Abstract
Tristetraprolin (TTP) is a nonclassical CCCH zinc finger protein that regulates inflammation. TTP targets AU-rich RNA sequences of cytokine mRNAs forming a TTP/mRNA complex. This complex is then degraded, switching off the inflammatory response. Cadmium, a known carcinogen, triggers proinflammatory effects, and there is evidence that Cd increases TTP expression in cells, suggesting that Zn-TTP may be a target for cadmium toxicity. We sought to determine whether Cd exchanges with Zn in the TTP active site and measure the effect of RNA binding on this exchange. A construct of TTP that contains the two CCCH domains (TTP-2D) was employed to investigate these interactions. A spin-filter ICP-MS experiment to quantify the metal that is bound to the ZF after metal exchange was performed, and it was determined that Cd exchanges with Zn in Zn2-TTP-2D and that Zn exchanges with Cd in Cd2-TTP-2D. A native ESI-MS experiment to identify the metal-ZF complexes formed after metal exchange was performed, and M-TTP-2D complexes with singular and double metal exchange were observed. Metal exchange was measured in both the absence and presence of TTP’s partner RNA, with retention of RNA binding. These data show that Cd can exchange with Zn in TTP without affecting function.
Graphical Abstract
INTRODUCTION
Zinc finger (ZF) proteins belong to a large family of proteins that utilize a combination of cysteine and/or histidine ligands to bind zinc (Zn) in a tetrahedral geometry.1–3 Upon zinc binding, ZFs fold into three-dimensional structures that function in recognition of DNA, RNA, or other proteins (Figure 1).1–7 ZFs are involved in a variety of functions, including transcriptional and translational regulation, and the role of the ZF is typically to bind to the DNA or RNA.1,2,8 ZFs are ubiquitous, particularly in eukaryotes, where it is estimated that 3–10% of all proteins are ZFs.2,5 There are multiple classes of ZFs which are defined based upon domain sequence, the number of cysteine and histidine residues (e.g., CCCC, CCCH, CCHH),2,6 and at least 13 different classes of ZFs are known.1,9,10
Figure 1.
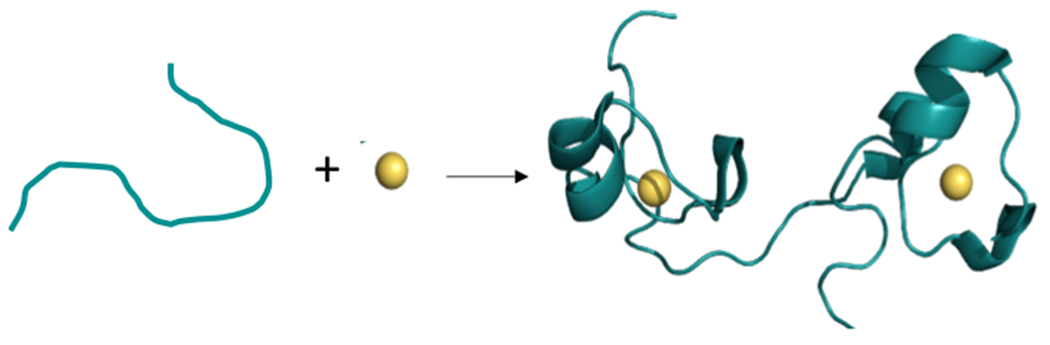
Cartoon of the effect of zinc binding to a ZF protein on structure. In the absence of zinc (left) the protein is unstructured, in the presence of zinc (right) the protein adopts secondary structure. ZF structure shown here is of Tis11d, a CCCH type ZF. (PDB = 1RGO, PyMOL).
Although zinc is the native cofactor for ZFs, the thiol rich nature of their sequences makes them targets for exogenous metals, including iron, copper, nickel, lead, cadmium, and gold.11–24 In some cases, these metals are upregulated during certain biological conditions (Fe, Cu), and in other cases, these metals are introduced exogenously from the environment (e.g., Ni, Pb, Cd) or delivered as therapeutics (e.g., Au). There is a body of biochemical work aimed at understanding how exogenous metals target ZFs.11–25 Much of this work has focused on determining how these exogenous metals directly bind to specific types of ZFs and elucidating the structural and functional consequences.11–25 In these studies, both short peptides that mirror the native ZF protein domain sequences and full length ZF proteins have been utilized, and experiments have involved direct titrations of metals with these ZF peptides and proteins in the apo-form.11–25 Collectively, these studies have provided fundamental insight into how exogenous metals can directly bind to apo-ZFs and affect structure and activity.11–25 In the cellular environment, zinc is typically coordinated to ZFs (Zn-ZF), and it is important to understand how exogenous metals interact with ZFs that have zinc coordinated. However, there are far fewer studies that examine how exogenous metals interact with Zn-ZFs.
One class of ZFs that has received increased attention in recent years are the Cys3His or CCCH type-ZFs.2,26 Proteins from this ZF class play roles in RNA regulation,26 and one of the best studied is tristetraprolin (TTP) which regulates inflammation.4,8,27 TTP contains two CCCH domains that, when zinc is bound, folds into a three-dimensional structure composed of loops and turns.28 TTP turns off the inflammatory response by regulating cytokines, e.g., TNF-α. Specifically, TTP binds to AU-rich sequences present in the 3′ untranslated region of cytokine mRNAs leading to degradation which shuts off the inflammatory response.4,27,29 Misregulation of TTP has been linked to a number of diseases, including rheumatoid arthritis (chronic inflammation), sepsis, and cancer.2,27,30–32 Factors that lead to misregulation of TTP are not well understood.
One potential factor that may promote misregulation of TTP is cadmium (Cd). Cd is a known environmental carcinogen, with major sources of exposure including waste from the industrial production of batteries and plastics, car exhaust, and cigarette smoke.33–42 The biological targets of Cd are not clear; however, cysteine rich proteins are potential targets because cadmium a “soft acid” and cysteine thiolates are a “soft bases.”43,44 There is published evidence for Cd binding to cysteine-rich proteins, including studies with several types of ZFs (e.g., TFIIIA, SP1, and XPA), as well as metallothioneins.44–56 Cd is known to have proinflammatory effects triggering inflammatory response and there is evidence that TTP levels are increased upon Cd exposure, suggesting that TTP may be a target of Cd toxicity.57–60
Our laboratory has previously investigated direct binding of Cd to a functional construct of TTP that contains the two CCCH ZF domains (TTP-2D).18 We reported that Cd binds to TTP-2D with nanomolar affinity forming a folded protein that binds to AU-rich RNA, much like its Zn-TTP-2D counterpart, albeit with greater sequence selectivity.18 These data inform on the ability of Cd to bind to apo-TTP; however, how Cd interacts with Zn-TTP-2D is not known. Cd may exchange with Zn and/or bind adventitiously and nonspecifically to TTP. Similarly, the effect of RNA binding on the interaction Cd with Zn-TTP (i.e., Zn-TTP/RNA) is not known.
Measuring the interactions and possible exchange of Cd with Zn in Zn-TTP-2D is challenging. The broad range of spectroscopic methods afforded to metalloproteins with open shell metal cofactors are not available to study Cd and Zn because of their d10 electron count. We have recently reported the application of two new approaches to measure the interaction of exogenous metals with TTP.12,25 The first approach is a spin filter/ICP-MS approach. In this approach, the exchange of the exogenous metal is tracked and quantified using ICP-MS.12 This method allows for the amount of metal associated with the protein to be quantified but does not provide information on the metal-protein complex. The second approach is a native ESI-MS approach in which the exogenous metal is titrated with Zn-TTP-2D in the absence and presence of RNA, and the metal-TTP complexes formed are identified.25 Although the ESI-MS method is qualitative–it identifies metal-TTP complexes that are formed– it complements the ICP-MS method that quantifies the metal content. We applied these two distinct approaches, along with a functional RNA binding assay, to determine whether Cd exchanges with Zn in Zn-TTP-2D and with Zn in TTP-2D bound to RNA (Zn-TTP-2D/RNA). From these approaches we were able to quantify the metals that associate with TTP-2D under different conditions, identify metal-ZF complexes that are formed during metal exchange, and elucidate the effects of metal exchange on RNA binding.
Materials and Methods.
Zinc chloride (ZnCl2), cadmium chloride (CdCl2), cadmium acetate dihydrate (Cd(OAc)2. 2H2O), ampicillin sodium salt, bovine serum albumin, 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES), urea, SP sepharose resin, trifluoracetic acid (TFA) (HPLC grade), ammonium acetate, and nonlabeled RNA oligonucleotide (5′UUUAUUUAUUU3′) were obtained from Sigma-Aldrich. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was purchased from Research Products International. BL21-DE3 competent cells were purchased from New England Biolabs. Luria–Bertani Lennox broth (LB) and dithiothreitol (DTT) were acquired from American Bio. Sodium chloride (NaCl), nitric acid (HNO3) (trace metal grade), and acetonitrile (HPLC grade) were obtained from Fisher Scientific. Zinc, cadmium, germanium, and scandium ICP-MS standards as well as zinc acetate dihydrate (Zn(OAc)2·2H2O) were purchased from Fluka analytical. Rhodium ICP-MS standard was obtained from VWR analytical. ICP-MS tuning solution was acquired from Agilent. MES was purchased from Amersco. EDTA-free protease inhibitor tablets were obtained from Thermo Scientific. A 0.5 mL portion of 3 kDa MWCO spin filters were purchased from (Amicon). 3′-fluorescein (F)-labeled PAGE-purified, deprotected, and desalted RNA oligonucleotide (5′UUUAUUUAUUU3′-F) was purchased from Dharmacon Research Inc. All metal free conical tubes used for ICP-MS sample preparation were purchased from VWR. PVDF 0.22 μm steriflip filters were purchased from Millipore.
TTP-2D Expression and Purification.
A pET-15b vector containing the TTP-2D gene fragment encoding for two zinc binding domains (SRYKTELCRTYSESGRCRYGAKCQ-FAHGLGELRQANRHPKYKTELCHKFYLQGRCPYGSRCHFIHNPTEDLAL) that was previously reported by our laboratory was utilized to obtain TTP-2D.11 Briefly, the TTP-2D vector was transformed into E. coli BL21-(DE3) competent cells and grown overnight in Luria–Bertani (LB) medium containing 100 μg/mL ampicillin at 37 °C. The following day, 15 mL of the overnight culture was added to 1 L of LB medium containing 100 μg/mL ampicillin and grown to mid log phase (≈ OD600 of 0.6 to 0.8) at 37 °C. Protein expression was induced by adding 1 mM isopropyl β-d1-thiogalactopyranoside (IPTG). The cultures were allowed to grow for 4 h post induction after which they were centrifuged at 7800g for 20 min at 4 °C. The cell pellets were then harvested and stored at −20 °C. Cell pellets were resuspended in 8 M urea and 10 mM MES pH 6 containing 10 mM dithiothreitol (DTT) and one EDTA-free protease inhibitor mini tablet. The resuspended cells were placed on ice and lysed using a Fisher Scientific Sonic Dismembrator model 100. Cellular debris were removed by centrifugation (17710 xg for 20 min at 4 °C). The bacterial supernatant was then poured into a SP-sepharose gravity column and rocked for 1 h at room temperature. A stepwise gradient from 0 to 2 M NaCl in 4 M urea and 10 mM MES pH 6 was applied. TTP-2D eluted in 4 M urea, 600 mM NaCl, and 10 mM MES pH 6 containing 10 mM DTT. Following elution, 25 mM DTT was added to the protein which was then incubated in a 56 °C water bath for 2 h to reduce any remaining disulfide bonds. The protein was then filtered through a steriflip 0.22 μm filter and purified using reverse-phase HPLC with a Symmetry C18 300 column on an Agilent 1200 series LC system using an acetonitrile:water gradient, in 0.1% trifluoroacetic acid. Apo-TTP-2D eluted at 32% acetonitrile and was lyophilized to dryness in a Coy anaerobic chamber (97% nitrogen and 3% hydrogen atmosphere) using a SpeedVac concentrator (Thermo Scientific). The purified protein was verified by SDS-PAGE (>95% pure) and MALDI-MS. All remaining protein manipulations were performed anaerobically to avoid cysteine oxidation.
ICP-MS ANALYSIS
For ICP-MS analysis, samples were diluted using trace metal grade HNO3 to a final concentration of 6% nitric acid. The samples were heated in an oven at 80 °C overnight, cooled to room temperature, and analyzed by ICP-MS. The concentrations of cadmium and zinc in protein samples were determined by injecting samples into an Agilent 7700x ICP-MS instrument (Agilent Technologies, Santa Clara, CA, USA) using the same instrument run conditions as previously reported.12 Cd111 and Zn66 concentrations in the samples were derived from an external calibration curve generated by a series of dilutions of atomic absorption standard (Fluka Analytical) prepared in the same 6% HNO3 matrix as the samples. Data analysis was performed using Agilent’s Mass Hunter software. All buffers are routinely analyzed for metal content using ICP-MS. Because Milli-Q water is employed to prepare all buffers, we do not typically observe the presence of metals above the lower limit of quantitation. If we do observe detectable levels of metal in the buffer, the buffer is discarded and remade.
Zn2-TTP-2D + CdCl2 and Cd2-TTP-2D + ZnCl2: Spin-filter ICP-MS Analysis.
A 30 μM solution of Zn2-TTP-2D in HEPES buffer (100 mM HEPES, 100 mM NaCl at pH 7.5) was prepared by adding 2 mol equiv of ZnCl2 to 30 μM apo-TTP-2D under anaerobic conditions. To this solution, either 2 or 4 mol equiv of CdCl2 was added using a stock solution of 15 mM CdCl2 in filtered deionized water. The protein solutions were then allowed to equilibrate by shaking at 300 rpm for 5 min at room temperature. The proteins were then transferred to 3 kDa MWCO spin filters (Amicon 500 Ultra −0.5 mL) and spun at 14000g for 30 min in a 40° fixed angle table-top centrifuge (Fisher Scientific) at 25 °C, per the manufacturer’s guidelines, to remove adventitiously bound metal ions. Spin filtration was repeated three times, after which the protein concentrate was recovered by inverting the spin filter into a fresh collection tube and centrifuging at 1000g for 2 min at room temperature. The resultant protein was diluted to 500 μL, and an aliquot was taken to measure protein concentration. The protein was unfolded by titration with HCl to reach a pH of 2, after which the concentration measured via UV–visible spectroscopy (A276; e = 8520 M−1cm−1)12 The metal content was measured via ICP-MS. To determine the percent metal associated with the protein, the measured concentration of each metal was divided by the measured concentration of protein and multiplied by 100.
The analogous experiment with Cd2-TTP-2D + ZnCl2 was performed using this same method. In addition to these protein samples, the following metal and protein stocks were also quantified by ICP-MS and serve as controls: 200 μM ZnCl2, 200 μM CdCl2, 30 μM apo-TTP-2D, 30 μM Zn (II)2-TTP-2D, and 30 μM Cd (II)2-TTP-2D. All measurements were performed in triplicate.
Zn2-TTP-2D + Cd(OAc)2-2H2O and Cd2-TTP-2D + Zn(OAc)2·2H2O: Native ESI-MS.
Zn2-TTP-2D samples (500 μL) were prepared by the addition of 2 mol equiv of Zn acetate dihydrate (Zn(OAc)2)·2H2O) to 10 μM apo-TTP-2D in 10 mM ammonium acetate (NH4OAc) buffer at pH 6.9 under anaerobic conditions. The samples were equilibrated for 5 min via shaking at 300 rpm and applied to an Amicon ultra 0.5 mL, 3 kDa MWCO centrifugation filter to remove any adventitious metal ions. The concentrated samples were then recovered by inverting each concentrator and centrifuging at 1000 g for 2 min at 25 °C into fresh collection tubes. Once centrifuged, NH4OAc buffer was added to each collection tube to bring the total sample volume to 500 μL. The protein concentration was measured independently by measuring unfolded apo-TTP-2D via UV–visible spectroscopy (A276; ε = 8520 M−1cm−1).12 Cd2-TTP-2D samples were prepared analogously. For metal exchange experiments, Cd acetate dihydrate (Cd(OAc)2·2H2O) (1 and 2 mol equiv) was added to Zn2-TTP-2D and Zn(OAc)2·2H2O(1 and 2 mol equiv) was added to Cd2-TTP-2D and allowed to equilibrate for 10 min. All samples were directly infused at a flow rate of 500–1000 nL/min into a Waters Synapt G2S mass spectrometer (Waters) with a nano-ESI interface coupled with a 15 μm diameter spray tip (Silicatip, New Objective, Inc., Woburn, MA, USA). The carrier solution was ammonium acetate (NH4OAc) at pH 6.9. The instrument parameters used for native analysis were as follows: source temperature: 50 C; capillary voltage, 2.0 kV; sample cone, 30 V; source offset, 0 V; trap collision energy, 4 V; trap gas flow, 2 mL/min; helium cell gas flow, 180 mL/min. Sodium iodide was used to calibrate over the range of m/z 100–3000. Ten mM Ammonium acetate and 1:1:1:1 water:methanol:acetonitrile:isopropanol rinses were used between each infusion. 6% HNO3 was used to rinse the syringe between injections. All mass spectra data were processed using Masslynx 4.1 software (Waters).
Simulations.
All m/z simulations were performed by enviPat Web 2.4.1 The theoretical chemical formula of TTP-2D is C374H575N117O105S6. The parameters used for simulations were: type, Gaussian; threshold, 0.01; fraction, 0.1; resolution, 50–70 ppm.61
Functional Effects of Cd Reactivity: Zn2-TTP-2D/RNA + CdCl2 and Cd2-TTP-2D/RNA + ZnCl2, Fluorescence Anisotropy.
The effect of Cd and Zn on Zn2-TTP-2D binding to the RNA target AAAUAAAUAAA and Cd2-TTP-2D binding to the RNA target AAAUAAAUAAA, respectively, was measured by fluorescence anisotropy (FA). All FA experiments were performed on an ISS K2 spectrofluorometer, configured in the L format. Upon the basis of full excitation and emission scans of the fluorescently labeled RNA probe, all FA experiments were performed using an excitation wavelength of 495 nm/2 nm and an emission wavelength of 523 nm/2 nm. Spectrosil far-UV quartz window fluorescence cuvettes (Strana Cells) were utilized for all measurements. For the experiments, the RNA probe (UUUAUUUAUUU-F; F=fluorescein) was suspended in 200 mM HEPES, 100 mM NaCl, 0.05 mg/mL bovine serum albumin (to avoid protein adherence to the cuvette) at pH 7.5. In a typical experiment, a solution of 10 nM UUUAUUUAUUU-F was titrated with M2-TTP-2D (M = Zn or Cd) to saturation, followed by the addition of MCl2 (M = Cd or Zn (II))(2, 4, 6, 8, and 10 mol equiv) and the fluorescence anisotropy (r) was recorded.
Zn2-TTP-2D/RNA + Cd(OAc)2·2H2O and Cd2-TTP-2D/RNA + Zn(OAc)2·2H2O: Native ESI-MS.
The Zn2-TTP-2D/UUUAUUUAUUU RNA complex was generated by adding 1 mol equiv of RNA oligomer (sequence 5′-UUUAUUUAUUU-3′, M.W.: 3352.0 Da, a custom oligomer from Sigma-Aldrich) to 10 μM Zn2-TTP-2D in 10 mM NH4OAc buffer at pH 6.9. The ESI-MS of the complex was obtained via direct infusion of the protein/RNA complex (500–1000 nL/min) into a Waters Synapt G2S mass spectrometer. Subsequently, 1 and 2 mol equiv of Cd(OAc)2·2H2O were then applied to the Zn2-TTP-2D/RNA complex and measured via ESI-MS using direct infusion. The analogous experiments were performed with Cd2-TTP-2D/UUUAUUUAUUU RNA plus Zn (OAc)2·2H2O. All samples were directly infused at a flow rate of 500–1000 nL/min into a Waters Synapt G2S mass spectrometer (Waters) with a nano-ESI interface coupled with a 30μm diameter spray tip (Silicatip, New Objective, Inc., Woburn, MA, USA). The instrument parameters used for native analysis were as follows: source temperature, 50 C; capillary voltage, 2.0 kV; sample cone, 30 V; source offset, 0 V; trap collision energy, 4 V; trap gas flow, 2 mL/min; helium cell gas flow, 180 mL/min. Sodium iodide was used to calibrate over the range of m/z 100–3000. Ten mM ammonium acetate and 1:1:1:1 water:methanol:acetonitrile:isopropanol rinses were used between each infusion. HNO3 (6%) was used to rinse the syringe between injections. All mass spectra were processed using Masslynx software 4.1 (Waters).
RESULTS
Preparation of Zn2-TTP-2D.
The construct of TTP used for these studies, TTP-2D, contains the two CCCH ZF domains. We have previously reported that when isolated in the apo-form, apo-TTP-2D binds zinc, folds and functions (binds RNA).11 TTP-2D is highly soluble, and for this work, we overexpressed the construct in E. coli and isolated it in the apo-state. Zn2-TTP-2D was prepared by adding 2 equiv of ZnCl2 to obtain the folded, functional Zn2-TTP-2D construct.
Exchange of Cd with Zn in Zn2-TTP-2D Measured by ICP-MS.
To determine whether Cd exchanges with Zn in Zn2-TTP-2D, a spin-filter ICP-MS assay developed in our laboratory was utilized.12 In this assay, the metal of interest is added to the metal-protein complex (e.g., Cd + Zn2-TTP-2D) at various concentrations, after which the protein is applied to a molecular weight cut off (MWCO) filter which separates the protein bound metal from nonprotein bound metal complexes. The identity and quantity of the metals that are isolated with the protein are measured by ICP-MS and the protein is quantified by UV–visible spectroscopy. The metal:ZF stoichiometry is then determined by relating the total metal concentration to the total protein concentration. (Figure 2).25 To measure Cd exchange with Zn-TTP-2D, either 2 or 4 equiv of Cd was added to Zn-TTP-2D. These stoichiometries were chosen because we had previously reported upper limit Kds for Zn and Cd binding directly to TTP in the low nM to pM regime and the addition of these equivalents should promote metal exchange.18 In the first experiment, 2 mol equiv of CdCl2 was added to Zn2-TTP-2D. The sample was allowed to equilibrate for 5 min at room temperature and then applied to a 3 kDa MWCO spin filter (3X). The isolated protein was analyzed for metal identity and content by ICP-MS. The analogous experiment with 4 mol equiv of CdCl2 was also performed. Cd exchange with Zn was observed in both experiments, with 52% of the isolated protein identified as Cd bound after two equivalents of Cd had been added and 74% after four equivalents added. (Table 1).
Figure 2.
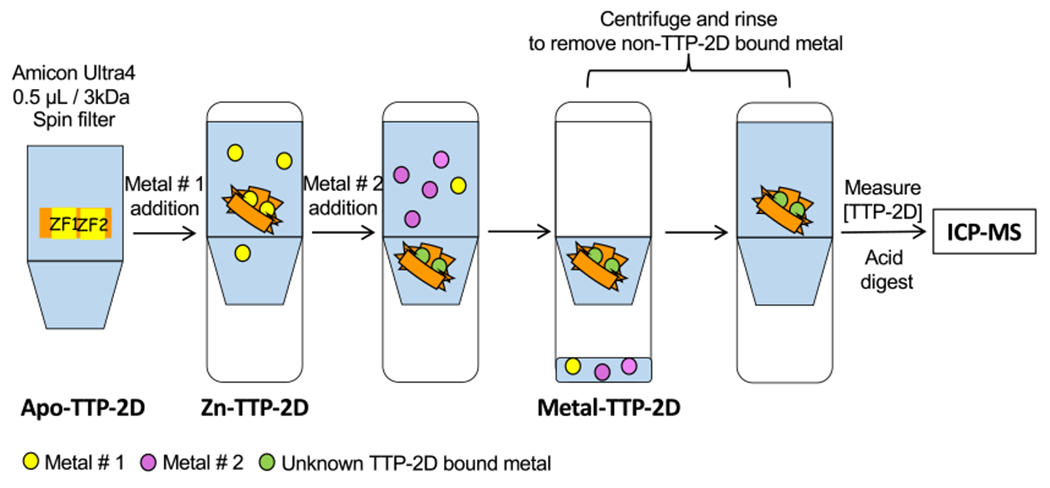
Schematic of the competitive metal binding TTP-2D spin filter method.
Table 1.
| Metal Equivalents Added | Zn2-TTP-2D + CdCl2 | Cd2-TTP-2D + ZnCl2 |
|---|---|---|
| 2 | 52 ± 0% Cd | 32 ± 0% Zn |
| 4 | 74% ± 4% Cd | 44% ± 1% Zn |
Exchange of Zn with Cd in Cd2-TTP-2D Measured by ICP-MS.
Zn supplementation is one of several therapeutic approaches used to minimize Cd toxicity.62 Therefore, in addition to determining whether Cd exchanges with Zn in Zn2-TTP-2D, we sought to determine if Zn exchanges with Cd2-TTP-2D. Following the spin filter-ICP-MS approach (vide supra), it was determined that when 2 equiv of Zn were added to Cd2-TTP-2D, 32% of the isolated protein contained Zn, and when 4 equiv were added, 44% of the isolated protein contained Zn (Table 1). These data support the exchange of Zn for Cd in TTP-2D.
Native ESI-MS to Identify M-TTP-2D Complexes Formed upon Exchange of Cd with Zn2-TTP-2D.
To identify the metal-TTP complexes that can form upon exchange of Cd with Zn, native-electrospray ionization mass spectrometry (native ESI-MS) was utilized. Native ESI-MS is gentle enough to detect the metal bound form of the ZF making it amenable to study this exchange.25,63–65 Initial studies focused obtaining the native ESI-MS of apo-TTP-2D. Here, a major peak with an m/z = 1430.8699 (z = 6+) was observed, which matches that previously reported at 1430.3560 (z = 6+).25 We then obtained the spectrum of Zn2-TTP-2D and observed a major peak at m/z = 1452.5217 (z = 6+), matching that previously reported for Zn2-TTP-2D (Figure 3a).25 With these data in hand, we performed experiments in which 1 and 2 equiv of Cd(OAc)2·2H2O were titrated with Zn2-TTP-2D and the ZF products were identified by native ESI-MS (Figure 3). Following the addition of 1 equiv of Cd(OAc)2·2H2O to Zn2-TTP-2D, peaks that best fit to Cd1Zn1-TTP-2D m/z at 1460.3499 (calc.1460.3377; z = 6+), and to Cd2-TTP-2D m/z at 1468.3495 (calc. 1468.3317; z = 6+) were observed in addition to Zn2-TTP-2D (Figure 3b). These peak assignments were confirmed via simulation (Figure 4). Upon the addition of 2 mol equiv of Cd(OAc)2·2H2O, the peaks that correspond to Zn2-TTP-2D and Cd1Zn1-TTP-2D were significantly reduced in intensity, and the peak that corresponds to Cd2-TTP-2D dominated the spectrum (Figure 3c). Taken together, these data are consistent with Cd displacing either a single or both Zn cofactors in TTP-2D and provide evidence that Cd exchanges with Zn in the Zn2-TTP-2D active site. The observation of the Cd1Zn1-TTP-2D species is of particular interest. We have previously reported that each domain of TTP-2D binds to Zn with similar affinity,11 and therefore one might expect to observe only Zn2-TTP-2D or Cd2-TTP-2D in the ESI-MS. We note that the affinities reported to Zn binding to TTP-2D were derived from a competition experiment with Co loaded TTP-2D, and the experiment performed here was an exchange of Cd with Zn loaded TTP-2D.
Figure 3.
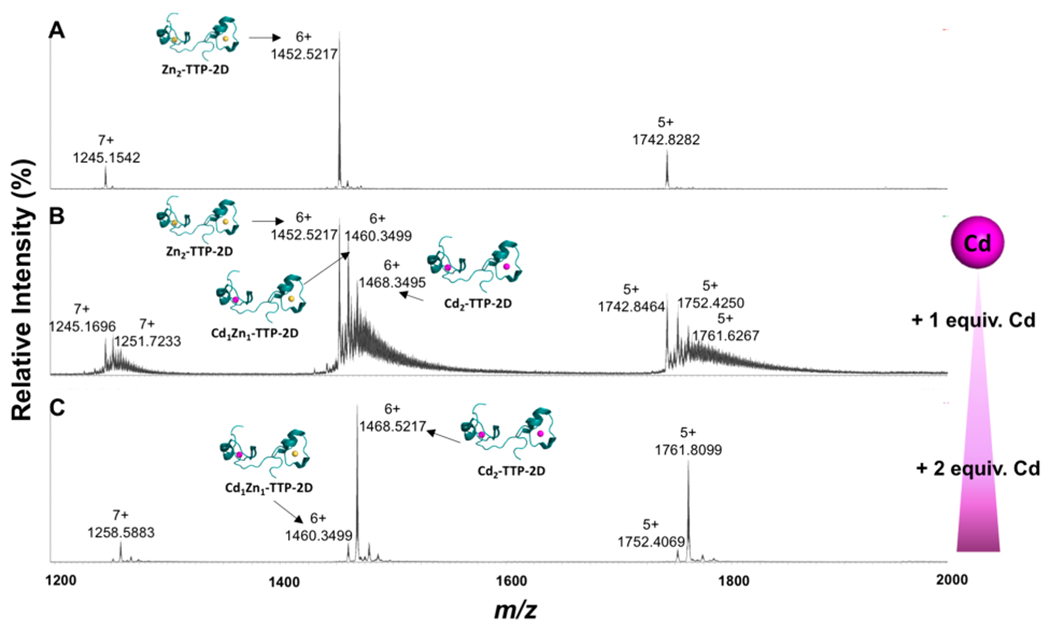
Change in the native nanoelectrospray mass spectrum of Zn2-TTP-2D (A) following the addition of 1 and 2 molar equivalents of Cd(OAc)2·2H2O to Zn2-TTP-2D (B to C) measured on a Waters Synapt-G2S mass spectrometer.
Figure 4.
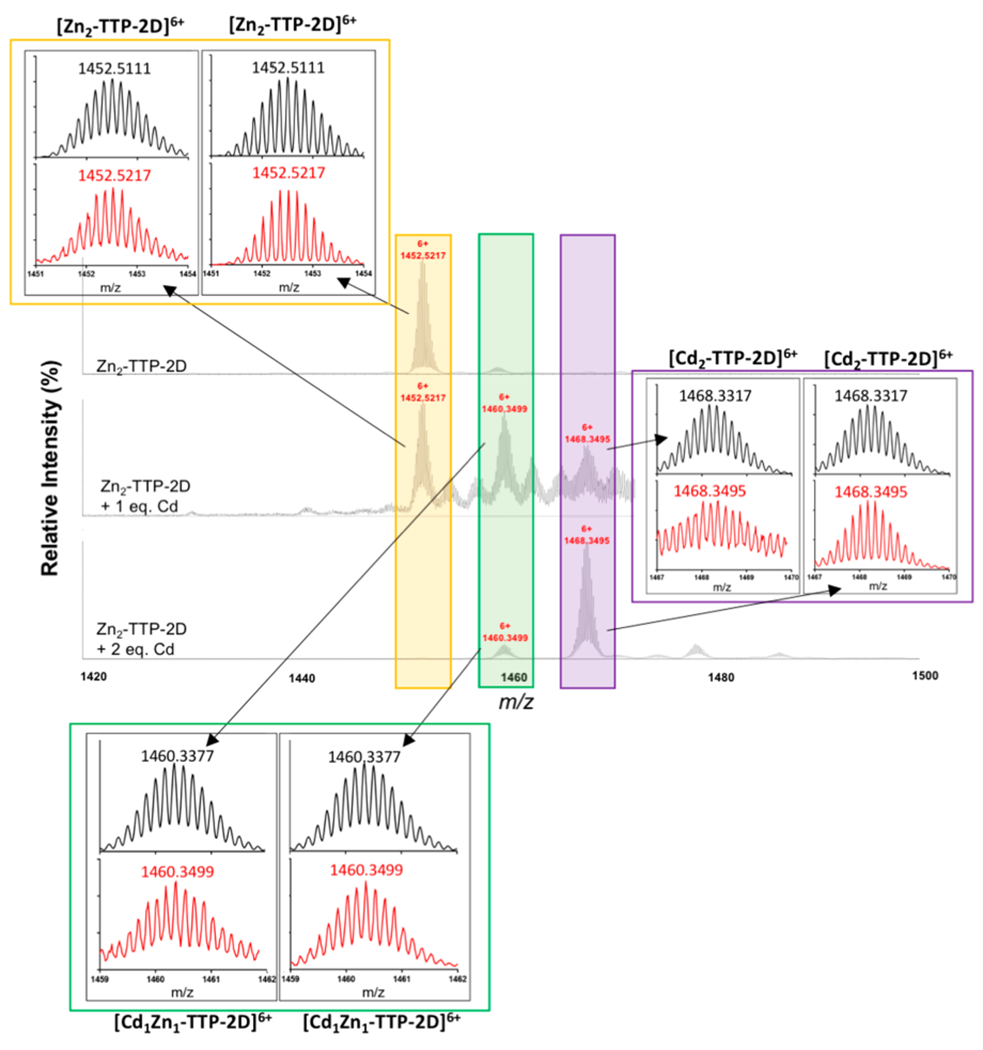
Comparison of the observed isotopic distribution pattern (red) of 6+ charged Zn2-TTP-2D (yellow inset), 6+ charged Cd1Zn1TTP-2D (green inset), and 6+ charged Cd2-TTP-2D (purple inset) ions with the simulated theoretical isotopic distribution (black) obtained with enviPat Web 2.4.
Native ESI-MS to Identify M-TTP-2D Complexes Formed upon Exchange of Zn with Cd2-TTP-2D.
The reverse experiment in which Zn was titrated with Cd2-TTP-2D was also performed. Here, 1 and 2 equiv of Zn(OAc)2·2H2O were titrated with Cd2-TTP-2D after which the native ESI-MS spectra were recorded (Figure 5). The peak at m/z = 1468.3458, indicative of Cd2-TTP-2D, was consistent with the calculated theoretical m/z value 1468.3317 (z = 6+) (Figure 5a). Following the addition of 1 mol equiv of Zn(OAc)2·2H2O, Cd2·TTP-2D (m/z = 1468.3625 (z = 6+)) and Cd1Zn1-TTP-2D (m/z = 1460.3629 (z = 6+)) signals were observed (Figure 5b). Upon the addition of 2 mol equiv of Zn(OAc)2·2H2O, increases in Cd1Zn1-TTP-2D relative signal intensity and decreases in Cd2-TTP-2D were detected. (Figure 5c). These data are consistent with the spin/filter ICP-MS results, and support the exchange of Zn with Cd bound in Cd2-TTP-2D.
Figure 5.
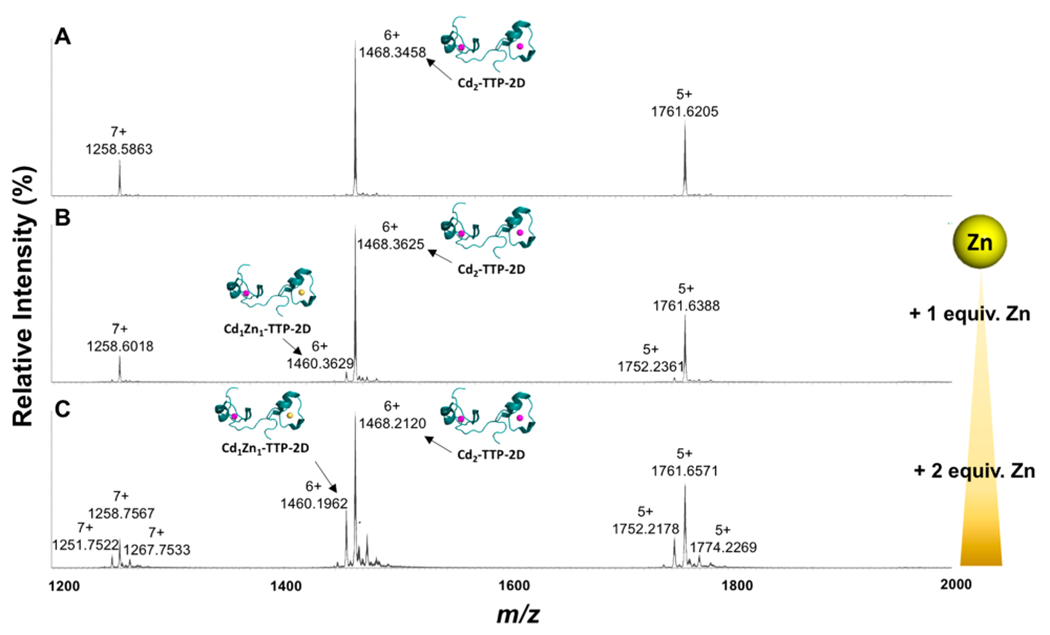
Change in the native nanoelectrospray mass spectrum of Cd2-TTP-2D (A) following the addition of 1 and 2 mol equiv of Zn(OAc)2·2H2O to Cd2-TTP-2D (B to C) measured on a Waters Synapt-G2S mass spectrometer.
Zn2-TTP-2D/RNA + Cd Monitored by Fluorescence Anisotropy (FA) and Native ESI-MS.
In cells, Zn2-TTP-2D binds to AU-rich RNA, and we sought to determine whether Cd exchanges with Zn2-TTP-2D when it is bound to RNA (Zn2-TTP-2D/RNA). We recently reported that RNA has a protective effect toward metal exchange with Zn2-TTP-2D in a study focused on how gold complexes interact with TTP,25 which we attributed to the RNA shielding the metal center. We hypothesized that RNA may also prevent Cd exchange with TTP via a similar shielding effect. To test this hypothesis, a competitive fluorescence anisotropy assay (FA), which probes protein/RNA binding, and a competitive native ESI-MS experiment, which identifies the metal-ZF-RNA complexes were performed.
The RNA target sequence recognized by TTP-2D is UUUAUUUAUUU, which corresponds to the 3′-untranslated region (3′UTR) of cytokine mRNA.8 We have previously reported that both Zn(II)-TTP-2D and Cd(II)-TTP-2D bind to this RNA sequence selectively and with high affinity (Kd = 16 ± 1 nM and 2.4 ± 0.2 nM, respectively) using FA.18 For the Cd exchange experiments, we titrated Zn2-TTP-2D with fluorescein-labeled RNA (UUUAUUUAUUU-F; F = fluorescein) and observed an increase in the FA signal (anisotropy, r) as the RNA was titrated, confirming Zn2-TTP-2D/RNA binding (Figure 6). Cd (4, 6, 8, and 10 equiv) was then titrated into the Zn2-TTP-2D/RNA complex, and the effect on FA monitored (Figure 6). No changes in fluorescence anisotropy (r) were observed. This finding suggests that either (1) the RNA protects the Zn bound to TTP from exchange or (2) the Cd is exchanging with Zn and RNA binding is retained. We have previously reported that Cd2-TTP-2D binds to the AAAUAAAUUUA RNA sequence with similar affinity that measured for Zn2-TTP-2D to RNA;18 therefore, we could not rule out either conclusion with the FA data alone (Figure 6 inset). Native ESI-MS was employed to determine if Cd exchanges with Zn when titrated with Zn2-TTP-2D/RNA. For these experiments, a 1:1 Zn2-TTP-2D/UUUAUUUAUUU-RNA complex was prepared, and the complexes formed were identified via ESI-MS. As Figure 7 shows, a peak at m/z = 2011.1294 (z = +6) indicative of the protein:RNA complex we identified. This peak is consistent with that we have previously reported (Figure 7).25 1 and 2 equiv of Cd(OAc)2·2H2O were then titrated into the Zn2-TTP2D/RNA complex and the resultant native ESI-MS spectra were obtained (Figure 7). Following the addition of a 1 equiv of Cd(OAc)2·2H2O, peaks indicative of Zn2-TTP-2D/RNA, Cd1Zn1-TTP-2D/RNA (m/z = 2019.3363 (z = +6)), and Cd2-TTP-2D/RNA (m/z = 2026.8152 (z = +6)) were observed (Figure 7b). Following the addition of 2 equiv of Cd(OAc)2·2H2O, peaks indicative of Cd1Zn1-TTP-2D/RNA and Cd2-TTP-2D/RNA were detected, with concomitant loss of the Zn2-TTP2D/RNA signal (Figure 7c). Taken together, these data reveal that Cd displaces Zn from the TTP-2D active site. Thus, we conclude that retention of RNA binding observed when Cd is titrated with the Zn2-TTP-2D/RNA complex involves Cd exchange with Zn with concurrent retention of RNA binding.
Figure 6.
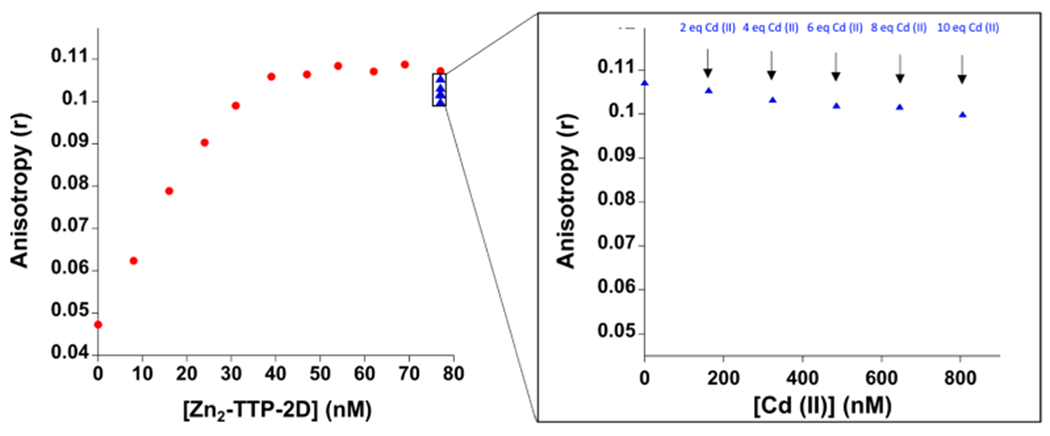
Plot of the change in anisotropy upon the addition of Zn2-TTP-2D (red) to the RNA oligonucleotide UUUAUUUAUUU-F (F = fluorescein) followed by (B) the addition of CdCl2. FA experiments were performed in 200 mM HEPES, 100 mM NaCl, at pH 7.5 via ISS multifrequency phase fluorometer (n = 3).
Figure 7.
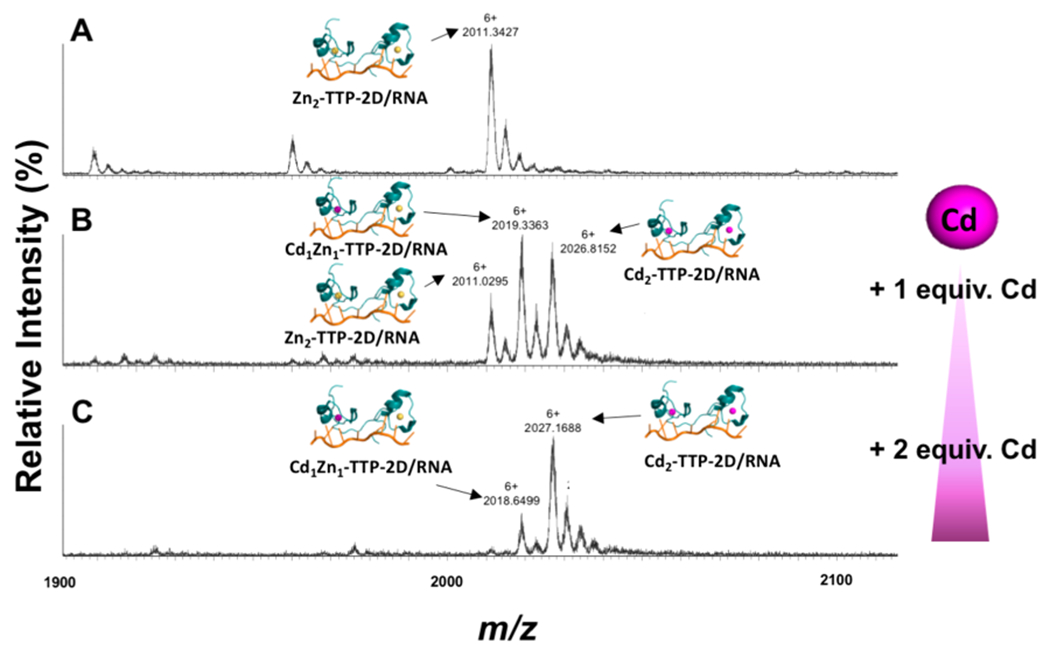
Change in the nanoelectrospray mass spectrum of Zn2-TTP-2D/RNA complex (A) following the addition of 1 and 2 mol equiv of Cd(OAc)2·2H2Oto Zn2-TTP-2D/RNA complex (B to C) measured on a Waters Synapt-G2S mass spectrometer.
DISCUSSION
The impetus for this work was the growing appreciation that toxic metals can target zinc finger proteins and the biological reports that TTP may be a cellular target for cadmium. We had previously reported that Cd can bind to apo-TTP-2D (a functional construct of TTP) with retention of RNA binding affinity and specificity to the sequence UUUAUUUAUUU.18 Although studies of metals binding to apo-ZFs are informative, a better mimic of cellular metal targeting is the zinc bound form of the ZF, as this is the form the exogenous metal likely encounters in the cell. A challenge in measuring metal exchange with Zn bound ZFs, particularly with exogenous metals that have filled d shells such as Cd, is the lack of a spectroscopic handle. We overcame this hurdle in this work by utilizing two MS approaches. The first was a spin-filter/ICP-MS approach which allowed us to quantify the metal exchange at the zinc site, and the second was a native ESI-MS approach that allowed us to identify the metal-TTP complexes that formed upon metal exchange. These two methods provide different information – ICP-MS is quantitative but only informs on the metal and native ESI-MS is qualitative but provides insight into metal–ligand complexes.
In the spin-filter/ICP-MS approach, a 3000 molecular weight cutoff filter was utilized to separate metal bound to protein (MW >9000) from low molecular weight metal salts. We then measured how much metal was associated with the protein using quantitative ICP-MS and determined the TTP-2D concentration using UV–visible spectroscopy (with a previously determined extinction coefficient). The ratio of [metal]/[protein] allowed us to determine the percent Zn and percent Cd associated with the protein. From these data, we observed that Cd can exchange with Zn in Zn-TTP-2D and vice versa suggesting that both exchanges are thermodynamically favorable. We previously utilized this approach for Cu exchange with Zn-TTP-2D,12 rigorously tested the assay to determine the number of time the spin filter must be spun (three) to separate all metal salts from metal-protein complexes, and followed that protocol here. We note that other approaches to follow metal exchange with ZFs prior to ICP-MS are also possible. These include size exclusion chromatography and chelex resin.66 Regardless of the exchange method, the coupling of metal exchange with ICP-MS offers a tool to measure metal exchange in ZF proteins that could potentially be applied to other metalloproteins.
The metal exchange results that we obtained for TTP suggest potential follow up studies. We observed greater exchange of Cd with Zn in Zn2-TTP-2D than Zn with Cd in Cd2-TTP-2D (Table 1) suggesting a thermodynamic preference for Cd exchange, and a future study could determine whether this difference is due to kinetics or thermodynamics.45,67,68 Similarly, the structural consequence of metal exchange are not known, and NMR approaches would provide insight.55,69,70
The native ESI-MS assay employed to identify the metal-TTP complexes that can form upon Cd and Zn exchange with Zn2-TTP-2D and Cd2-TTP-2D, respectively, provided information on the coordination complexes that can form upon metal exchange. These experiments involved titrating one and two equivalents of exogenous metal in order to determine if mixed metal species could be observed. We have previously reported that the affinity of zinc for both domains of TTP is equivalent.11 As such, we expected to observe mixed metal species when less than two equivalents of exogenous metal are added. We indeed saw mixed metal species: Cd1Zn1-TTP-2D was observed for exchange of both Cd with Zn2-TTP-2D and Zn with Cd2-TTP-2D (Figures 3 and 5). We did observe full exchange of Cd with Zn2-TTP-2D (i.e., Cd2-TTP-2D) but did not observe full exchange of Zn with Cd2-TTP-2D (i.e., Zn2-TTP-2D). This may be related to the lability of each metal, and future studies will address this question.
We also found that RNA does not prevent Cd exchange with Zn2-TTP-2D (by titrating Cd with Zn2-TTP-2D/RNA and monitoring by native ESI-MS). In contrast, we previously reported that AuIII(terpy) does not exchange with Zn in TTP-2D when it is bound to RNA, although it does exchange with Zn in the absence of RNA.25 This difference could be due to the nature of the metal complex: Cd is a salt, Cd(OAc)2, while Au is a complex, [AuIII(terpy)Cl]Cl2. It could also be due to the coordination properties of each metal: Cd, like Zn, can bind in a tetrahedral geometry, whereas AuIII(terpy)Cl is reduced to AuI upon coordination to apo-TTP-2D and favors a linear geometry.12
Overall, this work demonstrates the utility of combining a spin-filter-ICP-MS assay with native-ESI-MS titrations to follow metal exchange in metalloproteins, particularly those for which traditional spectroscopic approaches are not amenable (e.g., d10 metals). Of particular note is the ESI-MS monitored titration of metal exchange for a protein/RNA complex. To our knowledge, this is only the second example of such a titration and follows the first example, also performed by our laboratory, in which exchange with gold was monitored.25 The native ESI-MS approach is particularly appealing as it requires low concentration of samples and is mild enough to retain the intact metal-ZF complexes. This combined ICP/ESI MS approach has the potential to be applied to other systems to understand the effects of exogenous metal ions on protein metalation and function.
Cadmium is known to be linked to inflammation, and the impetus for examining the interactions of cadmium with TTP, which regulates inflammation, was to understand whether TTP is a target for Cd as part of its pro-inflammatory mechanism. The results presented here reveal that Cd does not affect TTP binding to its target RNA sequence (UUUAUUUAUUU) suggesting that TTP may not be a target for Cd. We note, however, that we have previously shown that Cd-TTP-2D binds with higher specificity to its target RNA when compared to other RNA sequences in contrast to Zn-TTP-2D which exhibits lower specificity.18 The greater specificity of Cd-TTP-2D for target RNA may alter the inflammatory response and be linked to the role of Cd in inflammation. Alternatively, the link between Cd and TTP may be kinetically driven. Our studies have focused on thermodynamics, and the kinetics of RNA binding by TTP have not been reported. Future studies both in vitro and in cells are needed to determine if TTP is a target for the inflammatory effect of cadmium.
ACKNOWLEDGMENTS
S.L.J.M. is grateful for the NSF (CHE-1708732) for support of this work. Additional support was provided by the University of Maryland School of Pharmacy Mass Spectrometry Center (SOP1841-IQB2014).
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.inorgchem.0c03808
The authors declare no competing financial interest.
Contributor Information
Joel E. P. Brandis, Department of Pharmaceutical Sciences, University of Maryland School of Pharmacy, Baltimore, Maryland 21201, United States
Stephanie M. Zalesak, Department of Pharmaceutical Sciences, University of Maryland School of Pharmacy, Baltimore, Maryland 21201, United States
Maureen A. Kane, Department of Pharmaceutical Sciences, University of Maryland School of Pharmacy, Baltimore, Maryland 21201, United States.
Sarah L. J. Michel, Department of Pharmaceutical Sciences, University of Maryland School of Pharmacy, Baltimore, Maryland 21201, United States
REFERENCES
- (1).Laity JH; Lee BM; Wright PE Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol 2001, 11 (1), 39–46. [DOI] [PubMed] [Google Scholar]
- (2).Lee SJ; Michel SL Structural metal sites in nonclassical zinc finger proteins involved in transcriptional and translational regulation. Acc. Chem. Res 2014, 47 (8), 2643–2650. [DOI] [PubMed] [Google Scholar]
- (3).Michalek JL; Besold AN; Michel SL Cysteine and histidine shuffling: mixing and matching cysteine and histidine residues in zinc finger proteins to afford different folds and function. Dalton Trans. 2011, 40 (47), 12619–32. [DOI] [PubMed] [Google Scholar]
- (4).Blackshear P Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans 2002, 30, 945–52. [DOI] [PubMed] [Google Scholar]
- (5).Andreini C; Banci L; Bertini I; Rosato A Counting the zincproteins encoded in the human genome. J. Proteome Res 2006, 5 (1), 196–201. [DOI] [PubMed] [Google Scholar]
- (6).Andreini C; Bertini I; Cavallaro G Minimal functional sites allow a classification of zinc sites in proteins. PLoS One 2011, 6 (10), No. e26325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Decaria L; Bertini I; Williams RJ Zinc proteomes, phylogenetics and evolution. Metallomics 2010, 2 (10), 706–9. [DOI] [PubMed] [Google Scholar]
- (8).Brooks SA; Blackshear PJ Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim. Biophys. Acta, Gene Regul. Mech 2013, 1829 (6-7), 666–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Klug A The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q. Rev. Biophys 2010, 43 (1), 1. [DOI] [PubMed] [Google Scholar]
- (10).Decaria L; Bertini I; Williams RJ Zinc proteomes, phylogenetics and evolution. Metallomics 2010, 2 (10), 706–709. [DOI] [PubMed] [Google Scholar]
- (11).di Targiani RC; Lee SJ; Wassink S; Michel SL Functional characterization of iron-substituted tristetraprolin-2D (TTP-2D, NUP475–2D): RNA binding affinity and selectivity. Biochemistry 2006, 45 (45), 13641 –13649. [DOI] [PubMed] [Google Scholar]
- (12).Shimberg GD; Ok K; Neu HM; Splan KE; Michel SL Cu (I) disrupts the structure and function of the nonclassical zinc finger protein tristetraprolin (TTP). Inorg. Chem 2017, 56 (12), 6838–6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Liang Y; Ewing PM; Laursen WJ; Tripp VT; Singh S; Splan KE Copper-binding properties of the BIR2 and BIR3 domains of the X-linked inhibitor of apoptosis protein. J. Inorg. Biochem 2014, 140, 104–10. [DOI] [PubMed] [Google Scholar]
- (14).Doku RT; Park G; Wheeler KE; Splan KE Spectroscopic characterization of copper(I) binding to apo and metal-reconstituted zinc finger peptides. JBIC, J. Biol. Inorg. Chem 2013, 18 (6), 669–78. [DOI] [PubMed] [Google Scholar]
- (15).Ghering AB; Jenkins LM; Schenck BL; Deo S; Mayer RA; Pikaart MJ; Omichinski JG; Godwin HA Spectroscopic and functional determination of the interaction of Pb2+ with GATA proteins. J. Am. Chem. Soc 2005, 127 (11), 3751–9. [DOI] [PubMed] [Google Scholar]
- (16).Neupane KP; Pecoraro VL Pb-207 NMR spectroscopy reveals that Pb(II) coordinates with glutathione (GSH) and tris cysteine zinc finger proteins in a PbS3 coordination environment. J. Inorg. Biochem 2011, 105 (8), 1030–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ordemann JM; Austin RN Lead neurotoxicity: exploring the potential impact of lead substitution in zinc-finger proteins on mental health. Metallomics 2016, 8 (6), 579–88. [DOI] [PubMed] [Google Scholar]
- (18).Michalek JL; Lee SJ; Michel SL Cadmium coordination to the zinc binding domains of the non-classical zinc finger protein Tristetraprolin affects RNA binding selectivity. J. Inorg. Biochem 2012, 112, 32–38. [DOI] [PubMed] [Google Scholar]
- (19).Padjasek M; Maciejczyk M; Nowakowski M; Kerber O; Pyrka M; Koźmiński W; Krȩźel A Metal Exchange in the Interprotein Zn(II) -Binding Site of the Rad50 Hook Domain: Structural Insights into Cd(II) -Induced DNA-Repair Inhibition. Chem. - Eur. J 2020, 26 (15), 3297–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).de Paiva REF; Du Z; Nakahata DH; Lima FA; Corbi PP; Farrell NP Gold-Catalyzed C-S Aryl-Group Transfer in Zinc Finger Proteins. Angew. Chem. Int. Ed 2018, 57 (30), 9305–9309. [DOI] [PubMed] [Google Scholar]
- (21).Wenzel MN; Meier-Menches SM; Williams TL; Ramisch E; Barone G; Casini A Selective targeting of PARP-1 zinc finger recognition domains with Au(iii) organometallics. Chem. Commun 2018, 54 (6), 611–614. [DOI] [PubMed] [Google Scholar]
- (22).Jacques A; Lebrun C; Casini A; Kieffer I; Proux O; Latour JM; Seneque O Reactivity of Cys4 zinc finger domains with gold(III) complexes: insights into the formation of “gold fingers. Inorg. Chem 2015, 54 (8), 4104–13. [DOI] [PubMed] [Google Scholar]
- (23).Franzman MA; Barrios AM Spectroscopic evidence for the formation of goldfingers. Inorg. Chem 2008, 47 (10), 3928–30. [DOI] [PubMed] [Google Scholar]
- (24).Nagaoka M; Kuwahara J; Sugiura Y Alteration of DNA binding specificity by nickel (II) substitution in three zinc (II) fingers of transcription factor Sp1. Biochem. Biophys. Res. Commun 1993, 194 (3), 1515–20. [DOI] [PubMed] [Google Scholar]
- (25).Ok K; Li W; Neu H; Batelu S; Stemmler T; Kane M; Michel S The Role of Gold in Inflammation and Tristetraprolin Activity. Chem. - Eur. J 2020, 26 (7), 1535–1547. [DOI] [PubMed] [Google Scholar]
- (26).Fu M; Blackshear PJ RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat. Rev. Immunol 2017, 17 (2), 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Sanduja S; Blanco FF; Young LE; Kaza V; Dixon DA The role of tristetraprolin in cancer and inflammation. Front. Biosci., Landmark Ed 2012, 17, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Hudson BP; Martinez-Yamout MA; Dyson HJ; Wright PE Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol 2004, 11 (3), 257–64. [DOI] [PubMed] [Google Scholar]
- (29).Maeda K; Akira S Regulation of mRNA stability by CCCHtype zinc-finger proteins in immune cells. Int. Immunol 2017, 29 (4), 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Brennan SE; Kuwano Y; Alkharouf N; Blackshear PJ; Gorospe M; Wilson GM The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009, 69 (12), 5168–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Suzuki T; Tsutsumi A; Suzuki H; Suzuki E; Sugihara M; Muraki Y; Hayashi T; Chino Y; Goto D; Matsumoto I Tristetraprolin (TTP) gene polymorphisms in patients with rheumatoid arthritis and healthy individuals. Mod. Rheumatol 2008, 18 (5), 472–479. [DOI] [PubMed] [Google Scholar]
- (32).Fairhurst A-M; Connolly JE; Hintz KA; Goulding NJ; Rassias AJ; Yeager MP; Rigby W; Wallace PK Regulation and localization of endogenous human tristetraprolin. Arthritis Res. Ther 2003, 5 (4), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Bernhard D; Rossmann A; Wick G Metals in cigarette smoke. IUBMB Life 2005, 57 (12), 805–809. [DOI] [PubMed] [Google Scholar]
- (34).Caruso RV; O’Connor RJ; Stephens WE; Cummings KM; Fong GT Toxic metal concentrations in cigarettes obtained from US smokers in 2009: results from the International Tobacco Control (ITC) United States survey cohort. Int. J. Environ. Res. Public Health 2014, 11 (1), 202–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Waisberg M; Joseph P; Hale B; Beyersmann D Molecular and cellular mechanisms of cadmium carcinogenesis. Toxicology 2003, 192 (2-3), 95–117. [DOI] [PubMed] [Google Scholar]
- (36).Richter P; Faroon O; Pappas RS Cadmium and Cadmium/Zinc Ratios and Tobacco-Related Morbidities. Int. J. Environ. Res. Public Health 2017, 14 (10), 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Hartwig A Cadmium and cancer. Met. Ions Life Sci 2013, 11, 491–507. [DOI] [PubMed] [Google Scholar]
- (38).Anwar S; Naz A; Ashraf MY; Malik A Evaluation of inorganic contaminants emitted from automobiles and dynamics in soil, dust, and vegetations from major highways in Pakistan. Environ. Sci. Pollut. Res 2020, 27 (26), 32494–32508. [DOI] [PubMed] [Google Scholar]
- (39).Brucker N; Moro A; Charao M; Bubols G; Nascimento S; Goethel G; Barth A; Prohmann AC; Rocha R; Moresco R; Sangoi M; Hausen BS; Saint’Pierre T; Gioda A; Duarte M; Castro I; Saldiva PH; Garcia SC Relationship between blood metals and inflammation in taxi drivers. Clin. Chim. Acta 2015, 444, 176–81. [DOI] [PubMed] [Google Scholar]
- (40).Gao P; Liu S; Ye W; Lin N; Meng P; Feng Y; Zhang Z; Cui F; Lu B; Xing B Assessment on the occupational exposure of urban public bus drivers to bioaccessible trace metals through resuspended fraction of settled bus dust. Sci. Total Environ 2015, 508, 37–45. [DOI] [PubMed] [Google Scholar]
- (41).Nazzal Y; Rosen MA; Al-Rawabdeh AM Assessment of metal pollution in urban road dusts from selected highways of the Greater Toronto Area in Canada. Environ. Monit. Assess 2013, 185 (2), 1847–58. [DOI] [PubMed] [Google Scholar]
- (42).Kim ND; Fergusson JE The concentrations, distribution and sources of cadmium, copper, lead and zinc in the atmosphere of an urban environment. Sci. Total Environ 1994, 144 (1-3), 179–89. [DOI] [PubMed] [Google Scholar]
- (43).Maret W; Moulis JM The bioinorganic chemistry of cadmium in the context of its toxicity. Met. Ions Life Sci 2013, 11, 1–29. [DOI] [PubMed] [Google Scholar]
- (44).Cotton FA; Wilkinson G; Mutillo C; Bochmann M Adv Inorg Chem. J. Chem. Educ 2000, 77 (3), 311. [Google Scholar]
- (45).Petering D; Huang M; Moteki S; Shaw III C Cadmium and lead interactions with transcription factor IIIA from Xenopus laevis: a model for zinc finger protein reactions with toxic metal ions and metallothionein. Mar. Environ. Res 2000, 50 (1-5), 89–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Gehrig PM; You C; Kägi JH; Hunziker PE; Dallinger R; Gruber C; Brouwer M Electrospray ionization mass spectrometry of zinc, cadmium, and copper metallothioneins: Evidence for metal-binding cooperativity. Protein Sci. 2000, 9 (2), 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Buchko GW; Hess NJ; Kennedy MA Cadmium mutagenicity and human nucleotide excision repair protein XPA: CD, EXAFS and 1H/15N-NMR spectroscopic studies on the zinc (II)-and cadmium (II)-associated minimal DNA-binding domain (M98-F219). Carcinogenesis 2000, 21 (5), 1051–1057. [DOI] [PubMed] [Google Scholar]
- (48).Razmiafshari M; Kao J; d’Avignon A; Zawia N NMR identification of heavy metal-binding sites in a synthetic zinc finger peptide: toxicological implications for the interactions of xenobiotic metals with zinc finger proteins. Toxicol. Appl. Pharmacol 2001, 172 (1), 1–10. [DOI] [PubMed] [Google Scholar]
- (49).Perinelli M; Tegoni M; Freisinger E Different Behavior of the Histidine Residue toward Cadmium and Zinc in a Cadmium-Specific Metallothionein from an Aquatic Fungus. Inorg. Chem 2020, 59 (23), 16988–16997. [DOI] [PubMed] [Google Scholar]
- (50).Krizek BA; Merkle DL; Berg JM Ligand variation and metal ion binding specificity in zinc finger peptides. Inorg. Chem 1993, 32 (6), 937–940. [Google Scholar]
- (51).Pinter TB; Stillman MJ Kinetics of zinc and cadmium exchanges between metallothionein and carbonic anhydrase. Biochemistry 2015, 54 (40), 6284–6293. [DOI] [PubMed] [Google Scholar]
- (52).Pinter TB; Irvine GW; Stillman MJ Domain selection in metallothionein 1a: Affinity-controlled mechanisms of zinc binding and cadmium exchange. Biochemistry 2015, 54 (32), 5006–5016. [DOI] [PubMed] [Google Scholar]
- (53).Ruiz C; Rodríguez AR Study of the additions of cadmium and zinc to the cadmium, zinc metallothioneins using differential pulse polarography Influence of the pH. Anal. Chim. Acta 1996, 325 (1-2), 43–51. [Google Scholar]
- (54).Malgieri G; Zaccaro L; Leone M; Bucci E; Esposito S; Baglivo I; Del Gatto A; Russo L; Scandurra R; Pedone PV Zinc to cadmium replacement in the A. thaliana SUPERMAN Cys2His2 zinc finger induces structural rearrangements of typical DNA base determinant positions. Biopolymers 2011, 95 (11), 801–810. [DOI] [PubMed] [Google Scholar]
- (55).Kowald GR; Sturzenbaum SR; Blindauer CA Earthworm Lumbricus rubellus MT-2: Metal Binding and Protein Folding of a True Cadmium-MT. Int. J. Mol. Sci 2016, 17 (1), 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Zeitoun-Ghandour S; Charnock JM; Hodson ME; Leszczyszyn OI; Blindauer CA; Sturzenbaum SR The two Caenorhabditis elegans metallothioneins (CeMT-1 and CeMT-2) discriminate between essential zinc and toxic cadmium. FEBS J. 2010, 277 (11), 2531–42. [DOI] [PubMed] [Google Scholar]
- (57).Freitas M; Fernandes E Zinc, cadmium and nickel increase the activation of NF-κB and the release of cytokines from THP-1 monocytic cells. Metallomics 2011, 3 (11), 1238–1243. [DOI] [PubMed] [Google Scholar]
- (58).Cox JN; Rahman MA; Bao S; Liu M; Wheeler SE; Knoell DL Cadmium attenuates the macrophage response to LPS through inhibition of the NF-κB pathway. Am. J. Physiol. Lung Cell Mol. Physiol 2016, 311 (4), L754–L765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Chin-Ju JH; Stapleton SR Early sensing and gene expression profiling under a low dose of cadmium exposure. Biochimie 2009, 91 (3), 329–343. [DOI] [PubMed] [Google Scholar]
- (60).Hartwig A Mechanisms in cadmium-induced carcinogenicity: recent insights. BioMetals 2010, 23 (5), 951–960. [DOI] [PubMed] [Google Scholar]
- (61).Loos M; Gerber C; Corona F; Hollender J; Singer H Accelerated isotope fine structure calculation using pruned transition trees. Anal. Chem 2015, 87 (11), 5738–5744. [DOI] [PubMed] [Google Scholar]
- (62).Bernhoft RA Cadmium toxicity and treatment. Sci. World J 2013, 2013, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Cassou CA; Sterling HJ; Susa AC; Williams ER Electrothermal supercharging in mass spectrometry and tandem mass spectrometry of native proteins. Anal. Chem 2013, 85 (1), 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Kaltashov IA; Bobst CE; Abzalimov RR Mass spectrometry-based methods to study protein architecture and dynamics. Protein Sci. 2013, 22 (5), 530–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Susa AC; Xia Z; Tang HY; Tainer JA; Williams ER Charging of proteins in native mass spectrometry. J. Am. Soc. Mass Spectrom 2017, 28 (2), 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Peroza EA; Kaabi AA; Meyer-Klaucke W; Wellenreuther G; Freisinger E The two distinctive metal ion binding domains of the wheat metallothionein Ec-1. J. Inorg. Biochem 2009, 103 (3), 342–53. [DOI] [PubMed] [Google Scholar]
- (67).Huestis J; Zhou X; Chen L; Feng C; Hudson LG; Liu KJ Kinetics and thermodynamics of zinc(II) and arsenic(III) binding to XPA and PARP-1 zinc finger peptides. J. Inorg. Biochem 2016, 163, 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Quintal SM; dePaula QA; Farrell NP Zinc finger proteins as templates for metal ion exchange and ligand reactivity. Chemical and biological consequences. Metallomics 2011, 3 (2), 121–39. [DOI] [PubMed] [Google Scholar]
- (69).Legge GB; Martinez-Yamout MA; Hambly DM; Trinh T; Lee BM; Dyson HJ; Wright PE ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module. J. Mol. Biol 2004, 343 (4), 1081–93. [DOI] [PubMed] [Google Scholar]
- (70).Loebus J; Peroza EA; Bluthgen N; Fox T; Meyer-Klaucke W; Zerbe O; Freisinger E Protein and metal cluster structure of the wheat metallothionein domain gamma-E(c)-1: the second part of the puzzle. J. Biol. Inorg. Chem 2011, 16 (5), 683–94. [DOI] [PubMed] [Google Scholar]


