Abstract
Human Immunodeficiency Virus type-1 (HIV-1) is the causative agent of AIDS. Its entry step is mediated by the envelope glycoprotein (Env). During the entry process, Env vastly changes its conformation. While non-liganded Env tends to have a closed structure, receptor-binding of Env opens its conformation, which leads to virus-cell membrane fusion. Single-molecule fluorescence resonance energy transfer (smFRET) imaging allows observation of these conformational changes on the virion surface. Nascent HIV-1 particles incorporate multiple host transmembrane proteins, some of which inhibit the entry process. The Env structure or its dynamics may determine the effectiveness of these antiviral mechanisms. Here, we review recent findings about the Env conformation changes on virus particles and inhibition of Env activities by virion-incorporated host transmembrane proteins.
Introduction
Human Immunodeficiency Virus type-1 (HIV-1) is the causative agent of Acquired Immunodeficiency Syndrome (AIDS). Currently over 38 million people in the world are living with HIV-1 (2020 https://www.who.int/news-room/fact-sheets/detail/hiv-aids). Combination antiretroviral therapy has allowed the control of viral loads in infected individuals and thereby the steady decrease of new HIV-1 infection and mortality. However, in the absence of effective vaccines and cure regimens, the emergence of drug-resistant mutant viruses and long-term effects of the current anti-HIV-1 drugs remain as major challenges to human health and society. The HIV-1 entry, mediated by the viral envelope glycoprotein Env, is one of the promising targets for the development of new antiretrovirals due to its importance in HIV-1 replication cycle. Detailed understanding of the native structure and dynamics of Env, the sole viral protein exposed on the surface of infectious virus particles, is intensely pursued to help the efforts to develop vaccines and small compounds that block HIV-1 entry.
Overview of HIV-1 Entry
HIV-1 is a retrovirus that infects CD4+ T cells [1], macrophages [2], and dendritic cells [3]. HIV-1 entry consists of attachment of a virus particle to target cells and subsequent fusion of viral and target cell membranes. Both virus attachment and fusion are driven by Env. Env is synthesized as a precursor protein, gp160 and processed into the surface glycoprotein gp120 and the transmembrane glycoprotein gp41. Heterodimers of gp120 and gp41 form the trimer, which is a functional unit on the surface of infectious virus particles [4]. Virus attachment is mediated by several cellular factors, but binding of gp120 to the host receptor CD4 is central to the attachment process on the surface of target cells [5]. The binding between gp120 and CD4 triggers conformational changes in Env that allow for a subsequent interaction of gp120 with coreceptors CCR5 or CXCR4 [6–8]. This interaction induces extension of gp41 and insertion of its N-terminal fusion peptide to the target cell membrane. Finally, gp41 refolds into a hairpin-like conformation in a six-helix bundle structure, leading to fusion of viral and host cell membranes [7–9].
Coreceptor usage determines HIV-1 cell tropism. In early studies, HIV-1 strains efficiently replicating in T cell lines and macrophages were called T cell-tropic and macrophage-tropic (M-tropic) strains, respectively. Later, it was found that the viruses that grow efficiently in T cell lines use CXCR4 as the coreceptor, whereas M-tropic viruses utilize CCR5. Thus, the classification of HIV-1 into two types, X4 T cell-tropic and R5 M-tropic viruses, had been commonly used. However, since CCR5-using viruses also grow in primary CD4+ T cells, and not all CCR5-dependent virus isolates efficiently infect macrophages, the binary link between coreceptor usage (CXCR4 versus CCR5) and cell tropism (T cell versus macrophage) is no longer accepted. Currently, HIV-1 is classified into three types: (i) R5 T cell-tropic, (ii) X4 T cell-tropic, and (iii) R5 M-tropic [10]. Most of HIV-1 isolated at early stages of infection is R5 T-tropic virus [11]. This type of viruses requires high levels of CD4 on the surface of target cells for fusion. Therefore, the major target of these viruses is CD4+ T cells but not macrophages, which express a low density of CD4 compared to CD4+ T cells [12]. X4 T cell-tropic viruses emerge at late stages of HIV-1 infection in a proportion of HIV-1-infected patients. This coreceptor switch correlates with disease progression and immune activation as evident from the increase in HLA-DR+ and HLA-DR+CD38+ populations in CD4+ T cells [13–16]. R5 M-tropic viruses are mainly detected in brain tissues and the cerebrospinal fluid [17,18]. These viruses are able to enter cells expressing a low density of CD4 on the surface (i.e. macrophages) efficiently [10]. To enable efficient entry into CD4low target cells, Env derived from these viruses binds to CD4 with a high affinity [19,20]. Thus, the adaptation to the low CD4 density determines the cell tropism of R5 viruses for macrophages versus primary CD4+ T cells. As for the choice of coreceptors (i.e. R5 versus X4), the gp120 V3 loop, which is exposed upon Env-CD4 binding [21,22] and forms a part of the interface in Env-coreceptor binding [23], is a major molecular determinant [24]. Mechanistic aspects of Env-coreceptor binding had been hampered due to the lack of structural information, but recent studies finally revealed the details of the gp120-coreceptor interactions [21,23,25,26].
HIV-1 fusion takes place at multiple sites [27]. Early studies suggested that HIV-1 entry takes place at the plasma membrane based on the observations, among others, that HIV-1 can fuse with target cells at neutral pH and that mutagenesis of the CD4 cytoplasmic tail that impairs ligand-induced internalization does not block HIV-1 infection [1,28,29]. However, more recent evidence obtained through inhibition of clathrin-mediated endocytosis, time-course analysis of surface accessibility of cell-associated viruses versus virus-cell fusion, and/or live cell imaging supports the contribution of virion endocytosis to HIV-1 infection [30–35]. Yet, whether endocytosis plays a role in productive HIV-1 entry into CD4+ T cells is still debated due to the contradictory results obtained with this cell type [30,35–37]. The cell activation status also seems to affect the frequency of virus fusion at endosomes in primary CD4+ T cells [30]. Macropinocytosis, a form of clathrin-independent endocytosis, has been shown to contribute to HIV-1 entry into macrophages [38,39], but the details remain to be elucidated [27].
Considering the importance of the HIV-1 entry step in virus replication cycle and hence in development of therapeutic and preventive strategies, it is not surprising that a tremendous amount of research has been performed on many aspects of HIV-1 entry. For more comprehensive understanding on HIV-1 entry, Env structures, and strategies to block the entry process, readers are directed to recent excellent reviews [5,40–42]. In the subsequent sections, we will highlight recent findings in two aspects of the entry process, the conformational dynamics of Env and the effects of virus-incorporated host proteins.
Conformational dynamics of Env
The Env trimer on the surface of infectious native virions is structurally flexible. Pre-fusion Env trimers are thought to adopt a closed structure, which, along with sequence variations and heavy glycosylation, is likely to allow HIV-1 to evade the host immune system because key functional regions of Env are hidden [43]. Once Env binds to CD4, the closed structure transitions toward an open conformation to expose the co-receptor binding site [21,22,44–47] (also see above). However, unliganded Env trimers also spontaneously transition between closed, open, and intermediate conformations.
Single-molecule fluorescence resonance energy transfer (smFRET) imaging performed using the total internal reflection fluorescence microscopy has allowed real-time monitoring of this conformational change of Env on the surface of native virions occurring in the milliseconds-to-seconds range [48–51]. Non-liganded Env on the native virions shows three distinct conformations, low-FRET (State 1), intermediate-FRET (State 3), and high-FRET (State 2) conformations (Figure 1) [48,52,53]. State 1 likely represents the pre-triggered closed conformation. State 2 is an intermediate conformation, which is also observed for the non-CD4-bound Env protomer in an asymmetric Env trimer where other protomer(s) is bound to CD4. State 3 is observed with the Env protomer bound to CD4 and hence likely to be in an open conformation.
Figure 1. Conformational dynamics of the Env trimer.
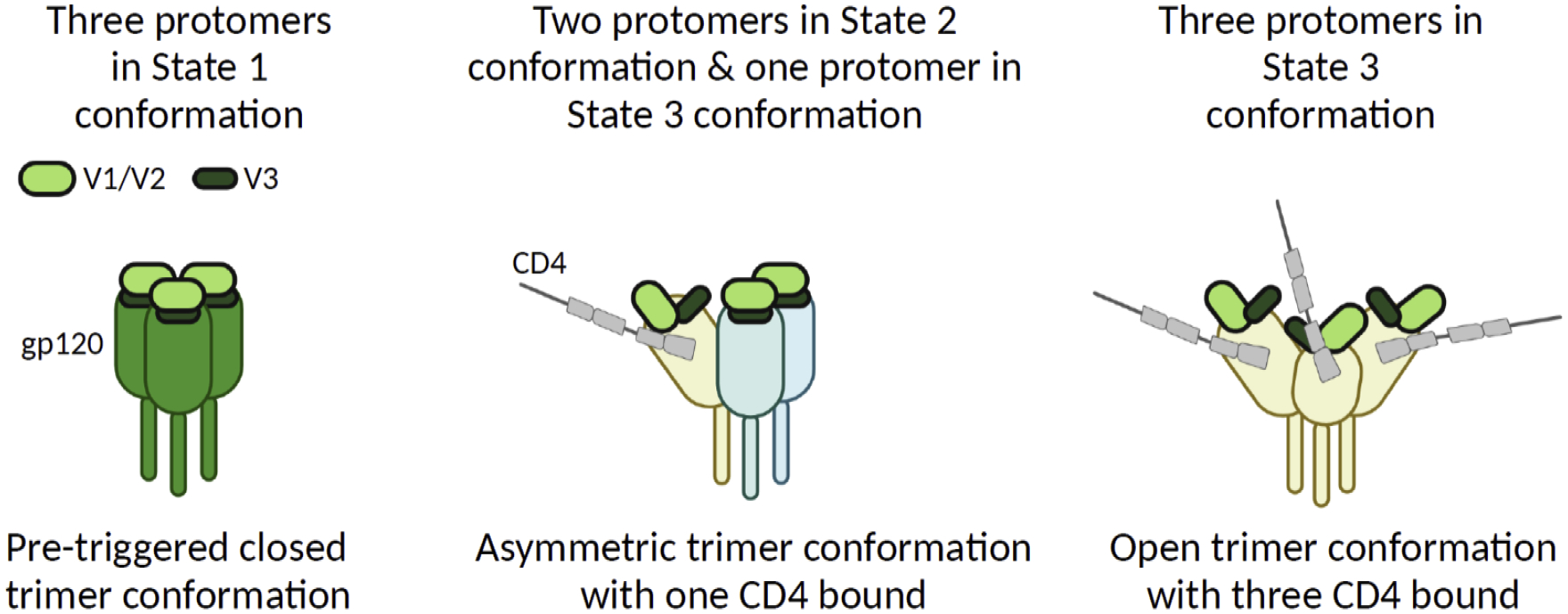
smFRET imaging revealed three conformational states of Env protomers (State 1, State 2, and State 3). State 1 Env is likely to represent a non-liganded conformation in pre-triggered trimers that have a closed structure. State 2 represents an intermediate conformation, which is observed for the non-CD4-bound Env protomer in an asymmetric Env trimer where other protomer(s) is bound to CD4. State 3 Env is observed with the Env protomer bound to CD4, which has an open conformation. In non-liganded Env, the V1/V2 loop masks the V3 loop that binds to coreceptors, CCR5 and CXCR4. Once CD4 binds to Env, the V3 loop is exposed. High-resolution structure of State 1 Env has not been determined. Created with BioRender.com.
Notably, non-liganded Env proteins of primary isolate strains JR-FL and BG505 display the signal for State 1 conformation more abundantly than Env of a lab-adapted HIV-1 strain NL4–3 [48,52]. Therefore, compared to the lab-adapted HIV-1 Env, Env trimers of the primary isolates likely spend a longer time in closed structures in the absence of ligands. The dynamic nature of Env structures has implications in development of vaccines aimed at eliciting broadly neutralizing antibodies (bNAbs). As noted in the tier classification of HIV-1 neutralization phenotypes [54], Env trimers that have open conformation as observed for lab adapted strains are readily blocked by non-bNAb antibodies (Tier 1A). In contrast, Env trimers of most circulating strains, which are the high priority target for bNAb vaccines, are associated with a predominantly closed conformation (Tier 2/3). Antibodies that neutralize Tier 1A viruses are often unable to neutralize Tier 2/3 viruses.
Transition of Env conformational states is likely to be mediated by various amino acid residues in gp120. Introducing an amino acid substitution in the V1/V2 loop or the β20-β21 region of JR-FL gp120 reduces State 1 conformation and increases State 2 and State 3 conformations [55,56]. These data in combination with the outcomes of other approaches indicate that the V1/V2 loops and the β20-β21 element, which are located at the trimer apex and near the CD4 contact site, respectively, play a role in maintaining the closed structure of Env. In addition, substitutions of specific amino acids in several regions of gp120 prevent transitioning of Env conformation from State 1 to State 2 and State 3 even when gp120 is bound to CD4 [57]. These and single particle cryoEM results identify the gp120 allosteric network that is involved in conformational changes of Env upon CD4 binding. The smFRET assay has also revealed the conformational states of Env that is bound to bNAbs or HIV-1 entry inhibitors [48,53]. Binding of Env with most of bNAbs stabilizes State 1 conformation despite the fact that the bNAbs recognize different epitopes. Similarly, BMS-626539, one of HIV-1 entry inhibitors, also increases State 1 conformation of Env. Therefore, stabilization of State 1 and/or inhibition of the transition of Env from State 1 to downstream conformations is a promising strategy for inhibition of HIV-1 entry.
Inhibition of HIV-1 entry by virion-incorporated host transmembrane proteins
In addition to Env, HIV-1 particles incorporate various host transmembrane proteins during the assembly at the plasma membrane. When incorporated into virions, some of the transmembrane proteins, such as ICAM-1, can promote HIV-1 entry through interactions with receptors on the surface of target cells [58]. HIV-1 also incorporates transmembrane proteins that inhibit the HIV-1 entry step. Earlier studies showed that tetraspanin proteins, which are efficiently incorporated into HIV-1 particles and inhibit cell-cell fusion [59,60], suppress post-virus attachment entry step(s) in a strain-specific manner [61]. Recent studies have identified an increasing number of virion-incorporated host transmembrane proteins that inhibit HIV-1 entry process (Figure 2), including serine incorporators (SERINCs) [62,63], interferon inducible transmembrane (IFITM) proteins [64–66], P-selectin glycoprotein ligand-1 (PSGL-1), and CD43 [67,68].
Figure 2. Inhibitory effect of antiviral proteins to HIV-1 entry steps.
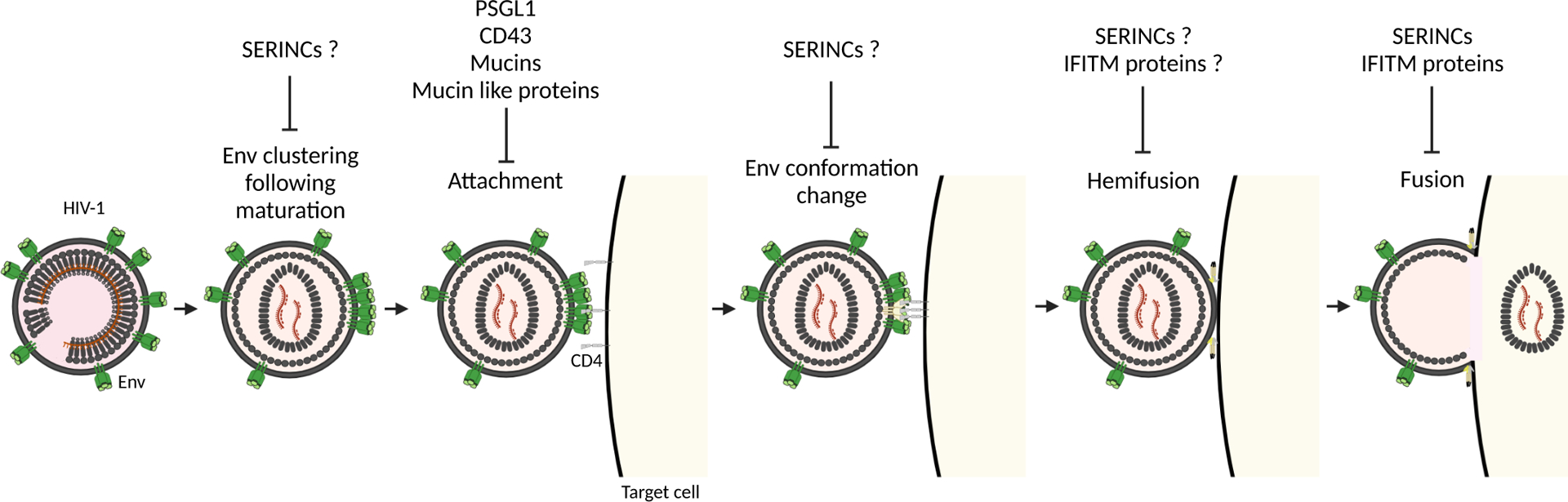
SERINCs and IFITM proteins prevent fusion, and PSGL-1, CD43, mucins, and mucin-like proteins inhibit virus attachment to target cells. For clarity, coreceptors are not shown. Created with BioRender.com.
SERINCs.
SERINCs are multipass transmembrane proteins. It was originally reported that these proteins play a role in serine incorporation and promoting phosphatidylserine (PS) and sphingomyelin biosynthesis [69]. At least SERINC3 and 5 are known to prevent HIV-1 infection, and SERINC5 has the stronger antiviral activity than SERINC3 [62,63]. SERINCs are counteracted by Nef, one of accessary proteins of HIV-1. In the absence of Nef, SERINCs are incorporated into progeny virions. The virion-incorporated SERINCs restrict HIV-1-cell fusion [62,63]. Although the exact molecular mechanism(s) by which SERINCs inhibit HIV-1 infection remains to be determined, SERINC5 reduces HIV-1 infectivity in a manner dependent on Env [62,63,70]. A correlation between the openness of Env trimer and the sensitivity to SERINCs (or Nef dependence) has been observed when Envs from different strains are compared [70–72]. Moreover, CD4, which induces open Env conformation (see above), was observed to sensitize a Tier 3 Env to SERINC5 [72]. Furthermore, the cytoplasmic tail (CT) of Env is required for the sensitivity to SERINC5 [73]. Upon deletion of the CT, which is known to alter Env conformation [74], HIV-1 is completely insensitive to SERINC5 [73]. Based on the sensitivity to neutralization by antibodies, the authors suggested that the CT-deleted Env may display a more closed conformation and thereby evade SERINC5-mediated inhibition [73]. However, the Env trimer propensity to adopt an open conformation as assessed by antibody neutralization does not appear to be the sole determinant for the SERINC sensitivity [75].
Approaches based on antibody binding also revealed that SERINC5 modifies the conformation of SERINC-sensitive Env on the surface of virions [76,77]. The changes in Env conformation include ones in gp41, perhaps leading to suppression of the fusion pore formation between viruses and target cells as well as spontaneous inactivation of Env [78]. Suppression of fusion pore formation by SERINCs is further examined in detail using the cryo-electron tomography of giant plasma membrane vesicles, which enabled observation of each step in the HIV-1 fusion process. This approach demonstrated that virion-incorporated SERINC3 and 5 stall HIV-1 fusion at the hemifusion and abnormal early fusion steps and inhibit opening of fusion pores [79]. Another line of investigation suggests a role for interactions between Env trimers; a 3D superresolution microscopy technique showed that SERINC5 blocks the formation of Env clusters on the surface of virus particles, which promotes efficient HIV-1 entry [80], without affecting Env incorporation [81]. The deletion of Env CT alters the distribution pattern of Env clusters [80], and Env clustering requires its interactions with cholesterol on the virus particles through the CT [82]. Therefore, considering that the CT-deleted Env is insensitive to SERINC5 [73], SERINC5 might affect the interactions of the Env CT with cholesterol directly or indirectly via changes in other lipids that interact with cholesterol. Nevertheless, the two restriction mechanisms, that is, the modification of Env conformation and the inhibition of Env clustering, are unlikely to require altering lipid composition of virus particles because lipid mass spectrometry revealed that SERINC5 does not affect steady-state lipid composition of virus-producing cells and virions [83]. However, SERINCs may still affect lipid clustering or inter-leaflet distribution in the viral envelop membrane. In addition, it is currently unknown whether these mechanisms postulated for the antiviral function of SERINC5 are mutually exclusive or not.
IFITMs.
IFITM proteins are small transmembrane proteins consisting of a transmembrane domain, a hydrophobic intra-membrane-associated domain, and an intervening highly conserved intracellular loop [84,85]. Among IFITM proteins, IFITM1, 2, and 3 inhibit HIV-1 infection [84–86]. Earlier studies suggested that these IFITM proteins modulate fluidity or rigidity of target cell membranes, resulting in inhibition of virus-cell fusion [87–90]. Notably, it has been reported that CXCR4- and CCR5-dependent HIV-1 strains show different sensitivity to IFITM1 versus IFITM2/3; CXCR4-dependent viruses are more sensitive to IFITM2/3, whereas CCR5-requiring viruses are more susceptible to IFITM1 [91]. Since IFITM proteins show different subcellular localization patterns [91–93], it was suggested that the site of coreceptor-triggered HIV-1 fusion might correlate with the sensitivity to each IFITM protein [91]. Of note, endocytosis inhibitors diminish the inhibitory effect of IFITM2 and 3 on infection of primary isolates, suggesting that the viruses utilize endocytosis for entry and that IFITM2 and IFITM3 could inhibit fusion to the endosomal membrane [91]. Transmitted/founder (T/F) viruses, which are isolated from recently infected patients and CCR5-dependent, are shown to be resistant to all IFITM proteins expressed in target cells [91,94,95]. However, the sensitivity of T/F viruses to restriction by IFITM proteins is a matter of debate; a more recent study showed that IFITM proteins in target cells inhibit infection of T/F viruses [96].
In addition to the role of IFITM proteins in target cells, IFITM proteins in virus-producing cells also attenuate HIV-1 fusion through inhibition of Env processing or when these proteins are incorporated into virions [64–66,97]. Therefore, IFITM proteins suppress HIV-1 fusion in either target cells or virus-producing cells. Similar to SERINC5 [70], Env, in particular the V3 loop, determines the sensitivity to IFITM3 [94]. However, it appears that IFITM3 and SERINC5 target Envs that may differ in sampling of conformations. When a panel of Env isolates is compared for sensitivities to a CD4 blocking antibody and soluble CD4 (sCD4), which inform about the Env-CD4 binding affinity, SERINC5-sensitive Envs tend to show stronger CD4 binding, whereas no such correlation is observed between the IFITM3 sensitivity and CD4 binding [98]. Moreover, upon binding to a CD4 mimetic that stabilizes the open conformation [99], an Env isolate resistant to both SERINC5 and IFITM3 is sensitized markedly to SERINC5 but only modestly, if any, to IFITM3 [98]. The mechanism(s) by which the presence of IFITM proteins in virus-producing cells and/or progeny virus particles inhibits entry of progeny virions remains to be determined. While a reduction in virion-associated Env levels upon IFITM3 expression in virus-producing cells has been observed in some studies [94,97,100,101], others did not detect a difference in Env incorporation using different experimental systems [64,66,91,102]. Furthermore, even in the study where reduced Env incorporation is observed, the impact of IFITM3 on virion infectivity cannot be fully explained by the Env quantity in virions [100,101]. Whether virion-incorporated IFITM proteins affect viral membrane rigidity, as is shown to occur in target cells [89,90,101] or whether they alter other properties of the envelope membrane is unknown. However, virion incorporation of IFITM3 increases the sensitivity of Env to several neutralizing antibodies [102], suggesting that IFITM3 directly or indirectly affects the Env conformation on the virus particle surface.
PSGL-1 and CD43.
PSGL-1 is a mucin-like type I transmembrane glycoprotein, and CD43 is a sialomucin type I transmembrane glycoprotein [103–105]. These proteins are primarily expressed on the surface of lymphocytes, mediating cell tethering and rolling through interactions with selectin family proteins to promote cell migration into inflamed tissue [106]. Both PSGL-1 and CD43 are also known to prevent cell-cell interactions via its extended extracellular domains estimated to be 45–50 nm long [107–110]. We previously showed that PSGL-1 and CD43 associate with HIV-1 structural protein Gag at the plasma membrane of virus-producing cells and get incorporated into nascent virions [111–113]. Later, both PSGL-1 and CD43 were identified as antiviral factors that reduce infectivity of progeny virions when it is expressed by virus-producing cells [114,115]. These proteins share some of characteristics of restriction factors, such as a signature of positive selection during primate evolution [114,115] and downregulation upon HIV-1 infection [67,68,115–117]. Therefore, PSGL-1 and CD43 could potentially be classified as restriction factors. More recently, Fu et al. and our group discovered that virion-incorporated PSGL-1 and CD43 inhibit the HIV-1 entry step [67,68]. These studies revealed that unlike SERINCs and IFITM proteins in virus particles, PSGL-1 and CD43 suppress virus entry at the step of virus attachment to target cells and that they inhibit the attachment regardless of molecules mediating virus-cell binding. This inhibition requires the intact extended extracellular domain at least for PSGL-1. Since these extracellular domains are longer than combined length of receptor and ligand pairs mediating HIV-1-cell binding, these results suggest that virion-incorporated PSGL-1 and CD43 create a physical barrier that sterically prevents HIV-1 from binding to target cells or inserting its fusion peptide into the target cell membrane. Indeed, the cryo electron tomography in the presence of fusion inhibitors revealed that the distance between a target cell and cell-attached HIV-1 by extended pre-hairpin intermediate Env is 15.6±2.8 nm [118], that is, ~3 fold shorter than the lengths of PSGL-1 and CD43. Extracellular domains of PSGL-1 and CD43 are highly O-glycosylated [104,105,119], and the O-glycosylation is thought to contribute to the maintenance of the extended structure [120]. Other mucins and mucin-like proteins, such as CD164, PODXL1, PODXL2, CD34, TMEM123, and MUC1, have a similar structure, that is, highly O-glycosylated extended extracellular domains. These proteins, collectively termed SHREK proteins, are also incorporated into progeny HIV-1 and reduce HIV-1 infectivity at least in overexpression experiments [121]. These observations support the possibility that virion-incorporated transmembrane proteins that have the elongated extracellular domains can sterically hinder virus attachment to target cells.
As an additional mechanism, PSGL-1-mediated inhibition of Env incorporation into nascent virions at the plasma membrane has been proposed based on the experiments in which 293T or Jurkat cells were used as virus-producing cells [67,122]. Consistent with this possibility, in virus-producing Jurkat cells, PSGL-1 interacts with Env gp41 and alters the localization of gp41. However, in another study in which HIV-1 is produced from PBMCs, amounts of virion-incorporated PSGL-1 do not inversely correlate with amounts of Env in the virions [123]. Therefore, whether PSGL-1 affects Env incorporation into virions in primary CD4+ T cells warrants future investigation. Likewise, another proposed antiviral mechanism in which PSGL-1 inhibits actin depolymerization in the virions, thereby reducing virion infectivity [122], awaits validation using primary CD4+ T cells. Finally, PSGL-1 in target cells was suggested restrict HIV-1 reverse transcription through inhibition of actin disassembly [115,122]. However, another group observed that PSGL-1 expression in target cells did not block any early HIV-1 replication step including reverse transcription [67]. Therefore, it remains to be determined which aspect of experimental conditions caused this discrepancy and to what extent PSGL-1 expressed in target cells inhibits reverse transcription under physiological conditions.
Concluding remarks
Understanding of conformational dynamics of Env on the virions is important not only for the development of neutralizing antibodies and antiretrovirals but also for elucidating the action of host-encoded antiviral proteins. The smFRET approach visualized real-time conformational dynamics of non-liganded and CD4-bound, antibody-bound, or small compound-bound Env trimer on the surface of native virions and defined the predominant conformation of the Env trimer in the milliseconds-to-seconds time scale. This technique identified the previously uncharacterized asymmetric intermediate state, key Env residues regulating the conformational transitioning of Env, and the conformational state of bNAb-bound or entry inhibitor-bound Env. The presence of asymmetric intermediate State 2 suggests that Env trimers more intricately rearrange the conformation upon receptor and coreceptor binding during HIV-1 entry than we previously imagined. Further understanding of detailed mechanism(s) regulating conformational changes within an Env protomer and between protomers, the latter using different approaches [124], will potentially allow us to develop new entry inhibitors and vaccine strategies. It is also important to determine the high-resolution structure of State 1 Env, since it may explain the mechanisms of inhibition by bNAbs and an entry inhibitor, which stabilize this state. Notably, high-resolution structures of gp120 obtained thus far are largely based on a soluble Env trimer engineered for stabilization (SOSIP) [21,22,44,125–131], which was found to be in the State 2 conformation [53].
Various virion-incorporated host transmembrane proteins that inhibit HIV-1 entry were recently identified. However, the molecular mechanism(s) by which these proteins suppress HIV-1 entry remain to be determined. For example, how SERINCs affect Env conformation and clustering is unclear. Whether IFITM proteins act on the behavior of Env trimers on the virus surface and whether they target Env in a specific conformation remain to be determined. Whether PSGL-1 and CD43 inhibit HIV-1 binding to target cells due to lengths or chemical properties of these proteins, for example, increased negative charge or heavy glycosylation, needs to be addressed. In addition, how these proteins get incorporated into infectious virus particles warrants future investigation. Elucidation of these mechanisms could offer insights into new therapeutic strategies.
Acknowledgements
We thank members of our laboratory for helpful discussions. This work is supported by N.I.H., grants R37 AI 071727 and R21 AI 148381 (to A.O.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Stein BS, Gowda SD, Lifson JD, Penhallow RC, Bensch KG, Engleman EG: pH-independent HIV entry into CD4-positive T cells via virus envelope fusion to the plasma membrane. Cell 1987, 49:659–668. [DOI] [PubMed] [Google Scholar]
- 2.Gartner S, Markovits P, Markovitz DM, Betts RF, Popovic M: Virus isolation from and identification of HTLV-III/LAV-producing cells in brain tissue from a patient with AIDS. JAMA 1986, 256:2365–2371. [PubMed] [Google Scholar]
- 3.Patterson S, Knight SC: Susceptibility of human peripheral blood dendritic cells to infection by human immunodeficiency virus. J Gen Virol 1987, 68:1177–1181. [DOI] [PubMed] [Google Scholar]
- 4.Wyatt R, Sodroski J: The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 1998, 280:1884–1888. [DOI] [PubMed] [Google Scholar]
- 5.Chen B: Molecular mechanism of HIV-1 entry. Trends Microbiol 2019, 27:878–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch ML, Earl PL, Fargnoli K, Picciafuoco S, Giombini F, Wong-Staal F, Franchini G: Identification of the fusion peptide of primate immunodeficiency viruses. Science 1989, 244:694–697. [DOI] [PubMed] [Google Scholar]
- 7.Berger EA, Murphy PM, Farber JM: Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol 1999, 17:657–700. [DOI] [PubMed] [Google Scholar]
- 8.Checkley MA, Luttge BG, Freed EO: HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol 2011, 410:582–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin M, Da LT: Refolding dynamics of gp41 from pre-fusion to pre-hairpin states during HIV-1 entry. J Chem Inf Model 2020, 60:162–174. [DOI] [PubMed] [Google Scholar]
- 10.Joseph SB, Swanstrom R: The evolution of HIV-1 entry phenotypes as a guide to changing target cells. J Leukoc Biol 2018, 103:421–431. [DOI] [PubMed] [Google Scholar]
- 11.Joseph SB, Swanstrom R, Kashuba AD, Cohen MS: Bottlenecks in HIV-1 transmission: insights from the study of founder viruses. Nat Rev Microbiol 2015, 13:414–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW: Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci U S A 1999, 96:5215–5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuitemaker H, Koot M, Kootstra NA, Dercksen MW, de Goede RE, van Steenwijk RP, Lange JM, Schattenkerk JK, Miedema F, Tersmette M: Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol 1992, 66:1354–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR: Change in coreceptor use correlates with disease progression in HIV-1—infected individuals. J Exp Med 1997, 185:621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng HK, Malnati MS, Plebani A, Siccardi AG, Littman DR et al. : In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med 1997, 3:1259–1265. [DOI] [PubMed] [Google Scholar]
- 16.Connell BJ, Hermans LE, Wensing AMJ, Schellens I, Schipper PJ, van Ham PM, de Jong D, Otto S, Mathe T, Moraba R et al. : Immune activation correlates with and predicts CXCR4 co-receptor tropism switch in HIV-1 infection. Sci Rep 2020, 10:15866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnell G, Joseph S, Spudich S, Price RW, Swanstrom R: HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog 2011, 7:e1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sturdevant CB, Joseph SB, Schnell G, Price RW, Swanstrom R, Spudich S: Compartmentalized replication of R5 T cell-tropic HIV-1 in the central nervous system early in the course of infection. PLoS Pathog 2015, 11:e1004720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beauparlant D, Rusert P, Magnus C, Kadelka C, Weber J, Uhr T, Zagordi O, Oberle C, Duenas-Decamp MJ, Clapham PR et al. : Delineating CD4 dependency of HIV-1: adaptation to infect low level CD4 expressing target cells widens cellular tropism but severely impacts on envelope functionality. PLoS Pathog 2017, 13:e1006255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quitadamo B, Peters PJ, Repik A, O’Connell O, Mou Z, Koch M, Somasundaran M, Brody R, Luzuriaga K, Wallace A et al. : HIV-1 R5 macrophage-tropic envelope glycoprotein trimers bind CD4 with high affinity, while the CD4 binding site on non-macrophage-tropic, T-tropic R5 envelopes is occluded. J Virol 2018, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.••.Wang H, Cohen AA, Galimidi RP, Gristick HB, Jensen GJ, Bjorkman PJ: Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proc Natl Acad Sci U S A 2016, 113:E7151–E7158 [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang et al. [21••] and Ozorowski et al. [22••] elucidated structural details of Env conformational changes upon CD4 binding, including repositioning of the V3 loop and the fusion peptide by analyzing SOSIP Env trimers using the cryo-electron microscopy.
- 22.••.Ozorowski G, Pallesen J, de Val N, Lyumkis D, Cottrell CA, Torres JL, Copps J, Stanfield RL, Cupo A, Pugach P et al. : Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 2017, 547:360–363 [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang et al. [21••] and Ozorowski et al. [22••] elucidated structural details of Env conformational changes upon CD4 binding, including repositioning of the V3 loop and the fusion peptide by analyzing SOSIP Env trimers using the cryo-electron microscopy.
- 23.••.Shaik MM, Peng H, Lu J, Rits-Volloch S, Xu C, Liao M, Chen B: Structural basis of coreceptor recognition by HIV-1 envelope spike. Nature 2019, 565:318–323 [DOI] [PMC free article] [PubMed] [Google Scholar]; This study revealed the first cryo-electron microscopy structure of a full-length gp120 bound to CD4 and CCR5.
- 24.Hartley O, Klasse PJ, Sattentau QJ, Moore JP: V3: HIV’s switch-hitter. AIDS Res Hum Retroviruses 2005, 21:171–189. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Barnes CO, Yang Z, Nussenzweig MC, Bjorkman PJ: Partially open HIV-1 envelope structures exhibit conformational changes relevant for coreceptor binding and fusion. Cell Host Microbe 2018, 24:579–592 e574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.•.Yang Z, Wang H, Liu AZ, Gristick HB, Bjorkman PJ: Asymmetric opening of HIV-1 Env bound to CD4 and a coreceptor-mimicking antibody. Nat Struct Mol Biol 2019, 26:1167–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]; This study revealed a cryo-electron microscopy structure of a CD4-bound Env trimer interacting with a tyrosine-sulfated coreceptor-mimicking antibody. This is the first paper that observed high-resolution Env trimer asymmetry.
- 27.Jakobsdottir GM, Iliopoulou M, Nolan R, Alvarez L, Compton AA, Padilla-Parra S: On the whereabouts of HIV-1 cellular entry and its fusion ports. Trends Mol Med 2017, 23:932–944. [DOI] [PubMed] [Google Scholar]
- 28.McClure MO, Marsh M, Weiss RA: Human immunodeficiency virus infection of CD4-bearing cells occurs by a pH-independent mechanism. EMBO J 1988, 7:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelchen-Matthews A, Clapham P, Marsh M: Role of CD4 endocytosis in human immunodeficiency virus infection. J Virol 1995, 69:8164–8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aggarwal A, Hitchen TL, Ootes L, McAllery S, Wong A, Nguyen K, McCluskey A, Robinson PJ, Turville SG: HIV infection is influenced by dynamin at 3 independent points in the viral life cycle. Traffic 2017, 18:392–410. [DOI] [PubMed] [Google Scholar]
- 31.Daecke J, Fackler OT, Dittmar MT, Krausslich HG: Involvement of clathrin-mediated endocytosis in human immunodeficiency virus type 1 entry. J Virol 2005, 79:1581–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB: HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 2009, 137:433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de la Vega M, Marin M, Kondo N, Miyauchi K, Kim Y, Epand RF, Epand RM, Melikyan GB: Inhibition of HIV-1 endocytosis allows lipid mixing at the plasma membrane, but not complete fusion. Retrovirology 2011, 8:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Kleist L, Stahlschmidt W, Bulut H, Gromova K, Puchkov D, Robertson MJ, MacGregor KA, Tomilin N, Pechstein A, Chau N et al. : Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell 2011, 146:471–484. [DOI] [PubMed] [Google Scholar]
- 35.Marin M, Kushnareva Y, Mason CS, Chanda SK, Melikyan GB: HIV-1 fusion with CD4+ T cells is promoted by proteins involved in endocytosis and intracellular membrane trafficking. Viruses 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones DM, Alvarez LA, Nolan R, Ferriz M, Sainz Urruela R, Massana-Munoz X, Novak-Kotzer H, Dustin ML, Padilla-Parra S: Dynamin-2 stabilizes the HIV-1 fusion pore with a low oligomeric state. Cell Rep 2017, 18:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herold N, Anders-Osswein M, Glass B, Eckhardt M, Muller B, Krausslich HG: HIV-1 entry in SupT1-R5, CEM-ss, and primary CD4+ T cells occurs at the plasma membrane and does not require endocytosis. J Virol 2014, 88:13956–13970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marechal V, Prevost MC, Petit C, Perret E, Heard JM, Schwartz O: Human immunodeficiency virus type 1 entry into macrophages mediated by macropinocytosis. J Virol 2001, 75:11166–11177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carter GC, Bernstone L, Baskaran D, James W: HIV-1 infects macrophages by exploiting an endocytic route dependent on dynamin, Rac1 and Pak1. Virology 2011, 409:234–250. [DOI] [PubMed] [Google Scholar]
- 40.Wang Q, Finzi A, Sodroski J: The conformational states of the HIV-1 envelope glycoproteins. Trends Microbiol 2020, 28:655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falkenhagen A, Joshi S: HIV entry and its inhibition by bifunctional antiviral proteins. Mol Ther Nucleic Acids 2018, 13:347–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao T, Cai Y, Chen B: HIV-1 entry and membrane fusion inhibitors. Viruses 2021, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson WE, Desrosiers RC: Viral persistence: HIV’s strategies of immune system evasion. Annu Rev Med 2002, 53:499–518. [DOI] [PubMed] [Google Scholar]
- 44.•.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP et al. : Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 2013, 342:1477–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper for the first time showed a crystal structure of a soluble cleaved Env trimer stabilized by mutations (BG505 SOSIP.664 gp140 trimer) in complex with a broadly neutralizing antibody.
- 45.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S: Molecular architecture of native HIV-1 gp120 trimers. Nature 2008, 455:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW et al. : Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc Natl Acad Sci U S A 2011, 108:11440–11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S: Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog 2012, 8:e1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.••.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB 3rd, Kwong PD et al. : Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 2014, 346:759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]; Munro et al. for the first time applied smFRET to observe conformation changes of Env trimer on virions and found that Env trimers are in three distinct states.
- 49.Munro JB, Mothes W: Structure and dynamics of the native HIV-1 Env trimer. J Virol 2015, 89:5752–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu M, Ma X, Mothes W: Illuminating the virus life cycle with single-molecule FRET imaging. Adv Virus Res 2019, 105:239–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu M: Single-molecule FRET imaging of virus spike-host interactions. Viruses 2021, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma X, Lu M, Gorman J, Terry DS, Hong X, Zhou Z, Zhao H, Altman RB, Arthos J, Blanchard SC et al. : HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. eLife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.••.Lu M, Ma X, Castillo-Menendez LR, Gorman J, Alsahafi N, Ermel U, Terry DS, Chambers M, Peng D, Zhang B et al. : Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 2019, 568:415–419 [DOI] [PMC free article] [PubMed] [Google Scholar]; The prefusion Env molecule is an important potential immunogen. This smFRET study revealed that the available high resolution structures of one type of engineered stabilized Env trimer may not represent the prefusion status.
- 54.Montefiori DC, Roederer M, Morris L, Seaman MS: Neutralization tiers of HIV-1. Curr Opin HIV AIDS 2018, 13:128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herschhorn A, Ma X, Gu C, Ventura JD, Castillo-Menendez L, Melillo B, Terry DS, Smith AB 3rd, Blanchard SC, Munro JB et al. : Release of gp120 restraints leads to an entry-competent intermediate state of the HIV-1 envelope glycoproteins. mBio 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herschhorn A, Gu C, Moraca F, Ma X, Farrell M, Smith AB 3rd, Pancera M, Kwong PD, Schon A, Freire E et al. : The beta20-beta21 of gp120 is a regulatory switch for HIV-1 Env conformational transitions. Nat Commun 2017, 8:1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.•.Henderson R, Lu M, Zhou Y, Mu Z, Parks R, Han Q, Hsu AL, Carter E, Blanchard SC, Edwards RJ et al. : Disruption of the HIV-1 envelope allosteric network blocks CD4-induced rearrangements. Nat Commun 2020, 11:520. [DOI] [PMC free article] [PubMed] [Google Scholar]; Henderson et al. identified critical amino acid residues in different regions of Env, which form an allosteric network that are important for a CD4-induced conformational change of Env.
- 58.Kondo N, Melikyan GB: Intercellular adhesion molecule 1 promotes HIV-1 attachment but not fusion to target cells. PLoS One 2012, 7:e44827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nydegger S, Khurana S, Krementsov DN, Foti M, Thali M: Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J Cell Biol 2006, 173:795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weng J, Krementsov DN, Khurana S, Roy NH, Thali M: Formation of syncytia is repressed by tetraspanins in human immunodeficiency virus type 1-producing cells. J Virol 2009, 83:7467–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sato K, Aoki J, Misawa N, Daikoku E, Sano K, Tanaka Y, Koyanagi Y: Modulation of human immunodeficiency virus type 1 infectivity through incorporation of tetraspanin proteins. J Virol 2008, 82:1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.••.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J et al. : HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 2015, 526:212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]; Rosa et al. as well as Usami et al. identified SERINC3 and 5 as a Nef-sensitive restriction factor that is incorporated into virions and reduces infectivity of progeny virions.
- 63.••.Usami Y, Wu Y, Gottlinger HG: SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 2015, 526:218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]; Usami et al. as well as Rosa et al. identified SERINC3 and 5 as a Nef-sensitive restriction factor that is incorporated into virions and reduces infectivity of progeny virions.
- 64.•.Compton AA, Bruel T, Porrot F, Mallet A, Sachse M, Euvrard M, Liang C, Casartelli N, Schwartz O: IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe 2014, 16:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrated that expression of IFITM proteins in virus-producing cells suppresses cell-to-cell transmission of HIV-1 for the first time and that virion-incorporated IFITM proteins inhibit HIV-1 fusion.
- 65.•.Tartour K, Appourchaux R, Gaillard J, Nguyen XN, Durand S, Turpin J, Beaumont E, Roch E, Berger G, Mahieux R et al. : IFITM proteins are incorporated onto HIV-1 virion particles and negatively imprint their infectivity. Retrovirology 2014, 11:103. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study also demonstrated that virion-incorporated IFITM proteins inhibit HIV-1 fusion. This is the first paper that reported that virion-incorporated IFITM proteins inhibit viral infection.
- 66.Tartour K, Nguyen XN, Appourchaux R, Assil S, Barateau V, Bloyet LM, Burlaud Gaillard J, Confort MP, Escudero-Perez B, Gruffat H et al. : Interference with the production of infectious viral particles and bimodal inhibition of replication are broadly conserved antiviral properties of IFITMs. PLoS Pathog 2017, 13: e1006610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.•.Fu Y, He S, Waheed AA, Dabbagh D, Zhou Z, Trinite B, Wang Z, Yu J, Wang D, Li F et al. : PSGL-1 restricts HIV-1 infectivity by blocking virus particle attachment to target cells. Proc Natl Acad Sci U S A 2020, 117:9537–9545 [DOI] [PMC free article] [PubMed] [Google Scholar]; This study found that virion-incorporated PSGL-1 reduces virion infectivity through preventing virus attachment to target cells. This and Ref. [68•] are the first papers that reported inhibition of virus attachment to target cells by a virion-incorporated host transmembrane protein.
- 68.•.Murakami T, Carmona N, Ono A: Virion-incorporated PSGL-1 and CD43 inhibit both cell-free infection and transinfection of HIV-1 by preventing virus-cell binding. Proc Natl Acad Sci U S A 2020, 117:8055–8063 [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrated that virion-incorporated PSGL-1 and CD43 inhibit both cell-free infection and trans-infection mediated by fibroblastic reticular cells by blocking virus-cell binding. This and Ref. [67•] are the first papers that identified virion-incorporated host transmembrane proteins that prevent HIV-1 from binding to cells.
- 69.Inuzuka M, Hayakawa M, Ingi T: Serinc, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J Biol Chem 2005, 280:35776–35783. [DOI] [PubMed] [Google Scholar]
- 70.Beitari S, Ding S, Pan Q, Finzi A, Liang C: Effect of HIV-1 Env on SERINC5 antagonism. J Virol 2017, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.•.Usami Y, Gottlinger H: HIV-1 Nef responsiveness is determined by Env variable regions involved in trimer association and correlates with neutralization sensitivity. Cell Rep 2013, 5:802–812 [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first paper that reported the relationship between Nef responsiveness, which was later attributed to SERINC sensitivity, and Env structure.
- 72.Zhang X, Shi J, Qiu X, Chai Q, Frabutt DA, Schwartz RC, Zheng YH: CD4 expression and Env conformation are critical for HIV-1 restriction by SERINC5. J Virol 2019, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haider T, Snetkov X, Jolly C: HIV envelope tail truncation confers resistance to SERINC5 restriction. Proc Natl Acad Sci U S A 2021, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen J, Kovacs JM, Peng H, Rits-Volloch S, Lu J, Park D, Zablowsky E, Seaman MS, Chen B: HIV-1 ENVELOPE. Effect of the cytoplasmic domain on antigenic characteristics of HIV-1 envelope glycoprotein. Science 2015, 349:191–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Angerstein AO, Stoneham CA, Ramirez PW, Guatelli JC, Vollbrecht T: Sensitivity to monoclonal antibody 447–52D and an open Env trimer conformation correlate poorly with inhibition of HIV-1 infectivity by SERINC5. Virology 2020, 548:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Featherstone A, Aiken C: SERINC5 inhibits HIV-1 infectivity by altering the conformation of gp120 on HIV-1 particles. J Virol 2020, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Staropoli I, Dufloo J, Ducher A, Commere PH, Sartori-Rupp A, Novault S, Bruel T, Lorin V, Mouquet H, Schwartz O et al. : Flow cytometry analysis of HIV-1 Env conformations at the surface of infected cells and virions: role of Nef, CD4, and SERINC5. J Virol 2020, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sood C, Marin M, Chande A, Pizzato M, Melikyan GB: SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins. J Biol Chem 2017, 292:6014–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ward AE, Kiessling V, Pornillos O, White JM, Ganser-Pornillos BK, Tamm LK: HIV-cell membrane fusion intermediates are restricted by serincs as revealed by cryo-electron and TIRF microscopy. J Biol Chem 2020, 295:15183–15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chojnacki J, Staudt T, Glass B, Bingen P, Engelhardt J, Anders M, Schneider J, Muller B, Hell SW, Krausslich HG: Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 2012, 338:524–528. [DOI] [PubMed] [Google Scholar]
- 81.•.Chen YC, Sood C, Marin M, Aaron J, Gratton E, Salaita K, Melikyan GB: Super-resolution fluorescence imaging reveals that serine incorporator protein 5 inhibits human immunodeficiency virus fusion by disrupting envelope glycoprotein clusters. ACS Nano 2020, 14:10929–10943 [DOI] [PMC free article] [PubMed] [Google Scholar]; This study applied a 3D superresolution microscopy technique to visualize the effect of SERINC5 on Env distribution on the surface of virions for the first time and discovered that SERINC5 blocks the clustering of Env that is required for efficient HIV-1 fusion on the surface of virions.
- 82.Nieto-Garai JA, Arboleya A, Otaegi S, Chojnacki J, Casas J, Fabrias G, Contreras FX, Krausslich HG, Lorizate M: Cholesterol in the viral membrane is a molecular switch governing HIV-1 Env clustering. Adv Sci (Weinh) 2021, 8:2003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trautz B, Wiedemann H, Luchtenborg C, Pierini V, Kranich J, Glass B, Krausslich HG, Brocker T, Pizzato M, Ruggieri A et al. : The host-cell restriction factor SERINC5 restricts HIV-1 infectivity without altering the lipid composition and organization of viral particles. J Biol Chem 2017, 292:13702–13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi G, Schwartz O, Compton AA: More than meets the I: the diverse antiviral and cellular functions of interferon-induced transmembrane proteins. Retrovirology 2017, 14:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marziali F, Cimarelli A: Membrane interference against HIV-1 by intrinsic antiviral factors: the case of IFITMs. Cells 2021, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.•.Lu J, Pan Q, Rong L, He W, Liu SL, Liang C: The IFITM proteins inhibit HIV-1 infection. J Virol 2011, 85:2126–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first paper that reported the inhibitory effect of IFITM proteins to HIV-1 infection.
- 87.Amini-Bavil-Olyaee S, Choi YJ, Lee JH, Shi M, Huang IC, Farzan M, Jung JU: The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 2013, 13:452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin TY, Chin CR, Everitt AR, Clare S, Perreira JM, Savidis G, Aker AM, John SP, Sarlah D, Carreira EM et al. : Amphotericin B increases influenza A virus infection by preventing IFITM3-mediated restriction. Cell Rep 2013, 5:895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB: IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog 2014, 10:e1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li K, Markosyan RM, Zheng YM, Golfetto O, Bungart B, Li M, Ding S, He Y, Liang C, Lee JC et al. : IFITM proteins restrict viral membrane hemifusion. PLoS Pathog 2013, 9:e1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Foster TL, Wilson H, Iyer SS, Coss K, Doores K, Smith S, Kellam P, Finzi A, Borrow P, Hahn BH et al. : Resistance of transmitted founder HIV-1 to IFITM-mediated restriction. Cell Host Microbe 2016, 20:429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jia R, Pan Q, Ding S, Rong L, Liu SL, Geng Y, Qiao W, Liang C: The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J Virol 2012, 86:13697–13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weston S, Czieso S, White IJ, Smith SE, Kellam P, Marsh M: A membrane topology model for human interferon inducible transmembrane protein 1. PLoS One 2014, 9:e104341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.•.Wang Y, Pan Q, Ding S, Wang Z, Yu J, Finzi A, Liu SL, Liang C: The V3 loop of HIV-1 Env determines viral susceptibility to IFITM3 impairment of viral infectivity. J Virol 2017, 91 [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang et al. identified the V3 loop of Env as a determinant of sensitivity to IFITM3.
- 95.Wu WL, Grotefend CR, Tsai MT, Wang YL, Radic V, Eoh H, Huang IC: Delta20 IFITM2 differentially restricts X4 and R5 HIV-1. Proc Natl Acad Sci U S A 2017, 114:7112–7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu J, Liu SL: The Inhibition of HIV-1 entry imposed by interferon inducible transmembrane proteins is independent of co-receptor usage. Viruses 2018, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu J, Li M, Wilkins J, Ding S, Swartz TH, Esposito AM, Zheng YM, Freed EO, Liang C, Chen BK et al. : IFITM proteins restrict HIV-1 infection by antagonizing the envelope glycoprotein. Cell Rep 2015, 13:145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Beitari S, Pan Q, Finzi A, Liang C: Differential pressures of SERINC5 and IFITM3 on HIV-1 envelope glycoprotein over the course of HIV-1 infection. J Virol 2020, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Richard J, Veillette M, Brassard N, Iyer SS, Roger M, Martin L, Pazgier M, Schon A, Freire E, Routy JP et al. : CD4 mimetics sensitize HIV-1-infected cells to ADCC. Proc Natl Acad Sci U S A 2015, 112:E2687–E2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ahi YS, Yimer D, Shi G, Majdoul S, Rahman K, Rein A, Compton AA: IFITM3 reduces retroviral envelope abundance and function and is counteracted by glycoGag. mBio 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rahman K, Coomer CA, Majdoul S, Ding SY, Padilla-Parra S, Compton AA: Homology-guided identification of a conserved motif linking the antiviral functions of IFITM3 to its oligomeric state. eLife 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Drouin A, Migraine J, Durand MA, Moreau A, Burlaud-Gaillard J, Beretta M, Roingeard P, Bouvin-Pley M, Braibant M: Escape of HIV-1 envelope glycoprotein from the restriction of infection by IFITM3. J Virol 2020, 95:e01994–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Carlsson SR, Fukuda M: Isolation and characterization of leukosialin, a major sialoglycoprotein on human leukocytes. J Biol Chem 1986, 261:12779–12786. [PubMed] [Google Scholar]
- 104.Norgard KE, Moore KL, Diaz S, Stults NL, Ushiyama S, McEver RP, Cummings RD, Varki A: Characterization of a specific ligand for P-selectin on myeloid cells. A minor glycoprotein with sialylated O-linked oligosaccharides. J Biol Chem 1993, 268:12764–12774. [PubMed] [Google Scholar]
- 105.Alon R, Rossiter H, Wang X, Springer TA, Kupper TS: Distinct cell surface ligands mediate T lymphocyte attachment and rolling on P and E selectin under physiological flow. J Cell Biol 1994, 127:1485–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zarbock A, Ley K, McEver RP, Hidalgo A: Leukocyte ligands for endothelial selectins: specialized glycoconjugates that mediate rolling and signaling under flow. Blood 2011, 118:6743–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cyster JG, Shotton DM, Williams AF: The dimensions of the T lymphocyte glycoprotein leukosialin and identification of linear protein epitopes that can be modified by glycosylation. EMBO J 1991, 10:893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Manjunath N, Correa M, Ardman M, Ardman B: Negative regulation of T-cell adhesion and activation by CD43. Nature 1995, 377:535–538. [DOI] [PubMed] [Google Scholar]
- 109.McEver RP, Moore KL, Cummings RD: Leukocyte trafficking mediated by selectin-carbohydrate interactions. J Biol Chem 1995, 270:11025–11028. [DOI] [PubMed] [Google Scholar]
- 110.Matsumoto M, Miyasaka M, Hirata T: P-selectin glycoprotein ligand-1 negatively regulates T-cell immune responses. J Immunol 2009, 183:7204–7211. [DOI] [PubMed] [Google Scholar]
- 111.Llewellyn GN, Hogue IB, Grover JR, Ono A: Nucleocapsid promotes localization of HIV-1 gag to uropods that participate in virological synapses between T cells. PLoS Pathog 2010, 6: e1001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Llewellyn GN, Grover JR, Olety B, Ono A: HIV-1 gag associates with specific uropod-directed microdomains in a manner dependent on its MA highly basic region. J Virol 2013, 87:6441–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Grover JR, Veatch SL, Ono A: Basic motifs target PSGL-1, CD43, and CD44 to plasma membrane sites where HIV-1 assembles. J Virol 2015, 89:454–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McLaren PJ, Gawanbacht A, Pyndiah N, Krapp C, Hotter D, Kluge SF, Gotz N, Heilmann J, Mack K, Sauter D et al. : Identification of potential HIV restriction factors by combining evolutionary genomic signatures with functional analyses. Retrovirology 2015, 12:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.•.Liu Y, Fu Y, Wang Q, Li M, Zhou Z, Dabbagh D, Fu C, Zhang H, Li S, Zhang T et al. : Proteomic profiling of HIV-1 infection of human CD4(+) T cells identifies PSGL-1 as an HIV restriction factor. Nat Microbiol 2019, 4:813–825 [DOI] [PubMed] [Google Scholar]; Liu et al. identified PSGL-1 as a restriction factor that inhibits HIV-1 reverse transcription when PSGL-1 is expressed by either target cells or virus-producing cells.
- 116.Haller C, Muller B, Fritz JV, Lamas-Murua M, Stolp B, Pujol FM, Keppler OT, Fackler OT: HIV-1 Nef and Vpu are functionally redundant broad-spectrum modulators of cell surface receptors, including tetraspanins. J Virol 2014, 88:14241–14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Matheson NJ, Sumner J, Wals K, Rapiteanu R, Weekes MP, Vigan R, Weinelt J, Schindler M, Antrobus R, Costa AS et al. : Cell surface proteomic map of HIV infection reveals antagonism of amino acid metabolism by Vpu and Nef. Cell Host Microbe 2015, 18:409–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.•.Ladinsky MS, Gnanapragasam PN, Yang Z, West AP, Kay MS, Bjorkman PJ: Electron tomography visualization of HIV-1 fusion with target cells using fusion inhibitors to trap the pre-hairpin intermediate. eLife 2020, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first paper that showed a structure of an extended pre-hairpin intermediate state of Env via a electron tomography and revealed that HIV-1 binds to target cells by 2–4 narrow spokes.
- 119.Clark MC, Baum LG: T cells modulate glycans on CD43 and CD45 during development and activation, signal regulation, and survival. Ann N Y Acad Sci 2012, 1253:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shogren R, Gerken TA, Jentoft N: Role of glycosylation on the conformation and chain dimensions of O-linked glycoproteins: light-scattering studies of ovine submaxillary mucin. Biochemistry 1989, 28:5525–5536. [DOI] [PubMed] [Google Scholar]
- 121.Dabbagh D, He S, Hetrick B, Chilin L, Andalibi A, Wu Y: Identification of the SHREK family of proteins as broad-spectrum host antiviral factors. Viruses 2021, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu Y, Song Y, Zhang S, Diao M, Huang S, Li S, Tan X: PSGL-1 inhibits HIV-1 infection by restricting actin dynamics and sequestering HIV envelope proteins. Cell Discov 2020, 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Burnie J, Persaud AT, Thaya L, Liu Q, Miao H, Grabinsky S, Norouzi V, Lusso P, Tang VA, Guzzo C: The P-selectin ligand PSGL-1 (CD162) is efficiently incorporated by primary HIV-1 isolates and can facilitate trans-infection. bioRxiv 2021. 2021.2006.2029.450454. [Google Scholar]
- 124.Stadtmueller BM, Bridges MD, Dam KM, Lerch MT, Huey-Tubman KE, Hubbell WL, Bjorkman PJ: DEER spectroscopy measurements reveal multiple conformations of HIV-1 SOSIP envelopes that show similarities with envelopes on native virions. Immunity 2018, 49:235–246 e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S: Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol 2013, 20:1352–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.•.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B et al. : Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 2013, 342:1484–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper for the first time showed a high-resolution cryo-EM structure of BG505 SOSIP.664 gp140 trimers in complex with a CD4-binding site-recognizing broadly neutralizing antibody.
- 127.•.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J et al. : A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 2013, 9:e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first paper that reported that a near-native antigenicity of BG505 SOSIP.664 gp140 trimers.
- 128.•.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G et al. : Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 2014, 514:455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]; This study determined the structure of an HIV-1 Env trimer captured in a mature closed state by antibodies and revealed the pre-fusion conformation of gp41.
- 129.•.Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S et al. : Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 2015, 22:522–531 [DOI] [PMC free article] [PubMed] [Google Scholar]; Kwon et al. for the first time revealed the crystal structure of the ligand-free Env trimer and showed by smFRET a possibility that a single-CD4-bound state of Env trimer exists during virus entry.
- 130.Gristick HB, von Boehmer L, West AP Jr, Schamber M, Gazumyan A, Golijanin J, Seaman MS, Fatkenheuer G, Klein F, Nussenzweig MC et al. : Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat Struct Mol Biol 2016, 23:906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lee JH, Ozorowski G, Ward AB: Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 2016, 351:1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]


