Significance
The syntrophic interaction of fermenting bacteria with methanogenic archaea is crucial for the globally relevant conversion of biomass into methane. Fifty years after the discovery of syntrophy, it has remained enigmatic how the oxidation of saturated fatty acid fermentation intermediates can be coupled to the thermodynamically extremely unfavorable reduction of CO2 to methane and how such a process can sustain growth of both syntrophic partners. Here, we provide biochemical evidence that heme b cofactors of a membrane-bound oxidoreductase and a modified quinone with perfectly fine-tuned redox potentials are the key players in this microbial process. Bioinformatics analyses suggest that the oxidoreductase plays a crucial role in lipid catabolism of the majority of prokaryotes.
Keywords: syntrophy, microbial methane formation, diheme oxidoreductase, methylmenaquinone, redox loop
Abstract
The microbial production of methane from organic matter is an essential process in the global carbon cycle and an important source of renewable energy. It involves the syntrophic interaction between methanogenic archaea and bacteria that convert primary fermentation products such as fatty acids to the methanogenic substrates acetate, H2, CO2, or formate. While the concept of syntrophic methane formation was developed half a century ago, the highly endergonic reduction of CO2 to methane by electrons derived from β-oxidation of saturated fatty acids has remained hypothetical. Here, we studied a previously noncharacterized membrane-bound oxidoreductase (EMO) from Syntrophus aciditrophicus containing two heme b cofactors and 8-methylmenaquinone as key redox components of the redox loop–driven reduction of CO2 by acyl–coenzyme A (CoA). Using solubilized EMO and proteoliposomes, we reconstituted the entire electron transfer chain from acyl-CoA to CO2 and identified the transfer from a high- to a low-potential heme b with perfectly adjusted midpoint potentials as key steps in syntrophic fatty acid oxidation. The results close our gap of knowledge in the conversion of biomass into methane and identify EMOs as key players of β-oxidation in (methyl)menaquinone-containing organisms.
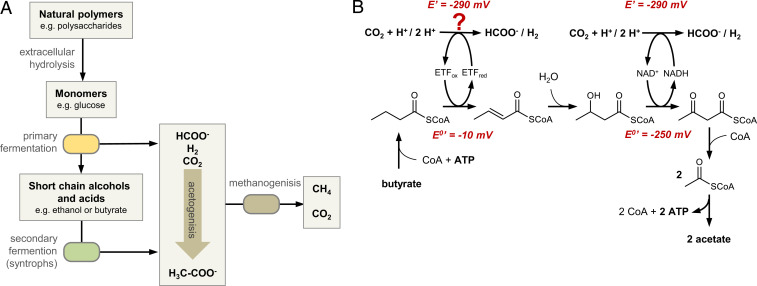
The microbial conversion of natural polymers into methane plays an important role in the global carbon cycle and accounts for more than one-half of all methane produced on Earth per year (1, 2). Methane is formed in anoxic environments, including marine and freshwater sediments, but also in biogas reactors of wastewater treatment plants and other engineered systems. A complex syntrophic association between fermenting bacteria and methanogenic archaea is involved in the degradation of biomass to CH4 and CO2. Primary fermenting bacteria hydrolyze complex polymers into monomers and degrade them mainly into short chain fatty acids (scFA) and alcohols. Secondary fermenting bacteria then oxidize these products to the methanogenic substrates acetate, H2, CO2, and formate that are finally converted into methane by hydrogenotrophic or acetotrophic archaea (Fig. 1A) (3). The reduction of CO2 to CH4 depends on interspecies electron transfer from secondary fermenting bacteria to methanogenic archaea usually via the diffusible low-potential carriers formate and/or H2 (3–5) or, as recently proposed, directly via nanowires (6).
Fig. 1.
Syntrophic degradation of organic matter to methane. (A) Major metabolic processes involving primary fermenters, secondary fermenters, and methanogenic archaea (for simpler presentation, acetogenic conversion of monomers is not depicted here). (B) Model for syntrophic β-oxidation of butyrate to two acetates coupled to the reduction of protons or CO2. The enzyme mediating electron transfer from reduced ETF to FDH has not been studied before and was assigned to noncharacterized DUF224 based on omics-based predictions.
The oxidation of scFA to acetate coupled to the reduction of H+ or CO2 is endergonic under standard conditions (+48 kJ/mol butyrate) but becomes clearly exergonic at an H2 partial pressure below 10 Pa (3, 7). On the other side, the H2 threshold partial pressure of hydrogenotrophic methanogenesis is around 8 Pa corresponding to E′(2H+/H2) ∼ −290 mV or around 10 µM formate, resulting in similar values for the CO2/formate redox couple (2). The syntrophic oxidation of the scFA model compound butyrate to acetate is accomplished by gram-positive Firmicutes (model organism Syntrophomonas wolfei) or gram-negative Deltaproteobacteria (model organism Syntrophus aciditrophicus) (3, 4, 7). It proceeds via two unequal β-oxidation steps (SI Appendix, Fig. 1) (8, 9): 1) Butyryl-CoA is oxidized to crotonyl-CoA by an acyl-CoA dehydrogenase (DH) (E°′ ∼ −10 mV) (10) with an electron-transferring flavoprotein (ETF) serving as electron acceptor, and 2) the 3-hydroxybutyryl-CoA formed by crotonase is subsequently oxidized to acetoacetyl-CoA by 3-hydroxybutyryl-CoA DH (ΔE°′ = −250 mV) (11) using NAD+ as acceptor. The reduction of H+ or CO2 by the NADH formed is feasible under syntrophic conditions, whereas butyryl-CoA oxidation coupled to H+ or CO2 reduction has to overcome a gap of ΔE ∼ −280 mV, giving ΔG°′ ∼ +54 kJ ⋅ mol–1 (3–5). Considering that only one ATP is gained via substrate-level phosphorylation, the energy metabolism of syntrophic butyrate oxidation has remained enigmatic.
Omics-based studies have led to the proposal of models for energy coupling processes during syntrophic scFA oxidation (12–16). The redox loop model is based on the identification of a putative membrane-bound gene product (DUF224) (14–16). It proposes that electrons are transferred from acyl-CoA via ETF, DUF224, and menaquinone (MK) to a membrane-bound formate dehydrogenase (FDH) or hydrogenase driven by the translocation of protons to the cytoplasm, resulting in two energetically unequal half reactions:
In agreement, a protonophore inhibited formation of H2 from butyrate in whole-cell suspension of S. wolfei (17), and MK was reported in S. wolfei and S. aciditrophicus (12, 17). In an alternative model, an electron-confurcating ETF couples endergonic reduction of NAD+ by ETFred to the exergonic reduction of NAD+ by reduced ferredoxin (Fdred–) (18). The NADH formed then may serve as an electron donor for a cytoplasmic FDH. Biochemical evidence for either of the two models is lacking.
Here, we study the missing membrane components that link fatty acid oxidation to CO2 reduction during syntrophic methane production. We provide biochemical evidence that a membrane-bound diheme oxidoreductase and a modified methylmenaquinone with perfectly adjusted redox potentials are the key players of this process. We further propose that related enzymes play a previously overlooked role in the lipid catabolism of the majority of microorganisms.
Results and Discussion
The Acyl-CoA DH/ETF System of S. aciditrophicus Supports a Redox Loop Model.
To shed light on the hypothetical processes involved in syntrophic fatty acid oxidation, we opted for cyclohexane carboxylate degradation in S. aciditrophicus as a model system because this bacterium uses the same enzyme inventory during growth under syntrophic or axenic conditions without substrate-specific gene regulation (12, 19, 20). The β-oxidation of the cyclohexane-carboxyl-CoA (CHCoA) intermediate is initiated by the dehydrogenation to cyclohex-1-ene-1-carboxyl-CoA (CHeneCoA) catalyzed by CHCoA DH (21) (SI Appendix, Fig. S1). We determined a midpoint potential of −89 mV of this reaction by titration in the presence of CHCoA DH and redox dyes (SI Appendix, Fig. S2).
The genome of S. aciditrophicus contains a single copy of etfAB genes (SYN_02636-7), and we enriched the corresponding ETF complex from wild-type cells (SI Appendix, Fig. S3A). It contained 0.95 ± 0.19 flavin adenine dinucleotide (FAD) and 1.1 ± 0.2 AMP per EtfAB heterodimer as determined by ultra-performance liquid chromatography. The oxidation of reduced ETF by CHeneCoA/CHCoA DH proceeded via the red semiquinone (SI Appendix, Fig. S3C) (Table 1). The presence of only one FAD, the only marginal NADH:iodonitrotetrazoliumchlorid oxidoreductase activity (≤0.5 nmol ⋅ min−1 ⋅ mg−1), and the one-electron reduction to the red semiquinone state are all typical properties of a nonelectron-bifurcating ETF (22). These results exclude the possibility that an electron-confurcating ETF drives the endergonic reduction of NAD+ by acyl-CoA to the exergonic reduction of NAD+ by Fdred–. Instead, they support a redox loop model involving a transmembrane electrochemical potential, generated by proton pumping ATP synthase or pyrophosphatase. To test the prerequisites for such a scenario, inner membrane vesicles of S. aciditrophicus cells were prepared for analyzing ΔpH by the pH-sensitive permeable dye 9-amino-6-chloro-2-methoxy acridine. Indeed, the ATP- or, to a minor extent, PPi-dependent formation of a ∆pH was observed, whereas quenching was negligible in the presence of ΔpH-uncoupling agents like nigericin (SI Appendix, Fig. S4).
Table 1.
Specific electron transfer activities in S. aciditrophicus
| Donor → acceptor | Preparation | Specific activity (nmol ⋅ min–1 ⋅ mg–1) |
| Direction: acyl-CoA → CO2 | ||
| CHCoA → ETF | CHCoA DH, ETF | 200* |
| CHCoA → EMO (high-potential heme b) | CHCoA DH, ETF, solubilized EMO | 32 ± 8* |
| EMO (low-potential heme b) → TMN | CHCoA DH, ETF, solubilized EMO | 84 ± 10† |
| CHCoA → EMO (high-potential heme b) | CHCoA DH, ETF, EMO proteoliposomes | 5.5 ± 1.0* |
| β-hydroxybutyryl-CoA → NAD+ | Soluble cell extract | 730 ± 50‡ |
| NADH → CO2 | Soluble cell extract | 23 ± 6‡ |
| Direction: formate/H2 → enoyl-CoA | ||
| Formate → DMN | Membrane protein fraction | 4,200 ± 400‡ |
| Formate → DMN | Solubilized mFDH | 8,000 ± 2,000§ |
| Formate → ETF | Membrane protein fraction, ETF | 16 ± 6‡ |
| Formate → CHeneCoA | Membrane protein fraction, ETF, CHCoA DH | 3.5 ± 1.5‡ |
| H2 → DMN | Membrane protein fraction | <1 |
| TMNH2 → ETF | Solubilized EMO | 260 ± 160† |
| Formate → NAD+ | Soluble protein fraction | 251 ± 36‡ |
| H2 → NAD+ | Soluble protein fraction | 290 ± 130‡ |
Activities are presented as mean value ± SD from at least three biological replicates.
CHCoA DH.
EMO.
Total protein.
mFDH.
Membrane-Bound and Cytoplasmic FDHs and Hydrogenases of S. aciditrophicus.
S. aciditrophicus may use formate and/or hydrogen as an interspecies electron carrier during the syntrophic conversion of fatty acids to methane (4). The genome contains genes encoding two versions of putative three-subunit, membrane-bound, molybdopterin-cofactor, and selenocysteine-containing FDH (SYN_00602-5 and SYN_00632-5, 63% sequence identity of catalytic α-subunits). Both contain a twin arginine translocation transport signal sequence in the α-subunit and four transmembrane helices in the γ-subunit. In contrast, no genes encoding membrane-bound NiFe-hydrogenases are found, whereas genes putatively encoding soluble, NADH-dependent versions of hydrogenases and FDHs exist (7). These findings suggest that CO2 serves as an exclusive acceptor in the predicted redox loop–mediated oxidation of acyl-CoA/ETF. However, this suggestion is in contrast to previous results where membrane-bound hydrogenase activities have been reported in S. aciditrophicus grown axenically and syntrophically (4). We therefore reinvestigated FDH and hydrogenase activities in the membrane fraction after two washing steps and in the soluble fractions using various electron acceptors. With the MK analog dimethylnaphthoquinone (DMN), we identified FDH but no hydrogenase activity in washed membrane preparations (Table 1). We solubilized and enriched membrane-bound FDHs (mFDHs) (SI Appendix, Fig. S3B) with a high specific activity of 8 ± 2 µmol ⋅ min–1 ⋅ mg–1, which is close to values reported for purified Escherichia coli FDH-N with ubiquinone as acceptor (12.2 µmol ⋅ min–1 ⋅ mg–1) (23). With NAD+ as acceptor, both hydrogenase and FDH activities were measured in the soluble fraction (Table 1). Soluble FDH also acted as CO2 reductase, thereby regenerating the NADH formed by β-hydroxybutyryl-CoA DH (Table 1 and Fig. 1).
In Vitro Electron Transfer from Formate to ETF and Enoyl-CoA.
To demonstrate the electronic connection of CHCoA DH to mFDH, we followed the formate-dependent reduction of ETF to the red semiquinone in the presence of washed membrane fraction by ultraviolet–visible (UV/vis) spectroscopy (Table 1). When CHCoA DH was added to this assay, we observed the formate-dependent reduction of CHeneCoA to CHCoA (SI Appendix, Fig. S3D). The activity was diminished by 60% in the presence of 60 µM of the quinone antagonist 2-n-heptyl-4-hydroxyquinoline-N-oxide (24), supporting the assumption that a quinone is involved in the electron transport between the redox couples enoyl-CoA/acyl-CoA and CO2/formate.
Methylmenaquinone Is the Only Quinone in S. aciditrophicus Synthesized from MK by a Radical S-Adenosyl Methionine (SAM) Methyltransferase.
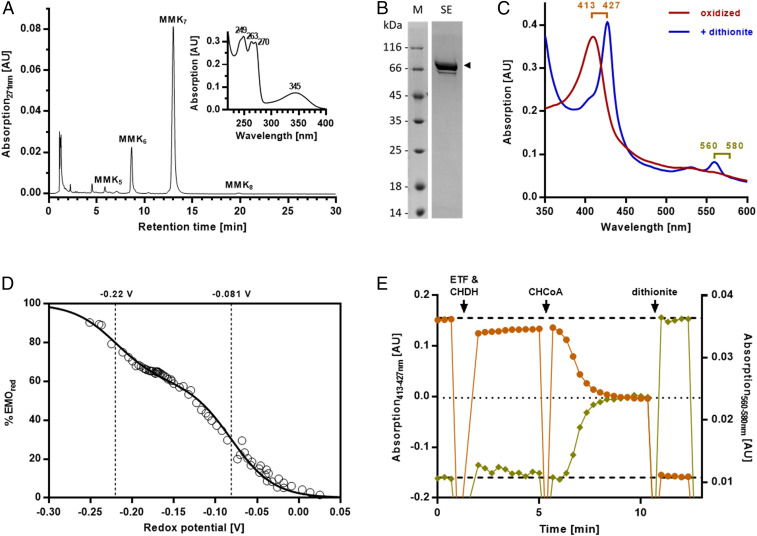
To identify the quinone involved in the suggested redox loop, we analyzed S. aciditrophicus membranes and identified species with UV/vis absorbance maxima at 345 nm characteristic for 8-methylmenaquinone (8-MMK) (25, 26) (Fig. 2A). MS analyses identified 8-MMK7 (with seven isoprenoid units) as the dominating form (SI Appendix, Fig. S5), whereas no MK was found. The formation of the low-potential 8-MMK (E°′ = −156 mV) from MK is catalyzed by a class C radical SAM methyltransferase designated MenK or MqnK (26). We identified a corresponding gene in S. aciditrophicus (SYN_01802), which was expressed in E. coli BL21(D3). The quinone profile of these cells showed an additional quinone that was identified as 8-MMK8, supporting the function of SYN_01802 as 8-MMK–forming methyltransferase (SI Appendix, Fig. S6).
Fig. 2.
Analysis of the quinone and EMO from S. aciditrophicus. (A) Ultra-performance liquid chromatography analysis of quinones with identical UV/vis spectra as 8-MKK (Inset). (B) Sodium dodecyl sulfate polyacrylamide gel electrophoresis of enrichment of EMO (the minor band below is most probably an EMO proteolysis product). M, mass standard; SE, size exclusion chromatography. (C) UV/vis spectra of solubilized EMO in the oxidized (as isolated) and dithionite-reduced form. (D) Redox titration of the heme b cofactors in the presence of redox dyes. Data points are fitted to two Nernst curves with n = 1 and midpoint potentials as indicated. (E) Reduction of solubilized EMO by CHCoA in the presence of ETF and CHCoA DH. Excess CHCoA reduced approximately one heme b equivalent. Light brown circles represent data points of differences in absorption 413 nm minus 427 nm and khaki diamonds of difference absorption 560 nm minus 580 nm.
The Membrane-Bound Diheme ETF:8-MMK Oxidoreductase Links Fatty Acid β-Oxidation to the 8-MMK Pool.
To unravel the function of DUF224, we solubilized and highly enriched a monomeric 75-kDa protein in the presence of detergents (Fig. 2B) that was identified as SYN_02638 by mass spectrometric analysis of tryptic peptides. The preparation showed a UV/vis spectrum typical for oxidized heme b (Fig. 2C). Metal cofactor analyses determined a 1.7-mol heme b and 12.4 ± 0.7 mol nonheme iron per mol protein, indicating the presence of two heme b and three [4Fe-4S] cofactors. Redox titration monitored by UV/vis spectroscopy resulted in a data set that readily fitted to two distinct Nernst curves (n = 1) with E°′ values of −81 and −220 mV. They are assigned to the one-electron redox transitions of a high- and a low-potential heme b (Fig. 2D). Solubilized SYN_02638 catalyzed the electron transfer from the soluble MMK analog 2,3,6-trimethylnaphthoquinol (TMNH2) to ETF (Table 1); we therefore refer to it as ETF:MMK oxidoreductase (EMO). In the presence of CHCoA DH and ETF, maximally half of the EMO heme b cofactors were reduced (Fig. 2E and Table 1). This finding fits to the redox potential of the CHeneCoA/CHCoA couple, which is sufficiently negative to reduce the high- but not the low-potential heme b. In the physiologically relevant forward direction, 50% of heme b cofactors of dithionite-reduced EMO were oxidized by equimolar 2,3,6-trimethylnaphthoquinone (TMN), suggesting that TMN accepts electrons only from the reduced low- but not from the reduced high-potential heme b. These results provide evidence that EMO is competent for electron transfer to/from 8-MMK/8-MMKH2 (low-potential heme b) and to/from ETFox/red (high-potential heme b). To study EMO function in a detergent-free environment, we prepared EMO-containing liposomes. Indeed, such liposomes catalyzed the CHCoA, CHCoA DH-, and ETF-dependent reduction of the high-potential heme b (SI Appendix, Fig. S7).
Bioinformatics analyses revealed a two-domain structure of EMO: 1) an N-terminal cytochrome b membrane domain belonging to the nitrate reductase γ-subunit (NarI) family with five transmembrane helices and 2) a C-terminal GlpC (anaerobic glycerol-3-phosphate DH subunit C) family domain with two typical Fd-like [4Fe-4S] cluster motifs and a cysteine-rich motif binding a further FeS cluster (SI Appendix, Fig. S8A). Using E. coli NarI as a template, we created a homology structure model of the membrane domain (SI Appendix, Fig. S9A) containing proximal and distal heme b binding sites. The latter site lacks one of the histidine ligands found in diheme-containing enzymes (27), and we propose that the resulting cavity serves as an 8-MMK binding site near the periplasm (SI Appendix, Fig. 9B). Intriguingly, EMO and mFDH (modeled accordingly) show complementary surface charges that suggest a possible interaction between EMO and mFDH (SI Appendix, Fig. 9C).
Biochemical Evidence-Based Model for Syntrophic CO2 Reduction Coupled to Fatty Acid Oxidation.
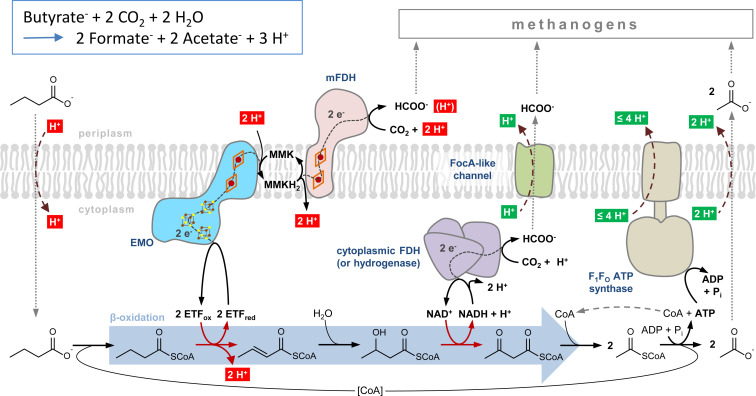
The identification of EMO as a key component in the endergonic electron transfer from acyl-CoA to CO2 closes the gap of knowledge in the conversion of biomass into methane. The syntrophic conversion of fatty acids to methanogenic substrates is often referred to as “life at thermodynamic equilibrium,” and syntrophic cultures have been frequently used to define the minimal free energy increment to sustain life (12, 28). In this context, the redox properties of EMO and the membranous electron carrier MMK act as a perfectly fine-tuned redox system. First, the replacement of MK by MMK divides the redox gap between acyl-CoA and CO2 into two energetically equal two-electron transfer reactions (∼27 kJ ⋅ mol–1). Second, ΔE between the high- and low-potential heme b differ by 140 mV, perfectly fitting to the anticipated membrane potential.
The functional characterization of EMO now provides a solid basis for a model of syntrophic CO2 reduction coupled to scFA oxidation (Fig. 3). Endergonic electron transfer from acyl-CoA to CO2 results in the net translocation of 4 H+ into the cytoplasm. Though transport was not studied here, we assume that the uptake of scFA growth substrates and export of the acetate end product will cotransport protons, respectively. Consumption of external acetate by methanogenic partners might facilitate diffusion-driven acetate export. The intracellularly formed formate is likely cotransported with a proton by a FocA-like formate channel (SYN_1200) (29). It is interesting to note that the SYN_1200 gene product shows high amino acid sequence identities (>80%) to FocA-like proteins from other syntrophs but also to those from methanogens (here referred to as FdhC) (30). Only one of the two acetyl-CoAs is available for ATP generation. The F0F1-ATP synthase from S. aciditrophicus may translocate three to four H+ per ATP across the cytoplasmic membrane. Consequently, the entire system allows for the net translocation of one to two H+ to the periplasm per butyrate oxidized. Assuming ΔG ∼−20 kJ ⋅ mol–1 per proton translocated, this value fits well to the theoretically calculated ΔG of −22 kJ ⋅ mol–1 for the conversion of butyrate to two acetates under syntrophic conditions (3).
Fig. 3.
Model of syntrophic scFA oxidation coupled to CO2 reduction in S. aciditrophicus. The model scFA butyrate is oxidized to two acetates coupled to the reduction of two CO2 to formate. Alternatively, the NADH formed by 3-hydroxybutyryl-CoA DH is regenerated by a soluble hydrogenase, and the H2 formed diffuses across the membrane without the involvement of a transporter. Protons that contribute to proton motive force (pmf) generation are shown in green; those that diminish pmf are shown in red. Note that transport of protons during uptake and excretion is based on assumptions and has not been studied in this work.
EMO Is a Key Enzyme in (M)MK-Containing Prokaryotes Competent for β-Oxidation of Lipids.
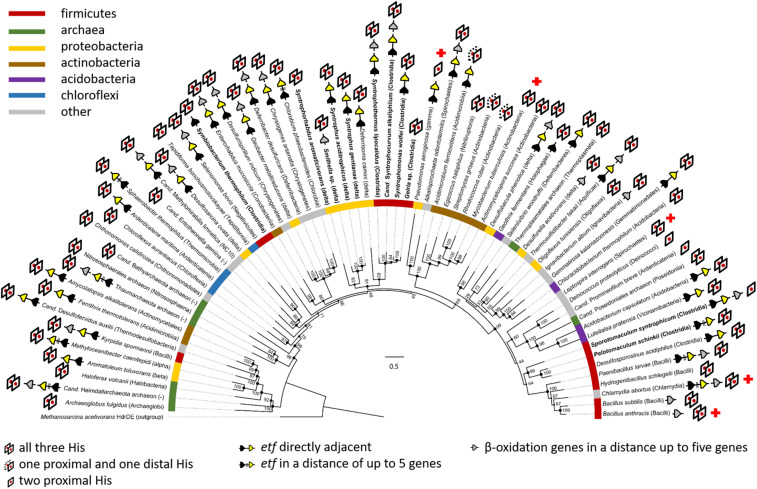
Endergonic electron transfer from acyl-CoA to (M)MK pools is a thermodynamic problem for all (M)MK-containing organisms growing with lipids. As the majority of prokaryotes contain (M)MK (31), we analyzed the occurrence of EMO-like proteins with the following criteria for reliable BLASTp (basic logical alignment search tool for proteins) application: 1) an E-value below 3e–60, 2) the presence of a NarI-like domain with five transmembrane helices and at least two conserved histidines involved in heme b binding, and 3) the presence of a GlpC-like domain. The analysis revealed that emo-like genes are highly abundant in the majority of bacterial and archaeal lineages (Fig. 4) and are often located in close vicinity to etfAB and genes encoding β-oxidation enzymes, supporting the general role of EMO in lipid catabolism. Mycobacterium tuberculosis, the causative agent of tuberculosis, contains MK as only quinone (32), and a single version of a putative emo gene was identified in the genome (Rv0338c, E-value 2e–151; SI Appendix, Fig. S8B). It is well established that the persistence of M. tuberculosis depends on host-derived fatty acids and cholesterol (33). Consequently, a wealth of studies have been carried out to identify inhibitors of lipid degradation (34) and MK biosynthesis (35) in M. tuberculosis. In light of this, we propose that the single copy of emo in M. tuberculosis is essential for pathogenicity, and its product serves as a previously overseen potential target for antimycobacterial drug development.
Fig. 4.
Phylogenetic analysis of emo genes in prokaryotes. EMO from S. aciditrophicus served as query sequence during BLASTp analysis. The presence of conserved His ligands of heme b and the genomic context with regard to etf and β-oxidation genes are indicated. Syntrophic bacteria are highlighted by bold typeface. The numbers refer to bootstrap values; the scale bar indicates the number of substitution per site. Red crosses mark selected pathogenic bacteria.
Materials and Methods
Detailed information on sources and detailed protocols for experimental procedures can be found in the SI Appendix. These include media and conditions for small- and large-scale cultivation of cells as well as the heterologous expression of genes and the enrichment of the gene products by chromatographic methods. Furthermore, it contains detailed descriptions of activity assays, quinone- and flavin-cofactor analyses, redox potential determinations, proton motif force determinations, and proteoliposome assays. The devices, programs, and protocols used for liquid chromatography–mass spectrometry analyses of proteins and quinone cofactors as well as for bioinformatics analyses are described.
Supplementary Material
Acknowledgments
We are grateful to Ivan Berg (Münster) and Thorsten Friedrich (Freiburg) for technical and editorial advice. This work was supported by the Deutsche Forschungsgemeinschaft (German Research Foundation) within CRC 1381, project identification #403222702, to M.B., S.V.A., and H.G.K.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.J.M. is a guest editor invited by the Editorial Board.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111682118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Evans P. N., et al., An evolving view of methane metabolism in the Archaea. Nat. Rev. Microbiol. 17, 219–232 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Thauer R. K., Kaster A. K., Seedorf H., Buckel W., Hedderich R., Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Stams A. J. M., Plugge C. M., Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Microbiol. 7, 568–577 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Sieber J. R., Le H. M., McInerney M. J., The importance of hydrogen and formate transfer for syntrophic fatty, aromatic and alicyclic metabolism. Environ. Microbiol. 16, 177–188 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Schink B., Montag D., Keller A., Müller N., Hydrogen or formate: Alternative key players in methanogenic degradation. Environ. Microbiol. Rep. 9, 189–202 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Walker D. J. F., et al., Syntrophus conductive pili demonstrate that common hydrogen-donating syntrophs can have a direct electron transfer option. ISME J. 14, 837–846 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sieber J. R., McInerney M. J., Gunsalus R. P., Genomic insights into syntrophy: The paradigm for anaerobic metabolic cooperation. Annu. Rev. Microbiol. 66, 429–452 (2012). [DOI] [PubMed] [Google Scholar]
- 8.McInerney M. J., Wofford N. Q., Enzymes involved in crotonate metabolism in Syntrophomonas woifei. Arch. Microbiol. 158, 344–349 (1992). [Google Scholar]
- 9.Wofford N. Q., Beaty P. S., McInerney M. J., Preparation of cell-free extracts and the enzymes involved in fatty acid metabolism in Syntrophomonas wolfei. J. Bacteriol. 167, 179–185 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stankovich M. T., Soltysik S., Regulation of the butyryl-CoA dehydrogenase by substrate and product binding. Biochemistry 26, 2627–2632 (1987). [DOI] [PubMed] [Google Scholar]
- 11.Gustafson W. G., Feinberg B. A., McFarland J. T., Energetics of beta-oxidation. Reduction potentials of general fatty acyl-CoA dehydrogenase, electron transfer flavoprotein, and fatty acyl-CoA substrates. J. Biol. Chem. 261, 7733–7741 (1986). [PubMed] [Google Scholar]
- 12.McInerney M. J., et al., The genome of Syntrophus aciditrophicus: Life at the thermodynamic limit of microbial growth. Proc. Natl. Acad. Sci. U.S.A. 104, 7600–7605 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sieber J. R., et al., The genome of Syntrophomonas wolfei: New insights into syntrophic metabolism and biohydrogen production. Environ. Microbiol. 12, 2289–2301 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Schmidt A., Müller N., Schink B., Schleheck D., A proteomic view at the biochemistry of syntrophic butyrate oxidation in Syntrophomonas wolfei. PLoS One 8, e56905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sieber J. R., et al., Proteomic analysis reveals metabolic and regulatory systems involved in the syntrophic and axenic lifestyle of Syntrophomonas wolfei. Front. Microbiol. 6, 115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crable B. R., et al., Membrane complexes of Syntrophomonas wolfei involved in syntrophic butyrate degradation and hydrogen formation. Front. Microbiol. 7, 1795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallrabenstein C., Schink B., Evidence of reversed electron transport in syntrophic butyrate or benzoate oxidation by Syntrophomonas wolfei and Syntrophus buswellii. Arch. Microbiol. 162, 136–142 (1994). [Google Scholar]
- 18.Boll M., Kung J. W., Ermler U., Martins B. M., Buckel W., Fermentative cyclohexane carboxylate formation in Syntrophus aciditrophicus. J. Mol. Microbiol. Biotechnol. 26, 165–179 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Mouttaki H., Nanny M. A., McInerney M. J., Use of benzoate as an electron acceptor by Syntrophus aciditrophicus grown in pure culture with crotonate. Environ. Microbiol. 10, 3265–3274 (2008). [DOI] [PubMed] [Google Scholar]
- 20.James K. L., et al., Syntrophus aciditrophicus uses the same enzymes in a reversible manner to degrade and synthesize aromatic and alicyclic acids. Environ. Microbiol. 21, 1833–1846 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kung J. W., Seifert J., von Bergen M., Boll M., Cyclohexanecarboxyl-coenzyme A (CoA) and cyclohex-1-ene-1-carboxyl-CoA dehydrogenases, two enzymes involved in the fermentation of benzoate and crotonate in Syntrophus aciditrophicus. J. Bacteriol. 195, 3193–3200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buckel W., Thauer R. K., Flavin-based electron bifurcation, a new mechanism of biological energy coupling. Chem. Rev. 118, 3862–3886 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Enoch H. G., Lester R. L., The role of a novel cytochrome b-containing nitrate reductase and quinone in the in vitro reconstruction of formate-nitrate reductase activity of E. coli. Biochem. Biophys. Res. Commun. 61, 1234–1241 (1974). [DOI] [PubMed] [Google Scholar]
- 24.Smirnova I. A., HOQNO interaction with cytochrome b in succinate:menaquinone oxidoreductase from Bacillus subtilis. Anal. Biochem. 217, 220–230 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Hein S., von Irmer J., Gallei M., Meusinger R., Simon J., Two dedicated class C radical S-adenosylmethionine methyltransferases concertedly catalyse the synthesis of 7,8-dimethylmenaquinone. Biochim. Biophys. Acta Bioenerg. 1859, 300–308 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Hein S., Klimmek O., Polly M., Kern M., Simon J., A class C radical S-adenosylmethionine methyltransferase synthesizes 8-methylmenaquinone. Mol. Microbiol. 104, 449–462 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Bertero M. G., et al., Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat. Struct. Biol. 10, 681–687 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Jackson B. E., McInerney M. J., Anaerobic microbial metabolism can proceed close to thermodynamic limits. Nature 415, 454–456 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Suppmann B., Sawers G., Isolation and characterization of hypophosphite–Resistant mutants of Escherichia coli: Identification of the FocA protein, encoded by the pfl operon, as a putative formate transporter. Mol. Microbiol. 11, 965–982 (1994). [DOI] [PubMed] [Google Scholar]
- 30.White W. B., Ferry J. G., Identification of formate dehydrogenase-specific mRNA species and nucleotide sequence of the fdhC gene of Methanobacterium formicicum. J. Bacteriol. 174, 4997–5004 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoepp-Cothenet B., et al., Menaquinone as pool quinone in a purple bacterium. Proc. Natl. Acad. Sci. U.S.A. 106, 8549–8554 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yassin A. F., Brzezinka H., Schaal K. P., Trüper H. G., Pulverer G., Menaquinone composition in the classification and identification of aerobic actinomycetes. Zentralblatt für Bakteriol. Mikrobiol. und Hyg. - Abt. 1 Orig. A 267, 339–356 (1988). [DOI] [PubMed] [Google Scholar]
- 33.Wilburn K. M., Fieweger R. A., VanderVen B. C., Cholesterol and fatty acids grease the wheels of Mycobacterium tuberculosis pathogenesis. Pathog. Dis. 76, fty021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baran M., et al., Development of small-molecule inhibitors of fatty acyl-AMP and fatty acyl-CoA ligases in Mycobacterium tuberculosis. Eur. J. Med. Chem. 201, 112408 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Debnath J., et al., Discovery of selective menaquinone biosynthesis inhibitors against Mycobacterium tuberculosis. J. Med. Chem. 55, 3739–3755 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.