Abstract
Large-scale molecular profiling studies in recent years have shown that central nervous system (CNS) tumors display a much greater heterogeneity in terms of molecularly distinct entities, cellular origins and genetic drivers than anticipated from histological assessment. DNA methylation profiling has emerged as a useful tool for robust tumor classification, providing new insights into these heterogeneous molecular classes. This is particularly true for rare CNS tumors with a broad morphological spectrum, which are not possible to assign as separate entities based on histological similarity alone. Here, we describe a molecularly distinct subset of predominantly pediatric CNS neoplasms (n = 60) that harbor PATZ1 fusions. The original histological diagnoses of these tumors covered a wide spectrum of tumor types and malignancy grades. While the single most common diagnosis was glioblastoma (GBM), clinical data of the PATZ1-fused tumors showed a better prognosis than typical GBM, despite frequent relapses. RNA sequencing revealed recurrent MN1:PATZ1 or EWSR1:PATZ1 fusions related to (often extensive) copy number variations on chromosome 22, where PATZ1 and the two fusion partners are located. These fusions have individually been reported in a number of glial/glioneuronal tumors, as well as extracranial sarcomas. We show here that they are more common than previously acknowledged, and together define a biologically distinct CNS tumor type with high expression of neural development markers such as PAX2, GATA2 and IGF2. Drug screening performed on the MN1:PATZ1 fusion-bearing KS-1 brain tumor cell line revealed preliminary candidates for further study. In summary, PATZ1 fusions define a molecular class of histologically polyphenotypic neuroepithelial tumors, which show an intermediate prognosis under current treatment regimens.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00401-021-02354-8.
Keywords: Brain tumor, Pediatric, Neurooncology, Neuroepithelial, PATZ1, EWSR1, MN1, Gene fusion
Introduction
Human central nervous system (CNS) tumors are an incredibly diverse set of neoplasms, reflecting the vast array of different temporo-spatially distinct stem/progenitor cells that are present (and which may undergo oncogenic transformation) at different stages of development. Recently, DNA methylation-based classification of CNS tumors has been shown to be a robust tool for molecular tumor classification, likely on the basis that individual tumor types maintain an epigenetic ‘memory’ of their distinct cell of origin [5]. Over the past years, many studies have confirmed the prognostic relevance of such profiling, and its ability to provide a molecular stratification with notable clinical and biological correlates across diverse CNS tumor entities (e.g. [5, 19, 31, 40]). Such an approach is particularly valuable for the identification of molecular subgroups displaying a broad morphological spectrum, which may not have previously been identified as a distinct entity due to their rarity or lack of obvious unifying features. This principle was recently demonstrated with the identification of four new molecular tumor types within the morphologically heterogeneous group previously referred to as CNS primitive neuroectodermal tumor (PNET) [46].
This substantial heterogeneity also extends to the glioma family. Until recently, malignant gliomas in children (pediatric high-grade glioma, pedHGG) were considered similar to their adult glioblastoma counterparts. The discovery of a variety of pediatric-enriched alterations, such as histone 3 mutations (K27M and G34R/V), PDGFRA alterations and others, has substantially increased our understanding of the molecular background of pedHGG [15, 32, 41, 55]. In parallel, multiple molecular studies have unravelled distinct (epi)-genetic subgroups of ‘histone wildtype’ pedHGG correlated with a variety of molecular and clinical features [19, 20, 26]. Recent studies on ‘HGG’ in infants have also shown that some of these tumors are biologically very different from those in older children, harbour recurrent, targetable gene fusions, and have a favourable prognosis despite histological features of malignancy [8]. Similarly, low-grade gliomas have also been found to be molecularly heterogeneous, with a number of subgroups showing consistent patterns of histological features, molecular profiles and genetic alterations (e.g. [10, 35, 36, 43, 57] and reviewed in [16]).
Interestingly, recent reports have identified a handful of pediatric brain tumors displaying a fusion of the PATZ1 gene with either MN1 or EWSR1 as a partner [1, 4, 6, 14, 35, 38, 42, 45] (summarized in Table 1, top panel). The histological diagnoses of these tumors included glial, glioneuronal and ‘polyphenotypic’ morphologies, hinting at some potential similarities but also variation in appearance, which may have prevented previous recognition as a defined entity. EWSR1:PATZ1 fusions have also been reported in a subset of sarcomas occurring across a wide range of ages, with a predilection for occurrence in the chest wall, showing substantial heterogeneity in their morphology and immunoprofile [3, 7, 27, 28, 33, 48, 52].
Table 1.
Summary of PATZ1-fused tumors previously reported in the literature
| Study | No. of cases | Fusion reported | Histopathologic features (ID in this series) |
|---|---|---|---|
| ‘CNS’ studies | |||
| Chadda et al. [6] | 1 | MN1:PATZ1 | Astroblastoma |
| Rossi et al. [38] | 1 | EWSR1:PATZ1 | Infantile Glioblastoma WHO Grade 4. Round monomorphous nuclei, cells with clear cytoplasm and oligodendroglia-like morphology, rich vascular network, microvascular proliferation, and microcysts |
| Lopez-Nunez et al. [23] | 1 | EWSR1:PATZ1 | Areas of cells with clear cytoplasm admixed with sheets of monotonous, round to spindled cells. “Malignant, poorly differentiated” |
| Burel-Vandenbos et al. [4] | 1 | MN1:PATZ1 | Malignant neuroepithelial tumor: hyperchromatic, polymorphous nuclei; clear cells, perivascular and stromal hyalinization, perivascular pseudorosettes, microcysts. (PATZ1-066) |
| Stichel et al. [45] | 3 |
EWSR1:PATZ1 MN1:PATZ1 |
Malignant neuroepithelial tumor with sarcomatous differentiation (PATZ1-014), glioneuronal tumor (PATZ-024), neuroepithelial neoplasia (PATZ-025) |
| Siegfried et al. [42] | 1 | EWSR1:PATZ1 | Low-grade glioneuronal tumor. Olig2 and synaptophysin positive cells with pleomorphic nuclei, vascular hyalinzation, pleomorphic clear cells (PATZ1-021)* |
| Alvarez-Breckenridge et al. [1] | 1 | EWSR1:PATZ1 | Glioneuronal tumor (no further details) |
| Johnson et al. [14] | 1 | EWSR1:PATZ1 | Low-grade glioma (no further details) |
| Qaddoumi et al. [35] | 1 | EWSR1:PATZ1 | BRAFV600E negative ganglioglioma (no further details) |
| ‘Sarcoma’ studies | |||
| Tsuda et al. [48] | 3 | EWSR1:PATZ1 | Round cell sarcomas |
| Pei et al. [33] | 1 | EWSR1:PATZ1 | GFAP positive and CD99 negative spinal intradural extramedullary tumor, eventually described as glioneuronal tumor. Monotonous spindle cells; abundant vasculature |
| Michal et al. [28] | 9 | EWSR1:PATZ1 | Spindle and round cell sarcomas |
| Bridge et al. [3] | 11 | EWSR1:PATZ1 | Morphologically variable; mostly undifferentiated or small round cell sarcomas, includes 4 CNS tumors |
| Chougule et al. [7] | 2 | EWSR1:PATZ1 | Spindle and round cell sarcomas |
| Watson et al. [52] | 5 | EWSR1:PATZ1 | Round cell sarcomas |
| Mastrangelo et al. [27] | 1 | EWSR1:PATZ1 | Small round cell tumor |
IHC immunohistochemistry
*PATZ-019 was also reported in Siegfried et al. [42], however, RNA-seq was only done on PATZ-021, revealing the EWSR1:PATZ1 rearrangement
Here, we describe a distinct molecular tumor type identified by investigation of a large cohort of DNA methylation and associated genomic profiling data. The morphological heterogeneity, marker expression and pathognomonic fusions of PATZ1 with either MN1 or EWSR1 in this group lead us to suggest a name of ‘neuroepithelial tumor with PATZ1 fusion’ (NET-PATZ1).
Materials and methods
Tumor tissue and clinical data
Our cohort comprises a total of 60 patients. All available clinical data are listed in Supplementary Table 1. Where not reported, patient sex was predicted using data from the DNA methylation array. Tumor samples and clinical data were provided by multiple international collaborating centers and collected at the Department of Neuropathology of the Heidelberg University Hospital (UKHD) and at the German Cancer Research Center (DKFZ, Heidelberg, Germany) with approval from the respective institutional review boards/ethics committees and informed consent from all patients or their guardians.
DNA methylation profiling and copy number variation calling
DNA methylation profiles on a genome-wide level were obtained using the Illumina Infinium HumanMethylation450 (450 k) or HumanMethylationEPIC (EPIC) BeadChip arrays according to the manufacturer’s instructions (Illumina, San Diego, USA). Raw data were generated at the Genomics and Proteomics Core Facility of the DKFZ, at the Neuropathology Department of the UKHD and at respective international contributing institutes, using both fresh-frozen and formalin-fixed paraffin-embedded (FFPE) tissue samples. All computational analyses were performed using R (http://www.R-project.org, R Development Core Team). Using the ‘conumee’ R package in Bioconductor (https://bioconductor.org/packages/release/bioc/html/conumee.html), the output data from the DNA methylation arrays were also used to investigate copy number alterations, which were assessed by manual inspection and in detail using IGV [37, 47]. The framework for data processing and downstream output after obtaining raw data from the methylation panels has been widely used and published elsewhere [5]. Summary copy number plots were generated using an in-house R-script (https://github.com/dstichel/CNsummaryplots).
Histopathology and immunohistochemistry
Confirmed histopathological diagnoses for the tumors included in this series were obtained through our international collaborators, as reviewed per the local pathologist and/or reference pathologists. Two experienced neuropathologists (F.S., A.Ko.) re-evaluated tumor haematoxylin–eosin (H&E) slides centrally available for n = 18 cases. Diagnostic criteria used were according to the 2016 WHO Classification of CNS tumors [24]. Features assessed included cell density, cytoplasm and nuclear pleomorphism, the number of mitotic figures per ten high-power fields (HPF; one HPF = 0.238 mm2), presence of microvascular proliferation and necrosis, and infiltrative nature. Immunohistochemical staining with Ki-67, GFAP, MAP2, NeuN, Olig2, Synaptophysin, S-100, CD34 and Vimentin was performed for a subset of tumors either in Heidelberg or at other contributing centers (n = 16). NG2 immunostaining was performed with NG2/CSPG4 (E3B3G) XP Rabbit monoclonal antibody (Cell Signaling, Danvers, Massachusetts, USA, 1:200).
Whole exome and gene panel sequencing
For n = 4 cases, whole exome sequencing (WES) data were generated for the INFORM study, from tumor and matched blood DNA, as previously described [54]. Gene panel sequencing data were available for n = 10 cases, including 7 cases for which normal blood DNA was also sequenced. The gene panel includes 130 commonly mutated genes in CNS neoplasms and has been described previously [39].
RNA sequencing analysis
RNA sequencing data (total n = 27) for the purpose of fusion detection was either generated by contributing sites (n = 8) or locally in Heidelberg. For samples sequenced in Heidelberg, sequencing was performed from FFPE (n = 8) on the NextSeq500 (Illumina) with 75 bp paired-end reads, at the Neuropathology Department of UKHD, as previously described [54]. For additional quantitative gene expression analysis, sequencing of high-quality RNA from frozen tissue (n = 11) was performed by the High Throughput Sequencing Unit of the Genomics and Proteomics Core Facility (GPCF) at the DKFZ using the Illumina HiSeq4000 or NovaSeq platform, with libraries prepared according to the manufacturer’s instructions using the TruSeq Stranded mRNA Library Prep Kit. Gene expression data analysis was performed using the online tool ‘R2: Genomics Analysis and Visualization Platform’ (https://hgserver2.amc.nl/cgi-bin/r2/main.cgi). Detection of gene fusions was performed using the Arriba analysis tool (https://github.com/suhrig/arriba).
Drug screening
The KS-1 cell line, from a 45-year-old female patient diagnosed with glioblastoma, was purchased from the Japanese Collection of Research Bioresources (JCRB) Cell Bank. RNA sequencing was performed to confirm the presence of the MN1:PATZ1 fusion. The cells were subjected to a drug screen at the Hopp Children’s Cancer Center, Heidelberg (KiTZ) Translational Drug Screening Unit (TDSU), with a library consisting of 74 widely used anti-cancer drugs. Cells were seeded at n = 750 per well in 384-well plates, pre-printed with drugs at 5 different concentrations, in duplicate per concentration. Cell Titer Glo (CTG2.0, Promega) viability readouts were performed on a FLUOstar OPTIMA plate reader (BMG Labtech) 72 h after cell seeding and quantified as residual metabolic activity. All cell viability readouts were normalized to the highest DMSO concentration used for the suspension of the respective drug. Validation experiments were carried out for five selected drugs. To that end, 1500 cells per well were seeded into a U-bottom 96-well plate. Drug concentrations used for validation experiments were selected individually per compound such that the IC50 could be confirmed and specified more precisely and the maximum plateau effect could be validated.
Results
DNA methylation profiling
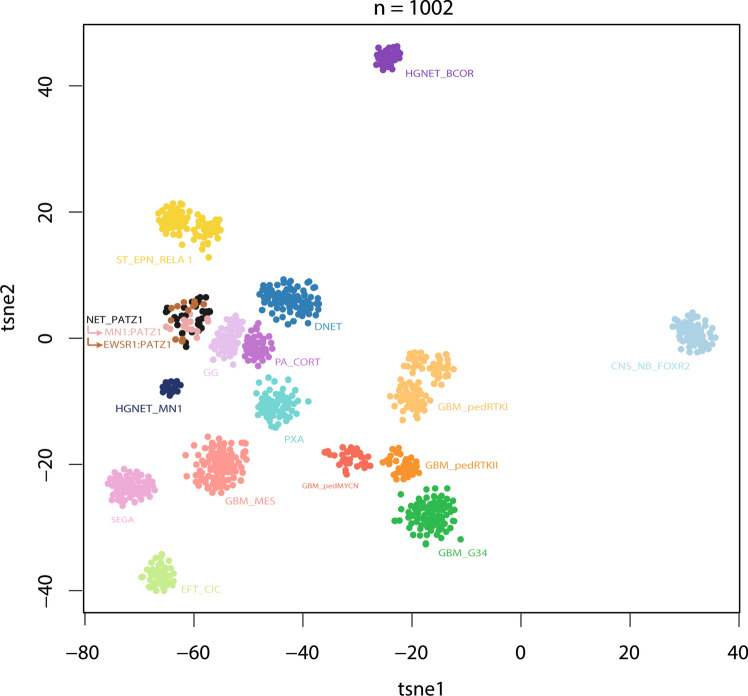
Routine diagnostic molecular profiling performed in Heidelberg in the context of the INFORM pipeline [54], the Molecular Neuropathology 2.0 or Pediatric Targeted Therapy 2.0 studies or otherwise in the Neuropathology Department of the UKHD [39, 45] revealed a small but recurring number of CNS tumors harboring fusions of the PATZ1 gene coupled to either MN1 or EWSR1 (see below). When further investigating the DNA methylation profile of these tumors within the context of a much larger reference database, they were found to form a distinct molecular cluster with several other tumors from other sources. Visualization of the genome-wide methylation pattern using t-distributed stochastic neighbor embedding (tSNE; Supplementary Fig. 1, online resource) and uniform manifold approximation and projection (UMAP; not shown) confirmed a common pattern with broad proximity to other molecular types of both low- and high-grade glial and glioneuronal tumors. A selected analysis of the tumors in this novel cluster (n = 60) compared with the genome-wide DNA methylation profiles of a reference cohort consisting of 15 other low- and high-grade glial and glioneuronal tumor types, confirmed a clearly distinct grouping (Fig. 1). No similarity was seen with the recently described HGNET_MN1 tumor type, which is characterized by MN1:BEND2 and MN1:CXXC5 (but not PATZ1) fusions. When assessed by the current Heidelberg Brain Tumor Classifier, which uses a random forest-based class prediction algorithm based on the output of the methylation analysis (v11b6; https://www.molecularneuropathology.org/mnp), tumors of this cluster scored poorly for all currently known entities (calibrated scores < 0.6), thus supporting the novel and distinct nature of this molecular type. Given their varied morphological appearance and defining fusions of PATZ1, as outlined further below, we provisionally suggest the term ‘neuroepithelial tumor with PATZ1 fusion’ to describe this molecular type.
Fig. 1.
t-distributed stochastic neighbor embedding (tSNE) clustering of DNA methylation patterns of 60 NET-PATZ1 tumors alongside 942 in-house reference samples representing 15 other low- and high-grade glial and glioneuronal tumor types, using the 10,000 most variably methylated probes. NET-PATZ1 forms a distinct ‘island’. CNS_NB_FOXR2 CNS neuroblastoma with FOXR2 activation; DNET dysembryoplastic neuroepithelial tumor; EFT_CIC CNS ewing sarcoma family tumor with CIC alteration; GBM_G34 glioblastoma, H3.3 G34 mutant; GBM_MES glioblastoma, subclass mesenchymal; GBM_pedMYCN pediatric-type glioblastoma, subclass MYCN; GBM_pedRTKI pediatric-type glioblastoma, subclass RTKI; GBM_pedRTKII pediatric-type glioblastoma, subclass RTK II; GG ganglioglioma; HGNET_BCOR CNS high-grade neuroepithelial tumor with BCOR alteration; HGNET_MN1 CNS high-grade neuroepithelial tumor with MN1 alteration; NET_PATZ1 neuroepithelial tumor with PATZ1 fusion; PA_CORT hemispheric pilocytic astrocytoma; PXA pleomorphic xanthoastrocytoma; SEGA subependymal giant cell astrocytoma; ST_EPN_RELA supratentorial ependymoma, RELA fused
Next-generation DNA and RNA sequencing analysis
Sequencing data of tumor DNA only (n = 3) or matched tumor-normal DNA pairs (n = 11), available through different platforms (gene panel or WES) revealed generally ‘quiet’ tumors at the level of point mutations or small insertions/deletions. Considering known CNS tumor-relevant genes, no mutations in genes such as IDH1/2, H3F3A, BRAF, or TP53 were detected. There were also no recurrent driver mutations found in the data, and no evidence for germline mutations in cancer predisposition genes in any of the cases with available germline sequencing data.
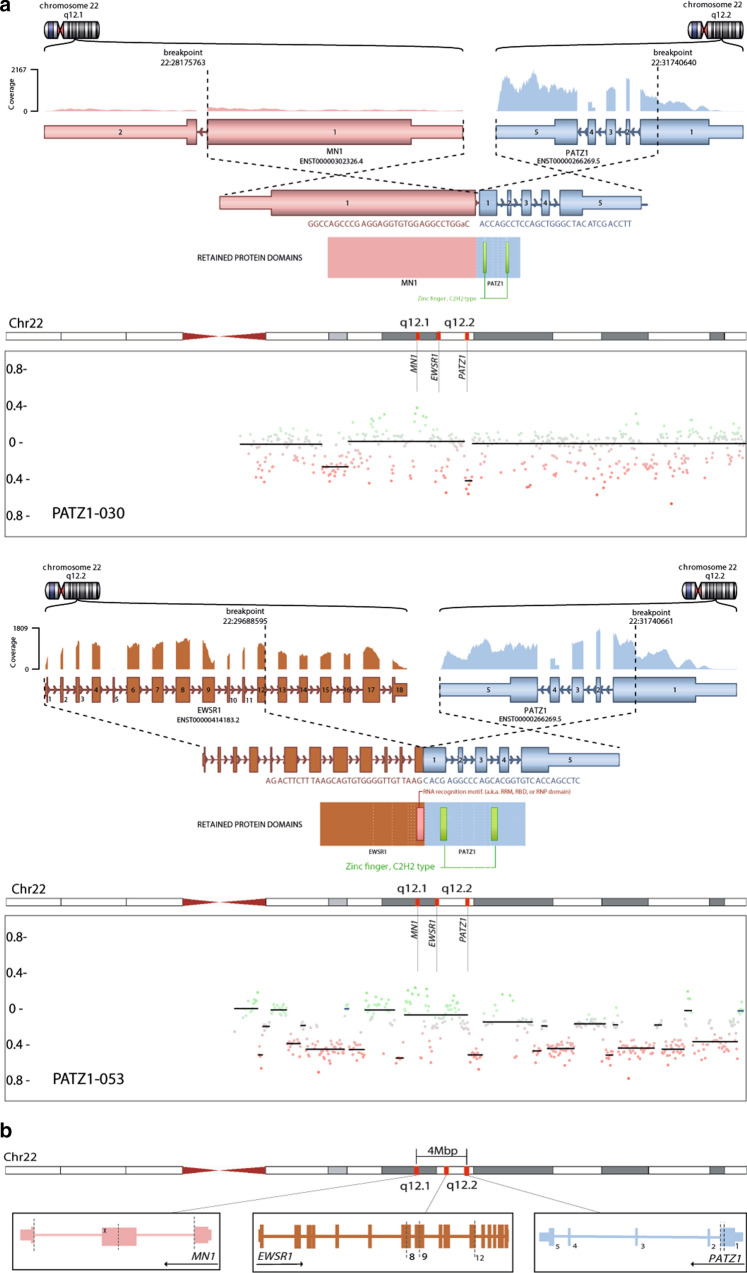
In contrast, the results of the RNA sequencing analysis were more informative. Supplementary Table 1 describes the RNA-seq fusion detection output for the 27 tumors with data available. Strikingly, in all cases where RNA-seq was conducted (27/27, 100%), an in-frame fusion gene involving PATZ1 was detected, with either MN1 or EWSR1 as the 5′ partner. This includes five of the aforementioned cases that have recently been previously published [4, 42, 45]. The two N′ terminal partners were roughly evenly distributed, with 13 EWSR1:PATZ1 fusions (48%) and 14 MN1:PATZ1 fusions identified (52%). The breakpoint within PATZ1 is unusual, with most fusion junctions occurring in the middle of exon 1 rather than at a splice junction. The remaining exons 2–5 of PATZ1 are also retained, conserving the zinc finger (C2H2 type) structure as part of the fusion product (Fig. 2). Most MN1:PATZ1 fusions showed a break at the end of MN1 exon 1. In some cases, however, an intronic breakpoint introduced an in-frame fusion with inclusion of an additional novel MN1 exon. The EWSR1:PATZ1 fusions harbor EWSR1 exons 1–8, 1–9 or 1–12. These findings are in line with the breakpoints detected in the previously reported CNS and sarcoma PATZ1 fusions [1, 14, 35, 52].
Fig. 2.
a Illustration of the PATZ1 fusion genes detected by RNA-seq for two selected cases; and corresponding copy number plots of Chromosome 22. The three involved genes MN1, EWSR1 and PATZ1 are marked. PATZ1-030 harbors an in-frame MN1:PATZ1 fusion, retaining an intronic pseudoexon (upper panel). PATZ1-053 demonstrates a variant fusion transcript juxtaposing Exon 12 of EWSR1 onto the usual partner 3′ sequence of PATZ1, in contrast to the more prevalent 5′ breakpoints observed in Exons 8 and 9 of EWSR1 (lower panel). b Schematic view of the loci of the genes involved in the fusions described (MN1, EWSR1, and PATZ1). Note the proximity of the three genes (all lie within approximately 4 Mbp). The dashed lines resemble the genomic breakpoints observed in the gene fusions. An intronic breakpoint observed in a subset of MN1_PATZ1 fusions introduces a novel pseudoexon (marked with x) whilst maintaining the reading frame. Isoforms illustrated: EWSR1 NM_005243; PATZ1 NM_0.13986.4; MN1 NM_002430.3
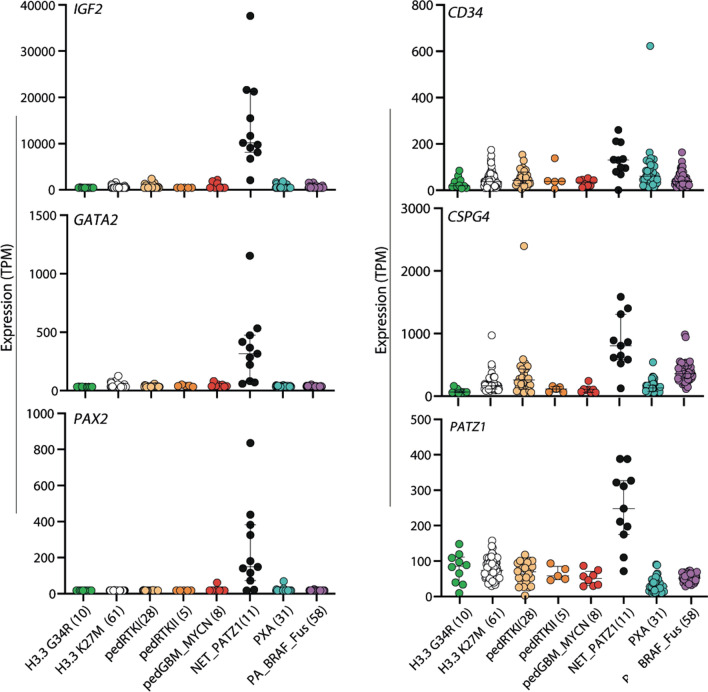
For RNA-seq data generated from high-quality frozen tumor tissue (n = 11), we also performed an exploratory differential gene expression analysis (with the caveat that the sample size is small). For this, we examined genes that were significantly differentially expressed between NET-PATZ1 and a combined reference cohort of other glioma subtypes (ANOVA test, p < 0.00001, corrected for multiple testing using FDR). This revealed a total of 964 upregulated transcripts in NET-PATZ1 compared with the reference samples, and 156 significantly down-regulated genes (Supplementary Table 2, online resource). The PATZ1 gene itself is more highly expressed in these cases when compared with other representative pediatric HGG or LGG tumors (Fig. 3). Additional candidates of potential interest within the top upregulated genes included IGF2, PAX2 and GATA2, with known roles in brain stem cell biology, differentiation and development [18, 49, 53, 58], although these data will need confirming in a larger series. Looking in a supervised way at expression of particular differentiation markers commonly used for immunohistochemical analysis of glial/glioneuronal tumors, we found low GFAP expression, and modest levels of OLIG2, NeuN (RBFOX3), MAP2 and synaptophysin (SYP). Expression of CD34 and CSPG4 (NG2) is higher than in the other comparison groups, opening possible avenues for diagnostic IHC staining. MKI67 levels as a surrogate for proliferation were higher than in pilocytic astrocytoma (PA), but lower than the GBM subgroups (Fig. 3, Supplementary Fig. 3, online resource). In contrast to reports on EWSR1:PATZ1-fused sarcoma [3, 7, 27, 52], there was no appreciable or distinguishing expression of CD99 or desmin (Supplementary Table 2; Supplementary Fig. 3, online resource) or other muscular markers such as MYOD1, myogenin, CALD1, or SOX10 in the present series (not shown).
Fig. 3.
Differential expression analysis between NET-PATZ1 and a reference cohort of other glioma subtypes; IGF2, GATA2, PAX2 and PATZ1 and are more highly expressed in NET-PATZ1 cases when compared with representative pediatric HGG or LGG tumors, while CD34 and NG2 (CSPG4) could represent potential IHC staining markers for NET-PATZ1. Gene expression values are shown as TPM (transcripts per kilobase million). Where relevant, bars indicate median and 1st/3rd Quartile. H3.3 G34R glioblastoma, IDH wildtype, H3.3 G34 mutant; H3.3 K27M diffuse midline glioma H3 K27M mutant: pedGBM_MYCN, glioblastoma, IDH wildtype, subclass MYCN; pedRTKI, glioblastoma, IDH wildtype, subclass RTK I; pedRTKII, glioblastoma, IDH wildtype, subclass RTK II; PXA pleomorphic xanthoastrocytoma; PA_BRAF_Fus pilocytic astrocytoma with BRAF fusion
Copy number alterations of chromosome 22 are frequent in NET-PATZ1
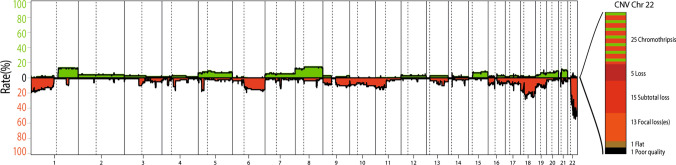
A summary of the copy number alterations identified in the combined cohort is given in Fig. 4. At the global level, the tumors are relatively ‘quiet’, with few recurrently altered regions. We excluded one tumor from the CNV analysis due to poor data quality (remaining n = 59). Chromosomal arms 1p (10/59, 17%), 1q (10/59, 17%) and 6q (10/59, 17%) and chromosome 18 (16/59, 27%) were most frequently affected at the level of broad changes. In comparison with data regarding PATZ1-fused sarcoma, the PATZ1-fused CNS tumors in this series were not enriched for homozygous deletions mapping to the CDKN2A/B locus (observed in only 2/59 cases, 3%) [3]. The most notable feature, however, is the presence of recurrent structural copy number variations on chromosome 22, seen in 98% of the cases (58/59). Among those alterations, the most frequently observed event was a chromothripsis-like, shattered pattern along chromosome 22 (25/59, 42%). Supplementary Fig. 2, online resource provides a closer look at the copy number variations observed on chromosome 22. These structural alterations are presumably driving the fusion formation through an intra-chromosomal arrangement, since EWSR1, MN1 and PATZ1 are all located on chromosome 22 (Fig. 2b). This high frequency of copy number alterations, including in cases where no RNA data were available, further supports the suggestion that PATZ1 fusion is likely a defining feature of this molecular tumor type.
Fig. 4.
Summary plot of copy number alterations in NET-PATZ1 and categorization of copy number alterations observed on chromosome 22, the most frequent being a dramatic shattering pattern (chromothripsis)
Clinical parameters and morphological aspects
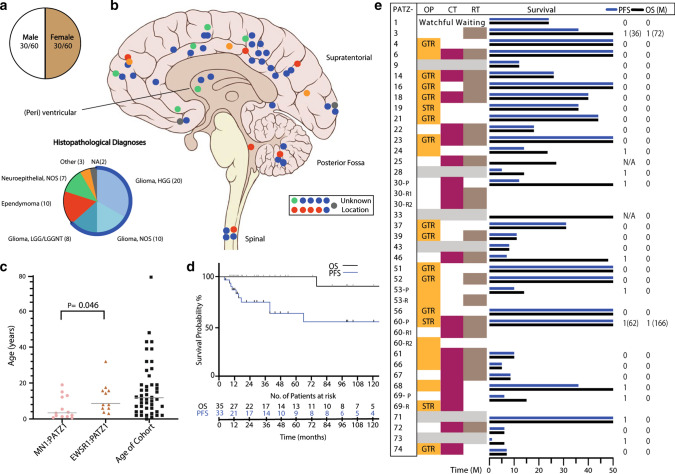
All clinical data available for our cohort are outlined in Fig. 5. More detailed descriptions are given in Supplementary Table 1, online resource. There is no sex-specific predominance observed in NET-PATZ1 (M:F = 1:1; Fig. 5a). Among the samples where the tumor location was known (n = 49), NET-PATZ1 most commonly showed a supratentorial manifestation (36/49 hemispheric, 4/49 peri- and intraventricular, total 40/49, 82%). Five of the 49 tumors (10%) were located in the posterior fossa, and 4/49 (8%) were spinal, including 2 metastatic lesions (Fig. 5b).
Fig. 5.
Clinical features of NET-PATZ1. a Patient sex distribution. b Distribution of tumor location. The institutional histopathological diagnoses of the series are also shown, representing a broad spectrum of mostly glial diagnoses. c Age distribution with the horizontal line representing median age of our cohort (11 years). MN1:PATZ1-fused tumors appear to be significantly enriched in younger ages (median = 3.5 years) vs EWSR1:PATZ1 (median = 8 years), (p value; 0.046, Student’s t test). d Clinical outcome in terms of OS and PFS of NET-PATZ1. e Overview of the different therapy protocols NET_PATZ1 patients within this cohort received. Where highlighted, the patients received that particular therapy modality, grey bars indicate unknown data. Different management as per primary vs relapse tumor is also shown. Note that some patients benefited from surgery (OP) alone. CT chemotherapy, RT radiation therapy, P primary, R (1/2) relapse 1 or relapse 2, GTR gross total resection, STR subtotal resection, 1 relapse/death, 0 censored. Detailed therapy protocols are listed in Supplementary Table 1, online resource
Figure 5c shows the age distribution for NET-PATZ1. The median age in our cohort was 11.0 years (range 0–80), with 74% of tumors occurring in patients under 18 years of age, indicating that NET-PATZ1 is primarily a childhood disease. Because some tumor samples were from recurrent tumors, and data about patient age could not always be confirmed to be the age at initial diagnosis, we analyzed the subgroup with definite age at primary diagnosis separately (n = 40), but this did not show any significant differences from the overall cohort (median age 9 years, p value; 0.42, Mann–Whitney U test). Interestingly, when looking deeper into cases with a confirmed fusion variant, MN1:PATZ1-fused tumors seem to manifest at a younger age (median = 3.5 years) vs EWSR1:PATZ1-fused tumors (median = 8 years; p value = 0.046, Student’s t test).
The original histopathological diagnoses of the cases described in this series were very diverse and variably polyphenotypic (Fig. 5b, Supplementary Table 1, online resource). The most common pathologic diagnoses described a high-grade astrocytic histology, including glioblastoma or anaplastic astrocytoma (high-grade glioma, HGG) in 20/58 annotated tumors (34%). Ten tumors (17%) had an institutional diagnosis of ependymal morphology, including two subependymomas. A further seven tumors (12%) had low-grade glial or glioneuronal diagnosis, and seven further tumors were broadly described as neuroepithelial. Across the morphological spectrum provided, five tumors were noted as additionally having a ‘sarcomatous’ or mesenchymal differentiation pattern on a glioneuronal/neuroepithelial background.
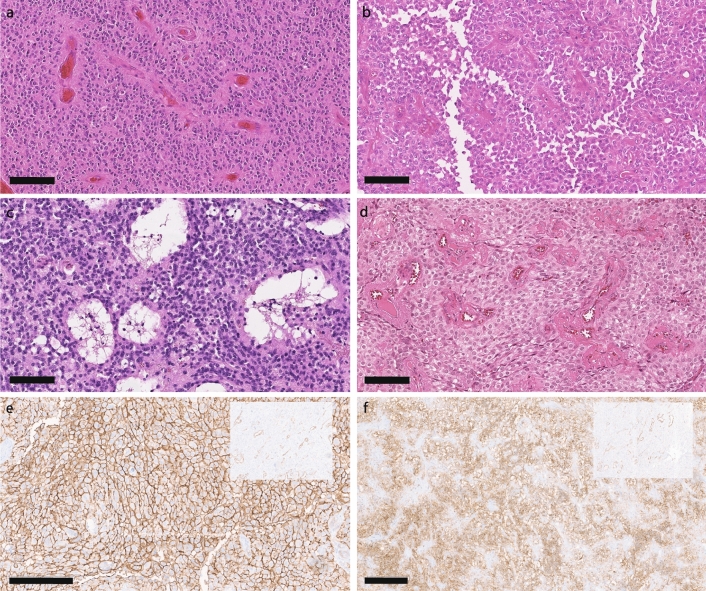
Given this very varied spectrum, we reassessed this novel tumor entity for any unifying histopathologic features. Figure 6 and supplementary Fig. 4, online resource show the extensively polyphenotypic morphology of NET-PATZ1. Histologically, hypercellularity was observed in almost all tumors (17/18, 94%). Nuclear morphology was sometimes, but not uniformly, irregular, with 8/18 tumors (57%) showing monomorphous nuclei with either large or small cells, arranged in clusters in two cases. Spindle-shaped cells were observed in 5/18 cases (28%). Mitoses were scarce for the centrally re-evaluated tumors, with only 1 tumor showing > 5 mitoses per 10 high-power fields (HPFs, one HPF = 0.238 mm2). However, most tumors (17/18, 94%) showed microvascular proliferation, with four tumor showing marked perivascular hyalinization. Necrosis was present in 33% of the cases (6/18), and was correlated with the presence of pseudorosettes and/or astroblastoma-like morphology. Details about our re-assessment are outlined in Table 2. A diffuse infiltration pattern was observed in 3/18 tumors (17%), with one tumor showing a cell cluster infiltration front. The clear biphasic and mesenchymal differentiation pattern of a subset of the tumors was also evident. However, within the five tumors showing sarcomatous or spindle cellular morphology, no correlation to a specific fusion partner (MN1/EWSR1) was observed.
Fig. 6.
Histology of NET-PATZ1 tumors. a PATZ1-012: perivascular pseudorosettes resembling ependymal morphology are observed. b PATZ1-040 showed an astroblastoma-like morphology. c PATZ1-025: high cellularity was consistently observed across almost all cases. Microcysts are here encircled by monomorphous nuclei. d PATZ1-039: in this tumor monomorphous nuclei with clear cell morphology along with perivascular pseudorosettes were encountered. e, f NG2 staining in PATZ-030 and PATZ-014, respectively, with negative control insets (IDH WT GBM). Thick scale bars represent 100 µm, thin scale bars represent 200 µm
Table 2.
Summary of the re-evaluated histopathologic findings for tumor samples with available material
| # | Detailed institutional histopathological diagnosis | Cell density | Cytoplasm and nuclei pleomorphism | Mitotic figures/10 HPF | Microvascular proliferation | Necrosis | Infiltration | Notes/fusion detected |
|---|---|---|---|---|---|---|---|---|
| 4 | Pleomorphic xanthoastrocytoma | High | Slightly pleomorphic, focally spindle cells | 0 | Hyalinized | No | EWSR1:PATZ1, perivascular pseudorosettes, biphasic differentiation | |
| 10 | Glioblastoma WHO grade 4 | High | Monomorphous nuclei, single giant cells | 2 | Few | No | Glial | |
| 12 | Anaplastic ependymoma WHO grade 3 | High | Monomorphous nuclei | 1 | Yes | Yes | Pseudorosettes | |
| 13 | N/A | Moderate | Monomorphous, small, arranged in groups/lobules | 0 | Hyalinized | No | Microcysts, biphasic differentiation | |
| 14 | Malignant NET with focal glial and sarcomatous differentiation | High | Small cell, partially spindle-shaped | 0 | No | No | MN1:PATZ1, biphasic differentiation | |
| 22 | Giant cell glioblastoma WHO grade 4 | High | Pleomorphic, small and large cells | 0 | Yes | No | MN1:PATZ1, biphasic differentiation | |
| 23 | Glioblastoma WHO grade 4 | High | Focally very pleomorphic, giant cells | 3 | Hyalinized | No | Diffuse | Focally spindle cells, biphasic differentiation |
| 24 | Low-grade glial/glioneuronal tumor | High | Pleomorphic, small and large cells | 0 | Yes | No | Diffuse | EWSR1:PATZ1 |
| 25 | Malignant neuroepithelial tumor | High | Monomorphous nuclei | 0 | Yes | No | EWSR1:PATZ1, microcysts | |
| 28 | Pleomorphic xanthoastrocytoma | High | Monomorphous, single giant cells, focally spindle cells | 0 | Few | No | Strong infiltration | MN1:PATZ1 |
| 30 | High-grade glioma | High | Pleomorphic, focally spindle-shaped, many apoptotic bodies | 8 | Yes | Yes | MN1:PATZ1, focally perivascular pseudorosettes, biphasic differentiation | |
| 39 | Anaplastic clear cell ependymoma WHO grade 2 | High | Monomorphous, clear cells | 0 | Yes | No | Ependymal, pseudorosettes | |
| 40 | Anaplastic ependymoma WHO grade 3 | High | Monomorphous nuclei | 0 | Yes | Yes | EWSR1:PATZ1, astroblastoma-like | |
| 46 | High-grade glioma | Moderate/high | Fibrillary, monomorphous nuclei | 0 | Yes | Yes | Diffuse | MN1:PATZ1, pseudorosettes, astroblastoma-like |
| 54 | Glioblastoma WHO grade 4 | High | Monomorphous, small clusters of cells | 1 | Yes | No | Clusters | Infiltration in small clusters of cells |
| 55 | N/A | High | Moderate nuclear pleomorphism | 1 | Yes | Yes | MN1:PATZ1, pseudorosettes | |
| 56 | Glioblastoma WHO grade 4; Gliosarcoma | High | Spindle-shaped cells | 0 | Yes | No | Sarcomatous differentiation | |
| 71 | Glioblastoma WHO grade 4 | High | Slightly pleomorphic, focally clear cells, prominent nucleoli | 5 | Hyalinized | Yes | EWSR1:PATZ1, perivascular pseudorosettes, focally spindle cells, biphasic differentiation |
Details about immunohistochemistry is listed in Supplementary Table 1
N/A not applicable
Insufficient unstained sections were available to perform a comprehensive immunohistochemical analysis for most cases. The limited available data suggest variable staining for OLIG2 and GFAP, similar to the gene expression data, which also outlined the potential role of NG2 for future investigation (Fig. 6e, f). We exploited this finding and found specific positive staining in all cases prospectively evaluated where material was available (n = 4/4). Immunoprofiling data from previously reported PATZ1-fused CNS tumors included in this series are summarized in Supplementary Table 1, online resource.
Overall, given the absence of clear uniform histopathological or immunophenotypic features to classify the tumors as being of glial/ependymal versus neural origin, we provisionally propose the non-specific term ‘neuroepithelial tumor’ (NET) within the NET-PATZ1 nomenclature. We anticipate that in time, this unspecific term may be replaced by a clearer description if new evidence on cellular origins emerges.
Patient outcomes may suggest an intermediate malignancy grade
Follow-up data were obtained for 35 patients, as listed in Supplementary Table 1, online resource, with a median follow-up period of 31 months (range 6–288 months). Eleven of the patients experienced a relapse during the follow-up period, including two patients with multiple relapses and spinal metastasis (PATZ-030, PATZ-068). Median PFS was 144 months. Only two of the patients died of their disease during the follow-up period, at 72 and 166 months after diagnosis (having relapsed at 36 and 62 months, respectively) (Fig. 5d). This clinical course is broadly compatible with the moderate MKI67 (the gene encoding Ki-67) expression seen in the RNA data, and the mostly rare mitoses observed histologically. The limited follow-up data and the heterogeneity of treatments applied (Fig. 5e, Supplementary Table 1, online resource) preclude a definitive statement regarding the malignancy grade of this group of tumors, or a precise estimate of their overall survival rates or median OS. Our initial data, however, suggest an intermediate grade and notably better outcome than expected for true ‘high-grade’ tumors—important given that a third of cases initially received a diagnosis of HGG. Also, it is notable that a subset of patients maintained a stable disease despite being treated with surgery alone.
Drug screening identifies potential candidates for future treatments
Screening of cell line molecular data in the Broad Institute’s Cancer Dependency Map portal (DepMap, https://depmap.org/portal/) revealed a CNS tumor cell line harboring an MN1:PATZ1 fusion. This line, KS-1, is derived from a 45-year-old female patient diagnosed with a glioblastoma. The line grows both adherently or in neurosphere conditions, and confirmatory RNA sequencing of the line validated the presence of the fusion. Although this long-term in vitro culture certainly will not recapitulate all features of the primary tumors, we nevertheless performed an initial drug screening to obtain some possible first hints at future therapeutic options. The drug screen was performed using the Hopp Children’s Cancer Center, Heidelberg (KiTZ) Translational Drug Screening Unit (TDSU) library. This library comprises 74 approved cancer drugs targeting a variety of different pathways. The output chemiluminescence intensities were fed into the web-based BREEZE drug screen analysis pipeline (https://breeze.fimm.fi); [34]), which calculates a drug-specific sensitivity (DSS) score for each drug [56]. DSS scores were also compared to the chemostatic effect on normal fetal astrocytes, to assess tumor specificity. Data analysis yielded five drugs that emerged as potential candidates due to their superior efficacy in tumor cells: paclitaxel, d-actinomycin, volasertib (PLK1 inhibitor), navitoclax (BCL-2/BCL-XL/BCL-w inhibitor) and I-BET-151 (a Bromodomain family inhibitor) (Supplementary Fig. 5a, online resource). These drugs, particularly paclitaxel, D-actinomycin and volasertib, showed similar effects when subsequently validated individually in a small scale tissue culture setting (Supplementary Fig. 5b, online resource), and may thus represent a starting point for future preclinical studies in this tumor type.
Discussion
Recent reports on a small number of pediatric brain tumors showing PATZ1 fusions, as well as the detection of further such cases through our ongoing molecular diagnostic studies, prompted us to investigate the properties of these cases in more detail. In this study, we identified a distinct DNA methylation-based cluster of 60 tumor samples, within which all tumor samples where RNA sequencing was conducted (n = 27) were found to harbour a PATZ1 fusion. We, therefore, believe it to be likely that these MN1:PATZ1 and EWSR1:PATZ1 fusions are pathognomonic for this novel tumor type.
Diagnosis and origins of NET-PATZ1
Our findings suggest that when integrating morphological aspects and molecular analysis, unusual glial, ependymal or glioneuronal histology along with clustered, focal chr22 alterations may together give diagnostic hints for NET-PATZ1. As with the sarcomas in which EWSR1:PATZ1 has been reported, the morphological variety, inconclusive immunoprofile and wide range of age at presentation are striking features that may account for the fact that these tumors have not been identified as a clear entity until now. When RNA sequencing cannot be performed to confirm presence of the fusion, methylation analysis and derived copy number data may be exploited as an alternative to establish the diagnosis of NET-PATZ1.
Exploring the differential diagnostic spectrum of this observation, the recently described HGNET-MN1 tumor type, which is characterized by alternative MN1 fusions (mostly MN1:BEND2 and MN1:CXXC5), manifests more frequently with an embryonal/astroblastoma histology and shows a clearly different DNA methylation profile [46]. The majority of NET-PATZ1 tumors presented with a histology extending along the glial or glioneuronal spectrum, but the scarce tissue available for immunohistochemistry together with marker gene expression analysis through RNA sequencing was inconclusive as to a clear lineage or distinct marker. Taken together with the mesenchymal phenotype noted in a subset of the cases in our cohort, we, therefore, advocate to provisionally call this tumor entity ‘neuroepithelial’ until a more concrete assessment of its likely origins can be made.
The observed overexpression of IGF2, PAX2 and GATA2 in NET-PATZ1 is also intriguing with respect to possible developmental origins and potential targets for therapeutic intervention. For example, GATA2 has roles in fate determination of neural progenitors into midbrain GABAergic neurons [17], while PAX2 is linked with development of the midbrain–hindbrain boundary and of GABAergic interneurons [11]. IGF2 has multiple roles in brain development and disease, as reviewed for example in [2, 22]. GATA2 has been reported to directly upregulate IGF2 in chemo-resistant prostate cancer [51], while IGF2 activity leads to PAX2 overexpression in Wilms’ tumor [13], suggesting a possible signalling axis in NET-PATZ1. Of particular interest will be a future comparison with PATZ1-fused tumors occurring in non-CNS sites, to investigate the possibility of a common neuroectodermal/neural crest progenitor cell that may also be of relevance in the formation of the previously described sarcomas (although markers of EWSR1:PATZ1 sarcomas such as CD99, desmin and myogenin were not found to be expressed in the CNS tumors). The histology and immunoprofile of EWSR1:PATZ1-fused sarcoma has also been described as incredibly polyphenotypic, but MN1:PATZ1-fused sarcomas have not yet been described.
An initial look at the survival data, with an average follow-up period of 31 months, indicates that NET-PATZ1 patients have a better overall survival than high-grade tumors (median OS not reached), despite experiencing several relapses (median PFS 144 months). This should prompt questioning of the nature of the GBM diagnoses in a subset of the patients included and could explain the observed survival pattern. This finding has consequences related to the aggressiveness and nature of the future therapy that is suitable for patients with tumors driven by this biology. However, more information is obviously needed to fully assess the malignancy of this tumor entity, particularly in the context of a uniform treatment protocol. Future retro- and prospective studies will, therefore, be critical for determining optimal clinical management.
Potential oncogenic mechanisms and the role of PATZ1
PATZ1 (POZ/BTB and AT Hook Containing Zinc Finger 1) is a transcription factor belonging to the POZ/BTB (Pox virus and Zinc finger/Broad-complex, tramtrack, and bric-à-brac) family and containing an AT-hook zinc finger domain [44]. It has been described as an important node as part of a network of transcription factors that maintain the “stemness” of embryonic stem cells [30], and a regulator of cellular reprogramming by inhibiting Pou5f1, depending on its expression levels. It has thus been assigned as both an activator and repressor of transcription, depending on the cellular context [25, 50].
MN1 and EWSR1 are both involved in fusions in other tumor entities. As described above, MN1 alteration is one of the molecular hallmarks of the recently described HGNET-MN1 tumor subentity, while EWSR1 has long been known to be involved in Ewing sarcoma, even though the exact factors underlying its oncogenicity remain to be fully understood [21]. EWSR1:PATZ1 and similar oncogenic EWSR1 fusions have been shown to cause globally altered transcriptional signatures [21, 52], and other EWSR1 fusions have been detected in primary intra-axial tumors [23]. Both MN1 and EWSR1 have transcriptional activator activity [12, 29].
In the fusion described here, the transactivating domain of MN1/EWSR1 is retained, and fused to the zinc finger domain of PATZ1. It, therefore, seems likely that the downstream consequences of the fusion are determined by the aberrant recruitment of a transactivating domain at binding sites determined by the PATZ1 binding domain, perhaps together with upregulated expression via the MN1/EWSR1 promoter, as seen, for example, with EWSR1:FLI1 fusions in Ewing sarcoma [9, 12]. The exact details of the oncogenic mechanisms of MN1:PATZ1 and EWSR1:PATZ1, however, are yet to be fully established and understood. Further studies will be needed to reveal the exact role of the fusion partners and the subsequent downstream effects in terms of global transcriptional deregulation.
Summary
In conclusion, we describe here ‘neuroepithelial tumor with PATZ1 fusion’ (NET-PATZ1)—a novel, molecularly distinct CNS tumor type with strikingly variable histopathologic morphology and heterogeneous multiphasic differentiation patterns. Being the sole molecular finding constant across all tumors analyzed, we postulate that the PATZ1 fusions are a key driver of tumor initiation. Preliminary indications suggest an intermediate prognosis, although further studies will be needed to confirm this and to investigate the detailed biology, cellular origins, treatment sensitivity and clinical course of these tumors. The recognition of NET-PATZ1 as a defined tumor type, however, is an important step towards conducting such analyses, and will hopefully provide a foundation for optimized clinical management in future.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file3. Supplementary Fig. 1 t-distributed stochastic neighbor embedding (tSNE) visualization of DNA methylation patterns for our in house cohort including more than 80,000 bulk tumor samples, the platform on which mnp is based (https://www.molecularneuropathology.org/mnp). NET_PATZ1 cluster close to a variety of glial tumors but form a distinct ‘island’, representing a distinct molecular tumor type (TIF 27774 kb)
Supplementary file4. Supplementary Fig. 2 Copy number status observed on chromosome 22 for the two selected cases in Fig. 2. Top panel: B-Allele frequency (BAF) in the tumor at SNP positions which are heterozygous in the germline. 2nd, 3rd and bottom panel: Rescaled tumor: germline coverage ratio, indicating copy-number gains or losses; tumor and germline coverage. MN1, EWSR1 and PATZ1 loci are indicated (TIF 26046 kb)
Supplementary file5. Supplementary Fig. 3 Expression of particular differentiation/proliferation markers: GFAP, MAP2, CD99, Desmin (DES) NeuN (RBFOX3): low, OLIG2 and Synaptophysin (SYP): modest. MKI67 levels were higher than in PA, but lower than the GBM subgroups (similar to PXA). Gene expression is shown as TPM (transcripts per kilobase million). H3.3 G34R, glioblastoma, IDH wildtype, H3.3 G34 mutant; H3.3 K27M, diffuse midline glioma H3 K27M mutant: pedGBM_MYCN, glioblastoma, IDH wildtype, subclass MYCN; pedRTKI, glioblastoma, IDH wildtype, subclass RTK I; pedRTKII, glioblastoma, IDH wildtype, subclass RTK II; PXA pleomorphic xanthoastrocytoma; PA_BRAF_Fus pilocytic astrocytoma with BRAF fusion (TIF 14413 kb)
Supplementary file6. Supplementary Fig. 4 a PATZ1-013: marked perivascular hyalinization and microcysts are seen together with monomorphous nuclei and small cells arranged in lobules. b PATZ1-056 displayed spindle-shaped cells with strong resemblance to a mesenchymal phenotype. c Immunohistochemistry of available sections for PATZ1-056 revealed very sparse staining with antibodies against GFAP and negative staining for Olig2, synaptophysin and MAP2, in keeping with the gene expression-based analysis. Scale bars represent 100 µm. Supplementary Table 1 includes more information about the staining patterns seen in NET_PATZ1 (TIF 39143 kb)
Supplementary file7. Supplementary Fig. 5 a Heatmap of the primary drug screen conducted on the KS-1 cell line. Drugs are ordered according to the drug specific sensitivity score (DSS) of the KS-1 cells. Area under the curve for drug efficacy (AUC) and IC50 values are also shown. Five potentially interesting candidate hits were selected based on high DSS and superior efficacy in KS-1 compared with astrocytes. b The five candidate hits were tested again on the KS-1 cells with a greater range of concentrations to confirm efficacy and obtain a more precise IC50 value. X indicates that the IC50 value measured for fetal astrocytes is higher than the highest concentration of drug applied (drug cytotoxic effect was not completely reached)(XLSX 9176 kb)
Acknowledgements
For technical support and expertise, we thank the German Cancer Research Centre (DKFZ) Genomics and Proteomics Core Facility, and Hai-Yen Nguyen, Laura Doerner & Moritz Leon Schalles (Department of Neuropathology, Institute of Pathology at the University Hospital Heidelberg). We gratefully acknowledge Teresa de Rojas for support in collecting clinical metadata. This work was supported by the German Childhood Cancer Foundation (“Neuropath 2.0—Increasing diagnostic accuracy in pediatric neurooncology”; DKS 2015.01), the Everest Centre for Low-Grade Paediatric Brain Tumour Research (The Brain Tumour Charity, UK; GN-000382), the German Federal Ministry of Education and Research (BMBF), and Cancéropôle Lyon Auvergne Rhône-Alpes (CLARA). DNA methylation profiling at NYU was in part supported by grants from the Friedberg Charitable Foundation, the Sohn Conference Foundation and the Making Headway Foundation (to M. Snuderl.)
Funding
Open Access funding enabled and organized by Projekt DEAL.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Felix Sahm and David T. W. Jones have contributed equally.
References
- 1.Alvarez-Breckenridge C, Miller JJ, Nayyar N, Gill CM, Kaneb A, D’Andrea M, et al. Clinical and radiographic response following targeting of BCAN-NTRK1 fusion in glioneuronal tumor. NPJ Precis Oncol. 2017;1:5. doi: 10.1038/s41698-017-0009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benarroch EE. Insulin-like growth factors in the brain and their potential clinical implications. Neurology. 2012;79:2148–2153. doi: 10.1212/WNL.0b013e3182752eef. [DOI] [PubMed] [Google Scholar]
- 3.Bridge JA, Sumegi J, Druta M, Bui MM, Henderson-Jackson E, Linos K, et al. Clinical, pathological, and genomic features of EWSR1-PATZ1 fusion sarcoma. Mod Pathol. 2019;32:1593–1604. doi: 10.1038/s41379-019-0301-1. [DOI] [PubMed] [Google Scholar]
- 4.Burel-Vandenbos F, Pierron G, Thomas C, Reynaud S, Gregoire V, Duhil de Benaze G, et al. A polyphenotypic malignant paediatric brain tumour presenting a MN1-PATZ1 fusion, no epigenetic similarities with CNS High-Grade Neuroepithelial Tumour with MN1 Alteration (CNS HGNET-MN1) and related to PATZ1-fused sarcomas. Neuropathol Appl Neurobiol. 2020 doi: 10.1111/nan.12626. [DOI] [PubMed] [Google Scholar]
- 5.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chadda KR, Holland K, Scoffings D, Dean A, Pickles JC, Behjati S, et al. A rare case of paediatric astroblastoma with concomitant MN1-GTSE1 and EWSR1-PATZ1 gene fusions altering management. Neuropathol Appl Neurobiol. 2021 doi: 10.1111/nan.12701. [DOI] [PubMed] [Google Scholar]
- 7.Chougule A, Taylor MS, Nardi V, Chebib I, Cote GM, Choy E, et al. Spindle and round cell sarcoma with EWSR1-PATZ1 gene fusion: a sarcoma with polyphenotypic differentiation. Am J Surg Pathol. 2019;43:220–228. doi: 10.1097/PAS.0000000000001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke M, Mackay A, Ismer B, Pickles JC, Tatevossian RG, Newman S, et al. Infant high-grade gliomas comprise multiple subgroups characterized by novel targetable gene fusions and favorable outcomes. Cancer Discov. 2020;10:942–963. doi: 10.1158/2159-8290.CD-19-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 10.Deng MY, Sill M, Chiang J, Schittenhelm J, Ebinger M, Schuhmann MU, et al. Molecularly defined diffuse leptomeningeal glioneuronal tumor (DLGNT) comprises two subgroups with distinct clinical and genetic features. Acta Neuropathol. 2018;136:239–253. doi: 10.1007/s00401-018-1865-4. [DOI] [PubMed] [Google Scholar]
- 11.Goode DK, Elgar G. The PAX258 gene subfamily: a comparative perspective. Dev Dyn. 2009;238:2951–2974. doi: 10.1002/dvdy.22146. [DOI] [PubMed] [Google Scholar]
- 12.Grünewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Álava E, Kovar H, et al. Ewing sarcoma. Nat Rev Dis Primers. 2018;4:5. doi: 10.1038/s41572-018-0003-x. [DOI] [PubMed] [Google Scholar]
- 13.Hu Q, Gao F, Tian W, Ruteshouser EC, Wang Y, Lazar A, et al. Wt1 ablation and Igf2 upregulation in mice result in Wilms tumors with elevated ERK1/2 phosphorylation. J Clin Investig. 2011;121:174–183. doi: 10.1172/JCI43772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson A, Severson E, Gay L, Vergilio JA, Elvin J, Suh J, et al. Comprehensive genomic profiling of 282 pediatric low- and high-grade gliomas reveals genomic drivers, tumor mutational burden, and hypermutation signatures. Oncologist. 2017;22:1478–1490. doi: 10.1634/theoncologist.2017-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones C, Baker SJ. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer. 2014 doi: 10.1038/nrc3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones DTW, Bandopadhayay P, Jabado N. The power of human cancer genetics as revealed by low-grade gliomas. Annu Rev Genet. 2019;53:483–503. doi: 10.1146/annurev-genet-120417-031642. [DOI] [PubMed] [Google Scholar]
- 17.Kala K, Haugas M, Lillevali K, Guimera J, Wurst W, Salminen M, et al. Gata2 is a tissue-specific post-mitotic selector gene for midbrain GABAergic neurons. Development. 2009;136:253–262. doi: 10.1242/dev.029900. [DOI] [PubMed] [Google Scholar]
- 18.Kala K, Haugas M, Lilleväli K, Guimera J, Wurst W, Salminen M, et al. Gata2 is a tissue-specific post-mitotic selector gene for midbrain GABAergic neurons. Development. 2009;136:253–262. doi: 10.1242/dev.029900. [DOI] [PubMed] [Google Scholar]
- 19.Korshunov A, Ryzhova M, Hovestadt V, Bender S, Sturm D, Capper D, et al. Integrated analysis of pediatric glioblastoma reveals a subset of biologically favorable tumors with associated molecular prognostic markers. Acta Neuropathol. 2015;129:669–678. doi: 10.1007/s00401-015-1405-4. [DOI] [PubMed] [Google Scholar]
- 20.Korshunov A, Schrimpf D, Ryzhova M, Sturm D, Chavez L, Hovestadt V, et al. H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol. 2017;134:507–516. doi: 10.1007/s00401-017-1710-1. [DOI] [PubMed] [Google Scholar]
- 21.Kovar H, Amatruda J, Brunet E, Burdach S, Cidre-Aranaz F, de Alava E, et al. The second European interdisciplinary Ewing sarcoma research summit–a joint effort to deconstructing the multiple layers of a complex disease. Oncotarget. 2016;7:8613–8624. doi: 10.18632/oncotarget.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Speder P, Brand AH. Control of brain development and homeostasis by local and systemic insulin signalling. Diabetes Obes Metab. 2014;16(Suppl 1):16–20. doi: 10.1111/dom.12337. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Nunez O, Cafferata B, Santi M, Ranganathan S, Pearce TM, Kulich SM, et al. The spectrum of rare central nervous system (CNS) tumors with EWSR1-non-ETS fusions: experience from three pediatric institutions with review of the literature. Brain Pathol. 2020 doi: 10.1111/bpa.12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 25.Ma H, Ow JR, Tan BCP, Goh Z, Feng B, Loh YH, et al. The dosage of Patz1 modulates reprogramming process. Sci Rep. 2014;4:7519. doi: 10.1038/srep07519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR, et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32(520–537):e525. doi: 10.1016/j.ccell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mastrangelo T, Modena P, Tornielli S, Bullrich F, Testi MA, Mezzelani A, et al. A novel zinc finger gene is fused to EWS in small round cell tumor. Oncogene. 2000;19:3799–3804. doi: 10.1038/sj.onc.1203762. [DOI] [PubMed] [Google Scholar]
- 28.Michal M, Rubin BP, Agaimy A, Kosemehmetoglu K, Rudzinski ER, Linos K, et al. EWSR1-PATZ1-rearranged sarcoma: a report of nine cases of spindle and round cell neoplasms with predilection for thoracoabdominal soft tissues and frequent expression of neural and skeletal muscle markers. Mod Pathol. 2020 doi: 10.1038/s41379-020-00684-8. [DOI] [PubMed] [Google Scholar]
- 29.Miyake N, Takahashi H, Nakamura K, Isidor B, Hiraki Y, Koshimizu E, et al. Gain-of-function MN1 truncation variants cause a recognizable syndrome with craniofacial and brain abnormalities. Am J Hum Genet. 2020;106:13–25. doi: 10.1016/j.ajhg.2019.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ow JR, Ma H, Jean A, Goh Z, Lee YH, Chong YM, et al. Patz1 regulates embryonic stem cell identity. Stem Cells Dev. 2014;23:1062–1073. doi: 10.1089/scd.2013.0430. [DOI] [PubMed] [Google Scholar]
- 31.Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27:728–743. doi: 10.1016/j.ccell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paugh BS, Zhu X, Qu C, Endersby R, Diaz AK, Zhang J, et al. Novel oncogenic PDGFRA mutations in pediatric high-grade gliomas. Cancer Res. 2013;73:6219–6229. doi: 10.1158/0008-5472.CAN-13-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pei J, Zhao X, Patchefsky AS, Flieder DB, Talarchek JN, Testa JR, et al. Clinical application of RNA sequencing in sarcoma diagnosis: an institutional experience. Medicine. 2019;98:e16031. doi: 10.1097/md.0000000000016031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Potdar S, Ianevski A, Mpindi JP, Bychkov D, Fiere C, Ianevski P, et al. Breeze: an integrated quality control and data analysis application for high-throughput drug screening. Bioinformatics. 2020;36:3602–3604. doi: 10.1093/bioinformatics/btaa138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qaddoumi I, Orisme W, Wen J, Santiago T, Gupta K, Dalton JD, et al. Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol. 2016;131:833–845. doi: 10.1007/s00401-016-1539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramkissoon LA, Horowitz PM, Craig JM, Ramkissoon SH, Rich BE, Schumacher SE, et al. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc Natl Acad Sci USA. 2013;110:8188–8193. doi: 10.1073/pnas.1300252110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossi S, Barresi S, Giovannoni I, Alesi V, Ciolfi A, Stefania Colafati G, et al. Expanding the spectrum of EWSR1-PATZ1 rearranged CNS tumors: an infantile case with leptomeningeal dissemination. Brain Pathol. 2020 doi: 10.1111/bpa.12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahm F, Schrimpf D, Jones DT, Meyer J, Kratz A, Reuss D, et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 2016;131:903–910. doi: 10.1007/s00401-015-1519-8. [DOI] [PubMed] [Google Scholar]
- 40.Sahm F, Schrimpf D, Stichel D, Jones DTW, Hielscher T, Schefzyk S, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;18:682–694. doi: 10.1016/s1470-2045(17)30155-9. [DOI] [PubMed] [Google Scholar]
- 41.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 42.Siegfried A, Rousseau A, Maurage CA, Pericart S, Nicaise Y, Escudie F, et al. EWSR1-PATZ1 gene fusion may define a new glioneuronal tumor entity. Brain Pathol. 2019;29:53–62. doi: 10.1111/bpa.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sievers P, Appay R, Schrimpf D, Stichel D, Reuss DE, Wefers AK, et al. Rosette-forming glioneuronal tumors share a distinct DNA methylation profile and mutations in FGFR1, with recurrent co-mutation of PIK3CA and NF1. Acta Neuropathol. 2019;138:497–504. doi: 10.1007/s00401-019-02038-4. [DOI] [PubMed] [Google Scholar]
- 44.Siggs O, Beutler B. The BTB-ZF transcription factors. Cell Cycle. 2012;11:3358–3369. doi: 10.4161/cc.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stichel D, Schrimpf D, Casalini B, Meyer J, Wefers AK, Sievers P, et al. Routine RNA sequencing of formalin-fixed paraffin-embedded specimens in neuropathology diagnostics identifies diagnostically and therapeutically relevant gene fusions. Acta Neuropathol. 2019;138:827–835. doi: 10.1007/s00401-019-02039-3. [DOI] [PubMed] [Google Scholar]
- 46.Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D, et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164:1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsuda Y, Zhang L, Meyers P, Tap WD, Healey JH, Antonescu CR. The clinical heterogeneity of round cell sarcomas with EWSR1/FUS gene fusions: impact of gene fusion type on clinical features and outcome. Genes Chromosomes Cancer. 2020;59:525–534. doi: 10.1002/gcc.22857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urbánek P, Fetka I, Meisler MH, Busslinger M. Cooperation of Pax2 and Pax5 in midbrain and cerebellum development. Proc Natl Acad Sci USA. 1997;94:5703–5708. doi: 10.1073/pnas.94.11.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valentino T, Palmieri D, Vitiello M, Pierantoni GM, Fusco A, Fedele M. PATZ1 interacts with p53 and regulates expression of p53-target genes enhancing apoptosis or cell survival based on the cellular context. Cell Death Dis. 2013;4:e963–e963. doi: 10.1038/cddis.2013.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vidal SJ, Rodriguez-Bravo V, Quinn SA, Rodriguez-Barrueco R, Lujambio A, Williams E, et al. A targetable GATA2-IGF2 axis confers aggressiveness in lethal prostate cancer. Cancer Cell. 2015;27:223–239. doi: 10.1016/j.ccell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watson S, Perrin V, Guillemot D, Reynaud S, Coindre JM, Karanian M, et al. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J Pathol. 2018;245:29–40. doi: 10.1002/path.5053. [DOI] [PubMed] [Google Scholar]
- 53.Willett RT, Greene LA. Gata2 is required for migration and differentiation of retinorecipient neurons in the superior colliculus. J Neurosci. 2011;31:4444–4455. doi: 10.1523/jneurosci.4616-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Worst BC, van Tilburg CM, Balasubramanian GP, Fiesel P, Witt R, Freitag A, et al. Next-generation personalised medicine for high-risk paediatric cancer patients - the INFORM pilot study. Eur J Cancer. 2016;65:91–101. doi: 10.1016/j.ejca.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 55.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yadav B, Pemovska T, Szwajda A, Kulesskiy E, Kontro M, Karjalainen R, et al. Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci Rep. 2014;4:5193. doi: 10.1038/srep05193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45:602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziegler AN, Feng Q, Chidambaram S, Testai JM, Kumari E, Rothbard DE, et al. Insulin-like Growth Factor II: an essential adult stem cell niche constituent in brain and intestine. Stem Cell Rep. 2019;12:816–830. doi: 10.1016/j.stemcr.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file3. Supplementary Fig. 1 t-distributed stochastic neighbor embedding (tSNE) visualization of DNA methylation patterns for our in house cohort including more than 80,000 bulk tumor samples, the platform on which mnp is based (https://www.molecularneuropathology.org/mnp). NET_PATZ1 cluster close to a variety of glial tumors but form a distinct ‘island’, representing a distinct molecular tumor type (TIF 27774 kb)
Supplementary file4. Supplementary Fig. 2 Copy number status observed on chromosome 22 for the two selected cases in Fig. 2. Top panel: B-Allele frequency (BAF) in the tumor at SNP positions which are heterozygous in the germline. 2nd, 3rd and bottom panel: Rescaled tumor: germline coverage ratio, indicating copy-number gains or losses; tumor and germline coverage. MN1, EWSR1 and PATZ1 loci are indicated (TIF 26046 kb)
Supplementary file5. Supplementary Fig. 3 Expression of particular differentiation/proliferation markers: GFAP, MAP2, CD99, Desmin (DES) NeuN (RBFOX3): low, OLIG2 and Synaptophysin (SYP): modest. MKI67 levels were higher than in PA, but lower than the GBM subgroups (similar to PXA). Gene expression is shown as TPM (transcripts per kilobase million). H3.3 G34R, glioblastoma, IDH wildtype, H3.3 G34 mutant; H3.3 K27M, diffuse midline glioma H3 K27M mutant: pedGBM_MYCN, glioblastoma, IDH wildtype, subclass MYCN; pedRTKI, glioblastoma, IDH wildtype, subclass RTK I; pedRTKII, glioblastoma, IDH wildtype, subclass RTK II; PXA pleomorphic xanthoastrocytoma; PA_BRAF_Fus pilocytic astrocytoma with BRAF fusion (TIF 14413 kb)
Supplementary file6. Supplementary Fig. 4 a PATZ1-013: marked perivascular hyalinization and microcysts are seen together with monomorphous nuclei and small cells arranged in lobules. b PATZ1-056 displayed spindle-shaped cells with strong resemblance to a mesenchymal phenotype. c Immunohistochemistry of available sections for PATZ1-056 revealed very sparse staining with antibodies against GFAP and negative staining for Olig2, synaptophysin and MAP2, in keeping with the gene expression-based analysis. Scale bars represent 100 µm. Supplementary Table 1 includes more information about the staining patterns seen in NET_PATZ1 (TIF 39143 kb)
Supplementary file7. Supplementary Fig. 5 a Heatmap of the primary drug screen conducted on the KS-1 cell line. Drugs are ordered according to the drug specific sensitivity score (DSS) of the KS-1 cells. Area under the curve for drug efficacy (AUC) and IC50 values are also shown. Five potentially interesting candidate hits were selected based on high DSS and superior efficacy in KS-1 compared with astrocytes. b The five candidate hits were tested again on the KS-1 cells with a greater range of concentrations to confirm efficacy and obtain a more precise IC50 value. X indicates that the IC50 value measured for fetal astrocytes is higher than the highest concentration of drug applied (drug cytotoxic effect was not completely reached)(XLSX 9176 kb)