Significance
Neurons have an exceptional capacity to grow axons and form synaptic circuits during development but not later life. In adults, the lack of circuit formation may support retention of skilled actions and memories but also limits regeneration and repair after injuries and in disorders. Research on developing and damaged neurons has revealed many molecules that help circuit formation and regeneration, and yet factors that could induce axon growth and synapse formation in adult brain neurons remain elusive. Here, we searched for such key molecules and find one that alone can induce complete circuit formation. After engineering a new circuit in adult mice, we also looked into its function and relevance for memories.
Keywords: mossy fiber, single-cell RNA-seq, Id2, circuit formation, adult brain rewiring
Abstract
Circuit formation in the central nervous system has been historically studied during development, after which cell-autonomous and nonautonomous wiring factors inactivate. In principle, balanced reactivation of such factors could enable further wiring in adults, but their relative contributions may be circuit dependent and are largely unknown. Here, we investigated hippocampal mossy fiber sprouting to gain insight into wiring mechanisms in mature circuits. We found that sole ectopic expression of Id2 in granule cells is capable of driving mossy fiber sprouting in healthy adult mouse and rat. Mice with the new mossy fiber circuit solved spatial problems equally well as controls but appeared to rely on local rather than global spatial cues. Our results demonstrate reprogrammed connectivity in mature neurons by one defined factor and an assembly of a new synaptic circuit in adult brain.
Connectivity is one of the most defining features of cellular identity in neurons of the brain and spinal cord. During development, cell surface receptors and neurotrophic gradients guide axon growth and formation of synaptic connections between neurons (1–3). After the developmental milieu dissipates, inhibitors of neurite growth are up-regulated (4, 5), and the axonal and dendritic structure of neurons become established. In adult brain, the lack of cell intrinsic axon growth, growth-supportive substrate, and chemoattraction hinders not only further wiring but also the attainment of therapeutic rewiring after injury (6). Currently, cell-autonomous mechanisms—including those mediated by transcription factors such as members of the CREB, HDAC, ID, KLF, SMAD, STAT, and SOX families—have been able to partially recapitulate circuit wiring (7, 8). However, it is thought that additional cell nonautonomous mechanisms are required to fully recapitulate circuit wiring (6). Nevertheless, other studies demonstrated that under mixed developmental-adult conditions, as seen after adult neurogenesis (9) and after in vivo glia/astrocyte to neuron conversion and stem cell grafting (10), long-range axonal wiring and synaptic integration can be achieved in adult circuits. This would suggest that some cell-autonomous mechanisms can overcome barriers to further circuit formation, but the nature of such permissive signals is not understood.
To explore wiring mechanisms in mature circuits of adult brain, we investigated axonal sprouting in the hippocampal mossy fiber (MF) system. Formed during normal development, naive MF axons originate from dentate gyrus granule cells (GCs) and target different cell types in hilus and CA3 (11). By contrast, during MF sprouting, which is typically studied in the context of temporal lobe epilepsies (TLE), GCs form new axons and synapses within the dentate and thereby create a new circuit on top of the developmentally established wiring scheme (12–14). MF sprouting is also inducible by different approaches, such as coarse mechanical (15, 16), electrical (17), and chemical induction (18), or by excessive activation of the mTOR pathway in postnatally generated GCs (19). However, these manipulations coinduce multiple other alterations in the network (e.g., cell death, GC dispersion, formation of aberrant GC dendrites, and changes in cell excitability), which thus far hindered understanding their relative contributions to TLE pathology (20) and of molecular mechanisms of MF sprouting (21–23). Regardless of its relation to TLE, MF sprouting involves all key stages of circuit assembly (i.e., axon growth, target cell specification, and synapse formation) and thus also represents a comprehensive model for studying wiring mechanisms in adult brain.
Our study aimed to elucidate transcriptional mechanisms behind MF sprouting and test if, analogous to reprogramming of differentiated cells (24), the connectivity of differentiated mature GCs can be reprogrammed via cell-autonomous genetic induction. Consistent with the assembly of an entirely new circuit, we hypothesized that MF sprouting was associated with broad transcriptomic changes in GCs. To recapitulate these changes in their entirety, we focused on transcription factors and regulators, whose activation may initiate the wiring process. Using a single-cell RNA-sequencing (RNA-seq) screen, we identified Id2, an inhibitor of transcription factors, as master regulator whose sole activation in GCs drove MF sprouting and formation of functional synapses. Mechanistically, activation of Id2 led to transcriptomic up-regulation of molecules in the JAK/STAT and interferon signaling pathways and controlled downstream expression of multiple wiring-related effectors. We furthermore tested if genetically induced MF sprouting generated hyperexcitability in the hippocampus, as posited by certain models of seizure generation, and investigated its consequences on learning and memory.
Results
Single-Cell Transcriptome of MF Sprouting.
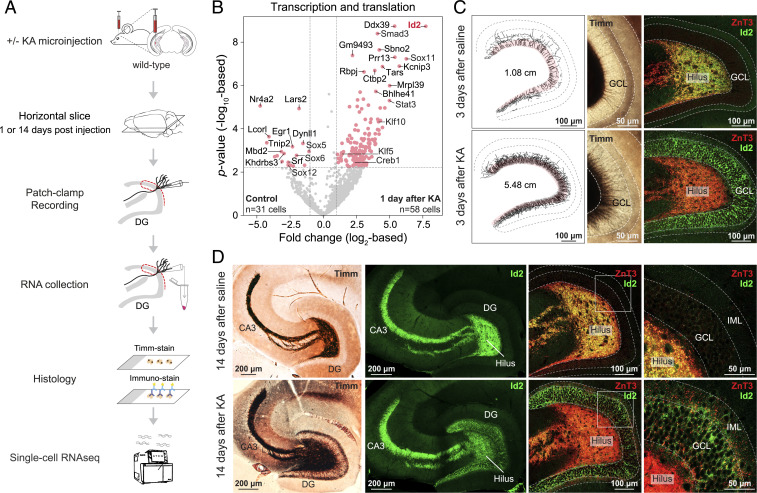
To begin, we used chemical induction (neuroexcitatory kainic acid [KA], microinjection into the hippocampus) in 2-mo-old mice to investigate the MF sprouting transcriptome. Using single-cell patch RNA-seq (25, 26), we sampled mature GCs 1 d (representing acute cellular response to induction) and 14 d after induction (27), by which time MF sprouting, but not epilepsy (28, 29), reliably develops (Fig. 1A). Transcriptomic analysis revealed up-regulated expression (FDR < 0.05) of several transcription regulators, including members of the ID (Id2), SMAD (Smad3), SOX (Sox11), STAT (Stat3, Stat1), HDAC (Hdac9), KLF (Klf10, Klf5), and CREB (Creb1) families (Fig. 1B and SI Appendix, Fig. S1). Of these, Id2, a developmentally active inhibitor of transcription factors, was a particular candidate of interest. Previously, Id2 was shown to have increased expression in GCs after status epilepticus (30), and separate studies have linked this gene to axon growth in cell and slice culture (31–33) and following spinal cord injury (34). Using immunostaining, we confirmed up-regulation of Id2 protein levels, which was apparent in few GCs already 1 d after KA (SI Appendix, Fig. S1G) but present in most GCs 3 d after KA (Fig. 1C and SI Appendix, Fig. S1H). This up-regulation persisted 14 d later (Fig. 1D). At first, the Id2 signal was present in nucleus (1 d) but later (3 d and after) become enriched in cytosol. This was consistent with a model in which Id2 is either sequestered to the cytosol by other factors (35–37) or itself binds and sequesters transcription factors to the cytosol (38). Notably, our analyses also revealed the presence of Id2 protein in MF axons in CA3, which was independent of its lack (in controls) or presence (after KA) in GC soma (Fig. 1D). While this may suggest a yet unrecognized role for Id2 in MF axons, since in this study our focus was on rewiring, we followed up on the up-regulation of Id2 that corresponded to sprouting.
Fig. 1.
Transcriptomic analysis of chemically induced MF sprouting. (A) Experimental design. (B) Volcano plot shows acute transcriptomic changes of transcription and translation-related molecules in single GCs 1 d after KA induction of MF sprouting. Red points denote differentially expressed genes (FDR < 0.05 and >2-fold change, or |log2FC| > 1 as in plot). (C) MF sprouting is already ongoing and Id2 is up-regulated in GCL 3 d after KA induction. Upper panels show saline-injected controls; lower panels show KA. From left to right: Timm’s staining–based MF axon tracings reconstructed from 80-µm-thick sections; Timm’s-stained brain slices at higher magnification; immunohistochemical staining of ZnT3 and Id2. (D) MF sprouting develops and Id2 remains up-regulated in GCL 14 d after KA. Upper panels show saline-injected controls; lower panels show KA. From left to right: Timm’s-stained hippocampal slices displaying the whole MF system; immunostaining of Id2 in the MF system (note the presence of Id2 in naive fibers in hilus and CA3); immunostaining of ZnT3 and Id2 in dentate gyrus shown at higher magnifications.
AAV-Id2 Induces Axon Growth in the Mature MF System.
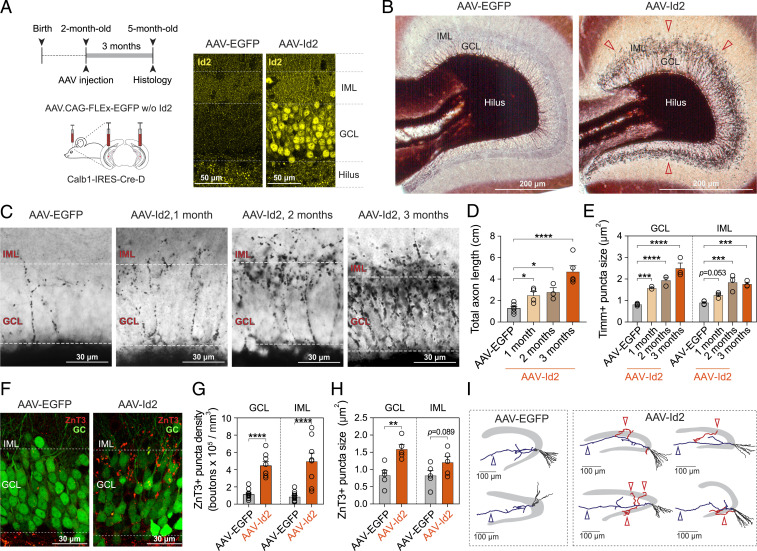
To test if Id2 played a role in MF sprouting, we cloned and ectopically expressed the Id2 gene selectively in GCs of the ventral hippocampus of adult Calb1-IRES2-Cre-D mice using Cre-dependent AAVDJ/8 virus (note that although AAV-Id2 was coinjected with AAV-EGFP to confirm the injection site by visualizing GCs, hereafter, we refer to this injection simply as AAV-Id2; for controls, only AAV-EGFP was used, in equal volume; Fig. 2A). One to three months after induction, histological analyses revealed newly formed MF axons targeting the GC and inner molecular layer (GCL and IML, respectively; Fig. 2 B and C and SI Appendix, Fig. S2). Quantification of axonal length and puncta further corroborated these observations. Both the measurement of MF axon length (AAV-EGFP: 1.3 ± 0.17 cm, n = 6 mice; AAV-Id2, 1 mo: 2.5 ± 0.34 cm, n = 4 mice, 2 mo: 2.8 ± 0.42 cm, n = 3 mice, 3 mo: 4.7 ± 0.56 cm, n = 5 mice; Fig. 2D) and puncta size (in GCL, AAV-EGFP: 0.82 ± 0.024 µm2, 737 puncta/4 mice; AAV-Id2: 1 mo: 1.6 ± 0.038 µm2, 691 puncta/3 mice, 2 mo: 1.9 ± 0.14 µm2, 853 puncta/3 mice, 3 mo: 2.5 ± 0.25 µm2, 712 puncta/3 mice; in IML, AAV-EGFP: 0.90 ± 0.044 µm2, 206 puncta/4 mice; AAV-Id2, 1 mo: 1.3 ± 0.076 µm2, 309 puncta/3 mice, 2 mo: 1.8 ± 0.22 µm2, 534 puncta/3 mice, 3 mo: 1.8 ± 0.081 µm2, 358 puncta/3 mice; Fig. 2E) revealed time-dependent increases after AAV-Id2 delivery. Because zinc transporter-3 (ZnT3) is a known marker of naive MFs (39), we tested its expression by immunostaining (Fig. 2F) and found both increased density (in GCL, AAV-EGFP: 1.1 ± 0.16 × 105 puncta/mm3, AAV-Id2: 4.5 ± 0.47 × 105 puncta/mm3; in IML, control: 0.86 ± 0.16 × 105 puncta/mm3; AAV-Id2: 5.0 ± 0.96 × 105 puncta/mm3; AAV-EGFP: 11 hippocampi from 6 mice, AAV-Id2: n = 8 hippocampi from 6 mice; Fig. 2G) and size (in GCL, AAV-EGFP: 0.83 ± 0.15 µm2; AAV-Id2: 1.6 ± 0.13 µm2; in IML, AAV-EGFP: 0.83 ± 0.15 µm2; AAV-Id2: 1.2 ± 0.17 µm2; AAV-EGFP: n = 5 hippocampi from 5 mice, AAV-Id2: n = 5 hippocampi from 5 mice; Fig. 2H) of ZnT3+ puncta 3 mo after AAV-Id2 induction. We also confirmed that AAV-Id2 induced rewiring at the single-cell level by morphological reconstruction of individual GCs, which revealed recurrent fibers in GCL/IML after AAV-Id2 induction without apparent reorganization of the cells’ developmentally established axonal and dendritic structure (Fig. 2I). In order to test if, in addition to its effect on anatomical rewiring, AAV-Id2 also changed action potential (AP) firing of GCs or their overall synaptic drive onto CA3, we performed electrophysiological characterization of these properties. These revealed that despite an increase in the cells’ input resistance, AAV-Id2 did not appear to change either current-pulse–induced AP firing properties or the cells’ developmentally established synaptic drive onto CA3 pyramidal cells (SI Appendix, Fig. S3).
Fig. 2.
AAV-delivered Id2 induces axon growth and target-specific rewiring in mature hippocampal GCs. (A) Experimental design. The Calb1-IRES-Cre-D transgenic line allows genetic access to dentate GCs via expression of Cre. Confocal images show confirmation of Id2 overexpression by immunostaining. (B) Timm’s staining shows the dentate gyrus 3 mo after Cre-dependent AAV-EGFP (control) and AAV-Id2 injections. After AAV-Id2, dark ring-like precipitation around GCL represents newly formed MFs (red arrowheads). (C) Timm’s stainings show MFs in GCL/IML 1, 2, and 3 mo after AAV-EGFP and AAV-Id2 injections. (D) Quantification of total axon length after AAV-EGFP and AAV-Id2 injections (one-way ANOVA, AAV-EGFP versus AAV-Id2, 1 mo, *P = 0.043; 2 mo, *P = 0.023; 3 mo, ****P < 0.0001). (E) Quantification of Timm’s positive puncta size in GCL/IML after AAV-EGFP and 1, 2, and 3 mo after AAV-Id2 injections (data points represent one individual; two-way ANOVA, GCL: AAV-EGFP versus AAV-Id2, 1 mo, ***P = 0.0003; 2 mo, ****P < 0.0001; 3 mo, ****P < 0.0001. IML: AAV-EGFP versus AAV-Id2, 1 mo, P = 0.053; 2 mo, ****P < 0.0001; 3 mo, ****P < 0.0001). (F) EGFP labeling of GCs (green) and ZnT3 staining of MF synapses (red) in GCL/IML 3 mo after AAV-EGFP and AAV-Id2 injections (note that AAV-Id2 was coinjected with AAV-EGFP to visualize GCs). (G) Quantification of ZnT3-positive puncta density in GCL/IML 3 mo after AAV-EGFP and AAV-Id2 injections (two-way ANOVA, GCL: AAV-EGFP versus AAV-Id2, ****P < 0.0001; IML: AAV-EGFP versus AAV-Id2, ****P < 0.0001). (H) Quantification of ZnT3-positive puncta size in GCL/IML 3 mo after AAV-EGFP and AAV-Id2 (two-way ANOVA, GCL: AAV-EGFP versus AAV-Id2, **P = 0.0022; IML: AAV-EGFP versus AAV-Id2, P = 0.089). (I) Reconstruction of single GCs 3 mo after AAV-EGFP and AAV-Id2 injections. After AAV-Id2, newly formed axons that extend into GCL and IML are shown in red. Original MF projections to CA3 are marked with blue arrowhead. CA3 is omitted for clarity.
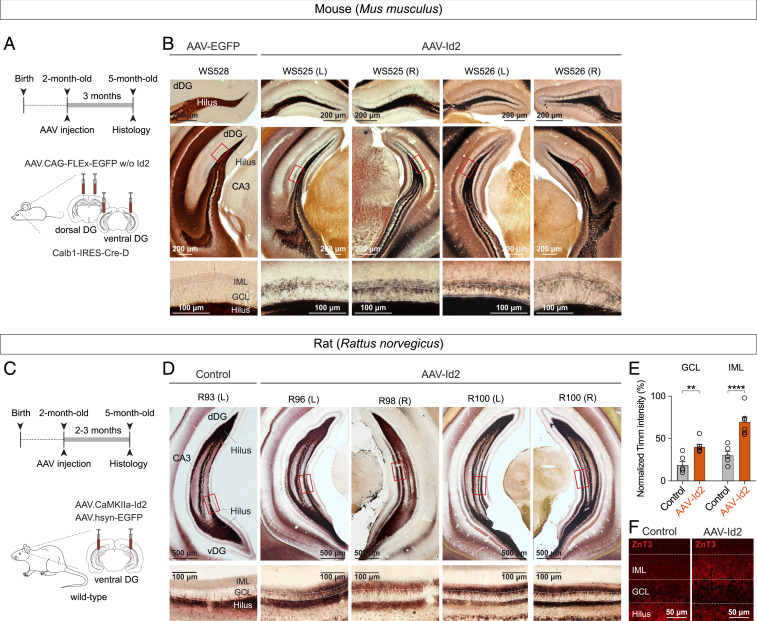
Since the ventral and dorsal hippocampus support different brain functions and display distinct molecular patterns (40–42) and left-right asymmetries in hippocampal function have also been recognized (43), in a next set of experiments, we tested if AAV-Id2 induced MF rewiring in hippocampal locations other than the ventral part. For this, we expressed AAV-Id2 in dorsal and ventral GCs, both in left and right hippocampi (Fig. 3A), which led to development of MF rewiring throughout the entire mouse hippocampus, independent of anatomical location (Fig. 3B and SI Appendix, Fig. S2A).
Fig. 3.
AAV-delivered Id2 induces MF rewiring throughout the mouse and rat hippocampus. (A) Experimental design showing Id2 overexpression in mouse dorsal and ventral hippocampus. (B) Images show Timm's-stained sections collected from different levels of dorsal hippocampus (bregma, –2.0 mm and –3.2 mm) after AAV-EGFP (control) and AAV-Id2 injections. Higher-magnification images at bottom show sprouting in GCL and IML in AAV-Id2 mice. (C) Experimental design showing Id2 overexpression in rat ventral hippocampus. (D) Example images of Timm staining in rats after Id2 overexpression. Coronal sections of rat ventral hippocampus (bregma, –6.2 mm) were collected from regions where AAV infection was confirmed by EGFP expression. Non-AAV-infected hippocampus was used as control. (E) Quantification of Timm’s staining intensity. Intensities were measured relative to signals in the hilus of the same sections (two-way ANOVA, GCL: Control versus AAV-Id2, **P = 0.0017; IML: Control versus AAV-Id2, ****P < 0.0001). (F) ZnT3 staining of MF synapses in GCL/IML 3 mo after AAV-Id2 injections.
Finally, in addition to mice, because MF sprouting has been extensively characterized in rats (12–18, 21, 22), we tested if Id2 had the ability to induce MF sprouting in this different species. For this, we injected Cre-independent AAV-Id2 into the hippocampus of wild-type rats. Similarly to observations in mouse, this manipulation revealed powerful MF rewiring (Fig. 3 C–F). Two to three months after injection, the relative intensity of Timm’s staining was significantly higher both in GCL and IML in AAV-Id2–injected dentate gyrus compared to controls (Control, GCL: 18.4 ± 4.3%, IML: 30.3 ± 4%, six hippocampi from three rats; AAV-Id2, GCL: 40.4 ± 2.8%, IML: 69.9 ± 5.5%, seven hippocampi from five rats; percentages represent signal intensity relative to hilus in the same sections). These observations demonstrate that AAV-Id2 can uniformly activate a GC wiring program in different hippocampal segments and species.
AAV-Id2 Induces Functional MF Synapse Formation.
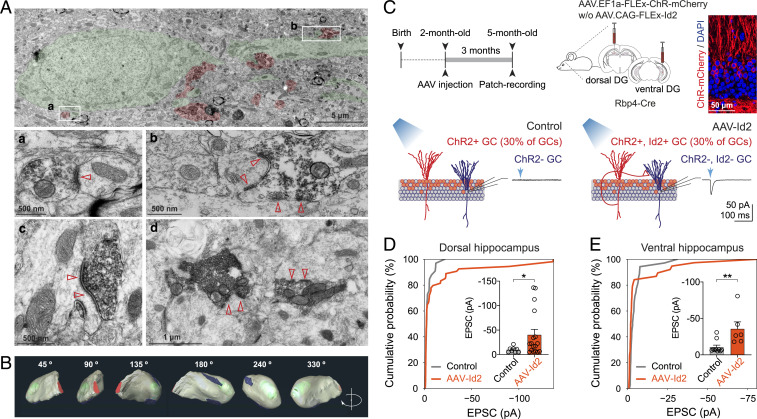
To test if genetically induced MF sprouting by AAV-Id2 involved formation of functional synapses, we labeled MF boutons by ZnT3 immunostaining and characterized them using electron microscopy. This showed that ZnT3+ boutons were formed on GC dendrites and spines, each bouton containing one or multiple release sites and abundant supply of synaptic vesicles (Fig. 4 A and B). To probe physiological transmission, we induced MF rewiring with AAV-Id2 while also expressing channel-rhodopsin (AAV-ChR) in GCs in vivo and prepared brain slices for electrophysiology 3 mo after induction. Because large conductance ChR currents would mask the comparably smaller synaptic currents, we restricted ChR expression and rewiring to a subset of GCs by using Rbp4-Cre transgenic mice, in which only ∼30% of GCs express Cre and thus Cre-dependent Id2 and ChR (Fig. 4C). In separate experiments in the dorsal and ventral hippocampus, we performed patch-clamp recordings from ChR nonexpressing cells. Independent of anatomical location, ChR activation by blue light evoked larger and more frequent excitatory synaptic events in slices after AAV-Id2 injection compared to controls (in dorsal hippocampus, Control: −9.9 ± 1.9 pA, AAV-Id2: −40 ± 11pA; Control, n = 8 out of 34 cells, from six mice; AAV-Id2, n = 17 out of 55 cells, from seven mice; Fig. 4D; in ventral hippocampus, Control: −11 ± 2.8 pA, AAV-Id2: −36 ± 9.7 pA; Control, n = 10 out of 36 cells, from six mice; AAV-Id2, n = 6 out of 39 cells, from seven mice; Fig. 4E). To summarize, AAV delivery of the single transcriptional regulator Id2 led to formation of a new MF circuit, including formation of functional synapses.
Fig. 4.
AAV-delivered Id2 induces functional synapse formation. (A) Electron microscopy images show ZnT3-positive boutons (red) on GC dendrite and spines (green) 3 mo after AAV-Id2. Insets a and b are shown enlarged, whereas c and d show additional examples that are not present in panel A. (B) 3D electron-tomographic reconstruction of a ZnT3-positive MF bouton located in the GCL/IML border 3 mo after AAV-Id2. Red: axon shaft, green: mitochondria, blue: synapse formed by this bouton terminal. (C) Experimental design and injection schedule to test physiological transmission after MF rewiring. In acute brain slices, patch-clamp recordings were made from ChR-negative GCs, while ChR-positive GCs were activated with blue light (∼30% of total GCs population in these experiments) 3 mo after AAV-ChR (Control) and AAV-Id2 (mixed with AAV-ChR) injections. (D) Cumulative probability plot shows the fraction of recorded cells versus light-evoked EPSC amplitude in GCs of dorsal hippocampus (recorded in 10 µM Gabazine; Kolmogorov-Smirnov test, P = 0.57). Inset shows light-evoked EPSC amplitudes (Mann–Whitney U test, *P = 0.048). (E) Cumulative probability plot shows the fraction of recorded cells versus light-evoked EPSC amplitude in GCs of ventral hippocampus (recorded in 10 µM Gabazine; Kolmogorov-Smirnov test, *P = 0.041). Inset shows light-evoked EPSC amplitudes (Mann–Whitney U test, **P = 0.0075).
The transcriptomic network controlled by Id2 in mature GCs.
Next, we sought to better understand the molecular mechanisms behind AAV-Id2–induced MF rewiring. Although Id2 has been extensively characterized for its role in transcriptional regulation (31, 38, 44), one previous study suggested that Id2 may directly contribute to axon growth and growth cone formation independently of transcriptional regulation (33). This function was dependent on Akt-mediated phosphorylation of the serine 14 site in Id2 (33). To test this possibility first, we virally expressed phosphorylation-ablated mutant Id2(S14A), AAV-Id2(S14A), in the ventral hippocampus. We hypothesized that MF rewiring would not develop using this mutant form if Akt/Id2 signaling was involved. However, AAV-Id2(S14A) still induced MF rewiring (SI Appendix, Fig. S4), suggesting that direct growth cone formation by Id2 was unlikely.
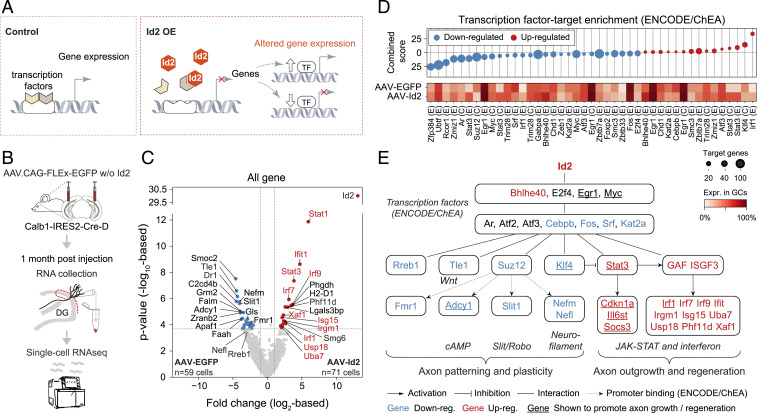
Second, we investigated transcriptomic changes induced by Id2. By directly binding transcription factors, Id2 inhibits their DNA binding and thereby their transcriptional activity (31, 44) (Fig. 5A). Consequently, increased expression of Id2 would feasibly lead to both up-regulated and down-regulated expression of genes, whose identity depends on the transcription factors that were inhibited by Id2. To study transcriptomic consequences, we sequenced single GCs 1 mo after AAV-Id2 induction (Fig. 5B), when growing axons were already apparent (Fig. 2C). Consistent with a role in transcriptional regulation, this revealed broad transcriptomic changes, which were dominated by members of the JAK-STAT (Stat1, Stat3, and Irf9) (45) and interferon signaling pathways (e.g., Irf1, Irf7, Irf9 Isg15, Usp18) (46) as well as by multiple other molecules that have been associated with axonal wiring (e.g., Tle1, Nefm, Slit1, Adcy1; Fig. 5C and SI Appendix, Fig. S5; see Discussion).
Fig. 5.
Single-cell transcriptomics reveal a comprehensive rewiring program induced by AAV-delivered Id2. (A) Drawing depicts transcriptional function of Id2. (B) Experimental strategy. (C) Volcano plot shows gene expression differences between AAV-EGFP (n = 59 cells) and AAV-Id2 (n = 71 cells)–delivered single GCs. Horizontal and vertical dashed lines show FDR = 0.05 and 2-fold change (|log2FC|>1), respectively. Gene names highlighted in red belong to the JAK-STAT and interferon pathways. (D) Enrichr transcription factor-target enrichment analysis based on 285 up-regulated (red) and 848 down-regulated genes (blue) that were differentially expressed (P < 0.05) between the AAV-EGFP or AAV-Id2 data sets. Identified transcription factors (E: Encode, C: ChEA) and their expression rate in GCs are shown in the bottom. Circle size represents the number of target genes present in the inputted data. (E) The gene regulatory network activated by AAV-Id2. Nodes represent molecules from C and D; edges represent interactions. GAF and ISGF3 refer to proteomic assembly of Stat1 homodimers and Stat1, Stat2, and Irf9, respectively (45).
We next asked what might be key mediators of the AAV-Id2–induced rewiring program, that is, molecules that interface Id2 and the aforementioned genes. Since Id2 inhibits the activity of transcription factors, without necessarily changing their expression, the found transcriptomic changes likely represent downstream effects and do not indicate upon which transcription factors Id2 directly acts. Therefore, we performed transcription factor-target enrichment analysis with the aim of identifying transcription factors that were either directly or indirectly inhibited or indirectly disinhibited by Id2. Using Enrichr (47, 48), we found 26 such factors whose known regulatory network matches the observed gene expression patterns and which were expressed in at least 30% of GCs (Fig. 5D). Together with this analysis, our results outlined a comprehensive transcriptomic model behind MF rewiring, in which Id2 exerts control over members of the wiring-related JAK-STAT, Wnt, cAMP, and Slit/Robo signaling pathways (Fig. 5E and SI Appendix, Figs. S5–S7).
Hippocampal brain dynamics after AAV-Id2–induced MF rewiring.
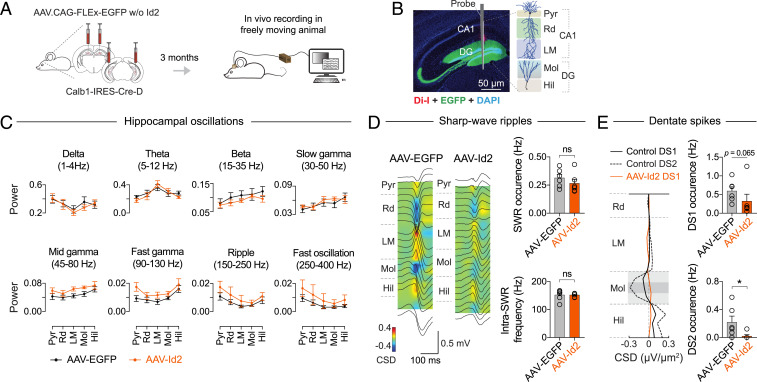
The ability to genetically induce MF sprouting allowed us to examine the network effects of this observed rewiring event. MF sprouting is observed in human TLE and is a hallmark in experimental TLE, but the question of whether it is a cause or consequence of seizures has been debated (20, 21). Thus, by performing multichannel silicon probe recordings in the hippocampus of freely moving mice, we looked to see if signatures of pathological brain dynamics (49) have developed 3 mo after AAV-Id2–induced MF rewiring (Fig. 6 A and B). In the 1 to 400 Hz range of local field potentials, our recordings did not register pathological oscillations or seizure-like activity (Fig. 6C). Specifically, theta, beta, slow and fast gamma, and ripple and fast ripple range oscillations remained intact. In addition, we analyzed CA1 sharp wave-ripple (SWR) and dentate spike (DS) events because their intrinsic frequency and occurrence, respectively, both increased in TLE (50, 51). Neither of these pathologies were present in our data. The occurrence (AAV-EGFP: 0.31 ± 0.033 Hz, n = 6 mice; AAV-Id2: 0.27 ± 0.037 Hz, n = 6 mice) and intrinsic frequency (AAV-EGFP: 152 ± 7.8 Hz, n = 6 mice; AAV-Id2: 152 ± 2.7 Hz, n = 6 mice) of SWRs were not different between AAV-EGFP and AAV-Id2–injected mice (Fig. 6D and SI Appendix, Fig. S8). By contrast, the occurrence of type-1 DS (DS1) decreased (AAV-EGFP: 0.59 ± 0.12 Hz, n = 6 mice; AAV-Id2: 0.32 ± 0.19 Hz, n = 6 mice; P = 0.06, Mann–Whitney U test), whereas type-2 DS (DS2) were selectively lost after AAV-Id2 delivery (AAV-EGFP: 0.22 ± 0.089 Hz, n = 6 mice; AAV-Id2: 0.021 ± 0.020 Hz, n = 6 mice; P = 0.015, Mann–Whitney U test, Fig. 6E and SI Appendix, Fig. S8). Taken together, intact oscillations and SWRs suggested that the network dynamics in AAV-Id2 mice are divergent from TLE, whereas the decrease in DS occurrence suggested that activity routing was still effectively altered in the dentate network.
Fig. 6.
Hippocampal dynamics after AAV-Id2–induced MF rewiring. (A) Experimental design. After AAV-EGFP and AAV-Id2 injections, each mouse was implanted with a linear silicon probe in dorsal hippocampus. Three months after AAV delivery, recordings were made from freely moving mice. (B) Histological image of a silicon probe track through the hippocampus (Pyr: pyramidal layer, Rd: radiatum, LM: lacunosum-moleculare, Mol: molecular layer, Hil: hilus). (C) Local field potential (LFP) power of delta, theta, beta, slow gamma, midgamma, fast gamma (all during locomotion), and ripple and fast ripple (during resting) range frequencies (AAV-EGFP, n = 7 mice; AAV-Id2, n = 6 mice). Neither pair-wise comparisons between the AAV-EGFP and AAV-Id2 groups revealed statistically significant (P < 0.05) differences using two-way ANOVA test. (D) Left: Regional distribution of currents associated with SWRs in ripple peak-triggered CSD maps; average LFP waveforms (black traces) are shown superimposed. Right: Quantification of ripple occurrence (Mann–Whitney U test, P = 0.31) and intraripple frequency in CA1 Pyr and Rd (Mann–Whitney U test, P = 0.45; AAV-EGFP, n = 6 mice; AAV-Id2, n = 6 mice). (E) Left: CSD profiles of DS1 and DS2. Right: Quantification of DS1 (Mann–Whitney U test, P = 0.065) and DS2 (Mann–Whitney U test, P = 0.015; AAV-EGFP, n = 6 mice; AAV-Id2, n = 6) occurrence.
Learning and memory after AAV-Id2–induced MF rewiring.
DS1 and DS2 events have been suggested to be triggered by population bursts of layer II stellate cells of the lateral entorhinal cortex (LEC) and medial entorhinal cortex (MEC), respectively (52). Since the LEC and MEC are proposed to support navigation based on local (“egocentric reference framework”) and global (“allocentric reference framework”) landmarks, respectively (53–55), and the dentate gyrus is critically involved in certain forms of object-related and spatial learning and memory (11, 41, 42), we hypothesized that MF sprouting-related network effects would manifest themselves during behavior. To test this hypothesis, we utilized eight different previously validated assays to phenotype mice 3 mo after bilateral, dorsal, and ventral AAV-EGFP or AAV-Id2 injections (see Methods for particulars of each assay).
Before the tests, the animals were subjected to light cycle inversion. To examine light cycle adaptation, locomotor activity, and freely moving behavior, we monitored each mouse in its home cage for 13 d and in a novel open-field environment for 20 min. In the home cage, mice in both groups adapted equally well to an inverted day-light cycle, and their overall activity level was not different (SI Appendix, Fig. S9A). In addition, in the open field, there was no discernible difference in total travel distance (AAV-EGFP: 66 ± 2.7 m, n = 12; AAV-Id2: 75 ± 4.4 m, n = 12) and time spent in center zone (AAV-EGFP: 29 ± 2.2%, n = 12; AAV-Id2: 25 ± 2.5%, n = 12) between the two groups (SI Appendix, Fig. S9B). Together, these suggest that AAV-Id2 mice did not display hyperactivity or anxiety-like behavior.
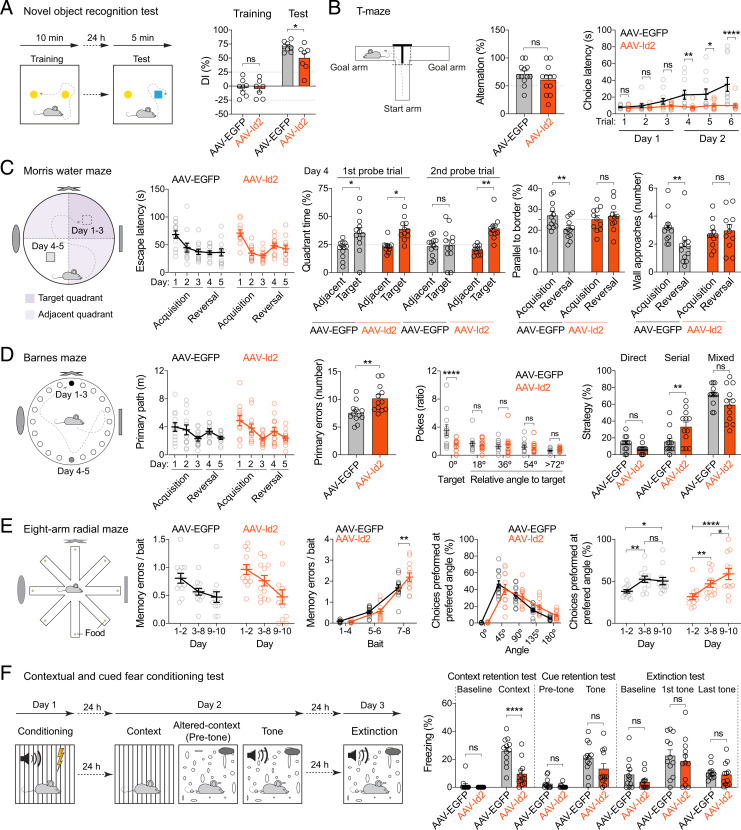
Next, we tested hippocampus-dependent long-term and short-term memory performance using novel object recognition and T-maze tests, which both take advantage of strong preference to novelty, whether object-related or environment-related, shown by rodents. The novel object recognition test showed that although both groups displayed preference to the novel object, AAV-Id2 mice did so to a lesser degree than controls (discrimination index [DI]; AAV-EGFP: 71 ± 3.2%, n = 8; AAV-Id2: 50 ± 8.7%, n = 7; mice with DI > 25% during training were excluded from analysis) (Fig. 7A and SI Appendix, Fig. S9C). In T-maze, alternation was above chance for both groups (AAV-EGFP: 71 ± 5.6%, n = 12; AAV-Id2: 61 ± 7.5%, n = 12) (Fig. 7B), which is also the expected outcome in untreated rodents. However, AAV-Id2 mice did not show a normally occurring increase in choice latencies across trials, and their latency to enter an arm remained significantly shorter than that of AAV-EGFP mice (at trial 6: AAV-EGFP: 35 ± 8.3 s, n = 11; AAV-Id2: 10 ± 2.2 s, n = 11) (Fig. 7B).
Fig. 7.
Learning and memory after AAV-Id2–induced MF rewiring. (Statistical tests are two-way ANOVA unless stated otherwise.) (A) Novel object recognition. Left to right: experiment design, DI (AAV-EGFP versus AAV-Id2, training P = 0.87, test *P = 0.021). (B) T-maze. Left to right: experiment design, alteration (Mann–Whitney U test, P = 0.33), and choice latency (AAV-EGFP versus AAV-Id2, trial 1, P = 0.87, trial 2, P = 0.66, trial 3, P = 0.31, trial 4, **P = 0.0048, trial 5, *P = 0.011, trial 6, ****P < 0.0001). (C) Morris water maze. Left to right: experiment design, escape latency [FDay (4, 84) = 13, P < 0.0001; FTreatment (1, 21) = 0.056, P = 0.82; FTreatment × Day (4, 84) = 1.4, P = 0.23], quadrant time (adjacent versus target, first probe trial, AAV-EGFP: *P = 0.017, AAV-Id2: *P = 0.014; second probe trial, AAV-EGFP: P = 0.82, AAV-Id2: **P = 0.0026), swim path length parallel to walls (acquisition versus reversal, AAV-EGFP: **P = 0.0025, AAV-Id2: P = 0.45), and number of wall approaches (acquisition versus reversal, AAV-EGFP: **P = 0.0079, AAV-Id2: P = 0.57). (D) Barnes maze. Left to right: experiment design, primary path length [FDay (4, 88) = 6.6, P = 0.0001; FTreatment (1, 22) = 0.4, P = 0.52; FTreatment × Day (4, 88) = 0.3, P = 0.85], primary errors (Mann–Whitney U test, **P = 0.0038), poke ratio in probe trial after acquisition (AAV-EGFP versus AAV-Id2, angle = 0°: ****P < 0.0001, angle = 18°: P = 0.52, angle = 36°: P = 0.54, angle = 54°: P = 0.98, angle > 72°: P = 0.72), and average strategy used during acquisition and reversal AAV-EGFP versus AAV-Id2: direct, P = 0.31, serial, **P = 0.0047, mixed P = 0.059). (E) Eight-arm radial maze. Left to right: experiment design, memory errors per consumed baits over days [FDay (2, 44) = 9.0, P = 0.0005; FTreatment (1, 22) = 2.8, P = 0.11; FTreatment × Day (2, 44) = 0.53, P = 0.59], memory errors per consumed bait (AAV-EGFP versus AAV-Id2: bait 1 to 4, P = 0.88, bait 5 to 6, P = 0.86, bait 7 to 8, **P = 0.0029), preferred angle [FAngle (2, 44) = 16, P < 0.0001; FTreatment (1, 22) = 0.056, P = 0.81; FTreatment × Angle (2, 44) = 2.6, P = 0.086], and choices performed at preferred angle (AAV-EGFP, days 1 to 2 versus 3 to 8, **P = 0.0071, days 1 to 2 versus 9 to 10, *P = 0.020, day 3 to 8 versus 9 to 10, P = 0.68; AAV-Id2, days 1 to 2 versus 3 to 8, **P = 0.0049, days 1 to 2 versus 9 to 10, ****P < 0.0001, days 3 to 8 versus 9 to 10, *P = 0.026). (F) Contextual and cued fear conditioning. Left to right: experiment design, freezing during context retention (AAV-EGFP versus AAV-Id2: baseline, P = 0.60, context, ****P < 0.0001), freezing during cue retention (AAV-EGFP versus AAV-Id2: pretone, P = 0.58, tone, P = 0.032, q = 0.064; does not meet FDR criterion), and freezing during extinction (AAV-EGFP versus AAV-Id2: baseline, P = 0.24, first tone, P = 0.38, last tone, P = 0.73).
To test spatial information-related memory performance, we used the Morris water, Barnes, and eight-arm radial maze assays, in which goal-oriented navigation is reinforced by aversive, natural, and positive stimuli, respectively. In the Morris water maze, indicative of successful spatial learning, escape latencies (Fig. 7C) and swim path lengths (SI Appendix, Fig. S9D) robustly decreased in both groups during the acquisition. Moreover, both groups displayed robust preference for the original target quadrant in the first probe trial of reversal learning, which was to test spatial retention (day 4; time in quadrant as percentage of total time, AAV-EGFP: target quadrant 36 ± 4.8%, adjacent quadrants 21 ± 2.2%; n = 12; AAV-Id2: target 39 ± 3.0%, adjacent 23 ± 1.6%, n = 12) (Fig. 7C). However, during the second probe trial of reversal learning (also on day 4), which was to test reversal learning itself, AAV-Id2, but not AAV-EGFP, mice still displayed a preference for the original target quadrant (AAV-EGFP: target quadrant 25 ± 4.5%, adjacent quadrants 24 ± 2.4%, n = 12; AAV-Id2: target 39 ± 3.1%, adjacent 21 ± 1.2%, n = 12) (Fig. 7C), suggesting spatial perseverance. Furthermore, AAV-Id2 mice reverted to a wall-oriented, nonspatial swimming strategy (Fig. 7C). Congruent with the observations in the Morris water maze, both groups learned the Barnes maze task (Fig. 7D and SI Appendix, Fig. S9E). However, AAV-Id2 mice made more errors in finding the escape chamber (primary errors, throughout all trials; AAV-EGFP: 7.6 ± 0.52, n = 12; AAV-Id2: 10 ± 0.69, n = 12) and did not show preference for the original target when the escape chamber was removed in the probe trial (poke ratio at original target, angle = 0°, AAV-EGFP: 3.6 ± 0.75, n = 12; AAV-Id2: 1.3 ± 0.28, n = 12). This was likely because AAV-Id2 mice more frequently adapted a serial search strategy (i.e., trying neighboring holes one after another) than AAV-EGFP mice (serial strategy, AAV-EGFP: 16 ± 4.1% of all tries, n = 12; AAV-Id2: 33 ± 5.7%, n = 12). In the eight-arm radial maze, again, both groups learned equally well to decrease their memory errors, which is entry to an arm where the bait was already consumed per consumed baits of the total of eight baits over days (test days 9 to 10, AAV-EGFP: 0.47 ± 0.095, n = 12; AAV-Id2: 0.47 ± 0.13, n = 12) (Fig. 7E). However, memory errors during collection of the last two baits were higher in the AAV-Id2 mice (averaged over all test days, AAV-EGFP: 1.7 ± 0.15, n = 12; AAV-Id2: 2.2 ± 0.2, n = 12; Fig. 7E), suggesting a buildup of memory load in these mice. As a preferred strategy, both groups tended to enter the neighboring arm after visiting one (angle = 45°) (Fig. 7E). However, AVV-Id2 mice, but not AAV-EGFP, robustly increased their choices at this preferred angle over days (AAV-EGFP: days 1 to 2 = 38 ± 2.0%, days 3 to 8 = 52 ± 3.7%, days 9 to 10 = 50 ± 4.3%, n = 12; AAV-Id2: days 1 to 2 = 32 ± 3.5%, days 3 to 8 = 47 ± 4.1%, days 9 to 10 = 59 ± 6.7%, n = 12) (Fig. 7E).
Finally, we used a cued and contextual fear conditioning test to assay associative learning. Here, while both groups showed freezing response during context retention test, the response was smaller in AAV-Id2 mice (AAV-EGFP: 12 ± 2.1%, n = 12; AAV-Id2: 3.1 ± 1.7%, n = 12) (Fig. 7F). By contrast, although the response to tone retention still appeared to be lower in AAV-Id2 mice, freezing responses during tone retention and extinction tests were not significantly different between the two groups (tone retention: AAV-EGFP: 21 ± 3.1%, n = 12; AAV-Id2: 13 ± 3.6%, n = 12; extinction test: first tone: AAV-EGFP: 23 ± 4.2%, n = 12; AAV-Id2: 19 ± 4.6%, n = 12, last tone: AAV-EGFP: 11 ± 1.4%, n = 12; AAV-Id2: 9.1 ± 2.4%, n = 12) (Fig. 7F).
Discussion
In this study, we systematically analyzed MF sprouting with the aim of understanding transcriptomic mechanisms that can facilitate axon growth and circuit formation in the adult brain. Our study design was motivated by the hypothesis that activation of certain cell-autonomous mechanisms may be sufficient to drive rewiring in the adult brain, where further axon growth is generally inhibited. Our results suggest three major conclusions, which have implications not only for circuit assembly in adult brain but also for TLE pathophysiology and information processing in the dentate gyrus.
Molecular Mechanisms of MF Sprouting.
In the developing nervous system, Id2 enhances cell proliferation and inhibits the activity of mainly basic helix–loop–helix (31) but also other transcription factors (44). Mechanistically, developmentally regulated degradation of Id2 has been linked to up-regulation of axon growth inhibitors (31), which suggested that activation of Id2 could counteract growth inhibition and enhance axon growth, but this prediction was only tested in culture systems and after spinal cord injury (31–34). In contrast to previous studies, we identified Id2 using an unbiased single-cell RNA-seq screen and interrogated consequences of Id2 activation in mature, uninjured neurons of healthy adult mice and rats. We found that the sole activation of Id2 in mature GCs is capable of driving MF sprouting and formation of functional synapses.
During our initial analyses, we found that during chemically induced MF sprouting, both Id2 mRNA and protein became enriched in GCs (Fig. 1). This suggested that Id2 may play a role in MF sprouting, which we tested by cell-autonomous activation of Id2 in GCs. We found that AAV-Id2 had the remarkable capacity to induce MF rewiring throughout the whole hippocampus with a circuit architecture that resembled previous descriptions of MF sprouting (e.g., refs. 12–14, 21) but without signs of GC death or layer dispersion (Figs. 2–4).
As a master regulator, Id2 activated a comprehensive transcriptomic program for rewiring. Because Id2 is an inhibitor of transcription factors, MF rewiring appeared to result from suppression of persistently active transcriptional programs, and consistent with this, we found activation and silencing of molecules whose function is related to circuit-level reorganization (Fig. 5). Up-regulated JAK-Stat3 has been implicated in promoting axon growth after injury in visual and spinal systems (56–59). While previous work has separately linked Id2 and Stat3 to axon growth, our results establish a link between the two whereby activation of Id2 promotes downstream expression of Stat3. Other up-regulated genes included Phf11d, a regulator of the transcription factor Bex1 (60), which facilitates axon regeneration (61).
Down-regulated molecules were also consistent with an increased capacity for axonal organization and circuit formation and included 1) Tle1, a corepressor of the axon patterning Wnt signaling (62, 63); 2) neurofilaments Nefl and Nefm, which determine mature axon structure and caliber (64); 3) Slit1, a regulator of developmental axon guidance and patterning (65), down-regulation of which may allow MF entry to the dentate gyrus; 4) Adcy1, loss of which led to developmental axon retraction arrest (66), exuberant axon branching in sensory areas (67, 68), and recovery after spinal cord injury (69); 5) Fmr1, modulator of local translation of synaptic proteins (70); and 6) Rreb1, a regulator of Wallerian axon degeneration after injury (71).
Delineating the sequential activation/inactivation and stepwise role of involved molecules will be important for understanding the MF rewiring process in detail. It remains possible that further aspects of MF rewiring could also be controlled independently of Id2 yet still cell autonomously. For example, AAV-Id2–induced MF sprouting developed over months (Fig. 2), which is similar to other observations (21), although slower than that is achieved by non-cell-specific chemical induction (Fig. 1). Whether the factor(s) controlling the speed of circuit formation are cell autonomous and can be separately identified and how synapse targeting can be reprogrammed to other cell types instead of GCs remain outstanding questions. With regard to other pathways that have been previously implicated in axon growth, regeneration, or MF sprouting in particular, our data from single GCs did not reveal Id2-induced transcriptomic changes in the TGF-beta/BMP-Smad (axon growth) (72–74), BDNF (MF sprouting) (21, 23), PTEN-mTOR (MF sprouting in postnatally generated GCs) (19, 22), and p38/JNK (in axon regeneration) (75) pathways (SI Appendix, Fig. S5). Some of these may be upstream of Id2 (e.g., TGF-beta/Smad2) (72), be controlled translationally, act independently of Id2, or simply not be involved in MF sprouting by mature GCs.
MF Sprouting and Hippocampal Brain Activity.
Our ability to genetically induce MF rewiring in GCs allowed us to examine consequent network effects in the context of two related but independent hypotheses. According to one hypothesis, MF sprouting may generate hyperexcitable network states. MF sprouting is observed in human TLE and is one of the hallmarks of chemically induced (which is nonspecific and broadly impacts different cell types) circuit alterations in experimental TLE (21). Whether MF sprouting is a cause or consequence of seizures has been debated (20). Thus, one particular question was whether pathological brain dynamics, such as in the epileptic brain, would appear after genetically induced MF rewiring. However, our in vivo electrophysiological recordings did not register pathological oscillations, SWRs, or seizure-like activity 3 mo after AAV-Id2 delivery (Fig. 6). By contrast, after chemically induced epilepsy, decreased theta and midgamma oscillations (49), and increased intra-SWR frequency (50) have been reported. Our results thus reveal network dynamics divergent from epilepsy and indicate that MF rewiring does not generate seizures (20).
According to another hypothesis, the dentate gyrus is presumed to process information about content (e.g., objects) and local spatial landmarks, delivered from the LEC, and about global spatial landmarks, delivered from the MEC (53–55). Our in vivo recordings in AAV-Id2 mice revealed a decrease in the occurrence of DS1 and a loss of DS2 events, which have been suggested to be triggered by population bursts in LEC and MEC, respectively (52). While circuit mechanisms behind DS events remain elusive, our data suggested that MF sprouting differentially interfered with the generation of DS1 and DS2 events. Consequently, information from LEC about content and local landmarks and from MEC about global spatial context of an experience may be differentially impacted by MF sprouting.
Learning and Memory after MF Sprouting.
By employing multiple different memory assays to evaluate object and spatial-related information processing, we found a recurrent schema in the behavior of AAV-Id2 animals that had MF sprouting. With regard to objects, we found that AAV-Id2 mice displayed preference to novel objects, although the preference was less than that of controls (Fig. 7). Notably, the decreased preference to novel objects correlated with decreased occurrence of LEC-linked DS1 events in the AAV-Id2 mice. These represent further divergence from TLE, as novel object preference was not altered (76, 77) and DS occurrence was increased in epileptic mice (51) (note that DS1 and DS2 were not separately analyzed by this later study). Nonetheless, these do not contradict our findings, which suggest that DS1 events may be related to recognition of novel objects. With regard to spatial information, AAV-Id2 mice did not display deficits in primary task performance, but they appeared to solve spatial problems differently from controls. Specifically, the Morris water, Barnes, and eight-arm radial maze assays showed that AAV-Id2 mice learn and perform well, without deficits in spatial retention (Fig. 7). Note that these findings represent yet another divergence from TLE, where the same assays revealed learning and memory impairments (49, 78–80). However, AAV-Id2 mice displayed a higher level of spatial perseverance and opted to nonspatial, wall-oriented swimming strategies during reversal learning in the Morris water maze and performed more serial trials at the expense of direct and mixed trials in both the Barnes maze and eight-arm maze, together indicating that AAV-Id2 mice were prone to use local landmark-based and possibly self-referential navigation-based strategies rather than relying on global spatial cues. Since MEC (global spatial information)–linked DS2 events were specifically lost in AAV-Id2 mice, our finding suggests that DS2 events are not required for solving primary spatial tasks per se but may facilitate the processing of global spatial cues that guide navigation to enhance performance. One interpretation of these findings that should also be considered is a potential confusion in pattern separation caused by recurrent redistribution of neural activity among GCs after MF sprouting. Pattern separation is one principal function performed by dentate gyrus (11, 81) thought to be controlled by young but not mature adult-born GCs (82). Since in these experiments the Calb1-IRES2-Cre-D line was used to deliver Id2 to GCs and the onset of Calb1 expression marks a transition of adult-born GCs into more mature states (83), it is unlikely that altered pattern separation by young adult-born GCs substantially contributed to our observations. Overall, our results are consistent with the hypothesis that MEC and LEC support navigation based on global and local cues, respectively (54, 55). Finally, we also found that AAV-Id2 mice, unlike controls, did not increase their choice latencies over subsequent trials in the T-maze. This phenotype could potentially be the result of an egocentric navigation strategy that utilizes left-right sequences with high precision but could also be interpreted as an inability to habituate (although our other data did not suggest this) or faster decision making, and therefore, its relationship to MF sprouting is currently less clear.
Summary.
Repair being a key objective, factors that facilitate circuit rewiring in the adult nervous system are typically studied in the context of pathology or injury. Our results provide evidence for cell-autonomous activation of axon growth and circuit formation in healthy mature neurons in the absence of developmental or injury signaling. More detailed understanding of how involved molecules contribute to this process will help to advance circuit engineering approaches that can induce axon growth and control target cell-specific circuit formation in the adult brain.
Methods
All mouse protocols and husbandry practices were approved by the Veterinary Office of Zürich Kanton. For comprehensive description of 1) animals, 2) plasmids and viruses, 3) stereotaxic injections, 4) single-cell RNA-seq and bioinformatics, 5) histology and neuroanatomy, 6) electron microscopy, 7) in vitro electrophysiology, 8) in vivo electrophysiology, 9) behavior, and 10) statistical analyses, see the SI Appendix.
Supplementary Material
Acknowledgments
This study was supported by the Swiss National Science Foundation grant (310030_188506 to C.F.), Dr. Eric Slack-Gyr-Stiftung award (to C.F.), Novartis Stiftung für medizinisch-biologische Forschung grant (to C.F.), 20017-1.2.1-NKP-2017-00002 grant (to C.V.), EFOP-3.6.2-16-2017-00008 grant (to C.V.), 20765-3/2018/FEKUTSTRAT grant (to C.V.), ERC Consolidator Grant (nanoAXON #772452 to J.S.), and University of Zürich Forschungskredit fellowship (to W.L.). We thank Drs. Jean-Charles Paterna and Melanie Rauch (Viral Vector Facility, University of Zürich/ETH Zürich) for discussions and virus production and the Functional Genomics Center Zürich for RNA-seq support.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108239118/-/DCSupplemental.
Data Availability
RNA-seq data have been deposited in National Center for Biotechnology Information Gene Expression Omnibus (GSE161619) (27).
References
- 1.Tessier-Lavigne M., Goodman C. S., The molecular biology of axon guidance. Science 274, 1123–1133 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Dickson B. J., Molecular mechanisms of axon guidance. Science 298, 1959–1964 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Sanes J. R., Zipursky S. L., Synaptic specificity, recognition molecules, and assembly of neuronal circuits. Cell 181, 536–556 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Yiu G., He Z., Glial inhibition of CNS axon regeneration. Nat. Rev. Neurosci. 7, 617–627 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwab M. E., Functions of Nogo proteins and their receptors in the nervous system. Nat. Rev. Neurosci. 11, 799–811 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Anderson M. A., et al., Required growth facilitators propel axon regeneration across complete spinal cord injury. Nature 561, 396–400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore D. L., Goldberg J. L., Multiple transcription factor families regulate axon growth and regeneration. Dev. Neurobiol. 71, 1186–1211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahar M., Cavalli V., Intrinsic mechanisms of neuronal axon regeneration. Nat. Rev. Neurosci. 19, 323–337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonçalves J. T., Schafer S. T., Gage F. H., Adult neurogenesis in the hippocampus: From stem cells to behavior. Cell 167, 897–914 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Barker R. A., Götz M., Parmar M., New approaches for brain repair-from rescue to reprogramming. Nature 557, 329–334 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Hainmueller T., Bartos M., Dentate gyrus circuits for encoding, retrieval and discrimination of episodic memories. Nat. Rev. Neurosci. 21, 153–168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wenzel H. J., Woolley C. S., Robbins C. A., Schwartzkroin P. A., Kainic acid-induced mossy fiber sprouting and synapse formation in the dentate gyrus of rats. Hippocampus 10, 244–260 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Cavazos J. E., Zhang P., Qazi R., Sutula T. P., Ultrastructural features of sprouted mossy fiber synapses in kindled and kainic acid-treated rats. J. Comp. Neurol. 458, 272–292 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Frotscher M., Jonas P., Sloviter R. S., Synapses formed by normal and abnormal hippocampal mossy fibers. Cell Tissue Res. 326, 361–367 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Laurberg S., Zimmer J., Lesion-induced sprouting of hippocampal mossy fiber collaterals to the fascia dentata in developing and adult rats. J. Comp. Neurol. 200, 433–459 (1981). [DOI] [PubMed] [Google Scholar]
- 16.Zimmer J., Gähwiler B. H., Growth of hippocampal mossy fibers: A lesion and coculture study of organotypic slice cultures. J. Comp. Neurol. 264, 1–13 (1987). [DOI] [PubMed] [Google Scholar]
- 17.Sutula T., He X. X., Cavazos J., Scott G., Synaptic reorganization in the hippocampus induced by abnormal functional activity. Science 239, 1147–1150 (1988). [DOI] [PubMed] [Google Scholar]
- 18.Tauck D. L., Nadler J. V., Evidence of functional mossy fiber sprouting in hippocampal formation of kainic acid-treated rats. J. Neurosci. 5, 1016–1022 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pun R. Y., et al., Excessive activation of mTOR in postnatally generated granule cells is sufficient to cause epilepsy. Neuron 75, 1022–1034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buckmaster P. S., Does mossy fiber sprouting give rise to the epileptic state? Adv. Exp. Med. Biol. 813, 161–168 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Buckmaster P. S., Mossy Fiber Sprouting in the Dentate Gyrus. Jasper’s Basic Mechanisms of the Epilepsies (National Center for Biotechnology Information (US), Bethesda, MD, ed. 4, 2012). [PubMed] [Google Scholar]
- 22.Godale C. M., Danzer S. C., Signaling pathways and cellular mechanisms regulating mossy fiber sprouting in the development of epilepsy. Front. Neurol. 9, 298 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koyama R., Ikegaya Y., The molecular and cellular mechanisms of axon guidance in mossy fiber sprouting. Front. Neurol. 9, 382 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi K., Yamanaka S., Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Földy C., et al., Single-cell RNAseq reveals cell adhesion molecule profiles in electrophysiologically defined neurons. Proc. Natl. Acad. Sci. U.S.A. 113, E5222–E5231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winterer J., et al., Single-cell RNA-Seq characterization of anatomically identified OLM interneurons in different transgenic mouse lines. Eur. J. Neurosci. 50, 3750–3771 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo W., Lukacsovich D., Que L., Winterer J., Földy C., Data from “Single-cell transcriptome of dentate gyrus granule cells after chemical and genetic induction.” NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE161619. Deposited 17 November 2020.
- 28.Ben-Ari Y., “Kainate and temporal lobe epilepsies: 3 decades of progress” in Jasper’s Basic Mechanisms of the Epilepsies, Noebels J. L., Avoli M., Rogawski M. A., Olsen R. W., Delgado-Escueta A. V., Eds. (National Center for Biotechnology Information, Bethesda, MD, ed. 4, 2012). [PubMed] [Google Scholar]
- 29.Jagirdar R., Drexel M., Bukovac A., Tasan R. O., Sperk G., Expression of class II histone deacetylases in two mouse models of temporal lobe epilepsy. J. Neurochem. 136, 717–730 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elliott R. C., Khademi S., Pleasure S. J., Parent J. M., Lowenstein D. H., Differential regulation of basic helix-loop-helix mRNAs in the dentate gyrus following status epilepticus. Neuroscience 106, 79–88 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Lasorella A., et al., Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature 442, 471–474 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Huang Z., et al., Inhibitor of DNA binding 2 promotes axonal growth through upregulation of Neurogenin2. Exp. Neurol. 320, 112966 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Ko H. R., et al., Akt1-inhibitor of DNA binding2 is essential for growth cone formation and axon growth and promotes central nervous system axon regeneration. eLife 5, e20799 10.7554/eLife.20799. (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu P., et al., Inhibitor of DNA binding 2 promotes sensory axonal growth after SCI. Exp. Neurol. 231, 38–44 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Li X., et al., Polycystin-1 and polycystin-2 regulate the cell cycle through the helix-loop-helix inhibitor Id2. Nat. Cell Biol. 7, 1202–1212 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Lasorella A., Iavarone A., The protein ENH is a cytoplasmic sequestration factor for Id2 in normal and tumor cells from the nervous system. Proc. Natl. Acad. Sci. U.S.A. 103, 4976–4981 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haenold R., et al., NF-κB controls axonal regeneration and degeneration through cell-specific balance of RelA and p50 in the adult CNS. J. Cell Sci. 127, 3052–3065 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Samanta J., Kessler J. A., Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development 131, 4131–4142 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Wenzel H. J., Cole T. B., Born D. E., Schwartzkroin P. A., Palmiter R. D., Ultrastructural localization of zinc transporter-3 (ZnT-3) to synaptic vesicle membranes within mossy fiber boutons in the hippocampus of mouse and monkey. Proc. Natl. Acad. Sci. U.S.A. 94, 12676–12681 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strange B. A., Witter M. P., Lein E. S., Moser E. I., Functional organization of the hippocampal longitudinal axis. Nat. Rev. Neurosci. 15, 655–669 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Kesner R. P., An analysis of the dentate gyrus function. Behav. Brain Res. 254, 1–7 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Kesner R. P., An analysis of dentate gyrus function (an update). Behav. Brain Res. 354, 84–91 (2018). [DOI] [PubMed] [Google Scholar]
- 43.El-Gaby M., Shipton O. A., Paulsen O., Synaptic plasticity and memory: New insights from hippocampal left-right asymmetries. Neuroscientist 21, 490–502 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Roschger C., Cabrele C., The Id-protein family in developmental and cancer-associated pathways. Cell Commun. Signal. 15, 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aaronson D. S., Horvath C. M., A road map for those who don’t know JAK-STAT. Science 296, 1653–1655 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Barrat F. J., Crow M. K., Ivashkiv L. B., Interferon target-gene expression and epigenomic signatures in health and disease. Nat. Immunol. 20, 1574–1583 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen E. Y., et al., Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuleshov M. V., et al., Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shuman T., et al., Breakdown of spatial coding and interneuron synchronization in epileptic mice. Nat. Neurosci. 23, 229–238 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marchionni I., Oberoi M., Soltesz I., Alexander A., Ripple-related firing of identified deep CA1 pyramidal cells in chronic temporal lobe epilepsy in mice. Epilepsia Open 4, 254–263 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flynn S. P., Barriere S., Scott R. C., Lenck-Santini P. P., Holmes G. L., Status epilepticus induced spontaneous dentate gyrus spikes: In vivo current source density analysis. PLoS One 10, e0132630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bragin A., Jandó G., Nádasdy Z., van Landeghem M., Buzsáki G., Dentate EEG spikes and associated interneuronal population bursts in the hippocampal hilar region of the rat. J. Neurophysiol. 73, 1691–1705 (1995). [DOI] [PubMed] [Google Scholar]
- 53.Knierim J. J., Neunuebel J. P., Deshmukh S. S., Functional correlates of the lateral and medial entorhinal cortex: Objects, path integration and local-global reference frames. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang C., et al., Egocentric coding of external items in the lateral entorhinal cortex. Science 362, 945–949 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernández-Ruiz A., et al., Gamma rhythm communication between entorhinal cortex and dentate gyrus neuronal assemblies. Science 372, eabf3119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bareyre F. M., et al., In vivo imaging reveals a phase-specific role of STAT3 during central and peripheral nervous system axon regeneration. Proc. Natl. Acad. Sci. U.S.A. 108, 6282–6287 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun F., et al., Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature 480, 372–375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pernet V., et al., Misguidance and modulation of axonal regeneration by Stat3 and Rho/ROCK signaling in the transparent optic nerve. Cell Death Dis. 4, e734 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo X., et al., Enhanced transcriptional activity and mitochondrial localization of STAT3 co-induce axon regrowth in the adult central nervous system. Cell Rep. 15, 398–410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Accornero F., et al., BEX1 is an RNA-dependent mediator of cardiomyopathy. Nat. Commun. 8, 1875 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khazaei M. R., et al., Bex1 is involved in the regeneration of axons after injury. J. Neurochem. 115, 910–920 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chodaparambil J. V., et al., Molecular functions of the TLE tetramerization domain in Wnt target gene repression. EMBO J. 33, 719–731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ciani L., Salinas P. C., WNTs in the vertebrate nervous system: From patterning to neuronal connectivity. Nat. Rev. Neurosci. 6, 351–362 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Fuchs E., Cleveland D. W., A structural scaffolding of intermediate filaments in health and disease. Science 279, 514–519 (1998). [DOI] [PubMed] [Google Scholar]
- 65.Brose K., Tessier-Lavigne M., Slit proteins: Key regulators of axon guidance, axonal branching, and cell migration. Curr. Opin. Neurobiol. 10, 95–102 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Nicol X., Muzerelle A., Rio J. P., Métin C., Gaspar P., Requirement of adenylate cyclase 1 for the ephrin-A5-dependent retraction of exuberant retinal axons. J. Neurosci. 26, 862–872 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iwasato T., et al., Cortical adenylyl cyclase 1 is required for thalamocortical synapse maturation and aspects of layer IV barrel development. J. Neurosci. 28, 5931–5943 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ravary A., et al., Adenylate cyclase 1 as a key actor in the refinement of retinal projection maps. J. Neurosci. 23, 2228–2238 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nait Taleb Ali H., et al., Lack of adenylate cyclase 1 (AC1): Consequences on corticospinal tract development and on locomotor recovery after spinal cord injury. Brain Res. 1549, 1–10 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Darnell J. C., et al., FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Farley J. E., et al., Transcription factor Pebbled/RREB1 regulates injury-induced axon degeneration. Proc. Natl. Acad. Sci. U.S.A. 115, 1358–1363 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stegmüller J., Huynh M. A., Yuan Z., Konishi Y., Bonni A., TGFbeta-Smad2 signaling regulates the Cdh1-APC/SnoN pathway of axonal morphogenesis. J. Neurosci. 28, 1961–1969 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yi J. J., Barnes A. P., Hand R., Polleux F., Ehlers M. D., TGF-beta signaling specifies axons during brain development. Cell 142, 144–157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhong J., Zou H., BMP signaling in axon regeneration. Curr. Opin. Neurobiol. 27, 127–134 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nix P., Hisamoto N., Matsumoto K., Bastiani M., Axon regeneration requires coordinate activation of p38 and JNK MAPK pathways. Proc. Natl. Acad. Sci. U.S.A. 108, 10738–10743 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zeidler Z., et al., Targeting the mouse ventral hippocampus in the intrahippocampal kainic acid model of temporal lobe epilepsy. eNeuro 5, ENEURO.0158-18.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bui A. D., et al., Dentate gyrus mossy cells control spontaneous convulsive seizures and spatial memory. Science 359, 787–790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mohajeri M. H., et al., The impact of genetic background on neurodegeneration and behavior in seizured mice. Genes Brain Behav. 3, 228–239 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Gröticke I., Hoffmann K., Löscher W., Behavioral alterations in a mouse model of temporal lobe epilepsy induced by intrahippocampal injection of kainate. Exp. Neurol. 213, 71–83 (2008). [DOI] [PubMed] [Google Scholar]
- 80.Van Den Herrewegen Y., et al., The Barnes maze task reveals specific impairment of spatial learning strategy in the intrahippocampal kainic acid model for temporal lobe epilepsy. Neurochem. Res. 44, 600–608 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Leutgeb J. K., Leutgeb S., Moser M. B., Moser E. I., Pattern separation in the dentate gyrus and CA3 of the hippocampus. Science 315, 961–966 (2007). [DOI] [PubMed] [Google Scholar]
- 82.Nakashiba T., et al., Young dentate granule cells mediate pattern separation, whereas old granule cells facilitate pattern completion. Cell 149, 188–201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brandt M. D., et al., Transient calretinin expression defines early postmitotic step of neuronal differentiation in adult hippocampal neurogenesis of mice. Mol. Cell. Neurosci. 24, 603–613 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in National Center for Biotechnology Information Gene Expression Omnibus (GSE161619) (27).