Abstract
Activation of transcription can occur by the facilitated recruitment of TFIID to promoters by gene-specific activators. To investigate the role of TFIIA in TFIID recruitment in vivo, we exploited a class of yeast TATA-binding protein (TBP) mutants that is activation and DNA binding defective. We found that co-overexpression of TOA1 and TOA2, the genes that encode yeast TFIIA, overcomes the activation defects caused by the TBP mutants. Using a genetic screen, we isolated a new class of TFIIA mutants and identified three regions on TFIIA that are likely to be involved in TBP recruitment or stabilization of the TBP-TATA complex in vivo. Amino acid replacements in only one of these regions enhance TFIIA-TBP-DNA complex formation in vitro, suggesting that the other regions are involved in regulatory interactions. To determine the relative importance of TFIIA in the regulation of different genes, we constructed yeast strains to conditionally deplete TFIIA levels prior to gene activation. While the activation of certain genes, such as INO1, was dramatically impaired by TFIIA depletion, activation of other genes, such as CUP1, was unaffected. These data suggest that TFIIA facilitates DNA binding by TBP in vivo, that TFIIA may be regulated by factors that target distinct regions of the protein, and that promoters vary significantly in the degree to which they require TFIIA for activation.
Initiation of mRNA synthesis in eukaryotes depends upon the controlled and coordinated activities of a large number of proteins. RNA polymerase II and six different general transcription factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) assemble into a transcriptionally competent preinitiation complex (PIC) at the core promoter elements of class II genes (47). Among the general transcription factors, TFIID plays a central role in initiation as it binds specifically to the TATA box through its component TATA-binding protein (TBP) and nucleates PIC assembly (47). Gene-specific transcriptional regulators typically bind upstream of the TATA box and, in concert with coactivators and corepressors, modulate the level of initiation by affecting the assembly or activity of the PIC. Although any step in the initiation reaction potentially may be regulated by the gene-specific factors, a growing body of data suggests that some transcriptional activators enhance the rate of initiation by recruiting TFIID to the core promoter or by recruiting the RNA polymerase II holoenzyme to the nascent PIC (20, 54, 65).
The importance of the TFIID recruitment step in gene regulation is supported by both genetic and biochemical studies. In the yeast Saccharomyces cerevisiae, the rate-limiting association of TBP with the TATA box can be accelerated by the acidic activator Gcn4 (32), and artificial recruitment of TBP via a heterologous DNA-binding domain can bypass the requirement for upstream activators (8, 31, 75). Two independent genetic screens for TBP mutants that exhibit transcription activation defects in vivo uncovered mutants with significantly reduced affinity for DNA in vitro (3, 37). More recently, analysis of transcription initiation and PIC assembly on immobilized DNA templates in yeast nuclear extracts showed that two different acidic activators can facilitate TFIID recruitment (56). In agreement with these studies of yeast, biochemical studies performed with purified mammalian and viral transcription factors have demonstrated activator-mediated enhancement of TFIID binding to TATA elements (38, 71) in a manner that leads to a stable conformation change in the developing PIC (9). Activator function in these assays is critically dependent upon the presence of TFIIA and the TBP-associated factors (TAFs), which, together with TBP, comprise TFIID.
TFIIA has multiple roles in RNA polymerase II transcription. In agreement with its description as an RNA polymerase II general transcription factor, TFIIA is required for basal transcription in reaction mixtures containing TFIID (12, 51, 69, 76). However, TFIIA does not stimulate basal transcription when TFIID is replaced with TBP (41, 59, 69), even though TFIIA can increase the affinity of TBP for TATA elements in vitro (25, 36, 72, 77). In addition to its involvement in basal transcription, TFIIA has an important role in the regulation of transcription. Together with a number of activators and coactivators, TFIIA stimulates formation of the TFIID-TATA complex, perhaps by forming a molecular bridge between the regulatory proteins and TFIID (27, 33, 38, 61, 62, 71). Consistent with this idea, TFIIA can interact directly with certain activators (33, 51) and with a TAF (76). Moreover, crystallographic studies have shown that TFIIA binds to TBP and contacts DNA upstream of the TATA box (16, 70). Finally, a genetic screen of yeast identified a TBP mutant that is impaired in its response to acidic activators and defective for its interaction with TFIIA (66).
In addition to its coactivator function, TFIIA can stimulate transcription by counteracting the effects of certain repressors that target TFIID (20). Some of these repressors, such as Mot1/ADI, inhibit transcription by disrupting the TFIID-TATA complex (4), while others, such as Dr1-DRAP1/NC2, and HMG1, block subsequent steps in PIC assembly (14, 26, 45). TFIIA can also compete with TAFs that inhibit the DNA-binding activity of TBP, providing a possible explanation for the requirement of TFIIA in basal transcription reactions programmed with TFIID (34, 50). Interestingly, the coactivation and antirepression activities of human TFIIA have been found to reside in separable domains of the protein, confirming that TFIIA can employ both mechanisms to enhance TFIID function (40).
In S. cerevisiae, TFIIA is composed of two subunits, a 32-kDa subunit encoded by the TOA1 gene and a 13.5-kDa subunit encoded by the TOA2 gene (55). Both genes are essential for cell viability and show striking similarity to the TFIIA-encoding genes of higher eukaryotes. Several mutational studies have been performed to identify residues in TFIIA important for its various functions. Deletion mutations in TOA1 and TOA2 have identified regions of TFIIA required for subunit-subunit interaction and for TBP and DNA binding (28). A search for conditional mutations yielded temperature-sensitive mutations in TOA1 that reduce formation of a TFIIA-TBP-DNA complex and mutations in TOA2 that do not impair TFIIA function in vitro (28). Site-directed mutagenesis of the human small-subunit gene and the yeast TOA2 gene demonstrated the importance of the TBP-TFIIA interaction for activation of specific genes in vitro and in vivo (48, 49).
Despite these genetic studies, our current understanding of TFIIA derives mostly from its properties in highly purified, reconstituted transcription systems. To begin to elucidate the function and regulation of TFIIA in vivo, we have analyzed the interaction of TFIIA with a class of TBP mutants that exhibits promoter-specific defects in activated transcription in vivo (3). One of the mutants in this class, TBP-N159D, which is encoded by the spt15-341 mutation, has an amino acid substitution at a position that directly contacts the TATA box. A second mutant in this class TBP-P109A, which is encoded by the spt15-328 mutation, has a proline-to-alanine substitution within a loop connecting two of the β strands that form the DNA-binding surface of TBP (3, 29, 30). Both of these proteins are severely compromised for DNA binding in vitro (1, 3). We have found that TFIIA overexpression strongly suppresses the transcriptional defects of the TBP mutants and that purified TFIIA restores DNA binding to the one mutant that was tested. Using a genetic screen, we have identified three regions of TFIIA that are important for TBP recruitment or TBP-TATA complex stabilization in vivo. Finally, using a method for conditionally expressing TOA1, we have depleted Toa1 and demonstrated that promoters vary significantly in the degree to which they require normal levels of TFIIA for activation. These results suggest that TFIIA coactivates transcription in a gene-specific manner in vivo by facilitating or stabilizing the interaction of TBP with the TATA box.
MATERIALS AND METHODS
Yeast strains and media.
Rich (YP), minimal (SD), synthetic complete (SC), 5-fluoroorotic acid, and inositol starvation media were prepared as described previously (58). YPGal and SCGal media contained 2% galactose. Solid YPGal and SCGal media also contained 1 mg of antimycin A/liter. Yeast transformants were selected on SC media lacking the appropriate nutrient.
With the exception of YJL1471 (a gift from C. S. Detweiler and J. J. Li), all strains used in this study are congenic to FY2, a GAL2+ ura3-52 derivative of S288C (73). Strains were constructed by standard genetic procedures (58) and are listed in Table 1. Strains containing the spt15 mutations were described previously (3). Plasmids pKA92 and pKA100 are derivatives of pRS314 (63) that contain the spt15-330 and spt15-338 mutations, respectively (3).
TABLE 1.
S. cerevisiae strains
| Strain | Genotype |
|---|---|
| KY214 | MATα spt15-328 his4-917δ lys2-173R2 leu2Δ1 ura3-52 trp1Δ63 |
| KY216 | MATα spt15Δ102::LEU2 his4-917δ lys2-173R2 leu2Δ1 ura3-52 trp1Δ63 [pKA92] |
| KY224 | MATα spt15Δ102::LEU2 his4-917δ lys2-173R2 leu2Δ1 ura3-52 trp1Δ63 [pKA100] |
| KY230 | MATα spt15-336 his4-917δ lys2-173R2 leu2Δ1 ura3-52 trp1Δ63 |
| KY231 | MATα spt15-341 his4-917δ lys2-173R2 leu2Δ1 ura3-52 trp1Δ63 |
| KY549 | MATa/MATα toa1Δ::LEU2/TOA1 lys2-128δ/lys2-128δ leu2Δ1/leu2Δ1 ura3-52/ura3-52 trp1Δ63/trp1Δ63 |
| KY550 | MATα toa1Δ::LEU2 lys2-128δ leu2Δ1 ura3-52 trp1Δ63 [pLQ55] |
| KY551 | MATα toa1Δ::LEU2 lys2-128δ leu2Δ1 ura3-52 trp1Δ63 [pLQ56] |
| KY552 | MATα toa1Δ::LEU2 lys2-128δ leu2Δ1 ura3-52 trp1Δ63 [pLQ57] |
| FY83 | MATa/MATα lys2-128δ/lys2-128δ leu2Δ1/leu2Δ1 ura3-52/ura3-52 trp1Δ63/trp1Δ63 |
| FY630 | MATα his4-917δ lys2-173R2 leu2Δ1 ura3-52 trp1Δ63 |
| YJL1471 | MATa leu2-3,112 ura3-52 trp1-289 bar1::LEU2 [pMET-(HA)3-CDC6] |
To introduce the toa1Δ100::LEU2 mutation into yeast, strain KY310 was transformed with the 3.7-kb SalI-PstI fragment of pLQ14. Precise replacement of one chromosomal copy of TOA1 was confirmed by tetrad analysis and Southern hybridization. To construct strains that were used in the Toa1 depletion experiments, strain KY549 was transformed with plasmid pLQ36 prior to sporulation and tetrad dissection. Segregants containing the toa1Δ100::LEU2 allele and pLQ36 were subsequently transformed with the TOA1 fusion plasmids. To evict pLQ36, Trp+ transformants were transferred by replica plating to 5-fluoroorotic acid media lacking methionine.
Plasmids.
Standard methods were used to construct plasmids, isolate DNA, and transform DNA into Escherichia coli (5). All plasmids used in yeast transformations were derived from the pRS series of shuttle vectors (10, 63). Plasmids recovered by glass bead lysis of yeast (23) were transformed into E. coli MH1 (19) cells for amplification.
Plasmids pLQ1, pLQ2, and pLQ3 used in the TFIIA overexpression experiments were derived from plasmid pSH346, a gift from Steve Hahn. To generate pLQ1, a 2.3-kb SalI-PstI fragment from pSH346 containing the wild-type TOA1 gene was inserted at the same sites in pRS425. The 2.0-kb PstI-PstI fragment from pSH346, which contains the wild-type TOA2 gene, was inserted at the PstI site of pRS425 or pLQ1 to give rise to pLQ2 or pLQ3, respectively. pLQ99 is a 2μm, URA3-marked plasmid that expresses both TOA1 and TOA2 and that was constructed by inserting the 4.1-kb XhoI-BamHI fragment of pLQ3 into the same sites of pRS426. pSB238, a gift from Steve Buratowski, is a pRS426 derivative containing the SUA7 gene (53). For TOA1 mutagenesis, plasmid pLQ21 was created by subcloning the 2.3-kb SalI-PstI fragment from pLQ1 into the same sites of pRS314. For TOA2 mutagenesis, pLQ23 was generated by insertion of the 2.0-kb PstI-PstI fragment from pLQ2 into the PstI site of pRS314.
Plasmids used for the in vitro transcription of TOA1 and TOA2 were constructed as follows. With pRS314 derivatives as templates, an MscI site was introduced by PCR 5 bp 5′ to the translation start codon of each TOA1 and TOA2 gene (28). MscI-BamHI (from the polylinker) fragments containing wild-type or mutant TOA1 genes were subcloned into the same sites of pCITE-4a (+) (Novagen). For the TOA2 derivatives, a SalI site was also introduced by PCR approximately 300 bp downstream of the translation stop codon. MscI-SalI fragments containing wild-type or TOA2 genes were subcloned into the same sites of pCITE-4a (+).
To generate a chromosomal deletion of TOA1, plasmid pLQ14 was constructed. First, the 2.3-kb SalI-PstI fragment from pLQ1 was inserted into the same sites of pUC18, generating pLQ6. Second, pLQ6 was used as a template in a PCR with divergent primers that annealed immediately adjacent to the ATG and TAA codons of TOA1. The primers incorporated XhoI restriction sites into the PCR product. Third, the 4.2-kb PCR product was cut with XhoI and self-ligated to create plasmid pLQ7. Fourth, a 2.2-kb SacI-XbaI fragment containing the LEU2 gene was excised from plasmid pGP43 (a gift from Greg Prelich), blunted with a Klenow fragment, and inserted into the XhoI (blunted) site of pLQ7 to create pLQ14.
The fusion genes used for Toa1 depletion experiments were derivatives of plasmids bc64, bc65, bc66, and bc67 (11). These plasmids contain UBI-lacI-SPT15 fusion genes under the control of the CUP1 promoter and differ in the encoded amino acid at the junction between ubiquitin and LacI. The four plasmids were cut with BamHI to remove the CUP1 promoter and UBI gene and religated to generate pLQ9, pLQ10, pLQ11, and pLQ12. TOA1 was amplified from plasmid pLQ6 with primers that incorporated an XbaI site at the ATG and a SalI site approximately 300 bp 3′ of the stop codon. The TOA1 open reading frame of the PCR product was sequenced with an ABI PRISM 377 sequencer. The 1.2-kb PCR products were digested with XbaI and SalI and then inserted at the same sites in pLQ9, pLQ10, pLQ11, and pLQ12 to replace the SPT15 gene. The BamHI-BamHI fragment containing the CUP1 promoter and UBI gene was inserted back into the newly made plasmids. Finally, to express TOA1 from the MET3 promoter, EcoRI-SalI fragments containing UBI-lacI-TOA1 fusion genes were excised from the CUP1 plasmids, made blunt by treatment with Klenow fragment, and inserted at the blunted EcoRI site of pRN500. Plasmid pRN500 is a derivative of pRS414 and contains the MET3 promoter (a gift from R. Nash). The resulting plasmids, pLQ54, pLQ55, pLQ56, and pLQ57, contain arginine, methionine, isoleucine, and tyrosine, respectively, at the junction between ubiquitin and LacI.
Isolation of TFIIA mutants.
The TOA1 and TOA2 genes were amplified from plasmids pLQ21 and pLQ23 by error-prone PCR (78). Primers that annealed to sequences on the pRS314 vector outside the polylinker were used. To generate a toa1 library, amplified fragments were digested with SalI and PstI and inserted into SalI-PstI-digested pRS314 (CEN ARS TRP1 vector). To generate a toa2 library, amplified fragments were cut with PstI and inserted into the PstI site of pRS314. Strain KY214 was transformed with the toa1 and toa2 mutant libraries, and transformants were replica plated to a series of media to test inositol auxotrophy, galactose utilization, and temperature sensitivity. To determine if the TOA1 mutations affected the four-helix bundle or β-sandwich region of the protein, in vivo mapping experiments were performed (46). All TOA1 mutations that were able to suppress the mutant TBP mapped to the β-sandwich coding region. DNA sequencing of this region revealed the nature of the TOA1 mutations. TOA2 mutations were identified by sequencing the entire TOA2 gene.
Saturation mutagenesis.
An oligonucleotide was designed to incorporate random changes at codon 254 of TOA1 and was used in a site-directed mutagenesis reaction (43) with plasmid pLQ21 to construct a plasmid library of toa1 mutations. Sequencing the DNA of 10 randomly chosen library clones showed that the mutagenesis frequency was approximately 40%. This library was then transformed into KY214 and screened for mutations that suppress the phenotypes conferred by spt15-328. Of 900 transformants, plasmids from 15 candidates were recovered, passaged through E. coli, and retransformed into KY214. Eleven plasmids that retained suppression were sequenced in the region encompassing codon 254 of TOA1.
RNA analysis.
Cells were grown at 30°C to 1 × 107 to 2 × 107 cells/ml in the appropriate media, and transcription of INO1, GAL1, GAL10, and CUP1 was induced as described in the figure legends (see also reference 3). RNA isolation and Northern hybridization analysis were performed by procedures described previously (3). DNA probes were derived from plasmids pJH310 (INO1) (22), pGAL1-GAL10 (EcoRI fragment 4812 [67] in pUC18), pCC69 (ACT1; a gift from F. Winston), pKA110 (GAL4) (1), pFW45 (HIS4) (74), and pLD2 (CUP1; a gift from M. Grunstein). Transcript levels were quantitated with a FUJIX BAS2000 phosphorimager with MacBAS, version 2.4, software.
Depletion of TFIIA with the MET3 promoter.
To analyze gene activation upon TFIIA depletion, yeast strains YJL1471 expressing (HA)3-CDC6 and KY552 expressing UBI-lacI-TOA1 (Y), in which the fusion genes are both under the control of the MET3 promoter, were grown at 30°C in the appropriate noninducing media lacking methionine to an optical density at 600 nm (OD600) of 0.3. To repress transcription of (HA)3-CDC6 and UBI-lacI-TOA1 (Y), l-methionine was added to a final concentration of 1 mM. To maintain cells in log-phase growth, the OD600 was measured every hour throughout the course of the experiment and cultures were repeatedly diluted to an OD600 of 0.3 with the appropriate media. The transcription of CDC6 and TOA1 was repressed by exposure to methionine for the times indicated in the figure legends so that similar percentages of cell survival were obtained. Subsequently, transcription of INO1, GAL1, GAL10, HIS4, and CUP1 was induced by adding the appropriate chemicals. Samples were taken for RNA preparation and Northern analysis. Percent viability was determined by spreading duplicate samples of cells from the depleted (with methionine) and nondepleted (without methionine) cultures on SC medium lacking methionine. To measure levels of the ubiquitin-LacI-Toa1 fusion protein, whole-cell extracts were prepared essentially as described previously (68). Protein extract from 0.25 OD600 unit of cells was loaded in each lane and analyzed by immunoblotting with rabbit polyclonal antisera directed against Toa1. Fusion Toa1 protein levels were quantitated on a Power Macintosh 7100/80 computer with the public-domain National Institutes of Health Image program and normalized to ribosomal protein L3 levels. During the course of our experiments, we noticed that the kinetics of Toa1 depletion in SD and SC media differed slightly. Therefore, to allow a meaningful comparison of the effects of TFIIA depletion at different promoters, control experiments were performed to establish the time needed to deplete Toa1 levels to less than 5% of undepleted levels in both media conditions.
In vitro DNase I footprinting assays and electrophoretic mobility shift assays.
A radiolabelled DNA probe containing the adenovirus major-late TATA box was prepared from plasmid pRW essentially as described previously (2). Wild-type TBP and TBP-N159D were purified from E. coli overexpression strains by standard chromatographic procedures (7, 52). The wild-type TBP used in all experiments was approximately 95% pure, and the TBP-N159D preparations used in DNase I footprinting and electrophoretic mobility shift assays were approximately 45 and 95% pure, respectively. Recombinant yeast TFIIA, which was approximately 70% pure, was a gift from Tony Imbalzano and Robert Kingston. In DNase I footprinting assays, wild-type TBP and TBP-N159D were incubated for 30 min at 30°C with <1 ng of the pRW probe in a reaction volume of 25 μl. Buffer conditions were as described previously (2). In the reaction mixtures that contained recombinant yeast TFIIA, 670 nM TFIIA was added before the incubation. This amount of TFIIA was found to be saturating for TFIIA-TBP-DNA ternary complex formation with both wild-type TBP and TBP-N159D. Samples were treated with DNase I and processed as described previously (2). In electrophoretic mobility shift assays, 15 nM recombinant yeast TBP and 2 μl of 1:100 dilutions of in vitro-translated TFIIA were incubated with the pRW probe for 30 min at 30°C under previously described conditions (13). TOA1 and TOA2 genes were transcribed in vitro with T7 RNA polymerase (Stratagene). Toa1 and Toa2 were cotranslated with a rabbit reticulocyte lysate system (Promega). [35S]methionine was added to a portion of each translation reaction mixture to compare amounts of protein made. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of these parallel reactions demonstrated that the electrophoretic mobility shift assay mixtures contained equivalent amounts of TFIIA (39). Electrophoretic mobility shift assays, carried out with unlabeled translation products, were analyzed on nondenaturing 4% polyacrylamide gels at room temperature (13).
RESULTS
Overexpression of TFIIA suppresses phenotypes caused by activation-defective TBP mutants.
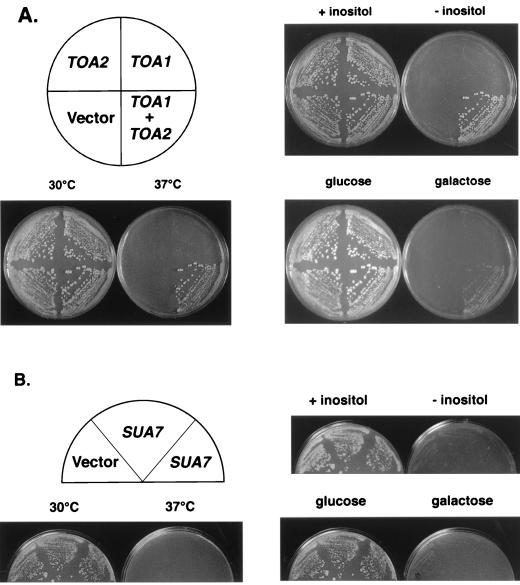
To investigate the importance of TFIIA in the recruitment of TBP to TATA elements in vivo, we have tested the ability of TFIIA to restore function to a class of TBP mutants that is activation and DNA binding defective (3). In our initial experiments, we asked whether overexpression of TFIIA could suppress the characteristic phenotypes, inositol auxotrophy (Ino−) and slow growth on galactose-containing media (Gal−), of five different TBP mutant strains in this class. High-copy-number plasmids expressing both TOA1 and TOA2 were transformed into the strains, and suppression was examined by replica plating the transformants to selective media lacking inositol or containing galactose. In addition, for experiments involving the temperature-sensitive (Ts−) spt15-328 mutation, transformants were analyzed for their ability to grow at 37°C. As shown in Fig. 1A and Table 2, the Ino−, Gal−, and Ts− phenotypes of the TBP mutant strains were significantly suppressed when both TOA1 and TOA2 were present on the 2μm plasmid. Specificity of suppression is indicated by the inability of TOA1 or TOA2 alone to reverse the mutant phenotypes (Fig. 1A) (39). Overexpression of TFIIA in these strains was confirmed by immunoblot analysis (39).
FIG. 1.
TFIIA overexpression suppresses the phenotypes conferred by TBP-P109A. (A) Yeast 2μm plasmids carrying TOA1, TOA2, or both genes were transformed into strain KY214. Transformants were replica plated to selective media to assay inositol auxotrophy, galactose utilization, or temperature sensitivity (SC). For the Ino and Ts phenotypes, plates were incubated at 30°C for 3 days. For the Gal phenotype, plates were incubated at 30°C for 4 days. (B) A yeast 2μm plasmid carrying SUA7 and a vector control were transformed into KY214 and replica plated to the indicated media. Plates were incubated at 30°C for 4 days to monitor the Ino and Ts phenotypes or for 5 days for the Gal phenotype.
TABLE 2.
Suppression of TBP mutants by TFIIA overexpressiona
| Amino acid change | Allele | Suppression of phenotype
|
||
|---|---|---|---|---|
| Ino | Gal | Ts | ||
| TBP-P109A | spt15-328 | +/− | +/−− | + |
| TBP-N159D | spt15-341 | −/+ | −−/+ | NA |
| TBP-V71A | spt15-330 | +/−− | −/+ | NA |
| TBP-F116Y | spt15-336 | +/−− | −/+ | NA |
| TBP-V161A | spt15-338 | +/−− | −−/+ | NA |
Strains containing the indicated spt15 mutations were transformed with 2μm plasmids (pLQ3 or pLQ99) expressing both TOA1 and TOA2. The Ino and Ts phenotypes were scored after 3 days of growth at 30°C, and the Gal phenotype was scored after 5 days. Very strong, strong, moderately strong, moderate, weak, and no suppression are indicated by the symbols +, +/−, +/−−, −/+, −−/+, and −, respectively. With the exception of TBP-V71A, which has not been tested, each of the TBP mutants has been shown to be DNA binding defective (3, 37, 57). A related mutant, TBP-V71E, also has greatly reduced affinity for DNA (57). NA, not applicable.
Previously, TFIIA and TFIIB have been shown to increase the affinity of wild-type TBP for the TATA box under suboptimal binding conditions (25). To test if TFIIB can also suppress the phenotypes caused by the mutant TBPs, SUA7, the gene that encodes TFIIB in S. cerevisiae, was overexpressed in spt15-328 and spt15-341 mutant strains. Unlike the strong suppression observed upon TFIIA overexpression, the high-copy-number SUA7 plasmid did not suppress the Ino−, Gal−, or Ts− phenotypes caused by TBP-P109A and TBP-N159D (Fig. 1B) (39). The presence of high levels of TFIIB in these strains was confirmed by immunoblot analysis (39). Together, these findings indicate that suppression of the activation- and DNA binding-defective TBP mutants is specific to TFIIA overexpression and suggest that TFIIA and TFIIB differ in their abilities to enhance the TBP-TATA interaction in vivo.
Overexpression of TFIIA restores transcriptional activation in the TBP mutant strains.
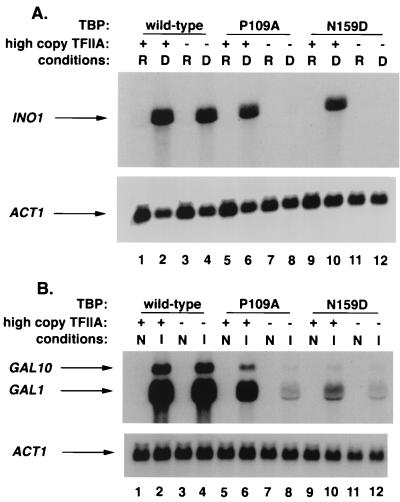
The inositol auxotrophy and galactose utilization defect caused by TBP-P109A and TBP-N159D correlate strongly with reduced transcriptional activation of the INO1, GAL1, and GAL10 genes (3). To determine whether TFIIA overexpression can rescue these activation defects in the TBP mutant strains, Northern analyses were performed. In wild-type strains, transcription of the INO1 gene is repressed by high levels of inositol and is dramatically induced when levels of inositol are low (Fig. 2A, lanes 3 and 4). However, in agreement with previous results (3), transcriptional activation of INO1 is severely impaired in strains that contain TBP-P109A or TBP-N159D (Fig. 2A, lanes 8 and 12). Whereas overexpression of TFIIA does not affect transcriptional activation of INO1 in strains containing wild-type TBP (Fig. 2A, lanes 1 and 2), overexpression of TFIIA in the TBP mutant strains restores activated transcription of INO1 to approximately 40 and 30% of wild-type levels for TBP-P109A and TBP-N159D, respectively (Fig. 2A, lanes 6 and 10). A similar result for the galactose-induced GAL1 and GAL10 genes was observed. For GAL1, overexpression of TFIIA in strains that contain TBP-P109A and TBP-N159D raises the activated-transcription levels to approximately 30 and 15%, respectively, of the levels observed in strains that contain wild-type TBP (Fig. 2B, lanes 6 and 10). These results reveal that suppression of the Ino− and Gal− phenotypes of the TBP mutant strains occurs at the transcriptional level. In addition, they demonstrate that the requirement for a transcriptional activator cannot be bypassed by an increase in TFIIA levels, as TFIIA overexpression does not lead to constitutive expression of INO1, GAL1, or GAL10. Instead, TFIIA apparently functions as a coactivator at these promoters.
FIG. 2.
TFIIA overexpression suppresses the INO1, GAL1, and GAL10 transcriptional defects caused by TBP-P109A and TBP-N159D. (A) Northern analysis of INO1 transcription in strains that overexpress TFIIA. Yeast strains containing wild-type TBP (KY300), TBP-P109A (KY214), or TBP-N159D (KY231) were transformed with plasmid pLQ3 (2μm, TOA1 and TOA2) or pRS425 (vector control) and grown in repressing (R; 200 μM inositol) and derepressing (D; 10 μM inositol) conditions. (B) Northern analysis of GAL1 and GAL10 transcription in strains that overexpress TFIIA. The same yeast strains used in panel A were grown in noninducing (N; 2% raffinose) and inducing (I; 2% raffinose plus 5% galactose) conditions. In each case, the filter used in the upper panel was stripped and probed for ACT1 message as a normalization control.
TFIIA restores DNA binding to TBP-N159D in vitro.
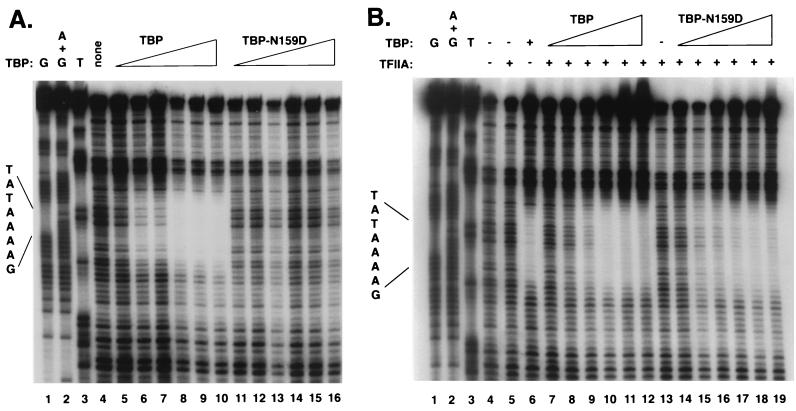
TBP-P109A and TBP-N159D are severely defective for binding to TATA elements in vitro, yet yeast strains that contain these mutant TBPs as the only source of TBP activity are viable (3). Therefore, in vivo, other factors most likely compensate for the extreme DNA-binding defects of these mutants. To determine whether TFIIA can restore TATA box binding to the mutant TBPs, we purified recombinant TBP-N159D and tested its ability to bind to the adenovirus major-late promoter (AdMLP) TATA box in the presence and absence of recombinant yeast TFIIA. In agreement with previous results from electrophoretic mobility shift assays (3), DNase I footprinting clearly demonstrates that, relative to wild-type TBP, TBP-N159D has greatly reduced affinity for the AdMLP TATA box. We detected no protection of the TATA box, even when the amount of TBP-N159D in the reaction mixture was 40-fold greater than the amount of wild-type TBP needed for half-maximal binding (Fig. 3A). However, in the presence of saturating amounts of yeast TFIIA, TBP-N159D protects the TATA region to levels similar to those observed with equivalent molar amounts of wild-type TBP (Fig. 3B). These results are in good agreement with an earlier study, which investigated the effects of TFIIA on a different set of TBP DNA-binding mutants (36). In conjunction with our genetic results on TFIIA overexpression, these findings demonstrate that TFIIA can overcome substantial impediments to TBP-TATA complex formation and support models in which TFIIA plays a role in TBP recruitment in vivo.
FIG. 3.
TFIIA increases the affinity of TBP-N159D for DNA in vitro. (A) DNase I footprinting assay of wild-type TBP and TBP-N159D binding to the AdMLP TATA box. Reaction mixtures for lanes 5 to 10 contained wild-type TBP, and reaction mixtures for lanes 11 to 16 contained TBP-N159D. Amounts were as follows: lanes 5 and 11, 7 nM; lanes 6 and 12, 15 nM; lanes 7 and 13, 30 nM; lanes 8 and 14, 60 nM; lanes 9 and 15, 120 nM; lanes 10 and 16, 240 nM. (B) DNase I footprinting assay of wild-type TBP and TBP-N159D binding to the AdMLP TATA box in the presence of TFIIA. Recombinant yeast TFIIA (670 nM) was added to the reaction mixtures in lanes 5 and 7 to 19. Reaction mixtures for lanes 6 to 12 contained wild-type TBP, and reaction mixtures for lanes 14 to 19 contained TBP-N159D. Amounts were as follows: lanes 7 and 14, 7 nM; lanes 8 and 15, 15 nM; lanes 9 and 16, 30 nM; lanes 10 and 17, 60 nM; lanes 6, 11, and 18, 120 nM; lanes 12 and 19, 240 nM. Lanes 1 to 3 in each panel contain the products of DNA sequencing reactions. In these assays, we did not detect a significant TFIIA-dependent increase in the affinity of wild-type TBP for DNA, presumably because conditions optimal for TBP-TATA complex formation were used (25).
Suppression of mutant TBPs by dominant mutations in TOA1 and TOA2.
The strong mutant phenotypes caused by TBP-P109A and TBP-N159D allowed us to perform a genetic screen for mutations in TOA1 and TOA2 that suppress these mutant TBPs and, therefore, might facilitate the TBP-TATA interaction in vivo. TOA1 and TOA2 were mutagenized by PCR, and amplified fragments were used to construct mutant toa1 and toa2 libraries. The libraries were transformed into a strain containing TBP-P109A, and transformants were screened for suppression of the Ino−, Gal−, and Ts− phenotypes. Since the endogenous wild-type copies of TOA1 and TOA2 were present in the strain, the suppressor mutations were expected to be dominant.
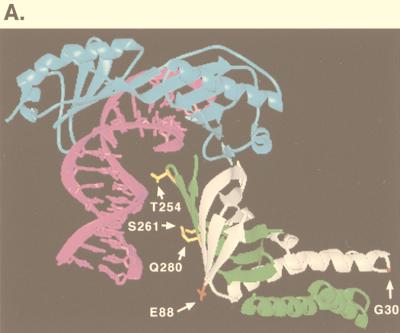
For the analysis of TOA1, a screen of 24,000 transformants yielded 29 candidates that carried plasmid-borne mutations responsible for suppression of TBP-P109A. Nineteen showed suppression of both the Ino− and Ts− phenotypes, while 10 exhibited suppression of only the Ino− phenotype. Since we have demonstrated that overexpression of TFIIA can reverse the mutant phenotypes conferred by TBP-P109A, TFIIA levels in these 29 candidates were assayed by immunoblotting. None of the 29 candidates exhibited TFIIA levels different from those of the wild type (39). An in vivo mapping strategy (46) was used to show that all of the TOA1 mutations affect the β-sandwich region of Toa1 (39). DNA sequencing revealed that among the 10 Ino+ candidates 6 encode the change S261R and four encode Q280R. Eleven Ino+ Ts+ candidates encode the change D226V, seven encode T254I, and one encodes T254K. On the TFIIA-TBP-DNA crystal structures, Thr254 and Ser261 lie within a region of Toa1 that contacts DNA (16, 70) (Fig. 4A). Gln280 occupies a position near Ser261, but on the adjacent β-strand, and may also be involved in contacting the promoter upstream of the TATA box (Fig. 4A). In contrast, Asp226 lies in a very acidic, solvent-exposed region of Toa1 that exhibits no direct contacts with TBP or DNA and, therefore, represents a possible site for regulatory interactions (15, 16). The degree of suppression caused by these amino acid changes in Toa1 is summarized in Table 3.
FIG. 4.
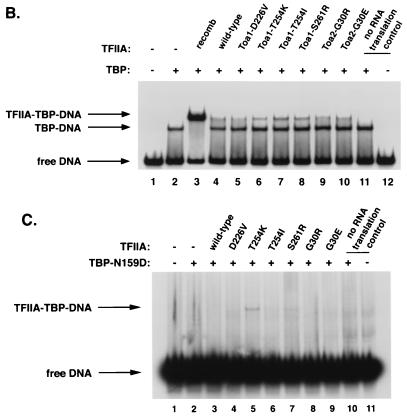
Effects of TOA1 and TOA2 mutations on TBP-TFIIA-DNA complex formation. (A) Locations of the amino acids altered by the dominant TOA1 and TOA2 mutations on the TFIIA-TBP-DNA crystal structure. Affected amino acids are highlighted in yellow for Toa1 and in red for Toa2. TBP, DNA, Toa1, and Toa2 are shown in blue, magenta, green, and white, respectively. The figure was adapted from reference 70. The position of Asp226 could not be accurately determined on this structure (70) and therefore is not indicated here. (B) Electrophoretic mobility shift assays were performed with recombinant wild-type TBP where indicated and a radiolabelled DNA probe containing the AdMLP TATA box. Prior to incubating the reaction mixture at 30°C for 30 min, recombinant yeast TFIIA (40 nM; lane 3) or in vitro-translated TFIIA (lanes 4 to 10) was added. Reaction mixtures for lanes 5 to 8 contained the indicated Toa1 mutant subunits cotranslated with wild-type Toa2, and reaction mixtures for lanes 9 and 10 contained the indicated Toa2 mutant subunits cotranslated with wild-type Toa1. Equivalent, but limiting, amounts of wild-type and mutant TFIIA were added to the reaction mixtures (see Materials and Methods). Lanes 11 and 12 contained an unprogrammed translation reaction mixture. (C) Electrophoretic mobility shift assays were performed with recombinant TBP-N159D and in vitro-translated TFIIA as described for panel B. A control reaction mixture containing TBP-N159D and recombinant TFIIA was loaded on the same gel to indicate the mobility of the TFIIA-TBP-DNA complex. The migration of this complex is indicated by an arrow.
TABLE 3.
Suppression of TBP mutants by dominant TOA1 and TOA2 mutationsa
| Protein | Suppression of indicated phenotype in strains containing:
|
||
|---|---|---|---|
| TBP-P109A
|
TBP-N159D, Ino | ||
| Ino | Ts | ||
| Wild-type Toa1 | − | − | − |
| Toa1-D226V | +/− | + | −− |
| Toa1-T254K | + | + | + |
| Toa1-T254I | + | + | + |
| Toa1-S261R | −/+ | − | −/+ |
| Toa1-Q280R | +/− | − | +/− |
| Wild-type Toa2 | − | − | − |
| Toa2-G30R | −−/+ | − | −−/+ |
| Toa2-G30E | −−/+ | − | −−/+ |
| Toa2-E88K | −−/+ | − | ND |
Strains containing spt15-328 (TBP-P109A) or spt15-341 (TBP-N159D), endogenous wild-type copies of TOA1 and TOA2, and the indicated TOA1 or TOA2 mutations were monitored for growth on media lacking inositol at 30°C. For spt15-328, growth of strains was also monitored at 37°C on SC media lacking tryptophan. Very strong, strong, moderate, weak, and no suppression after 3 days of incubation are indicated by +, +/−, −/+, −−/+, and −, respectively. −−, stronger Ino− phenotype caused by Toa1-D226V relative to wild-type Toa1 in the spt15-341 background. ND, not determined.
Because we recovered two different substitutions for Thr254 in our screen, we investigated the specificity of these changes by performing saturation mutagenesis. Codon 254 was randomized by site-directed mutagenesis. A plasmid library constructed from the mutagenesis products was transformed into a strain containing TBP-P109A. Of 900 transformants screened, 11 contained plasmid-encoded TOA1 mutations that were able to suppress the Ino− and/or Ts− phenotypes of the strain. Among the six strong Ino+ Ts+ TOA1 suppressors, five encode the T254K substitution and one encodes the T254I change. Among the five weak Ino+ suppressors, four change Thr254 to arginine and one encodes a glutamine at this position. These results indicate that only specific substitutions at residue 254 of Toa1, and not simple loss of a potential site of phosphorylation, suppress the activation- and DNA binding-defective TBP mutant.
To identify amino acids in Toa2 that are potentially important for recruitment of TBP to TATA elements in vivo, we constructed two independent toa2 mutant libraries and screened 26,000 transformants for suppression of TBP-P109A. After purification and retransformation, 11 candidates contained plasmid-encoded TOA2 mutations that partially reversed the Ino− phenotype conferred by TBP-P109A. The 11 mutant TOA2 genes were sequenced in their entirety. Five mutations change Gly30 to arginine, and four encode a glutamic acid at this position. Gly30 lies at the tip of the solvent-exposed four-helix bundle domain of TFIIA (16, 70) (Fig. 4A). The remaining two mutations encode the substitution E88K. As shown on the TFIIA-TBP-DNA crystal structure (Fig. 4A), Glu88 of Toa2 lies on the surface of TFIIA that contacts DNA, and the introduction of a lysine at this position may extend the TFIIA-DNA interaction upstream of the TATA box. Suppression of TBP-P109A by each of these TOA2 mutations is highly reproducible but noticeably weaker than suppression by the TOA1 mutations we identified (Table 3).
Plasmids expressing the dominantly acting TOA1 and TOA2 mutations were also transformed into a strain containing TBP-N159D. With one exception, the pattern and degree of suppression by the TOA1 and TOA2 mutations paralleled the results observed with TBP-P109A (Table 3). TOA1 mutations that altered residues on the TFIIA-DNA interface strongly suppressed the Ino− phenotype conferred by TBP-N159D, while the TOA2 mutations resulted in significantly weaker levels of suppression. Interestingly, the Toa1-D226V mutant, which strongly suppressed TBP-P109A, failed to suppress TBP-N159D. Indeed, this Toa1 mutant appeared to exert a dominant-negative effect on TBP-N159D, causing a stronger Ino− phenotype than that conferred by TBP-N159D alone. The basis for the differential behavior of Toa1-D226V in strains containing TBP-P109A and TBP-N159D is considered in the Discussion. All of the TOA1 and TOA2 mutations that were tested (i.e., all except Toa1-Q280R and Toa2-E88K) are sufficient to support viability in strains containing wild-type TBP and a disruption of the appropriate chromosomal TOA gene (39).
One explanation for the suppressive effects of the dominant TOA1 and TOA2 mutations, particularly those that introduce basic residues on the TFIIA-DNA interface, is that they stabilize or enhance formation of the TFIIA-TBP-DNA complex. To test this possibility, electrophoretic mobility shift assays were performed with in vitro-translated TFIIA, recombinant yeast TBP, and the AdMLP TATA probe. TFIIA mutant proteins were synthesized by cotranslating a mutant subunit (e.g., Toa1-D226V) with its wild-type counterpart (e.g., wild-type Toa2). Limiting, but equivalent (see Materials and Methods) (39), amounts of wild-type and mutant TFIIA proteins were added to the reaction mixtures so that any increase in complex formation could be detected. Toa1 mutants with changes at Thr254 and Ser261 exhibited increased formation or stability of TFIIA-TBP-DNA complexes containing wild-type TBP (Fig. 4B, lanes 6 to 8). Relative to wild-type TFIIA, these mutants gave rise to more discrete ternary complex bands and decreased dissociation in the native gel. In contrast, the Toa1-D226V, Toa2-G30R, and Toa2-G30E mutants supported TFIIA-TBP-DNA complex formation to approximately the same degree as wild-type TFIIA (Fig. 4B; compare lane 4 with lanes 5, 9, and 10). These results suggest that suppression by the TFIIA mutants with amino acid substitutions on the DNA-binding surface involves a direct effect on the assembly or stability of the TFIIA-TBP-DNA complex. To further test this hypothesis, similar assays were performed with recombinant TBP-N159D. With the limiting amounts of in vitro-translated TFIIA used in this experiment, wild-type TFIIA did not form detectable levels of a stable TFIIA-TBP-DNA complex (Fig. 4C, lane 3). In contrast, an equivalent amount of the Toa1-T254K mutant gave rise to a readily observable ternary complex (Fig. 4C, lane 5). The Toa1-T254I and Toa1-S261R derivatives supported only very low levels of complex formation in this assay (Fig. 4C, lanes 6 and 7), while the remaining mutants behaved similarly to wild-type TFIIA. Any significant differences among the latter mutants are masked by the overall instability of the TBP-N159D-containing complexes in the electrophoretic mobility shift assay. Nonetheless, the combined results of these experiments, together with the nature and locations of the amino acid changes, suggest that the dominant TOA1 and TOA2 mutations suppress the TBP defects by different mechanisms, one of which involves a direct effect on the assembly or stability of the TFIIA-TBP-DNA complex.
The requirement for TFIIA in transcriptional activation is promoter specific.
TBP-P109A and TBP-N159D exhibit promoter-specific defects in transcriptional activation (3). Activated transcription of certain genes, such as INO1, GAL1, and GAL10, is dramatically reduced in strains containing these TBP mutants, whereas activation of other genes, such as HIS3 and HIS4, is largely unaffected. These results suggest that transcriptional control at certain promoters is very sensitive to the stability of the TBP-TATA complex. Since TFIIA increases the affinity of TBP for DNA in vitro (25, 36, 72, 77) (see above) and since TFIIA overexpression restores activation to strains containing TBP-P109A or TBP-N159D, we reasoned that promoters may differ in the degree to which they require TFIIA for regulated transcription. Specifically, the properties of the TBP mutant strains suggested that activation of INO1, GAL1, and GAL10 may be more dependent upon the activity of TFIIA than activation of other promoters, such as HIS4. To test this hypothesis, we analyzed transcriptional activation from several highly regulated promoters in strains that contain wild-type TBP and reduced levels of TFIIA.
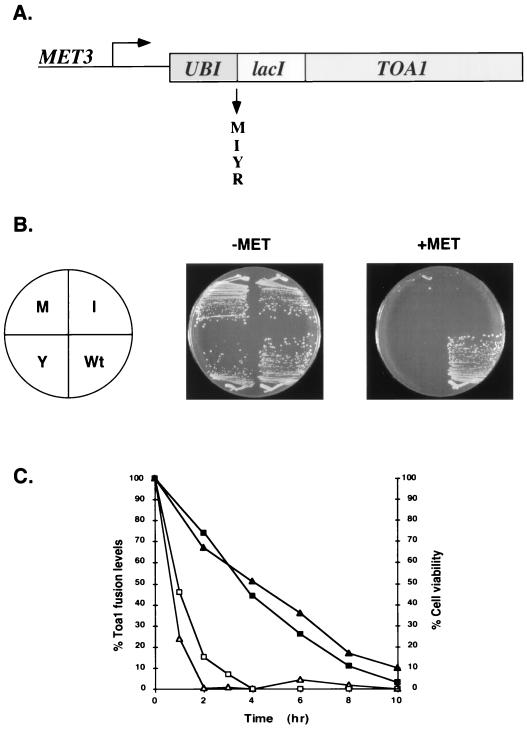
To conditionally deplete TFIIA in vivo, four different fusion genes that encode unstable versions of Toa1 were placed under the control of the methionine-repressible MET3 promoter (Fig. 5A). A related strategy was employed by Cormack and Struhl (11) for the depletion of TBP in yeast. The stabilities of the ubiquitin-LacI-Toa1 fusion proteins are governed by the junctional amino acid between ubiquitin and LacI (6). The predicted order of fusion protein stability is Met > Ile > Tyr > Arg. The four fusion genes were each introduced into a haploid strain in which the endogenous TOA1 gene was disrupted. Toa1 fusion proteins with Met, Ile, or Tyr at the junction between ubiquitin and LacI can support life in media lacking methionine (Fig. 5B), while a strain containing the Arg derivative as the only source of Toa1 was inviable under all conditions tested. Importantly, cells expressing the Met, Ile, or Tyr derivatives were inviable on media that contained methionine (Fig. 5B). In liquid cultures, the Tyr derivative exhibited the slowest growth rate upon transfer to media containing methionine (39). Therefore, a strain containing this fusion was chosen to perform the depletion experiments described below and will be referred to as the Toa1 depletion strain.
FIG. 5.
Depletion of TFIIA with the MET3 promoter. (A) Fusion genes comprised of UBI, lacI, and TOA1 are under the control of the MET3 promoter. The amino acid at the junction of ubiquitin and LacI is either M, I, Y, or R. (B) Three strains expressing, as their only source of Toa1, ubiquitin-LacI-Toa1 fusion proteins with the indicated junctional amino acids were streaked on plates lacking (−MET) or containing methionine (+MET) and were grown at 30°C for 3 days. A strain containing a wild-type (Wt) TOA1 gene is included for comparison. (C) Correlation between levels of the ubiquitin-LacI-Toa1 fusion protein and cell viability. KY552 expressing ubiquitin-LacI-Toa1(Y) was grown to early log phase in SC (triangles) or SD (squares) media lacking methionine. Methionine was added at zero time to repress transcription of the fusion gene. Samples were taken at the indicated times and assayed for cell viability (solid symbols) and fusion protein levels (open symbols). Fusion protein levels were quantitated from immunoblots. At each of the time points, an immunoreactive band with a mobility similar to that of recombinant Toa1 was observed (not shown). The levels of this protein did not change substantially over the time course. Confirmation that this protein represents a Toa1 derivative was not made. However, the reduction in cell viability detected upon methionine addition indicates that this protein cannot substitute for wild-type Toa1.
To investigate the requirement for TFIIA in the activation of various genes, we used the following strategy. First, the Toa1 depletion strain was grown to early log phase in media that lacked methionine and that was noninducing for the gene of interest. Next, methionine was added to one-half of the culture to deplete Toa1, and after a period of incubation, cells were transferred to conditions that were activating for gene expression. For each medium condition, control experiments were performed to determine the optimal amount of time needed for Toa1 depletion, which was measured by immunoblot analysis and cell viability (Fig. 5C).
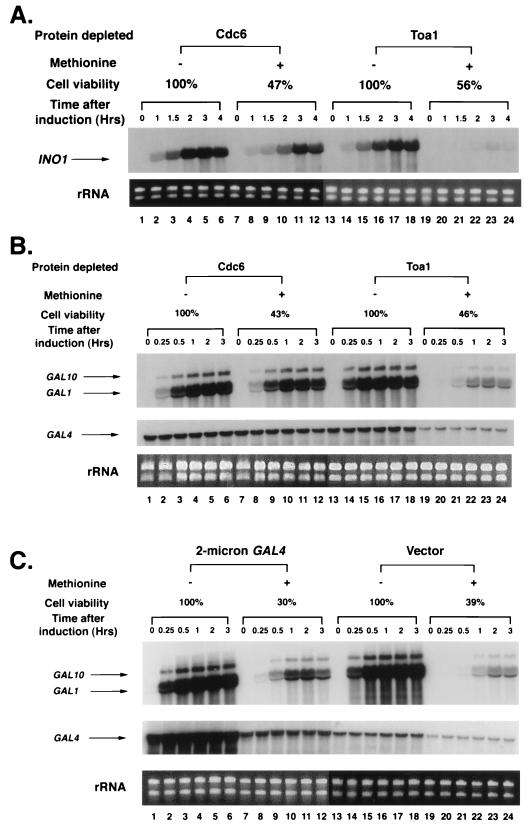
Our results show that INO1 activation is extremely sensitive to TFIIA levels (Fig. 6A). Following 3 h of exposure to methionine, a time sufficient to reduce Toa1 levels to approximately 5% of the nondepleted levels (Fig. 5C), cells were washed and transferred to media that lacked inositol but still contained methionine. Activation of INO1 transcription following the depletion period was significantly impaired. At the time of maximal INO1 induction, the culture that was depleted of Toa1 produced only 12% of the activated INO1 transcript levels that were observed in the absence of Toa1 depletion (Fig. 6A; compare lanes 17 and 23). To control for the effect of cell death in this and subsequent experiments, we analyzed transcriptional activation in a strain depleted of an essential protein, Cdc6, that is unrelated to RNA polymerase II transcription. In this control strain, transcription of CDC6, which encodes a protein required for DNA replication in yeast (21), is also directed by the MET3 promoter. Even when Cdc6 levels were reduced so that cell viability was less than that of the Toa1-depleted culture, INO1 activation was largely unaffected (Fig. 6A; compare lanes 5 and 11). Therefore, the striking decrease in INO1 activation observed in the Toa1-depleted strain is not due to an indirect effect of cell death caused by the loss of an essential protein. These findings suggest that TFIIA plays a critical role in INO1 activation even in the presence of a fully functional TBP.
FIG. 6.
TFIIA depletion impairs activation at INO1, GAL1, and GAL10. Northern analyses of INO1 (A) and GAL1-GAL10 (B) transcription following depletion of TFIIA are shown. Cdc6 and Toa1 depletion strains were grown to early log phase in media that lacked methionine and that was repressing (200 μM inositol for INO1) or noninducing (2% raffinose for GAL genes) for the gene of interest. Transcription of CDC6 and TOA1 was inhibited by the addition of 1 mM methionine where indicated, and the incubation was continued for times sufficient to achieve approximately equal levels of cell viability, as determined from control experiments. Subsequently, strains were transferred to derepressing media (lacking inositol for INO1 and 5% galactose for GAL1-GAL10). Times of exposure to methionine prior to gene activation were as follows for the Cdc6 and Toa1 depletion strains, respectively: 2 and 3 h for INO1 and 2 and 6 h for GAL1-GAL10. (C) Effect of GAL4 overexpression on GAL1-GAL10 induction. Strains carrying a GAL4 overexpression plasmid or control vector were grown as described for panel B except that the cultures were treated with methionine for 8 h prior to the addition of 5% galactose. Samples were taken for Northern analysis at the indicated times after gene induction. The zero-hour sample was removed prior to exposing cells to inducing conditions. RNA levels were normalized by measuring the A260 and by ethidium bromide staining of control gels to visualize rRNA.
We next analyzed the effect of TFIIA depletion on two other genes, GAL1 and GAL10, whose transcription is affected by the activation-defective TBP mutants. For these experiments, the Toa1 and Cdc6 depletion strains were first grown in raffinose media, and methionine was subsequently added to one-half of each culture for times sufficient to achieve comparable levels of cell death. For the Toa1 depletion strain, this treatment lowered Toa1 protein levels to less than 5% of nondepleted levels (Fig. 5C). GAL1 and GAL10 transcription was then induced by the addition of galactose. As shown in Fig. 6B, activation of these Gal4-regulated genes was significantly impaired under conditions of Toa1 depletion. The levels of GAL1 and GAL10 transcripts produced by the Toa1 depletion strain in the presence of methionine were 6 and 5%, respectively, of those produced in the absence of methionine at the time of maximal induction for the nondepleted strain (Fig. 6B; compare lanes 15 and 21). A reduction in GAL gene transcription was not observed upon depletion of Cdc6, indicating that the effect was not due to a loss in cell viability.
Activation of GAL1 and GAL10 is extremely sensitive to the amount of Gal4 in the cell (18). Therefore, the reduction in GAL gene activation observed upon Toa1 depletion could simply be a consequence of reduced activator levels. Indeed, Toa1 depletion resulted in a fourfold decrease in GAL4 message levels (Fig. 6B, middle; compare lanes 13 to 18 and lanes 19 to 24). To determine whether the impaired activation of GAL1 and GAL10 was due entirely to lowered Gal4 levels, we tested the effect of Toa1 depletion in strains that overexpressed Gal4 from a 2μm plasmid. In these strains, Toa1 depletion reduced GAL4 transcript levels as expected; however, the amount of GAL4 mRNA following depletion was still approximately twofold greater than that observed in a control strain that carried empty vector and that was not depleted of Toa1 (Fig. 6C; compare lanes 7 to 12 with lanes 13 to 18). When the Gal4-overexpressing strain was exposed to methionine to deplete Toa1 and then treated with galactose, transcription of GAL1 and GAL10 was still impaired (Fig. 6C; compare lanes 7 to 12 with lanes 13 to 18). These results suggest that lowered activator levels are not solely responsible for the GAL1-GAL10 activation defect and support a direct role for TFIIA in Gal4-mediated activation.
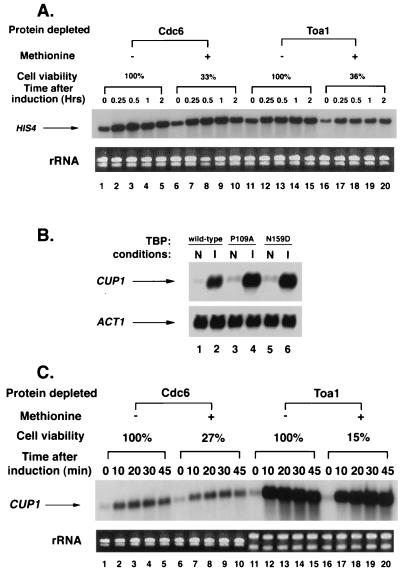
To determine the importance of TFIIA in Gcn4-mediated activation, we analyzed transcription of HIS4 under conditions of Toa1 depletion. To induce Gcn4 synthesis, 3-aminotriazole was added following the depletion of Toa1 (less than 5% of nondepleted levels; Fig. 5C). HIS4 stimulation with and without Toa1 depletion was by 2.3-fold and 2.4-fold, respectively (Fig. 7A, lanes 11 and 14 and lanes 16 and 19). These results suggest that activation of HIS4 is less sensitive than INO1, GAL1, and GAL10 to the levels of TFIIA in the cell. The mechanism that accounts for the decreased sensitivity of HIS4 to the TBP mutants may also lessen the dependence of this promoter on TFIIA.
FIG. 7.
TFIIA depletion does not impair activation of HIS4 or CUP1. (A) Northern analysis of HIS4 transcription following depletion of TFIIA. Cdc6 and Toa1 depletion strains were grown to early log phase in media that lacked methionine. Where indicated, transcription of CDC6 and TOA1 was inhibited by exposure of the cells to 1 mM methionine for 2.5 and 4 h, respectively. To induce HIS4 transcription, 10 mM 3-aminotriazole was added to each culture. Samples were taken at the indicated times prior to (zero-hour point) or following induction. (B) Northern analysis of CUP1 transcription in strains containing wild-type TBP (FY630) or the TBP mutants, TBP-P109A (KY214) and TBP-N159D (KY231). For induction, cells were treated with 100 μM CuSO4 for 30 min prior to harvesting. The filter shown in the upper panel was stripped and probed for ACT1 mRNA as a normalization control. (C) Northern analysis of CUP1 transcription following depletion of TFIIA. The experiment was conducted as described for panel A, except that cells were exposed to methionine for 2.5 and 6 h to achieve comparable levels of cell death for the Cdc6 and Toa1 depletion strains, respectively. CuSO4 (100 μM) was added to activate CUP1. For panels A and C, RNA levels were normalized by measuring the A260 and by ethidium bromide staining of control gels to visualize rRNA. In panel C, the two rRNA control gels were run for different periods of time.
Relative to INO1 and the GAL genes, HIS4 is only moderately induced by its activator, Gcn4. To find out whether these promoters differ in their requirement for TFIIA because they differ in their level of induction, we tested another highly activated promoter, CUP1, for its response to the TBP mutants and Toa1 depletion. Strikingly, activation of CUP1 by copper was unaffected by the TBP mutants, TBP-P109A and TBP-N159D (Fig. 7B). In addition, a reduction in Toa1 levels to less than 5% of nondepleted levels had no effect on the magnitude of CUP1 induction: with and without depletion of Toa1, the activation of CUP1 was by 17-fold and 13-fold, respectively (Fig. 7C, lanes 11 and 13 and lanes 16 and 18). These results demonstrate that at least one highly induced promoter is largely insensitive to the stability of the TBP-TATA complex or to TFIIA levels. These findings strongly suggest that the requirement for TFIIA in transcriptional activation varies at different promoters.
DISCUSSION
One important mechanism by which transcriptional activators stimulate gene expression is by accelerating the rate-limiting binding of TBP to the TATA box. Biochemical experiments have demonstrated a requirement for TFIIA and TAFs in communicating the activation signal to TBP. In this study, we sought to elucidate the role of TFIIA in transcriptional activation in vivo by exploiting the properties of a well-characterized class of TBP mutants (3). Three significant findings concerning the in vivo function of TFIIA have come from this work. First, we have demonstrated that TFIIA, when either overexpressed or altered by mutation, can restore function to the DNA binding- and activation-defective TBP mutants. These results strongly suggest that TFIIA facilitates the TBP-TATA interaction in vivo. Second, we have identified through a genetic screen three distinct regions on TFIIA that affect TBP recruitment or the stability of the TBP-TATA complex. Third, using a conditional expression system, we have demonstrated a promoter-specific involvement of TFIIA in transcriptional activation.
Together with the ability of TFIIA to restore DNA binding to TBP-N159D in vitro, our TFIIA overexpression results suggest that TFIIA assists in the recruitment of TBP to promoters in vivo. Although our data do not rule out an effect of TFIIA on TBP-TATA complex dissociation, a recruitment role for TFIIA is most consistent with previous biochemical studies that employed purified factors or yeast extracts (38, 56, 71). Our observation that overexpression of TFIIA causes an observable phenotype (i.e., suppression of the activation-defective TBP mutants) suggests that TFIIA may be limiting in vivo, at least at certain promoters. Interestingly, TFIIA overexpression also reverses the mutant phenotypes conferred by an unrelated TBP mutant, encoded by the spt15-21 allele, and by a mutation in the S. cerevisiae SPT3 gene (42). These mutations alter the interaction between TBP and the SAGA histone acetyltransferase complex (17). High-level expression of TFIIA most likely suppresses these mutations by assisting the TBP-TATA interaction in the context of a repressing chromatin structure. Therefore, TFIIA can lessen the effects of several types of impediments to TBP-TATA complex formation.
By searching for dominant mutations in TOA1 and TOA2 that suppress TBP-P109A, we have identified three different regions on TFIIA that may be involved in TFIID recruitment in vivo. Three of the TOA1 mutations, encoding the changes T254K, T254I, and S261R, alter residues along the TFIIA-DNA interface observed by crystallography (16, 70). Alanine scanning mutagenesis of charged residues near these positions caused a temperature-sensitive phenotype and a reduction in formation of a TFIIA-TBP-DNA complex in vitro (28). Crystallography of the TFIIA-TBP-TATA complex has revealed direct contacts between DNA and the Toa1 residues Thr252, Arg253, Lys255, Arg257, and Lys259 (16, 70). Two of these amino acids, Arg253 and Lys255, are in a position to make direct contacts with the phosphodiester backbone of the TATA box (70). The T254K and T254I substitutions, isolated in our mutant hunt, may create additional contacts between TFIIA and the TATA box. Such interactions might be especially important contributors to the overall stability of the TFIIA-TBP-DNA complex (72). Ser261 of Toa1 lies very near the basic amino acids that directly contact DNA upstream of the TATA box. Introduction of an arginine at this position might create a novel interaction between TFIIA and DNA. Two other mutations identified in our screen, encoding Toa1-Q280R and Toa2-E88K, also introduce basic amino acids in the proximity of DNA 5′ to the TATA box (Fig. 4A), and may further extend the TFIIA-DNA interface. In agreement with these predictions, TFIIA derivatives containing the Toa1-T254K, Toa1-T254I, and Toa1-S261R subunits increased the formation and/or stability of TFIIA-TBP-DNA complexes in vitro (Fig. 4B and C).
In contrast to the behavior of the Ser261 and Thr254 replacements in Toa1, we identified three other TFIIA mutants, Toa1-D226V, Toa2-G30E, and Toa2-G30R, that significantly affected the TBP mutant phenotypes in vivo but did not detectably enhance TFIIA-TBP-DNA complex formation in vitro. Analysis of a Toa1 mutant protein in which residues 217 to 240 had been deleted demonstrated that this region is important for interacting with TBP (28). However, on one of the two reported TFIIA-TBP-DNA crystal structures, aspartic acid 226 is not in direct contact with TBP but rather lies in a highly acidic, solvent-exposed region (15, 16). On the other crystal structure, the positions of Asp226 and adjacent amino acids could not be determined accurately, indicating flexibility in this region (70). Our results suggest that this region may represent an important interaction site for factors that regulate TFIIA activity. Interestingly, three serine residues near Asp226, including Ser225, have been recently implicated in phosphorylation of Toa1 (64). Phosphorylation of TFIIA increases its ability to support TFIIA-TBP-TATA complex formation (64). Our genetic studies demonstrated that the Toa1-D226V mutant strongly suppressed the TBP-P109A mutant but significantly enhanced the Ino− phenotype of the TBP-N159D mutant. The latter result is in excellent agreement with the proximity of Asp226 to the presumed sites of phosphorylation. If the D226V change interferes with phosphorylation of Toa1, then the activity of TFIIA in TBP recruitment will be reduced, further compromising TBP-TATA complex formation. The basis for suppression of TBP-P109A by Toa1-D226V is less easily explained by this model. By many other criteria, TBP-P109A and TBP-N159D behave similarly (3, 60). However, substitution of a critical proline residue in TBP that is important for DNA binding and that lies near a TFIIA contact point (16, 70) may significantly alter the conformation of TBP in such a way that it can be suppressed by the mutant TFIIA. This conclusion is consistent with the temperature-sensitive growth properties of TBP-P109A mutant strains.
Despite an extensive search for TOA2 mutations that suppress the TBP mutants, we identified nine mutations that change the same amino acid in the protein, Gly30. This residue lies at the tip of the four-helix bundle domain, within a turn that connects the two alpha helices of Toa2 (16, 70). A replacement of glycine at this position with either glutamic acid or arginine very likely affects the structure of the four-helix bundle domain. Previous studies demonstrated that the four-helix bundle domain is important for the coactivation properties of TFIIA in vitro (40). Our data provide strong support for this conclusion in vivo. Interestingly, alanine scanning mutagenesis of TOA2 generated only three temperature-sensitive alleles, two of which affect residues near glycine 30 (28). In agreement with the inability of the Toa2-G30E and Toa2-G30R proteins to enhance TFIIA-TBP-TATA complex formation in vitro (Fig. 4B and C), Kang et al. (28) concluded that the temperature-sensitive toa2 mutants were defective in a function other than TBP binding, TFIIA-TBP-TATA complex assembly, or subunit association.
The promoter-specific effects of the activation- and DNA binding-defective TBP mutants, coupled with the suppression of these mutants by high levels of TFIIA, suggested to us that the requirement for TFIIA may vary at different promoters. Indeed, depletion of TFIIA dramatically reduced activation of INO1, GAL1, and GAL10 but had no effect on activation of HIS4 or CUP1. These results complement those of Ozer et al. (49), who recently reported a temperature-sensitive toa2 mutation that reduced TBP binding and affected the transcription of a distinct set of genes, including some involved in cell cycle progression. A defect in GAL1 activation was also observed with this toa2 mutant; however, the magnitude of the effect, about twofold, was significantly smaller than what we observed by reducing TFIIA levels. This difference may be due to residual function of the Toa2 mutant protein under the conditions tested. This complexity, which is inherent to mutant analyses of essential genes, can be avoided by using depletion strategies such as that described here. In agreement with the results of Ozer et al. (49), we observed no effect on CUP1 activation following Toa1 depletion. Moreover, the TBP-P109A and TBP-N159D mutant proteins supported wild-type levels of CUP1 induction. These results are in apparent conflict with an earlier report of a TBP mutant, N2-1, which is unable to bind TFIIA in vitro and which causes a significant reduction in CUP1 activation (66). One explanation for this discrepancy is that N2-1 may be impaired for functions other than TFIIA binding and that these functions are critical for CUP1 activation. Interestingly, artificial recruitment of TFIIA by fusing N2-1 with Toa2 only partially rescued the GAL gene activation defect of the TBP mutant (66).
By depleting TFIIA to very low levels and then exposing the cells to conditions for gene activation, we have determined the relative need for TFIIA in the induction of various genes. Previous studies involving conditional depletion of TFIIA focused on genes that were constitutively transcribed and demonstrated a general requirement for TFIIA in maintaining the expression of these genes (28). By first activating INO1 and HIS4 and then depleting TFIIA levels, we have found a similar overall requirement for TFIIA in maintaining the transcription of these genes (39). Moreover, the rates at which these mRNAs decline following TFIIA depletion are similar. These results are in dramatic contrast to the promoter-specific effects in gene induction we have observed (Fig. 6 and 7). Together, our findings suggest that the requirement for TFIIA in the establishment of a preinitiation complex is highly variable; but once formed, the activated transcription complex remains TFIIA dependent to similar degrees at different genes. However, in the absence of more-direct biochemical support, our data can also be explained by a model in which the earliest rounds of transcription reinitiation at promoters such as INO1 are particularly dependent on TFIIA.
By regulating transcription in a gene-specific manner, TFIIA behaves similarly to several other generally acting transcription factors. In a recent genome-wide expression study, important components of the Srb-mediator, Swi-Snf, TFIID, and SAGA complexes were shown to affect only fractions of the RNA polymerase II-transcribed genes in yeast (24). What is the basis for the promoter specificity of TFIIA function? In keeping with the multiple functions of TFIIA, a number of causes are probable. The severity of the activation defect associated with certain TFIIA mutants depends significantly on the activator proteins involved (49). These mutants are also sensitive to differences in promoter structure in vivo (49). For some promoters, this effect of promoter structure most likely stems from the action of additional regulatory factors. For example, by selecting for genetic suppressors of TBP-P109A, we have implicated the Opi1 repressor and the Snf1 kinase pathway in the regulation of PIC assembly at INO1 (60). These findings suggest that TFIID recruitment may be impeded, directly or indirectly, by gene specific-repressor proteins or chromatin at INO1. The apparent lack of a TFIIA requirement at certain promoters, such as CUP1, may indicate that redundant mechanisms contribute to activation. CUP1 is a particularly interesting case because activation of this gene is unaffected by conditions that disfavor holoenzyme (35, 44) or TFIID recruitment (Fig. 7). Perhaps either mode of activation suffices for this promoter, or perhaps this promoter utilizes an entirely different mechanism of activation. Genetic studies on the RNA polymerase II general transcription factors should continue to elucidate the many and diverse strategies used for transcriptional activation in vivo.
ACKNOWLEDGMENTS
We thank the following individuals for the gifts of plasmids and strains: Cori Detweiler and Joachim Li, Steve Hahn, Steve Buratowski, Greg Prelich, Brendan Cormack and Kevin Struhl, Robert Nash, Michael Grunstein, and Fred Winston. We are very grateful to Jim Geiger for crystal structure coordinates and valuable discussions, to Steve Hahn for antisera against TFIIA, to Jonathan Warner for antisera against L3, and to Tony Imbalzano and Bob Kingston for recombinant yeast TFIIA. We thank Jeff Brodsky and members of his laboratory for technical assistance and Greg Prelich, Martin Schmidt, Peggy Shirra, and Fred Winston for helpful discussions and critical reading of the manuscript.
This work was supported by NIH grant GM52593 and by an NSF Career Development Award (MCB-9600955) to K.M.A. S.E.G. was supported by an NSF REU Award.
REFERENCES
- 1.Arndt, K. M. Unpublished data.
- 2.Arndt K M, Ricupero S L, Eisenmann D M, Winston F. Biochemical and genetic characterization of a yeast TFIID mutant that alters transcription in vivo and DNA binding in vitro. Mol Cell Biol. 1992;12:2372–2382. doi: 10.1128/mcb.12.5.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arndt K M, Ricupero-Hovasse S, Winston F. TBP mutants defective in activated transcription in vivo. EMBO J. 1995;14:1490–1497. doi: 10.1002/j.1460-2075.1995.tb07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auble D T, Hahn S. An ATP-dependent inhibitor of TBP binding to DNA. Genes Dev. 1993;7:844–856. doi: 10.1101/gad.7.5.844. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl, ed. 1988. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 6.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 7.Chasman, D., and R. Kornberg. Personal communication.
- 8.Chatterjee S, Struhl K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 9.Chi T, Carey M. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 10.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 11.Cormack B P, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 12.DeJong J, Bernstein R, Roeder R G. Human general transcription factor TFIIA: characterization of a cDNA encoding the small subunit and requirement for basal and activated transcription. Proc Natl Acad Sci USA. 1995;92:3313–3317. doi: 10.1073/pnas.92.8.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenmann D M, Arndt K M, Ricupero S L, Rooney J W, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 14.Ge H, Roeder R G. The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J Biol Chem. 1994;269:17136–17140. [PubMed] [Google Scholar]
- 15.Geiger, J. H. Personal communication.
- 16.Geiger J H, Hahn S, Lee S, Sigler P B. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 17.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 18.Griggs D W, Johnston M. Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc Natl Acad Sci USA. 1991;88:8597–8601. doi: 10.1073/pnas.88.19.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall M N, Hereford L, Herskowitz I. Targeting of E. coli β-galactosidase to the nucleus in yeast. Cell. 1984;36:1057–1065. doi: 10.1016/0092-8674(84)90055-2. [DOI] [PubMed] [Google Scholar]
- 20.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartwell L H. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976;104:803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch J P, Henry S A. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol Cell Biol. 1986;6:3320–3328. doi: 10.1128/mcb.6.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 24.Holstege F C P, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 25.Imbalzano A N, Zaret K S, Kingston R E. Transcription factor (TF) IIB and TFIIA can independently increase the affinity of the TATA-binding protein for DNA. J Biol Chem. 1994;269:8280–8286. [PubMed] [Google Scholar]
- 26.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 27.Kaiser K, Stelzer G, Meisterernst M. The coactivator p15(PC4) initiates transcriptional activation during TFIIA-TFIID-promoter complex formation. EMBO J. 1995;14:3520–3527. doi: 10.1002/j.1460-2075.1995.tb07358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang J J, Auble D T, Ranish J A, Hahn S. Analysis of the yeast transcription factor TFIIA: distinct functional regions and a polymerase II-specific role in basal and activated transcription. Mol Cell Biol. 1995;15:1234–1243. doi: 10.1128/mcb.15.3.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J L, Nikolov D B, Burley S K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 30.Kim Y, Geiger J H, Hahn S, Sigler P B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 31.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 32.Klein C, Struhl K. Increased recruitment of TATA-binding protein to the promoter by transcriptional activation domains in vivo. Science. 1994;266:280–282. doi: 10.1126/science.7939664. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi N, Boyer T G, Berk A J. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol Cell Biol. 1995;15:6465–6473. doi: 10.1128/mcb.15.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kokubo T, Swanson M J, Nishikawa J I, Hinnebusch A G, Nakatani Y. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol Cell Biol. 1998;18:1003–1012. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee D, Lis J T. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature. 1998;393:389–392. doi: 10.1038/30770. [DOI] [PubMed] [Google Scholar]
- 36.Lee D K, DeJong J, Hashimoto S, Horikoshi M, Roeder R G. TFIIA induces conformational changes in TFIID via interactions with the basic repeat. Mol Cell Biol. 1992;12:5189–5196. doi: 10.1128/mcb.12.11.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee M, Struhl K. Mutations on the DNA-binding surface of TBP can specifically impair the response to acidic activators in vivo. Mol Cell Biol. 1995;15:5461–5469. doi: 10.1128/mcb.15.10.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lieberman P M, Berk A J. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 39.Liu, Q., and K. M. Arndt. Unpublished data.
- 40.Ma D, Olave I, Merino A, Reinberg D. Separation of the transcriptional coactivator and antirepression functions of transcription factor IIA. Proc Natl Acad Sci USA. 1996;93:6583–6588. doi: 10.1073/pnas.93.13.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma D, Watanabe H, Mermelstein F, Admon A, Oguri K, Sun X, Wada T, Imai T, Shiroya T, Reinberg D, Handa H. Isolation of a cDNA encoding the largest subunit of TFIIA reveals functions important for activated transcription. Genes Dev. 1993;7:2246–2257. doi: 10.1101/gad.7.11.2246. [DOI] [PubMed] [Google Scholar]
- 42.Madison J M, Winston F. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:287–295. doi: 10.1128/mcb.17.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClary J A, Witney F, Geisselsoder J. Efficient site-directed in vitro mutagenesis using phagemid vectors. BioTechniques. 1989;7:282–289. [PubMed] [Google Scholar]
- 44.McNeil J B, Agah H, Bentley D. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev. 1998;12:2510–2521. doi: 10.1101/gad.12.16.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meisterernst M, Roeder R G. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 46.Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 47.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 48.Ozer J, Bolden A H, Lieberman P M. Transcription factor IIA mutations show activator-specific defects and reveal a IIA function distinct from stimulation of TBP-DNA binding. J Biol Chem. 1996;271:11182–11190. doi: 10.1074/jbc.271.19.11182. [DOI] [PubMed] [Google Scholar]
- 49.Ozer J, Lezina L E, Ewing J, Audi S, Lieberman P M. Association of transcription factor IIA with TATA binding protein is required for transcriptional activation of a subset of promoters and cell cycle progression in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:2559–2570. doi: 10.1128/mcb.18.5.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozer J, Mitsouras K, Zerby D, Carey M, Lieberman P M. Transcription factor IIA derepresses TATA-binding protein (TBP)-associated factor inhibition of TBP-DNA binding. J Biol Chem. 1998;273:14293–14300. doi: 10.1074/jbc.273.23.14293. [DOI] [PubMed] [Google Scholar]
- 51.Ozer J, Moore P A, Bolden A H, Lee A, Rosen C A, Lieberman P M. Molecular cloning of the small (γ) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev. 1994;8:2324–2335. doi: 10.1101/gad.8.19.2324. [DOI] [PubMed] [Google Scholar]
- 52.Petri V, Hsieh M, Brenowitz M. Thermodynamic and kinetic characterization of the binding of the TATA binding protein to the adenovirus E4 promoter. Biochemistry. 1995;34:9977–9984. doi: 10.1021/bi00031a020. [DOI] [PubMed] [Google Scholar]
- 53.Pinto I, Ware D E, Hampsey M. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell. 1992;68:977–988. doi: 10.1016/0092-8674(92)90040-j. [DOI] [PubMed] [Google Scholar]
- 54.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 55.Ranish J A, Lane W S, Hahn S. Isolation of two genes that encode subunits of the yeast transcription factor IIA. Science. 1992;255:1127–1129. doi: 10.1126/science.1546313. [DOI] [PubMed] [Google Scholar]
- 56.Ranish J A, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reddy P, Hahn S. Dominant negative mutations in yeast TFIID define a bipartite DNA-binding region. Cell. 1991;65:349–357. doi: 10.1016/0092-8674(91)90168-x. [DOI] [PubMed] [Google Scholar]
- 58.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 59.Sayre M H, Tschochner H, Kornberg R D. Reconstitution of transcription with five purified initiation factors and RNA polymerase II from Saccharomyces cerevisiae. J Biol Chem. 1992;267:23376–23382. [PubMed] [Google Scholar]
- 60.Shirra M K, Arndt K M. Evidence for the involvement of the Glc7-Reg1 phosphatase and the Snf1-Snf4 kinase in the regulation of INO1 transcription in Saccharomyces cerevisiae. Genetics. 1999;152:73–87. doi: 10.1093/genetics/152.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shykind B M, Kim J, Sharp P. Activation of the TFIID-TFIIA complex with HMG-2. Genes Dev. 1995;9:1354–1365. doi: 10.1101/gad.9.11.1354. [DOI] [PubMed] [Google Scholar]
- 62.Shykind B M, Kim J, Stewart L, Champoux J J, Sharp P A. Topoisomerase I enhances TFIID-TFIIA complex assembly during activation of transcription. Genes Dev. 1997;11:397–407. doi: 10.1101/gad.11.3.397. [DOI] [PubMed] [Google Scholar]
- 63.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Solow S P, Lezina L, Lieberman P M. Phosphorylation of TFIIA stimulates TATA binding protein-TATA interaction and contributes to maximal transcription and viability in yeast. Mol Cell Biol. 1999;19:2846–2852. doi: 10.1128/mcb.19.4.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stargell L A, Struhl K. Mechanisms of transcriptional activation in vivo: two steps forward. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- 66.Stargell L A, Struhl K. The TBP-TFIIA interaction in the response to acidic activators in vivo. Science. 1995;269:75–78. doi: 10.1126/science.7604282. [DOI] [PubMed] [Google Scholar]
- 67.St. John T P, Davis R W. The organization and transcription of the galactose gene cluster of Saccharomyces. J Mol Biol. 1981;152:285–315. doi: 10.1016/0022-2836(81)90244-8. [DOI] [PubMed] [Google Scholar]
- 68.Stolinski L A, Eisenmann D M, Arndt K M. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4490–4500. doi: 10.1128/mcb.17.8.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun X, Ma D, Sheldon M, Yeung K, Reinberg D. Reconstitution of human TFIIA activity from recombinant polypeptides: a role in TFIID-mediated transcription. Genes Dev. 1994;8:2336–2348. doi: 10.1101/gad.8.19.2336. [DOI] [PubMed] [Google Scholar]
- 70.Tan S, Hunziker Y, Sargent D F, Richmond T J. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 71.Wang W, Gralla J D, Carey M. The acidic activator GAL4-AH can stimulate polymerase II transcription by promoting assembly of a closed complex requiring TFIID and TFIIA. Genes Dev. 1992;6:1716–1727. doi: 10.1101/gad.6.9.1716. [DOI] [PubMed] [Google Scholar]
- 72.Weideman C A, Netter R C, Benjamin L R, McAllister J J, Schmiedekamp L A, Coleman R A, Pugh B F. Dynamic interplay of TFIIA, TBP and TATA DNA. J Mol Biol. 1997;271:61–75. doi: 10.1006/jmbi.1997.1152. [DOI] [PubMed] [Google Scholar]
- 73.Winston F, Dollard C, Ricupero-Hovasse S. Construction of a set of convenient S. cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 74.Winston F, Durbin K J, Fink G R. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell. 1984;39:675–682. doi: 10.1016/0092-8674(84)90474-4. [DOI] [PubMed] [Google Scholar]
- 75.Xiao H, Friesen J D, Lis J T. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yokomori K, Admon A, Goodrich J A, Chen J, Tjian R. Drosophila TFIIA-L is processed into two subunits that are associated with the TBP/TAF complex. Genes Dev. 1993;7:2235–2245. doi: 10.1101/gad.7.11.2235. [DOI] [PubMed] [Google Scholar]
- 77.Yokomori K, Zeidler M P, Chen J, Verrijzer C P, Mlodzik M, Tjian R. Drosophila TFIIA directs cooperative DNA binding with TBP and mediates transcriptional activation. Genes Dev. 1994;8:2313–2323. doi: 10.1101/gad.8.19.2313. [DOI] [PubMed] [Google Scholar]
- 78.Zhou Y, Zhang X, Ebright R H. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res. 1991;19:6052. doi: 10.1093/nar/19.21.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]