Abstract
Background
Drought stress has great negative effects on the plant growth and development. The tolerance of plants to such abiotic stress is triggered by complicated and multilayered signaling pathways to restore cellular homeostasis and to promote survival. The WRKY family is one of the largest transcription factor families in higher plants, and has been well recognized for the roles in regulating plants tolerance to abiotic and biotic stress. However, little is known about how the WRKY genes regulate drought resistance in cotton.
Results
In this work, we identified the WRKY transcription factor GhWRKY1-like from upland cotton as a positive regulator of tolerance to drought that directly manipulates abscisic acid (ABA) biosynthesis. Overexpression of GhWRKY1-like in Arabidopsis constitutively activated ABA biosynthesis genes, signaling genes, responsive genes and drought related maker genes, and led to enhanced tolerance to drought. Further analysis has shown that GhWRKY1-like can interact with “W-box” cis-elements of the promoters of AtNCED2, AtNCED5, AtNCED6 and AtNCED9 which are essential enzymes for ABA biosynthesis, and promotes the expression of those target genes.
Conclusions
In summary, our findings suggest that GhWRKY1-like may act as a positive regulator in Arabidopsis tolerance to drought via directly interacting with the promoters of AtNCED2, AtNCED5, AtNCED6 and AtNCED9 to promote ABA biosynthesis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-021-03238-5.
Keywords: Drought tolerance, ABA, Transcriptional regulation, WRKY, NCED
Background
Plants are constantly threatened by various biotic and abiotic stresses in the terrestrial phyllosphere, which cause massive yield and quality losses annually [1, 2]. Drought is a major factor restricting plant growth, survival and yield all over the world [3, 4]. Due to the never stopped co-evolution and mutual selection, plants have evolved a sophisticated surveillance system to perceive water deficit, and to launch prompt defense responses [5–7].
The plant hormone abscisic acid (ABA) is known as an essential phytohormone that positive regulates plant cellular adaptation to drought [8]. When plants are exposed to drought condition, elevation of ABA content is usually seen within a few minutes or hours, leading to stomatal closure and activation of drought responses related genes [9, 10]. ABA is produced from xanthophylls, the cleavage of cis-isomers of xanthophylls by 9-cis-epoxycarotenoid dioxygenases (NCED) into xanthoxin is the first committed step. A short-chain dehydrogenase/reductase encoded by ABA2, catalyzes the oxidation of xanthoxin to abscisic aldehyde which is then converted into ABA by an abscisic aldehyde oxidase encoded by AAO3 [11]. The ABA perception and signaling transduction consist of three families of proteins, including ABA receptors encoded by pyrabactin resistance 1/pyrabactin resistance 1-like/regulatory component of ABA receptor (PYR/PYL/RCAR) genes, negative regulators encoded by type 2C protein phosphatases (PP2Cs) genes, and positive regulators encoded by sucrose non-fermenting 1-related protein kinases [10, 12]. In the absence of ABA, PYR/PYL/RCARs are not bound to PP2Cs, resulting in high PP2C activity, thereby preventing the activation of SnRK2s. ABA perception leads to the conformational change of PYR/PYL/RCAR receptors, which bind to PP2Cs, thereby releasing phosphorylated SnRK2s for subsequent phosphorylation of ABA-responsive element (ABRE) binding factors (ABFs) to regulate transcription of ABA-responsive genes [13–15]. Although, the biosynthesis pathway, perception model and signaling transduction of ABA have been clearly identified, and already uncovered part of the components’ role in plant responses to drought stress, the factors that involved in transducing drought perception to ABA accumulation are unclear.
The WRKY transcription factors is one of the largest transcription factor families in plants [16, 17]. The WRKY family proteins are characterized by one or two highly conserved WRKYGQK heptapeptide at N-terminal and an atypical zinc finger-like motif at its C-terminal [18]. Generally, the WRKY family proteins are divided into 3 groups (I, II, and III) based on the number of the WRKY domains and the type of Zn-finger motif. The Group I WRKY proteins contain two WRKY domains with C2H2-type zinc finger; The Group II and III contain only one WRKY domain, with group II harboring the C2H2 zinc finger, while the group III harboring the C2HC-type zinc finger [18]. Since the first WRKY gene was cloned in sweet tomato, more members of WRKY family were isolated and widely reported to participate in plant response to biotic and abiotic stresses [19–23]. For example, overexpression of GhWRKY91 (from Gossypium hirsutum) enhances drought tolerance in Arabidopsis [24]. AtWRKY46 regulates development, stress and hormonal response by facilitating growth of lateral roots in salt stress through ABA signaling and auxin homeostasis in Arabidopsis [25]. Three Arabidopsis group III WRKY transcription factors, WRKY46, WRKY54, and WRKY70, are involved in both BR-regulated plant growth and drought response, the wrky46 wrky54 wrky70 triple mutant shows defects in BR-regulated growth,but is more tolerant to drought stress [26]. Overexpression of GhWRKY27a reduces tolerance to drought and resistance to Rhizoctonia solani infection in transgenic tobacco [27].
Cotton is one of the most important economic crops and is cultivated globally [10]. Cotton production is limited by various abiotic stresses, especially drought stress, which causes substantial loss of cotton yield [10]. Therefore, it is meaningful to elucidate the molecular mechanism how cotton copes with drought stress. Here, we identified a Group I WRKY transcription factor GhWRKY1-like, which was obviously up-regulated by mannitol, dehydration and NaCl treatment, overexpression WRKY enhances tolerance to drought stress with hyperaccumulated ABA content and activated ABA-dependent stress responses in Arabidopsis. Further experiments demonstrated that the key enzymes involved in ABA biosynthesis NCED3, NCED5, NCED6 and NCED9 were the direct target of GhWRKY1-like, and were significantly up regulated by GhWRKY1-like. Our results suggest that GhWRKY1-like may promotes plant tolerance to drought stress via directly manipulating ABA de novo biosynthesis.
Results
GhWRKY1-like is nuclear-localized protein and function as a transcription factor
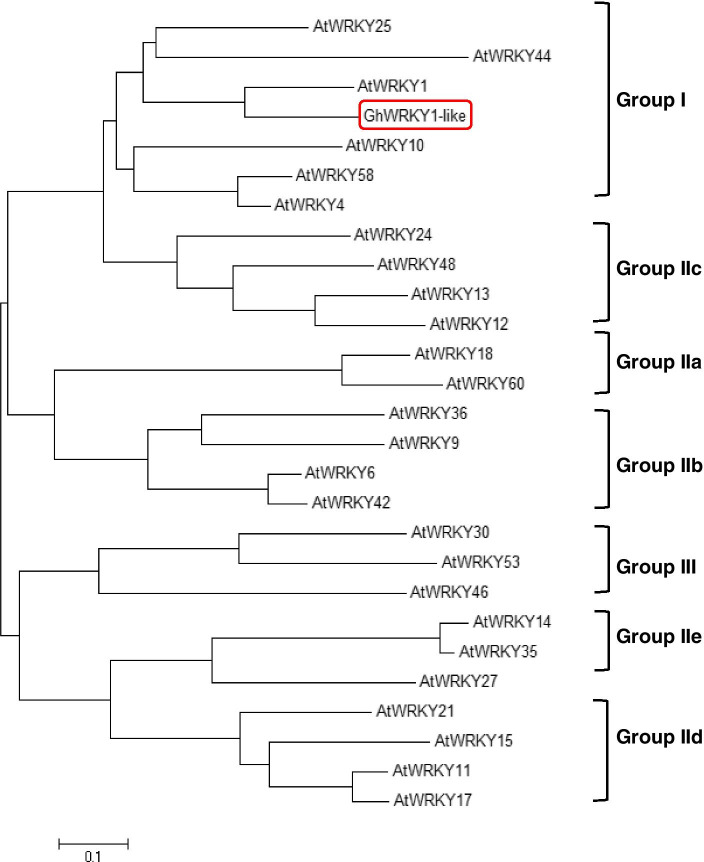
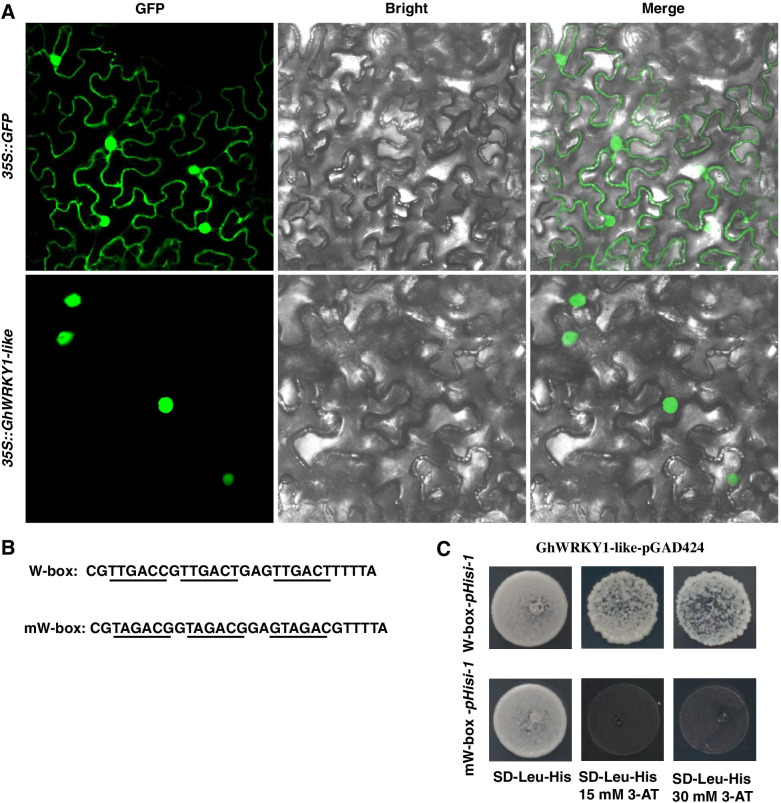
Previously, we have identified a WRKY transcription factor GhWRKY1-like as a positive regulator in cotton defense against Verticillium dahliae [28]. The GhWRKY1-like gene contains a complete open reading frame (ORF) of 1215 bp that encodes a protein with 404 amino acids. Alignment analysis of GhWRKY1-like protein sequence with its homologous sequences, including AtWRKY1, GmWRKY1, PtWRKY1 and TcWRKY1 was performed and the result showed that GhWRKY1-like contains two WRKY domain (WRKYGQK) and two C2H2 zinc finger (C-X4–5-CX22–23-H-X1-H) motif (Fig. S1). Phylogenetic analysis performed with the AtWRKY1 protein sequences showed that the closest orthologs of GhWRKY1-like is AtWRKY1, and belongs to group I WRKY transcription factor (Fig. 1). To analysis the subcellular localization of GhWRKY1-like, the protein of GhWRKY1-like was fused with C-terminal GFP protein, and transient expressed in Nicotiana benthamiana leaves. The result showed that the GhWRKY1-like-GFP fluorescence was seen mainly in the nuclei of cells, indicating GhWRKY1-like is a nuclear-localized protein (Fig. 2 A).
Fig. 1.
Phylogenetic relationship of WRKY proteins from Arabidopsis and Gossypium hirsutum. All the 27 WRKYs protein sequences were subjected to Clustal W using the neighbor-joining method in MEGA 6 and can be divided into three groups (I, II, III), and Group II is further divided into five subgroups (IIa, IIb, IIc, IId, IIe). All the WRKY proteins used for the phylogenetic tree are: AtWRKY1 (AEC05881), AtWRKY4 (NP_172849), AtWRKY6 (NP_564792), AtWRKY9 (AEE34757), AtWRKY10 (NP_175956), AtWRKY11 (NP_849559), AtWRKY12 (NP_566025), AtWRKY13 (NP_195651), AtWRKY14 (NP_564359), AtWRKY15 (NP_973521), AtWRKY17 (NP_565574), AtWRKY18 (NP_567882), AtWRKY21 (NP_565703), AtWRKY24 (NP_198972), AtWRKY25 (NP_180584), AtWRKY27 (NP_568777), AtWRKY30 (NP_568439), AtWRKY35 (AEC09027), AtWRKY36 (NP_564976), AtWRKY42 (NP_192354), AtWRKY44 (NP_181263), AtWRKY46 (AEC10690), AtWRKY48 (NP_199763), AtWRKY53 (NP_194112), AtWRKY58 (NP_186757), AtWRKY60 (NP_180072), and GhWRKY1-like (XP_016696352). At, Arabidopsis thaliana; Gh, Gossypium hirsutum
Fig. 2.
Characterization of GhWRKY1-like as a WRKY family transcriptional factor. A Subcellular localization of GhWRKY1-like protein. GFP fluorescence of 35S::GFP and 35S::GFP-GhWRKY1-like were observed in the tobacco epidermal cell using confocal microscope. B The sequence of the triple tandem repeats of the W-box and mutant W-box binding elements. C Yeast one-hybrid assay using the triple tandem repeats of the W-box and mW-box as bait. Yeast cells were grown on SD-Leu-His containing different concentrations of 3-amino-1,2,4-triazole (3-AT) for 3 to 5 d at 30 °C
Existing research suggests that WRKY transcription factor reprogram gene(s) expression level by directly binding to the W-box [(C/T)TGAC(C/T)] or W-box like [TGAC(C/T)] cis-regulatory elements in the promoter regions of downstream target genes [29]. Thus, we detected whether GhWRKY1-like protein has the ability to bind to the W-box sequence by yeast one-hybrid assay. The triple tandem repeat sequences of W-box (TTGACT) and triple tandem repeat sequences of mutant W-box (TAGACG) were inserted into the pHisi-1 vector to generate W-box-pHisi-1 and mW-box-pHisi-1 constructs, respectively (Fig. 2 B). The full length of GhWRKY1-like was amplified and inserted into pGAD424 vector to form a yeast effector vector GhWRKY1-like-pGAD424. The results confirmed the interaction between GhWRKY1-like-pGAD424 and W-box-pHisi-1, but not occurred between GhWRKY1-like-pGAD424 and mW-box-pHisi-1 (Fig. 2 C). Above findings implied that GhWRKY1-like was a WRKY transcription factor and could bind to the W-box elements of the target promoters to modulate expression of downstream genes.
Overexpression of GhWRKY1-like enhances transgenic plant sensitivity to ABA and tolerance to mannitol
Previous study has shown that GhWRKY1-like is ubiquitously expressed in several organs, and is significantly up-regulated by V. dahliae treatment. Furthermore, we found that GhWRKY1-like was obviously up-regulated by mannitol, dehydration and NaCl treatment in both roots and leaves in upland cotton cv YZ1 using reverse transcription quantitative PCR (RT-qPCR), but GhWRKY1-like showed no response to ABA treatment (Fig. S2A). The induce expression patter suggested that GhWRKY1-like might also involved in plant response to abiotic stress (Fig. S2B). To elucidate the putative biofunction of GhWRKY1-like in plant abiotic stress tolerance, the GhWRKY1-like was introduced into Arabidopsis thaliana for ectopic expression. According to the expression level, we choose one high expression line (OE-1) and one moderate expression line (OE-7) for further research (Fig. S2C). The T4 generation homozygous transgenic seeds of OE-1 and OE-7 was subjected to ABA and mannitol treatment to observe the sensitivity to ABA and mannitol by calculating the germination rate or green seedling rate (Fig. 3). The results showed that the germination rate or green seedling rate of GhWRKY1-like over-expression transgenic Arabidopsis lines were similar to that of WT under normal conditions, while compared with WT, the seed germination rate of the GhWRKY1-like over-expression transgenic Arabidopsis lines were significantly decreased with ABA treatment (Fig. 3 A), and the green seedling rate of GhWRKY1-like over-expression transgenic Arabidopsis lines were significantly increased with mannitol treatment (Fig. 3 B). These findings indicated that GhWRKY1-like may promotes plant drought tolerance via ABA signaling pathway.
Fig. 3.
Overexpression of GhWRKY1-like causes hypersensitivity to ABA-elicited seed germination and enhanced tolerance to mannitol treatment. A Green seedling rate of WT and GhWRKY1-like transgenic lines on 1/2 MS medium supplied with 0.6 μM ABA. Values are means ± SD; n = 6. Comparisons were performed with Student’s t test. **, P < 0.01. B Green seedling rate of WT and GhWRKY1-like transgenic lines on 1/2 MS medium supplied with 500 mM mannitol. Values are means ± SD; n = 6. Comparisons were performed with Student’s t test. **, P < 0.01
Overexpression of GhWRKY1-like enhances transgenic plant tolerance to drought
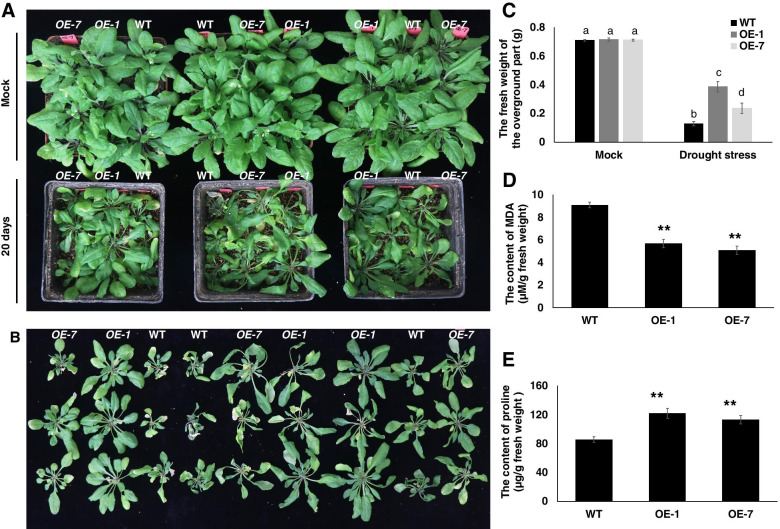
To further investigate how GhWRKY1-like functions in drought response during vegetative growth of the transgenic plants. Two-week-old vermiculite-grown (watered with Hogland medium) GhWRKY1-like transgenic Arabidopsis seedlings and WT were kept away from water for 20 days. As showed in Fig. 4 A, the GhWRKY1-like over-expression plants were much bigger than that of WT, and the performance of OE-1 line with higher GhWRKY1-like expression level was better than that of OE-7, which suggested that drought tolerance mediated by GhWRKY1-like was dosage-dependent (Fig. 4 A and B). The results of fresh weight of the transgenic lines and WT under normal conditions or water-withholding conditions also supported this conclusion (Fig. 4 C). Malondialdehyde (MDA) is one of the most important products of cell-membrane lipid peroxidation, and its production can aggravate the membrane damage. Therefore, MDA content is a common indicator to understand the degree of membrane lipid peroxidation and the potential capability of stress tolerance in plants [30]. The MDA content was obviously lower in GhWRKY1-like transgenic lines than that of WT (Fig. 4 D), which indicating GhWRKY1-like promotes Arabidopsis tolerance to drought by suppression MDA accumulation. Proline is a low molecular weight cyclic amino acid and is known to provide osmotic adjustments in plants under stressful environments. Proline equilibrates the intracellular redox homeostasis by stabilizing antioxidant system through osmotic adjustments and protecting the integrity of cell membranes [31]. Meanwhile, we detected the proline content in GhWRKY1-like transgenic lines and WT, and the result showed that proline was significantly hyperaccumulated in GhWRKY1-like over-expression lines than that of WT (Fig. 4 E), which indicated that GhWRKY1-like over-expression lines had stronger resistance capacity to drought stress.
Fig. 4.
Overexpression of GhWRKY1-like conferred drought tolerance in Arabidopsis. A The phenotypes of GhWRKY1-like transgenic lines tolerant to drought stress. B The size of WT and GhWRKY1-like transgenic lines after 20 days without watering. C The fresh weight of WT and GhWRKY1-like transgenic lines after 20 days without watering. Values are means ± SD; n = 18. In each column, values not followed by the same letters are significantly different based on Tukey’s multiple comparison test (P < 0.05). D The MAD content of WT and GhWRKY1-like transgenic lines after 20 days without watering. Values are means ± SD; n = 6. Comparisons were performed with Student’s t test. **, P < 0.01. E The proline content of WT and GhWRKY1-like transgenic lines after 20 days without watering. Values are means ± SD; n = 6. Comparisons were performed with Student’s t test. **, P < 0.01
GhWRKY1-like positively regulates ABA content and ABA-responsive genes in transgenic Arabidopsis
The phytohormone abscisic acid (ABA) plays an important role in plant development and adaption to biotic and abiotic stresses. Plants rapidly initiate ABA biosynthesis in various organs in response to water deficit. The activated ABA signaling pathway eventually leads to stomatal closure, osmoprotectants accumulation, phenotypical and physiological adaptations to enhance drought resistance [8]. The induced expression pattern showed that GhWRKY1-like had no response to ABA treatment (Fig. S2A and B), but the GhWRKY1-like over-expression lines were much sensitively to ABA treatment as indicated by lower seed germination rate (Fig. 3A). These findings implied that GhWRKY1-like may participate in ABA biosynthesis regulation rather than ABA signaling transduction. Thus, we detected the ABA content in GhWRKY1-like over-expression lines and WT. In accordance with expectation, the ABA content was obviously increased in GhWRKY1-like over-expression lines than that of WT in both normal conditions or drought conditions (Fig. 5A). The expression levels of genes involved in ABA biosynthesis also consistent with this conclusion (Fig. 5B). We also detected the ABA-responsive genes and drought-responsive genes in GhWRKY1-like over-expression lines and WT by RT-qPCR (Fig. 5C and D). The expression levels of these genes were consistently significantly up-regulated in the GhWRKY1-like over-expression lines (Fig. 5C and D). Above findings suggested that GhWRKY1-like was a positive regulator in Arabidopsis response to drought via manipulating ABA biosynthesis.
Fig. 5.
GhWRKY1-like positively regulates Arabidopsis tolerance to drought via manipulating ABA biosynthesis and ABA mediated drought responses. A The ABA content in WT and GhWRKY1-like transgenic lines in normal conditions and drought conditions. Values are means ± SD; n = 9. Comparisons were performed with Student’s t test. *, P < 0.05; **, P < 0.01. B Expression of genes involved in ABA biosynthesis and the ABA signaling pathway in WT and GhWRKY1-like transgenic lines after 10 days without watering. The values are normalized to AtACTIN2 and expressed as the means ± SD; n = 3. C Expression of genes involved in ABA responsive genes in WT and GhWRKY1-like transgenic lines after 10 days without watering. The values are normalized to AtACTIN2 and expressed as the means ± SD; n = 3. D Expression of genes involved in abiotic stress-responsive genes in WT and GhWRKY1-like transgenic lines after 10 days without watering. The values are normalized to AtACTIN2 and expressed as the means ± SD; n = 3
GhWRKY1-like promotes ABA biosynthesis by directly binding to the promoters of AtNCED2, AtNCED5, AtNCED6 and AtNCED9
To elucidated the mechanism(s) by which GhWRKY1-like regulates ABA biosynthesis, we cloned several promoters (about 2000 bp in the upstream of ATG) of ABA biosynthesis related genes to detect the interaction with GhWRKY1-like by yeast one hybrid. As shown in Fig. 6A, we identified that GhWRKY1-like could directly bind to the promoters of AtNCED2 (nine-cis-epoxycarotenoid dioxygenases, NCED; ProAtNCED2, 2434 bp in the upstream of ATG), AtNCED5 (ProAtNCED5, 2021 bp in the upstream of ATG), AtNCED6 (ProAtNCED6, 2278 bp in the upstream of ATG) and AtNCED9 (ProAtNCED9, 2314 bp in the upstream of ATG), but not interact with promoter of AtNCED3 (ProAtNCED3, 2200 bp in the upstream of ATG). Sequence analysis showed that there were at least one W-box in the promoter region of AtNCED2, AtNCED5, AtNCED6 and AtNCED9 (Fig. 6B). To elucidate the function of GhWRKY1-like (activator or repressor) for proAtNCED2, proAtNCED5, proAtNCED6 and proAtNCED9, a dual-luciferase reporter system was performed in Arabidopsis protoplasts (Fig. 6C). The ratio of LUC activity to the control REN (Renilla luciferase) activity or the LUC luminescence intensity was used to indicate the activity of promoters. As shown in Fig. 6D, compared with the control vector, the expression levels of proAtNCED2::LUC, proAtNCED2::LUC5, proAtNCED6::LUC and proAtNCED9::LUC were significantly activated in the present of GhWRKY1-like (Fig. 6D). These data suggested that AtNCED2, AtNCED5, AtNCED6 and AtNCED9 were the direct targets of GhWRKY1-like, and the accumulated ABA content in GhWRKY1-like over-expression lines might resulted from the activation of AtNCED2, AtNCED5, AtNCED6 and AtNCED9 expression.
Fig. 6.
GhWRKY1-like positively regulated the expression of ABA biosynthesis related genes AtNCED2, AtNCED5, AtNCED6 and AtNCED9. A Yeast one-hybrid assay showing the binding of GhWRKY1-like to the promoters of AtNCED2, AtNCED5, AtNCED6 and AtNCED9. Empty pHisi-1 vector was used as a negative control. Yeast cells were grown on SD-Leu-His containing different concentration different concentrations of 3-amino-1,2,4-triazole (3-AT) for 3 to 5 d at 30 °C. B Diagram of W-box positions in the promoters of AtNCED2, AtNCED5, AtNCED6 and AtNCED9. C Diagram of the effector and reporter constructs for the transactivation assay. The reporter carries a renilla luciferase gene under the control of the 35S promoter and a firefly luciferase reporter gene under the control of the promoter of AtNCED2, AtNCED5, AtNCED6 or AtNCED9. 35S, promoter of cauliflower mosaic virus 35S RNA gene; REN, Renilla luciferase; CaMV term, cauliflower mosaic virus terminator; LUC, firefly luciferase. D Translational changes of the reporters in the presence of GhWRKY1-like in Arabidopsis protoplasts. The LUC/REN activity ratios are the means ± SD; n = 3. Comparisons were performed with Student’s t test. *, P < 0.05; **, P < 0.01
Discussion
Global climate models predict a significant increase in intensity and frequency of hot and dry days, which is predicted to have a general negative effect on crop yield and quality [32, 33]. With increasing water scarcity and global population explosion, drought is emerging as a prominent constraint on crop production [10, 34]. Owing to the characteristics of the sessile lifestyle, plants have evolved a complex regulatory circuitry in response to drought stress. The plant hormone ABA regulates various physiological processes throughout plant life cycles and manipulated plant resistant responses to various abiotic stresses. The quickly accumulated ABA content is a typical plants’ response to water deficit and leads to stomatal closure and the expression of numerous stress-responsive genes [35, 36]. The mutant that impaired ABA synthesis or ABA signaling pathway always resulted in hypersensitivity to drought. For example, the AtNCED3 is known as an important enzyme for ABA accumulation during water deficit and the expression level is highly induced in the vascular tissues by drought stress, the knock out mutant of AtNCED3 (nced3–2) results in decreased ABA accumulation and is more sensitive to drought stress [37]. The aba2 mutant in Arabidopsis shows ABA-insensitive phenotype and reduced seed dormancy with high salt concentration, and reduced stomatal closure in response to a decrease in humidity [38]. The snrk2.2/3/6 triple mutant is totally abolished in ABA response and hypersensitive to water deficit [39, 40]. Although ABA biosynthesis and signaling pathway have been studied extensively, and nearly all enzymes involved in ABA synthesis have been cloned, the mechanism by which plants precisely regulate ABA accumulation (biosynthesis and degradation) after the perception of water deficit or osmotic stress is not well understood.
Actually, the basis for plants to establish effective defense responses depends on the precise expression reprogramming of stress-responsive genes including stress-induced hormones biosynthesis, signaling transduction and osmoprotectants metabolism related genes, and this regulatory circuitry is comprised of transcriptional activators and repressors [41, 42]. The WRKY faminly is one of the largest families of transcription regulators in plants, and have been well recognized for the roles in regulating abiotic and biotic stress tolerance. For instance, ectopic expression of GhWRKY33 enhances transgenic Arabidopsis drought sensitivity with down-regulated expression level of RD29A, DREB2A, ERD15, SOS2, ABI1 and RAB18 [43]. Interfamily expression of PbrWRKY53 from Pyrus betulaefolia in tobacco and Pyrus ussuriensis confers enhanced tolerance to drought stress in the transgenic plants, and exhibits better water status, less reactive oxygen species generation and higher levels of antioxidant enzyme activities and metabolites than the wild type [44]. Further research demonstrates that PbrWRKY53 can bind to the W-box element in the promoter region of PbrNCED1 to modulate stomatal aperture and ABA biosynthesis [44]. Conversely, the Arabidopsis wrky46wrky54wrky70 triple mutant shows enhanced tolerant to drought stress via modulating brassinosteroids-regulated plant growth and promoting expression levels of drought responsive genes, which indicates AtWRKY46, AtWRKY54 and AtWRKY70 are negative regulators of drought tolerance [26]. In our study, we found that GhWRKY1-like was significantly induced by mannitol, dehydration and Nacl treatment, but showed no response to ABA treatment (Fig. S2). Further research confirmed that over-expression GhWRKY1-like enhanced Arabidopsis transgenic lines resistance to drought and hyposensitive to ABA treatment accompanied with hyperaccumulated ABA content (Figs. 4 and 5). The yeast one hybrid assay and transient expression analysis demonstrated that GhWRKY1-like could bind to the W-box element in the promoter region of AtNCED2, AtNCED5, AtNCED6 and AtNCED9, and could promote the expression level of those genes involved in ABA biosynthesis (Fig. 6). According to present research in Arabidopsis, there are five genes encode the 9-cis-epoxycarotenoid dioxygenases and constitute a key step in the regulation of ABA biosynthesis [45]. AtNCED3 has been shown to play a major role in the regulation of ABA synthesis in response to water deficit [45], whereas AtNCED6 and AtNCED9 have been shown to be essential for the ABA production in the embryo and endosperm that imposes dormancy [46]; AtNCED2, AtNCED5, AtNCED9 have been shown to contribute to the thermoinhibition of germination by increasing ABA levels at high temperature [47]. A detailed phenotypic analysis of Atnced single, double and triple mutants generated from the combination of the Atnced5 mutation with Atnced3, Atnced6 and Atnced9 showed that AtNCED5 participates in the regulation of seed dormancy together with AtNCED6 and AtNCED9, and contributes with AtNCED3 to the increased ABA levels induced in vegetative tissues upon the onset of water deficit [11]. Although, GhWRKY1-like not directly binds to the promoter region of AtNCED3 which play a major role in the regulation of ABA synthesis in response to water deficit (Fig. 6A), GhWRKY1-like may indirectly manipulate AtNCED3 mediated drought responses via directly promoting the transcriptional levels of AtNCED5, AtNCED6 and AtNCED9 (Fig. 6A). Moreover, the expressions of the ABA signaling genes (ABI1, ABI2, ABI4, and ABI5), responsive genes (RD29A, COR15A, COR15B, and RD22) and stress-related marker genes (KIN1, LEA14, LEA76, and CER3) were significantly up-regulated in transgenic lines under drought stress (Fig. 5B).
Conclusion
Taken together, the results obtained in this study indicated that GhWRKY1-like played a positive role in drought tolerance by promoting ABA biosynthesis via directly regulating AtNCED2, AtNCED5, AtNCED6 and AtNCED9 expression, and thus holding a great potential in improving plant stress tolerance.
Methods
Plant material, growth conditions and treatments
Cotton (Gossypium hirsutum) cultivar “YZ1” provided by our lab was used to investigated the abiotic stresses induced expression pattern of GhWRKY1-like including 300 mM mannitol, 100 μM ABA, 200 mM NaCl and dehydration. Cotton seeds of YZ1 were germinated and cultivated with Hoagland solution [48] in a culture room until the three-leaf stage was reached [28]. Then, the seedings were moved to Hoagland solution containing 300 mM mannitol or 200 mM NaCl for mannitol and NaCl treatment. For ABA treatment, the seedings were moved to Hoagland solution containing 100 μM ABA and the leaves were also sprayed with this solution. For dehydration treatment, the whole plants were removed from the Hoagland solution, and placed on paper towels on a laboratory bench. For each treatment, the roots and leaves were harvested at 0, 3, 6, 12, 24, and 48 h, respectively. Samples were frozen in liquid nitrogen and stored at − 80 °C for further research.
Arabidopsis thaliana seeds (Col-0, obtained from the Arabidopsis Biological Resource Center (ABRC) through TAIR (www.arabidopsis.org) and kept in our lab) were firstly vernalized at 4 °C for 2 days, then sowing in soil and the seedlings were grown in a culture room at with a22 °C and 16 h light/8 h dark photoperiod [45]. For ABA and mannitol treatments, the Arabidopsis seeds were grown on the 1/2 Murashige and Skoog (MS) agar plate supplied with 0.6 μM ABA or 500 mM mannitol for 2 days at 4 °C, and them move to normal growth condition to observe the green seeding rate.
RAN extraction and expression analysis
Total RNA extraction, reversing transcription for the first strand cDNA synthesis and quantitative real-time PCR (qRT-PCR) was carried out according to our previous description [28]. The relative gene expression level was calculated by the 2–ΔΔCT method. GhUB7 and AtACTIN2 were used as an internal controls for cotton and Arabidopsis, respectively. The primer information was listed in Table S1.
GhWRKY1-like cloning, sequence analysis, vector construction and Arabidopsis transformation
The isolation and cloning of GhWRKY1-like open reading frame (ORF, Gh_D11G1536, https://cottonfgd.org) and the construction of GhWRKY1-like over-expression vector were performed as previously described [28], and the Agrobacterium-mediated transformation by floral dipping method was performed in Col-0 Arabidopsis to generate GhWRKY1-like over-expression lines [49]. The analysis of amino acid sequences alignment and phylogenetic relationship of the WRKY proteins were performed using ClustalX (http://www.clustal.org) and MEGA6 (http://www.megasoftware.net), respectively.
Subcellular localization of GhWRKY1-like protein
The ORF of GhWRKY1-like were amplified with GhWRKY1-like-GFP-F/R, and cloned into vector pMDC83 by BP and LR recombination reactions to generate the C-terminally fused GFP construct to determine the subcellular localization of GhWRKY1-like protein. The empty vector of pGWB452 with a N-terminally fused GFP was used as control. All vectors were transformed into Nicotiana benthamiana leaves via the Agrobacterium tumefaciens strain GV3101 [50] and the fluorescence of GFP were observed using a confocal microscope (Olympus FV1200) after infiltrated for 60 h. The primers used in this study are listed in Table S1.
Yeast one-hybird assay
The yeast one-hybird assay was conducted according to the manufacturer’s protocol (MATCHMAKER One-Hybrid System User Manual, Clontech). The ORF of GhWRKY1-like were amplified with GhWRKY1-like-AD-F/R and cloned into vector pGAD424 at EcoRI and BamH to generate GhWRKY1-like-pGAD424 construct as AD vector. Oligonucleotide sequences containing specific adaptor and the W-box or mW-box were synthesized and inserted into pHisi-1 vector at SacI to obtain W-box-pHisi-1 and mW-box-pHisi-1 as reporter vectors. The reporter vector was linearized with XhoI and co-transferred with GhWRKY1-like-pGAD424 into YM4271 yeast strain. The promoter region of AtNCED2 (AT4G18350, https://www.arabidopsis.org/), AtNCED3 (AT3G14440, https://www.arabidopsis.org/), AtNCED5 (AT1G30100, https://www.arabidopsis.org/), AtNCED6 (AT3G24220, https://www.arabidopsis.org/) and AtNCED9 (AT1G78390, https://www.arabidopsis.org/) were also amplified with the corresponding primers and inserted into pHisi-1 vector at SacI to obtain ProAtNCED2-pHisi-1, ProAtNCED3-pHisi-1, ProAtNCED5-pHisi-1, ProAtNCED6-pHisi-1 and ProAtNCED9-pHisi-1 as reporter vectors. The reporter vector was linearized with XhoI (ProAtNCED3-pHisi-1 and ProAtNCED9-pHisi-1 were linearized with AflII) and co-transferred with GhWRKY1-like-pGAD424 into YM4271 yeast strain. The yeast cells were cultured on synthetic SD-His-Leu medium with 15 mM or 30 mM 3-amino-1,2,4-triazole (3-AT) for 3 to 5 d at 30 °C to confirm the interaction between GhWRKY1-like and each promoter. The primers used in this study are listed in Table S1.
Drought treatment in transgenic Arabidopsis
For drought treatment, the seeds of WT and the T4 generation of GhWRKY1-like overexpression lines were sown in vermiculite for 3 weeks and watered with Hoagland solution. The drought stress were set as 20 days without watering. And the overground parts were collected for fresh weight, malondialdehyde (MAD) and proline content determination.
The determination of MAD and proline content
The measurement of MAD was performed using the thiobarbituric acid (TBA) method [51]. In brief, approximately 100 mg fresh samples were immediately homogenized in 1 mL 5% (w/v) trichloroacetic acid (TCA) solution using a Tissuelyser (Jingxin, Shanghai, China). After centrifugated at 4 °C for 10 min at 6000 rpm, 400 μL of the supernatant was taken out and added into 400 μL 10% TCA containing 0.67% (w/v) thiobarbituric acid (TBA) in a new tube, then the mixture was incubated in boiling water for 30 min. The mixture was centrifugated for 5 min at 12,000 rpm after cooling to room temperature and the OD450 nm, OD532 nm, and OD600nm of the supernatant were determined using a Multimode Plate Reader (PerkinElmer).
The content of free proline in plants was determined according to a reported method [52]. Briefly, 100 mg fresh samples were homogenized in 1 mL 3% (w/v) sulfosalicylic acid using a Tissuelyser (Jingxin, Shanghai, China), and incubated in boiling water for 10 min. After cooling to room temperature, the samples were centrifugated at 12,000 rpm for 5 min. 400 μL of the supernatant was taken out and mixed with 400 μL acid ninhydrin and 400 μL glacial acetic in a new tube, then mixture was incubated in a boiling water bath for 30 min. After centrifuged at 6000 rpm for 5 min, the proline content was measured at 520 nm using a Multimode Plate Reader (PerkinElmer).
The determination of ABA content
The quantitative analysis of the endogenous concentration of ABA was performed using an internal standard method as described previously [53]. 2H6ABA (OlChemIm, Olomouc, Czech Republic) was used as an internal standard.
Dual-luciferase reporter assays in Arabidopsis protoplasts
The promoter regions of AtNCED2, AtNCED5, AtNCED6 and AtNCED9 were amplified with the corresponding primers and inserted into pGreenII 0800 vector at HindIII and BamHI to obtain the ProAtNCED2-pGreenII 0800, ProAtNCED5- pGreenII 0800, ProAtNCED6- pGreenII 0800 and ProAtNCED9-pGreenII 0800 vectors as reporter constructs. The full length of GhWRKY1-like was amplified with GhWRKY1-like-SK-F/R and inserted into pGreenII 62-SK vector at PstI and BamHI to obtain GhWRKY1-like-62-SK construct as an effector; the pGreenII 62-SK vector without any DNA insertion was used as an empty vector control. The dual-luciferase reporter assays in Arabidopsis thaliana (Col-0) protoplasts were performed as described previously [54].
Supplementary Information
Additional file 1: Figure S1. Sequence and phylogenetic analysis of GhWRKY1-like. Sequence alignment of the amino acid sequence of GhWRKY1-like (XP_016696352) with AtWRKY1 (AEC05881), GmWRKY1 (XP_003518571), PtWRKY1 (XP_006375555), and TcWRKY1 (XP_007049283). Conserved WRKY domain and zinc finger motif are shown in red box and green box, respectively.
Additional file 2: Figure S2. The abiotic stresses including mannitol, ABA, dehydration and NaCl induced expression patterns of GhWRKY1-like.
Additional file 3: Table S1. Primers used in vectors constructed and qRT-PCR. The underlined sequences were the infusion sequences.
Acknowledgments
Not applicable.
Authors’ contributions
Q.H. and X.D. conceived and designed the experiments; Q.H. performed experiments for the gene biofunction assays and wrote the article; C.A., X.W. and Y.W. performed the drought tolerance assessment and the dual-luciferase report analysis; X.D. revised the article; all the authors discussed the results, revised the manuscript and approved submission of this work.
Funding
This work was supported by the National Natural Science Foundation of China (32001525) and the Natural Science Foundation of Hubei Province (2020CFB261) to Q.H., and the Natural Science Foundation of Hubei Province (2019CFA027) to X.D.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nelson R, Wiesner-Hanks T, Wisser R, Balint-Kurti P. Navigating complexity to breed disease-resistant crops. Nat Rev Genet. 2018;19(1):21–33. doi: 10.1038/nrg.2017.82. [DOI] [PubMed] [Google Scholar]
- 2.Avin-Wittenberg T. Autophagy and its role in plant abiotic stress management. Plant Cell Environ. 2019;42(3):1045–1053. doi: 10.1111/pce.13404. [DOI] [PubMed] [Google Scholar]
- 3.Dai AG. Increasing drought under global warming in observations and models (vol 3, pg 52, 2013) Nat Clim Chang. 2013;3(2):171. doi: 10.1038/nclimate1811. [DOI] [Google Scholar]
- 4.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167(2):313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Volaire F. A unified framework of plant adaptive strategies to drought: crossing scales and disciplines. Glob Chang Biol. 2018;24(7):2929–2938. doi: 10.1111/gcb.14062. [DOI] [PubMed] [Google Scholar]
- 7.Golldack D, Li C, Mohan H, Probst N. Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci. 2014;5:151. doi: 10.3389/fpls.2014.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida T, Mogami J, Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol. 2014;21:133–139. doi: 10.1016/j.pbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 9.Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci. 2014;5:170. doi: 10.3389/fpls.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li FJ, Li MY, Wang P, Cox KL, Duan LS, Dever JK, et al. Regulation of cotton (Gossypium hirsutum) drought responses by mitogen-activated protein (MAP) kinase cascade-mediated phosphorylation of GhWRKY59. New Phytol. 2017;215(4):1462–1475. doi: 10.1111/nph.14680. [DOI] [PubMed] [Google Scholar]
- 11.Frey A, Effroy D, Lefebvre V, Seo M, Perreau F, Berger A, et al. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 2012;70(3):501–512. doi: 10.1111/j.1365-313X.2011.04887.x. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, et al. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005;44(6):939–949. doi: 10.1111/j.1365-313X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- 13.Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15(7):395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Sussmilch FC, Atallah NM, Brodribb TJ, Banks JA, McAdam SAM. Abscisic acid (ABA) and key proteins in its perception and signaling pathways are ancient, but their roles have changed through time. Plant Signal Behav. 2017;12(9):e1365210. doi: 10.1080/15592324.2017.1365210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu FF, Xie Q. Ubiquitination modification precisely modulates the ABA signaling pathway in plants. Yi Chuan. 2017;39(8):692–706. doi: 10.16288/j.yczz.17-043. [DOI] [PubMed] [Google Scholar]
- 16.Barco B, Clay NK. Hierarchical and dynamic regulation of defense-responsive specialized metabolism by WRKY and MYB transcription factors. Front Plant Sci. 2020;10:1775. doi: 10.3389/fpls.2019.01775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schluttenhofer C, Yuan L. Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 2015;167(2):295–306. doi: 10.1104/pp.114.251769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15(5):247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol Gen Genet. 1994;244(6):563–571. doi: 10.1007/BF00282746. [DOI] [PubMed] [Google Scholar]
- 20.Tripathi P, Rabara RC, Rushton PJ. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta. 2014;239(2):255–266. doi: 10.1007/s00425-013-1985-y. [DOI] [PubMed] [Google Scholar]
- 21.Bakshi M, Oelmuller R. WRKY transcription factors: Jack of many trades in plants. Plant Signal Behav. 2014;9(2):e27700. doi: 10.4161/psb.27700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150(4):1648–1655. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai Y, Sunarti S, Kissoudis C, Visser RGF, van der Linden CG. The role of tomato WRKY genes in plant responses to combined abiotic and biotic stresses. Front Plant Sci. 2018;9:801. doi: 10.3389/fpls.2018.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu LJ, Ma Q, Zhang C, Wang CC, Wei HL, Wang HT, et al. The cotton GhWRKY91 transcription factor mediates leaf senescence and responses to drought stress in transgenic Arabidopsis thaliana. Front Plant Sci. 2019;10:1352. doi: 10.3389/fpls.2019.01352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding ZJ, Yan JY, Li CX, Li GX, Wu YR, Zheng SJ. Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 2015;84(1):56–69. doi: 10.1111/tpj.12958. [DOI] [PubMed] [Google Scholar]
- 26.Chen JN, Nolan TM, Ye HX, Zhang MC, Tong HN, Xin PY, et al. Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell. 2017;29(6):1425–1439. doi: 10.1105/tpc.17.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan Y, Jia HH, Wang F, Wang C, Liu SC, Guo XQ. Overexpression of GhWRKY27a reduces tolerance to drought stress and resistance to Rhizoctonia solani infection in transgenic Nicotiana benthamiana. Front Physiol. 2015;6:265. doi: 10.3389/fphys.2015.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Q, Xiao S, Wang X, Ao C, Zhang X, Zhu L. GhWRKY1-like enhances cotton resistance to Verticillium dahliae via an increase in defense-induced lignification and S monolignol content. Plant Sci. 2021;305:110833. doi: 10.1016/j.plantsci.2021.110833. [DOI] [PubMed] [Google Scholar]
- 29.Viana VE, Busanello C, da Maia LC, Pegoraro C, Costa de Oliveira A. Activation of rice WRKY transcription factors: an army of stress fighting soldiers? Curr Opin Plant Biol. 2018;45(Pt B):268–275. doi: 10.1016/j.pbi.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Tsikas D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: analytical and biological challenges. Anal Biochem. 2017;524:13–30. doi: 10.1016/j.ab.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 31.Fu Y, Ma H, Chen S, Gu T, Gong J. Control of proline accumulation under drought via a novel pathway comprising the histone methylase CAU1 and the transcription factor ANAC055. J Exp Bot. 2018;69(3):579–588. doi: 10.1093/jxb/erx419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawas LMF, Zuther E, Jagadish SK, Hincha DK. Molecular mechanisms of combined heat and drought stress resilience in cereals. Curr Opin Plant Biol. 2018;45(Pt B):212–217. doi: 10.1016/j.pbi.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 33.Bita CE, Gerats T. Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci. 2013;4:273. doi: 10.3389/fpls.2013.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu H, Xiong L. Genetic engineering and breeding of drought-resistant crops. Annu Rev Plant Biol. 2014;65:715–741. doi: 10.1146/annurev-arplant-050213-040000. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, et al. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015;38(1):35–49. doi: 10.1111/pce.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agurla S, Gahir S, Munemasa S, Murata Y, Raghavendra AS. Mechanism of stomatal closure in plants exposed to drought and cold stress. Adv Exp Med Biol. 2018;1081:215–232. doi: 10.1007/978-981-13-1244-1_12. [DOI] [PubMed] [Google Scholar]
- 37.Ruggiero B, Koiwa H, Manabe Y, Quist TM, Inan G, Saccardo F, et al. Uncoupling the effects of abscisic acid on plant growth and water relations. Analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in Arabidopsis. Plant Physiol. 2004;136(2):3134–3147. doi: 10.1104/pp.104.046169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez-Guzman M, Apostolova N, Belles JM, Barrero JM, Piqueras P, Ponce MR, et al. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell. 2002;14(8):1833–1846. doi: 10.1105/tpc.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujii H, Verslues PE, Zhu JK. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci U S A. 2011;108(4):1717–1722. doi: 10.1073/pnas.1018367108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009;50(12):2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- 41.Liu JG, Han X, Yang T, Cui WH, Wu AM, Fu CX, et al. Genome-wide transcriptional adaptation to salt stress in Populus. BMC Plant Biol. 2019;19(1):367. doi: 10.1186/s12870-019-1952-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu GY, Uan MY, Ai CR, Liu LJ, Zhuang E, Karapetyan S, et al. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature. 2017;545(7655):491–494. doi: 10.1038/nature22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang NN, Xu SW, Sun YL, Liu D, Zhou L, Li Y, et al. The cotton WRKY transcription factor (GhWRKY33) reduces transgenic Arabidopsis resistance to drought stress. Sci Rep. 2019;9(1):724. doi: 10.1038/s41598-018-37035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Yang TY, Lin ZK, Gu BJ, Xing CH, Zhao LY, et al. A WRKY transcription factor PbrWRKY53 from Pyrus betulaefolia is involved in drought tolerance and AsA accumulation. Plant Biotechnol J. 2019;17(9):1770–1787. doi: 10.1111/pbi.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato H, Takasaki H, Takahashi F, Suzuki T, Iuchi S, Mitsuda N, et al. Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc Natl Acad Sci U S A. 2018;115(47):11178–11187. doi: 10.1073/pnas.1811491115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, et al. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006;45(3):309–319. doi: 10.1111/j.1365-313X.2005.02622.x. [DOI] [PubMed] [Google Scholar]
- 47.Toh S, Imamura A, Watanabe A, Nakabayashi K, Okamoto M, Jikumaru Y, et al. High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 2008;146(3):1368–1385. doi: 10.1104/pp.107.113738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnon DI, Hoagland DR. Crop production in artificial solutions and soils with special reference to factors influencing yield and absorption of inorganic nutrients. Soil Sci. 1940;50:463–485. [Google Scholar]
- 49.Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc. 2006;1(2):641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 50.Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, et al. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008;146(2):368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heath RJ, Packer L. Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys. 1968;125:189–198. doi: 10.1016/0003-9861(68)90654-1. [DOI] [PubMed] [Google Scholar]
- 52.Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39(1):205–207. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
- 53.Liu HB, Li XH, Xiao JH, Wang SP. A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice-bacterium interaction. Plant Methods. 2012;8(1):2. doi: 10.1186/1746-4811-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Min L, Hu Q, Li YY, Xu J, Ma YZ, Zhu LF, et al. LEAFY COTYLEDON1-CASEIN KINASE I-TCP15-PHYTOCHROME INTERACTING FACTOR4 network regulates somatic embryogenesis by regulating Auxin homeostasis. Plant Physiol. 2015;169(4):2805–2821. doi: 10.1104/pp.15.01480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Sequence and phylogenetic analysis of GhWRKY1-like. Sequence alignment of the amino acid sequence of GhWRKY1-like (XP_016696352) with AtWRKY1 (AEC05881), GmWRKY1 (XP_003518571), PtWRKY1 (XP_006375555), and TcWRKY1 (XP_007049283). Conserved WRKY domain and zinc finger motif are shown in red box and green box, respectively.
Additional file 2: Figure S2. The abiotic stresses including mannitol, ABA, dehydration and NaCl induced expression patterns of GhWRKY1-like.
Additional file 3: Table S1. Primers used in vectors constructed and qRT-PCR. The underlined sequences were the infusion sequences.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.