Significance
The G4C2 repeat expansion in the C9orf72 gene is a major cause of frontotemporal dementia and amyotrophic lateral sclerosis. Unusual translation of the repeat sequence produces two highly toxic dipeptide repeat proteins (DPRs), PRX and GRX, which accumulate in the brain tissue of individuals with these diseases. Here, we show that PR and GR toxicity in both Caenorhabditis elegans and mammalian neurons depends on the E3 ubiquitin ligase adaptor SPOP. SPOP acts through bromodomain proteins to mediate dipeptide toxicity. SPOP inhibitors, which are currently being developed to treat SPOP-dependent renal cancer, also protect neurons against DPR toxicity. Our findings identify a highly conserved and “druggable” pathway that may represent a strategy for treating these currently incurable diseases.
Keywords: neurodegeneration, C. elegans, genetic screen, bromodomain proteins, proteasome
Abstract
A hexanucleotide repeat expansion in the C9orf72 gene is the most common cause of inherited amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Unconventional translation of the C9orf72 repeat produces dipeptide repeat proteins (DPRs). Previously, we showed that the DPRs PR50 and GR50 are highly toxic when expressed in Caenorhabditis elegans, and this toxicity depends on nuclear localization of the DPR. In an unbiased genome-wide RNA interference (RNAi) screen for suppressors of PR50 toxicity, we identified 12 genes that consistently suppressed either the developmental arrest and/or paralysis phenotype evoked by PR50 expression. All of these genes have vertebrate homologs, and 7 of 12 contain predicted nuclear localization signals. One of these genes was spop-1, the C. elegans homolog of SPOP, a nuclear localized E3 ubiquitin ligase adaptor only found in metazoans. SPOP is also required for GR50 toxicity and functions in a genetic pathway that includes cul-3, which is the canonical E3 ligase partner for SPOP. Genetic or pharmacological inhibition of SPOP in mammalian primary spinal cord motor neurons suppressed DPR toxicity without affecting DPR expression levels. Finally, we find that knockdown of bromodomain proteins in both C. elegans and mammalian neurons, which are known SPOP ubiquitination targets, suppresses the protective effect of SPOP inhibition. Together, these data suggest a model in which SPOP promotes the DPR-dependent ubiquitination and degradation of BRD proteins. We speculate the pharmacological manipulation of this pathway, which is currently underway for multiple cancer subtypes, could also represent an entry point for therapeutic intervention to treat C9orf72 FTD/ALS.
Mutations in C9orf72 are the most common monogenic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (the most common non-Alzheimer’s adult dementia) (1, 2). The mutation is an expansion of the microsatellite repeat 5′-GGGGCC-3′ in the intron between exons 1A and 1B. In unaffected individuals, the hexanucleotide repeat is reiterated 0 to 23 times (1, 2), while in ALS–FTD patients, the hexanucleotide repeat expansion (HRE) is considered to be pathogenic at >30 (2) or >60 repeats (3), although somatic expansion of the repeat can lead to hundreds or even thousands of repeats (1). Three nonmutually exclusive C9orf72 pathophysiological mechanisms have been proposed: 1) reduced abundance of the native C9ORF72 protein, 2) C9orf72 pre-mRNA containing the HRE adopts an imperfect hairpin structure that sequesters RNA binding proteins, and 3) the generation of dipeptide repeat proteins (DPRs) by repeat-associated non–AUG-dependent (RAN) translation (4) of the HRE-containing C9orf72 pre-mRNA (5).
RAN of the sense G4C2 sequence, as well as a disease-associated antisense G2C4 sequence, leads to the production of five DPRs—Gly-Ala (GA), Gly-Pro, Gly-Arg (GR), Pro-Ala (PA), and Pro-Arg (PR) (6). At autopsy, each of these DPRs is detected immunohistologically in C9orf72 FTD and ALS patients but not non-C9orf72 FTD or ALS patients. While DPR sites of expression and overall levels are highly variable, they are significantly more abundant within frontal and temporal lobe regions affected in FTD (7), suggesting a significant pathological role in this disease.
Of the five DPRs, two—GR and PR—are toxic in multiple cellular and animal model systems, although GA can also be toxic in some, but not all, settings (8–10). Pathophysiological processes linked to DPRs include the following: 1) perturbation of nucleocytoplasmic shuttling (11, 12), 2) impaired protein translation (13), 3) inhibition of proteasome function (14, 15), 4) defects in U2 snSNP-dependent splicing (16), 5) compromised mitochondrial function (17), and 6) perturbation of membraneless organelle dynamics (18, 19). In all of these studies, the key experimental approach involved the non-RAN expression of codon-varied DPRs, which allows experimental separation of G4C2 RNA toxicity from individual DPR toxicity. It is believed that these codon-varied DPR models accelerate the appearance of phenotypes that ordinarily emerge over decades in human C9orf72 patients (8, 13, 18–26).
Here, we describe an unbiased forward genetic screen for DPR modifiers in Caenorhabditis elegans. The genes identified in this screen included many known DPR modifiers, as well as several genes not previously associated with DPR toxicity. One PR50 suppressor was the E3 ubiquitin ligase adaptor Speckle-type Pox virus and Zinc Finger (POZ) domain Protein, or SPOP, which is a common genetic cause of prostate, endometrial, and renal cancer (27) but has never been linked to neurodegenerative disease. CRISPR-based introduction of SPOP missense alleles frequently found in prostate cancer also provided protection against PR50 toxicity. SPOP inhibition is a conserved mechanism to protect against DPR toxicity since either SPOP knockdown or an SPOP inhibitor in rat primary neurons also provided protection against DPR-induced neuron death. Finally, we find that the protective effects of SPOP inhibition depends on the function of bromodomain-containing proteins in both C. elegans and mammalian neurons.
Results
A Genome-Wide RNAi Screen for Suppressors of DPR Toxicity in C. elegans Identifies Highly Conserved Genes.
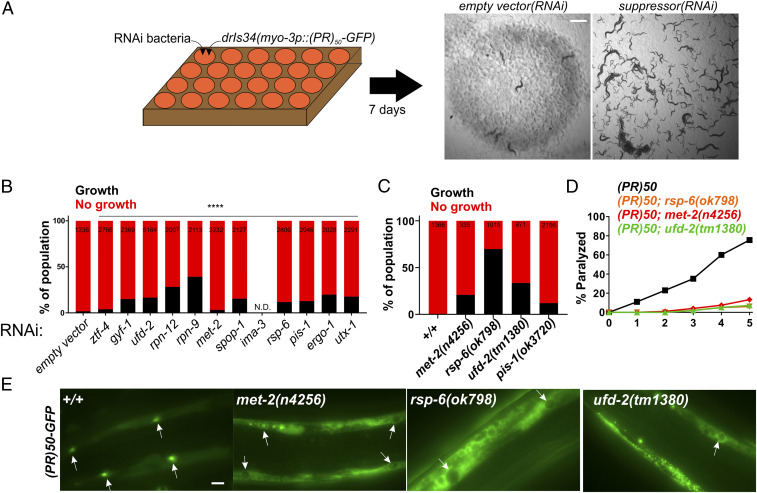
Expression of C9orf72 DPRs PR50 and GR50 are toxic and cause cellular and organismal toxicity in multiple systems (8, 10, 19, 20, 22, 28). Expression of these DPRs in C. elegans muscle cells lead to a completely penetrant larval developmental arrest and paralysis phenotype (21). To identify genes required for this phenotype, we performed a genome-wide RNA interference (RNAi) screen for suppressors of PR50-induced developmental arrest and/or paralysis (Fig. 1 A and B). We screened 15,865 individual gene knockdowns and identified 391 initial PR50 suppressors. Each hit was rescreened in six independent trials. In total, 12/391 hits showed suppression of the PR50 growth arrest in at least 4/6 of these trials (Table 1 and SI Appendix, Fig. S1). Knockdown of 83% of genes (10/12) also suppressed toxicity caused by GR50 in at least 1/2 age-dependent paralysis assays (Table 1 and SI Appendix, Fig. S2). Since loss of these genes suppresses toxicity, these genes are necessary for DPR toxicity. The genes identified as PR50 suppressors fell into three broad functional classes: 1) nuclear transport and RNA binding, 2) ubiquitin-mediated protein degradation, and 3) chromatin regulation (Table 1). Of these hits, 50% (6/12) encode homologs of genes previously identified as modifiers of or interactors with PR50 or GR50 in yeast, fly, or mammalian cells (10, 19, 20, 22), suggesting PR50 engages similar pathophysiological mechanisms in C. elegans as it does in other systems. In the list of PR50 suppressors, 75% (9/12) are conserved from yeast to humans, while 25% (3/12) of genes are absent from yeast but present in metazoan genomes. None of the genes are specific to C. elegans. Only one gene, ergo-1, appears to act via transgene suppression since knockdown reduces both GFP and RFP expression by >50% from a transgene expression control strain (SI Appendix, Fig. S3), which is consistent with previous reports of ergo-1(RNAi) acting as a transgene suppressor (29). The correlation between the level of mRNA knockdown and the degree of suppression of PR50 toxicity was poor (R2 = 0.03271; SI Appendix, Fig. S4), indicating no relationship between hit “strength” and the level of knockdown. A predicted nuclear localization signal or experimental evidence for nuclear localization is found in 75% (9/12) of genes, suggesting the encoded protein is localized to the nucleus where PR50 is required for toxicity (Table 1) (21). Genes annotated with “lethal” and/or “sterile” phenotypes made up 75% of the hits (9/12 genes), suggesting that traditional forward genetic screening for DPR suppressors would not have isolated these genes since these screens usually require mutants to be viable and fertile in order to be identified (Table 1). Therefore, our RNAi screen identified highly conserved and essential genes required for PR50 toxicity, many of which are likely to be localized to the nucleus, where PR50 is also required for toxicity.
Fig. 1.
A genome-wide RNAi screen for suppressors of PR50 toxicity in C. elegans. (A) RNAi screening strategy. ∼30 hypochlorite isolated eggs from the drIs34 (myo-3p::PR50-GFP; myo-3p::mCherry) strain are seeded onto 24-well RNAi plates. After 7 d, each well was visually examined and scored relative to the empty vector(RNAi) control, which exhibits no growth and strong paralysis. Suppressors are identified as wells in which worms continue to grow and/or regain motility (gfp(RNAi) shown as an example). (B) COPAS quantification of worm length (time-of-flight [TOF]) in RNAi suppressors of PR50. Worms with a TOF >200 were placed in the “Growth” group while those with TOF <200 were placed in the “No growth” group. Data are presented as the percentage of the total population in either the “Growth” or “No growth” groups. The size of each population (N) is indicated at the top of each bar. ****P < 0.0001, Fisher’s exact test versus empty vector(RNAi). (C) Quantification of the percentage of animals expressing PR50 that reach the L4 stage (“Growth”) after 72 h at 25 °C. N is indicated at the top of each bar. (D) Age-dependent paralysis assay was performed with homozygous viable mutations in genes identified in the RNAi screen. n = 100 for each genotype. For each genotype versus WT, P < 0.0001, log-rank test with Bonferroni correction. (E) Localization of PR50-GFP in either the WT or indicated homozygous mutant background. Arrows point to muscle nucleus. In WT, PR50-GFP is primarily localized to nuclei but in n4256, ok798, and tm1380, PR50-GFP is largely excluded from the nucleus and accumulates in nonnuclear compartments. (Scale bar, 10 microns.)
Table 1.
RNAi gene knockdowns that suppress myo-3p::PR50-GFP toxicity
| C. elegans | Yeast | Fly | Human | Description | Predicted or demonstrated NLS | Suppresses GR50? | Lethal/sterile |
| Nuclear transport/RNA binding | |||||||
| rsp-6 | MRD1 | X16* | SRSF3 † | RNA binding protein | Yes | Yes | Yes |
| ztf-4 | NGR1 | CG42458* | NCOA5/hnRNPC† | Contains RRM domain, similar to hnRNPC | Yes | Yes | Yes |
| gyf-1 | SYH1 | Gigyf | GIGYF † | Yeast homolog SYH1 binds proline-rich sequences and influences nuclear pore distribution | Yes | Yes | No |
| ima-3 | KAP122‡ | Kap-α3*,§ | KPNA3 † | One of three nuclear importin alpha subunits | No | No | Yes |
| ergo-1 | None | AGO1T1 | AGO3 | Production of endo-siRNAs | Yes | Yes | No |
| Ubiquitin-mediated protein degradation | |||||||
| spop-1 | None | roadkill | SPOP | Speckle-type POZ protein; no yeast homolog | Yes | Yes | No |
| ufd-2 | Ufd2 | UBE4B/ CG9934 | UBE4a | E3 and/or E4 ubiquitin ligase | Yes | Yes | No |
| rpn-12 | RPN12 | RPN12 | PSMD8 ¶ | nonATPase regulatory subunit of the 26S proteasome | No | Yes | No |
| rpn-9 | RPN9 | RPN9 | PSMD13 | nonATPase regulatory subunit of the 26S proteasome | No | Yes | Yes |
| Chromatin regulation | |||||||
| met-2 | MET2 | eggless | SETDB1 | Histone methyltransferase | Yes | Yes | Yes |
| utx-1 | CYC8 | Utx | UTX/UTY # | Histone demethylase | Yes | Yes | Yes |
| pis-1 | None | PTIP | PAXIP1 | Pax transcription factor interacting protein | Yes | No | Yes |
Enhanced the toxicity of GR50 in Drosophila model (19).
Interacts with both PR and GR in human cells (19).
Suppresses the toxicity of PR50 when overexpressed in yeast (10).
Enhanced the toxicity of PR25 in Drosophila model (22).
Gene is enriched or depleted in PR20-treated K562 cell CRISPR screen (20).
Gene is enriched or depleted in GR20-treated K562 cell CRISPR screen (20).
We obtained previously characterized mutant alleles that were viable and fertile for several of the genes identified in our screen. To validate our RNAi results, we crossed these mutants into the PR50 background. As we found for RNAi knockdown, loss-of-function mutants suppressed the developmental arrest phenotype and the postdevelopmental age-dependent paralysis phenotype caused by PR50 (Fig. 1C and D). We examined PR50 localization in each of these mutants and found that nuclear localization of the DPR was disrupted. In each mutant background, PR50-GFP was distributed throughout the nonnuclear compartments and was largely absent from either the nucleus or nucleolus (Fig. 1E). These data provide independent validation of the hits derived from our RNAi screen and suggest that the mechanism of suppression for some genes is associated with relocalization of PR50 out of the nucleus.
Loss-of-Function Mutations in the E3 Ubiquitin Ligase Adaptor SPOP Suppress PR50 and GR50 Toxicity.
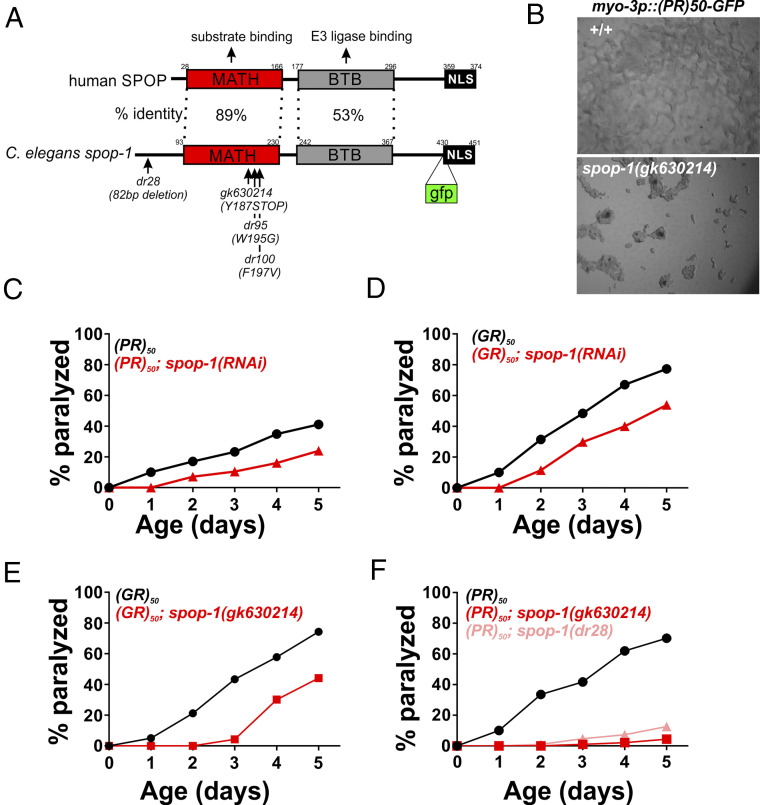
In addition to several previously identified DPR modulators (rsp-6/SRSF3, ima-3/KPNA3, ztf-4/hnRNPC, gyf-1/GIGYF, rpn-12/PSMD8, utx-1/UTX/UTY) (10, 19, 20, 22), our screen also identified several genes associated with regulation of the ubiquitin proteasome system. One such proteasome system gene was spop-1, the C. elegans homolog of human speckled-type POZ domain containing protein (SPOP) (Fig. 2 A and B). spop-1(RNAi) robustly suppressed the age-dependent toxicity of both PR50 and GR50 (Fig. 2 C and D). We obtained or generated via CRISPR/Cas9 mutations in spop-1 (gk630214, dr28) and crossed them into PR50- and GR50-expressing C. elegans. spop-1(gk630214) is a nonsense mutation early in the spop-1 coding sequence that truncates the protein within the MATH domain and is therefore likely a null allele (Fig. 2A). spop-1(dr28) is a CRISPR engineered 82-base pair deletion within exon 2 that deletes the start codon for isoform b and creates a reading-frame shift in the isoform a, also creating a likely null allele (Fig. 2A). Both spop-1 mutations recapitulated the RNAi phenotypes observed for both age-dependent paralysis (Fig. 2 E and F) and growth arrest (Fig. 2B). Additionally, spop-1(gk630214) suppressed the age-dependent toxicity of a motor neuron expressed GR50 transgene (SI Appendix, Fig. S5). Therefore, loss of spop-1 function suppresses C9orf72-associated DPR toxicity in multiple cellular contexts.
Fig. 2.
Loss-of-function mutations in the E3 ubiquitin ligase adaptor spop-1 suppress PR50 and GR50 toxicity in C. elegans. (A) Domain structure of the SPOP protein. The percent identity between the human and C. elegans domains are shown. The dr28 allele is a CRISPR-induced allele that deletes 82 bp within the second exon of spop-1. This shifts the reading frame of isoform 1 to encounter a premature stop codon and deletes that start ATG for isoform 2. Therefore, dr28 is a likely spop-1 null allele. The gk630214 allele is a spop-1 nonsense mutation isolated from the Million Mutation project. The gfp insertion site used to make the SPOP-1-GFP CRISPR allele is shown. (B) spop-1(gk630214) suppresses the developmental arrest phenotype of PR50-GFP. (C and D) spop-1(RNAi) suppresses the age-dependent paralysis phenotype of PR50-GFP (P = 0.000018) (C) and GR50-GFP (P = 0.0001) (D). (E and F) spop-1 alleles gk630214 and dr28 suppress PR50-GFP (P = 0.00000011) (E) and GR50-GFP (P < 0.0001) (F). For all paralysis assays, n = 100 animals for each genotype.
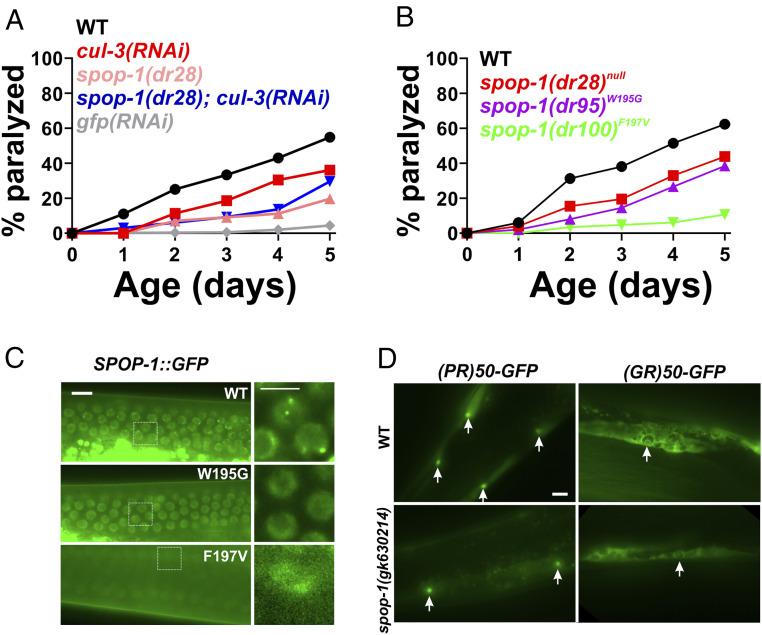
In mammals, SPOP functions as an adaptor protein that targets substrates for ubiquitination by cullin 3-type E3 ubiquitin ligases and subsequent proteasomal degradation (27). If spop-1 confers protection against DPR toxicity through its canonical function as an E3 ubiquitin ligase adaptor, then inhibition of the cullin 3 ubiquitin ligase should phenocopy spop-1 mutants and protect against DPR toxicity. Furthermore, this protective effective should exhibit a nonadditive gene interaction with spop-1 mutants. Since cul-3 mutants are not viable, we tested these predictions by performing cul-3(RNAi) in the wild-type and spop-1(dr28) mutant background. cul-3(RNAi) in a wild-type background provided significant protection against PR50 toxicity (Fig. 3A). While spop-1(dr28) also suppressed PR50 toxicity, cul-3(RNAi) in the spop-1(dr28) mutant did not provide additional suppression (Fig. 3A). This is unlikely to be due to a floor effect since gfp(RNAi) consistently suppresses PR50 toxicity to a greater extent than spop-1(dr28) (Fig. 3A). Together, these genetic data suggest that cul-3 functions in the same genetic pathway as spop-1 to mediate DPR toxicity in C. elegans.
Fig. 3.
Inhibition of a spop-1-cul-3 pathway protects against DPR toxicity without altering DPR levels or localization. (A) PR50 paralysis assay in the indicated genetic backgrounds. n = 100 animals per genotype. WT versus cul-3(RNAi), P = 0.0153; WT versus spop-1(dr28), P = 0.00000036; spop-1(dr28) versus spop-1(dr28); cul-3(RNAi), P = 0.4278. Log-rank test with Bonferroni correction for multiple comparisons. (B) PR50 paralysis assay in the indicated spop-1 genotypes. Mutations were introduced into the endogenous C. elegans spop-1 gene via CRISPR/Cas9. n = 100 animals per genotype. WT versus spop-1(dr28), P = 0.0059; WT versus spop-1(dr95), P = 0.0002; WT versus spop-1(dr100), P = 0.002. Log-rank test with Bonferroni correction for multiple comparisons. (C) Localization of a CRISPR-engineered SPOP-1-GFP fusion protein containing the indicated point mutations. Germline nuclei are shown because their high density facilitates observation of multiple instances of SPOP-1-GFP speckling. Similar speckling behavior is observed in somatic cells. (Scale bar, 10 microns.) Arrows in WT point to examples of SPOP-1-GFP nuclear localization exhibiting subnuclear speckles. (D) PR50-GFP and GR50-GFP expression in wild type or spop-1(gk630214). Images are exposure matched. Arrows point to muscle nuclei with nucleolar DPR enrichment, as was previously described for PR50-GFP and GR50-GFP localization in C. elegans (21). (Scale bar, 10 microns.)
Cancer-Causing Missense Mutations in SPOP Protect against PR50 Toxicity.
Genetic missense mutations in SPOP are a common cause of prostate cancer. The most common mutations, W131G and F133V, are loss-of-function alleles. These mutations are in the SPOP MATH domain and interfere with the ability of SPOP to interact with its substrates. If the role of SPOP in DPR toxicity is to target substrates for cul-3–dependent ubiquitination and degradation, then introduction of these mutations into C. elegans spop-1 is predicted to phenocopy the spop-1 null alleles and confer protection against PR50 toxicity. We used CRISPR/Cas9 to introduce each mutation into the endogenous spop-1 gene (W195G and F197V in C. elegans) in the PR50 background. We also knocked these mutations into a spop-1-gfp allele tagged at the endogenous locus with CRISPR/Cas9. Like the spop-1 null alleles, both missense mutations conferred protection against PR50 toxicity (Fig. 3B). While spop-1W195G-gfp exhibited normal abundance and nuclear localization, spop-1F197V-gfp showed a substantial decrease in the SPOP-1-GFP protein levels, suggesting the F197V mutation reduces SPOP-1 protein levels (Fig. 3C). Loss-of-function spop-1 alleles did not alter the levels or localization of the PR50 protein, suggesting spop-1 functions downstream of DPR nuclear localization to mediate toxicity (Fig. 3D). Together, these genetic data are consistent with the hypothesis that spop-1 mediates DPR toxicity via cul-3–dependent ubiquitination and degradation of one or more spop-1 substrates.
SPOP Is Required for DPR Toxicity in Mammalian Neurons.
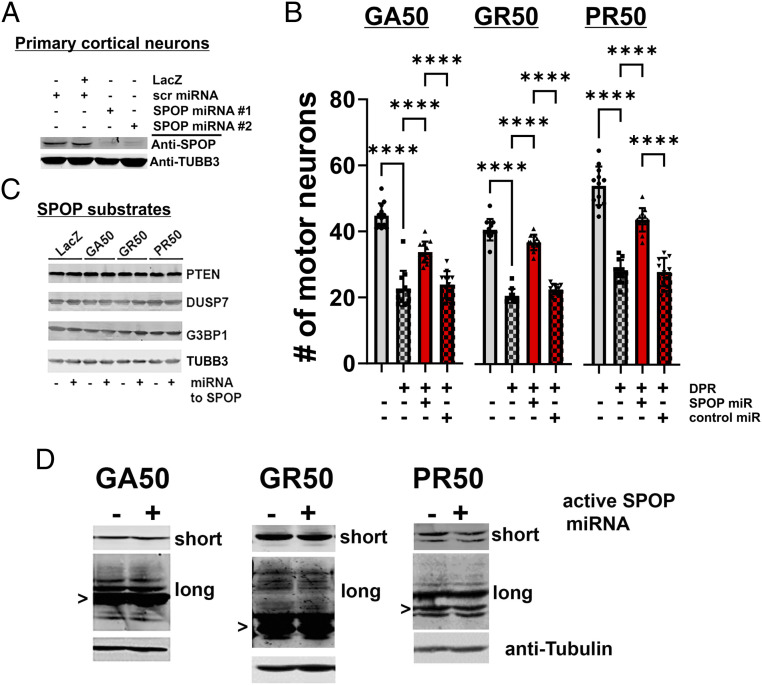
SPOP is highly conserved from C. elegans to humans (Fig. 2) but is absent from yeast. To determine if the role of SPOP in DPR toxicity is evolutionarily conserved, we utilized a rat spinal cord neuron culture system model of DPR toxicity. Embryonic rat spinal cord motor neurons were infected with recombinant herpes simplex virus (HSV) engineered to express microRNAs (miRs) targeting rat SPOP. In prior work, we show that both viruses express their cargo in >95% of neurons in a nontoxic manner (30, 31). Two independently derived SPOP miRs led to an ∼90% reduction in SPOP protein levels in pure neuronal cultures (Fig. 4A). After growing neurons on an astrocyte feeder layer for 2 wk, we coinfected our spinal cord neuronal cultures with two separate HSVs, one expressing a codon-optimized DPR (GA50, GR50, or PR50) and the second expressing either the SPOP miR or a scrambled sequence control miR. We found that expression of known toxic DPRs GA50, GR50, and PR50 caused significant neuronal toxicity and reduced the number of viable primary motor neurons ∼50% (Fig. 4B and SI Appendix, Fig. S6). Expression of a scrambled control sequence miR was neither toxic nor protected against the toxicity of PR50 (Fig. 4B and SI Appendix, Fig. S6). However, expression of the SPOP miR provided highly significant neuroprotection against the toxicity of all DPRs (Fig. 4B; Rescue relative to control: GA50, 50.0 ± 14.38%; GR50, 80.86% ± 11.75%; PR50, 60.26 ± 13.72%). Interestingly, while the SPOP miR robustly inhibited SPOP protein expression, the levels of several known SPOP ubiquitination substrates identified in cancer cells (e.g., PTEN, DUSP7, and G3BP1) were not altered by either PR50, GR50, or SPOP knockdown (Fig. 4C and SI Appendix, Fig. S7). Since these SPOP substrates have been largely characterized in renal, prostate, and endometrial cancer cell lines, these data present the possibility that neuronal SPOP functions may include the degradation of neuron-specific substrates. Finally, as we saw in C. elegans spop-1 mutants, SPOP knockdown in mammalian neurons did not alter the abundance of any of the toxic DPRs (either nominal monomers or higher molecular weight species; Fig. 4D and SI Appendix, Fig. S8). These data suggest that SPOP is an evolutionarily conserved factor broadly required for DPR toxicity from C. elegans to mammalian neurons and that it functions downstream of DPRs to engage mechanisms leading to cellular toxicity.
Fig. 4.
SPOP knockdown protects against DPR toxicity in mammalian primary neurons without reducing the abundance of known SPOP ubiquitination targets or DPRs themselves. (A) Cortical neuron cultures were infected with HSV engineered to express LacZ, a scrambled sequence miR or two different miRs targeting SPOP. Endogenous neuronal SPOP levels are reduced by targeted miRs. (B) DIV 14 spinal cord neuron cultures were coinfected with HSV engineered to express the indicated DPR and either the scrambled sequence miR (control, “cont”) or miR targeting SPOP. Control cultures were infected with HSV-LacZ. Cultures were processed for immunocytochemistry and motor neuron counts were obtained 5 d later. By ANOVA, DPRs lead to a statistically significant reduction in motor neuron number (compared with LacZ expressing cultures) and knockdown of SPOP leads to a statistically significant protection against SPR toxicity (e.g., GA50 group, F(3,44) = 72.49, P < 0.001; GR50 group, F(3,43) = 204.7, P < 0.0001; PR50 group, F(3,44) = 100.8, P < 0.0001). Pairwise comparisons utilized Tukey’s post hoc multiple comparisons test. ****P < 0.0001. (C) Cortical neuron cultures were infected with HSV engineered to express LacZ or the GA50, PR50, or GR50 DPRs. Simultaneously, cultures were infected with either the HSV engineered to express the scrambled sequence miR or SPOP miR. Then, 2 d later, before any neuron loss is detected, lysates were probed for known SPOP targets. No statistically significant group differences were detected in band intensities by ANOVA (SI Appendix, Fig. S7). (D) Lysates from experiments described in C were probed for GA, GR, or PR and imaged with short or long exposures (to see less abundant higher molecular weight species). Putative monomers are noted with “>.” No statistically significant group differences were detected in scan band intensities by ANOVA (SI Appendix, Fig. S8).
SPOP is strongly up-regulated in most clear cell renal carcinomas, and genetic inhibition of SPOP in these cancer cells attenuates their growth (32). As a result, Guo et al. performed a small molecule screen for SPOP inhibitors (33). One inhibitor, Compound 6b, blocks SPOP-substrate interactions with low micromolar affinity. We asked whether 6b was neuroprotective in our tissue culture model. Application of 6b had no effect on motor neuron survival in naive cultures (number of motor neurons: vehicle, 37.88 ± 2.497; Compound 6b, 36.06 ± 5.226; P = 0.2035, unpaired t test). However, 6b exhibited a statistically significant blunting of DPR toxicity in cultures that express GR50, GA50, or the exogenously provided PR20 peptide (SI Appendix, Fig. S9) (15, 20, 34). These observations suggest that the SPOP-CUL3 pathway we have defined may be amenable to small molecule modulation in mammals.
Bromodomain-Containing Proteins Are Required for SPOP Mutant Suppression of DPR Toxicity in C. elegans and Mammalian Neurons.
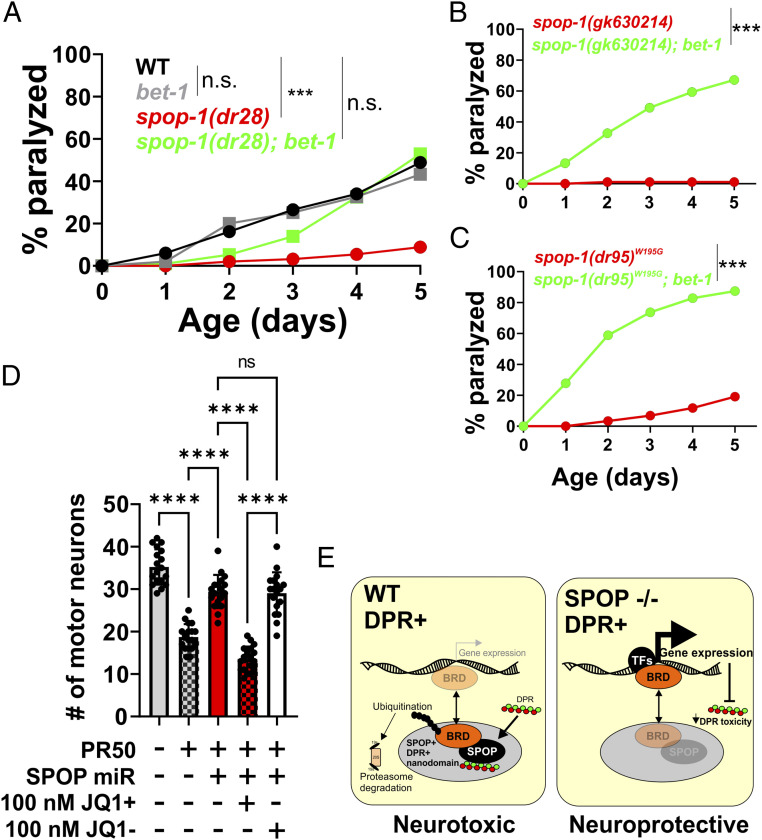
Our data suggest a model in which a conserved function of SPOP leads to the ubiquitin-dependent degradation of one or more substrates when toxic DPRs are present. In an SPOP mutant, this substrate(s) is no longer ubiquitinated and degraded, leading to increased protein levels and protection against DPR toxicity. A prediction of this model is that reducing the abundance of this substrate should reverse the protective effect of spop-1 mutants against DPRs. To identify such substrates, we performed a spop-1 mutant suppressor screen. In mammalian cancer cells, spop-1 targets >30 proteins for degradation (27). Eleven of these substrates have homologs in C. elegans. We performed feeding-based RNAi and paralysis assays for nine of these candidate SPOP substrates in both the wild type and spop-1(dr28) mutant. Only a single gene knockdown targeting the BRD2/3/4 homolog bet-1 suppressed spop-1(dr28) protection against DPR toxicity (Fig. 5). In wild-type animals, bet-1(RNAi) did not further enhance DPR toxicity (Fig. 5A). bet-1(RNAi) also suppressed the paralysis phenotypes of the spop-1 LOF allele gk630214 (Fig. 5B) and the spop-1 W195G missense allele (Fig. 5C). This indicates that the bet-1(RNAi) suppression of spop-1 is not allele specific. Rather, loss of bet-1 suppresses the phenotypic consequences associated with loss of spop-1.
Fig. 5.
spop-1 mutant protection against DPR toxicity requires the bromodomain protein bet-1. (A) Paralysis assay with the indicated genotypes. bet-1 null alleles are homozygous sterile (62). Therefore, bet-1(RNAi) was utilized. All genotypes include the drIs34 (myo-3p::3XFLAG-PR50-GFP) transgene. n = 100 animals per genotype. WT versus bet-(RNAi), P = 0.1187; WT versus spop-1(dr28), P < 0.0001; WT versus spop-1(dr28); bet-1(RNAi), P = 0.011; spop-1(dr28) versus spop-1(dr28); bet-1(RNAi), P = 0.0001, log-rank test with Bonferroni correction for multiple comparisons. (B) Paralysis assay in the indicated spop-1 allele ± bet-1(RNAi). n = 100 animals per genotype. ***P < 0.0001, log-rank test with Bonferroni correction for multiple comparisons. (C) Paralysis assay in the indicated spop-1 allele ± bet-1(RNAi). n = 100 animals per genotype. ***P < 0.0001, log-rank test with Bonferroni correction for multiple comparisons. (D) Inhibition of BRD proteins with the BET domain inhibitor JQ1+ following SPOP knockdown in mammalian neurons expressing PR50. JQ1− is an inactive enantiomer of JQ1+. One-way ANOVA, F(4,85) = 93.30, P < 0.0001. Tukey’s multiple comparisons test, ****P < 0.0001, not significant (n.s.), P = 0.9982. (E) Model for the function of SPOP and BRD proteins in C9orf72 DPR toxicity.
Small molecule BET bromodomain inhibitors such as JQ1+ have been developed as lead compounds for cancer chemotherapeutics (35). We employed it here to test if BET proteins are required for SPOP inhibition to protect against DPR toxicity, as it is in C. elegans. When applied to primary neuronal cultures, 100 nM JQ1+ (or the inactive enantiomer, JQ1−) was nontoxic and did not influence the toxicity of PR50 (SI Appendix, Fig. S10). Application of JQ1+ (but not JQ1−) suppressed the beneficial effects of knockdown of SPOP on PR50 toxicity (Fig. 5D). These results are consistent with the notion that the healthful effects of reducing SPOP on PR50 toxicity in mammalian neurons requires active BRD proteins. Taken together, these genetic data in both C. elegans and mammalian primary neurons are consistent with a model in which SPOP mediates the degradation of BET domain proteins and that stabilization of BET proteins protects against DPR toxicity (Fig. 5E).
Discussion
Aggregated, misfolded, and otherwise insoluble proteins, including DPRs, accumulate in the brains of individuals who bear the HRE in the C9orf72 locus, and these species are widely believed to be key contributors to disease pathophysiology. One potential driver of this process is direct inhibition of the proteasome by DPRs. Cryoelectron tomographic imaging of cells expressing GA DPRs shows that twisted GA filaments are decorated with proteasomes, which appear arrested in a nonfunctional state (14). Similarly, PR DPRs directly bind to purified proteasomes and inhibit degradation of ubiquitinated substrates (15). Here, we define components of a pathway that also ties the ubiquitin-proteasomal degradation pathway to DPR toxicity. In our unbiased RNAi screen, we discovered that the E3 ubiquitin ligase adaptor SPOP and its associated E3 ligase, cullin 3, are required for DPR toxicity. The beneficial effects of ablation of SPOP as well as introduction of mutations that inactivate its ability to present clients to the E3 ligase provide strong evidence that the loss of SPOP canonical function is a functionally important mechanism by which resistance to DPR toxicity is conferred.
Previously, we have shown that the toxicity of PR and GR DPRs are dependent upon nuclear localization (21). This aligns well with the fact that SPOP is also a predominantly nuclear protein and suggests that SPOP-dependent degradation of a nuclear protein(s) renders cells vulnerable to DPR toxicity. SPOP was originally identified in 1997 as a POZ domain containing protein that localizes to subnuclear speckles (36). Subsequent studies revealed that SPOP speckles have biophysical characteristics associated with proteins that undergo liquid–liquid phase separation (LLPS) (37). Moreover, SPOP nuclear speckles are highly dynamic and are enriched with known SPOP ubiquitination substrates (38). Given that PR and GR dipeptides can both undergo LLPS themselves and interfere with LLPS associated with other proteins (18, 19, 39), our discovery that SPOP is required for DPR toxicity suggests the possibility that alterations in the LLPS properties of SPOP speckles by PR50 could be the basis for DPR toxicity. These alterations could either occur through direct or indirect SPOP–DPR interactions. For example, DPRs could directly interact with SPOP and block its ability to colocalize with and degrade specific ubiquitination targets. Alternatively, DPRs could drive SPOP to inappropriately interact with and degrade targets that would not normally be degraded. Our data demonstrating that inhibition of the SPOP-substrate bet-1 suppresses the protective effect of SPOP inhibition on DPR toxicity supports the later hypothesis. Additional studies examining SPOP–DPR interactions, as well as the roles of other SPOP substrates in mammalian cells, are needed to further differentiate between these possibilities.
While SPOP has well-characterized links to nonneuronal diseases, such as prostate, endometrial, and renal cancer (32, 40, 41), our studies now link SPOP to a neurodegenerative disease. In prostate cancer, recurrent somatic missense mutations such as W131G and F133V occur in ∼10% of metastatic patients (42). These mutations lead to SPOP loss of function and stabilization of multiple SPOP substrates (43, 44). Many additional missense mutations are associated with SPOP-dependent cancer, including several gain-of-function mutations associated with endometrial cancer that enhance the degradation of SPOP substrates (45). A prediction of our findings is that such gain-of-function alleles may enhance DPR toxicity. The highly conserved nature of the SPOP protein from C. elegans to humans, combined with efficient CRISPR engineering in worms and highly penetrant DPR phenotypic assays, provides a unique opportunity to define SPOP structure–function relationships in SPOP-dependent DPR toxicity in ways that are not possible in other systems.
SPOP targets many proteins for degradation (46). In SPOP prostate cancer mutants, the ability to target and degrade these substrates is lost, which leads to their up-regulation (38). Up-regulation of several such SPOP substrates are associated with different aspects of the cancer phenotype (45, 47, 48). One important set of substrates are the Bromodomain and Extra-Terminal motif (BET)-containing proteins BRD2, BRD3, and BRD4. BET proteins are transcriptional coactivators that interact with acetylated histones and transcription factors to control gene expression (49). Our studies here show that BET proteins are also likely to play a role in SPOP-dependent DPR toxicity. We discovered that the protective effect of spop-1 mutants on DPR toxicity was suppressed by reducing the abundance of the C. elegans protein bet-1. In addition, pharmacological inhibition of BRD proteins blocks the beneficial effects of reduced SPOP on DPR toxicity in mammalian neurons. Our findings suggest a model in which DPRs lead to the inappropriate SPOP-dependent degradation of BRD proteins and possibly other substrates. Loss of SPOP leads to BRD protein up-regulation, which protects against DPR toxicity via alterations in gene expression. Inhibition of bet-1 in the spop-1 mutant suppresses these compensatory transcriptional responses and restores DPR toxicity. While our genetic studies are consistent with this model, additional cell biological and biochemical tests are needed to more rigorously test its predictions. Interestingly, in a recent small molecule screen, two BET inhibitors were identified as suppressors of exogenous PR20 toxicity (50). These observations differ from our findings, which show no effect of BET inhibitors on DPR toxicity when SPOP is present. The disparate results may be due to the differences in discovery platforms, cell types, read-out measures, redundant effects on multiple BRD domain proteins, and/or off-target drug effects. In this respect, genetic studies may be a more precise tool for investigating the role of BET proteins in DPR toxicity.
The primary goal of disease mechanism research is to find a druggable target. In this regard, SPOP is an attractive candidate. The SPOP inhibitor Compound 6b stops the growth of SPOP overexpressing ccRCC cells in culture and attenuates tumor growth in a mouse model. A recent study identified 6b derivatives that are even more potent (51), suggesting that the SPOP pathway is highly suitable for small molecule intervention. Given that genetic inhibition of the cancer-relevant functions of SPOP can also suppress DPR toxicity, we predict that 6b and its derivatives could also be a new and potentially viable therapeutic option for opposing DPR toxicity. Preclinical studies show that 6b is well tolerated and has no major side effects (33), although the blood–brain barrier permeability of this compound is not known. Future studies investigating 6b or other SPOP inhibitors could provide a novel entry point for new ALS/FTD therapies.
Conclusion
In conclusion, our unbiased genetic screening approach in C. elegans discovered that the SPOP ubiquitin ligase pathway is essential for C9orf72 DPR toxicity. Given that SPOP is a nuclear protein and DPR nuclear localization is required for toxicity, our findings provide additional evidence that the mechanism of DPR toxicity originates in the nucleus. Drugs targeting both SPOP and its relevant targets, such as BET domain proteins, are useful tools for treating cancer and may have utility as C9orf72 disease therapeutics.
Materials and Methods
C. elegans Strains, Culture, and Genome-Wide RNAi Screen.
Reference SI Appendix, Table S1 for a list of the strains utilized in this study. Unless otherwise indicated, animals were cultured at 20 °C. The genome-wide RNAi screen utilized the Ahringer feeding library (Source Bioscience) and was performed as previously described (52, 53). Briefly, 96-well plates containing ∼100 µl Luria Broth (LB) + 25 µg/mL carbenicillin liquid media were seeded with individual bacterial clones using a 96-well pin tool and grown for 18 to 24 h at 37 °C. Of this overnight culture, 20 µl was seeded into a single well of a 24-well plate containing nematode growth media (NGM) agar + 25 µg/mL carbenicillin + 1mM IPTG (Thermo Fisher Scientific) and incubated overnight at room temperature (∼20 °C). Prior to seeding on RNAi screening plates, the drIs34 strain is fed gfp(RNAi) bacteria to suppress the expression of PR50-GFP, which allows drIs34 animals to grow and reproduce. When fed empty vector(RNAi) or standard OP50, the drIs34 strain produces progeny that are growth arrested and paralyzed. This phenotype is 100% penetrant. Therefore, gfp(RNAi) serves as a positive control for the PR50-GFP suppression screen, and EV(RNAi) serves as a negative control. Approximately thirty synchronized L1 stage drIs34 worms were seeded into each well and incubated at 20 °C. After 7 d, each well was manually screened for RNAi clones that allowed further growth and/or restored mobility compared to the EV(RNAi) negative control. “Hits” from this primary screen were rescreened six times, and clones that were identified as “hits” in 4/6 rescreens were considered validated hits. During the screening process, the experimenters were blinded to the identity of the RNAi clone. RNAi clones from validated hits were sequenced to determine the identity of the gene targeted. These sequence-validated strains were frozen as glycerol stocks and used for all subsequent RNAi-based experiments.
Molecular Biology and CRISPR Methods.
All procedures involving recombinant or synthetic nucleic acid molecules and materials were approved by the University of Pittsburgh Institutional Biosafety Committee. The drIs34 (PR50) and drIs28 (GR50) transgenes were previously described (21). CRISPR/Cas9 engineering was performed according to the method of Ghanta et al. (54). For point mutations and deletion edits, worms were injected with a mixture of Cas9 (250 ng/µl [IDT]), tracrRNA (100 ng/µl [IDT]), guide RNA (56 ng/µl [IDT]), repair oligonucleotide (110 ng/µl [IDT]), and rol-6(su1006) marker plasmid (40 ng/µl). The Cas9, tracrRNA, and guide RNA was incubated at 37 °C for 10 min prior to the addition of the repair oligo and rol-6 plasmid. For generation of the spop-1-GFP CRISPR allele, a gfp PCR product was PCR amplified from pPD95.75 (Addgene) using primers containing 35 bp of overlap with the spop-1 gene immediately upstream of the predicted nuclear localization sequence. Guide RNAs, repair oligonucleotides, PCR primers, and genotyping primers used in this study are listed in SI Appendix, Table S2.
qPCR.
Approximately 20 gravid adult drIs34 worms grown on gfp(RNAi) plates were transferred to either empty vector(RNAi) or suppressor(RNAi) 10-cm RNAi plates (four plates per RNAi condition) and allowed to lay eggs for 24 h. After 7 d at 20 °C, animals from each plate were collected separately. Animals from one plate were analyzed for growth, as measured with the time-of-flight measurement on a COPAS Biosorter. For each of the three remaining plates, total RNA was collected (TRIzol/RNAeasy [Qiagen]), cDNA was synthesized from 200 ng total RNA (Superscript IV Vilo with EZ DNase [Invitrogen]), and gene expression was measured from 2 ng input RNA using PowerTrack SYBR green Master Mix (Applied Biosystems) on a CFX96 Real Time PCR system (BioRad). qPCR primers were designed with the GetPrimer Design Tool (https://gecftools.epfl.ch/getprime) (55).
Microscopy.
Low-magnification images were captured on an MZ16FA fluorescence stereo microscope equipped with a DFC345 FX camera and advanced fluorescence (AF) imaging software (Leica Microsystems). For high-magnification images, day 1 adult hermaphrodites were anesthetized in 10 mM levamisole and imaged within a silicon grease enclosed slide chamber. Animals were imaged on a wide-field DMI4000B inverted microscope with a 63× oil immersion lens. Images were captured with a DFC 340 FX digital camera using AF imaging software (Leica Microsystems). All images were captured with identical exposure times, and image brightness/contrast/intensity was adjusted for all images within a panel simultaneously within CorelDRAW X7.
Paralysis and Thrashing Assays.
DPR paralysis assays were performed as previously described (21, 56). For the thrashing assay quantification of the integrated GR50 motor neuron line, 10 L4 animals were picked onto 3-cm NGM plates spotted with OP50 (five plates per genotype). At each time point, animals were picked into a dish containing ∼1 mL M9 solution. After a 5 min of acclimation to liquid, the number of thrashes in 30 seconds was quantified. Each time the nose of the animals crossed the body midline was considered a thrash. At each time point, 10 worms per genotype were measured. At the completion of the assay, the thrashing rates for each genotype at each time point were normalized to the “Day 0” thrashing rate for that genotype in order to control for any baseline differences in thrashing. The experimenter counting the thrash rates was blinded to the genotype. Each experiment was performed two to three times, and the results from a single representative experiment are shown.
Mammalian Neuronal Cultures.
Primary neurons cultures were established according to previously described methods (15, 30, 31). Briefly, cortical astrocytes from newborn rat pups were plated on German Glass, acid-washed glass coverslips (18-mm round), grown in Minimal Essential Media (MEM) + 10% fetal bovine serum and 10% horse serum (HS), and when they achieved ∼60% confluency, dissociated embryonic (E) day 15 rat spinal cord neurons were added. The media is changed to astrocyte-conditioned MEM + 10% HS + glial derived neurotrophic factor, ciliary neurotrophic factor and cardiotropin-1 (all at 2 nM). AraC (10 μM) was then added to arrest astrocyte proliferation and washed out with subsequent biweekly media supplementation. After 14 d in vitro (DIV), recombinant HSV were added, and cultures were fixed 5 d later. The SPOP miR clone was designed using the BLOCK-IT miR RNAi design tool (SPOP accession no. NM_001100496.1; Thermo Fisher Scientific). Sense and antisense oligos for three distinct rat SPOP miRs beginning at SPOP cDNA base pairs 212, 239, and 336 were synthesized and cloned into the miR expression vector p1006 for packaging into HSV. Western blot validation experiments in HEK cells using overexpressed SPOP showed that the 336-miR was slightly less effective than the 212- or 239-miR. Therefore, only the 212-miR and 239-miRs were utilized. Viruses were generated by the Gene Delivery Technology Core at Massachusetts General Hospital.
For pure neuron biochemical studies, E17 rat cortex was dissociated and plated on plastic Petri dishes and maintained in astrocyte-conditioned, Neurobasal + B27 supplement. After 14 DIV, recombinant HSV was added and lysates prepared for Western blotting. Antibodies used were as follows: anti-GA (1:200; Millipore Sigma), anti-GR (kind gift of Q. Zhu/Cleveland laboratory, Ludwig Center for Cancer Research, San Diego, CA), anti-PR (kind gift of Y. Zhang/Petrucelli laboratory, Mayo Clinic, Jacksonville, FL), anti-SPOP (1:500; Proteintech), anti-tubulin (1:5,000; BioLegend), anti-PTEN (1:1,000; Cell Signaling Technology), anti-DUSP7 (1:1,000; Abcepta), and anti-G3BP1 (1:1,000; Cell Signaling Technology).
Motor Neuron Toxicity Assay.
Methodological details of the in vitro mammalian motor neuron toxicity assay have been reported previously (30, 31, 57, 58). In brief, coverslips containing paraformaldehyde fixed astrocyte/spinal cord cocultures were stained with SMI32 antibody—when restricted to neurons with a cell body diameter of 20 microns or greater, a validated marker of alpha motor neurons in this culture system (59). The number of motor neurons in three randomly selected 20× lower power field was determined and averaged. The average value of four coverslips was determined (experimental replicates). Determination of motor neuron number was obtained with the counter blinded to the experimental manipulation. The results from ≥3 independent experiments are presented.
Statistical Analysis.
Paralysis assays were analyzed using the Kaplan–Meier log-rank function with a Bonferroni correction for multiple comparisons [Online Application for Survival Analysis (OASIS; (60, 61)]. Comparisons of means were analyzed with either a two-tailed Student’s t test (two groups) or one- or two-way ANOVA (three or more groups) using the Tukey, Dunn’s, or Holm–Šidák posttest analysis as suggested in GraphPad Prism 9 (GraphPad Software, Inc.). P values of <0.05 were considered significant.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (NS094921 and NS096319 to T.L., NS05225 and NS087077 to R.G.K). Research in the R.G.K. laboratory is supported by the Heather Koster Family Charitable Fund and the Les Turner ALS Center. We thank the laboratory of Dr. Caiguang Yang (Shaghai Institute of Materia Medica, Chinese Academy of Sciences) for generously providing Compound 6b. We thank Ed Kipreos (University of Georgia) for providing the cul-3(RNAi) clone. Some strains were provided by the Caenorhabditis elegans Stock Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2104664118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.DeJesus-Hernandez M., et al., Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renton A. E., et al., A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gijselinck I., et al., A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: A gene identification study. Lancet Neurol. 11, 54–65 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Balendra R., Isaacs A. M., C9orf72-mediated ALS and FTD: Multiple pathways to disease. Nat. Rev. Neurol. 14, 544–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ling S. C., Polymenidou M., Cleveland D. W., Converging mechanisms in ALS and FTD: Disrupted RNA and protein homeostasis. Neuron 79, 416–438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zu T., et al., RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. U.S.A. 110, E4968–E4977 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson Y. S., et al., Brain distribution of dipeptide repeat proteins in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol. Commun. 2, 70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wen X., et al., Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84, 1213–1225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizielinska S., et al., C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345, 1192–1194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jovičić A., et al., Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci. 18, 1226–1229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang K., et al., The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freibaum B. D., et al., GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525, 129–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y. J., et al., Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat. Med. 24, 1136–1142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Q., et al., In situ structure of neuronal C9orf72 Poly-GA aggregates reveals proteasome recruitment. Cell 172, 696–705.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta R., et al., The proline/arginine dipeptide from hexanucleotide repeat expanded C9ORF72 inhibits the proteasome. eNeuro 4, ENEURO.0249-16.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin S., et al., Evidence that C9ORF72 dipeptide repeat proteins associate with U2 snRNP to cause Mis-splicing in ALS/FTD patients. Cell Rep. 19, 2244–2256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi S. Y., et al., C9ORF72-ALS/FTD-associated poly(GR) binds Atp5a1 and compromises mitochondrial function in vivo. Nat. Neurosci. 22, 851–862 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boeynaems S., et al., Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee K. H., et al., C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer N. J., et al., CRISPR-Cas9 screens in human cells and primary neurons identify modifiers of C9ORF72 dipeptide-repeat-protein toxicity. Nat. Genet. 50, 603–612 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudich P., et al., Nuclear localized C9orf72-associated arginine-containing dipeptides exhibit age-dependent toxicity in C. elegans. Hum. Mol. Genet. 26, 4916–4928 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boeynaems S., et al., Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci. Rep. 6, 20877 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y., et al., Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167, 789–802.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohki Y., et al., Glycine-alanine dipeptide repeat protein contributes to toxicity in a zebrafish model of C9orf72 associated neurodegeneration. Mol. Neurodegener. 12, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farg M. A., et al., C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum. Mol. Genet. 26, 4093–4094 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farg M. A., Konopka A., Soo K. Y., Ito D., Atkin J. D., The DNA damage response (DDR) is induced by the C9orf72 repeat expansion in amyotrophic lateral sclerosis. Hum. Mol. Genet. 26, 2882–2896 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Clark A., Burleson M., SPOP and cancer: A systematic review. Am. J. Cancer Res. 10, 704–726 (2020). [PMC free article] [PubMed] [Google Scholar]
- 28.Chai N., Gitler A. D., Yeast screen for modifiers of C9orf72 poly(glycine-arginine) dipeptide repeat toxicity. FEMS Yeast Res. 18, foy024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer S. E., et al., Multiple small RNA pathways regulate the silencing of repeated and foreign genes in C. elegans. Genes Dev. 27, 2678–2695 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jablonski A. M., et al., Loss of RAD-23 protects against models of motor neuron disease by enhancing mutant protein clearance. J. Neurosci. 35, 14286–14306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhai J., et al., Inhibition of cytohesins protects against genetic models of motor neuron disease. J. Neurosci. 35, 9088–9105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li G., et al., SPOP promotes tumorigenesis by acting as a key regulatory hub in kidney cancer. Cancer Cell 25, 455–468 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Z. Q., et al., Small-molecule targeting of E3 ligase adaptor SPOP in kidney cancer. Cancer Cell 30, 474–484 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Kwon I., et al., Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345, 1139–1145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilan O., et al., Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation. Science 368, 387–394 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagai Y., et al., Identification of a novel nuclear speckle-type protein, SPOP. FEBS Lett. 418, 23–26 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Marzahn M. R., et al., Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J. 35, 1254–1275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bouchard J. J., et al., Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell 72, 19–36.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White M. R., et al., C9orf72 Poly(PR) dipeptide repeats disturb biomolecular phase separation and disrupt nucleolar function. Mol. Cell 74, 713–728.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geng C., et al., Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc. Natl. Acad. Sci. U.S.A. 110, 6997–7002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barbieri C. E., et al., Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 44, 685–689 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blattner M., et al., SPOP mutations in prostate cancer across demographically diverse patient cohorts. Neoplasia 16, 14–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blattner M., et al., SPOP mutation drives prostate tumorigenesis in vivo through coordinate regulation of PI3K/mTOR and AR signaling. Cancer Cell 31, 436–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai X., et al., Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat. Med. 23, 1063–1071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janouskova H., et al., Opposing effects of cancer-type-specific SPOP mutants on BET protein degradation and sensitivity to BET inhibitors. Nat. Med. 23, 1046–1054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theurillat J. P., et al., Prostate cancer. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science 346, 85–89 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma J., et al., SPOP promotes ATF2 ubiquitination and degradation to suppress prostate cancer progression. J. Exp. Clin. Cancer Res. 37, 145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang P., et al., Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT-mTORC1 activation. Nat. Med. 23, 1055–1062 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujisawa T., Filippakopoulos P., Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat. Rev. Mol. Cell Biol. 18, 246–262 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Corman A., et al., A chemical screen identifies compounds limiting the toxicity of C9ORF72 dipeptide repeats. Cell Chem. Biol. 26, 235–243.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Dong Z., et al., Structure-activity relationship of SPOP inhibitors against kidney cancer. J. Med. Chem. 63, 4849–4866 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Rohlfing A. K., Miteva Y., Moronetti L., He L., Lamitina T., The Caenorhabditis elegans mucin-like protein OSM-8 negatively regulates osmosensitive physiology via the transmembrane protein PTR-23. PLoS Genet. 7, e1001267 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamitina T., Huang C. G., Strange K., Genome-wide RNAi screening identifies protein damage as a regulator of osmoprotective gene expression. Proc. Natl. Acad. Sci. U.S.A. 103, 12173–12178 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghanta K. S., Mello C. C., Melting dsDNA donor molecules greatly improves precision genome editing in Caenorhabditis elegans. Genetics 216, 643–650 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.David F. P., Rougemont J., Deplancke B., GETPrime 2.0: Gene- and transcript-specific qPCR primers for 13 species including polymorphisms. Nucleic Acids Res. 45 (D1), D56–D60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rudich P., Snoznik C., Puleo N., Lamitina T., Measuring RAN peptide toxicity in C. elegans. J. Vis. Exp. 158, 10.3791/61024 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McGurk L., et al., Poly(ADP-Ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell 71, 703–717.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta P., Uner O. E., Nayak S., Grant G. R., Kalb R. G., SAP97 regulates behavior and expression of schizophrenia risk enriched gene sets in mouse hippocampus. PLoS One 13, e0200477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mojsilovic-Petrovic J., et al., Protecting motor neurons from toxic insult by antagonism of adenosine A2a and Trk receptors. J. Neurosci. 26, 9250–9263 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han S. K., et al., OASIS 2: Online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 7, 56147–56152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J. S., et al., OASIS: Online application for the survival analysis of lifespan assays performed in aging research. PLoS One 6, e23525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shibata Y., Takeshita H., Sasakawa N., Sawa H., Double bromodomain protein BET-1 and MYST HATs establish and maintain stable cell fates in C. elegans. Development 137, 1045–1053 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.