Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19). Reports of new variants that potentially increase virulence and viral transmission, as well as reduce the efficacy of available vaccines, have recently emerged. In this study, we computationally analyzed the N439K, S477 N, and T478K variants for their ability to bind Angiotensin-converting enzyme 2 (ACE2). We used the protein-protein docking approach to explore whether the three variants displayed a higher binding affinity to the ACE2 receptor than the wild type. We found that these variants alter the hydrogen bonding network and the cluster of interactions. Additional salt bridges, hydrogen bonds, and a high number of non-bonded contacts (i.e., non-bonded interactions between atoms in the same molecule and those in other molecules) were observed only in the mutant complexes, allowing efficient binding to the ACE2 receptor. Furthermore, we used a 2.0-μs all-atoms simulation approach to detect differences in the structural dynamic features of the resulting protein complexes. Our findings revealed that the mutant complexes possessed stable dynamics, consistent with the global trend of mutations yielding variants with improved stability and enhanced affinity. Binding energy calculations based on molecular mechanics/generalized Born surface area (MM/GBSA) further revealed that electrostatic interactions principally increased net binding energies. The stability and binding energies of N439K, S477 N, and T478K variants were enhanced compared to the wild-type-ACE2 complex. The net binding energy of the systems was −31.86 kcal/mol for the wild-type-ACE2 complex, −67.85 kcal/mol for N439K, −69.82 kcal/mol for S477 N, and −69.64 kcal/mol for T478K. The current study provides a basis for exploring the enhanced binding abilities and structural features of SARS-CoV-2 variants to design novel therapeutics against the virus.
Keywords: SARS-CoV-2, Variants, Docking, Simulation, Free energy
Graphical abstract
1. Introduction
The frequent appearance of coronaviruses in the 21st century has caused unprecedented damage worldwide. The viruses that have emerged during the first two decades of the 21st century belong to the Orthocoronavirinae within the family Coronaviridae [1,2]. The devastating effects of the severe acute respiratory syndrome coronavirus (SARS-CoV-1), the Middle East respiratory syndrome-related coronavirus (MERS-CoV), and, more recently, the pandemic agent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have heavily impacted human health. Moreover, the current pandemic has affected the social fabric and economies worldwide [3,4]. The recently reported coronavirus species from the beta lineage are mostly pathogenic to humans [5]. The SARS-CoV-2 emerged in Wuhan, causing the coronavirus disease 2019 (COVID-19), which has evolved into a multiwave pandemic. New strains of the virus that are more transmissible and virulent were reported from different parts of the world. The newly emerged variants exhibit higher infectivity, contiguity, and causality [[5], [6], [7], [8], [9], [10]]. As of 08/29/2021, the number of cases reached 216,686,505, whereas the number of deaths reached 4,510,142 globally. Therefore, the COVID-19 case fatality ratio (CFR) is only 3%, which is comparatively lower than 10% in the case of SARS and 35% in the case of MERS [11]. The rapid spread of SARS-CoV-2 and the emergence of new variants present a severe threat to human health. Consequently, researchers around the world are exploiting different approaches to combat SARS-CoV-2, including the application of integrated multi-omics technologies to design novel and effective vaccines and drugs. The latter can be obtained by screening biological drug databases or repurposing old drugs to curtail the risks and threats associated with SARS-CoV-2 [[12], [13], [14]].
Deciphering the infection pathways using proteomics technologies may help relieve the threat of SARS-CoV-2 [15]. Therefore, knowledge of the SARS-CoV-2 proteome is essential to manage this virus [5,16]. Efforts to disclose the genomic organization of SARS-CoV-2 have revealed that the virus consists of sixteen nonstructural proteins (NSP1–NSP16) and four structural proteins (S protein, E protein, N protein, and M protein) [17,18]. The proteins encoded by SARS-CoV-2 are primarily associated with viral replication and infection, with the spike protein and proteases being primary targets for developing antiviral therapeutics [5,16,19]. The spike protein and angiotensin-converting enzyme (ACE2) receptors fuse to initiate the transmission and virulence of SARS-CoV-2 [[20], [21], [22]]. The S1 subunit of the spike protein binds to ACE2, while the S2 subunit further facilitates the fusion process by minimizing the distance between the viral spike protein and the host cell [23]. Next, the activated fusion peptide participates in critical processes, including deformation and membrane attachment [24]. Finally, the non-endosomal or endosomal pathways could be selected—individually or together—for entry into the host cell [25]. Therefore, the inhibition of the ACE2-RBD interaction is essential for controlling the infection caused by SARS-CoV-2. These treatments either stimulate the immune system to comply with infections or prevent infectious agents from adhering to the host receptor proteins [26].
The world is currently experiencing the third wave of the COVID-19 pandemic. During this prolonged occurrence, several variants harboring numerous mutations have surfaced that are more contagious, increase disease severity, and potentially evade the host immune response [[27], [28], [29], [30], [31]]. The RBD domain in VOCs may be an ideal target site for the development of novel antiviral therapies against SARS-CoV-2. The novel mutations N439K, S477 N, and T478K were recently reported to increase the binding affinity and impact the infectivity [32,33]. A detailed, systematic, and comprehensive investigation is essential to determine how the N439K, S477 N, and T478K substitutions affect the binding of the spike protein with the ACE2 receptor and initiate structural and functional changes. In the present study, we used different theoretical and computational methods such as protein-protein docking, molecular dynamics simulations, and binding free energy calculations to investigate the structural changes that (1) alter the binding between RDB and ACE2 receptor as a result of N439K, S477 N, and T478K mutations, and (2) increase the infectivity rates. This analysis will provide insights into the structural changes of the RBD domain, for application in future investigations and the development of therapeutics.
2. Methods
2.1. Data retrieval and mutants modeling
The RNA viruses are prone to mutations that lead to increasing or decreasing pathogenesis. In the case of SARS-CoV-2, the spike protein is prone to many mutations, which can cause disease, and is deemed as the primary therapeutic target. The literature reflected the structural and functional importance of the spike protein in human pathophysiology. Thus herein, we employed an in-silico mutagenesis strategy to model the structural variants and comprehend their effect on binding and pathogenesis. A recently described structure of the spike protein of SARS-CoV-2 was obtained from Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) using accession number 6M0J [34,35]. The 3D structures of mutations (N439K, S477 N, T478K), predicted to induce greater stability change and increase the binding affinity by many folds, were modeled using Modeller v15.2 embedded in Chimera software [36,37]. Furthermore, 3D structures were prepared, minimized, and the root mean square deviation (RMSD) differences were revealed by the superimposition of the mutants on the wild type. The mutated structures were validated through ProSA-web server [38], ERRAT [39] and VERIFY-3D [40].
2.2. Modeling the protein complexes
Consensus restrained docking of the spike RBD (wild mutant) and ACE2 cellular receptor was achieved through high ambiguity-driven protein-protein docking (HADDOCK) and HDOCK algorithms [41,42]. The interaction network, i.e., the salt-bridges, hydrogen interactions, and non-bonded contacts, were visualized through PDBsum [43].
2.3. Molecular dynamics simulation of the top complexes
Structure dynamics of the wild-type and mutants (N439K, S477 N, T478K) RBDs in complex with ACE2 were evaluated in a solvated system neutralized by counterions. The FF19SB force field in the assisted model building with energy refinement (AMBER20) simulation package was used to achieve all atoms molecular dynamics (MD) simulations [44,45]. The energy minimization procedure was being used to address the bad clashes in the protein complexes. For 6000 and 3000 cycles, the steepest descent algorithm and the conjugate gradient algorithm were used. The system was equilibrated at 1 atm constant pressure with weak restraint after 300 K heating. Furthermore, an all-atoms simulation executed for 500 ns each with the particle mesh Ewald algorithm (PME) and a cut-off distance 10.0 Å was used to treat the long-range electrostatic interactions. Bonds involving hydrogen atoms were treated by using SHAKE algorithm. A graphical processing unit (GPU) accelerated simulation was performed and MD trajectories were subjected to post-simulation analysis by using CPPTRAJ and PTRAJ packages [46].
2.4. Binding free energy calculations
The binding free energy is commonly used for the estimation of affinities of different macromolecular complexes, i.e., protein-protein/DNA/RNA [9,12,13]. To estimate the binding free energy of wild-type and mutants (N439K, S477 N, T478K) complexes, we used the molecular mechanics generalized Born surface area (MM/GBSA) approach, which was previously used by Abbas et al., 2021 to explore the binding differences of the wild-type and SARS-CoV-2 variants [10,47]. Employing the MM-GBSA.py script [48], electrostatic, van der Waals (vdW), solvation energy, and total free energy were estimated using the following equation.
| ΔG(bind) = ΔG(complex) − [ΔG(receptor) + ΔG(ligand)] |
The equation can be then used to determine the different components of the TBE.
| G = Gbonded + Gelectrostatic + Gvan der Waal + Gpolar + Gnon-polar |
3. Results and discussion
3.1. Structural modeling and evaluation
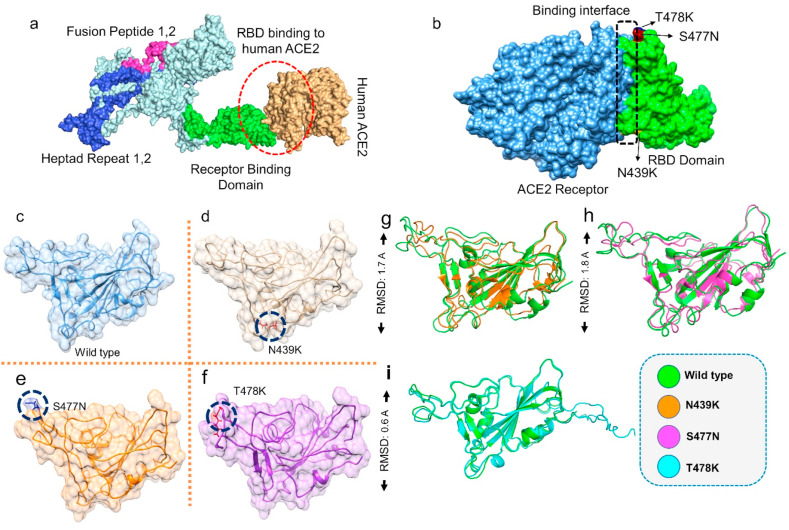
SARS-CoV-2, first reported in December 2019, is subject to frequent genetic changes that lead to variations in pathogenicity, infectivity, and treatment choices [9,10]. The spike protein's receptor-binding domain (RBD) upon attachment to the host receptor ACE2 elicits the pathogenesis pathway, is the most rapidly mutating protein, posing a continuous threat to coronavirus disease 2019 (COVID-19) containment and to lower vaccine and drug efficacy. Understanding these genetic variations, which may cause higher infectivity, can help to manage the increased risk of transmission and immune evasion [49]. The recently reported spike RBD mutations, which occur in higher frequency and are associated with higher infectivity, are N439K, S477 N, and T478K [31,32]. These mutations are structurally present at the binding interface between the human ACE2 receptor and the SARS-CoV-2 spike RBD domain (Fig. 1 a–b).
Fig. 1.
RBD variants modeling and the superimposition of RBD WT with mutants. (a) domain organization of the Spike protein, (b) binding interface of Spike RBD and ACE2 receptor, (c) wild type RBD (d) N439K mutant RBD (e) S477 N mutant RBD (f) T478K mutant RBD (g–i) Superimposed structure of RBD WT (green) with N439K (orange), S477 N (magenta), T478K (cyan), L84S (orange). The superimposition differences were shown as RMSD values.
To explore the interaction mechanism of the wild-type RBD and the mutants mentioned above with human ACE2, we used Chimera software to structurally model the N439K, S477 N, and T478K mutants (Fig. 1c–f). To visualize the effect of the generated mutants on the protein tertiary structure, we superimposed the wild-type protein on the mutants, and the RMSD values were recorded. The RMSD differences in each superimposed structure were significant, with 1.7 Å for N439K, 1.8 Å for S477 N, and 0.6 Å for T478K, indicating structural deviation, secondary structural element perturbation, and protein conformational variations in the mutant structures (Fig. 1g–i). The modeled mutated structures were also validated through ProSA-web and PROCHECK, which revealed that all the structures are modeled accurately with no topological error. The obtained results are shown in Table 1 .
Table 1.
Validation of the modeled 3D structures of the mutants.
| Mutant | ProSA-Web (Z-score) | ERRAT Quality Factor | VERIFY 3D | Output |
|---|---|---|---|---|
| N439K | −5.44 | 89.79% | 85.65% of the residues have averaged 3D-1D score≥0.2 | Pass |
| S477 N | −5.76 | 91.62% | 82.96% of the residues have averaged 3D-1D score≥0.2 | Pass |
| T478K | −5.64 | 88.57% | 90.87% of the residues have averaged 3D-1D score≥0.2 | Pass |
3.2. Interaction difference analysis using molecular docking
To understand the binding differences between the wild type and mutants, structural binding of the RBD with ACE2 was performed (Table 1). Comparative binding analysis of the wild type and mutants revealed key differences in the binding affinity and interaction network. To identify the underlying mechanism behind the increased infectivity of SARS-CoV-2 variants, we used the HDOCK server to perform molecular docking of the ACE2 receptor with the wild-type and mutant spike RBD domains. The predicted docking score of HDOCK for the wild-type ACE2 complex was −30,206 kcal/mol. Furthermore, the interaction interface analysis through PDBsum revealed that the ACE2–wild-type complex formed one salt bridge and 11 hydrogen bonds and the number of non-bonded contacts was 125. The hydrogen bonds that formed in the ACE2–WT complex were Gln493-Glu35, Lys417-Glu30, Thr500-Tyr41, Thr500-Asn330, Gln498-Gln42, Gln498-Lys353, Tyr449-Glu38, Gly496-Glu38, Gly496-Lys353, and Gly502-Lys353 (Fig. 2 a). The current findings are consistent with the previous reports that some important interactions, i.e., Lys417-Glu30, Tyr449-Glu38, Gly496-Glu38, and Gly496-Lys353, also conserved in this study [9,10,50]. However, the salt bridge was reported to be between Glu30 and Lys417, which is also reported by Abbas et al., in the recent findings on the new variants (B.1.1.7, B.1.351, P.1 and B.1.617) [9]. To compare the binding of T478K with the wild type and highlight the effect of this mutation on binding efficiency, we analyzed the binding network using the HDOCK server. Consequently, the binding network analysis revealed that the substituted T478K residue enhanced the binding of ACE2 to the spike RBD compared with the wild type by making one salt bridge, 14 hydrogen bonds, and 166 non-bonded contacts (Fig. 2b). Among the hydrogen bonds, Gln42–Gly446, Gln42–Tyr449, Ser19–Ala475, Tyr83–Asn487, Tyr83–Tyr489, Lys353–Gly496, Lys353–Gln498, Asn330–Thr500, Tyr41–Thr500, Glu38–Tyr449, Glu35–Gln493, Gln42-Gln498, Glu38–Gln498, Tyr41–Asn501 and Lys353–Gly502 residues were involved. Previously Abbas et al. concluded that the establishment of extra bonds, including salt bridges and hydrogen bonds, contributes to the higher binding and consequently the infectivity [9,10,33]. Another important mutation N439K in the spike RBD reported in a recent study revealed that this mutation is reported autonomously in multiple lineages and enhances the binding of spike RBD to the ACE2 receptor. It also confers resistance to several monoclonal antibodies and escapes some polyclonal responses [51]. We used the HDOCK online server to check the binding affinity of the N439K mutant to the ACE2 receptor. The interaction analysis of the complex using PDBsum, and a total of two salt bridges, 13 hydrogen bonds, and 169 non-bonded contacts were found. Salt bridges are the product of two non-covalent interactions, including ionic and hydrogen bonding. These interactions play a vital role in biological systems and are central to stabilizing docked conformers. Conversely, hydrogen bonding allows directional interactions and is important in protein-protein molecular recognition, structural integrity, and overall folding. Both electrostatic and hydrogen bonding contribute significantly to protein-protein interactions and the net stability of the complexes.
Fig. 2.
Docking complexes of wild-type and mutants spike protein with ACE2. (a) Represent the binding interface of the wild-type complex along with its stick representation of the key hydrogen interactions with ACE2. (b) The binding interface and stick representation of hydrogen bonding of the N439K mutant complex.
For instance, the increased electrostatic energy and hydrogen bonds were previously reported to enhance the affinity towards the host receptor ACE2 in other variants [10,50]. This variant shows a significant variation in the interaction pattern, with an additional salt bridge and two hydrogen bonds with the human receptor. This verifies that N439K establishes additional interactions and increases the binding affinity and infectivity compared with wild-type. Therefore, our results are similar to the experimental data in terms of the higher binding affinity and infectivity of the N439K mutant [51].
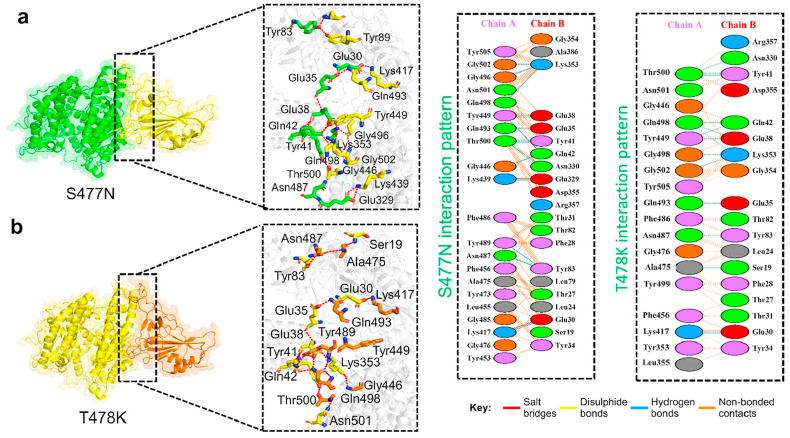
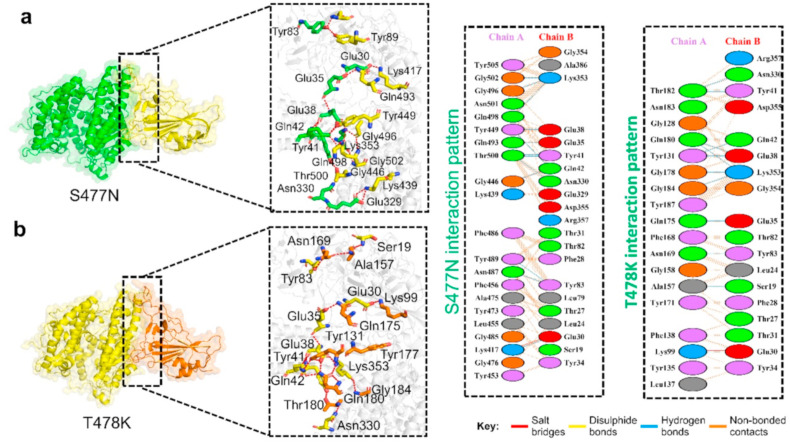
The hydrogen bonds involved in the ACE2–N439K complex include Glu32-Lys439, Ser19-Ala475, Tyr83-Asn487, Glu35-Gln493, Asn330-Thr500, Tyr41-Thr500, Gln42-Gly446, Gln42-Tyr449, Glu38-Tyr449, Lys353-Gln498, Lys353-Gly496, and Lys353-Gly502. The salt bridge was found between the Lys439–Glu329 and Glu30–Lys417 residues (Fig. 3 a). The Tyr83-Asn487 interaction is significantly conserved in various other variants, too [9,10,50]. Moreover, the cluster of interactions by Lys353 is important for the recognition and enhanced infectivity [52]. A similar approach was used for the analysis of the ACE2–S477 N using PDBsum, which revealed the formation of two salt bridges, 14 hydrogen bonds, and 181 non-bonded contacts, which is the highest bonding network among the above mutants and may be involved in higher infectivity. The hydrogen bonds of the ACE2–S477 N complex contained Glu30-Lys417, Tyr83-Asn487, Tyr83-Tyr489, Glu329-Lys439, Asn330-Thr500, Gln42-Tyr449, Tyr41-Thr500, Glu35-Gln493, Glu38-Tyr449, Lys353-Gln498, Lys353-Gly496, and Lys353-Gly502 residues. The salt bridge was found between Glu329–Lys439 and Glu30–Lys417, which are highly conserved [52]. Similarly, other essential interactions such as Glu30-Lys417, Tyr83-Asn487, Tyr83-Tyr489, and a cluster of interactions by Lys353 are necessary for the binding with ACE2 are conserved here. The interaction details are presented in Table 2 , while the bonding differences are presented in Table 3 .
Fig. 3.
Docking complexes of S477N and T478K mutants spike protein with ACE2. (a) Represent the binding interface of the S477 N complex along with its stick representation of the key hydrogen interactions with ACE2. (b) The binding interface and stick representation of hydrogen bonding of the T478K mutant complex.
Table 2.
Interaction difference between the wild type and mutant complexes.
| Complexes | Hydrogen Bonds | Salt Bridges |
|---|---|---|
| Wild Type | Gln493–Glu35, Lys417–Glu30, Thr500–Tyr41, Thr500–Asn330, Gln498–Gln42, Gln498–Lys353, Tyr449–Glu38, Gly496–Glu38, Gly496–Lys353, Gly502–Lys353 | Glu30–417 |
| N439K | Glu32-Lys439, Ser19-Ala475, Tyr83-Asn487, Glu35-Gln493, Asn330-Thr500, Tyr41-Thr500, Gln42-Gly446, Gln42-Tyr449, Glu38-Tyr449, Lys353-Gln498, Lys353-Gly496, Lys353-Gly502 | Lys439–Glu329, Glu30–Lys417 |
| S477N | Glu30-Lys417, Tyr83-Asn487, Tyr83-Tyr489, Glu329-Lys439, Asn330-Thr500, Gln42-Tyr449, Tyr41-Thr500, Glu35-Gln493, Glu38-Tyr449, Lys353-Gln498, Lys353-Gly496, Lys353-Gly502 | Lys439–Glu329, Glu30–Lys417 |
| T478K | Gln42–Gly446, Gln42–Tyr449, Ser19–Ala475, Tyr83–Asn487, Tyr83–Tyr489, Lys353–Gly496, Lys353–Gln498, Asn330–Thr500, Tyr41–Thr500, Glu38–Tyr449, Glu35–Gln493, Gln42–Gln498, Glu38–Gln498, Tyr41–Asn501, Lys353–Gly502 | Glu30–417 |
Table 3.
Bonding patterns of each of the RBD and ACE2 complexes.
| Complexes | Salt bridges | Disulfide bonds | hydrogen bonds | non-bonded contacts |
|---|---|---|---|---|
| Wild Type | 1 | 00 | 11 | 125 |
| S439K | 2 | 00 | 13 | 169 |
| S477 N | 2 | 00 | 14 | 181 |
| T478K | 1 | 00 | 14 | 166 |
3.3. Structural stability analysis
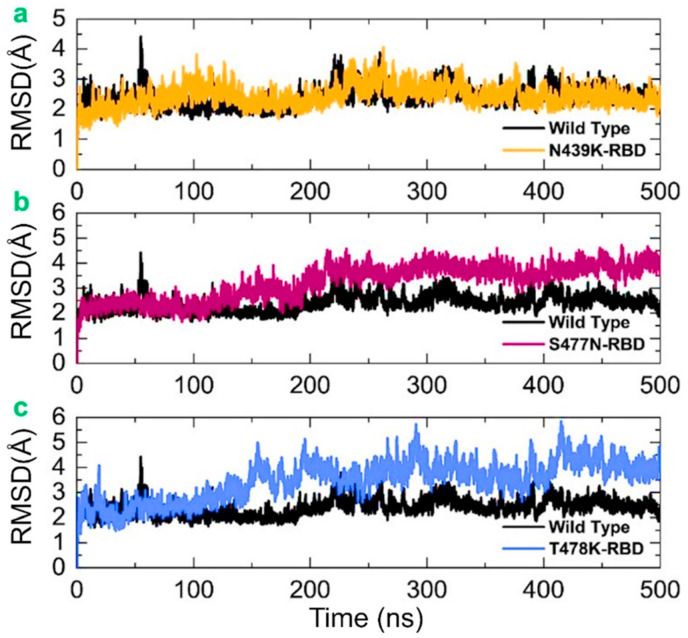
By estimating the thermodynamic state function as the RMSD, we evaluated the effect of the fixed amino acid changes identified in the RBD of the SARS-CoV-2 spike glycoprotein. The estimation of dynamic stability as the RMSD is a frequently used approach for the calculation of the differences in a protein's backbone from its initial structural conformation to its final position, demonstrating the stability of a system. The variations observed during the simulation time can be used to quantify the dynamic stability of a biological molecule regarding its conformation. A protein's stability is linked to the deviation experiences during simulation; a smaller number of deviations implies a more stable structure. In this study, dynamic stability was calculated as the RMSD from the carbon alpha backbone for the 500 ns trajectory of each complex. As shown in Fig. 4 , the wild-type system was equilibrated at 10 ns and attained stability at 2.0 Å. However, the system was sustained, and no notable convergence was observed except for a slight deviation of the RMSD at 55–60 ns during the simulation time, which reached up to 4.5 Å. After 60 ns, the RMSD value decreased back and remained stable at the level of 2.0 Å for the wild-type complex until 500 ns. However, a slight deviation was observed at 220–300 ns. Conversely, the N439K complex (spike–ACE2) was equilibrated at 5 ns and gained stability at 2.0 Å. The RMSD increased slightly and reached 3.0 Å for a short interval at 70–150 ns. Moreover, the N439K system remained dynamically more stable, and no notable deviation was observed until 230 ns. However, a slight deviation of the RMSD was observed at 230–270 ns during the simulation time, which reached up to 3.5 Å. Then, the RMSD decreased to 2.2 Å and followed the same pattern until the end of the simulation. The dynamic behavior of the wild type and N439K in terms of stability is comparable.
Fig. 4.
The figure represents the RMSDs of all the complexes. The RMSDs of the wild-type is shown in black colour while the other mutants are given in different colours. (a) show the comparative RMSD of the wild type and N439K, (b) show the comparative RMSD of the wild type and S477 N, while (c) show the comparative RMSD of the wild type and T478K.
The stability of the S477 N (ACE2–spike RBD) complex was also evaluated to demonstrate its dynamic behavior. As shown in Fig. 4, the RMSD value for the S477 N complex gained stability at 2.0 Å and remained steady until 100 ns. The system reached the equilibrium point at 5 ns. The RMSD gradually increased from 100 ns onwards. With a gradual increment in the RMSD after 100 ns, the average RMSD was 4.0 Å for the last 400 ns of simulation time. Conversely, the RMSD remained slightly higher for the S477 N than for the wild type, but the system exhibited a dynamically stable behavior, thus achieving stable binding of the mutant RBD. We also computed the stability of T478K (ACE2–spike RBD), which revealed that the system reached stability at 2.3 Å and remained steady until 100 ns. The RMSD scaled constantly during simulation until the end, reaching 4 Å. However, a slight deviation was observed at 220 ns, at which the RMSD increased a little. The wild-type system also experienced minor fluctuations in the RMSD values at 310 ns and 390–400 ns. Overall, the system remained stable, and the average RMSD was 2.1 Å. The RMSD of the N439K system also remained stable and experienced a slight deviation at 80–160 ns, 220–300 ns, and 360 ns time intervals. The mean RMSD was 2.1 Å, which is comparable with that of the wild type. The structure had stable dynamics and justified the previous finding that the mutations that increased the binding affinity also increased the stability, thus showing a strong correlation of the parameters. The S477 N also remained stable. The system was equilibrated at 10 ns, and the RMSD remained comparable with the wild type for a time period of 10–120 ns. During this time interval, the RMSD remained at 2.0 Å, but the RMSD gradually increased; this trend remained consistent until 500 ns. During the last 380 ns, the mean RMSD reached up to 3.5 Å. As shown in Fig. 4b, although the RMSD increased gradually, no significant deviation was found in the system. The evolution of the variant (S477 N) was stable, increasing the binding affinity. Moreover, the dynamics behavior of S477 N and T478K were comparable, as the RMSD of both systems increased after 100 ns, and the mean RMSD for the last 400 ns increased up to 4.0 Å (Fig. 4c). The fluctuating RMSD during simulation may be related to the opening or closing of the claw-like structure in ACE2, as reported by a previous study [22]. Our findings are also consistent with previous reports. As global RBD stability has been reported to contribute to ACE2-binding affinity, in this study, the mutants increased the stability of each system. Moreover, a strong relationship between RBD stability and affinity is corroborated by previous findings, in which mutations that increase structural stability and rigidity accompany upsurges in binding affinity. For example, other findings have reported that a destabilizing mutation C432D in the RBD lessens ACE2-assisted entry into the cell using a spike trimer. In recently reported mutations in the United Kingdom, South Africa, Brazil, and other countries, the stability also increased and claimed a stable evolution of the new variants [53]. Thus, our findings show that N439K, S477 N, and T478K have stable dynamics and have evolved stably, further increasing their unusual virulence. The RMSDs of all the complexes in triplicate are shown in Fig. 4.
3.4. Analysis of structural compactness
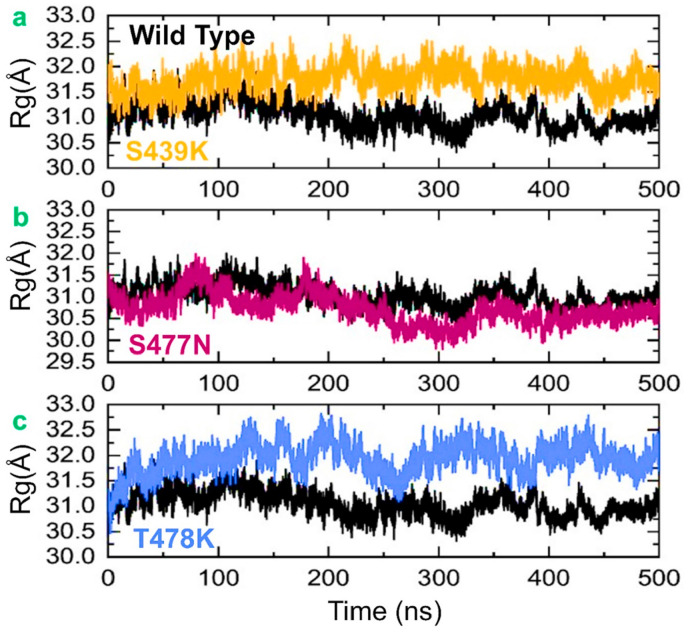
The radius of gyration (Rg) was employed for the measurement of protein structural compactness during simulation. This analysis helped to understand whether the interacting molecules got along with each other and whether the molecules remained in equilibrium and were not high-energy molecules that would push the system to be highly unstable. A higher Rg value is an indication of a highly unstable system, whereas a lower Rg implies a stable and equilibrium system. As shown in Fig. 5 , both the wild-type and mutant complexes followed the same pattern of Rg and underwent oscillation in the Rg values during the simulation time. The average Rg value was 31.0 Å for the wild type, 31.5 Å for N439K, 31.0 Å for S477 N, and 32.0 Å for T478K. During the entire simulation, the Rg values for the mutants N439K and T478K remained higher than those for the wild type, but the average value for the mutant S477 N was comparable with that for the wild type. The fluctuation of Rg value during the simulation was due to binding and unbinding of one or both ends of the spike receptor-binding domain, also reported by previous findings [9,10,50].
Fig. 5.
The figure represents the Rgs of all the complexes. The Rgs of the wild-type is shown in black colour while the other mutants are given in different colours. (a) show the comparative Rg of the wild type and N439K, (b) show the comparative Rg of the wild type and S477 N while (c) show the comparative Rg of the wild type and T478K.
3.5. Flexibility analysis of residues
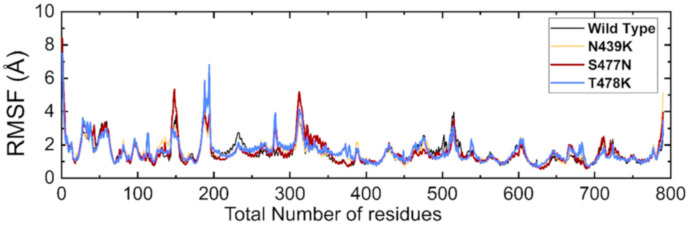
To give insights into the dynamic function relationship of protein motions caused by evolutionary divergence, the root mean square fluctuation (RMSF) value of C-alpha carbon was analyzed and compared (Fig. 6 ). Residual flexibility plays an imperative role in different biological processes, such as molecular recognition, macromolecular complex association, catalysis, and rigidity. The RMSF value visualizes the flexibility of the different regions. A lower RMSF value indicates a less flexible region, whereas a higher RMSF value indicates maximal movements in its average position during simulation. As shown in Fig. 6, regions 20–70, 140–160, 180–200, 280–320, 470–520, and 580–600 exhibited higher flexibility in all the complexes. In the case of the T478K variants, a higher fluctuation was observed in the 140–160 residues, which was not experienced by other systems. Surprisingly, the T478K mutant showed a higher fluctuation in the 180–200 amino acids. Fluctuation in this region is considerably higher than in other regions, and it is due to the distribution of three important loops vital for binding with ACE2, thus implicating the functional relevance of the mutant complexes. Three loops in the spike RBD domain, γ1 (474–485), γ2 (488–490), and γ3 (494–505), which are crucial for binding with ACE2, have higher fluctuation in the mutant systems but not in the wild type [9,10,22,35,50]. The residue Lys417 is essential for binding with ACE2, which shows limited fluctuation. These findings are similar to previous studies that reported ACE2–RBD dynamics [9,10]. The S477 N complex also showed an unusual fluctuation at 280–320, and no noticeable differences were observed in the other regions.
Fig. 6.
This figure represents the residual flexibility (RMSF) index of the wild-type and mutant complexes.
3.6. Hydrogen bonding analysis
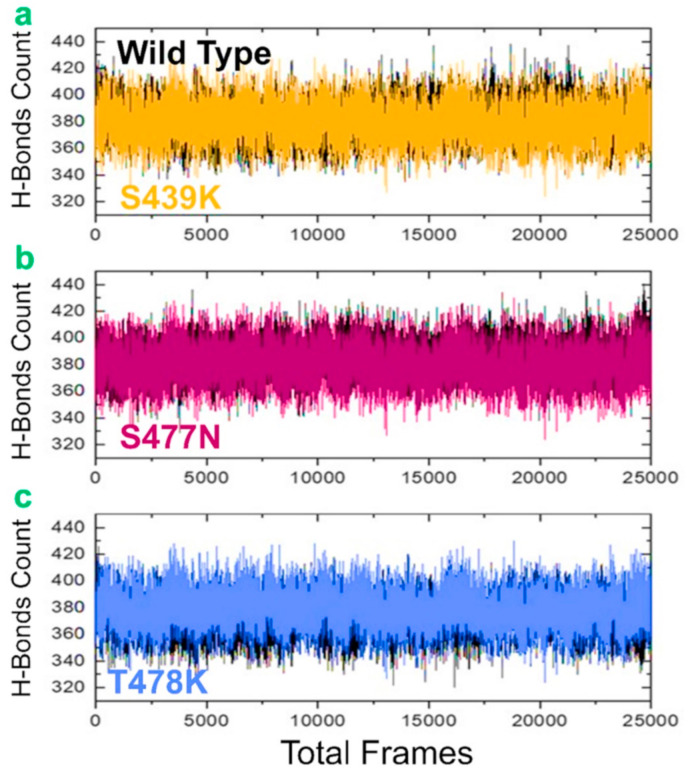
The hydrogen bonds were analyzed to verify the specificity of the wild-type and mutant complexes for the ACE2 receptor as a result of biological processes driven by hydrogen bonding. This is also important to derive stable connections that allow intermolecular contact for a longer duration and to carry out a functional role. The wild-type spike and variations are linked to the AC2 receptor by hundreds of hydrogen bonds in each frame of the MD simulations, as shown in the hydrogen bond graphs. The preceding MD simulation study was confirmed, and the systems were categorized as extremely stable. In each complex, the average hydrogen bonds were calculated. The average hydrogen bonds were 382 in the wild type, 387 in N439K, 385 in S477 N, and 386 in T478K (Fig. 7 ). These findings indicate that the mutations in the above-mentioned variants have changed their hydrogen-bonding contacts, suggesting that the increased binding affinity is associated with variations in the hydrogen bonding network.
Fig. 7.
The figure represents the total number of hydrogen bonds of all the complexes. The total number of hydrogen bonds of the wild-type is shown in black colour while the other mutants are given in different colours. (a) show the comparative total number of hydrogen bonds of the wild type and N439K, (b) show the comparative total number of hydrogen bonds of the wild type and S477 N while (c) show the comparative total number of hydrogen bonds of the wild type and T478K.
3.6.1. MM/GBSA based binding energies
The strength of intermolecular binding determined by docking studies is usually misleading, as the binding pose generated may result in false positives. Therefore, there is a need to conduct an in-depth atomic-level interaction energy analysis to assess the degree of binding between the receptor and the ligand molecule. The MM/GBSA is a popular post-simulation method because it is computationally less expensive, and the results are comparable with experimental data. Therefore, to determine the binding stability of the complexes and to highlight the strength of the hotspot residues in the binding, the MM/GBSA approach was used on the generated simulation trajectories. The MM/GBSA is mostly used to re-evaluate the docking conformation and affinities of the binding complexes. Due to the high credibility of this method, we analyzed the effect of the reported mutations in the RBD domain (N439K, S477 N, and T478K) on the interaction network with the human receptor ACE2. Table 4 shows the various energy components calculated using the MM/GBSA approach for the wild-type and mutant complexes. Free energy was computed using 25,000 structural frames. According to the MM/GBSA analysis, the reported mutations (N439K, S477 N, and T478K) enhanced the binding affinity toward the ACE2 receptor compared with the wild-type RBD complex. The total binding energies of the wild type and mutant complexes were −61.86, −67.85, −69.82, and −69.64 kcal/mol. These findings are consistent with prior research, which found that electrostatic interactions are the driving force behind greater binding and higher infectivity [9,54]. Furthermore, the total binding energy corroborated with the docking scores. The electrostatic, van der Waals, and other calculated components of the binding free energy are given in Table 4. This shows that the binding affinity toward the host cellular receptor increased but not significantly, thus altering infectivity.
Table 4.
MM-GBSA free energy calculation results derived from molecular dynamics simulations of both wild-type and mutants. All the energies in the table are calculated in kcal/mol.
| Complex | Wild Type | N439K | S477 N | T478K |
|---|---|---|---|---|
| vdW | −94.25 | −99.06 | −95.68 | −97.32 |
| Electrostatic | −591.47 | −630.22 | −680.26 | −641.29 |
| GB | 634.78 | 673.35 | 718.15 | 681.27 |
| SA | −10.92 | −11.92 | −12.03 | −12.3 |
| Total Binding energy | −61.86 | −67.85 | −69.82 | −69.64 |
vdW = Van Der Waal; GB = Generalized Born; SA = Surface Area.
4. Conclusions
In conclusion, the current study revealed the binding differences of some reported variants (N439K, S477 N, and T478K) of the SARS-CoV-2. Our results yield that N439K, S477 N, and T478K variants have a higher binding affinity towards the ACE2 receptor. This was inferred from the molecular docking data that predicted the formation of extra salt bridges, hydrogen bonds, and non-bonded contacts. These broad-spectrum interactions network enable the mutants to bind strongly to the ACE2 receptor, giving more structural stability and equilibrium of the systems. The MM/GBSA approach is a highly accepted endpoint technique in estimating binding free energies; the role of ligand-water interactions and protein-water interactions are often skipped. The binding energy estimated that enhanced binding of the mutants is the outcome of better electrostatic, van der Waals, and non-polar solvation energies that collectively contributed favorably to the net binding energy of the mutants. Conclusively this shows that the binding affinity towards the host cellular receptor is increased though not significantly and alters the infectivity.
Funding
Dong-Qing Wei is supported by grants from the Key Research Area Grant 2016YFA0501703 of the Ministry of Science and Technology of China, the National Science Foundation of China (Grant No. 32070662,61832019, 32030063), the Science and Technology Commission of Shanghai Municipality (Grant No.: 19430750600), the Natural Science Foundation of Henan Province (162300410060), as well as SJTU JiRLMDS Joint Research Fund and Joint Research Funds for Medical and Engineering and Scientific Research at Shanghai Jiao Tong University (YG2017ZD14). The computations were partially performed at the Pengcheng Lab. and the Center for High-Performance Computing, Shanghai Jiao Tong University.
Availability of data
All the data is available on RCSB, UniProt and any simulation data will be provided on demand.
Declaration of interest
Declared None.
Acknowledgments
The computations were partially performed at the Center for High-Performance Computing, Shanghai Jiao Tong University. We acknowledge their help.
References
- 1.Perlman S. Another decade, another coronavirus. N. Engl. J. Med. 2020;382:760–762. doi: 10.1056/NEJMe2001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019;382(2020):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawana A. [SARS, MERS and coronavirus infections], Nihon rinsho. Jpn. J. Clin. Med. 2016;74:1967–1972. [PubMed] [Google Scholar]
- 4.Zhu Z., Lian X., Su X., Wu W., Marraro G.A., Zeng Y. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir. Res. 2020;21:1–14. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison A.G., Lin T., Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;14:1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature microbiology. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Acter T., Uddin N., Das J., Akhter A., Choudhury T.R., Kim S. Evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as coronavirus disease 2019 (COVID-19) pandemic: a global health emergency. Sci. Total Environ. 2020;730:138996. doi: 10.1016/j.scitotenv.2020.138996. 138996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haque S.M., Ashwaq O., Sarief A., Azad John Mohamed A.K. A comprehensive review about SARS-CoV-2. Future Virol. 2020;15:625–648. doi: 10.2217/fvl-2020-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.-Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol. 2021;236:7045–7057. doi: 10.1002/jcp.30367. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.A. Khan, D.-Q. Wei, K. Kousar, J. Abubaker, S. Ahmad, J. Ali, F. Al-Mulla, S.S. Ali, N. Nizam-Uddin, A.M. Sayaf, Preliminary Structural Data Revealed that the SARS-CoV-2 B. 1.617 Variant's RBD Binds to ACE2 Receptor Stronger than the Wild Type to Enhance the Infectivity, ChemBioChem. [DOI] [PMC free article] [PubMed]
- 11.Day T., Gandon S., Lion S., Otto S.P. On the evolutionary epidemiology of SARS-CoV-2. Curr. Biol. 2020;30:R849–R857. doi: 10.1016/j.cub.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.A. Khan, W. Heng, Y. Wang, J. Qiu, X. Wei, S. Peng, S. Saleem, M. Khan, S.S. Ali, D.-Q. Wei, In Silico and in Vitro Evaluation of Kaempferol as a Potential Inhibitor of the SARS-CoV-2 Main Protease (3CLpro), Phytotherapy research: PTR. [DOI] [PMC free article] [PubMed]
- 13.Khan A., Khan M.T., Saleem S., Junaid M., Ali A., Ali S.S., Khan M., Wei D.-Q. Structural Insights into the mechanism of RNA recognition by the N-terminal RNA-binding domain of the SARS-CoV-2 nucleocapsid phosphoprotein. Comput. Struct. Biotechnol. J. 2020;18:2174–2184. doi: 10.1016/j.csbj.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan A., Khan S., Saleem S., Nizam-Uddin N., Mohammad A., Khan T., Ahmad S., Arshad M., Ali S.S., Suleman M. Immunogenomics guided design of immunomodulatory multi-epitope subunit vaccine against the SARS-CoV-2 new variants, and its validation through in silico cloning and immune simulation. Comput. Biol. Med. 2021:104420. doi: 10.1016/j.compbiomed.2021.104420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilal El Idrissi H. COVID-19: what you need to know. Gene reports. 2020;20:100756. doi: 10.1016/j.genrep.2020.100756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estola T. Coronaviruses, a new group of animal RNA viruses. Avian Dis. 1970;14:330–336. [PubMed] [Google Scholar]
- 17.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature microbiology. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voloch C.M., Silva F R.d., de Almeida L.G.P., Cardoso C.C., Brustolini O.J., Gerber A.L., Guimarães A.P.d.C., Mariani D., Costa R.M.d., Ferreira O.C., Cavalcanti A.C., Frauches T.S., de Mello C.M.B., Galliez R.M., Faffe D.S., Castiñeiras T.M.P.P., Tanuri A., de Vasconcelos A.T.R. medRxiv; 2020. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. 2020.2012.2023.20248598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahman H.S., Aziz M.S., Hussein R.H., Othman H.H., Salih Omer S.H., Khalid E.S., Abdulrahman N.A., Amin K., Abdullah R. The transmission modes and sources of COVID-19: a systematic review. International Journal of Surgery Open. 2020;26:125–136. doi: 10.1016/j.ijso.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spinello A., Saltalamacchia A., Magistrato A. Is the rigidity of SARS-CoV-2 spike receptor-binding motif the hallmark for its enhanced infectivity? Insights from all-atom simulations. J. Phys. Chem. Lett. 2020;11:4785–4790. doi: 10.1021/acs.jpclett.0c01148. [DOI] [PubMed] [Google Scholar]
- 21.Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microb. Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du L., Yang Y., Zhou Y., Lu L., Li F., Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin. Ther. Targets. 2017;21:131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alsaadi E.A., Jones I.M. Membrane binding proteins of coronaviruses. Future Virol. 2019;14:275. doi: 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosch B.J., Bartelink W., Rottier P.J. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 2008;82:8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Omolo C.A., Soni N., Fasiku V.O., Mackraj I., Govender T. Update on therapeutic approaches and emerging therapies for SARS-CoV-2 virus. Eur. J. Pharmacol. 2020;883:173348. doi: 10.1016/j.ejphar.2020.173348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng M.H., Krieger J.M., Kaynak B., Arditi M.A., Bahar I. bioRxiv; 2021. Impact of South African 501. V2 Variant on SARS-CoV-2 Spike Infectivity and Neutralization: A Structure-Based Computational Assessment. [Google Scholar]
- 28.Collier D., Meng B., Ferreira I., Datir R., Temperton N., Elmer A., Kingston N., Graves B., McCoy L., Smith K., Bradley, Thaventhiram J., Ceron-Gutierrez L., Barcenas-Morales G., Wills M., Doffinger R., Gupta R. medRxiv; 2021. Impact of SARS-CoV-2 B.1.1.7 Spike Variant on Neutralisation Potency of Sera from Individuals Vaccinated with Pfizer Vaccine BNT162b2. 2021.2001.2019.21249840. [Google Scholar]
- 29.Davies N.G., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J., Pearson C.A., Russell T.W., Tully D.C., Abbott S., Gimma A. medRxiv; 2020. Estimated Transmissibility and Severity of Novel SARS-CoV-2 Variant of Concern 202012/01 in England. [Google Scholar]
- 30.Kirby T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. The Lancet Respiratory Medicine. 2021;9:e20–e21. doi: 10.1016/S2213-2600(21)00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020;98:495. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R., Chen J., Gao K., Wei G.-W. Vaccine-escape and fast-growing mutations in the United Kingdom, the United States, Singapore, Spain, India, and other COVID-19-devastated countries. Genomics. 2021;113:2158–2170. doi: 10.1016/j.ygeno.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rezaei S., Sefidbakht Y., Uskoković V. Comparative molecular dynamics study of the receptor-binding domains in SARS-CoV-2 and SARS-CoV and the effects of mutations on the binding affinity. J. Biomol. Struct. Dyn. 2020:1–20. doi: 10.1080/07391102.2020.1860829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.B. Webb, A. Sali, Protein Structure Modeling with MODELLER, Structural Genomics, Springer2021, pp. 239-255.
- 37.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O. Dym, D. Eisenberg, T. Yeates, ERRAT, (2012).
- 40.R.A. Laskowski, M.W. MacArthur, D.S. Moss, J.M. Thornton, PROCHECK: a program to check the stereochemical quality of protein structures, J. Appl. Crystallogr., 26 (1993) 283-291.
- 41.Koukos P.I., Reau M.F., Bonvin A.M. bioRxiv; 2021. Shape-restrained Modelling of Protein-Small Molecule Complexes with HADDOCK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan Y., Tao H., He J., Huang S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020;15:1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- 43.Laskowski R.A. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:221–222. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salomon‐Ferrer R., Case D.A., Walker R.C. An overview of the Amber biomolecular simulation package. Wiley Interdisciplinary Reviews: Computational Molecular Science. 2013;3:198–210. [Google Scholar]
- 45.Salomon-Ferrer R., Götz A.W., Poole D., Le Grand S., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theor. Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 46.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theor. Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 47.Khan A., Gui J., Ahmad W., Haq I., Shahid M., Khan A.A., Shah A., Khan A., Ali L., Anwar Z. The SARS-CoV-2 B. 1.618 variant slightly alters the spike RBD–ACE2 binding affinity and is an antibody escaping variant: a computational structural perspective. RSC Adv. 2021;11:30132–30147. doi: 10.1039/d1ra04694b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan A., Khan T., Ali S., Aftab S., Wang Y., Qiankun W., Khan M., Suleman M., Ali S., Heng W. SARS-CoV-2 new variants: characteristic features and impact on the efficacy of different vaccines. Biomed. Pharmacother. 2021:112176. doi: 10.1016/j.biopha.2021.112176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan A., Gui J., Ahmad W., Haq I., Shahid M., Khan A.A., Shah A., Khan A., Ali L., Anwar Z., Safdar M., Abubaker J., Uddin N.N., Cao L., Wei D.-Q., Mohammad A. The SARS-CoV-2 B.1.618 variant slightly alters the spike RBD–ACE2 binding affinity and is an antibody escaping variant: a computational structural perspective. RSC Adv. 2021;11:30132–30147. doi: 10.1039/d1ra04694b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187. doi: 10.1016/j.cell.2021.01.037. e1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Andrade J., Gonçalves P.F.B., Netz P.A. Why does the novel coronavirus spike protein interact so strongly with the human ACE2? A thermodynamic answer. Chembiochem. 2021;22:865–875. doi: 10.1002/cbic.202000455. [DOI] [PubMed] [Google Scholar]
- 53.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310. doi: 10.1016/j.cell.2020.08.012. e1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hussain I., Pervaiz N., Khan A., Saleem S., Shireen H., Wei D.-Q., Labrie V., Bao Y., Abbasi A.A. Genes & Immunity; 2020. Evolutionary and Structural Analysis of SARS-CoV-2 Specific Evasion of Host Immunity; pp. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data is available on RCSB, UniProt and any simulation data will be provided on demand.