Abstract
Nuclear radiation and radioactive fallouts resulting from a nuclear weapon detonation or reactor accidents could result in injuries affecting multiple sensitive organs, defined as acute radiation syndrome (ARS). Rapid and early estimation of injuries to sensitive organs using markers of radiation response is critical for identifying individuals who could potentially exhibit ARS; however, there are currently no biodosimetry assays approved for human use. We developed a sensitive microRNA (miRNA)–based blood test for radiation dose reconstruction with ±0.5 Gy resolution at critical dose range. Radiation dose–dependent changes in miR-150-5p in blood were internally normalized by a miRNA, miR-23a-3p, that was nonresponsive to radiation. miR-23a-3p was not highly expressed in blood cells but was abundant in circulation and was released primarily from the lung. Our assay showed the capability for dose estimation within hours to 1 week after exposure using a drop of blood from mice. We tested this biodosimetry assay for estimation of absorbed ionizing radiation dose in mice of varying ages and after exposure to both improvised nuclear device (IND)–spectrum neutrons and gamma rays. Leukemia specimens from patients exposed to fractionated radiation showed depletion of miR-150-5p in blood. We bridged the exposure of these patients to fractionated radiation by comparing responses after fractionated versus single acute exposure in mice. Although validation in nonhuman primates is needed, this proof-of-concept study suggests the potential utility of this assay in radiation disaster management and clinical applications.
INTRODUCTION
With escalating tension in the international sociopolitical environment comes major concerns regarding the adequacy of preparedness for a mass casualty radiological event (1–3). Victims exposed to high doses of ionizing radiation develop acute radiation syndrome (ARS), which affects the hematopoietic and gastrointestinal system, and develop delayed effects of acute radiation exposure (DEARE), such as radiation pneumonitis, fibrosis, and multiorgan dysfunction (4, 5). In general, ARS follows a deterministic path, whereby dose effects have distinct clinical outcomes: Injuries resulting from <2 Gy gamma ray equivalent total body exposure exhibit mild symptoms; hematological effects are prominent at doses between 2 and 6 Gy; and gastrointestinal effects manifest at doses higher than 5 Gy, escalating to severity at higher doses (4–6). Injury to late-responding organs such as lung, heart, and kidney may not be evident for months or even years. In general, individuals exposed to 2 Gy and above to the whole body or substantial volume of the body need to be identified for treatment and follow-up. Early and rapid detection of ARS by accurate biological dose assessment will assist in effective medical management and proper allocation of resources and countermeasures (7). Over the years, a number of molecular, metabolomic, lipidomic, and protein markers exhibiting dose response have been identified (8–18); however, none have demonstrated adequate feasibility or capability for dose reconstruction in a heterogeneous population. As of now, there is no radiation biodosimetry assay or device approved by the U.S. Food and Drug Administration (FDA) for diagnostics in humans. Here, we report a two-microRNA–based scalable radiation biodosimetry (miR-RAD) assay that allows rapid and accurate estimation of absorbed ionizing radiation dose in pre-clinical studies.
MicroRNAs (miRNAs) are small noncoding RNA molecules of 18 to 24 nucleotides in length with a role in gene regulation (19). There are more than 2000 miRNAs reported from the human genome with cell type–specific signatures in their identity and abundance and can function as markers of diseases and organ responses (20). Multiple recent studies have reported the discovery and validation of circulating cell-free miRNAs as markers of radiation response (21–26). Evolutionarily conserved miR-150-5p detected in serum and plasma has been shown to exhibit dose response in rodent and nonhuman primate (NHP) models of total body irradiation (TBI) (21, 24, 25). Another evolutionarily conserved molecule, miR-23a-3p, was detected at amounts comparable to miR-150-5p in serum but was nonresponsive to radiation (21, 24). To explore the translational utility of these findings, we studied the response and the stability of miR-150-5p and miR-23a-3p to determine the feasibility of using these two miRNAs to assess radiation exposure at dose range and time points relevant to point-of-care and high-throughput radiation biodosimetry.
RESULTS
Peripheral blood mononuclear cells are the major source of radiation-sensitive miR-150-5p, and lungs are the major source of circulating radiation–nonresponsive miR-23a-3p
Our earlier mouse model studies identified radiation dose–dependent decrease in serum miR-150 (miR-150-5p), suggesting its potential utility as a radiation biodosimeter (24). Another abundant molecule detected in serum, miR-23a (miR-23a-3p), was found nonresponsive to radiation and could thus potentially serve as an endogenous normalizer. To identify the source and relative abundance of miR-150-5p and miR-23a-3p in circulation, we conducted miRNA profiling and comparative expression analysis in different tissues, cell types, and body fluids. Blood collected from healthy volunteers (leukopaks from American Red Cross) was fractionated and sorted for various cell subsets by immunomagnetic bead separation (schema in Fig. 1A). We analyzed the relative expression of miRNAs by nanoString-based profiling, an amplification-free hybridization-based direct digital counting method capable of parallel evaluation of more than 800 human miRNAs (Fig. 1B and fig. S1A). Comparative analysis of subsets of enriched mononuclear cells, such as mature T and B lymphocytes, natural killer (NK) cells, and hematopoietic stem cells (HSCs), revealed abundant yet varying expression of miR-150-5p, with T and B lymphocytes as well as HSCs showing high expression of miR-150-5p (Fig. 1B and fig. S1B) and low expression of miR-23a-3p (Fig. 1C and fig. S1B). Next, to identify the source of miR-23a-3p in circulation, we dissected several major organs (heart, liver, lung, kidney, small intestine, large intestine, spleen, brain, and bone marrow) from adult C57BL/6 mice and extracted total RNA for comparative expression analysis by quantitative reverse transcription polymerase chain reaction (qRT-PCR). We detected low cycle threshold (Ct) values for miR-23a-3p in RNA isolated from the lung, followed by the heart (Fig. 1, D and E, and fig. S1C), suggesting these organs as contributors of cell-free miR-23a-3p in circulation, with lung as the primary source. However, the release of miR-23a-3p into the circulation was not substantially altered even after a high dose of whole thorax irradiation, known to induce an acute inflammatory response, when followed up until 168 hours (day 7) (fig. S2, A and B). This further confirmed the abundant yet passive release of miR-23a-3p primarily from lungs to the circulation. The comparison of Ct values of both miRNAs in serum and exosomes purified from serum collected from healthy volunteers revealed the nonexosomal release of miR-23a-3p into the circulation (fig. S2, C and D). Collectively, these results suggested the feasibility of translating the physical dose to a quantifiable biologically absorbed dose based on differences in relative blood concentration of radiation-sensitive miR-150-5p using miR-23a-3p as an endogenous normalizer.
Fig. 1. PBMCs are the major source of radiation-sensitive miR-150-5p, and lungs are the major source of circulating radiation–nonresponsive miR-23a-3p.
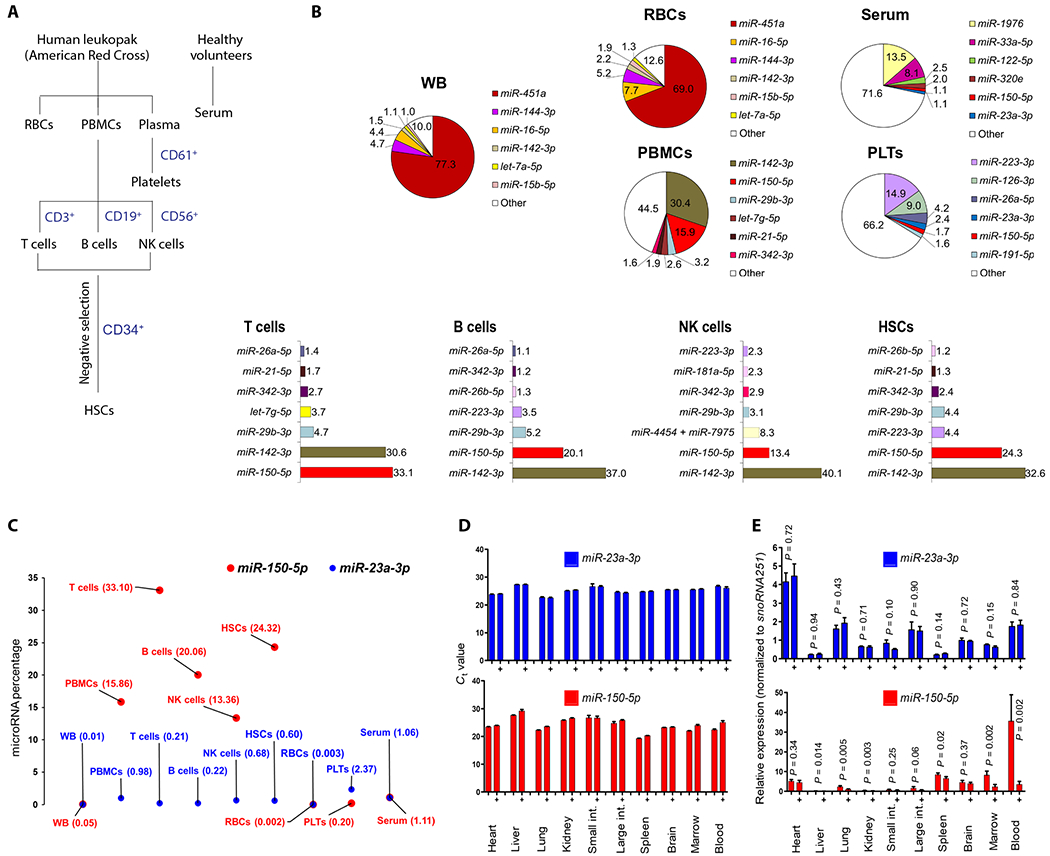
(A) Schematic showing separation of human blood cell subsets for miRNA profiling. (B) Pie chart showing top six or seven miRNAs expressed in whole blood (WB), serum, red blood cells (RBCs), platelets (PLTs), and PBMCs, detected by the nanoString nCounter assay. PBMCs were further separated into T cells, B cells, NK cells, and HSCs. Percentage is calculated by normalized count of each miRNA divided by mean of total counts of all miRNAs in a sample. (C) Scatterplot comparing percentage of miR-150-5p and miR-23a-3p between WB, PBMCs, T cells, B cells, NK cells, HSCs, RBCs, platelets, and serum detected by nanoString profiling. (D) Ct values of qRT-PCR assay obtained for miR-150-5p and miR-23a-3p across the major organs in control and 2 Gy irradiated (+) mice 24 hours after TBI. (E) Relative expression of miR-150-5p and miR-23a-3p normalized to snoRNA251 in various tissues of control and 2 Gy irradiated (+) mice (24 hours). Data are mean ± SD of n = 4 to 5 mice per group, statistical significance by unpaired t test.
Radiation response and kinetics of circulating miR-150-5p in bone marrow transplant patients
To investigate dose response in humans, we evaluated relative changes in serum concentration of miR-150-5p and miR-23a-3p in patients with acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), myelodysplastic syndrome (MDS), and mixed phenotype acute leukemia (MPAL) who underwent radiation-based myeloablative conditioning as a preparative regimen before HSC transplantation (HSCT). The standard ablative conditioning regimen for patients with leukemia used at many institutions, including The Ohio State University Comprehensive Cancer Center (OSUCCC), is 12 Gy (6 × 2 Gy) fractionated marrow-targeted TBI. About 4 days after completion of a 2 Gy BID (twice a day; with about 6-hour interval) radiation regimen allowed for complete marrow ablation before patients received stem cell transfusions. Analysis of blood samples collected at baseline and time points during and after marrow ablation and reconstitution (schema in Fig. 2, top) from patients enrolled in two clinical trials, NCT02122081 (OSU-13219 patients: n = 5 AML, n = 4 ALL, n = 2 MDS, and n = 1 MPAL; age, 20 to 70 years) and NCT01521039 (OSU-11002 patients: n = 7 ALL; age, 20 to 60 years), confirmed changes in the serum concentration of miR-150-5p after radiation with reference to miR-23a-3p. Consistent with the potential utility of circulating miR-150-5p as a readout of functional marrow, we observed depletion of normalized miR-150-5p (miR-150-5p/miR-23a-3p) expression through day −1 (pre-HSCT day), which then recovered to near baseline at week 4 (day +30) of stem cell transfusion in most patients (Fig. 2A). Analysis of serum samples collected at baseline and multiple time points during ablation and after reconstitution from an additional seven patients with AML, MDS, ALL, or MPAL confirmed the sensitivity of the miR-150-5p/miR-23a-3p assay for dose-response analysis (Fig. 2B). For additional validation, we used archived serum samples collected from seven patients with ALL enrolled in clinical trial NCT01521039 (OSU-11002) who underwent myeloablative TBI. As before, comparative analyses of miR-150-5p/miR-23a-3p values at baseline (before irradiation), day 0 (day of transplant), and week 4 (days 28 to 30) after transplant confirmed the robustness of these evolutionarily conserved molecules for providing a functional readout of marrow reconstitution after transplantation (Fig. 2C).
Fig. 2. Radiation dose response and kinetics of circulating miR-150-5p in bone marrow transplant (BMT) patients.
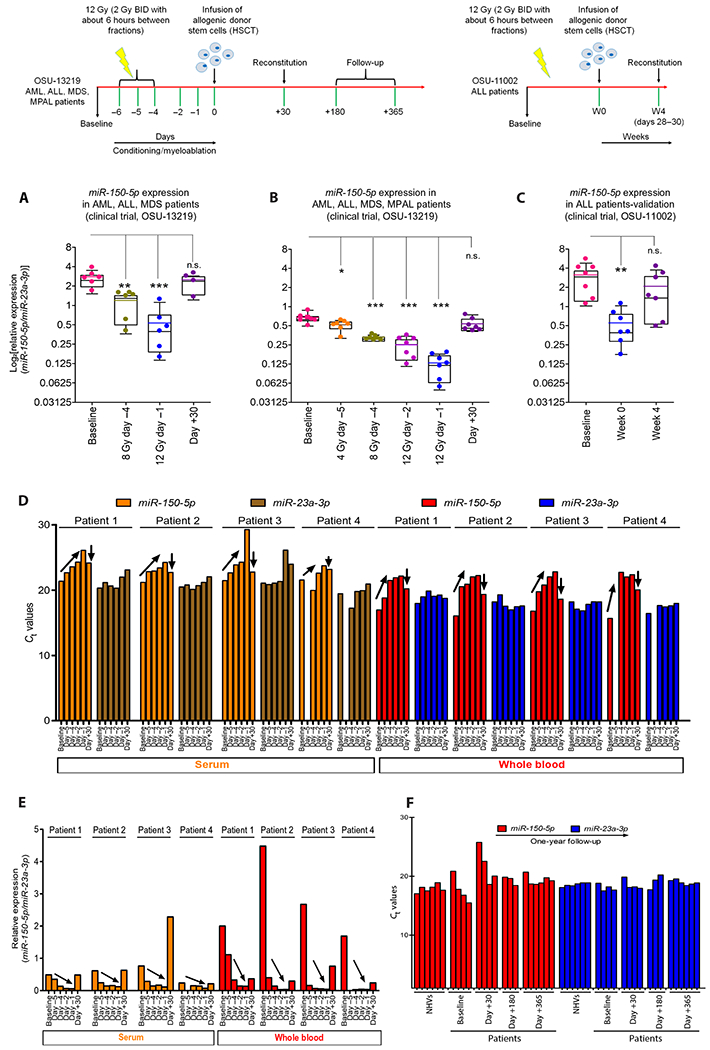
Top: Schematic of clinical trials OSU-13219 and OSU-11002 using radiation as a myeloablative conditioning regimen before allogeneic stem cell transplantation (HSCT), with sampling time points for miRNA analyses: OSU-13219 [baseline: day −5, 4 Gy (2 + 2 Gy); day −4, 8 Gy (2 + 2 + 2 + 2 Gy); day −2, 12 Gy (2 + 2 + 2 + 2 + 2 + 2 Gy); day −1 (before HSCT); and day +30] and OSU-11002 [baseline, week 0, and week 4 (days +28 to 30)]. (A) Dot-whisker plot shows depletion of miR-150-5p until day −1 and reconstitution at day +30 (n = 6 per time point except for day +30 with n = 4) in serum samples from OSU-13219 trial, and each dot represents patient sample. (B) For validation, more time points were added from newly enrolled patients (n = 7). (C) Dot-whisker plot depicts kinetics of miR-150-5p normalized to miR-23a-3p in cohort of patients from a different clinical trial, OSU-11002 (n = 7). For (A) to (C), the y axis represents miR-150-5p normalized to miR-23a-3p on a log2 scale. (D) Ct values of serum (orange and brown) and whole blood (red and blue) miR-150-5p and miR-23a-3p in four patients from OSU-13219 collected at baseline and at days−5, −4, −2, −1, and +30. Dose- and time-dependent depletion of miR-150-5p shown as an increase in Ct values until day −1 (upward-diagonal arrow) and reconstitution by day +30 (after HSCT) as a drop in Ct values. (E) Changes in normalized miR-150-5p in whole blood (red) and in serum (orange) in four patients as shown as ΔΔCt. (F) Ct values of miR-23a-3p (blue) and miR-150-5p (red) from qRT-PCR using RNA isolated from similar volumes of whole blood collected from normal healthy volunteers (NHVs) and patients (OSU-13219) at baseline and at days 30, 180, and 365 after HSCT; n = 6 for NHVs, n = 4 patients at baseline, n = 4 for day +30, n = 3 for day +180, and n = 6 for day +365. (A) to (D) were analyzed by a mixed-effect model with Bonferroni or Dunnett’s adjustment; *P < 0.05, **P < 0.01, and ***P < 0.001.
The comparable presence of miR-150-5p and miR-23a-3p in circulation prompted us to test dose response in whole blood and serum collected from patients with MDS, ALL, and AML (OSU-13219, n = 4). The data (Fig. 2D) confirmed dose- and time-dependent decrease in miR-150-5p after marrow-targeted irradiation (serum, P = 0.0079; whole blood, P < 0.0001) and increased concentration of miR-150-5p in blood on marrow reconstitution after transplantation. A steeper dose response was detected in whole blood RNA preparations compared to serum when translated as ΔΔCt values (Fig. 2E), indicating that whole blood provides higher sensitivity and translational feasibility for dose resolution and dose-response analysis. To further validate the utility of miR-23a-3p as a volume-independent internal normalizer, we evaluated its expression at various time points over a period of 365 days after HSCT. Ct values of miR-23a-3p in longitudinally collected blood samples from irradiated patients with leukemia were found comparable to that of blood collected from healthy volunteers (coefficient of variation, 2.15 to 3.26%), suggesting that the presence of miR-23a-3p in circulation was not confounded by leukemia load, myeloablative radiation, or after marrow reconstitution (Fig. 2F). We noted some variations in Ct values of miR-150-5p at baseline, potentially due to differences in blood cell counts, and at day +30 after transplant, perhaps due to the differences in marrow reconstitution (coefficient of variation, 4.12 to 12.9%). The amount of miR-150-5p was comparable in healthy volunteers and patients after complete remission as evaluated at day +180 and day +365.
We next investigated whether or not the analysis of both of these miRNAs was possible with a drop of blood (50 to 75 μl) collected by the finger-stick method. We recruited healthy volunteers (n = 21) for finger-stick blood collection and analyzed purified total RNA for relative expression of miR-150-5p and miR-23a-3p by qRT-PCR. We found nominal differences in the basal expression of miR-150-5p as expected in a heterogeneous human population, but without major confounding effects due to differences in age or gender (table S1 and fig. S3, A to D).
miR-RAD (miR-150-5p/miR-23a-3p assay) as a sensitive and robust radiation biodosimeter in mice in a dose range relevant for triage after radiological events
The radiation dose–dependent changes noted in ΔΔCt values of miR-150-5p expression normalized to miR-23a-3p prompted us to evaluate the dose response using a small volume of blood collected from mice exposed to various doses of TBI. Male and female C57BL/6 mice of varying ages were exposed to graded doses of 137Cs gamma rays, resulting in a dose range relevant to triage and dose estimation after radiological events. Total RNA extracted from a drop of blood collected by the submandibular method at multiple time points from the same or different animals was subjected to qRT-PCR for miR-150-5p and miR-23a-3p expression analysis. ΔΔCt values were calculated as fold change represented on a natural logarithmic scale and shown as miR-RAD response. Figure 3 (A and B) shows miR-RAD and lymphocytes depletion kinetics observed in blood collected at 24 and 48 hours from 3-month-old female mice exposed to gamma rays at 0.5 Gy increments until 4 Gy and then 2 Gy increments until 10 Gy. The observed differences in miR-RAD allowed dose resolution ±0.5 Gy in the range of 0.5 to 3.5 Gy; however, resolvability decreased at doses beyond 4 Gy. As expected, the miR-RAD response showed a strong correlation with lymphocyte and white blood cell (WBC) depletion kinetics (Pearson correlation, ρ > −0.8, P < 0.0001) (Fig. 3C). We observed a similar dose response of miR-150-5p with decreasing lymphocyte number and a correlation of miR-RAD with lymphocyte and WBC depletion in 3-month-old male mice (Fig. 3, D to F), suggesting consistent response irrespective of gender. As in humans, whole blood RNA preparations produced a steeper dose-response slope compared to those of serum (−6.67 versus −0.06, P = 0.0016), demonstrating that the analysis of whole blood was more sensitive (fig. S4, A to D).
Fig. 3. miR-RAD (miR-150-5p/miR-23a-3p assay) as a sensitive and robust radiation biodosimeter in mice in a dose range relevant for triage after radiological events.
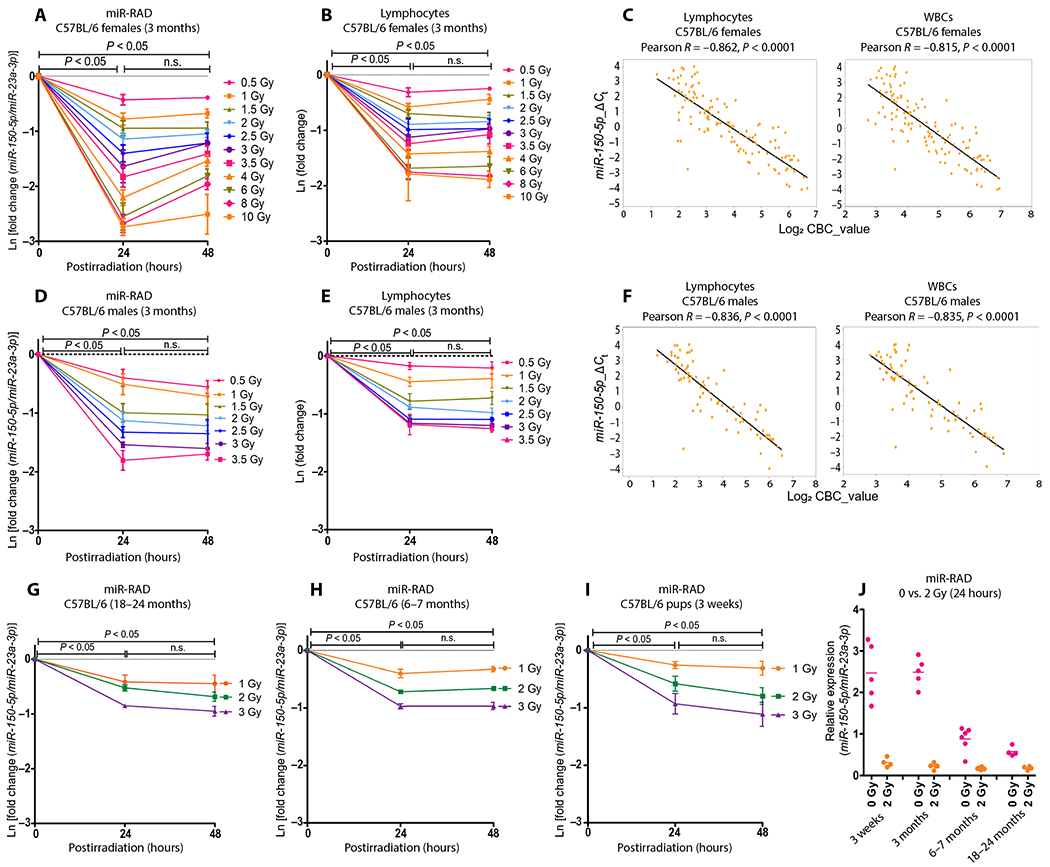
(A) Dose-time response in 3-month-old C57BL/6 female mice after total body exposure to 137Cs gamma rays (0.94 Gy/min) at a broad dose range (0.5 to 10 Gy) at 24 and 48 hours after TBI. miR-150-5p expression normalized to endogenous miR-23a-3p is plotted as fold expression on a natural logarithmic scale Ln[fold change (miR-150-5p/miR-23a-3p)] and (B) corresponding lymphocyte depletion kinetics, where the y axis is represented as natural logarithmic scale Ln(fold change). (C) Pearson correlation between miR-RAD and lymphocytes or white blood cells (WBCs), where the y axis represents ΔCt (miR-150-5p/miR-23a-3p) and the x axis represents the log2 of lymphocytes or WBC counts. (D) miR-RAD response in 3-month-old C57BL/6 male mice in the dose range 0.5 to 3.5 Gy at 24 and 48 hours after TBI, (E) corresponding lymphocyte depletion kinetics, and (F) Pearson correlation graphs between miR-RAD and lymphocytes or WBCs. (G) miR-RAD response of miR-150-5p normalized to miR-23a-3p at 1, 2, and 3 Gy doses in 18-to 24-month-old C57BL/6 (n = 4), (H) 6- to 7-month-old male and female C57BL/6 mice (n = 6), and (I) 3-week-old C57BL/6 mice (n = 4). (J) Dot plot shows the expression of miR-150-5p normalized with miR-23a-3p in mice with varying ages at baseline and 24 hours after 2 Gy TBI (n = 4 to 5). (A) and (B), (D) and (E), and (G) to (I) are mean ± SD and analyzed by two-way ANOVA with Bonferroni adjustment.
Because 2 Gy exposure has been shown to result in hematopoietic repression and is defined as a cut point for triage (27), we focused on the response in the 1 to 3 Gy range by including additional animals consisting of older (Fig. 3G), middle-aged (Fig. 3H), and pediatric (Fig. 3I) cohorts, modeling a heterogeneous population. Parallel analysis of miR-RAD response, lymphocyte depletion kinetics, and neutrophil to lymphocyte ratio (NLR) in animals of 3 weeks, 3 months, 6 to 7 months, and 18 to 24 months of age (comparable to 9-, 18- to 20-, 30- to 35-, and 55- to 80-year-old humans, respectively) at dose range (0.5 to 10 Gy or 1 to 3 Gy) and at time points (6 to 168 hours) showed robust responses (figs. S5 to S7). Irrespective of mouse age, the dose-dependent depletion observed at 24 hours stabilized at 48 and 96 hours with partial recovery at 168 hours (fig. S5, A to E). As in humans, some variations were noted in the basal values of miR-150-5p/miR-23a-3p, particularly when comparing animals of varying ages, potentially attributable to a decline in lymphocyte count with age (28). However, after exposure to an even lower dose (2 Gy as shown in Fig. 3J), signals in the exposed groups tightened, suggesting that age or other chronic conditions affecting lymphocyte number are not major confounders affecting dose response.
To evaluate the potential of the assay for dose-response analysis in immunocompromised populations, we measured the expression of miR-150-5p in athymic nude mice (NCr-nu/nu) carrying a mutation in Foxn1 (hence lacking T cells). Consistent with the finding that lymphocytes highly expressed miR-150-5p, the depletion of miR-150-5p was found to be steeper with a lower recovery in athymic nude mice in comparison to immunocompetent mice (fig. S8, A and B). In addition, the miR-RAD readings were not significantly confounded in animals exposed to bacterial endotoxin lipopolysaccharide (LPS) when tested at 48 hours after exposure (P = 0.282) (fig. S8C), although we did observe a decline in expression at 24 hours correlating with low lymphocyte count before they increase in number (29). As shown earlier, miR-150-5p expression provides a readout of functional marrow, and animals that received partial body radiation (upper versus lower body, about 60% versus 40% marrow exposure) exhibited a response correlating with the percent of marrow exposed (fig. S8D). The miR-150-5p expression in partially DNA repair–deficient (parp1−/−) mice in comparison to wild type in the same genetic background did not show notable variations at early time points after exposure to 2 Gy (24 to 96 hours) (fig. S8E), although parp1−/− mice showed poor recovery of miR-150-5p at 168 hours (P = 0.002), possibly due to DNA repair deficiency and increased apoptosis.
Utility of miR-RAD for evaluation of response after high-LET and low-LET radiation exposure in mice
The detonation of an improvised nuclear device (IND) is expected to expose victims to both high-LET (linear energy transfer) radiation, such as neutrons, and low-LET gamma rays, with varying biological effects. To evaluate the relative biological effectiveness for IND-spectrum neutrons, we directly compared the amounts of miR-150-5p in blood collected from mice at 24, 48, 96, and 168 hours after exposure to varying doses of total body gamma rays (0.5 to 8 Gy) versus IND-spectrum neutrons (0.1 to 2 Gy). Animals were exposed to neutrons with beam energies of 0.2 to 9 MeV, expected to mimic the environment about 1.5 km from the epicenter following the detonation of a Hiroshima-like explosion using the Columbia IND-spectrum Neutron Facility (CINF) (30). Parallel analysis of samples collected from animals exposed to neutrons versus gamma rays allowed comparison of dose response with time at a range of doses (Fig. 4, A and B). The response curves generated by plotting Ln[fold change (miR-150-5p/miR-23a-3p)] against dose showed an overall radiation quality dependence (fig. S9A). Exposure to even lower doses of neutrons induced a greater decrease in miR-150-5p and a greater increase in cell killing, as evident from complete blood count (CBC) analysis, compared to gamma rays (Fig. 4, C and D, and fig. S9, B to F). Head-to-head comparison of miR-RAD values in mice exposed to neutrons and gamma rays allowed us to identify the gamma dose equivalent of neutrons causing similar biological responses. miR-RAD response was evident even after 0.1 Gy neutron exposure (the lowest dose tested) and was resolvable in a range of 0.1 to 1.5 Gy. The responses in this range were comparable to that achieved with gamma rays in 0.5 to 4 Gy dose range (Fig. 4, E to H). We plotted log10[fold change (miR-RAD)] to extrapolate the relative biological effectiveness, which was in the range of 1.5 to 3.2 for 1 to 3 Gy gamma rays, with a tendency to decrease at the higher dose range.
Fig. 4. Comparison of miR-RAD and CBC in mice exposed to IND-spectrum neutrons and gamma rays.
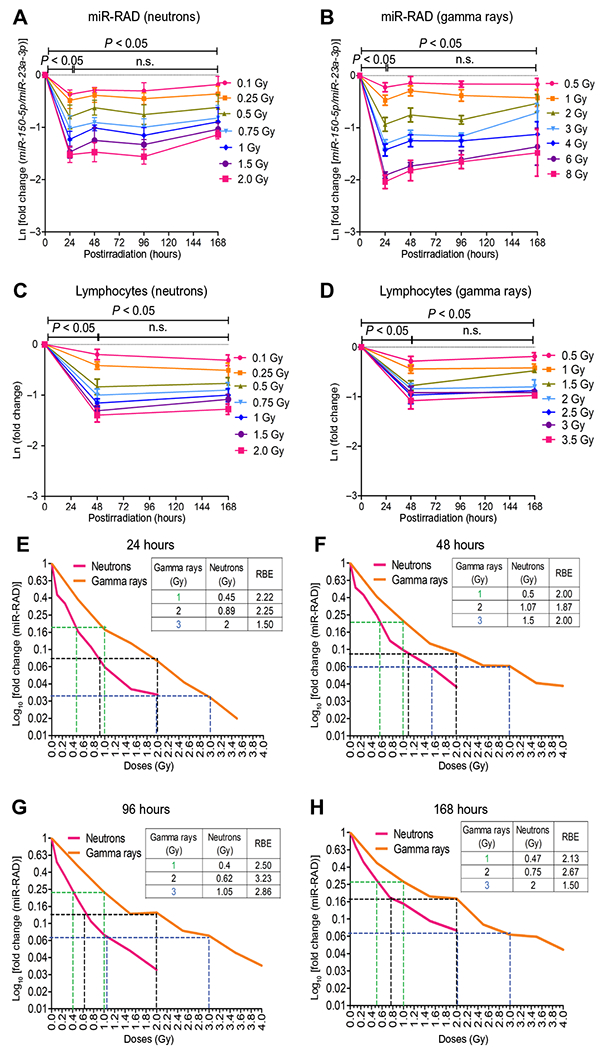
(A and B) Longitudinal evaluation of miR-150-5p normalized to miR-23a-3p at days 1 to 7 in 3-month-old C57BL/6 female mice after total body exposure to neutron (sham, 0.1, 0.25, 0.5, 0.75, 1.0, 1.5, or 2.0 Gy) or gamma rays (sham, 0.5, 1, 2, 3, 4, 6, or 8 Gy). miR-RAD response is plotted as natural logarithmic scale Ln[fold change (miR-150-5p/miR-23a-3p)]; n = 6 for neutrons or n = 5 for gamma rays. Corresponding lymphocyte depletion kinetics are shown in (C) and (D). (E to H) Relative biological effectiveness (RBE) of neutron to 1, 2, and 3 Gy gamma dose calculated at 24, 48, 96, and 168 hours plotted as log10[fold change (miR-RAD)]. (A) to (D) are mean ± SD and analyzed by two-way ANOVA with Bonferroni adjustment.
Development of dose estimation algorithm, dose reconstruction, and validation
To develop a practical biodosimetry assay that allows in vitro dose reconstruction from samples of unknown exposure, miR-RAD response calculated after varying acute single doses in mice (n = 8 to 10/dose) was used for developing algorithms by fitting in the experimental data points using the goodness of fit method. During mathematical modeling, instead of using traditional ΔΔCt values defined as 2^-(CtmiR-150-5p – CtmiR-23a-3p), we used the normalized ΔΔCt, defined as 2^-[(CtmiR-150-5p - CtmiR-23a-3p)/CtmiR-23a-3p], which reduced the mean uncertainty error at broader dose range by fold of 17.44 ± 2.30 at 24 hours, 17.77 ± 4.74 at 48 hours, 16.11 ± 3.79 at 96 hours, and 21.12 ± 3.76 at 168 hours (table S4), allowing better accuracy in dose prediction. We constructed separate plots for 24, 48, 96, and 168 hours of absorbed doses using normalized ΔΔCt, values that allowed further simplifying the mathematical relationship into a polynomial formula (Fig. 5A). Although we generated algorithms at each time point, the 48-hour algorithm best fitted to all time points except that of 168 hours. We validated our observations by conducting a single-blinded study in mice (n = 5 to 10 per dose for each time point) and extrapolated the results in the form of linear regression models (Fig. 5, B to F). The slopes showed values of 0.54 (24 hours), 1.03 (48 hours), 0.95 (96 hours), and 0.69 (168 hours), showing an overall agreement between the actual dose and the estimated dose (lack of fit, P >0.05). Combined analysis of estimated doses from all time points (24 to 168 hours) showed better accuracy in dose prediction at lower dose range (covering the 2 Gy triage cutoff), where the exposed dose differed from the estimated dose in the range of −0.5 to 0.5 Gy; however, the estimated dose deviated more at a higher dose range (4 to 10 Gy) (Fig. 5, B to G; fig. S10; and table S5). In addition, the 48- and 168-hour algorithms generated based on the response in mice after gamma ray exposure allowed estimation of the neutron dose with comparable biological effects (fig. S11).
Fig. 5. Development of dose estimation algorithm, dose reconstruction, and validation.

(A) Dose estimation algorithms developed for each time point (green, blue, magenta, and turquoise for 24, 48, 96, and 168 hours after gamma irradiation, respectively) by plotting normalized ΔΔCt data points from 3-month-old C57BL/6 female mice. (B) Linear regression plots of absorbed dose estimated versus actual doses at 24 hours, (C) 48 hours, and (D) 96 hours generated using 48-hour algorithm and (E) 168 hours using 168-hour algorithm. For each plot, dose estimated from 5 to 10 blinded samples is given on the y axis, while actual exposed dose is given on the x axis. (F) Combined plot with data pooled from 24, 48, 96, and 168 hours (20 to 40 data points per dose). Lack-of-fit test P > 0.05 and R2 > 0.8. (G) Box plots show the deviations between the actual dose and the estimated dose (combined n = 20 to 40 data points per dose). (H) ROC curves demonstrate AUC values >0.9 at each time point for actual absorbed dose >2 or >6 Gy.
We tested the performance of the miR-RAD by using receiver operating characteristic (ROC) analysis on the basis of the estimated absorbed radiation dose. The sensitivity and specificity of the assay were determined by setting cutoff values at 2 and 6 Gy. These cutoffs were previously proposed as relevant for effective clinical decision-making as the type of clinical manifestations of ARS is expected to differ on exposure to >2 Gy versus >6 Gy, and therefore, treatment is expected to differ (for example, monitoring, supportive care, growth factors, or blood transfusion) (31). The sensitivity of miR-RAD for identifying the subject exposed with 2 Gy was 95.9% (211/220), indicating high predictability of absorbed dose above 2 Gy, and the specificity was 89% (160/180), indicating accuracy of the miR-RAD assay at ≤2 Gy. Sensitivity at 6 Gy was 55% (33/60) with specificity of 94.4% (321/340), indicating high probability of accurate dose reconstruction when exposed to ≤6 Gy. ROC curves generated for doses >2 and >6 Gy showed that the area under the curve (AUC) were 0.97 and 0.93, respectively (fig. S12). Assessment of miR-RAD responses across time points from 24 to 168 hours independently showed AUC greater than 0.90 at both 2 and 6 Gy cutoff (Fig. 5H and table S6). In summary, the data show the capability of miR-RAD assay in distinguishing 2 Gy exposed versus unexposed individuals and dose estimation in a dose range relevant for triage and potential clinical decision-making.
Comparison of dose response after acute single versus fractionated irradiation in mice and humans
To test miR-RAD adaptability for dose estimation after acute exposure in humans, we evaluated the dose response in mice after fractionated versus acute single exposure and further compared with the changes observed in mice with that in humans after fractionated radiation. Twelve-week-old female C57BL/6 mice were subjected to TBI following a 2 Gy BID regimen for 3 days (total cumulative dose of 12 Gy), mimicking a myeloablative conditioning regimen in patients as part of preparation for HSCT. A drop of blood was collected at baseline and at days 1, 2, 4, 6, and 7 (reference 0 hours to first fraction, schema in Fig. 6, top), corresponding to the time points collected from patients. In addition to the 2 Gy BID exposure group, we included another cohort of mice that received 1 Gy BID exposure (cumulative dose of 6 Gy) for evaluation of dose response and kinetics. Comparative analysis of changes in miR-150-5p normalized to miR-23a-3p measured (unconverted ΔΔCt values) at time points after exposure to various single doses of irradiation, 1 Gy and 2 Gy BID fractionated irradiation in mice, 2 Gy BID fractionated irradiation in patients with leukemia, and baselines in healthy volunteers is shown in Fig. 6A. As expected, due to partial recovery by sublethal DNA damage repair between fractions, the depletion kinetics of miR-150-5p normalized to miR-23a-3p with cumulative fractionated irradiation was lower in comparison to single doses. For example, ΔΔCt values in 2 Gy single-dosed mice were comparable to that in mice receiving 1 Gy BID fractionated doses to a cumulative dose of 6 Gy, and ΔΔCt values in 3 and 4 Gy single-dosed mice were similar to 2 Gy BID fractionated dose mice that received a cumulative dose of 12 Gy. Direct comparison of ΔΔCt values in mice and humans showed a conserved pattern of depletion of miR-150-5p, albeit with variations in kinetics potentially attributable to differences in radiation sensitivity or to differences in body size and kinetics of hematopoiesis and bone marrow reconstitution (32). Next, to estimate the single-dose equivalents of cumulative fractionated irradiation in mice and humans, we used a dose reconstruction algorithm developed for retrospective estimation of acute dose based on the normalized ΔΔCt values in mice at 48 hours (Fig. 6B). The estimated single-dose equivalent of 2 Gy BID fractionated exposure in mice and humans at various time points with fold variations is compiled in Fig. 6C. We computed the estimated single-dose equivalent of 12 Gy fractionated irradiation as 4.63 ± 0.77 Gy and 3.76 ± 1.47 Gy for mouse and 7.32 ± 1.83 Gy and 7.38 ± 1.67 Gy for humans on days 4 and 6, respectively. Baseline corrections considering the differences in baseline values in healthy volunteers (0.40 ± 0.32 Gy) and patients with leukemia (0.90 ± 0.85 Gy) will need to be factored in for accurate dose estimation; however, our bridging studies in mice show the potential adaptability of miR-RAD for dose estimation in humans after single-dose exposure.
Fig. 6. Comparison of dose response after acute single versus fractionated irradiation in mice and humans.
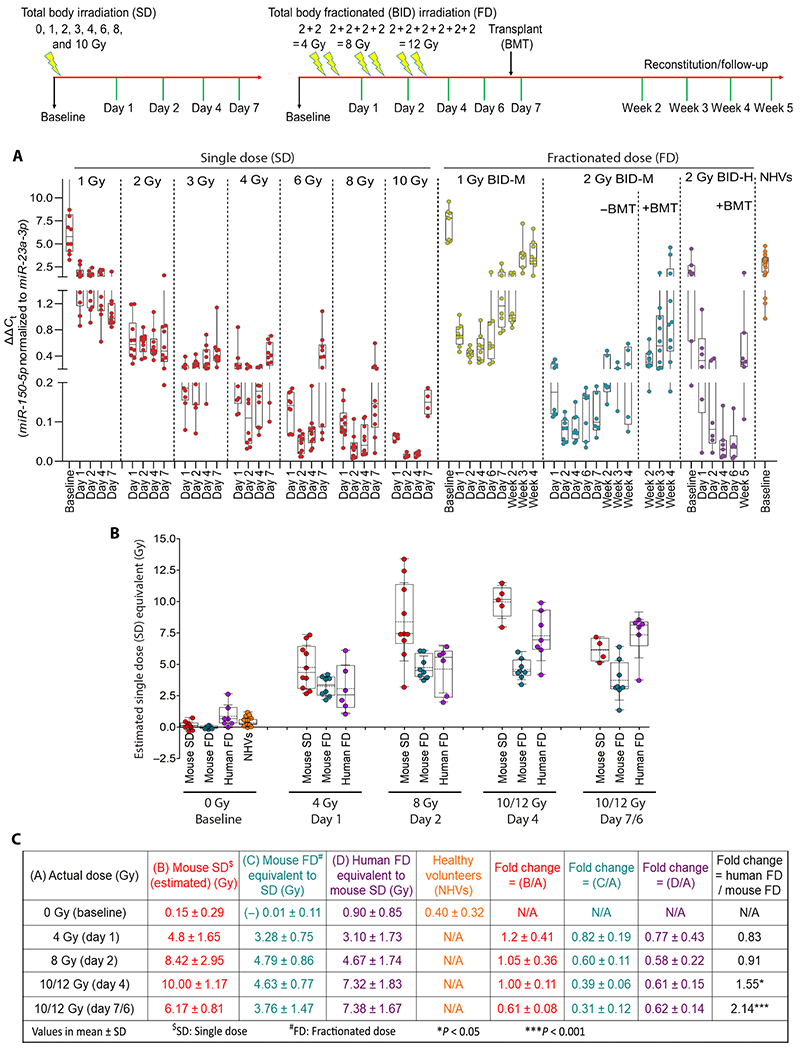
Top: Schema represents single dose (SD) and fractionated irradiation (FD) and sampling time points. (A) Dot-whisker plot represents ΔΔCt values (miR-150-5p normalized to miR-23a-3p) comparing baseline, dose, and time-dependent changes after single dose (n = 5 to 10) and fractionated irradiation 1 Gy BID (n = 8); 2 Gy BID (n = 3 to 8); 2 Gy BID + BMT (Transplant) (n = 8 to 10) in mice; and 2 Gy BID + BMT in patients with leukemia (n = 6 to 7) and NHVs (n = 21). (B) Dot-whisker plot represents the estimated mouse single-dose equivalent dose for various groups using 48-hour dose reconstruction algorithm developed. (C) Summary of estimated single-dose equivalents of fractionated irradiation in various groups with fold changes and kinetics.
DISCUSSION
The peripheral blood cell–based methods such as the dicentric chromosome assay (DCA) and cytokinesis-block micronucleus (CBMN) assay developed decades ago are labor intensive and time consuming; hence, they are not optimal for high-throughput practical radiation biodosimetry (33–38). Over the years, several radiation dose–responsive metabolites, lipids, proteins, and mRNAs have been identified as putative biomarkers, however, with less than appreciable sensitivity and robustness across a broad dose range, and the responses reported for many are limited to a narrow analytical range (10, 11, 39–42). The major impediment to productization of a marker panel–based radiation biodosimetry assay is probably the absence or the inadequacy of internal normalizers that are critical for in vitro dose reconstruction. The radiation dose–dependent depletion of blood cells, the primary source of both responders and normalizers, contributes to variations in dose estimation. Readings can vary due to genetics and underlying conditions, contributing to differences in DNA damage response and cell cycle checkpoint activation, resulting in changes in the expression of markers detected in peripheral blood. Thus, despite the investment of a large amount of resources over a decade and more, the translational utility of the above panels for developing a practical radiation biodosimetry test meeting the overwhelming demand is not evident (43, 44).
Using complementary screening platforms such as nanoString nCounter assay, qRT-PCR, and next-generation sequencing–based approaches, we and others have identified the dose-dependent depletion of serum or plasma expression of miR-150-5p in rodent and NHP models (21, 24, 25). Expanding on these studies, molecular profiling of blood cell subsets performed in this study allowed us to identify the primary source of circulating miR-150-5p as T and B lymphocytes, which are among the most radiation-sensitive cells. The translational utility of these findings was enhanced by our discovery of miR-23a-3p as a robust and abundant internal normalizer. Lungs, being highly vascularized organ with high miR-23a-3p expression, passively release miR-23a-3p into the circulation and provide an opportunity for internal normalization of radiation-sensitive markers. The constant release of miR-23a-3p from nonblood cells into circulation is not substantially affected by age or chronic conditions, nor it is affected by acute radiation exposure or injury to various sensitive organs (21, 24, 45). Although there are reports on the involvement of miR-23a-3p in pathogenesis (46, 47), our study showed abundance of cell-free circulating miR-23a-3p in blood that was not responsive to radiation injury, allowing a volume-independent internal normalization. A direct comparison of miRNAs purified from serum and serum-derived exosomes showed that the majority of miR-23a-3p detected in circulation was nonexosomal and therefore likely to be released as protein-bound. Use of such a normalizer with biochemical properties similar to that of the analyte contributes to the practicality and robustness of a diagnostic assay as this minimizes errors during sampling and processing.
The characteristics and performance of the miR-RAD assay we developed in mice align with the guidelines and stipulations set forth by the FDA for developing a radiation biodosimetry assay for triage, clinical decision-making, and follow-up after radiological events (44, 48). The assay is capable of gauging absorbed radiation dose at a lower dose range of 0.5 to 3.5 Gy with ±0.5 Gy resolution, ±2 Gy until 8 Gy gamma rays, and its equivalent of IND-spectrum neutrons. The dose reconstruction algorithms developed based on miR-150-5p depletion normalized with miR-23a-3p in mouse models are capable of estimating the absorbed dose at various time points relevant for triage, particularly until day 7 and follow-up during the recovery phase. Comparison of miR-RAD response after acute versus fractionated irradiation in mice and that in patients with leukemia exposed to fractionated radiation provides reasonable assurance that miR-RAD is translatable for dose estimation in humans after acute exposure. In mouse models, we have shown that miR-RAD is capable of distinguishing 2 Gy exposed from controls in a heterogeneous population at 6 to 168 hours after exposure. Radiobiological studies have shown that a total body exposure of ≥2 Gy results in hematological repression (49, 50). Therefore, 2 Gy resolvability at and after 24 hours is emphasized by federal and international agencies as the cut point for triage in radiological events (27). Our assay may be particularly useful for screening victims exposed in the range of 1 to 3 Gy who do not show overt symptoms during early times after exposure. As lymphocytes are highly sensitive to radiation, the miR-RAD signals converge even with low doses that naturally reduce the error rate in dose estimation. The sensitivity, resolvability, and favorable kinetics of the miRNA marker–based assay, together with clinical signs, may help in the management of victims involved in radiation accidents.
In conclusion, the data presented here demonstrate proof of concept for development of a practical biodosimetry assay with potentially broad clinical applications. If validated, this may help to provide qualitative information and quantitative output and facilitate triage and clinical decision-making at different stages after mass casualty radiological events. Sampling can be minimally invasive using finger-stick collection, stabilized in a generic lysis reagent without the need for refrigeration, and shipped at ambient conditions. The assay can be adapted for an automated high-throughput platform using a multitube matrix coupled with a simpler dose reconstruction module. Beyond its utility in nuclear and radiological disaster preparedness and management, the sensitivity of the assay should allow clinical decision-making and therapy guiding in radiotherapy patients where overdosing and underdosing are of major concerns.
The assay is neither approved nor ready for human use and there are limitations to our analysis. These include the lack of analysis of specimens collected from higher animal species, such as NHPs, that are closer to humans. Although miR-150-5p and miR-23a-3p are evolutionarily conserved (with 100% sequence similarity between primates and rodents) (25), and dose response has been shown in specimens collected from patients on radiotherapy, variations in assay sensitivity and differences in kinetics of radiation-sensitive marker response between rodents and primates are expected. A clinical trial directly evaluating responses in healthy humans exposed to varying acute single doses of radiation is ethically not possible. Therefore, pivotal validation and additional bridging studies using NHPs following the FDA animal rule (48, 51) are required for potential future regulatory approval. Another limitation of our study is the lack of dose-response analysis after mixed neutron and gamma radiation exposure. The neutron irradiation used in the current study contained about 18% gamma rays (30); however, the exposure from nuclear events is predicted to contain a higher percentage of gamma rays. Furthermore, the interactions between different types of damage elicited by low-LET versus high-LET radiations could be modifying the dose responses. Thus, follow-up studies modeling multiple mixed neutron and gamma ray radiation exposures in NHPs are also needed for fine-tuning and calibration of the two-miRNA–based radiation biodosimetry assay.
MATERIALS AND METHODS
Study design
The goal of the study was to develop a blood miRNA-based diagnostic assay for the retrospective evaluation of absorbed ionizing radiation with a view to triage and clinical decision-making after mass casualty radiological or nuclear events. This study used male and female mice of varying ages categorized into pediatric, young adult, middle-aged, and geriatric groups. We used an acceptable testing window with longitudinal follow-ups (from 6 to 168 hours after exposure with 24, 48, and 96 hours as intermediary time points) for detection of acute effects. Serum or whole blood (one to two drops) collected directly into a generic lysis reagent were used for total RNA extraction and analysis using commercially available reagents and machineries. In addition to total body exposure models, we used different testing scenarios such as partial body irradiation (PBI), immunocompromised, and immune-challenged, and DNA repair–deficient mouse models to assess robustness of the assay to various confounders. More than 2000 samples collected from mice were processed for dose-response analyses, dose reconstruction, and assay validation. Sample size, groups, and replicates for each animal study are summarized in table S2. Blinded studies were performed to validate the accuracy of prediction algorithms, where the principal investigator concealed sample identification from other investigators while processing the samples. Values were then slotted into the respective equations for calculating estimated doses and were eventually compared to the actual radiation doses. Specimens collected from human patients on radiotherapy and healthy volunteers with informed consent were used to establish assay feasibility in humans, including archived serum specimens from patients enrolled in a completed clinical trial (NCT01521039) and whole blood and serum collected from another recruiting clinical trial (NCT02122081). Patients underwent radiation-based myeloablative conditioning before HSCT and blood samples were collected before, after, or concurrently with fractionated radiation, as shown in the schema in Fig. 2 and table S3. The feasibility of the finger-stick blood test was evaluated using specimens collected from healthy volunteers.
Animal experiments
Animal studies were carried out in compliance with the guidelines approved by Institutional Animal Care and Use Committee (IACUC) of The Ohio State University (OSU) under protocol #2011A00000029. Mus musculus (C57BL/6, NCr-nu/nu, 129S1, and 129S-parp1tm1Zqw/J) of 6 to 10 weeks purchased from Charles River or The Jackson Laboratory were acclimatized for 1 to 3 weeks before the start of experiments. Naturally aged C57BL/6 mice housed in ULAR (University Laboratory Animal Resources) at OSU and pups (3 weeks old) derived by in-house breeding were used to evaluate responses in geriatric and pediatric populations, respectively. Mice were maintained under a 12-hour light/dark cycle with standard NIH31 diet and water ad libitum.
Radiation exposure
Animals, stratified according to age and gender, were placed in a RadDisk (Braintree Scientific Inc.) for TBI. Animals were exposed to TBI doses using a Gammacell 40 Exactor (137Cesium source, Best Theratronics) at a dose rate of 94 cGy/min. The dosimetry of Gammacell was done annually by Best Theratronics using Fricke dosimetry system, and the accuracy reported was within 3% of uncertainty. The LD50/30 for 10-week-old C57BL/6J mice under our laboratory conditions with minimal supportive care was determined as 7.7 Gy. Nonanesthetized animals were exposed to different doses of TBI, and sham-exposed animals were used as controls. Three-month-old female and male C57BL/6 mice were exposed to a low-dose range (0 to 4 Gy) in 0.5 Gy increments and to high-dose of 6 Gy (sublethal, partially ablative dose), 8 Gy (LD70/30), or 10 Gy (supralethal dose). Blood collection time points were 6, 24, 48, 96, and 168 hours after exposure. The geriatric (18 to 24 months old), middle-aged (6 to 7 months old), and pediatric (3 weeks old) groups of mice were exposed to 1, 2, or 3 Gy TBI. A group of 3-month-old female C57BL/6 mice was treated (intraperitoneal injection) with LPS (5 mg/kg; Escherichia coli 055:B5, Sigma-Aldrich) with or without radiation to mimic individuals with acute inflammatory responses. For region-targeted PBI, mice anesthetized using ketamine/xylazine and partial body were exposed to 160-kV x-rays using RS2000 Biological Research Irradiator (Rad Source Technologies). nanoDot Optically Stimulated Luminescence (OSL) detectors (Landauer) were used for radiation dosimetry under partial body exposure conditions. Custom-cut lead (Pb) shields were used to protect partial body, allowing targeted dose delivery to upper body (UBI; ~60% marrow), lower body (LBI; ~40% marrow), or the whole thorax. Animals were flipped halfway to minimize variations in dose delivery due to attenuation.
For high-LET irradiation, we used a neutron irradiator developed by the Radiological Research Accelerator Facility (RARAF) at CINF mimicking the neutron spectra from an IND at relevant distances (for example, 1.5 km) from the epicenter of an event (30). Three-month-old C57BL/6 females were exposed to varying neutron doses (sham, 0.1, 0.25, 0.5, 0.75, 1.0, 1.5, or 2.0 Gy). Dosimetry and mouse irradiation were performed as described previously (30, 52, 53). Dose rates of neutrons were as follows: 0.6 Gy/hour (0.1 Gy), 1.5 Gy/hour (0.25, 0.5, and 1 Gy), or 2.25 Gy/hour (0.75, 1.5, and 2 Gy) with 18% gamma photons. After irradiation at RARAF, animals were transported and maintained at the OSU animal quarantine facility until completion of a 7-day blood collection.
For studies bridging mice and humans, C57BL/6 female 3-month-old mice were subjected to fractionated TBI (gamma rays) following 1 Gy BID (with 6-hour gap, 3 days, total of 6 Gy) or 2 Gy BID (3 days, total of 12 Gy), with unirradiated mice used as baseline controls. Two batches of mice were used in each group to reduce repeated bleeding from the same animal. A batch of mice receiving 2 Gy BID was subjected to transplantation on day 7. Recipient mice received whole bone marrow cell suspension (three parts) mixed with spleen cells (one part) via tail vein injection (~20 million cells per mouse). Bone marrow and spleen cells were obtained from sex- and age-matched donors. Sampling was done on days 0, 1, 2, 4, 6, and 7 (before bone marrow/spleen cells infusion) and after transplant weeks 2, 3, and 4 for the analysis for miR-150-5p and miR-23a-3p.
Mouse blood and tissue sampling for RNA analysis
The submandibular bleeding method was primarily used for collecting blood drops (25 to 75 μl) directly into 5 volume or more of QIAzol lysis reagent and used for miRNA isolation with an miRNeasy kit (QIAGEN). For serum collection, ~200 μl of blood was collected into gold BD Microtainer blood collection tubes (BD Diagnostics) and allowed to clot for 30 min, then centrifuged at 10,000g for 10 min at room temperature on a tabletop centrifuge. Serum samples were stored at −80°C until use. For tissue miRNA expression analysis, C57BL/6 mice were euthanized by asphyxiation with CO2 followed by cervical dislocation. Transcardial perfusion with phosphate-buffered saline (PBS) was performed until organs (liver, heart, lung, kidney, small intestine, large intestine, brain, and spleen) were cleared of excess blood. RNA was extracted using a QIAGEN RNA isolation kit.
Human blood sampling
Blood specimens from healthy volunteers and patients with AML, ALL, MPAL, or MDS were obtained after consent under protocols approved by the Institutional Review Board (IRB) (protocols 2016C0032, 2011C0055, 2014C0014, and 2015C0124) at OSU. Specimens from patients with leukemia enrolled in clinical trials NCT02122081 (OSU-13219) and NCT01521039 (OSU-11002) were used. Blood was collected into BD SST II Plus Vacutainer tubes for serum separation and into ACD (acid citrate dextrose) vacutainer tubes for whole blood RNA isolation. Samples were collected before the start of the conditioning regimen (baseline); on days −5, −4, −2, and −1 (before stem cell infusion); and thereafter at days +30, +180, and +365 after allogeneic HSCT. Serum was separated by centrifugation at 650g for 10 min at room temperature and aliquoted as 250 μl in multiple vials. For finger-prick blood collection, fingers were cleaned using an alcohol swab and then punctured using a sterile and disposable lancet. Blood drops (75 to 150 μl) were collected into EDTA tubes (Microvette 100K3E) by gently squeezing the finger. About 500 μl of QIAzol lysis reagent was added to the blood collection tubes with 75 to 150 μl of blood, and half of the lysate (250 to 300 μl per patient/healthy volunteer) was processed for RNA isolation.
RNA isolation, TaqMan advanced cDNA synthesis, and qRT-PCR
RNA from whole blood or serum was extracted using a commercially available miRNA purification kit (primarily miRNeasy Kit, QIAGEN) following the manufacturer’s protocol. A portion of the purified/concentrated RNA was used for complementary DNA (cDNA) synthesis and preamplification following the manufacturer’s protocol (TaqMan Advanced miRNA cDNA Synthesis Kit, Thermo Fisher Scientific Inc.). cDNA was quantitatively analyzed using TaqMan Advanced probes, Hsa-miR-150-5p-FAM (assay ID: 477918_mir-5′-UCUCCCAACCCUUGUACCAGUG-3′) and Hsa-miR-23a-3p-FAM (assay ID: 478532_mir-5′-AUCACAUUGCCAGGGAUUUCC-3′), on a 7900HT Fast Real-Time PCR system (Applied Biosystems) or QuantStudio 3 Real-Time PCR system (Applied Biosystems). The epMotion automated liquid handling system (epMotion 5075) was used for routine pipetting of cDNA samples into 384-well plates for real-time PCR to eliminate manual pipetting errors and maximize the efficiency and reproducibility. The results were analyzed according to [2^-(Ct of miR-150-5p – Ct of miR-23a-3p)] method.
Blood cell sorting by magnetic immunobeads and miRNA expression profiling
Human blood leukopaks (American Red Cross) were processed for isolation of peripheral blood mononuclear cells (PBMCs), red blood cells (RBCs), and platelet enriched plasma using Ficoll (Ficoll-Paque PLUS, GE Healthcare, #17-1440-02) density gradient method as described earlier (54). The top plasma layer was washed twice with PBS and sorted for platelets using human CD61 magnetic beads (MACS Miltenyi Biotec, #130-051-101). The middle PBMC layer was washed twice and mixed with RBC lysis buffer for removal of residual erythrocytes. Washed PBMCs were selected for CD3+ (T lymphocytes; MACS Miltenyi Biotec, #130-050-101), CD19+ (B lymphocytes; MACS Miltenyi Biotec, #130-050-301), and CD56+ (NK cells; MACS Miltenyi Biotec, #130-050-401) using the magnetic bead column separation method. The negatively selected cells thus obtained were further separated for CD34+ cells using a Diamond CD34 Isolation Kit for human HSCs (MACS Miltenyi Biotec, #130-094-531) as described by the manufacturer. Five nanograms (whole blood and RBC) or 100 ng (PBMCs and cell subsets) of total RNA isolated using an miRNeasy kit (QIAGEN) was used for human miRNA profiling on nCounter assay (NanoString Technologies Inc.). Obtained miRNA profiling data were normalized to the mean of the top 100 miRNAs using nSolver software. miRNA profiling was done in triplicate using total RNA from blood cell subsets purified from three different leukopaks and serum from three healthy volunteers.
Statistical analysis
nCounter human miRNA panel (NanoString Technologies) was used for miRNA expression analysis following the manufacturer’s protocol. The output RCC files were used for biological normalization of miRNAs using geometric means of top 100 miRNAs on nSolver software (NanoString Technologies). For qRT-PCR data, fold changes of miR-150-5p (normalized to miR-23a-3p) were validated by analysis of variance (ANOVA) or by a mixed-effect model compared to baseline control, with multiplicities adjusted by Bonferroni or Dunnett’s method. Graphs were generated using Prism 5 software (GraphPad). The figures for miR-RAD kinetics were constructed by transforming the fold change into natural log (Ln) as the y axis against the postirradiation time points (x axis). The normality assumption for the difference between whole blood and serum ΔΔCt values from cohort of mice and human volunteers was confirmed by using Kolmogorov-Smirnov method (P > 0.15) and unpaired t tests to the difference between irradiated and nonirradiated groups. Slopes of whole blood and serum were modeled and compared by using ANOVA to demonstrate the assay sensitivity. The association between the miR-150-5p/miR-23a-3p and CBC values was assessed using Pearson correlation method and visualized by plotting ΔCt (CtmiR-150-5p – CtmiR-23a-3p) against lymphocytes/WBC count using Minitab v.17 software (Minitab Inc.). Linear regression modeling was used to evaluate the reliability of dose prediction using normalized ΔΔCt values of miR-150-5p and miR-23a-3p for each time point. Intercept of zero and slope of 1 indicate good agreement between calculated and actual doses. Model fit was validated by lack-of-fit tests. Regression analysis was conducted using SAS v.14 (SAS Inc.), and respective figures were generated in Minitab v.17. All values are expressed as mean ± SD. P < 0.05 was considered statistically significant, and n.s. as not significant. We performed ROC curve analysis to determine the specificity and sensitivity of the miR-RAD assay in predicting exposure dose >0.5, >2.0, and >6.0 Gy. All ROC analyses were generated using SAS 9.4 (SAS Inc.).
Supplementary Material
Fig. S1. miR-150-5p and miR-23a-3p expression in human blood cell subsets, serum, and mouse tissues.
Fig. S2. Normalizer miR-23a-3p is unaltered after exposure to high doses of whole thorax irradiation in mice and is secreted in nonexosomal fashion.
Fig. S3. Finger-stick assay benchmarking according to gender and age at baseline in humans.
Fig. S4. Comparison of dose response and sensitivity in mouse blood versus serum.
Fig. S5. Longitudinal evaluation of dose response in pediatric, young-adult, middle-aged, and geriatric mice.
Fig. S6. CBC in mice of different ages after exposure to a broader dose range.
Fig. S7. Comparison of miR-RAD with lymphocyte depletion and NLR.
Fig. S8. Dose response in immunocompetent, immunocompromised, immune-challenged, partial body exposure, and DNA repair–deficient mice.
Fig. S9. miR-RAD with CBC analysis after neutron irradiation in mice.
Fig. S10. An overview of the accuracy of miR-RAD in a broad dose range.
Fig. S11. Relative biological effectiveness of neutron with miR-RAD endpoint.
Fig. S12. ROC curves for absorbed dose >2 or >6 Gy using miR-RAD.
Table S1. Baseline Ct values of miR-150-5p and miR-23a-3p in finger-stick blood from healthy volunteers and venous blood from patients with leukemia.
Table S2. Overview of mouse samples including number of animals and sampling time points.
Table S3. Overview of human samples used for dose-response evaluation and controls.
Table S4. Comparison of uncertainty error with ΔΔCt versus normalized ΔΔCt in mice.
Table S5. Tabulation of actual dose versus estimated dose in mice.
Table S6. Tabulation of assay sensitivity and specificity at 0.5, 2, and 6 Gy cutoff.
Data file S1. Primary data.
Acknowledgments:
We thank patients and volunteers for donating blood; veterinary staff at the Ohio State University Laboratory Animal Resources (OSU-ULAR) for animal care; and S. Khan, I. Manring, S. Camilli, and N. R. Jacob for technical or editorial assistance. We thank Y. Efebera, S. Devine, R. Garzon, P. Ranganathan, and P. Kumchala for assistance in retrieving archived specimens from clinical trial NCT01521039. Neutron exposure was a paid service performed at CINF under the supervision of G. Garty and B. Ponnaiya.
Funding:
This study was supported, in part, by an Investigator-Initiated Research Award (IIRA) from United States Defense for Health Affairs through the Peer Reviewed Medical Research Program (award no. W81XWH-15-2-0054), National Aeronautics and Space Administration (NASA) (award no. 80NSSC18K1691), and Oakridge Associated Universities (ORAU)–Directed Research and Development fund to N.K.J. and, in part, with intramural funds from OSUCCC. Multiple shared resources used at the Institution were supported by National Cancer Institute (NCI) grant P30CA016058.
Footnotes
Competing interests: N.K.J. and A.C. are coinventors on U.S. patents 9,890,428 and 10,407,726 (titled “miRNA biomarkers for radiation biodosimetry”) and U.S. patents 10,131,952 and 10,513,736 (titled “miRNA biomarkers for monitoring bone marrow reconstitution”), issued to the Ohio State Innovation Foundation (OSIF). These patents are under license from OSIF to an outside party. All other authors declare that they have no competing interests.
Data and materials availability: microRNA profiles from this study are available in the GEO (Gene Expression Omnibus) database repository under accession number GSE116880. Raw data from figures are provided in data file S1.
REFERENCES AND NOTES
- 1.Coleman CN, Koerner JF, Biodosimetry: Medicine, science, and systems to support the medical decision-maker following a large scale nuclear or radiation incident. Radiat. Prot. Dosimetry 172, 38–46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coleman CN, Sullivan JM, Bader JL, Murrain-Hill P, Koerner JF, Garrett AL, Weinstock DM, Case C Jr., Hrdina C, Adams SA, Whitcomb RC, Graeden E, Shankman R, Lant T, Maidment BW, Hatchett RC, Public health and medical preparedness for a nuclear detonation: The nuclear incident medical enterprise. Health Phys. 108, 149–160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swartz HM, Williams BB, Flood AB, Overview of the principles and practice of biodosimetry. Radiat. Environ. Biophys 53, 221–232 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dainiak N, Medical management of acute radiation syndrome and associated infections in a high-casualty incident. J. Radiat. Res 59, ii54–ii64 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waselenko JK, MacVittie TJ, Blakely WF, Pesik N, Wiley AL, Dickerson WE, Tsu H, Confer DL, Coleman CN, Seed T, Lowry P, Armitage JO, Dainiak N; Strategic National Stockpile Radiation Working Group, Medical management of the acute radiation syndrome: Recommendations of the Strategic National Stockpile Radiation Working Group. Ann. Intern. Med 140, 1037–1051 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Dörr H, Meineke V, Acute radiation syndrome caused by accidental radiation exposure—Therapeutic principles. BMC Med. 9, 126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan JM, Prasanna PG, Grace MB, Wathen LK, Wallace RL, Koerner JF, Coleman CN, Assessment of biodosimetry methods for a mass-casualty radiological incident: Medical response and management considerations. Health Phys. 105, 540–554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blakely WF, Ossetrova NI, Whitnall MH, Sandgren DJ, Krivokrysenko VI, Shakhov A, Feinstein E, Multiple parameter radiation injury assessment using a nonhuman primate radiation model-biodosimetry applications. Health Phys. 98, 153–159 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Laiakis EC, Pannkuk EL, Chauthe SK, Wang YW, Lian M, Mak TD, Barker CA, Astarita G, Fornace AJ Jr., A Serum small molecule biosignature of radiation exposure from total body irradiated patients. J. Proteome Res 16, 3805–3815 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucas J, Dressman HK, Suchindran S, Nakamura M, Chao NJ, Himburg H, Minor K, Phillips G, Ross J, Abedi M, Terbrueggen R, Chute JP, A translatable predictor of human radiation exposure. PLOS ONE 9, e107897 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ossetrova NI, Blakely WF, Multiple blood-proteins approach for early-response exposure assessment using an in vivo murine radiation model. Int. J. Radiat. Biol 85, 837–850 (2009). [PubMed] [Google Scholar]
- 12.Pannkuk EL, Laiakis EC, Singh VK, Fornace AJ, Lipidomic signatures of nonhuman primates with radiation-induced hematopoietic syndrome. Sci. Rep 7, 9777 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redon CE, Nakamura AJ, Gouliaeva K, Rahman A, Blakely WF, Bonner WM, The use of gamma-H2AX as a biodosimeter for total-body radiation exposure in non-human primates. PLOS ONE 5, e15544 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tucker JD, Grever WE, Joiner MC, Konski AA, Thomas RA, Smolinski JM, Divine GW, Auner GW, Gene expression-based detection of radiation exposure in mice after treatment with granulocyte colony-stimulating factor and lipopolysaccharide. Radiat. Res 177, 209–219 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ Jr., Gonzalez FJ, Idle JR, Radiation metabolomics. 2. Dose- and time-dependent urinary excretion of deaminated purines and pyrimidines after sublethal gamma-radiation exposure in mice. Radiat. Res 172, 42–57 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul S, Kleiman NJ, Amundson SA, Transcriptomic responses in mouse blood during the first week after in vivo gamma irradiation. Sci. Rep 9, 18364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ossetrova NI, Stanton P, Krasnopolsky K, Ismail M, Doreswamy A, Hieber KP, Biomarkers for radiation biodosimetry and injury assessment after mixed-field (neutron and gamma) radiation in the mouse total-body irradiation model. Health Phys. 115, 727–742 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Port M, Majewski M, Herodin F, Valente M, Drouet M, Forcheron F, Tichy A, Sirak I, Zavrelova A, Malkova A, Becker BV, Veit DA, Waldeck S, Badie C, O’Brien G, Christiansen H, Wichmann J, Eder M, Beutel G, Vachelova J, Doucha-Senf S, Abend M, Validating baboon ex vivo and in vivo radiation-related gene expression with corresponding human data. Radiat. Res 189, 389–398 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Bartel DP, MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Hammond SM, An overview of microRNAs. Adv. Drug Deliv. Rev 87, 3–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Acharya SS, Fendler W, Watson J, Hamilton A, Pan Y, Gaudiano E, Moskwa P, Bhanja P, Saha S, Guha C, Parmar K, Chowdhury D, Serum microRNAs are early indicators of survival after radiation-induced hematopoietic injury. Sci. Transl. Med 7, 287ra269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aryankalayil MJ, Chopra S, Makinde A, Eke I, Levin J, Shankavaram U, MacMillan L, Vanpouille-Box C, Demaria S, Coleman CN, Microarray analysis of miRNA expression profiles following whole body irradiation in a mouse model. Biomarkers 23, 689–703 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fendler W, Malachowska B, Meghani K, Konstantinopoulos PA, Guha C, Singh VK, Chowdhury D, Evolutionarily conserved serum microRNAs predict radiation-induced fatality in nonhuman primates. Sci. Transl. Med 9, eaal2408 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacob NK, Cooley JV, Yee TN, Jacob J, Alder H, Wickramasinghe P, Maclean KH, Chakravarti A, Identification of sensitive serum microRNA biomarkers for radiation biodosimetry. PLOS ONE 8, e57603 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menon N, Rogers CJ, Lukaszewicz AI, Axtelle J, Yadav M, Song F, Chakravarti A, Jacob NK, Detection of acute radiation sickness: A feasibility study in non-human primates circulating miRNAs for triage in radiological events. PLOS ONE 11, e0167333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao F, Liu P, Narayanan J, Yang M, Fish BL, Liu Y, Liang M, Jacobs ER, Medhora M, Changes in miRNA in the lung and whole blood after whole thorax irradiation in rats. Sci. Rep 7, 44132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiCarlo AL, Maher C, Hick JL, Hanfling D, Dainiak N, Chao N, Bader JL, Coleman CN, Weinstock DM, Radiation injury after a nuclear detonation: Medical consequences and the need for scarce resources allocation. Disaster Med. Public Health Prep 5, S32–S44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moehrle BM, Geiger H, Aging of hematopoietic stem cells: DNA damage and mutations? Exp. Hematol 44, 895–901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pecaut MJ, Gridley DS, Radiation and secondary immune response to lipopolysaccharide. In Vivo 22, 423–434 (2008). [PubMed] [Google Scholar]
- 30.Xu Y, Randers-Pehrson G, Turner HC, Marino SA, Geard CR, Brenner DJ, Garty G, Accelerator-based biological irradiation facility simulating neutron exposure from an improvised nuclear device. Radiat. Res 184, 404–410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coleman CN, Weinstock DM, Casagrande R, Hick JL, Bader JL, Chang F, Nemhauser JB, Knebel AR, Triage and treatment tools for use in a scarce resources-crisis standards of care setting after a nuclear detonation. Disaster Med. Public Health Prep 5 (suppl. 1), S111–S121 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Sykes SM, Scadden DT, Modeling human hematopoietic stem cell biology in the mouse. Semin. Hematol 50, 92–100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ainsbury EA, Bakhanova E, Barquinero JF, Brai M, Chumak V, Correcher V, Darroudi F, Fattibene P, Gruel G, Guclu I, Horn S, Jaworska A, Kulka U, Lindholm C, Lloyd D, Longo A, Marrale M, Gil OM, Oestreicher U, Pajic J, Rakic B, Romm H, Trompier F, Veronese I, Voisin P, Vral A, Whitehouse CA, Wieser A, Woda C, Wojcik A, Rothkamm K, Review of retrospective dosimetry techniques for external ionising radiation exposures. Radiat. Prot. Dosimetry 147, 573–592 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Depuydt J, Baeyens A, Barnard S, Beinke C, Benedek A, Beukes P, Buraczewska I, Darroudi F, De Sanctis S, Dominguez I, Monteiro Gil O, Hadjidekova V, Kis E, Kulka U, Lista F, Lumniczky K, M’Kacher R, Moquet J, Obreja D, Oestreicher U, Pajic J, Pastor N, Popova L, Regalbuto E, Ricoul M, Sabatier L, Slabbert J, Sommer S, Testa A, Thierens H, Wojcik A, Vral A, RENEB intercomparison exercises analyzing micronuclei (Cytokinesis-block Micronucleus Assay). Int. J. Radiat. Biol 93, 36–47 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Dolphin GW, Bolton D, Humphreys DL, Speight DL, Stradling GN, Biological and physical dosimetry after a radiation accident. Nature 227, 165 (1970). [DOI] [PubMed] [Google Scholar]
- 36.Fenech M, Morley AA, Measurement of micronuclei in lymphocytes. Mutat. Res 147, 29–36 (1985). [DOI] [PubMed] [Google Scholar]
- 37.Rothkamm K, Beinke C, Romm H, Badie C, Balagurunathan Y, Barnard S, Bernard N, Boulay-Greene H, Brengues M, De Amicis A, De Sanctis S, Greither R, Herodin F, Jones A, Kabacik S, Knie T, Kulka U, Lista F, Martigne P, Missel A, Moquet J, Oestreicher U, Peinnequin A, Poyot T, Roessler U, Scherthan H, Terbrueggen B, Thierens H, Valente M, Vral A, Zenhausern F, Meineke V, Braselmann H, Abend M, Comparison of established and emerging biodosimetry assays. Radiat. Res 180, 111–119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulka U, Wojcik A, Di Giorgio M, Wilkins R, Suto Y, Jang S, Quing-Jie L, Jiaxiang L, Ainsbury E, Woda C, Roy L, Li C, Lloyd D, Carr Z, Biodosimetry and biodosimetry networks for managing radiation emergency. Radiat. Prot. Dosimetry 182, 128–138 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Lacombe J, Sima C, Amundson SA, Zenhausern F, Candidate gene biodosimetry markers of exposure to external ionizing radiation in human blood: A systematic review. PLOS ONE 13, e0198851 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu T-P, Hsu Y-Y, Lai L-C, Tsai M-H, Chuang EY, Identification of gene expression biomarkers for predicting radiation exposure. Sci. Rep 4, 6293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pannkuk EL, Fornace AJ Jr., Laiakis EC, Metabolomic applications in radiation biodosimetry: Exploring radiation effects through small molecules. Int. J. Radiat. Biol 93, 1151–1176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ Jr., Gonzalez FJ, Idle JR, Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat. Res 170, 1–14 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Homer MJ, Raulli R, DiCarlo-Cohen AL, Esker J, Hrdina C, Maidment BW, Moyer B, Rios C, Macchiarini F, Prasanna PG, Wathen L, United States Department of Health and Human Services biodosimetry and radiological/nuclear medical countermeasure programs. Radiat. Prot. Dosimetry 171, 85–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sproull MT, Camphausen KA, Koblentz GD, Biodosimetry: A future tool for medical management of radiological emergencies. Health Secur. 15, 599–610 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, Dahlsveen IK, Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 59, S1–S6 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Guo W, Wang H, Yang Y, Guo S, Zhang W, Liu Y, Yi X, Ma J, Zhao T, Liu L, Jian Z, Liu L, Wang G, Gao T, Shi Q, Li C, Down-regulated miR-23a contributes to the metastasis of cutaneous melanoma by promoting autophagy. Theranostics 7, 2231–2249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hatzl S, Geiger O, Kuepper MK, Caraffini V, Seime T, Furlan T, Nussbaumer E, Wieser R, Pichler M, Scheideler M, Nowek K, Jongen-Lavrencic M, Quehenberger F, Wölfler A, Troppmair J, Sill H, Zebisch A, Increased expression of miR-23a mediates a loss of expression in the RAF kinase inhibitor protein RKIP. Cancer Res. 76, 3644–3654 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.U.S. Food and Drug Administration, Radiation Biodosimetry Medical Countermeasure Devices Guidance for Industry and Food and Drug Administration Staff (U.S. Food and Drug Administration, 2016); https://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/UCM427866.pdf. [Google Scholar]
- 49.Plett PA, Sampson CH, Chua HL, Joshi M, Booth C, Gough A, Johnson CS, Katz BP, Farese AM, Parker J, MacVittie TJ, Orschell CM, Establishing a murine model of the hematopoietic syndrome of the acute radiation syndrome. Health Phys. 103, 343–355 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh VK, Newman VL, Berg AN, MacVittie TJ, Animal models for acute radiation syndrome drug discovery. Expert Opin. Drug Discovery 10, 497–517 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Singh VK, Newman VL, Romaine PL, Hauer-Jensen M, Pollard HB, Use of biomarkers for assessing radiation injury and efficacy of countermeasures. Expert Rev. Mol. Diagn 16, 65–81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Y, Randers-Pehrson G, Marino SA, Garty G, Harken A, Brenner DJ, Broad energy range neutron spectroscopy using a liquid scintillator and a proportional counter: Application to a neutron spectrum similar to that from an improvised nuclear device. Nucl. Instrum. Methods Phys. Res. A 794, 234–239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laiakis EC, Wang YW, Young EF, Harken AD, Xu Y, Smilenov L, Garty GY, Brenner DJ, Fornace AJ Jr., Metabolic dysregulation after neutron exposures expected from an improvised nuclear device. Radiat. Res 188, 21–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yadav M, Song F, Huang J, Chakravarti A, Jacob NK, Ocimum flavone Orientin as a countermeasure for thrombocytopenia. Sci. Rep 8, 5075 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. miR-150-5p and miR-23a-3p expression in human blood cell subsets, serum, and mouse tissues.
Fig. S2. Normalizer miR-23a-3p is unaltered after exposure to high doses of whole thorax irradiation in mice and is secreted in nonexosomal fashion.
Fig. S3. Finger-stick assay benchmarking according to gender and age at baseline in humans.
Fig. S4. Comparison of dose response and sensitivity in mouse blood versus serum.
Fig. S5. Longitudinal evaluation of dose response in pediatric, young-adult, middle-aged, and geriatric mice.
Fig. S6. CBC in mice of different ages after exposure to a broader dose range.
Fig. S7. Comparison of miR-RAD with lymphocyte depletion and NLR.
Fig. S8. Dose response in immunocompetent, immunocompromised, immune-challenged, partial body exposure, and DNA repair–deficient mice.
Fig. S9. miR-RAD with CBC analysis after neutron irradiation in mice.
Fig. S10. An overview of the accuracy of miR-RAD in a broad dose range.
Fig. S11. Relative biological effectiveness of neutron with miR-RAD endpoint.
Fig. S12. ROC curves for absorbed dose >2 or >6 Gy using miR-RAD.
Table S1. Baseline Ct values of miR-150-5p and miR-23a-3p in finger-stick blood from healthy volunteers and venous blood from patients with leukemia.
Table S2. Overview of mouse samples including number of animals and sampling time points.
Table S3. Overview of human samples used for dose-response evaluation and controls.
Table S4. Comparison of uncertainty error with ΔΔCt versus normalized ΔΔCt in mice.
Table S5. Tabulation of actual dose versus estimated dose in mice.
Table S6. Tabulation of assay sensitivity and specificity at 0.5, 2, and 6 Gy cutoff.
Data file S1. Primary data.


