Abstract
Secretion of cellular components across the plasma membrane is an essential process that enables organisms to interact with their environments. Production of extracellular vesicles in bacteria is a well-documented but poorly understood process. Outer membrane vesicles (OMVs) are produced in gram-negative bacteria by blebbing of the outer membrane. In addition to their roles in pathogenesis, cell-to-cell communication, and stress responses, OMVs play important roles in immunomodulation and the establishment and balance of the gut microbiota. In this review, we discuss the multiple roles of OMVs and the current knowledge of OMV biogenesis. We also discuss the growing and promising biotechnological applications of OMV.
Keywords: OMV, Secretion system, pathogenesis, vaccines, commensalism
1. Introduction
Secretion of cellular components across the plasma membrane is an essential process that occurs in all life-forms, enabling organisms to interact with their environments. One way cells accomplish this is by secreting vesicles, which are spherical, nanosized structures derived from lipid membranes of the cell surface (21, 39). Production of extracellular vesicles is a well-documented process that takes place in gram-positive and gram-negative bacteria. Outer membrane vesicles (OMVs), which are released from the cell envelope of gram-negative bacteria, have been studied for more than 50 years. However, little is known about OMV biogenesis. In this review, we discuss the current knowledge of the biogenesis of OMVs in gram-negative bacteria, consider their roles in bacterium-host interactions, and discuss their biotechnological applications. Finally, we briefly address some of the reasons why some researchers remain skeptical about the physiological roles of OMVs.
1.1. OMV Secretion and Its Discovery
OMV production was first observed in 1965 in an auxotrophic Escherichia coli strain that released significant amounts of cell-free lipopolysaccharides (LPS) under lysine-limiting growth conditions (17). Later, Knox and collaborators showed by electron microscopy that these secreted cell-free LPS elements were part of membrane structures and proposed that these vesicles were derived from the outer membrane (OM) (87). Rothfield & Pearlman-Kothencz (124) followed up these observations and showed that chloramphenicol exposure and amino acid starvation promote the secretion of these OM blebs in E. coli. Subsequent studies reported the observation and isolation of OMVs from different gram-negative bacteria, like Veillonella parvula (106), Vibrio cholerae (29), and Salmonella typhimurium (124). Despite the increasing evidence of OMV production by bacteria, OMVs were considered mere growth artifacts or cell lysis by-products for several years. Later, OMVs were observed in cerebrospinal fluid samples from patients with acute meningitis, suggesting that OMVs were not generated only in lab conditions (42). Since then, the biogenesis of OMVs and their roles have gained interest.
OMVs range between 20 and 300 nm and function as a versatile secretion and transport mechanism for bacterial cells (64). OMV composition in several species has been described and includes lipids, LPS and OM proteins as well as encapsulated periplasmic content. The presence of cytoplasmic elements, like DNA and RNA, has also been reported, but it is unclear how these elements are transported into the periplasm to be packed into OMVs (18, 37). Analysis of OM and OMV fractions from different microorganisms revealed a distinct enrichment of proteins and lipids in each fraction (1, 45, 63, 66, 72, 78, 93, 105, 121, 131). These findings favor the hypotheses that bacteria possess specific sorting mechanisms and that OMV formation is a directed process and not the result of cell lysis.
2. OMV Roles
OMVs have been implicated in an array of physiological processes, including intracellular and extracellular communication, quorum sensing, horizontal gene transfer, interbacterial killing, toxin delivery, polysaccharide hydrolysis, and stress responses (64, 67, 102, 129) (Figure 1).
Figure 1.
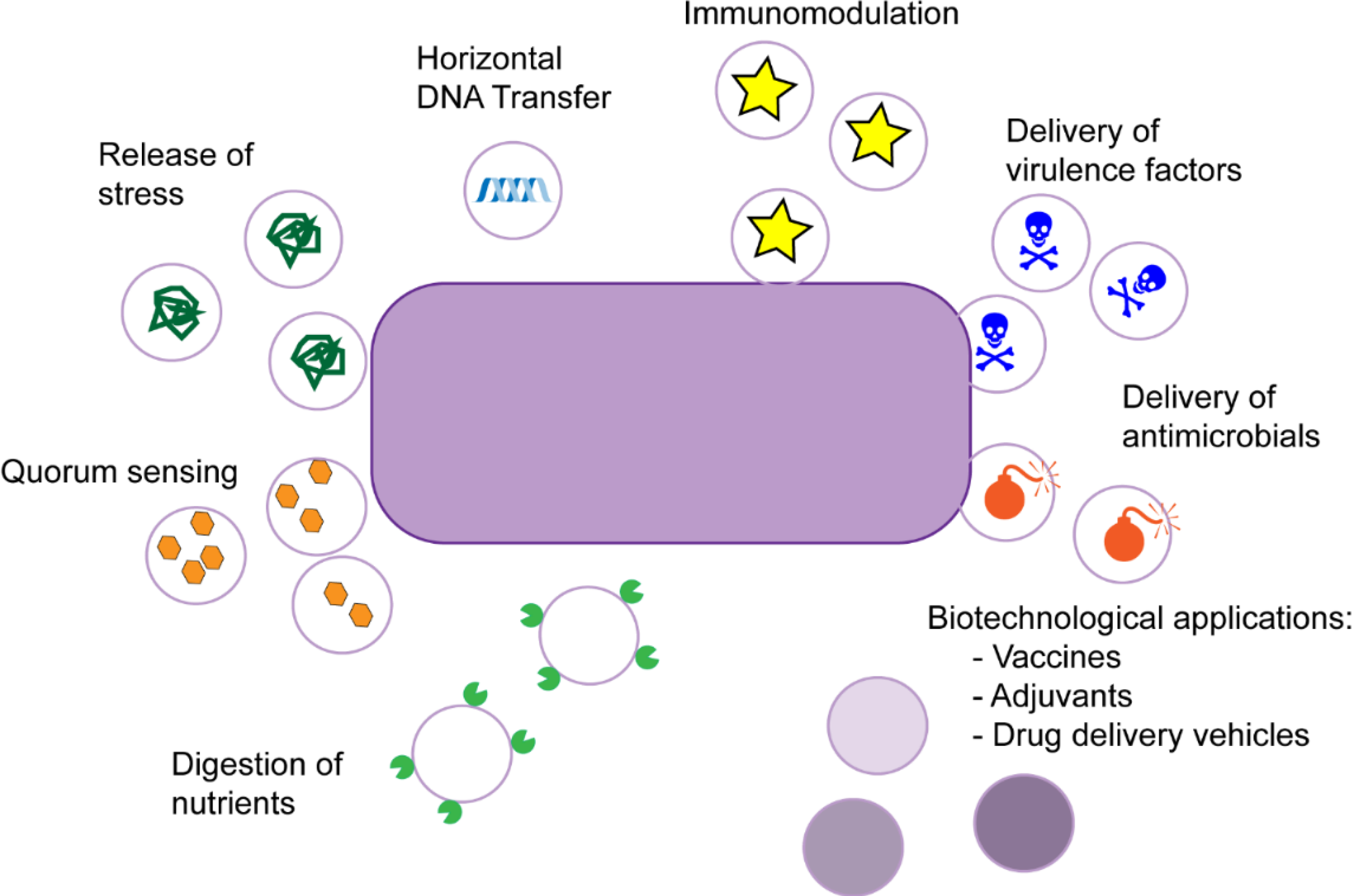
Different roles of outer membrane vesicles in Gram-negative bacteria. Roles of outer membrane vesicles in gram-negative bacteria. OMVs, which are released from the cell envelope, range between 20 and −300 nm and function as a versatile secretion and transport mechanism. OMV roles include intracellular and extracellular communication, quorum sensing, horizontal gene transfer, interbacterial killing, toxin delivery, nutrient hydrolysis, and stress responses.
2.1. OMVs in Pathogenesis
OMVs have been linked to pathogenesis. They can serve as long-distance delivery vehicles that can promote host colonization and immune evasion. They carry an array of immunogenic molecules, such as LPS, flagellin, and peptidoglycan, which stimulate the host immune system through Toll-like receptors (TLRs) (13, 23a). OMV-associated molecules include virulence factors related to adherence, invasion, antimicrobial resistance, and modulation of host immunity. To highlight the role of OMV in pathogenesis, we discuss examples in further detail below.
Enterotoxigenic E. coli (ETEC) is an important diarrheal pathogen that secretes several toxins, including heat-labile enterotoxin (LT) (72, 144). This toxin disrupts the electrolyte balance in the gut endothelium and has been detected in OMVs (72, 86). ETEC OMVs can also deliver the pore-forming cytotoxic protein cytolysin A (ClyA) (143). ClyA incorporated into OMVs has higher cytotoxicity compared to the purified toxin in a mammalian cell model (143). This finding has been attributed to an effect of the redox environment within the vesicles that allows ClyA oligomerization (143). Cytotoxic necrotizing factor type 1 (CNF1) is a virulence factor produced by uropathogenic E. coli contained in OMVs (88). Once this toxin reaches the host cells, it alters actin cytoskeleton and promotes bacterial invasion of endothelial cells of the blood–brain barrier.
Early studies in the human dental pathogen Porphyromonas gingivalis also linked OMV secretion to pathogenesis. The main virulence factors of P. gingivalis, gingipains, are enriched in OMVs, which contributes to impairment of host cell function (66, 142). Moreover, hemagglutinins and heat shock proteins, which are mainly involved in host cell attachment and invasion, are also secreted as OMV cargo (10a). Consequently, OMV production increases bacterial adherence to host cells, stimulating bacterial aggregation and leading to the formation of dental plaque (46, 63, 80).
Pseudomonas aeruginosa is an opportunistic pathogen that causes nosocomial infections mainly in immunocompromised patients (97). Its OMVs carry multiple virulence factors, including degradative and pore-forming molecules such as peptidoglycan hydrolase, phospholipase C, alkaline phosphatase, protease, elastase, and hemolysin (34, 78). Additionally, P. aeruginosa OMVs facilitate bacterial competition during infection, being able to kill gram-negative and gram-positive competitors in cocultures (79). Furthermore, P. aeruginosa elicits a potent destructive inflammatory response via combined sensing of both LPS and protein components (48). In vitro assays carried out in macrophages showed that OMVs led to a significant increase of MIP-2, TNF-α, IL-1, and IL-6 transcriptional levels compared to exposure to LPS alone.
S. typhimurium, the leading cause of gastroenteritis, also exploits OMVs to transport virulence factors. This bacterium is able to produce OMVs even during its intracellular life, secreting OMVs packed with cytolethal distending toxin inside infected epithelial cells (65). Interestingly, OMV biogenesis in S. typhimurium inside infected macrophages is triggered by the two-component system PhoPQ (44).
V. cholerae, the causative agent of cholera, produces cholera toxin (CT) as its main virulence factor (28). Although CT is primarily secreted by the type II secretion system (T2SS), OMVs serve as a secondary mechanism by which CT is secreted (28). In addition to CT, several other virulence factors have been linked to OMVs, such as the pore-forming toxin Vibrio cytolysin (117) as well as various serine proteases and metalloproteases (58, 123). These compounds were found to cause cytotoxicity and induce an inflammatory response in host cells (108). Moreover, this bacterium can alter the immunogenicity of OMV cargo by decreasing the expression of virulence factors through quorum sensing (16).
2.2. OMVs in Commensalism
Gut microbiota–derived vesicles are an emerging topic of study and were first reported less than ten years ago (81, 130). The functionality of microbiota-derived OMVs varies greatly depending on the species releasing them. Proteomic studies on OMVs from gut commensal strains suggest that OMV-associated proteins contribute to modulating host immunity, promote host colonization, and act as public goods by degrading various carbon sources in the gut (1, 45, 119, 147).
Bacteroides is well known for secreting large amounts of OMVs and actively contributing to gut symbiosis (69, 130). These bacteria secrete glycosylases and proteases, which degrade complex polysaccharides and mucins (45, 69, 119). The generated degradation products can then be utilized as nutrient sources by any other members of the gut microbiota (45, 69, 119). Thus, OMVs are considered public goods: Once OMVs are released, any member of the gut microbiota can benefit from their degradative activity. However, not all the OMV cargo fulfills this altruistic role. Recently, a family of peptide toxins that has broad-spectrum activity against Bacteroides has been found as OMV cargo. The genes encoding these toxins are widely distributed throughout the human gut microbiota (30, 36).
OMVs from commensal bacteria can also modulate the host innate immune system. For example, Bacteroides fragilis polysaccharide A triggers TLR-mediated signaling that attenuates host immune responses and promotes commensal gut colonization (103, 125). The constant and controlled immune stimulation mediated by OMVs from commensals in the gut is linked to intestinal health (23a, 107). This cross talk between microbiota and host cells constitutes a key process in maintaining gut homeostasis.
2.3. Production of OMVs by Plant Pathogens
OMVs have been investigated primarily in human pathogens and commensals. However, recent studies show that OMVs produced by plant pathogens perform similar functions (73, 84, 134). Plant pathogens deliver virulence factors, such as T2SS effectors and xylanase, as OMV cargo (73, 111, 134). The innate immune system of plants recognizes and responds to purified OMVs from plant pathogens (7). Thus, when Arabidopsis thaliana seedlings were incubated with OMVs purified from several Xanthomonas species, three defense responses (defense gene activation, reactive oxygen species burst, and medium alkalinization) were modulated (7). Still, it remains unexplored whether OMVs play a role in plant symbiosis or in interkingdom cell-to-cell cross talk.
2.4. Other OMV Roles
OMV production is increased when cells are subjected to physical or chemical stress (3, 62, 96, 104). E. coli mutants lacking the proteases DegS and DegP exhibited a hypervesiculating phenotype, and the amount of vesicles released correlated with the level of protein accumulated in the cell envelope (104). Furthermore, the increased vesiculation enhanced bacterial survival upon challenge with stressing agents. These observations led to the hypothesis that vesicle overproduction is linked to the maintenance of the cell envelope (104, 136). Temperature is another factor that can modulate the amount of OMVs generated. In vitro, Serratia marcescens produced significant amounts of vesicles at 22°C or 30°C, whereas it produced negligible quantities at 37°C (130). Moreover, inactivation of the synthesis of the enterobacterial common antigen resulted in hypervesiculation in this strain, and this hypervesiculating phenotype was reverted upon inactivation of the response regulator RcsB (130). Nutrient limitations have been also associated as a trigger for OMV production. For example, under sulphate-depleted conditions, Neisseria meningitidis increased its OMV biogenesis, while Haemophilus influenzae, V. cholerae, and E. coli increased OMV production under iron-limited conditions (57, 121).
OMVs can also act as decoys to confront and attenuate antibiotic activity. E. coli OMVs contributed to protection against the membrane-targeting antibiotics colistin and melittin (91). The protective effect was not limited to E. coli, as these purified E. coli OMVs provided protection against those antibiotics to P. aeruginosa and Acinetobacter radioresistens strains (91). However, the protective effect of the OMVs could not be extended to other antibiotics, like ciprofloxacin, streptomycin, and trimethoprim, indicating that OMVs appear to protect the bacterial community mainly against antibiotics targeting the membranes (91), possibly by sequestering and reducing the availability of the antibiotic. P. aeruginosa and other bacteria also secrete antibiotic-degrading enzymes like β-lactamases in OMVs (35, 59). Similarly, OMVs secreted by V. cholerae acted as decoys to increase resistance to antimicrobial peptides, such as polymyxin B and LL-37 (43). Furthermore, OMVs act as decoys during phage infections. OMVs irreversibly bind the phage, highly reducing their ability to infect the bacterial cell (96, 101).
Recent studies have suggested that OMVs can harbor DNA and different types of RNAs, mostly noncoding RNAs (18, 37, 60, 133). Intriguingly, many of these sequences align to intergenic noncoding regions of human DNA (27). The delivery of bacterial RNA through OMVs could exert epigenetic changes in host transcription (27). However, it remains unclear how these RNA molecules reach the periplasm prior to being packed inside OMVs.
OMVs are also pivotal in bacterial communication. Certain bacteria can regulate gene expression in response to quorum sensing. P. aeruginosa is able to secrete quinolone signal compounds [Pseudomonas quinolone signal (PQS)] through OMVs, which provide a protective environment to these highly hydrophobic molecules (102). PQS simultaneously stimulates OMV production by intercalating into the OM, generating a positive-feedback loop (51). As PQS is found at high concentrations in OMVs, the fusion of a single vesicle with a bacterial cell is enough to trigger the quorum sensing response. Hydrophobic signal molecules related to quorum sensing were also detected as OMV cargo in other microorganisms (20), thus suggesting a novel OMV-based mechanism for hydrophobic signal molecule trafficking.
3. Regulation and Biogenesis of Bacterial OMVs
Bacterial OMVs have been studied extensively for decades. However, researchers have not yet elucidated a definitive or universal mechanism to explain OMV production. Here, we address several biogenesis mechanisms proposed to explain how OMVs are formed (Figure 2).
Figure 2.
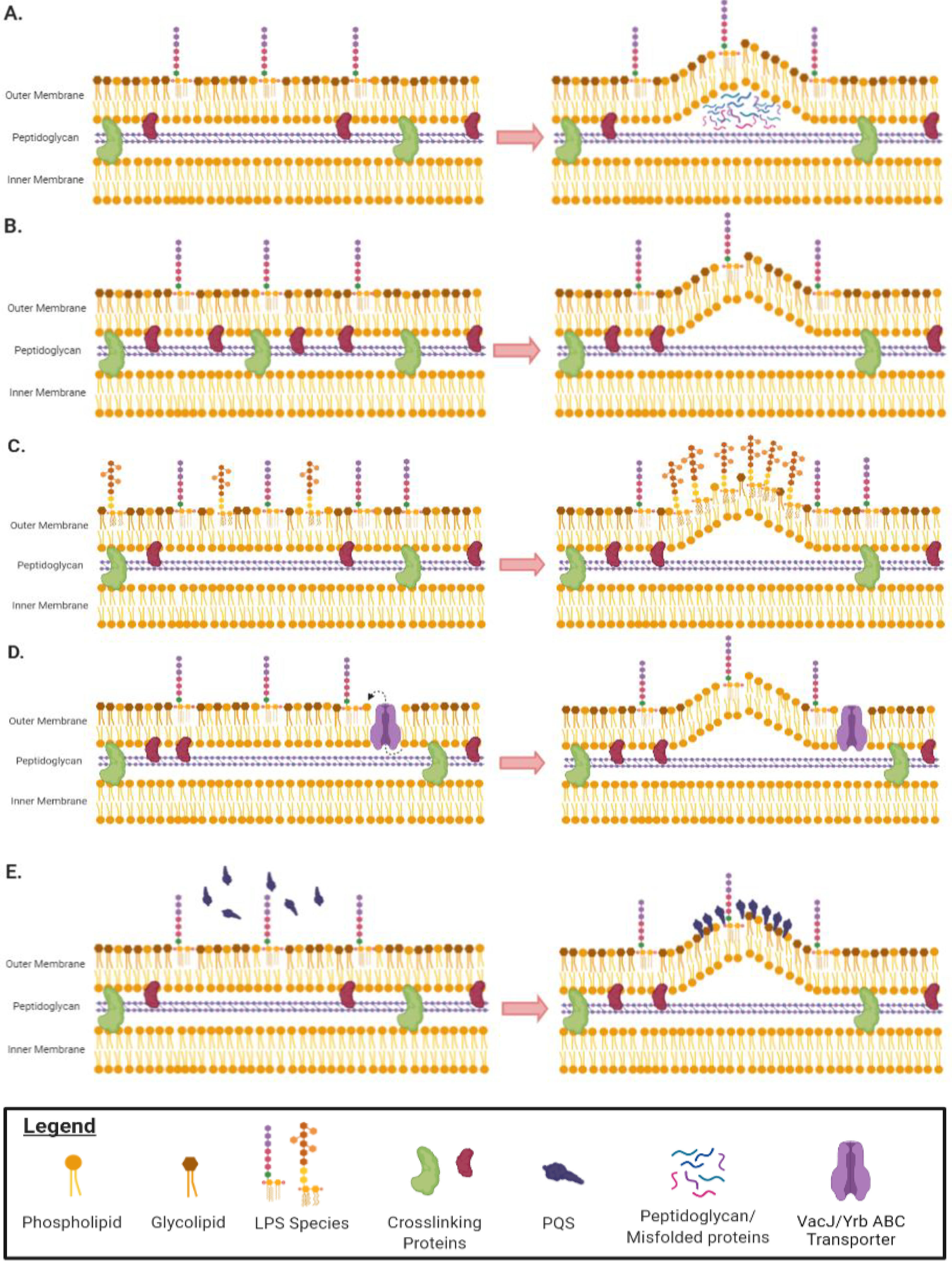
Mechanisms of OMV production in gram-negative bacteria. (a) Curvature of the OM is induced by the local accumulation of peptidoglycan fragments or misfolded proteins in the periplasm. (b) Removal of proteins anchoring the OM to the underlying peptidoglycan increases OM fluidity, enabling the membrane to bend and form vesicles. (c) Owing to charge repulsion, local enrichment of LPS species with anionic charges induces curvature of the OM and subsequent vesicle formation. Enrichment in deacylated LPS species also promotes membrane curvature because lipid A deacylation changes the shape of LPS from cylinder-like to an inverted cone. (d) The VacJ/Yrb ABC transporter is involved in retrograde transport of phospholipids from the OM. Downregulation of this transporter leads to the accumulation of phospholipids in the outer leaflet of the OM. This causes the outer leaflet to expand rapidly compared to the inner leaflet, which leads to membrane curvature and vesicle formation. (e) Once PQS is produced, it is secreted from the cell and subsequently intercalated into the outer leaflet of the OM owing to its interaction with LPS and phospholipids. Insertion of PQS into the OM causes outer leaflet expansion that increases OMV production. Abbreviations: ABC, ATP-binding cassette; LPS, lipopolysaccharide; OM, outer membrane; OMV, OM vesicle; PQS, Pseudomonas quinolone signal.
3.1. Membrane Cross-Links and OMV Production
One of the earliest models for OMV formation linked OMV biogenesis to a decrease in OM–peptidoglycan cross-links at the site of vesicle formation (Figure 2b). Braun’s lipoprotein (Lpp), outer membrane protein A (OmpA), and components of the Tol–Pal complex are OM proteins found to be involved in this process. Lpp is an abundant OM protein in some bacteria that acts as a molecular staple, linking the OM to the peptidoglycan layer. Inactivation of lpp results in increased OMV production in P. aeruginosa, E. coli, and S. typhimurium (12, 145). However, lpp mutants have defects in the integrity of their OMs, so it is difficult to distinguish OMV production from cell damage.
OmpA is another OM protein that associates noncovalently with the underlying peptidoglycan layer. OmpA is a common component of OMVs (113, 139, 145). Regulation of OmpA impacts OMV production, and its deletion induces hypervesiculation in many bacterial species (113, 139, 145). In V. cholerae, the small noncoding RNA VrrA is suggested to modulate OMV production by acting as a negative regulator of OmpA. vrrA mutants were found to display a hypervesiculation phenotype likely due to decreased OM–peptidoglycan interactions (135).
The Tol–Pal complex spans the inner membrane and OM of gram-negative bacteria and consists of five proteins, TolA, TolB, TolQ, TolR, and Pal (56). TolA, TolQ, and TolR form a complex in the inner membrane, whereas TolB is a periplasmic protein that interacts with Lpp, OmpA, and Pal (146). Pal is localized to the inner leaflet of the OM, where it interacts directly with the peptidoglycan layer and promotes membrane stability (56, 146). The Tol–Pal complex is proposed to have many functions; however, its best-characterized role is in cell division (56, 146). Tol–Pal proteins localize at invagination sites during cell division, and inactivation of the genes encoding these proteins leaves the cell unable to divide (56, 146). However, Tol–Pal mutants displayed increased bleb formation rates around invagination sites. It has been reported that the Tol–Pal complex is involved in producing OMVs at sites of cell division and that these OMVs are distinct from those generated at other sites on the cell (40).
3.2. Periplasmic Accumulation and OMV Production
It has been proposed that accumulation of periplasmic contents can induce vesicle formation (Figure 2a). For example, peptidoglycan fragments liberated in the periplasm during growth could exert turgor pressure on the OM, leading to vesicle production (149). Studies in P. gingivalis demonstrated that mutants lacking an autolysin displayed increased OMV production. Researchers explained this phenotype by claiming that the absence of the autolysin activity prevented P. gingivalis from degrading periplasmic peptidoglycan fragments, which accumulated in the periplasms, and were therefore expelled via OMVs (68). Misfolded proteins might also induce OMV production by exerting force on the membrane of bacteria with mutations in envelope stress pathways, since these cells were unable to degrade the proteins (128). This claim was supported by the fact that mutants grown at lower temperatures (30°C or 34°C), which cause less protein misfolding, produced OMVs at comparable rates to wild-type cells (104).
3.3. Lipopolysaccharide Remodeling and OMV Production
Several reports have shown that altering LPS content impacts OMV production (Figure 2c). P. aeruginosa produces two LPS types containing different O-polysaccharides, the A-band LPS (neutral charge) and the B-band LPS (anionic charge). Remarkably, only B-band LPS has been detected in the vesicles (78). This finding has led to the hypothesis that OMVs are generated in regions where B-band LPS is more abundant and that the OM bends to alleviate the charge repulsion between them (78). Mutants that only produce B-band LPS were found to produce more OMVs than wild-type organisms and mutants only able to produce A-band LPS (114). However, the OMVs secreted by the wild-type and these mutant strains are different sizes, which indicates that the overproduction of OMVs by strains producing only the B-band LPS could be the result of an envelope stress–coping mechanism (110).
P. gingivalis also synthesizes two different O-antigen chains, the A-LPS (anionic charge) and the O-LPS (neutral charge). Contrary to LPS in P. aeruginosa, both LPS types are packed into P. gingivalis OMVs (66). Mutations affecting A-LPS synthesis or attachment of both O-antigens onto the lipid A core did not affect OMV biogenesis, and all bacteria with LPS mutations produced OMVs comparable in size to wild-type OMVs (66). Even though P. gingivalis packs both types of LPS into OMVs, the lipid A compositions of the OM and OMVs are different (66). The lipid A sorted into OMVs is deacylated compared to those in the OM, suggesting that OMVs are generated in specific OM regions because of compartmentalization or remodeling of the OM (67). In this model, lipid A deacylation alters the configuration of LPS, increasing the membrane curvature and consequently inducing OMV production. When the lipid A deacylase PagL was overexpressed in S. typhimurium, deacylated lipid A was preferentially packed into OMVs, which subsequently increased OMV production (44). PagL is tightly regulated and is expressed inside macrophages (112). Upon macrophage infection, PagL is required for OMV formation by intracellular S. typhimurium (44). Given that most lipid A–modification enzymes, such as PagL, are not expressed in the lab, which is where samples are extracted for lipid A analysis, it is possible that other bacteria produce OMVs with deacylated lipid A during infection.
3.4. OMV Production and the Bilayer-Couple Model
The bilayer-couple model describes a mechanism where membrane curvature is initiated by the insertion of biomolecules into the outer leaflet of the OM (Figure 2d,e). This is proposed to cause the outer leaflet to expand faster than the inner leaflet, resulting in blebbing of the OM and formation of OMVs (126). This model has been extensively studied in P. aeruginosa (Figure 2e); however, a similar mechanism involving VacJ/Yrb ATP-binding cassette (ABC) transport has been investigated in other species (Figure 2d).
As previously mentioned, the molecule PQS is enriched in OMVs produced by P. aeruginosa, and reduced OMV production was found in PQS-deficient mutants (102). Subsequent studies found that PQS stimulates OMV production when it is secreted from the cell and intercalated into the OM (51). PQS insertion causes the OM to expand rapidly, resulting in vesicle formation (126) (Figure 2e). This mechanism appears to be limited to P. aeruginosa and related species that produce PQS (71).
V. cholerae OMVs are enriched in phospholipids and carry large amount of enzymes associated with phospholipid biosynthesis (121). It has been proposed that OMVs are formed as a result of accumulation of phospholipids in the OM, forcing blebbing of the membrane (121). The VacJ/Yrb ABC transport system is involved in the carriage of phospholipids from the OM (99). Disruption of this system promotes OMV overproduction (99, 121). Similar results have been reported for Chromobacterium violaceum, where VacJ/Yrb mutants hypervesiculated (10). Loss of VacJ/Yrb causes an accumulation of phospholipids in the outer leaflet of the OM, which leads to OM expansion and, subsequently, vesicle formation (121) (Figure 2d). Accordingly, OMVs from VacJ/Yrb mutants contain twice as many phospholipids as wild-type OMVs (121). In addition, during iron-limited conditions, the VacJ/Yrb ABC transporter is downregulated by the ferric uptake regulator, causing a hypervesiculation phenotype in H. influenzae, V. cholerae, and E. coli (121). Moreover, regulation of VacJ/Yrb modulated OMV production in the presence of the bile salt sodium taurocholate (38, 49). Since the VacJ/Yrb pathway has been shown to impact OMV production under various conditions and in various organisms, this mechanism of OMV biogenesis could serve as a general way to stimulate OMV production.
3.5. Proposed OMV Cargo Selection Mechanisms
Enrichment of specific protein and lipid cargo is an essential characteristic of OMVs. Vesicles from pathogenic bacteria like P. aeruginosa, V. cholerae, P. gingivalis, and ETEC have been shown to selectively pack virulence factors (19, 72, 78). OMVs from commensal Bacteroides are enriched with glycoside hydrolases that enable the degradation of environmental polysaccharides (45). Similarly, those from the predatory bacterium Myxococcus xanthus are enriched in acidic hydrolases and alkaline phosphatases, which aid the microbe in killing other environmental bacteria (11). The observed OMV cargo selection requires extensive OM compartmentalization to generate patches in the OM regions from which OMVs are secreted. In P. gingivalis, intact LPS is required to achieve proper cargo selection (66). Mutant strains deficient in the synthesis of A-LPS produced OMVs with aberrant cargo, packing a few additional proteins. Interactions of proteins with A-LPS could shape the OM by partitioning proteins into OMVs or excluding them from OMVs (66). This interaction may be direct, in which case the OMV-specific proteins could have a domain to recognize and interact with the O-antigen, promoting compartmentalization of the OM. Alternatively, the interaction could be mediated by a yet unknown sorting factor, which would recruit the protein to the O-antigen-enriched region in the OMV. This proposed mechanism has an uncanny resemblance to the role of galectin in protein sorting in exosomes (41). In agreement with the sorting model, B. fragilis and Bacteroides thetaiotaomicron utilize conserved protein sequences to specifically direct proteins to OMVs (45). The OMVs of these organisms are enriched in a large subset of OMV-unique proteins, which were found to be acidic lipoproteins (negatively charged), while the majority of the OM proteins, β-barrel proteins, are positively charged (45). In Bacteroidetes, lipoproteins enriched in OMVs were found to contain a conserved, negatively charged amino acid motif known as the LES (lipoprotein export signal) domain (140). This domain is required for surface exposure of lipoproteins in Bacteroidetes (94). When it was mutated, lipoproteins were less efficiently packaged into OMVs and were retained in the OM fraction (140). Although many studies have analyzed the OMV proteomes from different bacteria, this is the first time that a specific signal was shown to be linked to an OMV protein-sorting mechanism (140).
4. Mechanisms of OMV Entry into Host Cells
OMVs are long-distance delivery systems; however, the molecular mechanisms underlying OMV internalization and cargo delivery into host cells are not fully understood. Two pathways have been proposed: (a) direct cargo delivery via fusion to host cell membranes and (b) OMV internalization via the endocytic pathway (19, 53, 86, 115). During direct cargo delivery, OMV fusion to host cell lipid rafts induces actin remodeling to allow OMV soluble cargo to diffuse directly into the host cytoplasm. This is the mechanism proposed for P. aeruginosa, Legionella pneumophila, and Aggregatibacter actinomycetemcomitans OMVs (19, 75a, 122). Alternatively, delivery of OMVs is facilitated by diverse mechanisms of endocytosis, including clathrin-mediated endocytosis, caveola-mediated endocytosis, and clathrin- and caveola-independent endocytosis. After OMVs have bound to lipid rafts, they are internalized through one of many endocytic pathways (19, 53, 86, 116). The pathway employed is organism-, or even strain-dependent (19, 53, 86, 116). For example, OMVs derived from Helicobacter pylori, E. coli O157, non-O157 Enterohemorrhagic E. coli (EHEC), and nonpathogenic E. coli strains are internalized by clathrin-mediated endocytosis (15, 49a, 92), whereas the internalization of OMVs from ETEC, V. cholerae, P. aeruginosa, and A. actinomycetemcomitans depends on caveola-mediated endocytosis (19, 28, 82, 86, 122). Finally, P. gingivalis OMVs are internalized via clathrin- and caveola-independent endocytosis (53). Upon entry, the OMV can follow multiple fates inside the host trafficking network. ETEC OMVs containing LT accumulate in nonacidified compartments (86), whereas P. gingivalis and E. coli O157 OMVs and nonpathogenic OMVs are routed to the early endosome, followed by sorting to lysosomal compartments (14, 53, 75).
OMV size and cargo could determine which mechanism(s) of entry into the host cells is employed (76, 83, 104, 137). OMVs from pathogenic E. coli strains are internalized approximately 30% more rapidly and efficiently than those of nonpathogenic E. coli strains (116). Similarly, OMVs produced and secreted by mutant strains lacking one virulence factor, such as LT, Cif, or gingipains, cannot fuse with or enter host cells (19, 53, 86). Furthermore, a reduced OMV entry ratio was observed in strains lacking O-antigen (116). Smaller OMVs preferentially enter epithelial cells via caveola-mediated endocytosis (137). These findings suggest that the kinetics of entry are cargo dependent; proteins and/or LPS carried by OMVs probably interacts with or bind to receptors present on the lipids rafts that promote OMV attachment and entry (19, 53, 86, 116).
5. OMVs as a Platform for Biotechnological Applications
In recent years, OMVs have gained attention as a platform for biotechnological applications. Such applications take advantage of the fact that OMVs contain bacterium-derived antigens and multiple pathogen-associated molecular patterns (PAMPs) and can modulate the immune system. The best-known biotechnological application is the use of OMVs as vaccines (141)(106a). However, other applications have been proposed, such as drug-delivery systems (95).
5.1. OMVs as Vaccines
By mimicking a pathogen but not causing the related diseases, vaccines induce humoral (antibody-mediated) and/or cellular (immune cell– and cytokine-mediated) immunity and dramatically decrease infection and illness rates. As OMVs are non-replicative entities that mimic immunogenic properties of the producing bacteria, they are an attractive vaccine platform. Further advantages of OMVs are their size, which enables their entry into lymph vessels and uptake by antigen-presenting cells (APCs) (48). In addition, OMVs have natural adjuvant properties that strongly stimulate the innate and, more importantly, the adaptive immune responses (47, 141). The high stability of OMVs upon exposure to high temperatures and several chemical treatments further points to them as attractive vaccine candidates (4). Two types of OMV-based vaccines have been described: natural OMV and bioengineered OMVs.
5.1.1. Naturally secreted OMV-based vaccines.
The vaccines against N. meningitidis group B (MenB) are the most representative and successful OMV-based vaccines developed to date. In the last 25 years, these vaccines have been developed and employed to combat outbreaks in various countries (70) (see the sidebar titled Overview of OMV-Based MenB Vaccines). OMV-based vaccines against V. cholerae, S. typhimurium, and Shigella flexneri have been investigated in animal models, but none has progressed to clinical trials (2, 23, 127).
SIDEBAR 1: OVERVIEW OF OMV-BASED MenB VACCINES.
For most N. meningitidis serogroups, the available vaccines consist of CPS coupled to a carrier protein. However, the serogroup B capsule is homologous to molecular structures present in the human brain, making it impossible to produce a glycoconjugate vaccine for this serogroup (70). Thus, OM proteins such as PorA were considered as vaccine candidates. PorA, the main immunogenic protein in N. meningitidis and also present in OMVs, is highly variable between strains (70). To overcome the problem of strain specificity, a novel MenB vaccine using OMVs derived from bioengineered strains expressing multiple PorA variants has been developed in the Netherlands (70, 141). Most available OMV-based MenB vaccines (VA-MENGOC-BC, MenBvac and MeNZB) succeeded in combating specific outbreaks of MenB-caused meningitis in Cuba, Norway, and New Zealand, with an efficacy of at least 70% (70). In 2016, the BEXSERO vaccine was been approved for human use by the European Medicines Agency and the FDA as this vaccine provides protection against endemic disease (70).
OMVs have the intrinsic capacity to act as adjuvants. The immunogenic properties of OMV cargo lead to protective mucosal and systemic bactericidal antibody responses. OMVs can easily be phagocytized and processed by APCs, including dendritic cells (48). Then, the OMV-delivered antigens are presented by APCs to CD4+ T cells, leading to the generation of antigen-specific B cell responses (47, 141)(100, 106a). LPS, a major component of OMVs, is a potent activator of immune cells, such as monocytes/macrophages. Specific recognition by the TLR4/MD2 receptor on these cells triggers NF-κB- and IRF3-dependent gene expression (98, 100). However, this inflammatory activation can also result in high vaccine reactogenicity. In addition to LPS, OMVs carry lipoproteins that are recognized by TLR2 and also modulate and activate innate immunity (89, 100).
Despite a few successful examples, some disadvantages of vaccines based on naturally released OMVs need to be overcome before they are widely employed. The main concern is safety. OMVs contain lipid A, an endotoxic component of LPS that can provoke a severe, even lethal, inflammatory response in the host (112). Furthermore, OMV cargo varies between strains, which may in some cases limit their applicability to a specific subset of strains. Finally, many strains secrete low amounts of OMVs, which means that large volumes of pathogenic bacteria are needed. In the last decade, bioengineering of OMV-producing bacteria has substantially progressed to address these concerns.
5.1.2. Bioengineered OMV–based vaccines.
Bioengineered OMV–carrying, heterologous recombinant proteins expressed in lab strains have shown potential. One strategy to effectively direct heterologous antigens to the surface of the OMV is to fuse them with native OMV proteins. Many groups have selected ClyA from E. coli as the fusion partner to deliver exogenous proteins to OMVs (31, 55, 143). For example, ClyA has been fused to green fluorescent protein (GFP), to the ectodomain of the influenza A matrix protein 2 (M2), and to the domain 4 moiety of Bacillus anthracis protective antigen (31, 55, 120). As an alternative, antigens can be overexpressed and directed to the periplasmic space, where they will be entrapped and packed into the OMV lumen (85, 109). Several proteins from Streptococcus have been fused to the signal peptide of E. coli OmpA, and once in the periplasm, they were successfully packed into OMVs (50). A similar approach was employed to generate E. coli OMVs carrying the Chlamydia muridarum protein HtrA (9). Even though these proteins were not surface exposed, they were able to elicit antigen-specific antibody responses (9, 50, 109). The IgG antibodies generated had excellent functional activity in terms of bacterial opsonophagocytic killing and protection in murine lethal infectious challenge assays (50, 109).
Capsular polysaccharides (CPS) and LPS, which decorate the cell surfaces of pathogenic bacteria, are also good vaccine antigens. Unfortunately, vaccines consisting solely of polysaccharides typically promote T cell–independent immune responses, which do not include IgM-to-IgG class switch and fail to generate immunological memory (6). A common strategy to trigger immunological memory is to covalently couple the polysaccharide to a carrier. Capsule and O-antigen biosynthesis gene clusters from pathogenic bacteria can be transferred into E. coli lab strains. These glycoengineering techniques can be employed to display the glycans of choice as recombinant LPS in nonpathogenic bacteria. The glycoengineered OMVs can be purified and employed directly as conjugate vaccines. Streptococcus pneumoniae capsule (Sp-CPS) gene cluster was expressed in E. coli. There, Sp-CPS was attached to the lipid A core and displayed on the bacterial surface and glycoengineered OMVs (118). These glycoengineered OMVs raised specific IgG antibodies against Sp-CPS in immunized mice and were shown effective in opsonophagocytosis assays (118). In another example, the Francisella tularensis O-antigen polysaccharide gene cluster was also successfully expressed in E. coli to produce glycoengineered OMVs decorated with F. tularensis O-antigen (32). Mice immunized with these F. tularensis glycoengineered OMVs were protected against lethal challenge with several strains of F. tularensis (32). Furthermore, to highlight the versatility of this vaccine technology, glycosyltransferases and the enzymes required for the synthesis of nucleotide-activated sugars from various organisms can be expressed in E. coli cells to produce customized glycoengineered OMVs (138).
5.1.3. OMV-based vaccine detoxification alternatives.
Several strategies have been developed to obtain OMVs with a low level of LPS toxicity. The first method is to reduce LPS content by treating purified OMVs with detergents, such as deoxycholate and polyoxyethylene 10 oleyl ether (Brij-96) (70, 141). However, this approach has the disadvantage of a loss of lipoproteins, which are TLR agonists, thus reducing the adjuvant properties of OMVs (100). A second detoxification method is based on the fact that less-acylated lipid A has reduced toxicity (132). Monophosphoryl lipid A is a safe and effective vaccine adjuvant that is approved by the US Food and Drug Administration for human use (25). Some naturally occurring Neisseria strains produce penta-acylated LPS due to a mutation in the lipid A biosynthesis lauroyl acyltransferase, encoded by lpxL1. Their secreted OMVs do not need the detergent extraction step because of low endotoxic activity of the penta-acylated lipid A (52). Therefore, by modifying genes responsible for lipid A synthesis, it is possible to obtain genetically detoxified strains and, consequently, detoxified OMVs (100, 112). Inactivation of the genes encoding lipid A acyltransferases, such as msbA, msbB, lpxL1, and lpxM, and/or overexpression of genes encoding lipid A deacyltransferases, such as pagP, results in lipid A lacking an acyl chain and the concomitant reduced activation of TLR4/MD2 (5, 44, 74, 112). An additional fine-tuning step can be accomplished by introducing lipid A phosphatases, such as LpxE, to generate monophosphorylated penta-acylated lipid A (32, 112). Another alternative is to engineer nonpathogenic bacteria that naturally produce OMVs that only pack monophosphorylated penta-acylated lipid A. Bacteroides OMVs contain only monophosphorylated penta-acylated lipid A moieties, and these organisms can be engineered to deliver antigens and drugs (24, 45). A balanced response that generates sufficient adjuvant activity but prevents harsh side effects can be achieved by adopting any of the methods described above.
5.2. OMVs as Cancer Immunotherapy Agents
In the last decades, multiple preclinical and clinical studies involving cancer vaccines have been performed. Unfortunately, these vaccines have had an overwhelmingly low rate of efficacy (less than 5%) (148). However, promising results for the use of liposomes have been reported lately (90). Considering the immunomodulatory role of OMVs, they can be engineered to express cancer-specific epitopes or to carry small noncoding RNAs (61, 148). Furthermore, OMVs induced a durable antitumor immune response that inhibited tumor growth in multiple tumor models (33, 148). The caveat, again, is the toxic effect of LPS. However, some of the detoxification methods mentioned above are promising and worthy of exploration to improve the cancer therapies available.
6. Skepticism about OMVs and their Physiological Relevance
OMVs were described for the first time in the 1960s (17, 87). However, for several reasons, a number of scientists are still skeptical about their biological relevance. The story of OMVs resembles that of eukaryotic extracellular vesicles, or exosomes, which have also been a matter of controversy. Many concerns stem from the fact that despite more than 50 years of research, we have yet to elucidate the mechanism(s) for their formation. Pieces of the puzzle regarding OMV biogenesis have been uncovered in different species, however, and none of the working models has been confirmed to be universal.
It has been proposed that DNA and RNA are packed inside OMVs (18, 37, 133). None of the current models for OMV biogenesis specifically accounts for the packing of nucleic acids, as these molecules would need to be translocated into the periplasm and across the peptidoglycan layer to be included in an OMV. On the contrary, RNA and DNA are released upon cell lysis and could easily contaminate OMV preparations. A feasible explanation for how DNA or RNA is transported into the bacterial periplasm would support the biological function of these molecules in OMVs.
Multiple mutant strains presenting hypervesiculating phenotypes have been described (104, 116, 139, 145). Although some of these mutations appear to regulate OMV formation, many mutations also affect membrane stability. Consequently, the material produced by these strains that is considered OMVs may result from lysis or membrane destabilization. No mutants unable to produce OMVs have been identified. Those who believe in the relevance of OMVs explain this by postulating the essentiality of OMV biogenesis. Because vesicles formed as by-products of lysis have the same composition as the membrane from which they are derived, the fact that protein and lipid cargo selection has been shown argues against the notion that OMVs are simply bacterial debris. Many secreted proteins, such as flagellins, pilins, and T3SS effectors, may aggregate and copurify with OMVs. Therefore, claims of cargo selection have to be considered with caution when these are the proteins that are presumably enriched n OMV preparations. In any case, cargo selection has been conclusively demonstrated in a few bacterial species, with proteomics indicating that, at least in these species, OMVs do not result from lysis (19, 45, 66, 72, 78, 140, 142).
Numerous functions have been proposed for OMVs. OMVs, when reported, are purified from bacteria cultured in large volumes under standard lab conditions with regard to media, temperature, and aeration (28, 48, 92, 96a, 108, 133). In some studies, OMVs were obtained from up to 1.5 L of culture and concentrated by a factor in the hundreds before their biological properties were tested. Although this might suggest that OMVs are produced in negligible amounts, OMV biogenesis is likely stimulated in vivo. However, the results of these experiments have to be carefully interpreted, as the composition of OMVs produced during infection can be very different from that of OMVs produced in culturing media.
Part of the problem is that the vesicle biogenesis process is very difficult to investigate. Bona fide OMVs and lysis-derived vesicle-like structures are extremely difficult to differentiate by biophysical and biochemical methods. We recognize the limitations of current studies involving the biogenesis and functions of OMVs. It is possible that some of the published works contain artifacts. However, there is substantial evidence to assert that at least some bacteria under particular conditions produce bona fide OMVs as a result of an orchestrated process. Given recent technical advances in mass spectrometry and microscopy, it will be possible to investigate the vesiculation process in relevant clinical strains and in infection models to decipher OMV biogenesis mechanisms and establish their true biological significance.
SUMMARY POINTS.
Outer membrane vesicles (OMVs) constitute a universal secretion system that is found in all gram-negative bacteria and that results from a directed and selective cellular process.
OMV composition can be differentiated from that of the outer membrane, as OMVs are enriched with a specific subset of proteins and lipids.
At least five mechanisms have been proposed to explain OMV formation.
OMVs can have offensive and defensive roles in bacterium–bacterium and bacterium–host interactions.
Engineered OMVs carrying customized cargo and detoxified lipopolysaccharide could be used to improve current vesicle-based vaccines and drug-delivery platforms.
FUTURE ISSUES.
Is there a universal mechanism for OMV biogenesis?
How is protein cargo selected?
What is the composition of the cargo of OMVs secreted in vivo?
ACKNOWLEDGMENTS
We thank Juvenal Lopez for critical reading of the manuscript. This work was supported by National Institute of Allergy and Infectious Diseases grant R21AI151873, awarded to M.F.F, and a National Institutes of Health training program in “Cellular and Molecular Biology” awarded to E.J.P.
Footnotes
DISCLOSURE STATEMENT
M.F.F has a patent granted for the use of glycoengineered OMVs as vaccines (US9526775B2, “Glycoengineered outer membrane vesicles and use thereof as vaccines”). All other authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Aguilera L, Toloza L, Giménez R, Odena A, Oliveira E, et al. 2014. Proteomic analysis of outer membrane vesicles from the probiotic strain Escherichia coli Nissle 1917. Proteomics 14:222–29 [DOI] [PubMed] [Google Scholar]
- 2.Alaniz RC, Deatherage BL, Lara JC, Cookson BT. 2007. Membrane vesicles are immunogenic facsimiles of Salmonella typhimurium that potently activate dendritic cells, prime B and T cell responses, and stimulate protective immunity in vivo. J. Immunol. 179:7692–701 [DOI] [PubMed] [Google Scholar]
- 3.Allan ND, Beveridge TJ. 2003. Gentamicin delivery to Burkholderia cepacia group IIIa strains via membrane vesicles from Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 47:2962–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arigita C, Jiskoot W, Westdijk J, van Ingen C, Hennink WE, et al. 2004. Stability of mono- and trivalent meningococcal outer membrane vesicle vaccines. Vaccine 22:629–42 [DOI] [PubMed] [Google Scholar]
- 5.Asensio CJ, Gaillard ME, Moreno G, Bottero D, Zurita E, et al. 2011. Outer membrane vesicles obtained from Bordetella pertussis Tohama expressing the lipid A deacylase PagL as a novel acellular vaccine candidate. Vaccine 29:1649–56 [DOI] [PubMed] [Google Scholar]
- 6.Avci FY, Kasper DL. 2010. How bacterial carbohydrates influence the adaptive immune system. Annu. Rev. Immunol 28:107–30 [DOI] [PubMed] [Google Scholar]
- 7.Bahar O, Mordukhovich G, Luu DD, Schwessinger B, Daudi A, et al. 2016. Bacterial outer membrane vesicles induce plant immune responses. Mol. Plant-Microbe Interact. 29:374–84 [DOI] [PubMed] [Google Scholar]
- 8. Deleted in proof, duplication.
- 9.Bartolini E, Ianni E, Frigimelica E, Petracca R, Galli G, et al. 2013. Recombinant outer membrane vesicles carrying Chlamydia muridarum HtrA induce antibodies that neutralize chlamydial infection in vitro. J. Extracell. Vesicles 2. 10.3402/jev.v2i0.20181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batista JH, Leal FC, Fukuda TTH, Alcoforado Diniz J, Almeida F, et al. 2020. Interplay between two quorum sensing-regulated pathways, violacein biosynthesis and VacJ/Yrb, dictates outer membrane vesicle biogenesis in Chromobacterium violaceum. Environ. Microbiol 22:2432–42 [DOI] [PubMed] [Google Scholar]
- 10a.Bélanger M, Kozarov E, Song H, Whitlock J, Progulske-Fox A. 2012. Both the unique and repeat regions of the Porphyromonas gingivalis hemagglutin A are involved in adhesion and invasion of host cells. Anaerobe 18:128–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berleman JE, Allen S, Danielewicz MA, Remis JP, Gorur A, et al. 2014. The lethal cargo of Myxococcus xanthus outer membrane vesicles. Front. Microbiol 5:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernadac A, Gavioli M, Lazzaroni JC, Raina S, Lloubès R. 1998. Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol 180:4872–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bielaszewska M, Marejková M, Bauwens A, Kunsmann-Prokscha L, Mellmann A, Karch H. 2018. Enterohemorrhagic Escherichia coli O157 outer membrane vesicles induce interleukin 8 production in human intestinal epithelial cells by signaling via Toll-like receptors TLR4 and TLR5 and activation of the nuclear factor NF-κB. Int. J. Med. Microbiol 308:882–89 [DOI] [PubMed] [Google Scholar]
- 14.Bielaszewska M, Rüter C, Bauwens A, Greune L, Jarosch KA, et al. 2017. Host cell interactions of outer membrane vesicle-associated virulence factors of enterohemorrhagic Escherichia coli O157: intracellular delivery, trafficking and mechanisms of cell injury. PLOS Pathog. 13:e1006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bielaszewska M, Rüter C, Kunsmann L, Greune L, Bauwens A, et al. 2013. Enterohemorrhagic Escherichia coli hemolysin employs outer membrane vesicles to target mitochondria and cause endothelial and epithelial apoptosis. PLOS Pathog. 9:e1003797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bielig H, Rompikuntal PK, Dongre M, Zurek B, Lindmark B, et al. 2011. NOD-like receptor activation by outer membrane vesicles from Vibrio cholerae non-O1 non-O139 strains is modulated by the quorum-sensing regulator HapR. Infect. Immun 79:1418–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bishop DG, Work E. 1965. An extracellular glycolipid produced by Escherichia coli grown under lysine-limiting conditions. Biochem. J 96:567–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blenkiron C, Simonov D, Muthukaruppan A, Tsai P, Dauros P, et al. 2016. Uropathogenic Escherichia coli releases extracellular vesicles that are associated with RNA. PLOS ONE 11:e0160440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O’Toole GA, Stanton BA. 2009. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLOS Pathog. 5:e1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brameyer S, Plener L, Müller A, Klingl A, Wanner G, Jung K. 2018. Outer membrane vesicles facilitate trafficking of the hydrophobic signaling molecule CAI-1 between Vibrio harveyi cells. J. Bacteriol 200:e00740–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown L, Wolf JM, Prados-Rosales R, Casadevall A. 2015. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol 13:620–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deleted in proof. Moved to Reference 10a.
- 23.Camacho AI, de Souza J, Sánchez-Gómez S, Pardo-Ros M, Irache JM, Gamazo C. 2011. Mucosal immunization with Shigella flexneri outer membrane vesicles induced protection in mice. Vaccine 29:8222–29 [DOI] [PubMed] [Google Scholar]
- 23a.Cañas MA, Fábrega MJ, Giménez R, Badia J, Baldomà L. 2018. Outer membrane vesicles from probiotic and commensal Escherichia coli activate NOD1-mediated immune responses in intestinal epithelial cells. Front. Microbiol 9:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carvalho AL, Fonseca S, Miquel-Clopés A, Cross K, Kok KS, et al. 2019. Bioengineering commensal bacteria-derived outer membrane vesicles for delivery of biologics to the gastrointestinal and respiratory tract. J. Extracell. Vesicles 8:1632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casella CR, Mitchell TC. 2008. Putting endotoxin to work for us: monophosphoryl lipid A as a safe and effective vaccine adjuvant. Cell Mol. Life Sci 65:3231–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deleted in proof. Moved to Reference 23a.
- 27.Celluzzi A, Masotti A. 2016. How our other genome controls our epi-genome. Trends Microbiol. 24:777–87 [DOI] [PubMed] [Google Scholar]
- 28.Chatterjee D, Chaudhuri K. 2011. Association of cholera toxin with Vibrio cholerae outer membrane vesicles which are internalized by human intestinal epithelial cells. FEBS Lett. 585:1357–62 [DOI] [PubMed] [Google Scholar]
- 29.Chatterjee SN, Das J. 1967. Electron microscopic observations on the excretion of cell-wall material by Vibrio cholerae. J. Gen. Microbiol 49:1–11 [DOI] [PubMed] [Google Scholar]
- 30.Chatzidaki-Livanis M, Coyne MJ, Comstock LE. 2014. An antimicrobial protein of the gut symbiont Bacteroides fragilis with a MACPF domain of host immune proteins. Mol. Microbiol 94:1361–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen DJ, Osterrieder N, Metzger SM, Buckles E, Doody AM, et al. 2010. Delivery of foreign antigens by engineered outer membrane vesicle vaccines. PNAS 107:3099–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Valentine JL, Huang CJ, Endicott CE, Moeller TD, et al. 2016. Outer membrane vesicles displaying engineered glycotopes elicit protective antibodies. PNAS 113:E3609–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Q, Bai H, Wu W, Huang G, Li Y, et al. 2020. Bioengineering bacterial vesicle-coated polymeric nanomedicine for enhanced cancer immunotherapy and metastasis prevention. Nano Lett. 20:11–21 [DOI] [PubMed] [Google Scholar]
- 34.Choi DS, Kim DK, Choi SJ, Lee J, Choi JP, et al. 2011. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics 11:3424–29 [DOI] [PubMed] [Google Scholar]
- 35.Ciofu O, Beveridge TJ, Kadurugamuwa J, Walther-Rasmussen J, Høiby N. 2000. Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother 45:9–13 [DOI] [PubMed] [Google Scholar]
- 36.Coyne MJ, Béchon N, Matano LM, McEneany VL, Chatzidaki-Livanis M, Comstock LE. 2019. A family of anti-Bacteroidales peptide toxins wide-spread in the human gut microbiota. Nat. Commun 10:3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dauros-Singorenko P, Blenkiron C, Phillips A, Swift S. 2018. The functional RNA cargo of bacterial membrane vesicles. FEMS Microbiol. Lett 365. 10.1093/femsle/fny023 [DOI] [PubMed] [Google Scholar]
- 38.Davies C, Taylor AJ, Elmi A, Winter J, Liaw J, et al. 2019. Sodium taurocholate stimulates Campylobacter jejuni outer membrane vesicle production via down-regulation of the maintenance of lipid asymmetry pathway. Front. Cell Infect. Microbiol 9:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deatherage BL, Cookson BT. 2012. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect. Immun 80:1948–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deatherage BL, Lara JC, Bergsbaken T, Rassoulian Barrett SL, Lara S, Cookson BT. 2009. Biogenesis of bacterial membrane vesicles. Mol. Microbiol 72:1395–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delacour D, Greb C, Koch A, Salomonsson E, Leffler H, et al. 2007. Apical sorting by galectin-3-dependent glycoprotein clustering. Traffic 8:379–88 [DOI] [PubMed] [Google Scholar]
- 42.DeVoe IW, Gilchrist JE. 1975. Pili on meningococci from primary cultures of nasopharyngeal carriers and cerebrospinal fluid of patients with acute disease. J. Exp. Med 141:297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duperthuy M, Sjöström AE, Sabharwal D, Damghani F, Uhlin BE, Wai SN. 2013. Role of the Vibrio cholerae matrix protein Bap1 in cross-resistance to antimicrobial peptides. PLOS Pathog. 9:e1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elhenawy W, Bording-Jorgensen M, Valguarnera E, Haurat MF, Wine E, Feldman MF. 2016. LPS remodeling triggers formation of outer membrane vesicles in Salmonella. mBio 7:e00940–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elhenawy W, Debelyy MO, Feldman MF. 2014. Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio 5:e00909–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ellen RP, Grove DA. 1989. Bacteroides gingivalis vesicles bind to and aggregate Actinomyces viscosus. Infect. Immun 57:1618–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ellis TN, Kuehn MJ. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev 74:81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellis TN, Leiman SA, Kuehn MJ. 2010. Naturally produced outer membrane vesicles from Pseudomonas aeruginosa elicit a potent innate immune response via combined sensing of both lipopolysaccharide and protein components. Infect. Immun. 78:3822–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elmi A, Dorey A, Watson E, Jagatia H, Inglis NF, et al. 2018. The bile salt sodium taurocholate induces Campylobacter jejuni outer membrane vesicle production and increases OMV-associated proteolytic activity. Cell. Microbiol 20. 10.1111/cmi.12814 [DOI] [PubMed] [Google Scholar]
- 49a.Fábrega MJ, Aguilera L, Giménez R, Varela E, Alexandra Cañas M, et al. 2016. Activation of immune and defense responses in the intestinal mucosa by outer membrane vesicles of commensal and probiotic Escherichia coli strains. Front. Microbiol 7:705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fantappiè L, de Santis M, Chiarot E, Carboni F, Bensi G, et al. 2014. Antibody-mediated immunity induced by engineered Escherichia coli OMVs carrying heterologous antigens in their lumen. J. Extracell. Vesicles 3. 10.3402/jev.v3.24015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Florez C, Raab JE, Cooke AC, Schertzer JW. 2017. Membrane distribution of the Pseudomonas quinolone signal modulates outer membrane vesicle production in Pseudomonas aeruginosa. mBio 8:e01034–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fransen F, Heckenberg SG, Hamstra HJ, Feller M, Boog CJ, et al. 2009. Naturally occurring lipid A mutants in Neisseria meningitidis from patients with invasive meningococcal disease are associated with reduced coagulopathy. PLOS Pathog. 5:e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Furuta N, Tsuda K, Omori H, Yoshimori T, Yoshimura F, Amano A. 2009. Porphyromonas gingivalis outer membrane vesicles enter human epithelial cells via an endocytic pathway and are sorted to lysosomal compartments. Infect. Immun 77:4187–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Deleted in proof. Moved to Reference 49a.
- 55.Galen JE, Zhao L, Chinchilla M, Wang JY, Pasetti MF, et al. 2004. Adaptation of the endogenous Salmonella enterica serovar Typhi clyA-encoded hemolysin for antigen export enhances the immunogenicity of anthrax protective antigen domain 4 expressed by the attenuated live-vector vaccine strain CVD 908-htrA. Infect. Immun 72:7096–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. 2007. The trans-envelope Tol–Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol. Microbiol 63:1008–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerritzen MJH, Martens DE, Uittenbogaard JP, Wijffels RH, Stork M. 2019. Sulfate depletion triggers overproduction of phospholipids and the release of outer membrane vesicles by Neisseria meningitidis. Sci. Rep 9:4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghosh A, Saha DR, Hoque KM, Asakuna M, Yamasaki S, et al. 2006. Enterotoxigenicity of mature 45-kilodalton and processed 35-kilodalton forms of hemagglutinin protease purified from a cholera toxin gene-negative Vibrio cholerae non-O1, non-O139 strain. Infect. Immun 74:2937–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.González LJ, Bahr G, Nakashige TG, Nolan EM, Bonomo RA, Vila AJ. 2016. Membrane anchoring stabilizes and favors secretion of New Delhi metallo-β-lactamase. Nat. Chem. Biol 12:516–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.González Plaza JJ. 2018. Small RNAs in cell-to-cell communications during bacterial infection. FEMS Microbiol. Lett 365. 10.1093/femsle/fny024 [DOI] [PubMed] [Google Scholar]
- 61.Grandi A, Fantappiè L, Irene C, Valensin S, Tomasi M, et al. 2018. Vaccination with a FAT1-derived B cell epitope combined with tumor-specific B and T cell epitopes elicits additive protection in cancer mouse models. Front. Oncol 8:481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grenier D, Bélanger M. 1991. Protective effect of Porphyromonas gingivalis outer membrane vesicles against bactericidal activity of human serum. Infect. Immun 59:3004–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grenier D, Mayrand D. 1987. Functional characterization of extracellular vesicles produced by Bacteroides gingivalis. Infect. Immun 55:111–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guerrero-Mandujano A, Hernández-Cortez C, Ibarra JA, Castro-Escarpulli G. 2017. The outer membrane vesicles: secretion system type zero. Traffic 18:425–32 [DOI] [PubMed] [Google Scholar]
- 65.Guidi R, Levi L, Rouf SF, Puiac S, Rhen M, Frisan T. 2013. Salmonella enterica delivers its genotoxin through outer membrane vesicles secreted from infected cells. Cell Microbiol 15:2034–50 [DOI] [PubMed] [Google Scholar]
- 66.Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, et al. 2011. Selective sorting of cargo proteins into bacterial membrane vesicles. J. Biol. Chem 286:1269–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haurat MF, Elhenawy W, Feldman MF. 2015. Prokaryotic membrane vesicles: new insights on biogenesis and biological roles. Biol. Chem 396:95–109 [DOI] [PubMed] [Google Scholar]
- 68.Hayashi J, Hamada N, Kuramitsu HK. 2002. The autolysin of Porphyromonas gingivalis is involved in outer membrane vesicle release. FEMS Microbiol. Lett 216:217–22 [DOI] [PubMed] [Google Scholar]
- 69.Hickey CA, Kuhn KA, Donermeyer DL, Porter NT, Jin C, et al. 2015. Colitogenic Bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase-dependent manner via outer membrane vesicles. Cell Host Microbe 17:672–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holst J, Oster P, Arnold R, Tatley MV, Næss LM, et al. 2013. Vaccines against meningococcal serogroup B disease containing outer membrane vesicles (OMV): lessons from past programs and implications for the future. Hum. Vaccin. Immunother 9:1241–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horspool AM, Schertzer JW. 2018. Reciprocal cross-species induction of outer membrane vesicle biogenesis via secreted factors. Sci. Rep 8:9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Horstman AL, Kuehn MJ. 2000. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem 275:12489–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ionescu M, Zaini PA, Baccari C, Tran S, da Silva AM, Lindow SE. 2014. Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. PNAS 111:E3910–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Irene C, Fantappiè L, Caproni E, Zerbini F, Anesi A, et al. 2019. Bacterial outer membrane vesicles engineered with lipidated antigens as a platform for Staphylococcus aureus vaccine. PNAS 116:21780–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Irving AT, Mimuro H, Kufer TA, Lo C, Wheeler R, et al. 2014. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe 15:623–35 [DOI] [PubMed] [Google Scholar]
- 75a.Jäger J, Keese S, Roessle M, Steinert M, Schromm AB. 2015. Fusion of Legionella pneumophila outer membrane vesicles with eukaryotic membrane systems is a mechanism to deliver pathogen factors to host cell membranes. Cell Microbiol. 17:607–20 [DOI] [PubMed] [Google Scholar]
- 76.Jones EJ, Booth C, Fonseca S, Parker A, Cross K, et al. 2020. The uptake, trafficking, and biodistribution of Bacteroides thetaiotaomicron generated outer membrane vesicles. Front. Microbiol. 11:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Deleted in proof. Moved to 75a.
- 78.Kadurugamuwa JL, Beveridge TJ. 1995. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol 177:3998–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kadurugamuwa JL, Beveridge TJ. 1996. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J. Bacteriol 178:2767–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kamaguchi A, Nakayama K, Ichiyama S, Nakamura R, Watanabe T, et al. 2003. Effect of Porphyromonas gingivalis vesicles on coaggregation of Staphylococcus aureus to oral microorganisms. Curr. Microbiol 47:485–91 [DOI] [PubMed] [Google Scholar]
- 81.Kang CS, Ban M, Choi EJ, Moon HG, Jeon JS, et al. 2013. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLOS ONE 8:e76520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaparakis M, Turnbull L, Carneiro L, Firth S, Coleman HA, et al. 2010. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol. 12:372–85 [DOI] [PubMed] [Google Scholar]
- 83.Kaparakis-Liaskos M, Ferrero RL. 2015. Immune modulation by bacterial outer membrane vesicles. Nat. Rev. Immunol 15:375–87 [DOI] [PubMed] [Google Scholar]
- 84.Katsir L, Bahar O. 2017. Bacterial outer membrane vesicles at the plant–pathogen interface. PLOS Pathog. 13:e1006306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kesty NC, Kuehn MJ. 2004. Incorporation of heterologous outer membrane and periplasmic proteins into Escherichia coli outer membrane vesicles. J. Biol. Chem 279:2069–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ. 2004. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 23:4538–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Knox KW, Vesk M, Work E. 1966. Relation between excreted lipopolysaccharide complexes and surface structures of a lysine-limited culture of Escherichia coli. J. Bacteriol 92:1206–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kouokam JC, Wai SN, Fällman M, Dobrindt U, Hacker J, Uhlin BE. 2006. Active cytotoxic necrotizing factor 1 associated with outer membrane vesicles from uropathogenic Escherichia coli. Infect. Immun 74:2022–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kovacs-Simon A, Titball RW, Michell SL. 2011. Lipoproteins of bacterial pathogens. Infect. Immun 79:548–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kranz LM, Diken M, Haas H, Kreiter S, Loquai C, et al. 2016. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534:396–401 [DOI] [PubMed] [Google Scholar]
- 91.Kulkarni HM, Nagaraj R, Jagannadham MV. 2015. Protective role of E. coli outer membrane vesicles against antibiotics. Microbiol. Res 181:1–7 [DOI] [PubMed] [Google Scholar]
- 92.Kunsmann L, Rüter C, Bauwens A, Greune L, Glüder M, et al. 2015. Virulence from vesicles: novel mechanisms of host cell injury by Escherichia coli O104:H4 outbreak strain. Sci. Rep 5:13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lappann M, Otto A, Becher D, Vogel U. 2013. Comparative proteome analysis of spontaneous outer membrane vesicles and purified outer membranes of Neisseria meningitidis. J. Bacteriol 195:4425–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lauber F, Cornelis GR, Renzi F. 2016. Identification of a new lipoprotein export signal in Gram-negative bacteria. mBio 7:e01232–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li M, Zhou H, Yang C, Wu Y, Zhou X, et al. 2020. Bacterial outer membrane vesicles as a platform for biomedical applications: an update. J. Control. Release 323:253–68 [DOI] [PubMed] [Google Scholar]
- 96.Loeb MR. 1974. Bacteriophage T4-mediated release of envelope components from Escherichia coli. J. Virol 13:631–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96a.Losier TT, Akuma M, McKee-Muir OC, LeBlond ND, et al.2019. AMPK Promotes Xenophagy through Priming of Autophagic Kinases upon Detection of Bacterial Outer Membrane Vesicles. Cell Reports 26:2150–2165.e5 [DOI] [PubMed] [Google Scholar]
- 97.Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051–60 [DOI] [PubMed] [Google Scholar]
- 98.Maisonneuve C, Bertholet S, Philpott DJ, De Gregorio E. 2014. Unleashing the potential of NOD- and Toll-like agonists as vaccine adjuvants. PNAS 111:12294–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Malinverni JC, Silhavy TJ. 2009. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. PNAS 106:8009–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mancini F, Rossi O, Necchi F, Micoli F. 2020. OMV vaccines and the role of TLR agonists in immune response. Int. J. Mol. Sci 21:4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Manning AJ, Kuehn MJ. 2011. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 11:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mashburn LM, Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422–25 [DOI] [PubMed] [Google Scholar]
- 103.Mazmanian SK, Round JL, Kasper DL. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620–25 [DOI] [PubMed] [Google Scholar]
- 104.McBroom AJ, Kuehn MJ. 2007. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol 63:545–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McCaig WD, Loving CL, Hughes HR, Brockmeier SL. 2016. Characterization and vaccine potential of outer membrane vesicles produced by Haemophilus parasuis. PLOS ONE 11:e0149132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mergenhagen SE, Bladen HA, Hsu KC. 1966. Electron microscopic localization of endotoxic lipopolysaccharide in gram-negative organisms. Ann. N. Y. Acad. Sci 133:279–91 [DOI] [PubMed] [Google Scholar]
- 106a.Micoli F, MacLennan CA. 2020. Outer membrane vesicle vaccines. Semin Immunol. 50:101433. [DOI] [PubMed] [Google Scholar]
- 107.Molina-Tijeras JA, Gálvez J, Rodríguez-Cabezas ME. 2019. The immunomodulatory properties of extracellular vesicles derived from probiotics: a novel approach for the management of gastrointestinal diseases. Nutrients 11:1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mondal A, Tapader R, Chatterjee NS, Ghosh A, Sinha R, et al. 2016. Cytotoxic and inflammatory responses induced by outer membrane vesicle-associated biologically active proteases from Vibrio cholerae. Infect. Immun 84:1478–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Muralinath M, Kuehn MJ, Roland KL, Curtiss R. 2011. Immunization with Salmonella enterica serovar Typhimurium-derived outer membrane vesicles delivering the pneumococcal protein PspA confers protection against challenge with Streptococcus pneumoniae. Infect. Immun 79:887–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Murphy K, Park AJ, Hao Y, Brewer D, Lam JS, Khursigara CM. 2014. Influence of O polysaccharides on biofilm development and outer membrane vesicle biogenesis in Pseudomonas aeruginosa PAO1. J. Bacteriol 196:1306–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nascimento R, Gouran H, Chakraborty S, Gillespie HW, Almeida-Souza HO, et al. 2016. The type II secreted lipase/esterase LesA is a key virulence factor required for Xylella fastidiosa pathogenesis in grapevines. Sci. Rep 6:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Needham BD, Carroll SM, Giles DK, Georgiou G, Whiteley M, Trent MS. 2013. Modulating the innate immune response by combinatorial engineering of endotoxin. PNAS 110:1464–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nevermann J, Silva A, Otero C, Oyarzún DP, Barrera B, et al. 2019. Identification of genes involved in biogenesis of outer membrane vesicles (OMVs) in Salmonella enterica serovar Typhi. Front. Microbiol 10:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nguyen TT, Saxena A, Beveridge TJ. 2003. Effect of surface lipopolysaccharide on the nature of membrane vesicles liberated from the Gram-negative bacterium Pseudomonas aeruginosa. J. Electron. Microsc 52:465–69 [DOI] [PubMed] [Google Scholar]
- 115.O’Donoghue EJ, Krachler AM. 2016. Mechanisms of outer membrane vesicle entry into host cells. Cell Microbiol. 18:1508–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.O’Donoghue EJ, Sirisaengtaksin N, Browning DF, Bielska E, Hadis M, et al. 2017. Lipopolysaccharide structure impacts the entry kinetics of bacterial outer membrane vesicles into host cells. PLOS Pathog. 13:e1006760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Olivier V, Haines GK, Tan Y, Satchell KJ. 2007. Hemolysin and the multifunctional autoprocessing RTX toxin are virulence factors during intestinal infection of mice with Vibrio cholerae El Tor O1 strains. Infect. Immun 75:5035–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Price NL, Goyette-Desjardins G, Nothaft H, Valguarnera E, Szymanski CM, et al. 2016. Glycoengineered outer membrane vesicles: a novel platform for bacterial vaccines. Sci. Rep 6:24931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rakoff-Nahoum S, Coyne MJ, Comstock LE. 2014. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr. Biol 24:40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rappazzo CG, Watkins HC, Guarino CM, Chau A, Lopez JL, et al. 2016. Recombinant M2e outer membrane vesicle vaccines protect against lethal influenza A challenge in BALB/c mice. Vaccine 34:1252–58 [DOI] [PubMed] [Google Scholar]
- 121.Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, et al. 2016. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun 7:10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rompikuntal PK, Thay B, Khan MK, Alanko J, Penttinen AM, et al. 2012. Perinuclear localization of internalized outer membrane vesicles carrying active cytolethal distending toxin from Aggregatibacter actinomycetemcomitans. Infect. Immun 80:31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rompikuntal PK, Vdovikova S, Duperthuy M, Johnson TL, Åhlund M, et al. 2015. Outer membrane vesicle-mediated export of processed PrtV protease from Vibrio cholerae. PLOS ONE 10:e0134098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rothfield L, Pearlman-Kothencz M. 1969. Synthesis and assembly of bacterial membrane components: a lipopolysaccharide-phospholipid-protein complex excreted by living bacteria. J. Mol. Biol 44:477–92 [DOI] [PubMed] [Google Scholar]
- 125.Round JL, Lee SM, Li J, Tran G, Jabri B, et al. 2011. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332:974–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schertzer JW, Whiteley M. 2012. A bilayer-couple model of bacterial outer membrane vesicle biogenesis. mBio 3:e00297–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schild S, Nelson EJ, Camilli A. 2008. Immunization with Vibrio cholerae outer membrane vesicles induces protective immunity in mice. Infect. Immun 76:4554–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schwechheimer C, Kuehn MJ. 2013. Synthetic effect between envelope stress and lack of outer membrane vesicle production in Escherichia coli. J. Bacteriol 195:4161–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol 13:605–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. 2012. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12:509–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sidhu VK, Vorhölter FJ, Niehaus K, Watt SA. 2008. Analysis of outer membrane vesicle associated proteins isolated from the plant pathogenic bacterium Xanthomonas campestris pv. campestris. BMC Microbiol. 8:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Simpson BW, Trent MS. 2019. Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol 17:403–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sjöström AE, Sandblad L, Uhlin BE, Wai SN. 2015. Membrane vesicle-mediated release of bacterial RNA. Sci. Rep 5:15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Solé M, Scheibner F, Hoffmeister A-K, Hartmann N, Hause G, et al. 2015. Xanthomonas campestris pv. vesicatoria secretes proteases and xylanases via the Xps type II secretion system and outer membrane vesicles. J. Bacteriol 197:2879–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Song T, Mika F, Lindmark B, Liu Z, Schild S, et al. 2008. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol. Microbiol 70:100–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Strauch KL, Johnson K, Beckwith J. 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol 171:2689–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Turner L, Bitto NJ, Steer DL, Lo C, D’Costa K, et al. 2018. Outer membrane vesicle size determines their mechanisms of host cell entry and protein content. Front. Immunol 9:1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Valentine JL, Chen L, Perregaux EC, Weyant KB, Rosenthal JA, et al. 2016. Immunization with outer membrane vesicles displaying designer glycotopes yields class-switched, glycan-specific antibodies. Cell Chem. Biol. 23:655–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Valeru SP, Shanan S, Alossimi H, Saeed A, Sandström G, Abd H. 2014. Lack of outer membrane protein A enhances the release of outer membrane vesicles and survival of Vibrio cholerae and suppresses viability of Acanthamoeba castellanii. Int. J. Microbiol 2014:610190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Valguarnera E, Scott NE, Azimzadeh P, Feldman MF. 2018. Surface exposure and packing of lipoproteins into outer membrane vesicles are coupled processes in Bacteroides. mSphere 3:e00559–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.van der Pol L, Stork M, van der Ley P. 2015. Outer membrane vesicles as platform vaccine technology. Biotechnol. J 10:1689–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Veith PD, Chen YY, Gorasia DG, Chen D, Glew MD, et al. 2014. Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J. Proteome Res 13:2420–32 [DOI] [PubMed] [Google Scholar]
- 143.Wai SN, Lindmark B, Söderblom T, Takade A, Westermark M, et al. 2003. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115:25–35 [DOI] [PubMed] [Google Scholar]
- 144.Wai SN, Takade A, Amako K. 1995. The release of outer membrane vesicles from the strains of enterotoxigenic Escherichia coli. Microbiol. Immunol 39:451–56 [DOI] [PubMed] [Google Scholar]
- 145.Wessel AK, Liew J, Kwon T, Marcotte EM, Whiteley M. 2013. Role of Pseudomonas aeruginosa peptidoglycan-associated outer membrane proteins in vesicle formation. J. Bacteriol 195:213–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yeh YC, Comolli LR, Downing KH, Shapiro L, McAdams HH. 2010. The Caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J. Bacteriol 192:4847–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zakharzhevskaya NB, Vanyushkina AA, Altukhov IA, Shavarda AL, Butenko IO, et al. 2017. Outer membrane vesicles secreted by pathogenic and nonpathogenic Bacteroides fragilis represent different metabolic activities. Sci. Rep 7:5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang Y, Fang Z, Li R, Huang X, Liu Q. 2019. Design of outer membrane vesicles as cancer vaccines: a new toolkit for cancer therapy. Cancers 11:1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhou L, Srisatjaluk R, Justus DE, Doyle RJ. 1998. On the origin of membrane vesicles in gram-negative bacteria. FEMS Microbiol. Lett 163:223–28 [DOI] [PubMed] [Google Scholar]


