Abstract
muREC2/RAD51L1 is a radiation-inducible gene that regulates cell cycle progression. To elucidate the biological function of muREC2/RAD51L1, the gene was disrupted in embryonic stem cells by homologous recombination. Mice heterozygous for muREC2/RAD51L1 appear normal and fertile; however, no homozygous pups were born after interbreeding of heterozygous mice. Timed pregnancy studies showed that homozygous mutant embryos were severely retarded in growth as early as ca. 5 days gestation (E5.5) and were completely resorbed by E8.5. Mutant blastocyst outgrowth was also severely impaired in a double-knockout embryo, but embryonic development did progress further in a p53-null background. These results suggest that muREC2/RAD51L1 plays a role in cell proliferation and early embryonic development, perhaps through interaction with p53.
In the past few years, an increasing number of genes in the recA/RAD51 recombination-repair family have been cloned, including REC2/RAD51L1, RAD51C, RAD51D, R51H3, XRCC2, and XRCC3 (1, 3, 4, 24, 25, 31, 32). Among them, REC2/RAD51L1, which was first cloned in our lab and subsequently at two other labs, encodes a 350-amino-acid protein exhibiting significant homology to the Escherichia coli RECA and Ustilago maydis REC2 and RAD51 genes (25, 26). The regions of similarity include the nucleotide-binding A and B motifs and a DNA binding domain. Overexpression of hREC2/RAD51L1 in Chinese hamster ovary (CHO) cells causes a G1 delay in the cell cycle and hypersensitivity to UV irradiation (10). Overexpression of hREC2/RAD51L1 in T cells of transgenic mice results in partial blockage of T-cell differentiation and hypersensitivity of T cells to ionizing radiation (27a).
Although the human Rec2/Rad51L1 protein has not been shown to catalyze recombinase reactions such as DNA pairing and strand transfer, amino acid alignment classifies the gene as a RAD51 ortholog. Some similarities in function have been noted, however. The REC2/RAD51L1 gene is induced by both ionizing radiation (25) and UV radiation (22). The hsRad51 protein appears to be recruited to the nucleus in response to DNA damage (8) and has been shown to interact with p53 directly, suggesting a role in cell cycle regulation and perhaps apoptosis (2, 28). Finally, disruption of both the muRAD51 gene and the muREC2/RAD51L1 gene results in early embryonic lethality (16, 34) (see below). The role of muRec2/Rad51L1 in DNA repair may not involve direct interaction with the damaged site. Thus far, we have been unable to demonstrate a significant level of DNA recognition by this protein (24a), and no ATP-hydrolytic activity has been observed (6a). Furthermore, recent results indicate that this gene may form fusion products upon translocation with an HMGC1 gene in leiomyoma. Potentiating the response to DNA damage may involve an indirect association with the proteins that regulate the cell cycle. Thus, in terms of DNA repair, the properties of this homolog of Rad51 are likely to be very different from those of the prototype protein.
To investigate the biological function of muREC2/RAD51L1 and create a mouse model to study DNA repair mechanisms, we disrupted muREC2/RAD51L1 in embryonic stem (ES) cells via homologous recombination. These experiments resulted in the creation of mice bearing a single allelic copy of the mREC2/RAD51L1 gene. We were unable to obtain a homozygous knockout mouse because the targeting process resulted in early embryonic lethality. Heterozygous mice appear normal after 12 months, while homozygous mutants die during early embryonic development. Analysis of mutant embryos in vivo and in vitro indicated that REC2/RAD51L1 is essential for cell proliferation. Although breeding of REC2/RAD51L1-heterozygous mice with p53-knockout mice failed to generate double-knockout pups, the double-mutant embryos survived longer, indicating a partial rescue by p53. In this report, we outline the details of our efforts.
MATERIALS AND METHODS
Construction of targeting vectors.
A human REC2/RAD51L1 cDNA probe (an XbaI-KpnI fragment from the pT3T7 plasmid) was used to screen a 129/sv mouse genomic library (Stratagene), and a 16-kb fragment containing exons and 2 was isolated. Genomic subfragments from the fragment were subcloned into pBluescript SK(+) (Stratagene), and the restriction map and the intron-exon boundaries were determined by direct sequencing and restriction site mapping. A 3.6-kb EcoRI fragment containing exon 2 was used to make the targeting vectors. A dicistronic β-galactosidase–neomycin (β-geo) cassette containing the picornavirus internal ribosome entry sequence (IRES) and a splice receptor (kindly provided by P. Mountford, Agricultural and Food Research Council Centre for Genome Research, University of Edinburgh, Edinburgh, Scotland) was inserted into a unique StuI site on exon 2 (21). The insertion and correct orientation were confirmed by direct sequencing. To construct the hygromycin targeting vector, a phosphoglycerate kinase-hygromycin cassette (kindly provided by S. M. A. Swagemakers, Erasmus University, Rotterdam, The Netherlands) was inserted at the same StuI site on exon 2. The vector was linearized with XhoI before electroporation into ES cells.
Transfection and analysis of ES cells.
J1 ES cells were cultured as described elsewhere (15), and 25 μg of linearized IRES targeting vector was electroporated into 2 × 107 ES cells. G418 (Gibco-BRL) was added 24 h later to a final concentration of 250 μg/ml (active substance). After 7 to 8 days of selection, individual clones were picked and expanded. DNA was prepared and analyzed by Southern blotting with probe B, which hybridizes outside of the targeting vector (Fig. 1). Correct integration was confirmed by using probe A, which localizes on the 5′ side of the gene. Single integration events were tested by probing the same blots with a neomycin resistance gene (neo) probe.
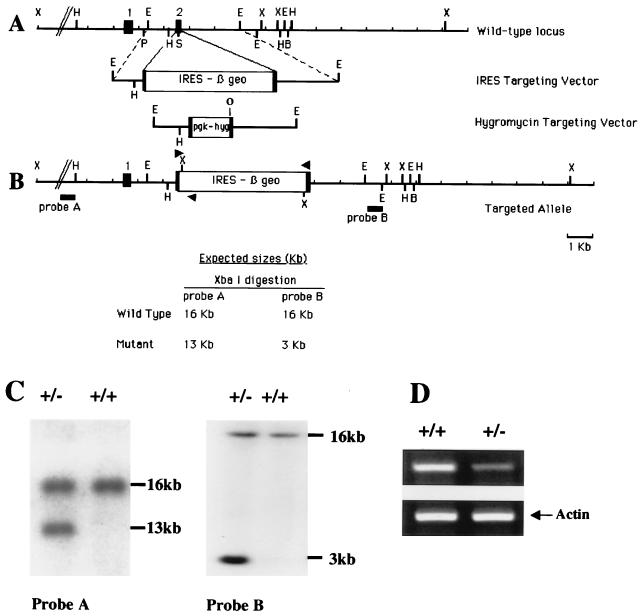
FIG. 1.
Targeted disruption of the muREC2/RAD51L1 gene. (A) Genomic structure of muREC2/RAD51L1 and targeting vectors. Exons 1 and 2 (closed boxes) encode the nuclear localization signal and part of the putative zinc finger motif. The IRES targeting vector was constructed by inserting an IRES–β-geo cassette into exon 2 at the StuI site (21). A hygromycin targeting vector was also constructed by inserting a PGK-hygromycin cassette (pgk-hyg) into the same StuI site to target the other allele (5). The insertion and correct orientation of the cassette were confirmed by direct sequencing. Abbreviations for restriction sites are as follows: X, XbaI; O, EcoRV; H, HindIII; P, PstI; E, EcoRI; S, StuI; and B, BamHI. (B) Targeted allele and expected size determined by Southern blotting. Upon XbaI digestion, probe A generates one 16-kb wild-type band and a 13-kb mutant band. Probe B generates one 16-kb wild-type band and a 3-kb mutant band. Arrowheads indicate primers used for genotyping. (C) Southern blot analysis of targeted ES cell clones. DNA from ES cells was digested with XbaI. Initial screening was performed with probe B and then confirmed by using probe A. (D) Semiquantitative RT-PCR of mouse thymus RNA. Total RNA were extracted from the thymuses of a muREC2/RAD51L1-heterozygous mouse and a wild-type mouse (6 to 8 weeks old). RT-PCR was performed with a pair of primers amplifying the entire muREC2/RAD51L1 cDNA (see Materials and Methods). The level of expression in heterozygous mice was roughly 50% of that in the wild-type mouse. Actin was used as an internal control for normalization.
Generation of chimeric mice and REC2/RAD51L1-heterozygous mice.
Two independent clones heterozygous for REC2/RAD51L1 were microinjected into C57BL/6 blastocysts at the Thomas Jefferson University Transgenic Facility by standard procedures (12). Integrated blastocysts were implanted into pseudopregnant (CBA × C57BL/6) F1 foster mothers. Chimeric mice, identified by their Agouti coat color, were mated with C57BL/6 mice (The Jackson Laboratory, Bar Harbor, Maine). Germ line transmission was confirmed by the presence of Agouti coat color in the F1 animals. All Agouti offspring were genotyped by PCR with three primers: E2F (5′-CTT TTA GCA CTT TTT AAG TCT CTC-3′), E2R (5′-GTT TGC ATT TGC GGG GCA CAG-3′), and IRES4 (5′-GTA TCT TAT ACA CGT GGC TTT TG-3′). E2F and E2R will amplify the wild-type allele (118 bp), while E2F and IRES4 will amplify the mutant allele (500 bp). PCR was performed for 35 cycles of 94°C for 1 min, 56°C for 30 s, and 68°C for 30 s. For reverse transcription (RT)-PCR analyses, total cellular RNA was isolated from the thymuses of heterozygous and wild-type mice by using an Ultraspec RNA isolation system (Biotecx). Semiquantitative RT-PCR was carried out with a pair of primers that amplifies the entire muREC2/RAD51L1 cDNA, F1 (5′-CGA AAT GAT CTC TTC CTC CAA AGA-3′) and F2 (5′-GAG CAG CAA GAA ACT AAG ACG AG-3′). To verify the existence of a fusion protein consisting of muRec2/Rad51L1 and the IRES cassette, RT-PCR was performed on total RNA from homozygous (8-day gestation [E8.5] REC2/RAD51L1−/− p53−/−]), heterozygous (thymus), and wild-type (thymus) embryos, using primers derived from exon 1 and the IRES cassette, MuREC2A (5′-ATG AGC AGC AAG AAA CTA AGA CGA-3′) and IRES4 (5′-GTA TCT TAT ACA CGT GGC TTT TG-3′), respectively.
Breeding and genotyping.
To genotype the pups resulting from breeding of chimeric and heterozygous muREC2/RAD51L1 animals, genomic DNA was extracted from the tail tips of 2-week-old mice by using a QIAamp tissue kit (Qiagen). To genotype by PCR, three primers were used: E2F, E2R, and IRES4. E2F-E2R will amplify the wild-type band (118 bp), and E2F-IRES4 will amplify the mutant band (500 bp). Proper insertion was confirmed by Southern blot analyses of isolated genomic DNA. The probes and conditions used were the same as those employed in the initial screening in ES cells.
The p53-knockout mice (13) were obtained from The Jackson Laboratory, and mice heterozygous for both muREC2/RAD51L1 and p53 were bred to generate double-knockout mice. For genotyping, four primers were used: OIMR013 (5′-CTT GGG TGG AGA GGC TAT TC-3′), OIMR014 (5′-AGG TGA GAT GAC AGG AGA TC-3′), OIMR336 (5′-ATA GGT CGG CGG TTC AT-3′), and OIMR337 (5′-CCC GAG TAT CTG GAA GAC AG-3′). The optimized protocol for PCR was provided by Carol Cutler-Linder of The Jackson Laboratory.
Histological analysis.
Timed pregnancies were carried out after mating muREC2/RAD51L1-heterozygous mice and muREC2/RAD51L1+/− p53+/− mice. Uteri from E5.5, E6.5, E7.5, and E8.5 pregnancies were isolated in ice-cold phosphate-buffered saline. Decidua were dissected, fixed overnight in 4% paraformaldehyde, processed, and embedded in paraffin. Sections were cut at a thickness of 5 mm, mounted, stained with hematoxylin and eosin (H&E), and photographed under an Olympus IX50 microscope. To genotype the sections, embryonic tissues were microdissected out from unstained paraffin sections. The tissues were lysed, and DNA was extracted for PCR. The sets of primers that were used for genotyping of pups were employed.
Culture of blastocyst outgrowths.
Pregnant females from heterozygous intercrosses were sacrificed at E3.5, and blastocysts were collected by flushing the uteri (12). Blastocysts were cultured individually in Dulbecco’s modified Eagle medium supplemented with 20% fetal bovine serum (HyClone) in 24-well plates at 37°C in a 5% CO2 incubator. The outgrowths were examined daily and photographed to monitor their development for 8 to 10 days. Finally, they were lysed and genotyped by PCR with primers E2F, E2R, and IRES4 (see above).
Generation of double-knockout muREC2/RAD51L1 ES cell lines.
Two approaches were taken to generate double-knockout ES cell lines. One was selection of heterozygous ES cells by using high concentrations of G418 (20). Briefly, one correctly targeted ES cell line (E16) was plated on a 6-mm-diameter plate at a density of 106 cells/plate. The cells were selected with G418 at four different concentrations (0, 1.2 mg/ml, 1.6 mg/ml, and 3.2 mg/ml) for 7 to 8 days. Clones which survived the highest concentration of G418 (3.2 mg/ml) were picked, expanded, and screened by Southern blotting with probe B. The other strategy involved retargeting the second allele by using a hygromycin vector. The hygromycin vector was linearized by XhoI digestion, and 25 μg of vector was electroporated into heterozygous clones (E16). Hygromycin was added at 200 μg/ml for a 7- to 8-day selection. Resistant clones were picked and expanded. Genomic DNA were extracted for screening by Southern blot analysis. The DNA were digested with XbaI-EcoRV and probed with probe B (Fig. 1).
RESULTS
Targeted disruption of the muREC2/RAD51L1 gene.
A 16-kb genomic fragment containing exons 1 and 2 of the muREC2/RAD51L1 gene was isolated by screening a lambda phage library from a 129/sv strain mouse genomic library. The genomic structure was determined by direct sequencing and restriction site mapping (Fig. 1A). To disrupt the muREC2/RAD51L1 gene, a 3.6-kb EcoRI fragment containing exon 2 was cloned into pBluescript SK(+). A dicistronic cassette containing an IRES and β-geo was inserted into a unique StuI site in exon 2 (21). The major advantages of this vector are its high targeting efficiency and its ability to cointegrate with a histochemically detectable reporter. The use of IRES–β-geo was particularly appropriate in our case because by RT-PCR, muREC2/RAD51L1 was found to be actively transcribed in ES cells (data not shown). The insertion introduced a stop codon, resulting in a shift of all three reading frames, thus eliminating 83% of the protein product. The truncated protein lacks important functional domains such as the A and B box and DNA binding domains, therefore rendering it nonfunctional. The IRES targeting vector was linearized and electroporated into J1 ES cells (15). After G418 selection, 46 clones were picked and expanded. The initial screening was done with a 500-bp EcoRI fragment (probe B) outside the vector. DNA was extracted from ES clones and digested with XbaI. Wild-type clones will produce only one 16-kb band, while heterozygous clones will generate one mutant band (3 kb) and the wild-type band (16 kb). Among the 46 clones screened, 28 were found to contain the disrupted genotype (56.5%). Southern blot analysis, using probe A on the 5′ end (which will produce a 13-kb mutant band and the 16-kb wild-type band), was used to confirm the targeted event (Fig. 1B and C). The blots were also hybridized with a neo probe to confirm that only one integration had occurred (data not shown). The high targeting frequency (56.5%) is due to selection for expression of the promoterless neo cassette; this eliminates insertions into introns (random integrations), thus resulting in considerable enrichment for homologous targeting events.
Phenotype of muREC2/RAD51L1-heterozygous mice.
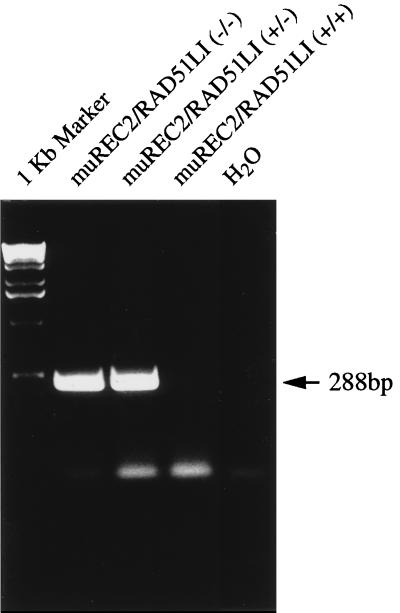
Two targeted ES clones were injected into C57BL/6 blastocysts, and seven high-percentage chimeric mice were obtained (five males and two females). They were mated with C57BL/6 mice for germ line transmission of the mutant allele. All of the males and one of the females exhibited germ line transmission, with one male giving 100% transmission as judged by the Agouti coat color of its offspring. Heterozygous mice were phenotypically normal and fertile, with no tumors or other abnormalities observed, for up to 16 months. To confirm that one allele had indeed been deleted, Northern and Western blotting was performed on mRNA and protein from thymuses of both heterozygous and wild-type mice. No signals were seen due to the fact that muREC2/RAD51L1 is expressed at very low levels in normal tissues. However, semiquantitative RT-PCR showed that only half of the wild-type level of mRNA was being expressed in the thymuses of heterozygous mice (Fig. 1D). To verify the fusion between exon 2 and IRES cassette, we designed a pair of primers. The forward primer is on exon 1, and the reverse primer on an IRES-selectable marker. Total RNA was extracted from homozygous embryos, heterozygous mice, and wild-type mice, and RT-PCR was performed. The results showed that the homozygous and heterozygous mice contained the fusion transcript while the wild type did not (Fig. 2).
FIG. 2.
RT-PCR for determination of the fusion protein. RT-PCR was performed with one primer on exon 1 and another primer on the IRES cassette. Only heterozygous and homozygous samples produced the fusion product (288 bp).
muREC2/RAD51L1−/− results in early embryonic lethality.
Heterozygous mice were interbred, and of the 228 pups genotyped by PCR, 160 were muREC2/RAD51L1+/− and 68 were wild type, producing a heterozygous:wild-type ratio of 2.35 to 1. No viable muREC2/RAD51L1−/− pups were identified, indicating that homozygosity of the muREC2/RAD51L1 mutation results in embryonic lethality (see Table 1).
TABLE 1.
Genotypic and phenotypic analysis of the progeny resulting from intercrosses of muREC2/RAD51L1-heterozygous micea
| Stage | Phenotype
|
muREC2/RAD51L1 genotype
|
Total no. of mice analyzed | |||
|---|---|---|---|---|---|---|
| Normal | Abnormal | +/+ | +/− | −/− | ||
| E3.5 | 11 | 4 | 4 | 7 | 4 | 15 |
| E4.5 | 15 | 5 | 5 | 10 | 5 | 20 |
| E5.5 | 31 | 9 | 9 | 22 | 9 | 40 |
| E6.5 | 24 | 8 | 7 | 17 | 8 | 32 |
| E7.5 | 22 | 6 | 7 | 15 | 6 | 28 |
| E8.5 to E13.5 | 26 | 9b | 8 | 18 | 0 | 35 |
| Pup (2 wks) | 228 | 0 | 68 | 160 | 0 | 228 |
Pups (2 weeks old) were genotyped by Southern blotting with probe B as shown in Fig. 1C. The embryos were collected from E3.5 to E13.5. DNA was extracted from whole embryos or yolk sacs. For stage 7.5, we were able to genotype only six of the embryos directly. Three primers, E2F, E2R, and IRES4, were used for genotyping, as described in Materials and Methods.
Embryos were completely resorbed; hence, genotyping could not be carried out.
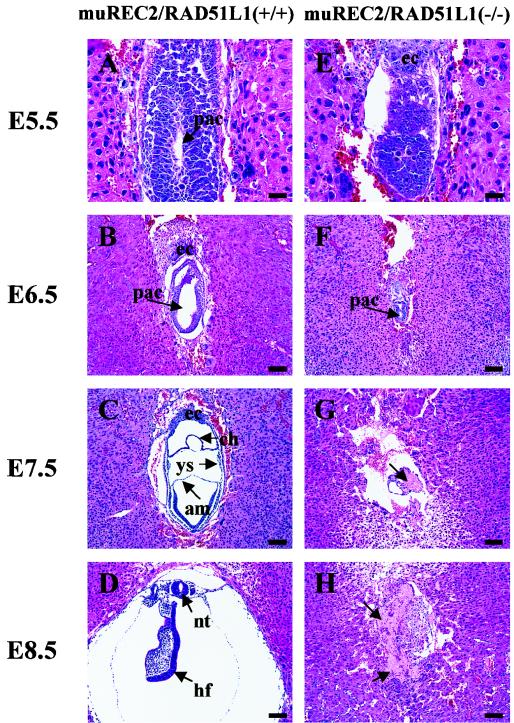
To pinpoint the differences between wild-type and muREC2/RAD51L1−/− mutant embryos, we next examined the histology of embryos between implantation and gastrulation. Intact decidual swellings obtained between E5.5 and E8.5 from muREC2/RAD51L1+/− intercross litters were fixed, sectioned, and stained with H&E. Following implantation (E4.5 to E5.5), abnormalities that distinguished normal conceptuses from muREC2/RAD51L1−/− conceptuses could be readily observed. Wild-type and heterozygous embryos showed normal growth and elongation of the egg cylinder, which contains both embryonic and extraembryonic ectoderm and distinct proamniotic cavities (Fig. 3A). In contrast, muREC2/RAD51L1−/− embryos were smaller in size and failed to form proamniotic cavities (Fig. 3E), although they did display embryonic and extraembryonic tissues. By E6.5, wild-type embryos are almost ready for gastrulation, with the egg cylinders nearly filling the yolk sac cavity. Elongated proamniotic and distinct exocolomic cavities are also well developed (Fig. 3B). By comparison, the mutant embryos were 75% smaller than that of the wild type, cells were disorganized and started to degenerate and presumptive areas where proamniotic cavities might develop were barely visible (Fig. 3F). By the time wild-type embryos undergo gastrulation (E7.5), the mesoderm develops concomitantly with the formation of three distinctive cavities: the amniotic cavity, yolk sac, and chorionic cavity (Fig. 3C). At this stage, mutant embryonic tissues almost completely disappeared and resorption was evident (Fig. 3G). At E8.5, wild-type embryos increase tremendously in size and exhibit formation of more structures, e.g., the neural tube and head fold (Fig. 3D). At this stage, mutant embryos were dead and were resorbed completely (Fig. 3H). This type of comparison was used to generate the data displayed in Table 1.
FIG. 3.
Histological sections of wild-type and muREC2/RAD51L1−/− embryos grown in utero. Sagittal sections are shown. All sections were stained with H&E. (A to D) Wild-type embryos; (E to H) muREC2/RAD51L1−/− embryos. (A) E5.5 wild-type embryo (early egg cylinder stage). Note the appearance of the proamniotic cavity and the clearly differentiated embryonic and extraembryonic ectoderm. (B) E6.5 wild-type embryo (egg cylinder stage). Note the formation of an exocoelomic cavity and enlargement of the proamniotic cavity (pac). (C) E7.5 wild-type embryo. The three germ layers are apparent. The yolk sac (ys), chorion (ch), and amnion (am) are clearly seen. (D) E8.5 wild-type embryo. More structures are formed (neural tube [nt], head fold [hf], etc.). (E) E5.5 mutant embryo. The embryonic region is reduced. No proamniotic cavity is seen. (F) E6.5 mutant embryo. There is evidence of a great loss of embryonic tissue and narrowing of the proamniotic cavity. (G) E7.5 mutant embryo. Only traces of embryonic tissue are left. Resorption is evident (indicated by the arrow). (H) E8.5 mutant embryo. The embryo is completely resorbed. The arrows indicate the pac. ec, ectoplacental cone. Bars, 100 μm.
muREC2/RAD51L1−/− blastocysts have a growth disadvantage in culture.
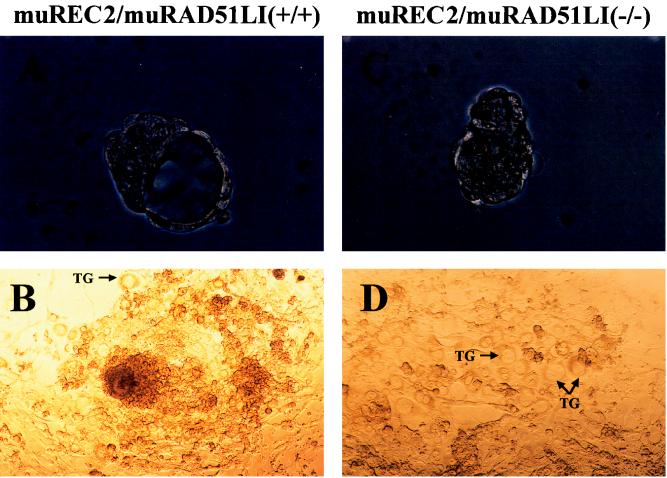
Blastocysts are composed of two cell types, pluripotent cells in the inner cell mass (ICM) and trophectoderm cells. When blastocysts are cultured in vitro, cells from the ICM proliferate rapidly while cells from the trophectoderm remain relatively quiescent. Blastocysts (E3.5) from progeny of heterozygous intercrosses were isolated by uterine flushing and photographed before and after in vitro culture. Eleven blastocysts which formed a well-developed epiblast surrounded by epithelial cells and very few trophoblastic giant cells (Fig. 4A and B) were found to have either the wild-type or a heterozygous genotype (Fig. 5). Four appeared smaller and lacked blastocoel cavities (Table 1; Fig. 4C). After 7 days in culture, those blastocysts showed an impaired outgrowth characterized by a smaller epiblast outgrowth surrounded by a monolayer of trophoblastic giant cells (Fig. 4D) or no epiblast outgrowth at all; they were genotyped as muREC2/RAD51L1−/− (Fig. 5). All of these results are consistent with the above-described in vivo observations of growth retardation in homozygous embryos.
FIG. 4.
Outgrowth of wild-type and muREC2/RAD51L1−/− blastocysts in vitro. The wild-type blastocyst is larger and has an intact blastocoel cavity (A), while the mutant blastocyst is smaller and lacks the blastocoel cavity (C). After 7 days in culture, the wild-type blastocyst grew into a huge mass characterized by rapid proliferation of the ICM surrounded by ectodermal cells. (B) Very few trophoblastic giant cells (TG) were seen (B); however, the growth of mutant blastocyst was greatly retarded. (D) There were no apparent epiblast outgrowth and few ectodermal cells; however, trophoblastic giant cells were abundant. Magnifications: ×66 (A and C) and ×33 (B and D).
FIG. 5.
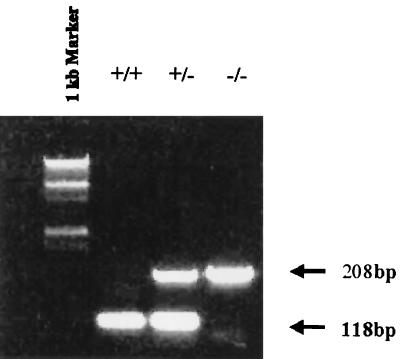
PCR genotyping of blastocysts and embryos resulting from muREC2/RAD51L1 interbreeding. DNA were extracted from blastocysts and embryos ranging from E4.5 to E13.5. PCRs were performed with probes E2F, E2R, and IRES4 (see Materials and Methods). Wild-type samples (+/+) produced only one 118-bp band, homozygous samples (−/−) produced one 208-bp band, while heterozygous samples (+/−) generated both bands. The marker lane contains a 1-kb DNA ladder.
muREC2/RAD51L1−/− cells fail to proliferate in vitro.
To generate muREC2/RAD51L1 double-knockout ES cell lines, we first attempted to target the other allele by using a hygromycin vector. A hygromycin vector was constructed by inserting a PGK-hygromycin cassette into the StuI site of exon 2 (Fig. 1A). The vector was linearized with XhoI and transfected into a heterozygous ES cell clone (E16), and a total of 100 clones were picked and expanded. Genomic DNA were digested with XbaI-EcoRV and probed with probe B. Wild-type clones produced one 16-kb band, while mutant clones generated a novel 3.3-kb band (Fig. 6A). Southern blotting showed that six of them were correctly targeted. None of them were homozygous. As an alternative, we selected heterozygous ES cells under conditions of elevated G418 concentration (20). One heterozygous ES clone (E16) was selected with various concentrations of G418. Forty-eight clones survived at the highest concentration (3.2 mg/ml). Southern blot analyses with probe B revealed that none of the clones were homozygous (Fig. 6B). The failure to obtain any homozygous ES cells is quite statistically significant (P < 0.01), which strongly suggests that the muREC2/RAD51L1 gene is required for the viability of ES cells. These data, coupled with the observation that no homozygous mutant cells were generated by retargeting or by increasing selective pressure, make a strong case for the RAD51L1 gene being indispensable for cell growth.
FIG. 6.
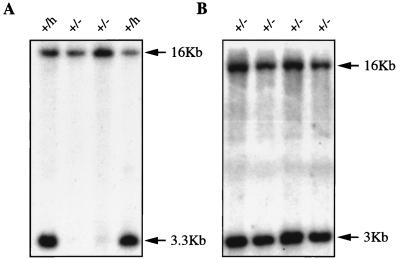
Southern blots of products of hygromycin retargeting and G418 selection. (A) Hygromycin retargeting. DNA was digested with XbaI and EcoRV and then probed with probe B. Nontargeted clones (+/−) produced one 16-kb band (middle two lanes). Heterozygous clones (+/h) generated one 16-kb wild-type band and one 3.3-kb mutant band (two outside lanes) because the hygromycin cassette introduced a new EcoRV site. (B) G418 selection. DNA was digested with XbaI and then probed with probe B (Fig. 1). All of the clones produced two bands (16-kb wild-type and 3-kb mutant bands), indicating that they were still heterozygous.
Early embryonic lethality is partially rescued by p53.
p53 plays a pivotal role in cell cycle control and apoptosis, and the embryonic lethality caused by mRAD51, BRCA1, and BRCA2 is partially rescued while mdm2-deficient mice are completely rescued in a p53-null background (9, 16, 17, 19). To explore the possibility of rescue by p53, muREC2/RAD51L1+/− mice were crossed with p53-knockout mice. One hundred seventy pups from double-heterozygous crosses (muREC2/RAD51L1+/− p53+/− × muREC2/RAD51L1+/− p53+/− were genotyped by PCR. None of them were homozygous for muREC2/RAD51L1, indicating that the lethal phenotype could not be rescued completely in a p53-null background. Twenty E7.5, 40 E8.5, and 30 E9.5 embryos from progeny of double-heterozygous crosses were isolated and genotyped by PCR. Interestingly, one E7.5 embryo, two E8.5 embryos, and one E9.5 embryo were homozygous for both muREC2/RAD51L1 and p53 (Table 2). No homozygous mutant muREC2/RAD51L1 mice were identified in either a wild-type or heterozygous p53 background, indicating that p53 heterozygosity was insufficient to rescue the embryonic-lethal phenotype. Double-mutant embryos appeared grossly smaller than their normal counterparts (Fig. 7). Histological studies of one E7.5 double mutant showed that some normal structures were formed—for example, chorion cavities; however, structures such as yolk sacs and amniotic cavities were missing. Cells in the embryos also looked abnormal (Fig. 7B). Compared with the muREC2/RAD51L1−/− embryos in a wild-type p53 or p53-heterozygous background, double mutants demonstrated significantly advanced proliferation and development.
TABLE 2.
Genotyping of progeny resulting from crosses between muREC2/RAD51L1+/− p53+/− and muREC2/RAD51L1+/− p53+/− mice
| Stage | Genotype
|
Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild type
|
muREC2/RAD51L1 knockout
|
p53 null
|
Double null muREC2/RAD51L1−/−p53−/− | |||||||
| muREC2/ RAD51L1+/+p53+/+ | muREC2/ RAD51L1+/+p53+/− | muREC2/ RAD51L1+/−p53+/+ | muREC2/ RAD51L1+/−p53+/− | muREC2/ RAD51L1−/−p53+/− | muREC2/ RAD51L1−/−p53+/+ | muREC2/ RAD51L1+/+p53−/− | muREC2/ RAD51L1+/−p53−/− | |||
| E7.5 | 3 | 4 | 3 | 4 | 0 | 0 | 3 | 2 | 1 | 20 |
| E8.5 | 6 | 8 | 6 | 5 | 0 | 0 | 7 | 6 | 2 | 40 |
| E9.5 | 5 | 6 | 4 | 4 | 0 | 0 | 6 | 4 | 1 | 30 |
| Pup | 34 | 24 | 28 | 30 | 0 | 0 | 26 | 28 | 0 | 170 |
FIG. 7.
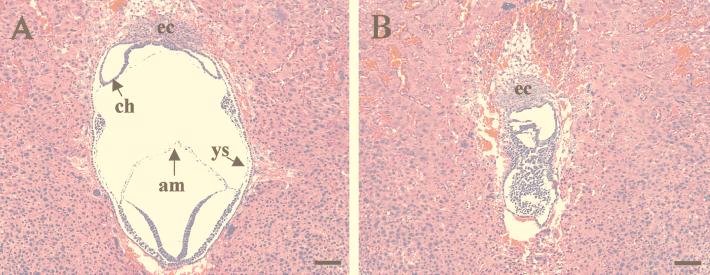
Histological sections of muREC2/RAD51L1+/− p53+/− (A) and muREC2/RAD51L1−/− p53−/− (B) embryos grown in utero to E7.5. Sagittal sections are shown. All sections were stained with H&E. (A) In double-heterozygous embryos, typical three chambers were observed, with formation of the chorion (ch), yolk sac (ys), and amnion (am). (B) The double-mutant embryo was smaller in size, and the chambers were not distinguishable. Although the ectoplacental cone (ec) is present, other structures cannot be identified, and embryonic cells looked morphologically abnormal. Bars, 100 μm.
DISCUSSION
In this study, we have generated a muREC2/RAD51L1 knockout by homologous recombination. Heterozygous mice were viable and looked normal; however, interbreeding of heterozygous mice failed to generate viable pups, indicating embryonic lethality. Timed pregnancies showed that mutant embryos died at approximately E5.5 to E6.5, and blastocyst outgrowth was also hindered in mutant embryos. Interestingly, muREC2/RAD51L1 mutant embryos survived longer (E8.5 to E9.5) and developed further in a p53-mutant background.
These findings show striking parallels with previous work on RAD51-, BRCA1-, and BRCA2-knockout mice. Both RAD51- and BRCA1-mutant embryos die before E7.5, while BRCA2-mutant embryos survive longer (E8.5 to E9.5) (9, 16, 17, 23, 30, 33). All of them exhibit reduced cellular proliferation, and the early lethality can be partially rescued by p53.
The phenotypic similarities shared by RAD51, BRCA1, BRCA2, and REC2/RAD51L1 mutants strongly suggest that these genes act together in some of the most important processes in a cell, e.g., DNA repair, transcription, and cell cycle control. Efficient repair of DNA damage is crucial to maintaining the integrity of the genome and, thereby, the survival of the cell or organism. Our current understanding of double-strand break (DSB) DNA repair is derived primarily from studies of bacteria and yeast. In Saccharomyces cerevisiae, Rad51 (a homolog of RecA) has been shown to interact with Rad52, Rad55, and replication protein A (6, 11, 18). Rad55 in turn interacts with Rad57, and a Rad55-Rad57 complex exhibits ATPase activity and promotes strand exchange mediated by Rad51 (14, 29). It is believed that together replication protein A, Rad52, Rad51, Rad55, and Rad57 assemble at the site of a DSB, forming a huge complex called a recombinosome which pulls the two ends of the DSB together and repairs the damage (11).
At the heart of this process is the activity of the RAD51 gene product. It is believed that Rad51 acts to conjoin the DNA during the repair event, providing a recombinational aspect of the process. Although this activity is crucial for repair of damaged DNA, the fact that it is essential for early embryonic life is somewhat puzzling. Our data suggest that the RAD51 analogue REC2/RAD51L1 is also an essential gene, since homozygous knockouts exhibit an embryonic-lethal phenotype. Interestingly, we have not been able to detect in vitro recombination activities, similar to those of Rad51 (29), mediated by Rec2/Rad51L1. Since hsRec2/Rad51L1 has been shown to interact directly with a related member of the Rad51 family, Rad51C (4), and Rad51C interacts with Rad51 (as judged by the yeast two-hybrid system), the loss of hsRec2/Rad51L1 may, in turn, lead to dysfunctional-complex formation. The activity of such a complex is likely to be similar to that of the recombinosome of S. cerevisiae (7) (see also reference 27 and references cited therein) which is responsible for DNA repair. Hence, it is possible that hsRec2/Rad51L1 performs a regulatory function within a complex that directs the repair of damaged DNA or monitors the accuracy of DNA replication events. If the complex were not properly formed, the cell would lack the capacity to monitor replication errors and, by extension, enable the propagation of genetic mutations. The activity of this protein may also be distinct from that of the so-called complex. In fact, Thacker (32) does not include this protein as part of the group of repair proteins (Rad51, Rad52, etc.) that act at the site of damage. He speculates that the Rec2/Rad51L1 protein could act more as a regulator of the repair process, and our data align with this notion. Since the overexpression of hRac2/hRad51L1 has been shown to reduce the cell cycle rate (10), we speculate that the protein functions at the level of DNA replication, expanding the window of time in which repair can take place. Close examination of the data of Havre et al. (10) data reveals that, in fact, S phase becomes elongated as a function of increased levels of this protein. This hypothesis is supported by recent evidence that disruption of another mouse RAD51-like gene, RAD51d, causes an early-embryonic-lethal phenotype (26a). We have recently demonstrated that hRec2/Rad51L1 is a protein kinase (9a), an activity that fits well with such a proposed role.
ACKNOWLEDGMENTS
We thank Peter Mountford for sending us the IRES cassette and Sigrid Swagemakers for the pPGK-Hygro vector. We are grateful to members of the Kmiec laboratory for helpful discussions and to Thomas Knudson and Leslie Lock (Thomas Jefferson University) for evaluation of embryonic development stages.
This work was supported by NIH grant R01 HL-58563-01A1.
REFERENCES
- 1.Albala J S, Thelen M P, Prange C, Fan W, Christensen M, Thompson L H, Lennon G G. Identification of a novel human RAD51 homolog, RAD51B. Genomics. 1997;46:476–479. doi: 10.1006/geno.1997.5062. [DOI] [PubMed] [Google Scholar]
- 2.Buchhop S, Gibson M K, Wang X W, Wagner P, Sturzbecher H W, Harris C C. Interaction of p53 with the human Rad51 protein. Nucleic Acids Res. 1997;25:3868–3874. doi: 10.1093/nar/25.19.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cartwright R, Dunn A M, Simpson P J, Tambini C E, Thacker J. Isolation of novel human and mouse genes of the recA/RAD51 recombination-repair gene family. Nucleic Acids Res. 1998;26:1653–1659. doi: 10.1093/nar/26.7.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dosanjh M K, Collins D W, Fan W, Lennon G G, Albala J S, Shen Z, Schild D. Isolation and characterization of RAD51C, a new human member of the RAD51 family of related genes. Nucleic Acids Res. 1998;26:1179–1184. doi: 10.1093/nar/26.5.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Essers J, Hendriks R W, Swagemakers S M, Troelstra C, de Wit J, Bootsma D, Hoeijmakers J H, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 6.Firmenich A A, Elias-Arnanz M, Berg P. A novel allele of Saccharomyces cerevisiae RFA1 that is deficient in recombination and repair and suppressible by RAD52. Mol Cell Biol. 1995;15:1620–1631. doi: 10.1128/mcb.15.3.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Gamper, H. Unpublished observations.
- 7.Golub E I, Kovalenko O V, Gupta R C, Ward D C, Radding C M. Interaction of human recombination proteins Rad51 and Rad54. Nucleic Acids Res. 1997;25:4106–4110. doi: 10.1093/nar/25.20.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haaf T, Golub E I, Reddy G, Radding C M, Ward D C. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci USA. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hakem R, de la Pompa J L, Elia A, Potter J, Mak T W. Partial rescue of Brca1 (5-6) early embryonic lethality by p53 or p21 null mutation. Nat Genet. 1997;16:298–302. doi: 10.1038/ng0797-298. [DOI] [PubMed] [Google Scholar]
- 9a.Havre, P., M. Rice, R. Ramos, and E. B. Kmiec. HsRec2/Rad51L1, a protein influencing cell cycle progression, has protein kinase activity. Exp. Cell Res., in press. [DOI] [PubMed]
- 10.Havre P A, Rice M C, Nöe A, Kmiec E B. The human REC2/RAD51B gene acts as a DNA damage sensor by inducing G1 delay and hypersensitivity to UV irradiation. Cancer Res. 1998;58:4733–4739. [PubMed] [Google Scholar]
- 11.Hays S L, Firmenich A A, Berg P. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc Natl Acad Sci USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the mouse embryo. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 13.Jacks T, Remington L, Williams B O, Schmitt E M, Halachmi S, Bronson R T, Weinberg R A. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 14.Johnson R D, Symington L S. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol Cell Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 16.Lim D S, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludwig T, Chapman D L, Papaioannou V E, Efstratiadis A. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 18.Milne G T, Weaver D T. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including RAD51 and RAD52. Genes Dev. 1993;7:1755–1765. doi: 10.1101/gad.7.9.1755. [DOI] [PubMed] [Google Scholar]
- 19.Montes de Oca Luna R, Wagner D S, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 20.Mortensen R M, Conner D A, Chao S, Geisterfer-Lowrance A A T, Seidman J G. Production of homozygous mutant ES cells with a single targeting construct. Mol Cell Biol. 1992;12:2391–2395. doi: 10.1128/mcb.12.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mountford P, Zevnik B, Duwel A, Nichols J, Li M, Dani C, Robertson M, Chambers I, Smith A. Dicistronic targeting constructs: reporters and modifiers of mammalian gene expression. Proc Natl Acad Sci USA. 1994;91:4303–4307. doi: 10.1073/pnas.91.10.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng L, Rice M C, Kmiec E B. Analysis of the human RAD51L1 promoter region and its activation by UV light. Genomics. 1998;54:529–541. doi: 10.1006/geno.1998.5536. [DOI] [PubMed] [Google Scholar]
- 23.Pentchev P G, Vanier M T, Suzuki K, Patterson M C. The metabolic and molecular basis of inherited diseases. Philadelphia, Pa: McGraw-Hill; 1995. [Google Scholar]
- 24.Pittman D L, Weinberg L R, Schimenti J C. Identification, characterization, and genetic mapping of Rad51d, a new mouse and human RAD51/RecA-related gene. Genomics. 1998;49:103–111. doi: 10.1006/geno.1998.5226. [DOI] [PubMed] [Google Scholar]
- 24a.Rice, M. Unpublished observations.
- 25.Rice M C, Smith S T, Bullrich F, Havre P A, Kmiec E B. Isolation of human and mouse genes based on homology to REC2, a recombinational repair gene from the fungus Ustilago maydis. Proc Natl Acad Sci USA. 1997;94:7417–7422. doi: 10.1073/pnas.94.14.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubin B P, Ferguson D O, Holloman W K. Structure of REC2, a recombinational repair gene of Ustilago maydis, and its function in homologous recombination between plasmid and chromosomal sequences. Mol Cell Biol. 1994;14:6287–6296. doi: 10.1128/mcb.14.9.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Schimenti, J. Personal communication.
- 27.Shen Z, Pardington-Purtymun P E, Comeaux J C, Moyzis R K, Chen D J. UBL1, a human ubiquitin-like protein associating with human RAD51/RAD52 proteins. Genomics. 1996;36:271–279. doi: 10.1006/geno.1996.0462. [DOI] [PubMed] [Google Scholar]
- 27a.Shu, Z., et al. Submitted for publication.
- 28.Sturzbecher H W, Donzelmann B, Henning W, Knippschild U, Buchhop S. p53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction. EMBO J. 1996;15:1992–2002. [PMC free article] [PubMed] [Google Scholar]
- 29.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki A, de la Pompa J L, Hakem R, Elia A, Yoshida R, Mo R, Nishina H, Chuang T, Wakeham A, Itie A, Koo W, Billia P, Ho A, Fukumoto M, Hui C C, Mak T W. Brca2 is required for embryonic cellular proliferation in the mouse. Genes Dev. 1997;11:1242–1252. doi: 10.1101/gad.11.10.1242. [DOI] [PubMed] [Google Scholar]
- 31.Tebbs R S, Zhao Y, Tucker J D, Scheerer J B, Siciliano M J, Hwang M, Liu N, Legerski R J, Thompson L H. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc Natl Acad Sci USA. 1995;92:6354–6358. doi: 10.1073/pnas.92.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thacker J. A surfeit of RAD51-like genes? Trends Genet. 1999;15:166–168. doi: 10.1016/s0168-9525(99)01733-3. [DOI] [PubMed] [Google Scholar]
- 33.Thompson A, Zhang Y, Kamen D, Jackson C W, Cardiff R D, Ravid K. Deregulated expression of c-myc in megakaryocytes of transgenic mice increases megakaryopoiesis and decreases polyploidization. J Biol Chem. 1996;271:22976–22982. doi: 10.1074/jbc.271.38.22976. [DOI] [PubMed] [Google Scholar]
- 34.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]