Abstract
Background
ARDS is a devastating syndrome with heterogeneous subtypes, but few causal biomarkers have been identified.
Research Question
Would multistage Mendelian randomization identify new causal protein biomarkers for ARDS 28-day mortality?
Study Design and Methods
Three hundred moderate to severe ARDS patients were selected randomly from the Molecular Epidemiology of ARDS cohort for proteomics analysis. Orthogonal projections to latent structures discriminant analysis was applied to detect the association between proteins and ARDS 28-day mortality. Candidate proteins were analyzed using generalized summary data-based Mendelian randomization (GSMR). Protein quantitative trait summary statistics were retrieved from the Efficiency and safety of varying the frequency of whole blood donation (INTERVAL) study (n = 2,504), and a genome-wide association study for ARDS was conducted from the Identification of SNPs Predisposing to Altered Acute Lung Injury Risk (iSPAAR) consortium study (n = 534). Causal mediation analysis detected the role of platelet count in mediating the effect of protein on ARDS prognosis.
Results
Plasma insulin-like growth factor binding protein 7 (IGFBP7) moderately increased ARDS 28-day mortality (OR, 1.11; 95% CI, 1.04-1.19; P = .002) per log2 increase. GSMR analysis coupled with four other Mendelian randomization methods revealed IGFBP7 as a causal biomarker for ARDS 28-day mortality (OR, 2.61; 95% CI, 1.33-5.13; P = .005). Causal mediation analysis indicated that the association between IGFBP7 and ARDS 28-day mortality is mediated by platelet count (OR, 1.03; 95% CI, 1.02-1.04; P = .01).
Interpretation
We identified plasma IGFBP7 as a novel causal protein involved in the pathogenesis of ARDS 28-day mortality and platelet function in ARDS, a topic for further experimental and clinical investigation.
Key Words: acute respiratory distress syndrome, biomarkers, insulin-like growth factor binding protein 7, mediation analysis, Mendelian randomization analysis
Abbreviations: ANG-2, angiopoietin 2; ARDSNet, ARDS Clinical Trials Network; AUC, area under the receiver operating characteristic curve; GSMR, generalized summary data-based Mendelian randomization; GWAS, genome-wide association study; IGFBP7, insulin-like growth factor binding protein 7; INTERVAL, Efficiency and safety of varying the frequency of whole blood donation; IV, instrumental variable; MAF, minor allele frequency; MEARDS, Molecular Epidemiology of ARDS; NB, net benefit; SNP, single nucleotide polymorphism; sRAGE, soluble receptor for advanced glycation end-products; t-PA, tissue-type plasminogen activator; vWF, von Willebrand factor; WPB, Weibel-Palade body
Take-home Points.
Study Question: Would multistage Mendelian randomization identify new causal protein biomarkers for ARDS 28-day mortality?
Results: Plasma IGFBP7 moderately increased ARDS 28-day mortality, and the association between IGFBP7 and ARDS 28-day mortality is mediated by platelet count.
Interpretation: Plasma IGFBP7 may involve in the pathogenesis of ARDS 28-day mortality and platelet function in ARDS, a topic for further experimental and clinical investigation.
ARDS is characterized by acute, diffuse inflammatory lung injury and affects nearly 200,000 patients annually in the United States.1,2 The mortality rate ranges from 35% to 46% despite decades of research into effective treatments.3 A promising pharmacotherapy for ARDS is neuromuscular blockade, but the efficacy of this approach has been brought into question. The latest clinical trial shows that in a cohort of moderate to severe ARDS patients, the addition of early continuous neuromuscular blockade with concomitant deep sedation did not result in lower mortality than a usual-care approach to mechanical ventilation.4 Because ARDS has heterogeneous lung pathophysiologic characteristics, identifying clinical and biological features to discriminate ARDS patients into subphenotypes may be helpful to categorize those who might be more responsive to different therapies.5
Proteomics technology applied to investigate the pathogenesis and progression of ARDS resulted in the identification of many plasma protein biomarkers. Plasma tumor necrosis factor-α, IL-1β, IL-6, and IL-8 levels were reported as significantly higher in patients dying of ARDS.6 Similarly, soluble receptor for advanced glycation end-products (sRAGE) in plasma independently is associated with death in ARDS patients, mainly because of its critical role in inflammatory reactions.7 Other evidence supports that a high level of angiopoietin 2 (ANG-2) leads to increased microvascular permeability in critically ill patients with ARDS and is associated with worse outcome.8 Although these studies provide promise, insufficient causal validity means biomarkers seldom are incorporated into clinical practice for ARDS treatment. Thus, although many potential biomarkers have been identified by observational studies,9 a gap in understanding their clinical applicability exists. Theoretically, causal biomarkers independent of confounding factors have more potential in pharmacologic responses to a therapeutic intervention.
A potential way to identify causal biomarkers independent of confounders is through Mendelian randomization. Mendelian randomization studies often are described as naturally occurring randomized controlled trials in which genetic factors are assigned randomly by nature,10 with this random assortment of genetic variants used to infer causal relationships between exposures and outcomes.11 In the context of ARDS, Mendelian randomization analysis linked two causal protein biomarkers (ANG-2 and sRAGE) with development of sepsis-associated ARDS.12,13 We hypothesized that additional plasma proteins can serve as causal biomarkers for ARDS. To identify markers that ultimately may have clinical usefulness, we used generalized summary data-based Mendelian randomization (GSMR) with protein quantitative trait loci data from the Efficiency and safety of varying the frequency of whole blood donation (INTERVAL) study and genome-wide association study (GWAS) data from the Identification of SNPs Predisposing to Altered Acute Lung Injury Risk (iSPAAR) consortium study to infer the causal association between protein levels and ARDS 28-day mortality.14,15
Methods
Study Populations and Plasma Samples
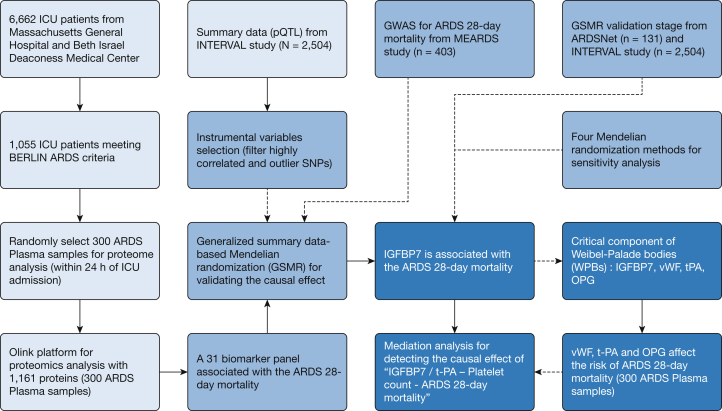
Study design is illustrated in Figure 1. ARDS patients were enrolled at the ICUs of Massachusetts General Hospital and Beth Israel Deaconess Medical Center as part of the Molecular Epidemiology of ARDS (MEARDS) cohort between 1998 and 2014. Detailed information on this cohort was published previously.16,17 Patients were eligible for the study if they did not have any of the following: age younger than 18 years, HIV infection, diffuse alveolar hemorrhage, chronic lung diseases other than COPD or asthma, directive to withhold intubation, immunosuppression not secondary to corticosteroid, treatment with granulocyte colony-stimulating factor, cytotoxic therapy, or solid organ or bone marrow transplant. We collected demographics, medical history, vital signs, hematologic characteristics, and biochemical indicators and performed frequent arterial blood gas analysis and chest radiography within 24 h of admission. All plasma samples also were collected within the 24 h of ICU admission, which were used for proteomics profiling. Presence of ARDS was adjudicated in accordance with Berlin criteria requiring that chest radiograph and oxygenation criteria be met on the same calendar day while invasively ventilated.18 Detailed biospecimen collection information was described previously,19 and 300 plasma samples (obtained from the MEARDS cohort) were quantified precisely by the Olink platform, including 13 panels (1,161 proteins) followed by quality control.20 The quality control process was sample control and variable control.21,22 All proteins were log2 transformed to obey normal distribution, and values of less than the detection limit of the assay were replaced by the lower limit of detection. Samples were removed if the control sample deviated more than ± 0.3 from the median value of all samples on the run. The batch effect in Olink data was adjusted by sva package from R version 3.6.1 software (R Foundation for Statistical Computing).23
Figure 1.
Study design flowchart. ARDSNet = ARDS Clinical Trials Network; GSMR = generalized summary data-based Mendelian randomization; GWAS = genome-wide association study; IGFBP7 = insulin-like growth factor binding protein 7; INTERVAL = Efficiency and safety of varying the frequency of whole blood donation; MEARDS = Molecular Epidemiology of ARDS; OPG = osteoprotegerin; OPLS-DA = orthogonal projections to latent structures discriminant analysis; pQTL = protein quantitative trait loci; SNP = single nucleotide polymorphism; t-PA = tissue-type plasminogen activator; vWF = von Willebrand factor; WPB = Weibel-Palade body.
GWAS Data Source and Quality Control
GWAS data for ARDS were obtained from the Identification of SNPs Predisposing to Altered Acute Lung Injury Risk (iSPAAR) consortium study. iSPAAR samples were from the MEARDS study enrolled at the Massachusetts General Hospital and Beth Israel Deaconess Medical Center and from the ARDS Clinical Trials Network (ARDSNet). In the current study, we selected ARDS cases with complete clinical information, including: status at 28 days, age, sex, ethnicity, BMI, Acute Physiology and Chronic Health Evaluation III score, vasopressors within 24 h, prednisone within 21 days, smoking status, alcohol abuse, and predisposing conditions such as sepsis, pneumonia, multiple trauma, or aspiration. Clinical variables were adjusted in GWAS multilogistic regression, Mendelian randomization, and causal mediation analyses. GWAS data and quality control procedures are described in e-Appendix 1.
Summary statistics of protein quantitative trait loci were obtained from the INTERVAL study, which analyzed 2,994 proteins via the SOMAScan assay from 3,301 European people.15 Selection of proteins on the platform reflects both the availability of purified protein targets and a focus on proteins suspected to be involved in the pathophysiologic characteristics of human disease.
Mendelian Randomization Analysis
We conducted Mendelian randomization analysis to infer the causal effect of genetically predicted proteins on ARDS 28-day mortality using GSMR. All single nucleotide polymorphisms (SNPs) significantly associated with each protein (P < 5 × 10-8) were selected as candidates for genetic instrumental variables (IVs). The effects of those SNPs on ARDS 28-day mortality risk were calculated separately from MEARDS and ARDSNet. Summarized results of the two parts were integrated separately with the INTERVAL study to evaluate the causal effect of the proteins on ARDS 28-day mortality. High linkage disequilibrium (r2 > 0.1) SNPs were removed in the IVs. Then, pleiotropic SNPs were removed by Heterogeneity in dependent instrument (HEIDI)-outlier detection methods with a P < .05 / (number of IVs).14 Using all markers on the genotyping platform, we performed principal component analysis to identify 10 principal components allowing for adjustment of genetic population stratification. Four Mendelian randomization methods—sample median, penalized weighted median, inverse-variance weighted, and robust inverse-variance weighted—were used to perform sensitivity analysis.24, 25, 26, 27 The statistical power of Mendelian randomization in this study was calculated by mRnd, an online tool calculating the statistical power given the chosen sample size, type I error (α value), causal effect of protein on ARDS 28-day mortality, and the proportion of death within 28 days.28
Statistical analysis is described in e-Appendix 1.
Results
Baseline characteristics of patients are described in Table 1. Ninety-nine ARDS patients died within 28 days (33.00%), which is similar to the mortality reported in a previous systematic review (33.53%).29 Briefly, patients of older age, with low BMI, high Acute Physiology and Chronic Health Evaluation III score, low platelet count, low Pao2/Fio2, high creatinine, high bilirubin, sepsis, or bacteremia had higher 28-day mortality.
Table 1.
Description of ARDS Patient Characteristics
| Baseline Characteristics | Survival Within 28 Days (n = 201) | Nonsurvival Within 28 Days (n = 99) | All Patients (N = 300) | P Value |
|---|---|---|---|---|
| Age, y | 53.71 ± 18.77 | 68.27 ± 15.50 | 58.51 ± 19.01 | 1.33 × 10-11 |
| Sex | ||||
| Female | 78 (38.81) | 36 (36.36) | 114 (38.00) | .77 |
| Ethnicity | ||||
| White | 190 (94.53) | 95 (95.96) | 285 (95.00) | .69 |
| BMI, kg/m2 | 29.18 ± 8.78 | 26.76 ± 7.58 | 28.39 ± 8.47 | .01 |
| APACHE III score | 71.71 ± 21.02 | 92.26 ± 20.46 | 78.49 ± 22.94 | 5.10 × 10-14 |
| Vasopressin within 24 h | ||||
| Yes | 137 (67.18) | 73 (73.74) | 210 | .39 |
| Prednisone within 21 d | ||||
| Yes | 20 (9.95) | 15 (15.15) | 35 | .25 |
| Smoking status | ||||
| Current | 68 (33.83) | 21 (21.21) | 89 (29.67) | .06 |
| Former | 44 (21.89) | 36 (36.36) | 80 (26.67) | |
| Alcohol abuse | ||||
| Yes | 25 (12.44) | 16 (16.16) | 41 (13.67) | .48 |
| ARDS risk factors | ||||
| Sepsis | 172 (85.57) | 94 (94.95) | 266 (88.67) | .03 |
| Septic shock | 126 (62.69) | 74 (74.75) | 200 (66.67) | .05 |
| Pneumonia | 81 (40.30) | 38 (38.38) | 119 (39.67) | .84 |
| Bacteremia | 25 (12.44) | 27 (27.27) | 52 (17.33) | .002 |
| Multiple fractures | 13 (6.47) | 1 (1.01) | 14 (4.67) | .07 |
| Aspiration | 17 (8.46) | 11 (11.11) | 28 (9.33) | .59 |
| Pulmonary contusion | 10 (4.98) | 2 (2.02) | 12 (4.00) | .36 |
| Platelet count, 109/L | 228.56 ± 147.22 | 167.26 ± 123.43 | 226.01 ± 208.36 | .006 |
| Pao2/Fio2 | 109.04 ± 42.44 | 98.08 ± 37.83 | 105.43 ± 41.24 | .02 |
| Creatinine, mg/dL | 1.53 ± 1.23 | 1.95 ± 1.33 | 1.67 ± 1.28 | .01 |
| Total bilirubin, mg/dL | 0.90 (0.20-9.07) | 1.55 (0.20-21.11) | 1.00 (0.20-16.25) | .004 |
| Ventilator-free days | 24 (15.35-27.83) | 22 (2.48-27.53) | 19 (0.00-27.00) | < 2.20 × 10-16 |
Data are presented at No. (%), mean ± SD, or median (95% CI), unless otherwise indicated. Bold represents P value < .05. APACHE = Acute Physiology and Chronic Health Evaluation.
Heterogeneous Proteins in Mortality and Survivor Groups
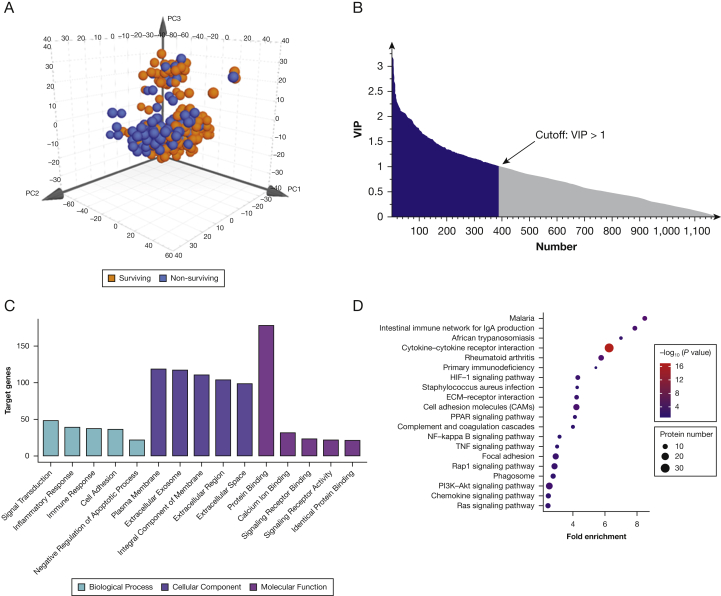
Overall, plasma protein levels differentiated between patients who died and those who survived, with R2Y at 0.60 and Q2 at 0.50 (Fig 2A). Three hundred ninety-three proteins with variable influence on projection of more than 1.00 were considered potential biomarkers (Fig 2B).30 The function of potential biomarkers was identified by gene ontology annotation and Kyoto Encyclopedia of Genes and Genomes pathway analysis. Gene ontology analysis categorized potential biomarkers into several essential biological processes, including inflammatory response, immune response, and cell adhesion, which are all critical processes in ARDS (Fig 2C). Kyoto Encyclopedia of Genes and Genomes analysis associated those 393 proteins mainly with 20 signaling pathways (Fig 2D). Two hundred thirty-seven of 393 potential biomarkers were identified as associating with ARDS mortality by single logistic regression without adjustment for covariates (e-Fig 1A, 1C, e-Table 1), and 31 of 237 were confirmed by multilogistic regression analysis with adjustment for critical covariates (e-Fig 1B, 1D, e-Table 2). Statistically significant protein biomarkers separately demonstrated a considerable discrimination ability, with areas under the receiver operating characteristic curve (AUCs) ranging from 0.64 to 0.74 (e-Fig 2). Predictive ability, evaluated by AUC, for the clinical variables (critical covariates) combination was 0.81 and increased to 0.92 with the contribution of 31 proteins (e-Fig 3A). Protein-protein interaction analysis showed that the 31 proteins participated in platelet-related, extracellular matrix-related, and proteinaceous extracellular matrix-related pathways critical in ARDS development and prognosis (e-Fig 3B).
Figure 2.
Orthogonal projection to latent structure-discriminant analysis (OPLS-DA) and protein pathway analysis. A, OPLS-DA score plots comparing deaths to survival within 28 days. Plasma protein levels differentiated between patients who died and those who survived, with R2Y at 0.60 and Q2 at 0.50. B, Feature selection results based on variable importance in projection (VIP). Three hundred ninety-three proteins with VIP of more than 1.00 were considered potential biomarkers. C, Gene ontology analysis for potential biomarkers. D, Kyoto Encyclopedia of Genes and Genomes analysis for potential biomarkers. ECM = extracellular matrix; HIF = hypoxia-inducible factor; NF = nuclear factor; PI3K-Akt = phosphatidylinositol 3-kinase/protein kinase B; PPAR = peroxisome proliferator-activated receptors; TNF = tumor necrosis factor.
Mendelian Randomization Analysis to Infer the Causal Effect of Proteins on ARDS 28-Day Outcome
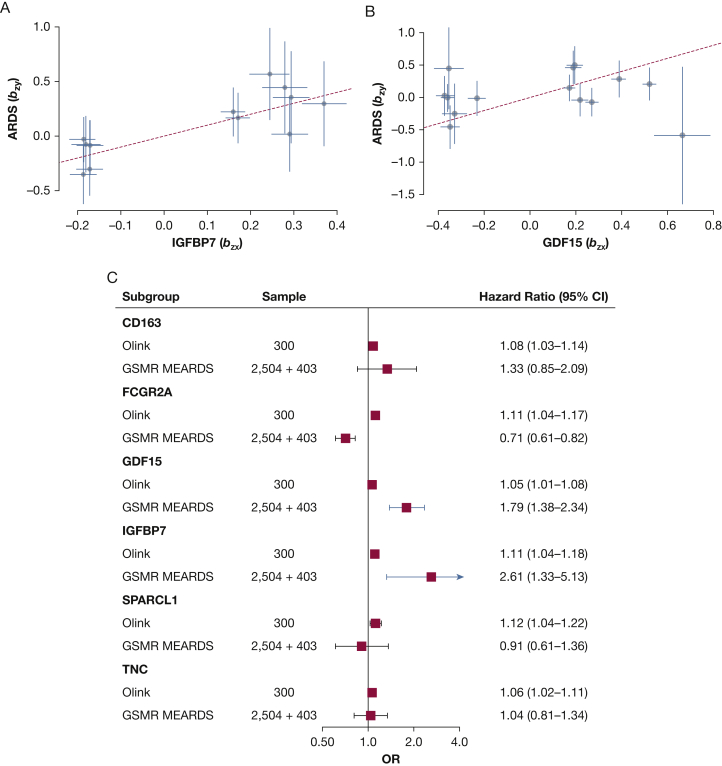
In total, 12 of the 31 proteins (CD163, FCGR2A, GDF15, IGFBP7, INHBC, MET, PAPPER, PROC, SIGLEC10, SPARCL1, VCAM, and TNC) were identified from the INTERVAL study, and the candidate IVs are described in e-Tables 3-14. Mendelian randomization analysis in MEARDS showed that two genetically predicted proteins (predicted by SNPs without confounder interference) significantly increased the risk for ARDS 28-day mortality: IGFBP7 (OR, 2.61; 95% CI, 1.33-5.13; P = .005) and GDF15 (OR, 1.80; 95% CI, 1.38-2.34; P = 1.61 × 10-5) (Fig 3 In contrast, one genetically predicted protein decreased the risk of ARDS 28-day mortality: FCGR2A (OR, 0.71; 95% CI, 0.61-0.82; P = 6.07 × 10-6). However, its effect on ARDS 28-day mortality was opposite to the proteomics analysis (Fig 3C), so FCGR2A was excluded from further analysis. Selected IVs, used in GSMR, are detailed in e-Tables 15 and 16 according to studies. Mendelian randomization analysis was applied in an independent ARDS GWAS data set from ARDSNet to replicate the results from MEARDS; analysis revealed that genetically predicted IGFBP7 significantly elevated the risk of ARDS 28-day mortality (OR, 7.61; 95% CI, 2.01-28.86; P = .003) (e-Fig 4). The statistical power of IGFBP7 detection in this study was calculated as 0.81 (sample sizeMEARDS = 403; α = 0.05; KProportion of 28-day death = 0.3; ORMendelian randomization = 2.61). However, genetically predicted GDF15 in ARDSNet exhibited an opposite effect with the Mendelian randomization results in MEARDS, although the effect was significant (OR, 0.32; 95% CI, 0.19-0.53; P = 1.02 × 10-5). Further, the four summary data-based Mendelian randomization methods used in sensitivity analysis support the adverse effect of IGFBP7 on ARDS 28-day mortality (e-Fig 5). Because most ARDS cases originate from sepsis, we also conducted a subgroup analysis restricted to sepsis patients. Consistently, multivariate logistic regression showed that IGFBP7 was associated with ARDS 28-day mortality (OR, 2.32; 95% CI, 1.05-1.19; P = .002) in plasma analysis and was associated causally with ARDS 28-day mortality (ORMEARDS = 2.23; 95% CI, 1.10-4.51; P = .02; ORARDSNet = 6.69; 95% CI, 2.06-21.67; P = .002) in GSMR analysis.
Figure 3.
Generalized summary data-based Mendelian randomization (GSMR) analysis for ARDS 28-day mortality. Relationship between the effect size estimates on proteins (x-axis) and the effect size estimates on ARDS 28-day mortality (y-axis) for all single nucleotide polymorphisms (SNPs) that served as instrumental variables. The 95% CIs for the estimated SNP effect sizes on proteins are shown as vertical orange lines, whereas the 95% CIs for the estimated SNP effect sizes on ARDS 28-day mortality are shown as horizontal orange lines. A, Insulin-like growth factor binding protein 7 (IGFBP7). B, Growth differentiation factor 15 (GDF15). C, Plasma proteins and GSMR analysis from the Molecular Epidemiology of ARDS (MEARDS) studies.
Causal Mediation Analysis for IGFBP7 on ARDS openbrace28-Day Outcome
Many indices at ICU admission reflect the severity of ARDS, including platelet count, Pao2/Fio2, bilirubin, and creatinine. Platelet count is believed to contribute to ARDS pathogenesis and prognosis through platelet involvement in the inflammatory response and disseminated intravascular coagulation.31,32 We previously showed that early-stage thrombocytopenia in critically ill patients is associated with the development of mortality in ARDS.33 Previous studies also examined the relationship between ARDS mortality and markers such as Pao2/Fio2, bilirubin, and creatinine.34, 35, 36 We therefore conducted a causal mediation analysis to identify the mechanism by which protein biomarkers relate to ARDS 28-day mortality via explanatory variables or mediators (eg, platelet count).37,38
IGFBP7 is a critical component of Weibel-Palade bodies (WPBs) along with von Willebrand factor (vWF), tissue-type plasminogen activator (t-PA), and osteoprotegerin.39, 40, 41, 42 The negative associations between each of vWF and t-PA and ARDS 28-day mortality also are replicated in this study (ORvWF = 1.06 [95% CI, 1.01-1.11; P = .02]; ORt-PA = 1.08 [95% CI, 1.04-1.13; P = 3.77 × 10-4]). Causal mediation analysis showed that the effects of IGFBP7 and t-PA on ARDS 28-day mortality were mediated significantly by platelet count, with average causal mediated effects up to an OR of 1.03 (95% CI, 1.02-1.04; mediated proportion, 23.67%; P = .01) for IGFBP7 and an OR of 1.02 (95% CI, 1.01-1.03; mediated proportion, 17.20%; P = .046) for t-PA (e-Table 17).
Clinical Application
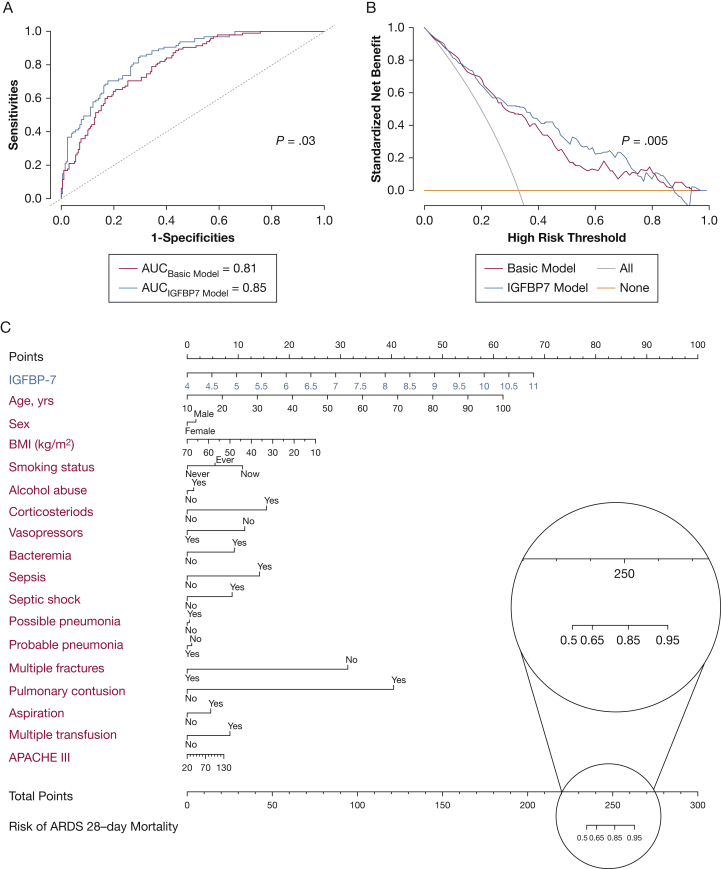
Clinical decision-making analysis revealed that the receiver operating characteristic curve of IGFBP7 was significantly different from clinical features and increased the AUC to 0.85 (P = .03) (Fig 4A). The receiver operating characteristic curve evaluates an overall discernment ability (sensitivity and specificity), but is insufficient in accounting for the clinical usefulness of a specific model.43 The decision curve analysis allows threshold probability to vary to examine whether the IGFBP7 model is superior to another at a certain range of threshold with respect to net benefits (NBs).44 The trapezoidal numerical integration method confirmed that the NBs of the IGFBP7 model were superior to those of the basic model (NBIGFBP7 model = 0.35 vs NBbasic model = 0.26; P = .005), crossing a wide range of threshold from 0.25 to 0.73 (Fig 4B).
Figure 4.
Clinical application of insulin-like growth factor binding protein-7 (IGFBP7). A, Receiver operating characteristic (ROC) curve, which was used to evaluate the performance of IGFBP7 and clinical variables for ARDS 28-day mortality prediction in plasma samples. B, Decision curve analysis. Comparison of standard net benefits among treat all (gray line), treat none (black line), IGFBP7 model (red line), and basic model (green line) groups. C, Nomogram constructed with clinical findings (green) and IGFBP7 (red) for ARDS 28-day mortality risk. The probability of each predictor can be converted into the points axis in the top of the nomogram. The summary of the points of each predictor corresponded to the total points at the bottom of the nomogram. After adding the points of each predictor in the total points axis, a patient’s probability of mortality can be found at the bottom of the nomogram. For example, if a patient’s score was 250, the ARDS 28-day mortality risk probability corresponds to 0.85. APACHE = Acute Physiology and Chronic Health Evaluation; AUC = area under the receiver operating characteristic curve.
To apply IGFBP7 easily in clinical practice, we combined clinical information and log2-transformed IGFBP7 of patients to develop a nomogram, making intuitive graphical and individualized predictions. A weighted score calculated using all predictors was used to predict 28-day mortality (Fig 4C). Discrimination and calibration methods were applied in both the discovery and validation phases. The c-index was calculated as 0.80, referring to a relatively good prediction of the nomogram. Calibration plot, drawing from a 1,000-time bootstrap for correcting overfitting, showed high-level accordance between predicted and actual probability (e-Fig 6).
Discussion
Our comprehensive study incorporating both plasma proteomic analysis and two rounds of Mendelian randomization analysis demonstrated that genetically predicted IGFBP7 causally effected 28-day mortality in ARDS. We determined that plasma IGFBP7 associated with ARDS 28-day mortality partially mediated by platelet count at ICU admission. To the best of our knowledge, this is the first causal association study on ARDS 28-day mortality outcomes leveraging GWAS, proteomics, and novel causal inference methods combined.
In recent years, clinical research groups have performed clinical studies assessing protein biomarkers in ARDS. Calfee et al45,46 used updated plasma biomarkers and proceeded to define a detailed endotype (hyperinflammatory and hypoinflammatory) of ARDS that may vary on the individual level. They described different ARDS subphenotypes and biomarker panels that may help clinicians to select patients who may benefit from different therapeutic strategies.47 A large biomarker study of 1,341 patients enrolled in the Protocolized Care of Early Septic Shock trial found that endothelial cell permeability and hemostasis-related proteins were associated with increased mortality.48 Unlike targeted proteomics study mentioned above, untargeted proteomics analysis in ARDS to date typically have examined a small number of samples.49, 50, 51, 52 Given the small sample size and potential environmental confounders, many proteomic biomarkers processed in other studies likely contributed to the heterogeneous conclusions. Causal biomarker studies benefit from interpretation of clinical outcomes, because disentangling correlation from causation is important in ARDS. For example, ANG-2 is an established biomarker of endothelial activation and permeability that is associated strongly with ARDS risk and outcome, but is limited in causality. Genetic variants exhibiting cis-protein quantitative trait loci with plasma ANG-2 were associated with ARDS risk from sepsis, and the risk is statistically determined by genetically predicted ANG-2 levels,53 emphasizing the causal role of ANG-2 in inducing sepsis-associated ARDS. A Mendelian randomization approach confirmed that plasma sRAGE acts as a genetically regulated causal intermediate in sepsis-associated ARDS.12 Subsequently, multiple clinical trials (clinicaltrials.gov Identifiers: NCT01600651, NCT00811629, NCT02070536, and NCT01270295) validated the usefulness of sRAGE in predicting ARDS development at ICU admission. Treatments based on ANG-2 and sRAGE have not been reported, and small sample sizes may limit clinical trial options. We used GSMR analysis in this study, a summary data Mendelian randomization framework, to expand current study sample size, resulting in higher power. One of the attractive features of our study is that the causal association between IGFBP7 and ARDS 28-day mortality risk is unlikely to be confounded with environmental factors and should be validated in follow-up clinical trials.
Previous studies strongly support that IGFBP7 plays a mechanistic role in pulmonary inflammatory response both in mammal models and population studies. For example, IGFBP7 levels were upregulated in experimental radiation-induced lung injury models54 and were displayed synchronously in zinc chloride smoke inhalation lung injury models, suggesting IGFBP7 derangement with pulmonary inflammation.55 A population-based retrospective study demonstrated that circulating IGFBP7 levels were increased during acute exacerbation of COPD and were reduced after convalescence, indicating the potential prognostic value of IGFBP7 for pulmonary inflammation.56 Given its comparatively low affinity in a comparison with the insulin-like growth factor binding protein family, IGFBP7 functions not only as an insulin-like growth factor binding protein, but also as a direct growth-suppressing factor with an insulin-like growth factor-independent action similar to that of insulin-like growth factor binding protein 3.57,58 Previous studies found that insulin-like growth factor 1, one of the IGFBP7 binding proteins, coupled with insulin-like growth factor binding protein 3 is involved in the pathogenesis of lung injury and is associated independently with ARDS risk and mortality.52,59,60
WPBs, storage granules of endothelial cells, play a dual role in hemostasis and inflammation by capturing platelets during the onset of primary hemostasis.61,62 IGFBP7 is a critical component of WPBs, along with vWF, t-PA, and osteoprotegerin.39, 40, 41, 42 The dysregulation of vWF and t-PA affect the development and prognosis of ARDS, mainly by regulating inflammatory response.63,64 The negative associations between each of vWF and t-PA and ARDS 28-day mortality also are replicated in this study. Carboxy-terminal D4-C1-C2-C3-CK domains (vWF variants) are needed for cotargeting of IGFBP7 to pseudo-WPBs, which suggests that the binding of IGFBP7 to vWF is required for its targeting to WPBs.39 However, little evidence indicates a potential interaction relationship between t-PA and IGFBP7. Platelets are believed to make an important contribution to ARDS among critically ill patients acting in conjunction with fibrinogen to mediate endothelial damage through multiple signal transduction pathways.65 Our results indicate that the effects of IGFBP7 and t-PA on ARDS 28-day mortality were mediated partially by platelet count at ICU admission. Because of the physiological function of WPBs on platelets, theoretically, platelet count has the capacity to be an intermediator between IGFBP7 and ARDS 28-day mortality.
Our study has several strengths. First, the study design is based on both plasma proteomic detection and multistage causal inference analysis, which bolsters reliability. Second, previous simulation studies have proven that GSMR outperforms existing summary data-based Mendelian randomization methods because GSMR leverages power from multiple genetic variants, accounting for linkage disequilibrium between variants and for pleotropic effects.25,66,67 Sensitivity analysis with the other four Mendelian randomization methods in both the MEARDS and ARDSNet studies also supports the adverse effect of IGFBP7 on ARDS 28-day mortality and further validates the robustness of the GSMR results. Third, the IV was selected based on a strict genome-wide significance threshold (P < 5 × 10-8). Fourth, in the current study, we restricted the causal inference to non-Hispanic White people to minimize the population stratification bias, which is the most important confounder in genetic studies. However, we acknowledge that this is a limitation because recent studies showed that acute lung injury mortality differs among different races and ethnicities, indicating the importance of studying a diverse population.68 Future studies with non-European populations are needed to replicate our findings. We also acknowledge some other limitations. First, the GWAS sample size arguably is small, decreasing the power of causal protein biomarker detection. Theoretically, more causal protein biomarkers will be discovered with an expansion of genotype ARDS cases. Second, our study did not elucidate underlying biological mechanisms linking these biomarkers to ARDS. Finally, although our study detected IGFBP7 as a causal protein biomarker by Mendelian randomization, the clinical benefits (AUCs, nomogram, and decision curve analysis) require external validation.
Interpretation
This study linking plasma IGFBP7 as a novel causal protein involved in the pathogenesis of ARDS 28-day mortality and in platelet function in ARDS provides a foundation for further experimental and clinical investigation of the mechanisms and usefulness of this biomarker.
Acknowledgments
Author contributions: D. C. C., Z. Z., and X. D. contributed to the study design. N. J. M., A. M. A., L. S., M. D., D. N., B. T. T., and P. T. contributed to data collection. X. D., H. H., and L. L. performed statistical analysis and interpretation. X. D. drafted the manuscript. Z. Z., A. M. A., Y. W., N. J. M., B. T. T., R. Z., and F. C. revised the manuscript. All authors contributed to critical revision of the manuscript and approved its final version. D. C. C. and Z. Z. supervised the study. All authors revised the manuscript and approved of the final manuscript.
Financial/nonfinancial disclosures: None declared.
Other contributions: The authors thank the international INTERVAL project for protein quantitative trait loci, sharing the GWAS data from European populations, and all the study participants and research staff for their contributions and commitment to the study.
Additional information: The e-Appendix, e-Figures, and e-Tables can be found in the Supplemental Materials section of the online article.
Footnotes
Drs D. C. Christiani and Z. Zhu are senior authors who supervised the study.
FUNDING/SUPPORT: This study was supported by the National Institutes of Health [Grants R01HL060710, R56HL134356, and ES000002 to D. C. C.], State’s Key Project of Research and Development Program [Grant 2016YFE0204900 to F. C.], National Natural Science Foundation of China [Grants 81530088 and 81473070 to F. C., 81402764 to Y. W., and 81402763 to R. Z.]), and China Scholarship Council [Grant 201906090239 to X. D.]. The funding source, the National Institutes of Health, had not role in the design, execution, or reporting of the study.
Supplementary Data
References
- 1.Fan E., Brodie D., Slutsky A.S. Acute respiratory distress syndrome: advances in diagnosis and treatment. JAMA. 2018;319:698–710. doi: 10.1001/jama.2017.21907. [DOI] [PubMed] [Google Scholar]
- 2.Elliott C.G., Rasmusson B.Y., Crapo R.O. Prediction of pulmonary function abnormalities after adult respiratory distress syndrome (ARDS) Am Rev Respir Dis. 1987;135:634–638. doi: 10.1164/arrd.1987.135.3.634. [DOI] [PubMed] [Google Scholar]
- 3.Bellani G., Laffey J.G., Pham T. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA. 2016;315:788–800. doi: 10.1001/jama.2016.0291. [DOI] [PubMed] [Google Scholar]
- 4.Moss M., Huang D.T., Brower R.G. Early Neuromuscular blockade in the acute respiratory distress syndrome. N Engl J Med. 2019;380:1997–2008. doi: 10.1056/NEJMoa1901686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laffey J.G., Kavanagh B.P. Fifty years of research in ARDS. Insight into acute respiratory distress syndrome. From models to patients. Am J Respir Crit Care Med. 2017;196:18–28. doi: 10.1164/rccm.201612-2415CI. [DOI] [PubMed] [Google Scholar]
- 6.Headley A.S., Tolley E., Meduri GU. Infections and the inflammatory response in acute respiratory distress syndrome. Chest. 1997;111:1306–1321. doi: 10.1378/chest.111.5.1306. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura T., Sato E., Fujiwara N. Increased levels of soluble receptor for advanced glycation end products (sRAGE) and high mobility group box 1 (HMGB1) are associated with death in patients with acute respiratory distress syndrome. Clin Biochem. 2011;44:601–604. doi: 10.1016/j.clinbiochem.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 8.Gallagher D.C., Parikh S.M., Balonov K. Circulating angiopoietin 2 correlates with mortality in a surgical population with acute lung injury/adult respiratory distress syndrome. Shock. 2008;29:656–661. doi: 10.1097/shk.0b013e31815dd92f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y.L., Chen W., Chen L.Y. Systemic and bronchoalveolar cytokines as predictors of in-hospital mortality in severe community-acquired pneumonia. J Crit Care. 2010;25 doi: 10.1016/j.jcrc.2009.05.002. 176.e7-13. [DOI] [PubMed] [Google Scholar]
- 10.Kamstrup P.R., Tybjaerg-Hansen A., Steffensen R. Genetically elevated lipoprotein(a) and increased risk of myocardial infarction. JAMA. 2009;301:2331–2339. doi: 10.1001/jama.2009.801. [DOI] [PubMed] [Google Scholar]
- 11.Smith G.D., Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 12.Jones T.K., Feng R., Kerchberger V.E. Plasma sRAGE acts as a genetically regulated causal intermediate in sepsis-associated acute respiratory distress syndrome. Am J Respir Crit Care Med. 2020;201:47–56. doi: 10.1164/rccm.201810-2033OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reilly J.P., Wang F., Jones T.K. Plasma angiopoietin-2 as a potential causal marker in sepsis-associated ARDS development: evidence from Mendelian randomization and mediation analysis. Intensive Care Med. 2018;44:1849–1858. doi: 10.1007/s00134-018-5328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Z., Zheng Z., Zhang F. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9:224. doi: 10.1038/s41467-017-02317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun B.B., Maranville J.C., Peters J.E. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh C.C., David S., Zhang R. Gene control of tyrosine kinase TIE2 and vascular manifestations of infections. Proc Natl Acad Sci U S A. 2016;113:2472–2477. doi: 10.1073/pnas.1519467113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang R., Wang Z., Tejera P. Late-onset moderate to severe acute respiratory distress syndrome is associated with shorter survival and higher mortality: a two-stage association study. Intensive Care Med. 2017;43:399–407. doi: 10.1007/s00134-016-4638-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferguson N.D., Fan E., Camporota L. The Berlin definition of ARDS: an expanded rationale, justification, and supplementary material. Intensive Care Med. 2012;38:1573–1582. doi: 10.1007/s00134-012-2682-1. [DOI] [PubMed] [Google Scholar]
- 19.Zhu Z., Liang L., Zhang R. Whole blood microRNA markers are associated with acute respiratory distress syndrome. Intensive Care Med Exp. 2017;5:38. doi: 10.1186/s40635-017-0155-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Assarsson E., Lundberg M., Holmquist G. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9 doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chua W., Purmah Y., Cardoso V.R. Data-driven discovery and validation of circulating blood-based biomarkers associated with prevalent atrial fibrillation. Eur Heart J. 2019;40:1268–1276. doi: 10.1093/eurheartj/ehy815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreira J.P., Verdonschot J., Collier T. Proteomic bioprofiles and mechanistic pathways of progression to heart failure. Circ Heart Fail. 2019;12 doi: 10.1161/CIRCHEARTFAILURE.118.005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leek J.T., Johnson W.E., Parker H.S. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowden J., Davey Smith G., Haycock P.C. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess S., Butterworth A., Thompson S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehret G.B., Munroe P.B., Rice K.M. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greco M.F., Minelli C., Sheehan N.A. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med. 2015;34:2926–2940. doi: 10.1002/sim.6522. [DOI] [PubMed] [Google Scholar]
- 28.Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol. 2014;43:922–929. doi: 10.1093/ije/dyu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zambon M., Vincent J.L. Mortality rates for patients with acute lung injury/ARDS have decreased over time. Chest. 2008;133:1120–1127. doi: 10.1378/chest.07-2134. [DOI] [PubMed] [Google Scholar]
- 30.Fan Y., Li Y., Chen Y. Comprehensive metabolomic characterization of coronary artery diseases. J Am Coll Cardiol. 2016;68:1281–1293. doi: 10.1016/j.jacc.2016.06.044. [DOI] [PubMed] [Google Scholar]
- 31.Reilly J.P., Christie J.D. Linking genetics to ARDS pathogenesis: the role of the platelet. Chest. 2015;147:585–586. doi: 10.1378/chest.14-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J., Sheng L., Wang S. Analysis of clinical risk factors associated with the prognosis of severe multiple-trauma patients with acute lung injury. J Emerg Med. 2012;43:407–412. doi: 10.1016/j.jemermed.2009.05.024. [DOI] [PubMed] [Google Scholar]
- 33.Wang T., Liu Z., Wang Z. Thrombocytopenia is associated with acute respiratory distress syndrome mortality: an international study. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu L., Ye Q.F., Wan Q.Q. Mortality predictors in acute respiratory distress syndrome renal transplant recipients with ESKAPE/rESKAPE pneumonia. Transplant Proc. 2015;47:2450–2455. doi: 10.1016/j.transproceed.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Lazzeri C., Bonizzoli M., Cianchi G. Bilirubin in the early course of venovenous extracorporeal membrane oxygenation support for refractory ARDS. J Artif Organs. 2018;21:61–67. doi: 10.1007/s10047-017-0979-0. [DOI] [PubMed] [Google Scholar]
- 36.Rice T.W., Wheeler A.P., Bernard G.R. Comparison of the SpO2/Fio2 ratio and the Pao2/Fio2 ratio in patients with acute lung injury or ARDS. Chest. 2007;132:410–417. doi: 10.1378/chest.07-0617. [DOI] [PubMed] [Google Scholar]
- 37.Dong X., Zhang R., He J. Trans-omics biomarker model improves prognostic prediction accuracy for early-stage lung adenocarcinoma. Aging (Albany, NY) 2019;11:6312–6335. doi: 10.18632/aging.102189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanderweele T.J., Vansteelandt S. Odds ratios for mediation analysis for a dichotomous outcome. Am J Epidemiol. 2010;172:1339–1348. doi: 10.1093/aje/kwq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Breevoort D., van Agtmaal E.L., Dragt B.S. Proteomic screen identifies IGFBP7 as a novel component of endothelial cell-specific Weibel-Palade bodies. J Proteome Res. 2012;11:2925–2936. doi: 10.1021/pr300010r. [DOI] [PubMed] [Google Scholar]
- 40.Huber D., Cramer E.M., Kaufmann J.E. Tissue-type plasminogen activator (t-PA) is stored in Weibel-Palade bodies in human endothelial cells both in vitro and in vivo. Blood. 2002;99:3637–3645. doi: 10.1182/blood.v99.10.3637. [DOI] [PubMed] [Google Scholar]
- 41.Shahbazi S., Lenting P.J., Fribourg C. Characterization of the interaction between von Willebrand factor and osteoprotegerin. J Thromb Haemost. 2007;5:1956–1962. doi: 10.1111/j.1538-7836.2007.02681.x. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y., Li T., Zhang R. Poor sleep quality and late-life depression among the elderly in urban communities in Liaoning, China: a moderated mediation analysis. Arch Gerontol Geriatr. 2018;79:158–163. doi: 10.1016/j.archger.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Fitzgerald M., Saville B.R., Lewis R.J. Decision curve analysis. JAMA. 2015;313:409–410. doi: 10.1001/jama.2015.37. [DOI] [PubMed] [Google Scholar]
- 44.Vickers A.J., Van Calster B., Steyerberg E.W. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ. 2016;352:i6. doi: 10.1136/bmj.i6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calfee C.S., Delucchi K., Parsons P.E. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2:611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calfee C.S., Delucchi K.L., Sinha P. Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: secondary analysis of a randomised controlled trial. Lancet Respir Med. 2018;6:691–698. doi: 10.1016/S2213-2600(18)30177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reilly J.P., Calfee C.S., Christie J.D. Acute respiratory distress syndrome phenotypes. Semin Respir Crit Care Med. 2019;40:19–30. doi: 10.1055/s-0039-1684049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou P.C., Filbin M.R., Wang H. Endothelial permeability and hemostasis in septic shock: results from the ProCESS trial. Chest. 2017;152:22–31. doi: 10.1016/j.chest.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhargava M., Becker T.L., Viken K.J. Proteomic profiles in acute respiratory distress syndrome differentiates survivors from non-survivors. PLoS One. 2014;9 doi: 10.1371/journal.pone.0109713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang D.W., Hayashi S., Gharib S.A. Proteomic and computational analysis of bronchoalveolar proteins during the course of the acute respiratory distress syndrome. Am J Respir Crit Care Med. 2008;178:701–709. doi: 10.1164/rccm.200712-1895OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X., Shan Q., Jiang L. Quantitative proteomic analysis by iTRAQ for identification of candidate biomarkers in plasma from acute respiratory distress syndrome patients. Biochem Biophys Res Commun. 2013;441:1–6. doi: 10.1016/j.bbrc.2013.09.027. [DOI] [PubMed] [Google Scholar]
- 52.Schnapp L.M., Donohoe S., Chen J. Mining the acute respiratory distress syndrome proteome: identification of the insulin-like growth factor (IGF)/IGF-binding protein-3 pathway in acute lung injury. Am J Pathol. 2006;169:86–95. doi: 10.2353/ajpath.2006.050612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tammela T., Sanchez-Rivera F.J., Cetinbas N.M. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature. 2017;545:355–359. doi: 10.1038/nature22334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong Y., Lin Z., Lin X. IGFBP7 contributes to epithelial-mesenchymal transition of HPAEpiC cells in response to radiation. J Cell Biochem. 2019;120:12500–12507. doi: 10.1002/jcb.28516. [DOI] [PubMed] [Google Scholar]
- 55.Xie X., Zhao J., Xie L. Identification of differentially expressed proteins in the injured lung from zinc chloride smoke inhalation based on proteomics analysis. Respir Res. 2019;20:36. doi: 10.1186/s12931-019-0995-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruan W., Wu M., Shi L. Serum levels of IGFBP7 are elevated during acute exacerbation in COPD patients. Int J Chron Obstruct Pulmon Dis. 2017;12:1775–1780. doi: 10.2147/COPD.S132652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oh Y. IGF-independent regulation of breast cancer growth by IGF binding proteins. Breast Cancer Res Treat. 1998;47:283–293. doi: 10.1023/a:1005911319432. [DOI] [PubMed] [Google Scholar]
- 58.Kim H.S., Nagalla S.R., Oh Y. Identification of a family of low-affinity insulin-like growth factor binding proteins (IGFBPs): characterization of connective tissue growth factor as a member of the IGFBP superfamily. Proc Natl Acad Sci U S A. 1997;94:12981–12986. doi: 10.1073/pnas.94.24.12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahasic A.M., Zhai R., Su L. IGF1 and IGFBP3 in acute respiratory distress syndrome. Eur J Endocrinol. 2012;166:121–129. doi: 10.1530/EJE-11-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahasic A.M., Tejera P., Wei Y. Predictors of circulating insulin-like growth factor-1 and insulin-like growth factor-binding protein-3 in critical illness. Crit Care Med. 2015;43:2651–2659. doi: 10.1097/CCM.0000000000001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang X., Halvorsen K., Zhang C.Z. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science. 2009;324:1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valentijn K.M., Eikenboom J. Weibel-Palade bodies: a window to von Willebrand disease. J Thromb Haemost. 2013;11:581–592. doi: 10.1111/jth.12160. [DOI] [PubMed] [Google Scholar]
- 63.Ware L.B., Eisner M.D., Thompson B.T. Significance of von Willebrand factor in septic and nonseptic patients with acute lung injury. Am J Respir Crit Care Med. 2004;170:766–772. doi: 10.1164/rccm.200310-1434OC. [DOI] [PubMed] [Google Scholar]
- 64.Grau G.E., de Moerloose P., Bulla O. Haemostatic properties of human pulmonary and cerebral microvascular endothelial cells. Thromb Haemost. 1997;77:585–590. [PubMed] [Google Scholar]
- 65.Wei Y., Wang Z., Su L. Platelet count mediates the contribution of a genetic variant in LRRC16A to ARDS risk. Chest. 2015;147:607–617. doi: 10.1378/chest.14-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bowden J., Davey Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burgess S., Dudbridge F., Thompson S.G. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016;35:1880–1906. doi: 10.1002/sim.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bime C., Poongkunran C., Borgstrom M. Racial differences in mortality from severe acute respiratory failure in the United States, 2008-2012. Ann Am Thorac Soc. 2016;13:2184–2189. doi: 10.1513/AnnalsATS.201605-359OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.