Abstract
A ubiquitous problem in aggregating data across different experimental and observational data sources is a lack of software infrastructure that enables flexible and extensible standardization of data and metadata. To address this challenge, we developed HDMF, a hierarchical data modeling framework for modern science data standards. With HDMF, we separate the process of data standardization into three main components: (1) data modeling and specification, (2) data I/O and storage, and (3) data interaction and data APIs. To enable standards to support the complex requirements and varying use cases throughout the data life cycle, HDMF provides object mapping infrastructure to insulate and integrate these various components. This approach supports the flexible development of data standards and extensions, optimized storage backends, and data APIs, while allowing the other components of the data standards ecosystem to remain stable. To meet the demands of modern, large-scale science data, HDMF provides advanced data I/O functionality for iterative data write, lazy data load, and parallel I/O. It also supports optimization of data storage via support for chunking, compression, linking, and modular data storage. We demonstrate the application of HDMF in practice to design NWB 2.0 [13], a modern data standard for collaborative science across the neurophysiology community.
Index Terms—: data standards, data modeling, data formats, HDF5, neurophysiology
I. Introduction
As technological advances continue to accelerate the volumes and variety of data being produced across scientific communities, data engineers and scientists must grapple with the arduous task of managing their data. A subtask to this broader challenge is the curation and organization of complex data. Within expansive scientific communities, this challenge is exacerbated by the idiosyncrasies of experimental design, leading to inconsistent and/or insufficient documentation, which in turn makes data difficult or impossible to interpret and share. A common solution to this problem is the adoption of a data schema, a formal description of the structure of data.
Proper data schemas ensure data completeness, allow for data to be archived, and facilitate tool development against a standard data structure. Despite the benefit to scientific communities, efforts to establish standards often fail. Diverse analysis tools and storage needs drive conflicting needs in data storage and API requirements, hindering the development and community-wide adoption of a common standard. Here, we present HDMF, a framework that addresses these problems by separating data standardization into three components: Data Specification, Data Storage, and Data Interaction. By creating interfaces between these components, we have created a modular system that allows users to easily modify these components without altering the others. We demonstrate the utility of HDMF by evaluating its use in the development of NWB, a data standard for storing and sharing neurophysiology data across the systems neuroscience community.
In this work, we first present an assessment of the state of the field (Sec. II) and common requirements for data standards (Sec. III). Motivated by these requirements, we then describe HDMF, a novel software architecture for hierarchical data standards that addresses key limitations of the current state of the art (Sec. IV). Finally, we assess this novel architecture and demonstrate its utility in building NWB 2.0, a community data standard for collaborative systems neuroscience (Sec. V).
II. Related Work
A. File Formats
File formats used in the scientific community vary broadly, ranging from: 1) basic formats that explicitly specify how data is laid out and formatted in binary or text data files (e.g., CSV, flat binary, etc.), 2) text files based on language standards, e.g., the Extensible Markup Language (XML) [3], JavaScript Object Notation (JSON) [6], or YAML [2], to 3) self-describing array-based formats and libraries, e.g., HDF5 [19], NetCDF [10], or Zarr [21]. While basic, explicit formats are common, they often suffer from a lack of portability, scalability and rigor in specification, and as such are not compliant with FAIR [20] data principles. Text-based standards, e.g., JSON, (in combination with character-encoding schema, e.g., ASCII or Unicode) are popular for standardizing documents for data exchange, particularly for relatively small, structured documents; however, they are impractical for storage and exchange of large, scientific data arrays. For storing large scientific data, self-describing, array-based formats, e.g., HDF5, have gained wide popularity in the scientific community.
B. Data Standards
Building on file formats, data standards specify the organization of data and metadata to enable standardized exchange, access, and use and to facilitate reuse, integration, and preservation of data assets. Tool-oriented data standards, e.g., VizSchema [16], NIX [17], XDMF [4], propose to bridge the gap between general-purpose, self-describing formats and the need for standardized tools via additional lightweight, low-level schema to further standardize the description of the low-level data organization to facilitate data exchange and tool development. However, such tool-oriented standards are still fairly low-level and often do not consider semantics of the data critical for applications.
Conversely, application-oriented data standards, such as NeXus [7], BRAINformat [11], or CXIDB [8] among many others, provide community-specific solutions that focus on the semantic organization of data for target applications. However, the approaches and tools developed do not generalize to other scientific communities.
III. Requirements
Data standards are central to all aspects of the data life-cycle, from data acquisition, pre-processing, and analysis to data publication, preservation, and reuse. A data standard must be flexible enough to accommodate the diversity of data types and metadata that arise throughout this cycle. Additionally, different use cases often emphasize different requirements. This leads to broad, and sometimes conflicting, requirements for data standards. Broadly speaking, requirements are driven by properties of and constraints on the data that need to be stored.
The 5 V’s of Big Data—volume, velocity, variety, value, and veracity—emphasize properties of the data itself, leading to strict requirements with regard to performance and flexibility of data standards. A data standard should be flexible enough to allow for the diversity of data generated by the scientific community. At the same time, it must not hinder scientists from storing and accessing large volumes of data. Providing this functionality requires that APIs for data access, I/O, and specification be decoupled in such a way that these attributes of big data can be properly addressed. For example, a standard should not be tied to a storage format that gives poor I/O performance nor a format that hinders interactive exploration.
FAIR data principles—findable, accessible, interoperable, reusable—aim to assist humans and machines in the discovery, access, and integration of data [20]. In contrast to the 5 Vs, FAIR focuses on requirements for reusability of data. Achieving FAIRness goals requires that data and metadata be properly curated and documented, and that data and metadata be properly associated, both onerous tasks. To ensure that researchers adhere to these principles, APIs for data access, I/O, and specification should simplify the process for including necessary documentation, have data documentation as a core capability, and facilitate machine and human readability.
IV. Software Architecture
The HDMF software architecture (see Fig. 1) consists of four main components:
specification interfaces (blue) for creation and use of the data standard schema (orange),
data storage and I/O interfaces for read/write (green),
front-end data containers defining the user API (purple), and
a flexible object mapping API to insulate and integrate the different system components (red).
Fig. 1:
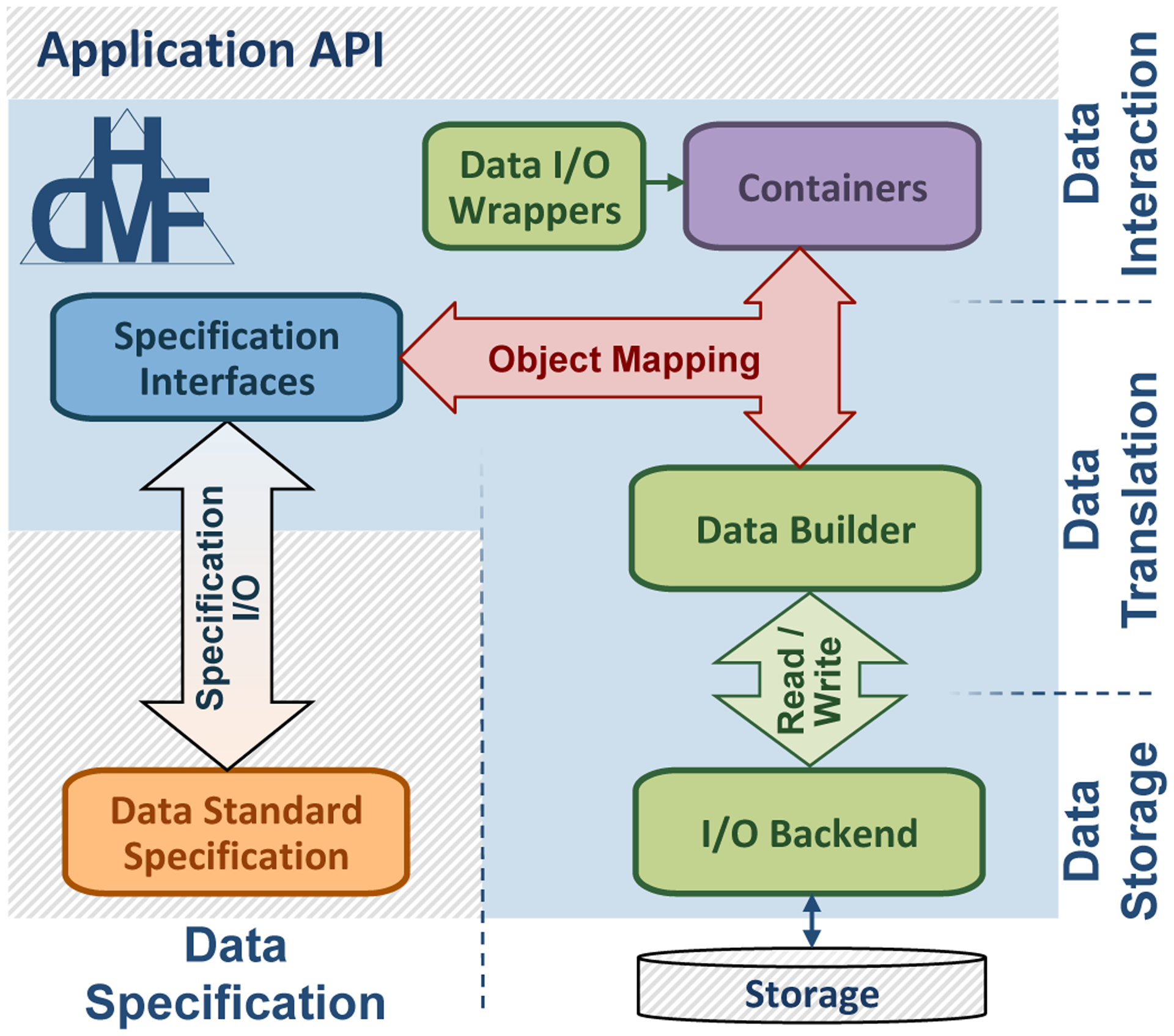
Software architecture of HDMF (blue box). HDMF is designed to facilitate the creation of formal data standards (orange), advanced application APIs (top), and flexible integration with modern data storage (bottom right).
Through its modular architecture, HDMF supports:
specification and sharing of data standards (Sec. IV-A),
advanced data storage, including multiple different storage backends (Sec. IV-B),
design of easy-to-use user APIs (Sec. IV-C),
flexible mapping between the specification, user APIs, and storage to insulate and integrate the various aspects of the system (Sec. IV-D), and
advanced I/O features to optimize data storage and I/O (Sec. IV-E).
HDMF is implemented in Python 3 (with 2.7.x support) and can be easily installed via the popular PIP and Conda package managers. All sources and documentation for HDMF are available online via GitHub at https://hdmf-dev.github.io/ using a BSD-style open source license.
A. Format Specification
A data standard describes the rules by which data is described and stored. In order to share, exchange, and understand scientific data, standards need to facilitate human and programmatic interpretation and formal verification. While use of text-based documents to describe standards is common, this approach is not scalable to increasingly complex data and impedes formal verification. To address this challenge, HDMF defines a formal language for specification of hierarchical data standards, i.e., a schema for defining hierarchical data schemas. This specification language enables the formal definition, programmatic interpretation, and verification of scientific data standards, as well as efficient sharing and versioning of standards documents.
1). Primitives:
The HDMF specification language supports the following main primitives for defining data standards. A Group (similar to a folder) defines a collection of objects (subgroups, datasets, and links). A Dataset defines an n-dimensional array with associated data type and dimensions. An Attribute defines a small metadata dataset associated with a group or dataset. Attributes and datasets may store i) basic data types, e.g., strings (ASCII, UTF-8) or numeric types (float, int, uint, bool, etc.), ii) flat compound data types similar to structs, iii) special types, e.g., ISO 8061 [5] datetime string, and iv) references to other objects.
2). Defining Reusable Data Types:
To enable the modular specification and reuse of data standard components, HDMF supports the concept of data types. A group or dataset may be assigned a data type to support referencing and reuse of the types elsewhere in the specification. Types here are similar to the concept of a class in object-oriented programming. All components (groups, datasets, attributes, links) in a standard specification must have either a unique type or unique name within a type specification, ensuring a unique mapping between objects on disk to components of a standard specification. HDMF enables reuse of types through inheritance and composition via the keys: i) data_type_inc to include an existing type and ii) data_type_def to define a new type (Tab. I).
TABLE I:
Defining and reusing types in a format specification.
| data_type_inc | data_type_def | Description |
|---|---|---|
| not set | not set | Define a dataset/group without a type (the name must be fixed) |
| not set | set | Define a new type |
| set | not set | Include an existing type (i.e., composition) |
| set | set | Define a new type that inherits components of an existing type |
Using the HDMF specification language, a data standard schema typically defines a collection and organization of reusable types. This approach supports modular creation and extension of data standards through definition of new data types, while allowing new types to build on existing ones through inheritance and composition.
3). Data Referencing:
In addition to the primitives described earlier, HDMF also supports the specification of Links to other types in the specification. Object references are similar to links but are stored as values of datasets or attributes (rather than objects inside groups). Using datasets of object references allows for efficient storage of large collections of references. Object references, like links, may point to datasets or groups with an assigned type. Finally, region references are similar to object references, but in contrast point to regions (i.e., select subsets) of datasets with an assigned type.
4). Namespaces:
The concept of a namespace is used to collect all specifications corresponding to a data standard, to document high-level concepts, and to insulate standard specifications. Within a namespace, specified type names must be unique, while the same type name may be used in different namespaces. The namespace further documents metadata about a standard, e.g., the authors, description, and version. Using the concept of a namespace makes it easy to create new data standards, avoids collisions between standards and format extensions, and facilitates sharing, versioning, and dissemination of standards.
5). Specification API:
HDMF provides a specification API for building and maintaining data type specifications. The specification API lets users progressively and programmatically build up data types and add them to namespaces. As these data types are definitions for groups and datasets, the specification API provides corresponding classes (i.e. GroupSpec and DatasetSpec) for building up these primitive objects into data type specifications. Additionally, the specification API provides AttributeSpec, LinkSpec, and RefSpec for specifying attributes, links, and references, respectively. After building up data type specifications and adding them to a namespace, users can serialize the namespace to YAML or JSON, which can be stored and used elsewhere.
6). Tools:
To validate files against a data standard, HDMF provides a built-in validator. Further, HDMF provides dedicated tools to automatically generate reStructuredText documentation and figures from a format specification.
B. Storage and I/O
The role of data I/O is to translate data primitives to and from storage. In practice, requirements of the storage backend vary depending on the particular use case, e.g., data archiving and publication emphasize FAIR principles, whereas data pipelines place more stringent demands on performance. Recognizing that no single storage modality can meet all requirements optimally, HDMF provides an abstract I/O interface to support integration of new I/O backends. The HDMF I/O interface consists of two main components: 1) Builders to describe data primitives (Sec. IV-B1) and 2) the HDMF I/O interface for reading/writing builders from/to storage (Sec. IV-B2). Leveraging this general design, HDMF implements a reference storage backend based on HDF5 (Sec. IV-B3), while providing the flexibility to add additional backends (see Sec. V-B).
1). Builder:
Builders define intermediary objects for I/O and represent the main data primitives. HDMF defines a corresponding builder class for each primitive from the specification language, i.e., GroupBuilder, DatasetBuilder, LinkBuilder, ReferenceBuilder, RegionBuilder, etc. Builders provide a format-independent description of data objects for storage.
2). HDMF I/O:
HDMFIO provides a general, abstract interface for creating new I/O backends. The role of the backend I/O is then to translate (i.e., read/write) builders to and from storage. The interface that new I/O backends need to implement consists of five main functions: the __init__(…) function to initialize the storage backend, the open() and close() functions to open and close the file (or connection to the storage, e.g., a database), the write_builder(builder) function to translate a given builder to storage, and the read_builder() function to load data from storage.
3). HDF5 I/O:
HDMF provides HDF5IO, a reference implementation of a storage backend based on HDF5. As a concrete implementation of the abstract HDMFIO class, HDF5IO depends only on the Builders. As such, HDF5IO is agnostic to the data format specification and front-end API and supports read/write of any data format specified via the HDMF specification language. HDF5 was chosen as the main storage backend for HDMF because it meets a broad range of the fundamental requirements of scientific data standards. In particular, HDF5 is self-describing, portable, extensible, optimized for storage and I/O of large scientific data, and is supported by many programming languages and analysis tools.
C. User APIs
The role of the user API is to enable users to efficiently interact with data in memory and to provide usable interfaces for building user applications. HDMF uses an object-oriented approach toward user interfaces in which each type in the format specification is represented by a corresponding frontend container class.
1). Containers:
Containers are Python object representations of the data specified in a data type specification. Containers are endowed with an object ID and pointers to other containers to facilitate internal tracking of relationships, e.g. parent-child relationships. Apart from this, the base Container class provides no other functionality to the user to working with instances. Instead, it provides a base for users to add functionality associated with more specialized data types. The base Container class is generated from the metaclass ExtenderMeta, which provides decorators for designating routines to be used for programmatically defining behavior and functionality of Container subclasses.
2). Dynamic Containers:
HDMF supports the dynamic generation of container classes to represent data types defined in a format specification. This functionality enables HDMF to read and write arbitrary data standards and extensions based on the format specification alone, i.e., without requiring users to write custom Container classes. Using dynamic containers allows users to easily prototype, evaluate, and share new data types and their definitions.
3). Data Validation:
Ensuring compliance of data with the format specification is central to data standardization. This includes checking of data compliance at run-time during data generation as well as tools for post-hoc validation of files.
As a dynamically-typed language, Python does not enforce data types and only recently added support for type hints (which are also not enforced). To address this challenge, HDMF defines docval; a function decorator that allows documentation and type declaration for inputs and outputs of a function. Using docval, functions and methods can enforce variable types and generate standardized Sphinx-style docstrings for comprehensive API documentation.
HDMF also includes a user-friendly tool for post-hoc validation. Using the validator, a file can be checked for compliance to a given format specification to identify errors, such as missing objects or additional components in a file that are not governed by the given standard.
D. Object Mapping
The role of the object mapping (Fig 1, red) is to insulate and integrate the specification, storage, and user API components of HDMF. Using the format specification, the object mapping translates between front-end containers and data builders for I/O. Fig. 2 provides an overview of the main components of the object mapping API, consisting of the Object Mapper classes, the Type Map, and the Build Manager.
Fig. 2:

Overview of the HDMF object mapping API. Object Mappers map between containers and builders while the Type Map maintains the mapping between data types and containers and containers and object mappers. Finally, the Build Manager manages the mapping process to construct containers and builders and memoizes builders and containers.
An Object Mapper maintains the mapping between frontend API container attributes and the data type specification components. Its role is to translate data containers using the format specification to builders for I/O and vice versa. For each type in the data standard, there exists a corresponding Container and ObjectMapper class. In many cases, the default ObjectMapper can be used, which implements a default mapping for: 1) objects in the specification to constructor arguments of the Container class and 2) Container attributes to specification objects. These mappings can be customized by creating a custom class that inherits from the default ObjectMapper. To optimize storage and improve usability of the data API, the representation of data in memory as part of the container can differ from the organization of the data in a file. In this case, a direct mapping between Container attributes, Container constructor arguments, and specification data types may not be possible. To address this challenge, custom functions can be designated for setting Container constructor arguments or retrieving Container attributes.
The role of the Type Map then is to map between types in the format specification, Container classes, and Object Mapper classes. Using TypeMap instance methods, users specify which data type specification corresponds to a defined Container class and which Container class corresponds to a defined ObjectMapper class. By maintaining these mappings, a Type Map is then able to convert between all data types to and from their respective Container classes.
Finally, the BuildManager is responsible for memoizing Builders and Containers. To ensure a one-to-one correspondence between in-memory Container objects and stored data (as represented by Builder objects), the BuildManager maintains a map between Builder objects and Container objects, and only builds a Builder (from a Container) or constructs a Container (from a Builder) once, thereby maintaining data integrity.
E. Advanced Data I/O
Due to the large size of many experimental and observational datasets, efficient data read and write is essential. HDMF includes optional classes and functions that provide access to advanced I/O features for each storage backend. Our primary storage system, HDF5, supports several advanced I/O features:
1). Lazy Data Load:
HDMF uses lazy data load, i.e., while HDMF constructs the full container hierarchy on read, the actual data from large arrays is loaded on request. This allows users to efficiently explore data files even if the data is too large to fit into main memory.
2). Data I/O Wrappers:
Arrays, such as numpy ndarrays, can be wrapped using the HDMF DataIO class to define perdataset, backend-specific settings for write. To enable standard use of wrapped arrays, DataIO exposes the normal interface of the array. Using the concept of array wrappers allows HDMF to preserve the decoupling of the front-end API from the data storage backend, while providing users flexible control of advanced per-dataset storage optimization.
To optimize dataset I/O and storage when using HDF5 as the storage backend, HDMF provides the H5DataIO wrapper, which extends DataIO, and enables per-dataset chunking and I/O filters. Rather than storing an n-dimensional array as a contiguous block, chunking allows the user to split the data into sub-blocks (chunks) of a specified shape. By aligning chunks with typical read/write operations, chunking allows the user to accelerate I/O and optimize data storage. With chunking enabled, HDF5 also supports a range of I/O filters, e.g., gzip for compression. Chunking and I/O filters are applied transparently, i.e., to the user the data appears as a regular n-dimensional array independent of the storage options used.
3). Iterative Data Write:
In practice, data is often not readily available in memory, e.g., data may be acquired continuously over time or may simply be too large to fit into main memory. To address this challenge, HDMF supports writing of dataset iteratively, one data chunk at a time. A data chunk, represented by the DataChunk class, consists of a block of data and a selection describing the location of data in the target dataset. The AbstractDataChunkIterator class then defines the interface for iterating over data chunks. Users may define their own data chunk iterator classes, enabling arbitrary division of arrays into chunks and ordering of chunks in the iteration. HDMF provides a DataChunkIterator class, which implements the common case of iterating over an arbitrary dimension of an n-dimensional array. DataChunkIterator also supports wrapping of arbitrary Python iterators and generators and allows buffering of values to collect data in larger chunks for I/O.
4). Parallel I/O:
The ability to access (read/write) data in parallel is paramount to enable science to efficiently utilize modern high-performance and cloud-based parallel compute resources and enable analysis of the ever-growing data volumes. HDMF supports the use of the Message Passing Interface (MPI) standard for parallel I/O via HDF5. In practice, experimental and observational data consists of a complex collection of small metadata (i.e, few MB to GB), with the bulk of the data volume appearing in a few large data arrays (e.g., raw recordings). In practice, parallel write is most appropriate for populating the largest arrays, while creation of the metadata structure is usually simpler and more efficient in serial. With HDMF, therefore, the initial structure of the file is created on a single node, and bulk arrays are subsequently populated in parallel. This use pattern has the advantage that it allows the user to maintain their workflow for creating files while providing explicit, fine-grained control over the parallel write for optimization. Similarly, on read, the object hierarchy is constructed on all ranks, while the actual data is loaded lazily in parallel on request.
5). Append:
As scientific data is being processed and analyzed, a common need is to append (i.e., add new components) to a file. HDMF automatically records for all builders and containers whether they have been written or modified. On write, this allows the I/O backend to skip builders that have already been written previously and append only new builders.
6). Modular Data Storage:
Separating data into different files according to a researcher’s needs is essential for efficient exploratory analysis of scientific data, as well as productionlevel processing of data. To facilitate this, HDMF allows users to reference objects stored in different files. By default, backend sources are automatically detected and corresponding external links are formed across files. For cases where this default functionality is not sufficient, the H5DataIO wrapper allows users to explicitly link to objects.
V. Evaluation
We demonstrate the application of HDMF in practice to design the NWB 2.0 [13] neurophysiology data standard. Developed as part of the US NIH BRAIN Initiative, the NWB data standard supports a broad range of neurophysiology data modalities, including extracellular and intracellular electrophysiology, optical physiology, behavior, stimulus, and experiment data and metadata. HDMF has been used to both design NWB 2.0 as well as to implement PyNWB, the Python reference API for NWB. Following the same basic steps as in the previous section, we first discuss the use of HDMF to specify the NWB data standard (Sec. V-A). Next, we demonstrate the use of the HDMF I/O layer to integrate Zarr as an alternate storage backend and show its application to store NWB files from a broad range of neurophysiology applications (Sec. V-B). We then discuss how HDMF facilitates the design of advanced user APIs (here, PyNWB; Sec. V-C). Finally, we demonstrate how HDMF facilitates the creation and use of format extensions (Sec. V-D) and show the application of the advanced data I/O features of HDMF to optimize storage and I/O of neurophysiology data (Sec. V-E).
A. Format Specification
Neurophysiology data consists of a wide range of data types, including recordings of electrical activity from brain areas over time, microscopy images of fluorescent activity from a brain areas over time, external stimuli, and behavioral measurements under different experimental conditions. A common theme among these various types of recordings is that they represent time series recordings in combination with complex metadata to describe experiments, data acquisition hardware, and annotations of features (e.g., regions of interest in an image) and events (e.g., neural spikes, experimental epochs, etc.). Because HDMF supports the reuse of data types through inheritance and composition, NWB defines a base TimeSeries data type consisting of a dataset of timestamps in seconds (or staring time and sampling rate for regularly sampled data), a dataset of measured traces, the unit of measurement, and a name and description of the data. TimeSeries then serves as the base type for more specialized time series types that extend or refine TimeSeries, such as ElectricalSeries for voltage data over time, ImageSeries for image data over time, SpatialSeries for positional data over time, among others. By supporting reusable and extensible data types, HDMF allows the NWB standard and its extensions to be described succinctly and modularly. In addition, NWB specifies generic types for column-based tables, ragged arrays, and several other specialized metadata types. On top of these modular types, NWB defines a hierarchical structure for organizing these types within a file. In total, the NWB standard defines 68 different types, which describe the vast majority of data and metadata used in neurophysiology experiments (see [13]).
NWB uses links to specify associations between data types. For example, the DecompositionSeries type, which represents the results of a spectral analysis of a time series, contains a link to the source time series of the analysis. A link can also point to a data type stored in an external file, which is often used for separating the results of different analyses from the raw data while maintaining the relationships between the results and the source data. As such, HDMF links facilitate documenting relationships and provenance while avoiding data duplication.
To model relationships between data and metadata and to annotate subsets of data, NWB uses object references. For example, neurophysiology experiments often consist of time series where only data at selected epochs of time are important for analysis, e.g. the electrical activity of a neuron during a one second period immediately following presentation of stimuli. These epochs are stored as a dataset of object references to time series and indices into the time series data. HDMF object references provide an efficient way to specify the large datasets of metadata that are often required to understand neurophysiology data.
Because HDMF supports storage of format specifications as YAML/JSON text files, the NWB schema is version controlled and hosted publicly on GitHub [14]. Sphinx-based documentation for the standard is then automatically generated from the schema using HDMF documentation tools, and is version controlled and integrated with continuous documentation web services with minimal customization required. These features make updating the NWB schema and its documentation to a new version simple and transparent, while maintaining a detailed history of previous versions.
B. Data Storage
In practice, different storage formats are optimal for different applications, uses, and compute environments. As such, it is critical that users can use data standards across storage modalities. Using the HDMF abstract storage API, we demonstrate the integration of Zarr’s DirectoryStore with HDMF by defining the ZarrIO class (see Appendix F). The goal of this evaluation is to demonstrate feasibility and practical use of multiple storage backends via HDMF. However, it is beyond the scope of this manuscript to evaluate and compare HDF5 and Zarr.
To integrate Zarr with HDMF and PyNWB, we only had to define new classes defining the backend, i.e., no modifications to the frontend container classes or the format specification were needed. As this example demonstrates, using the HDMF abstract storage API facilitates the integration of new storage backends and allows us to make new storage options broadly accessible to any application format built using HDMF.
Table II illustrates the use of the Zarr backend in practice. We converted four NWB files from the Allen Institute for Brain Science and one from the BouchardLab at LBNL from HDF5 to Zarr. We chose these datasets as they are very complex and allow us to cover a wide range of data sizes and NWB use cases, i.e., extracellular electrophysiology (D1, D4), intracellular electrophysiology (D2), as well as optical physiology and behavior (D3) (see also Appendix G).
TABLE II:
Overview of four NWB neurophysiology data files (top rows) stored in i) HDF5 without compression and with gzip compression enabled for all datasets, ii) Zarr, and iii) Zarr-C with automatic chunking enabled for all datasets.
| (D1) Allen EPhys |
(D2) Allen ICEphys |
(D3) Allen OPhys |
(D4) NSDS EPhys |
||
|---|---|---|---|---|---|
| Content | #Groups | 30 | 142 | 41 | 33 |
| #Datasets | 49 | 604 | 83 | 69 | |
| #Attributes | 251 | 1565 | 408 | 283 | |
| #Links | 8 | 127 | 12 | 6 | |
| HDF5 | #Files | 1 | 1 | 1 | 1 |
| Size | 705MB | 168MB | 1,569MB | 57,934MB | |
| Size (gzip=4) | 533MB | 49MB | 704MB | 49,057MB | |
| Zarr | #Folders | 79 | 731 | 124 | 102 |
| #Files | 198 | 2045 | 320 | 263 | |
| Size | 696MB | 165MB | 1565MB | 57,925MB | |
| Zarr-C | #Folders | 79 | 731 | 124 | 102 |
| #Files | 622 | 2383 | 1153 | 12,799 | |
| Size | 709MB | 167MB | 1584MB | 59,579MB |
As an example, Fig. 3a shows the conversion of dataset D2 from HDF5 to Zarr. Fig. 3b then shows a simple analysis to compare the same recording from the HDF5 and Zarr file, illustrating that both indeed store the same data (Fig. 3c). Importantly, as the front-end API is decoupled from the storage backend, we can use the same code to access and analyze the data, independent of whether it is stored via Zarr or HDF5.
Fig. 3:
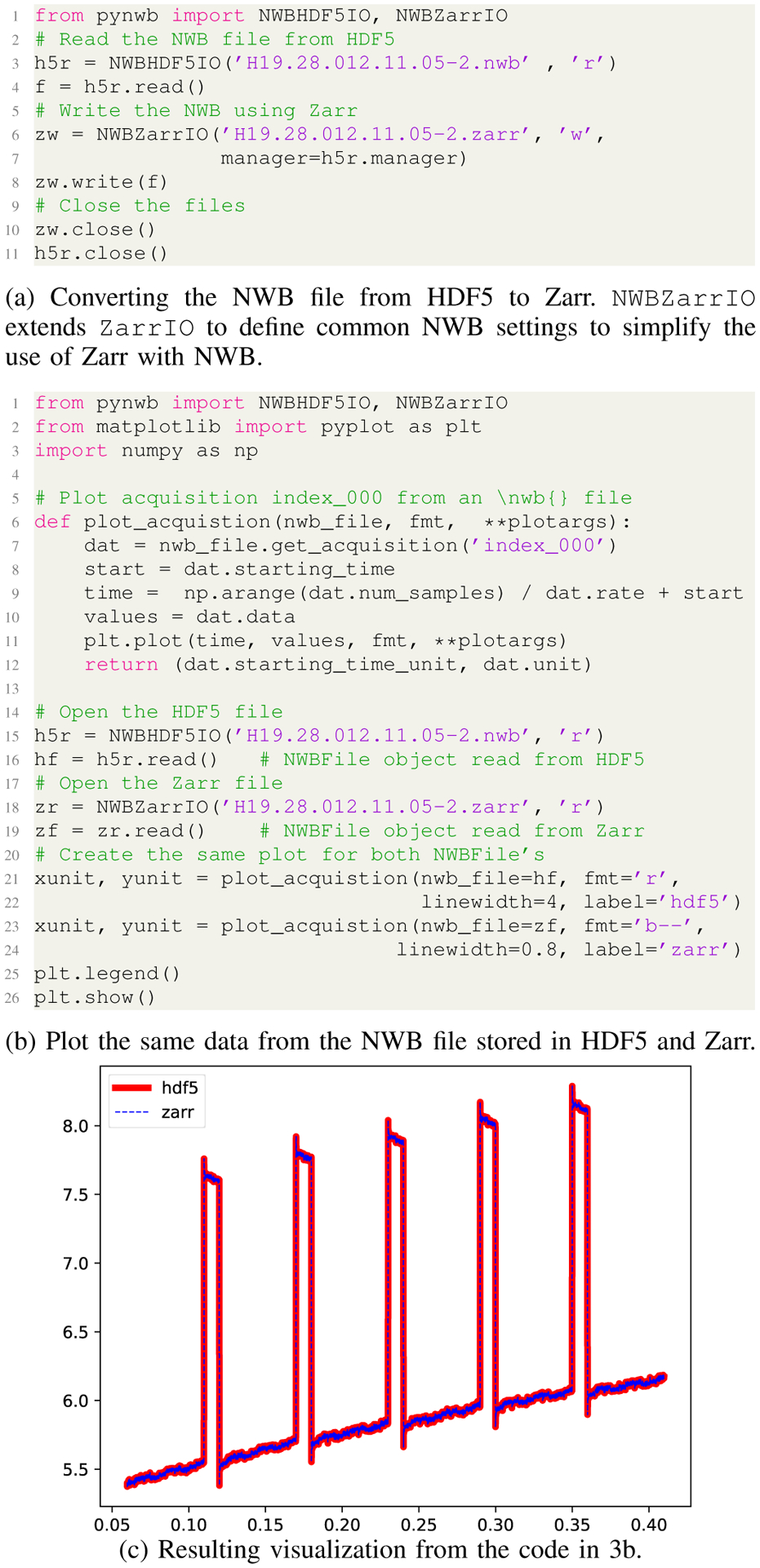
Example showing: (a) convert of dataset D2 (Tab. II) from HDF5 to Zarr and (b,c) use of the two files for analysis.
C. Data API
Building on top of HDMF, the PyNWB Python package is used for reading and writing NWB files. PyNWB primarily consists of a set of HDMF Container classes and ObjectMappers to represent and map NWB types. PyNWB provides a modified base Container class, NWBContainer, upon which the rest of the API is built. NWBContainer uses HDMFs ExtenderMeta metaclass decorators for defining functions that autogenerate setter and getter methods from parameterized macros, creating uniform functionality across the API and simplifying integration of new types. To ensure compliance of constructor arguments with the schema, PyNWB uses docval. Three main classes that exercise additional functionality of HDMF are NWBFile, TimeSeries, and DynamicTable.
1). NWBFile:
The NWBFile data type is the top-level type in the NWB standard, which defines the hierarchical organization of all data types in a file. These data types, in the form of Containers, are added to the NWBFile object through automatically generated instance methods. These methods are generated using the parameterized macros provided by the base NWBContainer class.
Another notable feature of the NWBFile Container is the copy method, which allows users to create a shallow copy of an NWBFile Container. Leveraging the modular storage capabilities of HDMF, this copy method allows users to easily create an interface file, which does not store any large data itself, but contains external links to all objects in the original file. This is useful for storing the results of exploratory data analysis separate from the raw data.
2). TimeSeries:
The TimeSeries Container represents the base TimeSeries type of the NWB data standard, which allows us to define a consistent base interface to all time series data in NWB. Neurophysiology experiments often use the same time axis for all time series data. To facilitate reuse of the same timestamps across different time series data, the timestamps of a TimeSeries Container can be defined as another TimeSeries object. To resolve this sharing, a custom ObjectMapper is used to override the base behavior for retrieving the timestamps from a given TimeSeries Container. This is done using decorators provided by ObjectMapper class in HDMF.
3). DynamicTable:
The DynamicTable data type is a group that represents tabular data, where each column is stored as a dataset. Using different types of datasets (i.e. unique data types), a DynamicTable can store one-to-one and one-to-many relationships. The details for populating and working with these different types of datasets are all managed by custom add and retrieval instance methods of the DynamicTable Container class, thereby shielding the user from details of the format specification.
As the DynamicTable data type does not predefine specific columns, users can extend DynamicTable to provide a stable specification for certain types of tabular data. To simplify development of corresponding DynamicTable Container subclasses, the DynamicTable Container uses directives of the ExtenderMeta metaclass in HDMF to automatically define structure of the Container class.
D. Extensions
Scientific data standards, by necessity, evolve slower than scientific experiments, but rather emerge from common needs across experiments. As a result, there is often a gap between common practices supported by scientific standards and the data needs of bleeding-edge science experiments. To address this challenge, HDMF enables users to extend data standards, enabling the integration of new data types while facilitating use (and reuse) of best practice and existing standard components.
To demonstrate the creation and use of format extensions in practice, we show the extension of the NWB data standard for electrocorticography (ECoG) data. ECoG uses electrodes placed directly on the exposed surface of the brain to record electrical activity from the cerebral cortex (Fig. 4 left). To enable localization of electrodes on the brain surface, we create a format extension to store a triangle mesh describing the cortical surface of the brain (Fig. 4 right). Similar to how one would typically write data to disk, we use the HDMF specification API to specify all data objects (groups, datasets, and attributes) and data types for our extensions (Fig. 5). We then specify a new namespace and export our extensions. The result is a collection of YAML files that describe our extensions (see Appendix A).
Fig. 4:

Left: Visualization of the cortical surface of the brain showing the location of the electrodes of the ECoG recording device. Right: Format extension for storing a triangle mesh of the cortical surface.
Fig. 5:

Example illustrating the creation of an extension for storing a mesh describing the cortical surface of the brain.
Using the HDMF load_namespace method to load our extension and get_class method to automatically create a Python class to represent our new Surface data type, we can then then immediately write and read data using our extension (Fig. 6). The ability to read/write extension data purely based on the specification supports fast prototyping, evaluation, sharing, and persistence of extensions and data. To enable users to define custom functionality for extensions, e.g., to facilitate specialized queries and visualizations, HDMF supports the creation of custom container classes.
Fig. 6:

Write (top) and read (bottom) ECoG extension data.
E. Data I/O
Next, we discuss the impact of HDMF’s advanced data I/O features for lazy data load, compression, iterative write, parallel I/O, append, and modular storage in practice.
1). Lazy Data Load:
Lazy data load enables us to efficiently read large data files, while avoiding loading the whole file into main memory. To demonstrate the impact of lazy data load, we evaluate the performance and memory usage for reading the NWB files from Table II. Here, file read includes opening the HDF5 file and subsequent lazy read to construct all builders and containers. We performed the tests on a 2017 MacBook Pro using the profiling scripts shown in Appendix C. We repeated the file read 100 times for each file and report the mean times: D1) 0.16s, D2) 0.92s, D3) 0.31s, and D4) 0.22s. For memory usage, we observe that instantiating the NWBHDF5IO object requires ≈ 0.3MB for all files. The actual read then requires: D1) 4.1MB, D2) 11.9MB, D3) 4.7MB, and D4) 3.5MB. We observe that data read requires only a few MB and less than one second in all cases. Here, read performance and memory usage depend mainly on the number of objects in the file, rather than file size. In fact, D2 as the smallest file overall but with the most objects, requires the most time and memory on initial read.
2). Chunking:
Chunking (and I/O filters, e.g., compression) allow us to optimize data layout for storage, read, and write. To illustrate the impact of chunking on read performance, we use as an example a dataset from file D4, which stores the frequency decomposition of an ECoG recording. The dataset consists of 916,385 timesteps for 128 electrodes and 54 frequency bands. We store the data as a single binary block as well as using (32×128×54) chunks. We evaluate performance for reading random blocks in time consisting of 512 consecutive time steps. We observe a mean read time of 0.179s without chunking and 0.012s with chunking, i.e., a ≈ 15× speed-up (see Appendix D). Design of optimal data layouts is a research area in itself and we refer the interested reader to the literature for details [1], [9], [12], [15], [18].
3). Compression:
NWB uses GZIP for compression. GZIP is available with all HDF5 deployments, ensuring that files are readable across compute systems. As shown in Table II, using GZIP we see compression ratios of 1.32×, 3.43×, 2.23×, and 1.18× for the four NWB files, respectively. Compression and chunking are applied transparently by HDF5 on a per-chunk basis. This ensures that we only need to de/compress chunks that we actually need and it allows users to interact with files the same way, independent of the storage optimizations used.
4). Iterative Data Write:
Iterative data write allows us to optimize memory usage and I/O for large data, e.g., to avoid loading all data at once into memory during data import (Fig. 7a) and support streaming write during acquisition (Fig. 7b). By combining the iterative write approach with chunking and compression, we can further optimize both storage and I/O of sparse data and data with missing data blocks (Fig. 7c).
Fig. 7:

Example applications of iterative data write.
A common example in neurophysiology experiments is intervals of invalid observations, e.g., due to changes in the experiment. Using iterative data write allows us to write only blocks of valid observations to a file, and in turn reduce the cost for I/O. To illustrate this process, we implemented a Python iterator that yields a set of random values for valid timesteps and None for invalid times. For write, we then wrap the iterator using HDMFs DataChunkIterator, which in turn collects the data into data chunks for iterative write, while automatically omitting write of invalid chunks (see Appendix E). When using chunking in HDF5, chunks are allocated in the file when written to. Hence, chunks of the array that contain only invalid observations are never allocated. In our example, the full array has a size of 2569.42MB while only 1233.51MB of the total data are valid. The resulting NWB file in turn has a size of just 1239.07MB. In addition, iterative write can help to greatly reduce memory cost, since we only need to hold the chunks relevant for the current write in memory, rather than the full array. In our example, memory usage during write was only 6.6MB.
5). Append:
The process for appending to a file in HDMF consists of: 1) reading the file in append mode, 2) adding new containers to the file, and finally 3) writing the file as usual. Using this simple process allows us to easily add, e.g., results from data processing pipelines, to an existing data file. See Appendix B for a code example.
6). Modular Data Storage:
HDMF’s support for modular storage, enables us to easily separate data from different acquisition, processing, and analysis stages across individual files. This approach is useful in practice to facilitate data management, avoid repeated file updates, and manage file sizes. At the same time, links are resolved transparently, enabling convenient access to all relevant data via a single file.
VI. Conclusion
Creating data standards is as much a social challenge as it is a technical challenge. With stakeholders ranging from application scientists to data managers, analysts, software developers, administrators, and the broader public, it is critical that we enable stakeholders to focus on the data challenges that matter most to them while limiting conflict and facilitating collaboration. HDMF addresses this challenge by clearly defining and insulating data specification, storage, and interaction as the core technical components of the data standardization process. At the same time, HDMF supports the integration of these core components via its sophisticated data mapping capabilities. HDMF facilitates, in this way, the creation, expansion, and technical evolution of data standards while simultaneously shielding and enabling collaboration between stakeholders. The successful use of HDMF in developing NWB 2.0, a standard for diverse neurophysiology data, suggests that it may be suitable for addressing analogous problems in other experimental and observational sciences.
In the future, we plan to enhance the specification language and API of HDMF to support complex data constraints to define dimensions scales, dependencies between datasets (e.g., alignment of shape), and mutually exclusive groups of attributes and datasets. We also plan to further expand the integration of HDMF with common Python analysis tools.
Acknowledgments
The authors thank Nicholas Cain, Nile Graddis, Lydia Ng, and Thomas Braun for providing us with pre-release data from the Allen Institute for Brain Science. We thank Max Dougherty for providing us with the NSDS dataset. We thank the NWB Executive Board, Technical Advisory Board, and the whole NWB user and developer community for their support and enthusiasm in developing and promoting the NWB data standard.
This work was sponsored by the Kavli foundation. Research reported in this publication was supported by the National Institute of Mental Health of the National Institutes of Health under Award Number R24MH116922 to O. Rübel and by the Simons Foundation for the Global Brain grant 521921 to L. Frank. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
APPENDIX A
ECOG Extension Example
Sources for the ECoG extension used in the paper are available at: https://github.com/bendichter/nwbext_ecog
Fig. A.1:

Source YAML files with the complete specification for the ECoG extension used in Sec. V-D.
APPENDIX B
Appending To an existing file
Fig. B.1:

Example illustrating the creation of an example NWB file (top) and process for appending a new SpatialSeries container to an existing file using HDMF (bottom).
APPENDIX C
Profiling Lazy Data Load
All tests for lazy data load were performed on a MacBook Pro with macOS 10.14.3, a 4-core, 3.1 GHz Intel Core i7 processor, a 1TB SSD hardrive, and 16GB of main memory.
A. Profiling Memory Usage for Lazy Data Load
Fig. C.1:

Evaluating the memory usage for lazy data load for the files listed in Tab. II.
B. Profiling Time for Lazy Data Load
Fig. C.2:

Evaluating read time for lazy data load for the files listed in Tab. II.
APPENDIX D
Profiling Chunking Performance
Fig. D.1:

Code used to test chunking performance
Fig. D.2:

Timing results for reading blocks in time with and without chunking.
APPENDIX E
Iterative Data Write
Fig. E.1:

Code used to test iterative data write performance
Fig. E.2:

Memory profiling results for iterative data write
Fig. E.3:

File size comparison for writing of sparse data
APPENDIX F
ZarrIO Implementation Details
Similar to HDF5, Zarr allows us to directly map groups, datasets, and attributes from a format specification to corresponding storage types. Specifically, groups are mapped to folders on the filesystem, datasets are mapped to folders storing a flat binary file for each array chunk, and attributes are stored as JSON files. The latter requires that we take particular care when creating attributes to ensure the data is JSON serializable.
As Zarr does not natively support links and references, we define the ZarrReference class to store the path of the source file and referenced object. On write, we then determine the required paths for each reference and serialize the ZarrReference objects via JSON. To identify and resolve reference on read, we define the reserved attribute zarr_type.
To support per-dataset I/O options (e.g, for chunking), we define ZarrDataIO. Like H5DataIO for HDF5, ZarrDataIO implements the HDMF DataIO class to wrap arrays for write to define I/O parameters.
Finally, NWBZarrIO extends our generic ZarrIO backend to setup the build manager and namespace for NWB.
APPENDIX G
Resources
All software and the majority of the data files used in the manuscript are available online. Here we summarize these resources.
A. Software
All software described in the manuscript is available online:
- ZarrIO: The Zarr I/O backend for HDMF and PyNWB are available online as part of the following pull requests:
B. Data
Datasets D1, D2, and D3 (see Tab. II) are available online from the Allen Institute for Brain Science at: http://download.alleninstitute.org/informatics-archive/prerelease/. Here we used the following files from this collection:
(D1): ecephys_session_785402239.nwb is a passive viewing extracellular electrophysiology dataset,
(D2): H19.28.012.11.05–2.nwb is an intracellular in-vitro electrophysiology dataset,
(D3) behavior_ophys_session_783928214.nwb is a visual behavior calcium imaging dataset.
Dataset D4 refers to R70_B9.nwb from the Neural Systems and Data Science Lab (NSDS) led by Kristofer Bouchard at Lawrence Berkeley National Laboratory https://bouchardlab.lbl.gov/. D4 is not available publicly yet.
In the case of (D2) as available online, compression is used for several datasets to reduce size. To gather data sizes without compression we used the h5repack tool available with the HDF5 library to remove compression from all datasets via h5repack -f NONE. To illustrate the potential impact of compression on file size we then used the h5repack to apply GZIP compression to all datasets via h5repack -f GZIP=4. This approach allows us to assess the expected impact of compression on file size. Using h5repack provides us with a convenient tool to test compression settings for existing HDF5 files. In practice, when generating new data files, users will typically use HDMF directly to specify I/O filters on a per-dataset basis, which has the advantage that it allows us to optimize storage layout independently for each dataset.
Footnotes
Publisher's Disclaimer: Legal Disclaimer
Publisher's Disclaimer: This document was prepared as an account of work sponsored by the United States Government. While this document is believed to contain correct information, neither the United States Government nor any agency thereof, nor the Regents of the University of California, nor any of their employees, makes any warranty, express or implied, or assumes any legal responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by its trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States Government or any agency thereof, or the Regents of the University of California. The views and opinions of authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof or the Regents of the University of California
References
- [1].Behzad B, Byna S, Prabhat, and Snir M. Optimizing i/o performance of hpc applications with autotuning. ACM Trans. Parallel Comput, 5(4):15:1–15:27, March. 2019. [Google Scholar]
- [2].Ben-Kiki O, Evans C, and Ingerson B. Yaml ain’t markup language (yaml) version 1.2. yaml. org, Tech. Rep, page 23, October 2009. [Google Scholar]
- [3].Bray T, Paoli J, Sperberg-McQueen C, Maler E, and Yergeau F. Extensible markup language (xml), 2008. [URL] http://www.w3.org/TR/2008/REC-xml-20081126/.
- [4].Clarke J and Mark E. Enhancements to the extensible data model and format (xdmf). In DoD High Performance Computing Modernization Program Users Group Conference, 2007, pages 322–327, June 2007. [Google Scholar]
- [5].Date and time format - ISO 8601 - An internationally accepted way to represent dates and times using numbers, 2019. [Google Scholar]
- [6].JSON: JavaScript Object Notation, 1999 – 2015. [URL] http://json.org/.
- [7].Klosowski P, Koennecke M, Tischler J, and Osborn R. Nexus: A common format for the exchange of neutron and synchroton data. Physica B: Condensed Matter, 241:151–153, 1997. [Google Scholar]
- [8].Maia FR. The coherent x-ray imaging data bank. Nature methods, 9(9):854–855, 2012. [DOI] [PubMed] [Google Scholar]
- [9].Nam B and Sussman A. Improving access to multi-dimensional self-describing scientific datasets. In CCGrid 2003. 3rd IEEE/ACM International Symposium on Cluster Computing and the Grid, 2003. Proceedings, pages 172–179, May 2003. [Google Scholar]
- [10].Rew R and Davis G. NetCDF: an interface for scientific data access. Computer Graphics and Applications, IEEE, 10(4):76–82, July 1990. [Google Scholar]
- [11].Rübel O, Dougherty M, Prabhat, Denes P, Conant D, Chang EF, and Bouchard K. Methods for specifying scientific data standards and modeling relationships with applications to neuroscience. Frontiers in Neuroinformatics, 10:48, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rübel O, Greiner A, Cholia S, Louie K, Bethel EW, Northen TR, and Bowen BP. Openmsi: A high-performance web-based platform for mass spectrometry imaging. Analytical Chemistry, 85(21):10354–10361, 2013. [DOI] [PubMed] [Google Scholar]
- [13].Rübel O, Tritt A, Dichter B, Braun T, Cain N, Clack N, Davidson TJ, Dougherty M, Fillion-Robin J-C, Graddis N, Grauer M, Kiggins JT, Niu L, Ozturk D, Schroeder W, Soltesz I, Sommer FT, Svoboda K, Lydia N, Frank LM, and Bouchard K. NWB:N 2.0: An Accessible Data Standard for Neurophysiology. bioRxiv, 2019. [Google Scholar]
- [14].Rübel O, Tritt A, and et al. NWB:N Format Specification V 2.0.1, July 2019. https://nwb-schema.readthedocs.io/en/latest/index.html [2019–0729].
- [15].Sarawagi S and Stonebraker M. Efficient organization of large multidimensional arrays. In Proceedings of 1994 IEEE 10th International Conference on Data Engineering, pages 328–336, February 1994. [Google Scholar]
- [16].Shasharina S, Cary JR, Veitzer S, Hamill P, Kruger S, Durant M, and Alexander DA. VizSchema–Visualization Interface for Scientific Data. In IADIS International Conference, Computer Graphics, Visualization, Computer Vision and Image Processing, page 49, 2009. [Google Scholar]
- [17].Stoewer A, Kellner CJ, and Grewe J. NIX, 2019. [URL] https://github.com/G-Node/nix/wiki.
- [18].Tang H, Byna S, Harenberg S, Zou X, Zhang W, Wu K, Dong B, Rubel O, Bouchard K, Klasky S, and Samatova NF. Usage pattern-¨ driven dynamic data layout reorganization. In 2016 16th IEEE/ACM International Symposium on Cluster, Cloud and Grid Computing (CCGrid), pages 356–365, May 2016. [Google Scholar]
- [19].The HDF Group. Hierarchical Data Format, version 5, 1997–2015. [URL] http://www.hdfgroup.org/HDF5/.
- [20].Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE, et al. The fair guiding principles for scientific data management and stewardship. Scientific data, 3, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dev Zarr. Zarr v. 2.3.2, 2019. [URL] https://zarr.readthedocs.io.


