Abstract
Broadly neutralizing antibodies (bNAbs) block infection by genetically diverse hepatitis C virus (HCV) isolates by targeting relatively conserved epitopes on the HCV envelope glycoproteins, E1 and E2. Many amino acid substitutions conferring resistance to these bNAbs have been characterized, identifying multiple mechanisms of bNAb escape. Some resistance substitutions follow the expected mechanism of directly disrupting targeted epitopes. Interestingly, other resistance substitutions fall in E2 domains distant from bNAb-targeted epitopes. These substitutions, which can confer broad resistance to multiple bNAbs, act by less clearly defined mechanisms. Some modulate binding of HCV to cell surface receptors, while others may induce conformational changes in the E2 protein. In this review, we discuss mechanisms of HCV bNAb resistance and implications for HCV vaccine development.
Keywords: Hepatitis C virus, broadly neutralizing antibody, antibody, escape, resistance
Introduction
One of the major challenges to the development of a successful hepatitis C virus (HCV) vaccine is the genetic diversity of the virus. There are seven well-described HCV genotypes that exhibit approximately 30% inter-genotypic amino acid variation in their envelope genes (E1 and E2). Each genotype has many subtypes that differ at approximately 15% of their E1E2 amino acids, and viral strains within the same subtype differ by up to 10% [1–3]. In addition to viral diversity at a population level, the virus’ high rate of replication with an error-prone polymerase, coupled with immune pressure, leads to rapid viral evolution within infected individuals [4–10]. Therefore, an effective vaccine must induce a broad immune response capable of neutralizing diverse HCV isolates.
Broadly neutralizing antibodies (bNAbs) target relatively conserved epitopes in the HCV envelope glycoproteins, E1 and E2. Early development of bNAbs has been associated with spontaneous clearance of HCV infection in humans [11–14], and infusion of bNAbs is protective against HCV infection in animal models [15–18]. However, some HCV strains resist even the most broadly neutralizing antibodies described to date. In this article, we will review studies of antibody selective pressure in vivo and in vitro that have been instrumental in elucidating how bNAb resistance arises and how it affects E2 function. Some resistance substitutions follow the expected mechanism of directly disrupting targeted epitopes, affecting only the bNAb targeting that epitope. Interestingly, other resistance substitutions are distant from targeted epitopes and act by less clearly defined mechanisms, either by modulating binding of HCV to cell surface receptors CD81 or scavenger receptor B1 (SR-B1), or perhaps by inducing conformational changes in the E2 protein. These substitutions can confer broad resistance to multiple bNAbs targeting distinct antigenic sites.
Antigenic sites targeted by bNAbs
Although this article is not intended as an exhaustive review of HCV bNAbs or HCV envelope protein structural biology [19–22], it is useful to consider antigenic sites targeted by bNAbs and the structure of HCV E2 when discussing mechanisms of bNAb resistance. Of the human mAbs with cross-reactivity against multiple HCV strains that have been isolated to date. the majority have binding residues falling within three overlapping antigenic sites on E2 or E1E2 [19, 21, 23, 24]. Many potent bNAbs (e.g., AR3A, HEPC3, HEPC74, HC-1 and HC84.26) target overlapping conformational epitopes spanning the front layer (amino acids 424–459, numbering based on reference strain H77) and CD81 binding loop (amino acids 519–535) of E2, at sites that have been designated antigenic Domain B, Domain D, antigenic region 3 (AR3) or ‘epitope II' [15, 17, 25–29]. In this review, these bNAbs will be referred to as ‘front layer bNAbs’. Other bNAbs such as HC33.1 and HCV1 target a conserved continuous epitope spanning amino acids 412–423 of the CD81 binding site of E2, which is referred to as Antigenic Domain E, AS412 or ‘epitope I’ [27, 30–34]. In this review, these bNAbs will be referred to as ‘AS412 bNAbs’. Binding epitopes of bNAbs AR4A, AR5A, and HEPC111/130 are poorly defined, but these mAbs recognize intact E1E2 heterodimers, with dependence on residues in the stem of E2 [17, 28, 35, 36]. In this review, these bNAbs will be referred to as ‘E1E2 bNAbs’.
Thus far, 17 structures of E2 in complex with Fabs of various monoclonal antibodies (mAbs) have been characterized [37–42]. The structure of H77 (genotype 1a) or HK6a (genotype 6a) E2 ‘core’ (E2 with various deletions and truncations) in complex with various front layer bNAbs revealed that E2 consists of a central β sandwich flanked by front (residues 421 to 459) and back layers (residues 597 to 645) [37, 40]. The structures of near full-length genotype 1a and genotype 1b E2 ectodomains complexed with bNAbs HEPC3 or HEPC74 showed a disulfide-stabilized β-turn motif at the tip of heavy-chain complementarity-determining region 3 (CDRH3) of the bNAbs that is essential for high affinity binding to the front layer of E2. These structures also revealed that the AS412 region of E2 is flexible, with different conformations in each structure [39]. The back layer, β sandwich core, and front layer structures of genotype 1a, 1b, and 6a E2 structures were very similar, indicating that the structure of E2 is largely conserved despite very extensive genetic variation. Different conformations of β sandwich loop were noted among two Fab-E2 complexes that utilize the identical strain of E2 ectodomain, suggesting that that region of E2 is flexible [39]. Several studies using electron microscopy or hydrogen—deuterium exchange have also suggested that the front layer of E2 may be conformationally flexible [38, 43–45]. A recent publication of structures of bNAbs HC1AM or 212.1.1 in complex with truncated E2 core constructs suggests that the front layer of E2 may also adopt an alternative conformation with exposure of several back-layer residues that were buried in prior structures [41].
Resistance to bNAbs targeting the front layer and CD81 binding loop of E2
A remarkable variety of substitutions have been identified that can confer resistance to front layer bNAbs (Table 1 and Figure 1). These substitutions fall either within or distant from targeted epitopes. Some substitutions abrogate bNAb binding while others appear to act at post-binding steps. Many substitutions carry significant fitness cost, while others may enhance viral entry.
Table 1.
bNAb resistance substitutions.
| bNAb target | Representative bNAbs | Resistance substitutions* | References |
|---|---|---|---|
|
| |||
| Front layer and CD81 binding loop of E2 (aa 424–459, 519535) | AR3C, CBH-2, HC84.26, HC-11, HC84.22, CBH-5, HC-2, HEPC3, HEPC74 | D431G, A439E, 438FZ, L438F/N434D, L438F/T435A | [46, 47, 50] |
| F442L/I, D431E/A | [25, 48, 50] | ||
| F442Y | [47] | ||
| S501N/V506A | [49] | ||
| I538V, Q546L, T563V | [48] | ||
| F447L, S458G, R478C | [51] | ||
| N434D, H444Y, N445H, R461L, F465Y, A466D, A475T, P498S, D533N, D610H | [50] | ||
| AS412 of E2 (aa 412–423) | HC33.1, F:V1, AP33 | N417S/T, N415K/S/D | [56–59] |
| H386R/N415D | [58] | ||
| E1E2 complex** | AR4A, AR5A | L665S/S680T, L665W/S | [60] |
| I696T/N | [61] | ||
| Multiple E2 and E1E2 sites*** | AR3A, CBH-5, HC84.22, HC33.4, AR4A | L403F, V438L | [62] |
| R424S | [63] | ||
| V400A, G401S, L403F, T404S, V414I, D431E, S453P | [65] | ||
|
| |||
Amino acid position numbering based on reference strain H77
Epitopes poorly defined
Targeted sites include front layer, AS412, and E1E2 complex
Figure 1.
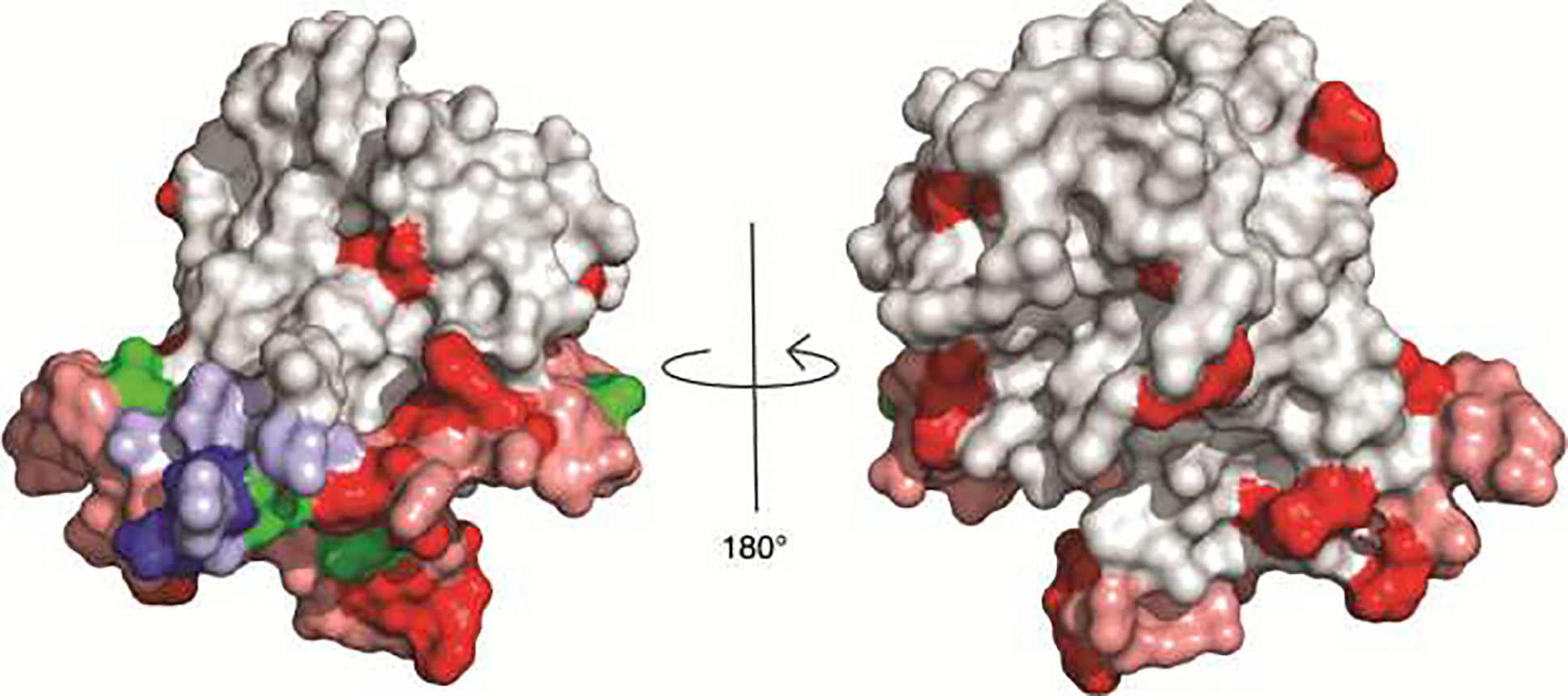
Location of resistance substitutions on the E2 ectodomain structure [PDB 6MEI] [39]. E2 is gray, with the front layer (aa 424–459) and CD81 binding loop (aa 519–535) shaded in pink and AS412 (aa 412–423) shaded in light blue. Location of substitutions conferring resistance to (i) bNAbs targeting front layer/CD81 binding loop (red), (ii) bNAbs targeting AS412 (blue), or (iii) multiple bNAbs targeting distinct antigenic sites (green) are displayed on the surface of E2. V506 (front layer bNAb resistance) is not visible because it is buried within the structure. Positions of resistance substitutions for E1E2 complex bNAbs (L665, I696) as well some substitutions conferring resistance to multiple bNAbs (V400, G401, L403, T404) are not shown because they are truncated or unresolved in the crystal structure.
In many cases, resistance to front layer bNAbs develops as a result of substitutions within targeted epitopes. Propagating JFH-1 replication competent cell culture virus (HCVcc) under increasing concentrations of front layer bNAbs CBH-2, HC-1, or HC-11 generated escape mutations at E2 front layer residues 425–443 that had varying degrees of viral fitness loss and loss of CD81 binding [46]. By culturing HVR1-deleted J6/JFH1 in the presence of AR3A, Velázquez-Moctezuma et al. identified a D431G substitution that resulted in decreased infectivity and decreased AR3A neutralization and binding of HVR1-deleted J6/JFH1. While no effect in AR3A sensitivity of J6/JFH1 was observed, this substitution conferred increased AR4A and AR5A neutralization sensitivity of J6/JFH1. With a similar approach, the authors also identified a F442Y substitution that conferred AR3A resistance to HVR1-deleted H77/JFH1 and reduced viral fitness [47]. In another study, Keck et al. compared sequences of sensitive and resistant isolates to identify D431E/A substitutions that conferred resistance to some front layer bNAbs [25]. Our group analyzed neutralization and amino acid sequences of a panel of HCV E1E2 clones isolated from individuals infected with genotype 1 HCV, also finding that D431E/A substitutions can confer resistance to front layer bNAbs, including CBH-2, AR3C, and HC84.22, without significant viral fitness cost. In the same study, we identified F442L/I substitutions conferring resistance to front layer bNAbs HC84.22 and HC84.26. Interestingly, introduction of F442I by site directed mutagenesis significantly reduced entry of HCV pseudoparticles (HCVpp) generated with this E1E2, but other HC84.26-resistant E1E2 clones with naturally-ocurring F442I substitutions had normal function, indicating that fitness cost of some bNAb resistance mutations is dependent on the E2 strain sequence [48].
Substitutions distant from targeted epitopes also appear to be a major mechanism of resistance to front layer bNAbs. A study of sequential E1E2 isolates from a chronically infected person found that one variant was resistant to neutralization by a panel of bNAbs targeting overlapping front layer epitopes. Substitutions in this variant at E1E2 residues 501 and 506 led to escape by reducing antibody binding. These changes also reduced E1E2 binding to CD81. Since these changes were distant from targeted epitopes, they presumably induced a structural change in conformational front layer epitopes that disrupted antibody and CD81 binding to E2 [49]. A study from our group also identified I538V, Q546L, and T563V substitutions in the central β-sheet of E2 that conferred broad resistance to front layer bNAbs as well as immune plasma by reducing bNAb binding to E2. These substitutions fell in a domain of E2 that was distant from bNAb binding sites, indicating that, like the substitutions identified by Keck et al., these substitutions must disrupt bNAb binding by causing conformational changes in E2 [48].
Another recent study by our group investigated mechanisms of bNAb resistance during spontaneous clearance of HCV infection. HCVpp bearing E1E2 amplified from longitudinal plasma isolates of two subjects who ultimately cleared HCV infection were progressively more resistant to neutralization and binding by autologous plasma and monoclonal autologous bNAbs HEPC3 and HEPC74. Of the resistance substitutions that arose in E2, N434D, H444Y, P498S, and D533N fell at or near HEPC3 binding residues, while L438I, F442I, and N445H fell at or near HEPC74 binding residues. In contrast, resistance substitutions R461L, F465Y, A466D, A475T, and D610H fell in hypervariable region 2 (HVR2) or the back layer of E2, distant from binding epitopes of either bNAb. These substitutions also resulted in significantly reduced E2 binding to CD81, progressive loss if viral fitness, and spontaneous clearance of infection [50].
Another case of viral escape was observed in viruses isolated during liver graft infection after transplant. Substitutions at positions 447, 458, and 478 in the front layer and HVR2 of E2 conferred resistance to antibodies in autologous pretransplant serum, as well as a panel of front layer bNAbs, including CBH-2, CBH-5, and HC-1. In contrast with the studies described above, these resistance substitutions enhanced rather than abrogated E2 binding to CD81, and they did not reduce bNAb binding to E1E2. Rather, they appeared to inhibit neutralization at a post-binding step [51].
Resistance to bNAbs targeting AS412
Another mechanism of HCV escape from bNAbs is the masking of targeted epitopes by glycosylation, which is known as glycan shielding [52–55]. Using in vitro HCVcc resistance selection studies, resistance substitutions conferring resistance to bNAbs HCV1 (human) and AP33 (rodent-derived) at asparagine 417 were identified. These substitutions caused an N-linked glycosylation shift from N417 to N415 [56]. In contrast, AS412 bNAb HC33.1 (isolated from a yeast display human antibody library) retained the ability to neutralize E2 with N415 glycan. The crystal structure of E2 region 412–423 peptide in complex with this mAb showed that HC33.1 recognizes a more extended conformation of the AS412 epitope than HCV1 or AP33, which might explain why N417 or N415 glycans do not interfere with HC33.1 binding [57]. A third study examining resistance mutants generated in vitro under the selective pressure of AS412-specific MAb24 determined that N415D, N417S and H386R/N415D substitutions resulted in resistance. Interestingly, these substitutions increased sensitivity to bNAbs targeting epitopes outside of the AS412 region, suggesting that a glycosylation shift in AS412 may have led to change in E2 structure, increasing sensitivity to neutralization by non-AS412 bNAbs [58]. Another study identified AS412 resistance substitions arising in vivo. HCV-infected patients receiving a liver transplant were treated in a clinical trial with AS412-specific bNAb HCV1. All treated patients experienced delayed viral rebound with virus with substitutions at N415 or N417 positions within the AS412 epitope [59]. Together, these studies demonstrate that glycosylation shifts within the AS412 epitope are a major mechanism of resistance to AS412 bNAbs.
Resistance to bNAbs targeting the E1E2 complex
AR4-, and AR5-specific bNAbs (‘E1E2 bNAbs’) bind epitopes on the E1E2 heterodimer surface and potently neutralize multiple HCV genotypes [17, 36]. Taking advantage of the fact that deletion of hypervariable region 1 (HVR1) renders HCV isolates more sensitive to antibody neutralization, escape mutants for AR5A were generated by passaging chimeric HVR1-deleted H77/JFH1 (genotype 1a) and J6/JFH1 (genotype 2a) HCVcc in the presence of increasing concentrations of the bNAb. For both strains, resistance substitutions at residue L665 (in the stem region of E2) caused loss of mAb binding with no decrease in viral replication competence. When these mutations were introduced into other genotypes 2–6 viruses, a reduction in AR5A sensitivity was observed, highlighting the importance of this residue in AR5A resistance across diverse viral isolates [60]. In a similar study, passage of HCVcc with bNAb AR4A induced only low level resistance substitutions in HVR1-deleted H77/JFH1 or J6/JFH1 HCVcc, suggesting that AR4A has a higher barrier to resistance than AR5A [61]. Notably, substitutions I696T or I696N in J6/JFH1 conferred AR4A resistance and loss of viral fitness in an isolate-dependent manner. Overall, substitutions in the stem region of E2 can confer resistance to E1E2-specific bNAbs, but some bNAbs in this class appear to have a high barrier to resistance.
Broad resistance to multiple bNAbs targeting distinct antigenic sites
A striking feature of HCV is the influence of substitutions distant from bNAb binding epitopes on bNAb resistance. As we have described, some of these substitutions can influence sensitivity to groups of bNAbs targeting a single antigenic site. Perhaps even more importantly, some substitutions confer resistance to multiple bNAbs targeting distinct antigenic sites. Recent studies have begun to elucidate possible mechanisms of this phenomenon, which include modulation of HCV receptor binding and temperature-dependent conformational changes of HCV envelope proteins (i.e. envelope ‘breathing’).
E1E2 variants with resistance to both HC33.4 (an AS412 bNAb) and AR4A (an E1E2 bNAb) were identified using a large panel of HCVpp generated using natural E1E2 isolates. Known critical binding residues for both bNAbs were conserved across the panel, but substitutions L403F or L438V, both distant from either bNAb binding epitope, conferred increased resistance or sensivity, respectively, to neutralization by both bNAbs. These substitutions had no effect on binding of either bNAb to E1E2, and instead increased or decreased neutralization resistance by enhancing (L403F) or reducing (L438V) E2 binding to HCV co-receptor SR-B1 [62]. In another study, polymorphism R424S conferred broad resistance to neutralization of Bole1a, a computationally derived, ancestral genotype 1a HCV strain, by both polyclonal HCV sera and diverse array of bNAbs targeting multiple antigenic sites (CBH-5, HC84.22, HC84.26, HC33.4, AR3A and AR4A). Interestingly, HCVcc carrying S424 showed lower replicative fitness than the R424 variant, suggesting that bNAb selection favors persistence of S424 variants, while greater replicative fitness favors persistence of R424 variants [63].
Prentoe et al. found that removal of glycans from E1E2 rendered HCVcc more sensitive to neutralization by AR3A (front layer bNAb), AR4A, AR5A (E1E2 bNAbs) and J6.36 (an HVR1-specific bNAb), even if the glycans did not fall near bNAb targeted epitopes. Interestingly, deletion of HVR1 rendered HCVcc neutralization sensitive regardless of whether glycans were present or deleted. Temperature-dependent neutralization experiments revealed that the removal of glycans or the absence of HVR1 appeared to destabilize E1E2, making it more globally neutralization sensitive. Removal of glycans or deletion of HVR1 also decreased SR-B1 dependency of HCVcc, leading the authors to suggest that E1E2 binding to SR-B1 may precipitate a conformational shift from a ‘closed’ to an 'open' conformation that is also precipitated by glycan or HVR1 deletion [64]. Another study by the same group used similar temperature-dependent neutralization experiments to show that polymorphisms at position 400–404 in HVR1 and 414, 431, and 453 in E2 can confer resistance to multiple bNAbs by modulating this shift between closed and open E2 conformations [65]. X-ray crystallographic studies with E2 variants possessing such polymorphisms are still needed provide a molecular basis for the existence of multiple E2 conformations.
Conclusions
HCV resistance to bNAbs is driven by substitutions within targeted epitopes, but also by amino acid changes distant from binding epitopes that likely induce conformational changes in E1E2 or alter E1E2 affinity for cell surface receptors. Some resistance substitutions (in particular substitutions in the E2 front layer region) have a significant fitness cost for the virus by decreasing the capacity of E2 to bind to CD81 and/or SR-B1, while others appear to have minimal impact on E2 function. Understanding these mechanisms of bNAb resistance is critical for HCV vaccine development. First, more work is needed to identify the bNAbs for which resistance substitutions consistently induce the greatest fitness cost to the virus, since these are the bNAbs that are most desirable for vaccine induction. Structural differences between monomeric, soluble E2 (sE2), full-length E2 in complex with E1, and E1E2 on replication competent virus or virus-like particles should be carefully considered in structure—function analyses, since antibody resistance and receptor binding effects of some substitutions may not be fully recapitulated by sE2 [65, 66]. Second, inclusion of both bNAb sensitive and resistant antigens in a vaccine might broaden the vaccine-induced antibody response. Third, multiple studies have shown that some combinations of bNAbs are synergistic, so a vaccine may need to simultaneously induce bNAbs targeting multiple distinct epitopes in order to limit immune escape [67–69]. Finally, more work is needed to understand the different conformational states that may be tolerated by functional E1E2 in vivo, so that bNAbs targeting these different states can be identified, and potentially antigens mimicking these states could be stabilized and included in a vaccine. Structural studies with E2 and E1E2 sensitive and resistant variants in complex with bNAbs will further aid in immunogen stabilization efforts. Overall, efforts to date have greatly advanced our understanding of HCV bNAb resistance, but very important work still remains.
Acknowledgements
This work was supported in part by by the National Institutes of Health grant R01 AI127469 (to JRB) and U.S. NIH grant K99 AI153465 (to AIF).
Footnotes
Declarations of Interests
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Smith DB, et al. , Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology, 2014. 59(1): p. 318–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bukh J, The history of hepatitis C virus (HCV): Basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control. J Hepatol, 2016. 65(1 Suppl): p. S2–S21. [DOI] [PubMed] [Google Scholar]
- [3].Smith DB, et al. , Proposed revision to the taxonomy of the genus Pestivirus, family Flaviviridae. J Gen Virol, 2017. 98(8): p. 2106–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Martell M, et al. , Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. Journal of Virology, 1992. 66: p. 3225–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Farci P, Bukh J, and Purcell RH, The quasispecies of hepatitis C virus and the host immune response. Springer Semin.Immunopathol, 1997. 19: p. 5–26. [DOI] [PubMed] [Google Scholar]
- [6].Forns X, Purcell RH, and Bukh J, Quasispecies in viral persistence and pathogenesis of hepatitis C virus. Trends Microbiol, 1999. 7(10): p. 402–10. [DOI] [PubMed] [Google Scholar]
- [7].Liu L, et al. , Acceleration of hepatitis C virus envelope evolution in humans is consistent with progressive humoral immune selection during the transition from acute to chronic infection. J Virol, 2010. 84: p. 5067–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cox AL, et al. , Cellular immune selection with hepatitis C virus persistence in humans. J Exp.Med, 2005. 201: p. 1741–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Erickson AL, et al. , Hepatitis C virus-specific CTL responses in the liver of chimpanzees with acute and chronic hepatitis C. J.Immunol, 1993. 151: p. 4189–4199. [PubMed] [Google Scholar]
- [10].Timm J, et al. , CD8 epitope escape and reversion in acute HCV infection. J.Exp.Med, 2004. 200: p. 1593–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Logvinoff C, et al. , Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc.Natl.Acad.Sci.U.S.A, 2004. 101: p. 10149–10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pestka JM, et al. , Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc.Natl.Acad.Sci.U.S.A, 2007. 104: p. 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bailey JR, et al. , Constraints on viral evolution during chronic hepatitis C virus infection arising from a common-source exposure. J Virol, 2012. 86(23): p. 12582–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Osburn WO, et al. , Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology, 2014. 59(6): p. 2140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Law M, et al. , Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat.Med, 2008. 14: p. 25–27. [DOI] [PubMed] [Google Scholar]
- [16].Keck ZY, et al. , Affinity maturation of a broadly neutralizing human monoclonal antibody that prevents acute hepatitis C virus infection in mice. Hepatology, 2016. 64(6): p. 1922–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Giang E, et al. , Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc.Natl.Acad.Sci.U.S.A, 2012. 109: p. 6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Morin TJ, et al. , Human Monoclonal Antibody HCV1 Effectively Prevents and Treats HCV Infection in Chimpanzees. PLoS.Pathog., 2012. 8: p. e1002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Freedman H, et al. , Structure and Function of the Hepatitis C Virus Envelope Glycoproteins E1 and E2: Antiviral and Vaccine Targets. ACS Infect Dis, 2016. 2(11): p. 749–762. [DOI] [PubMed] [Google Scholar]
- [20].Fuerst TR, et al. , Designing a B Cell-Based Vaccine against a Highly Variable Hepatitis C Virus. Front Microbiol, 2017. 8: p. 2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kinchen VJ, Cox AL, and Bailey JR, Can Broadly Neutralizing Monoclonal Antibodies Lead to a Hepatitis C Virus Vaccine? Trends Microbiol, 2018. 26(10): p. 854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bailey JR, Barnes E, and Cox AL, Approaches, Progress, and Challenges to Hepatitis C Vaccine Development. Gastroenterology, 2019. 156(2): p. 418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kong L, et al. , Capitalizing on knowledge of hepatitis C virus neutralizing epitopes for rational vaccine design. Curr Opin Virol, 2015. 11: p. 148–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Keck ML, et al. , Mapping Determinants of Virus Neutralization and Viral Escape for Rational Design of a Hepatitis C Virus Vaccine. Front Immunol, 2018. 9: p. 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Keck ZY, et al. , A point mutation leading to hepatitis C virus escape from neutralization by a monoclonal antibody to a conserved conformational epitope. J.Virol, 2008. 82: p. 6067–6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Keck ZY, et al. , Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV e2 with resistance to neutralization escape in a genotype 2a isolate. PLoS.Pathog., 2012. 8: p. e1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pierce BG, et al. , Global mapping of antibody recognition of the hepatitis C virus E2 glycoprotein: Implications for vaccine design. Proc Natl Acad Sci U S A, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gopal R, et al. , Probing the antigenicity of hepatitis C virus envelope glycoprotein complex by high-throughput mutagenesis. PLoS Pathog, 2017. 13(12): p. e1006735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bailey JR, et al. , Broadly neutralizing antibodies with few somatic mutations and hepatitis C virus clearance. JCI Insight, 2017. 2(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Flint M, et al. , Functional analysis of cell surface-expressed hepatitis C virus E2 glycoprotein. J Virol, 1999. 73(8): p. 6782–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tarr AW, et al. , Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology, 2006. 43: p. 592–601. [DOI] [PubMed] [Google Scholar]
- [32].Broering TJ, et al. , Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J.Virol, 2009. 83: p. 12473–12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sabo MC, et al. , Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step. J.Virol, 2011. 85: p. 7005–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Keck Z, et al. , Cooperativity in virus neutralization by human monoclonal antibodies to two adjacent regions located at the amino terminus of hepatitis C virus E2 glycoprotein. J.Virol, 2013. 87: p. 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].de Jong YP, et al. , Broadly neutralizing antibodies abrogate established hepatitis C virus infection. Sci Transl Med, 2014. 6(254): p. 254ra129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Colbert MD, et al. , Broadly Neutralizing Antibodies Targeting New Sites of Vulnerability in Hepatitis C Virus E1E2. J Virol, 2019. 93(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kong L, et al. , Hepatitis C virus E2 envelope glycoprotein core structure. Science, 2013. 342(6162): p. 1090–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Khan AG, et al. , Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature, 2014. 509(7500): p. 381–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Flyak AI, et al. , HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design. Cell Host Microbe, 2018. 24(5): p. 703–716 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tzarum N, et al. , Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1–69 antibodies. Sci Adv, 2019. 5(1): p. eaav1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tzarum N, et al. , An alternate conformation of HCV E2 neutralizing face as an additional vaccine target. Sci Adv, 2020. 6(30): p. eabb5642. * nAbs encoded by the VH1–69 germline were used to identify a new conformation of E2 susceptible to neutralization, referred to as ‘B conformation’. This conformation involves the exposure of back-layer residues Y613 and W616 for direct antibody interaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Flyak AI, et al. , An ultralong CDRH2 in HCV neutralizing antibody demonstrates structural plasticity of antibodies against E2 glycoprotein. Elife, 2020. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kong L, et al. , Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc.Natl.Acad.Sci.U.S.A, 2012. 109: p. 9499–9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Meola A, et al. , Structural flexibility of a conserved antigenic region in hepatitis C virus glycoprotein E2 recognized by broadly neutralizing antibodies. J Virol, 2015. 89(4): p. 2170–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kong L, et al. , Structural flexibility at a major conserved antibody target on hepatitis C virus E2 antigen. Proc Natl Acad Sci U S A, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Keck ZY, et al. , Mapping a region of hepatitis C virus E2 that is responsible for escape from neutralizing antibodies and a core CD81-binding region that does not tolerate neutralization escape mutations. J.Virol, 2011. 85: p. 10451–10463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Velázquez-Moctezuma R, et al. , Hepatitis C Virus Escape Studies of Human Antibody AR3A Reveal a High Barrier to Resistance and Novel Insights on Viral Antibody Evasion Mechanisms. J Virol, 2019. 93(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Bailey JR, et al. , Naturally selected hepatitis C virus polymorphisms confer broad neutralizing antibody resistance. J Clin Invest, 2015. 125(1): p. 437–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Keck ZY, et al. , Mutations in hepatitis C virus E2 located outside the CD81 binding sites lead to escape from broadly neutralizing antibodies but compromise virus infectivity. J.Virol, 2009. 83: p. 6149–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kinchen VJ, et al. , Broadly Neutralizing Antibody Mediated Clearance of Human Hepatitis C Virus Infection. Cell Host Microbe, 2018. 24(5): p. 717–730 e5. ** Using E1E2 sequences amplified from longitudinal plasma isolates of two subjects who clear HCV infection, the authors found that autologous plasma IgG and bNAbs were able to neutralize autologous strains. Naturally selected substitutions in E2 were found to confer partial resistance and sometimes complete escape from autologous bNAbs and also caused a progressive loss of fitness of the longitudinal natural E1E2 strains by a reduction in entry and in binding affinity for CD81 and SR-B1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Fofana I, et al. , Mutations that alter use of hepatitis C virus cell entry factors mediate escape from neutralizing antibodies. Gastroenterology, 2012. 143: p. 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Helle F, et al. , The neutralizing activity of anti-hepatitis C virus antibodies is modulated by specific glycans on the E2 envelope protein. J.Virol, 2007. 81: p. 8101–8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Goffard A, et al. , Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J Virol, 2005. 79(13): p. 8400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Falkowska E, et al. , Hepatitis C virus envelope glycoprotein E2 glycans modulate entry, CD81 binding, and neutralization. J.Virol, 2007. 81: p. 8072–8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Helle F, Duverlie G, and Dubuisson J, The hepatitis C virus glycan shield and evasion of the humoral immune response. Viruses., 2011. 3: p. 1909–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pantua H, et al. , Glycan shifting on hepatitis C virus (HCV) E2 glycoprotein is a mechanism for escape from broadly neutralizing antibodies. J Mol Biol, 2013. 425(11): p. 1899–1914. [DOI] [PubMed] [Google Scholar]
- [57].Li Y, et al. , Structural basis for penetration of the glycan shield of hepatitis C virus E2 glycoprotein by a broadly neutralizing human antibody. J Biol Chem, 2015. 290(16): p. 10117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gu J, et al. , Escape of Hepatitis C Virus from Epitope I Neutralization Increases Sensitivity of Other Neutralization Epitopes. J Virol, 2018. 92(9). * This study determined that resistance mutations to bNAbs targeting the 412-to-423 region increased the sensitivity to neutralization by bNAbs targeting epitopes outside this region, indicating an effective vaccine should elicit bNAbs directed toward the 412-to-423 region of E2 as well as additional epitopes of E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chung RT, et al. , Human monoclonal antibody MBL-HCV1 delays HCV viral rebound following liver transplantation: a randomized controlled study. Am J Transplant, 2013. 13(4): p. 1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Velázquez-Moctezuma R, et al. , Applying antibody-sensitive hypervariable region 1-deleted hepatitis C virus to the study of escape pathways of neutralizing human monoclonal antibody AR5A. PLoS Pathog, 2017. 13(2): p. e1006214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Velázquez-Moctezuma R, et al. , Hepatitis C Virus-Escape Studies for Human Monoclonal Antibody AR4A Reveal Isolate-Specific Resistance and a High Barrier to Resistance. J Infect Dis, 2019. 219(1): p. 68–79. * Taking advantage of the fact that deletion of HVR1 renders HCV isolates more sensitive to antibody neutralization, the authors generated escape mutants for AR4A in H77/JFH1 and J6/JFH1 HCVcc. Only low level resistance substitutions were induced in J6/JFH1, suggesting that AR4A has a high barrier to resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].El-Diwany R, et al. , Extra-epitopic hepatitis C virus polymorphisms confer resistance to broadly neutralizing antibodies by modulating binding to scavenger receptor B1. PLoS Pathog, 2017. 13(2): p. e1006235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wasilewski LN, et al. , A Hepatitis C Virus Envelope Polymorphism Confers Resistance to Neutralization by Polyclonal Sera and Broadly Neutralizing Monoclonal Antibodies. J Virol, 2016. 90(7): p. 3773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Prentoe J, et al. , Hypervariable region 1 and N-linked glycans of hepatitis C regulate virion neutralization by modulating envelope conformations. Proc Natl Acad Sci U S A, 2019. 116(20): p. 10039–10047. ** With temperature-dependent neutralization assays the authors determine that removal of glycans and HVR1 destabilized a closed, difficult to neutralize, envelope conformation. They suggest that SR-B1 may play a role in transitions from closed to open conformations during entry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Augestad EH, et al. , Global and local envelope protein dynamics of hepatitis C virus determine broad antibody sensitivity. Sci Adv, 2020. 6(35): p. eabb5938. ** Through temperature-dependent neutralization experiments they determined that HVRI and AS412 have a role in regulating the E1E2 conformational state involving a theoretical neutralization-sensitive “open” state of E1E2, favored at high temperatures, and a neutralization-resistant “closed” state of E1E2, favored at low temperatures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Owsianka A, et al. , Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J Gen Virol, 2001. 82(Pt 8): p. 1877–1883. [DOI] [PubMed] [Google Scholar]
- [67].Carlsen TH, et al. , Breadth of neutralization and synergy of clinically relevant human monoclonal antibodies against HCV genotypes 1a, 1b, 2a, 2b, 2c, and 3a. Hepatology, 2014. 60(5): p. 1551–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Mankowski MC, et al. , Synergistic anti-HCV broadly neutralizing human monoclonal antibodies with independent mechanisms. Proc Natl Acad Sci U S A, 2018. 115(1): p. E82–E91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kinchen VJ, et al. , Plasma deconvolution identifies broadly neutralizing antibodies associated with hepatitis C virus clearance. J Clin Invest, 2019. 130. [DOI] [PMC free article] [PubMed] [Google Scholar]


