Significance
Methionine metabolism is dramatically affected by age and restricting methionine consumption extends the lifespan across different species. However, we have limited information on the activity of different branches of methionine metabolism during aging. Furthermore, how tissue-specific perturbation of methionine metabolism affects aging is not known. Here, by feeding 13C5-methionine to Drosophila and tracing its fate in different branches of the methionine metabolism, we were able to identify impairments of methionine flux associated with aging. In addition, to deplete methionine in a tissue-specific manner, we created transgenic flies that express Methioninase. Strikingly, Methioninase expression even in a single tissue can dramatically extend the health- and lifespan.
Keywords: methionine restriction, Methioninase, aging, 13C-Methionine labeling, Alzheimer’s disease
Abstract
Loss of metabolic homeostasis is a hallmark of aging and is characterized by dramatic metabolic reprogramming. To analyze how the fate of labeled methionine is altered during aging, we applied 13C5-Methionine labeling to Drosophila and demonstrated significant changes in the activity of different branches of the methionine metabolism as flies age. We further tested whether targeted degradation of methionine metabolism components would “reset” methionine metabolism flux and extend the fly lifespan. Specifically, we created transgenic flies with inducible expression of Methioninase, a bacterial enzyme capable of degrading methionine and revealed methionine requirements for normal maintenance of lifespan. We also demonstrated that microbiota-derived methionine is an alternative and important source in addition to food-derived methionine. In this genetic model of methionine restriction (MetR), we also demonstrate that either whole-body or tissue-specific Methioninase expression can dramatically extend Drosophila health- and lifespan and exerts physiological effects associated with MetR. Interestingly, while previous dietary MetR extended lifespan in flies only in low amino acid conditions, MetR from Methioninase expression extends lifespan independently of amino acid levels in the food. Finally, because impairment of the methionine metabolism has been previously associated with the development of Alzheimer’s disease, we compared methionine metabolism reprogramming between aging flies and a Drosophila model relevant to Alzheimer’s disease, and found that overexpression of human Tau caused methionine metabolism flux reprogramming similar to the changes found in aged flies. Altogether, our study highlights Methioninase as a potential agent for health- and lifespan extension.
Aging is the primary risk factor for many major human pathologies (1). Age-dependent metabolic reprogramming has been noted in different organisms (2), including worms (3), mice (4), and humans (5–7). Moreover, we have previously demonstrated that metabolism in general, and methionine metabolism in particular, is perturbed during aging in Drosophila (8). In addition, methionine metabolism is altered in the tissues of several long-lived species, such as naked mole rats (9), in long-lived mutants, such as flies selected for delayed reproductive senescence (8, 10), and in long-lived Ames mice (11).
Although alterations in levels of specific metabolites suggest that the activity of the methionine metabolism pathway is affected, it is difficult to determine if the flux via methionine metabolism is up- or down-regulated. For example, deficiencies of enzymes involved in methionine metabolism (MAT, CBS, GNMT, AHCY) lead to methionine and homocysteine elevation (hypermethioninemias and hyperhomocysteinemias), and elevated levels of methionine and homocysteine reflect a disruption of flux in methionine metabolism (12). Additionally, metabolite changes do not explain how the flux is reprogrammed between different branches of methionine metabolism, as it is possible that the absolute levels of metabolites do not change if metabolites get processed via different routes.
Impairment of methionine metabolism flux results in the accumulation of detrimental metabolites belonging to the methionine cycle, such as S-adenosylhomocysteine (SAH) and homocysteine, and leads to different pathological manifestations (13). Lifespan can be extended by up-regulating the clearance of these metabolites via activation of methionine metabolism flux. In fact, methionine restriction (MetR) extends lifespan in yeast, flies, rodents, and human diploid fibroblasts (14–17) and exerts beneficial effects on metabolic health and inflammatory responses (18–20). Moreover, lifespan in worms and flies can be extended through manipulation of different enzymes, either belonging to the methionine metabolism pathway or those that affect the levels of methionine metabolism metabolites. Some of these manipulations include the overexpression of Cbs (21) and Gnmt (22, 23), and the down-regulation of SamS (24) and AhcyL1/AhcyL2 (8). In addition, manipulation of methionine metabolism in just one tissue is sufficient for lifespan extension (8, 21, 22).
Restoring age-reprogrammed activity of methionine metabolism can be an attractive option for the extension of health- and lifespan. However, studies relating to MetR have the following challenges: 1) MetR does not decrease levels of methionine equally across different organs; 2) it is impossible to study tissue-specific effects of MetR via manipulations of methionine levels in the food; and 3) the activation of methionine metabolism enzymes is difficult to achieve due to a lack of small-molecule activators. To solve these problems, we created transgenic flies carrying the enzyme Methioninase, which allows for the rapid, inducible, and tissue-specific degradation of methionine.
l-Methionine α-deamino-γ-mercaptomethane-lyase (Methioninase) is a bacterial enzyme that is capable of degrading methionine to ammonia, α-ketobutyrate, and methanthiol. Because methionine dependency was attributed to various cancers, recombinant Methioninase (rMetase) has been tested in various cancer models in vitro and in vivo (25, 26). Methioninase has also entered several clinical trials in humans (26–28). Based on this, Methioninase may provide an alternative option to dietary MetR, since it can be expressed in a tissue-specific manner, and its recombinant form can be used in humans.
To understand how methionine metabolism is reprogrammed with age or neurodegeneration, we supplemented flies with a labeled 13C5-methionine tracer and estimated its fate between different branches of methionine metabolism. We further created transgenic flies with inducible expression of Methioninase (genetic MetR) and performed metabolomics profiling to demonstrate that the effect was similar to the effects of MetR caused by the depletion of methionine from fly food (dietary MetR). It has been previously shown that dietary MetR extends the lifespan in flies only in low amino acid conditions (15). We demonstrate that either whole-body or tissue-specific expression of Methioninase (genetic MetR) can extend Drosophila lifespan without lowering levels of amino acids in the food. Altogether, our studies offer a strategy for restoring age-dependent defects related to impaired methionine metabolism that has strong potential for lifespan extension and treatment of neurodegenerative diseases in humans.
Results
Age-Dependent Reprogramming of Methionine Metabolism Flux.
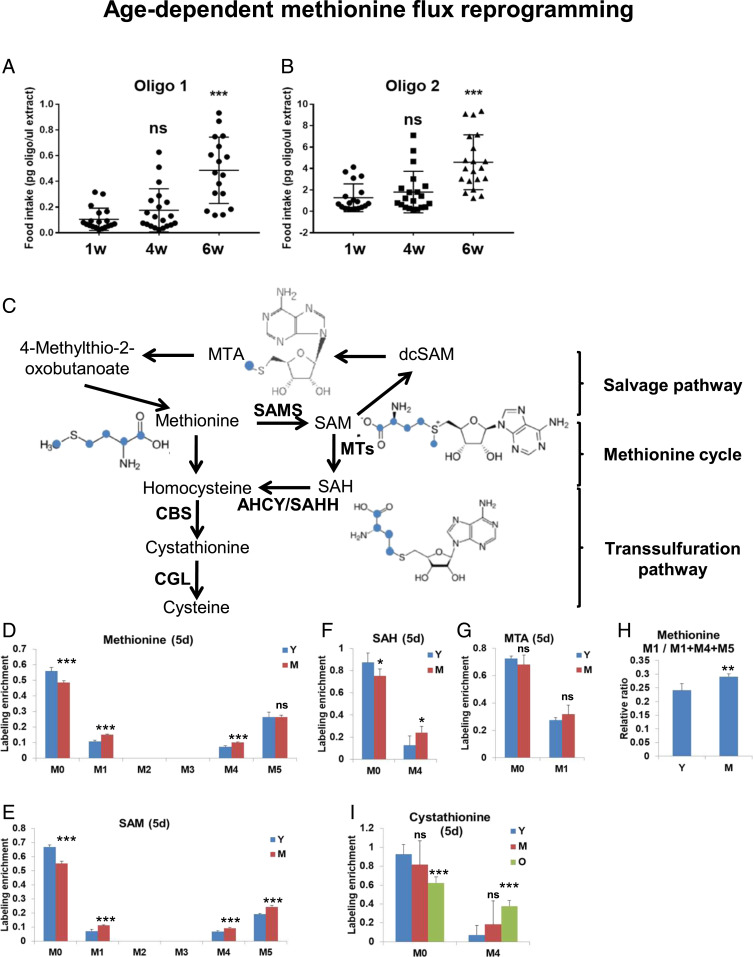
We previously demonstrated that steady-state levels of several metabolites in the methionine metabolism pathway are altered with age in Drosophila (8). In agreement with this, increased levels of plasma homocysteine (one of the metabolites in the methionine cycle) have been recognized as a risk factor for a number of human diseases (29). Although alterations in levels of particular metabolites may suggest altered pathway activity, increased levels of a specific metabolite can be attributed to either an increase or decrease in flux through a metabolic pathway. 13C-metabolic flux analysis is a common approach used to determine cellular metabolic flux (30). It is based on feeding cells/organisms with carbon isotope-labeled nutrients and measuring the labeling patterns of intracellular metabolites (31–33). To estimate how age affects the distribution of methionine via different branches of the methionine metabolism pathway, we implemented a method to feed flies with stable isotope-labeled methionine. We then coupled the approach with mass spectrometry analysis to determine the relative activities of different branches of methionine metabolism. We first fed young male (1 wk) wild-type OreR flies with 1 mM of labeled 13C5-methionine tracer in chemically defined food (lacking endogenous methionine) that has been previously used for lifespan studies related to MetR (15) for different periods of time (3 h, 12 h, 2 d, 5 d, and 10 d) to optimize the best timing for labeling (SI Appendix, Fig. S1). Because alterations in food uptake may significantly affect the interpretation of results, we compared food uptake in young (1 wk), middle (4 wk), and old (7 wk) age male flies using two different oligonucleotides and quantitating their uptake by qPCR, as it has been previously described (34).
Although we did not observe differences between young and middle age flies, we found dramatic differences in older flies (Fig. 1 A and B). Because food consumption and processing are dramatically different at this stage, and because flies actively die at this age, we decided to evaluate differences in the fate of labeled methionine in young and middle-age flies. We supplemented young (1 wk) and middle-age (4 wk) male flies with labeled 13C5-methionine for 1, 2, and 5 d and analyzed the distribution of labeled methionine to methionine metabolism metabolites by mass spectrometry (SI Appendix, Fig. S2). Methionine metabolism can be broken into three parts: the methionine cycle, the transsulfuration pathway, and the salvage cycle (Fig. 1C). Labeled methionine enters the methionine cycle from chemically defined food (labeled with 5 carbons, M5). Methionine is first converted to S-adenosyl-l-methionine (SAM) (M5), and after SAM donates its methyl group to acceptor molecules, it is converted to SAH (M4). SAH is further converted to homocysteine, and homocysteine can be either remethylated to form methionine (M4) to be retained in the methylation cycle, or it can be irreversibly converted to cysteine via the transsulfuration pathway and thus withdrawn from the methylation cycle (Fig. 1C). Methionine (M1) can also be regenerated from SAM via the methionine salvage pathway.
Fig. 1.
Age-dependent changes of the fate of labeled methionine. (A and B) Food consumption in 1-, 4-, and 6-wk-old male flies measured by qPCR using two different oligonucleotides. Twenty biological replicates per age. (C) Scheme of methionine metabolism. Blue color marks the labeled carbons. Relative labeling enrichment for methionine (D), SAM (E), SAH (F), and MTA (G) in whole male flies fed with 1 mM of labeled 13C5-methionine tracer in chemically defined food (lacking endogenous methionine) for 5 d. M0–M12 mark the number of labeled carbons. Labeling enrichment is the proportion of a particular labeled metabolite form among all measured isoforms. Note that although multiple isoforms of the same metabolite with different numbers of labeled carbons were measured, many of them are not present in Drosophila cells, and they are not displayed on the graph. Five biological replicates per condition. Means ± SD. Y and M symbols mark young (1 wk) and medium-age (4 wk) male flies *P < 0.05, **P < 0.01, ***P < 0.001. (H) The enrichment ratio (M1/M4+M5+M1) between methionine (M1) produced from the salvage pathway and total labeled methionine (M4+M5+M1) reflects the activity of the salvage (MTA cycle) pathway. Means ± SD; **P < 0.01. (I) Relative labeling enrichment for cystathionine. Means ± SD, ***P < 0.001.
Consistent with the observation that there were no differences in food uptake between 1- and 4-wk-old flies, we did not observe any differences in the amount of food-derived methionine (M5) (Fig. 1D). Interestingly, we found increased levels of SAM (M5) (Fig. 1E) and SAH (M4) (Fig. 1F), both of which are produced from food-derived methionine (M5), potentially indicating a decreased ability to process SAH via the methionine cycle. Although there was no difference in the levels of MTA (M1), an intermediate metabolite in the salvage cycle (Fig. 1G), we used the enrichment ratio (M1/M4+M5+M1) between methionine (M1) produced from the salvage pathway and total labeled methionine as an estimate of the activity via the salvage pathway. The methionine salvage pathway, or 5′-methylthioadenosine (MTA) cycle, regenerates methionine from SAM and is responsible for the production of polyamines (35, 36). The proportion of methionine (M1) produced from the salvage pathway increased in 4-wk-old flies (Fig. 1H), potentially reflecting a higher need for polyamine production. We did not observe a significant difference in the labeling of cystathionine (M4) between 1- and 4-wk-old flies (Fig. 1I). Interestingly, we observed a greater accumulation of food-derived methionine (M5) and its products, SAM (M5) and SAH (M4), in 7-wk-old flies compared to young flies, as expected according to the increased food consumption (SI Appendix, Fig. S2). Similarly, there was an increased level of labeled cystathionine (M4) in 7-wk-old flies (Fig. 1I), suggesting increased transsulfuration flux in old flies. This may be triggered either by increased requirements for the degradation of the excess of methionine and the necessity to withdraw methionine from the methionine cycle or by increased requirements for the production of cysteine-derived antioxidants (glutathione and taurine). In conclusion, our data suggest the age-dependent alterations in the activity of different branches of the methionine metabolism.
Methioninase Is a Genetic Tool for MetR.
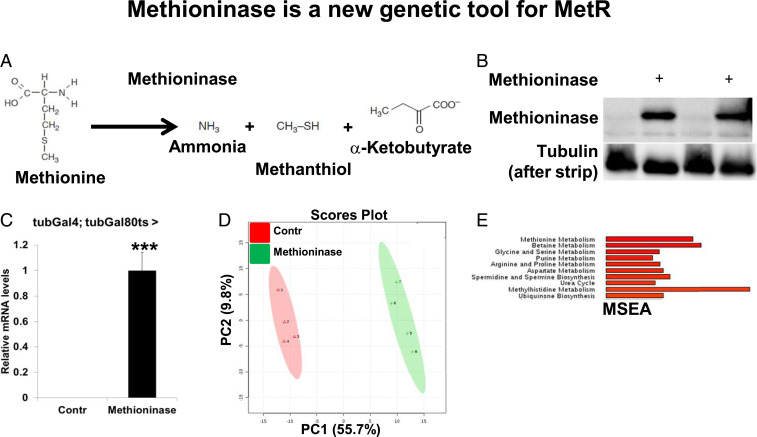
We hypothesized that degrading methionine and removing downstream metabolites ubiquitously or in a tissue-specific manner would “reset” methionine metabolism by removing any inhibitory feedback regulation from the abnormal accumulation of intermediary metabolites and can extend lifespan. Although dietary MetR can be used to deplete methionine and its downstream metabolites, it causes tissue-specific effects on the levels of methionine in different organs and cannot be applied at a tissue-specific level. To create a system for efficient methionine depletion without altering food composition and to study tissue-specific effects of MetR, we employed a bacterial enzyme, Methioninase, which degrades methionine to ammonia, α-ketobutyrate, and methanthiol (Fig. 2A). We cloned the Methioninase gene from Pseudomonas putida into the pW10roe Drosophila expression vector and created transgenic flies. To confirm the expression of Methioninase in flies, we ubiquitously expressed Methioninase in adults using the tubulin-Gal4,tubulin-Gal80ts temperature-inducible system (37). Gal80ts is active at 18 °C and represses Gal4, while at 29 °C, Gal80ts is inactivated, allowing for the Gal4-dependent expression of Methioninase. Flies were grown at 18 °C and switched to 29 °C after eclosion to induce expression of Methioninase for 10 d. As a control, we used the same line that we had injected with the UAS-Methioninase construct to produce transgenic UAS-Methioninase flies. We confirmed expression of Methioninase in flies using both immunoblot analysis (Fig. 2B) and qRT-PCR (Fig. 2C). To test the functional consequences of Methioninase expression, we performed metabolomic profiling of either control flies or flies that expressed Methioninase for 10 d (SI Appendix, Fig. S3A). Principal component analysis (PCA) of the measured metabolites clearly distinguished between control flies and flies expressing Methioninase (Fig. 2D). We identified 123 metabolites that were significantly changed in flies expressing Methioninase. Among these metabolites were multiple metabolites belonging to methionine metabolism: methionine, SAH, SAM, MTA, methionine sulfoxide, cysteine, and cystathionine (SI Appendix, Fig. S3A).
Fig. 2.
Methioninase is a genetic tool for MetR. (A) Scheme of methionine degradation by Methioninase. (B) Immunoblot analysis of Methioninase and tubulin in tubulin-Gal80ts,tubulin-Gal4 flies > control flies or flies expressing Methioninase (in duplicates). (C) Relative mRNA levels of Methioninase in tubulin-Gal80ts,tubulin-Gal4 > control flies or flies expressing Methioninase. Four biological replicates per condition. Means ± SD, ***P < 0.001. (D) PCA of tubulin-Gal80ts,tubulin-Gal4 > control flies, or flies expressing Methioninase. (E) MSEA of the metabolites that changed significantly in either tubulin-Gal80ts,tubulin-Gal4 > control flies or flies expressing Methioninase.
In an effort to understand which metabolic pathways are among the most significantly changed by Methioninase expression, we applied metabolite set enrichment analysis (MSEA). As expected, among the top five most significantly altered metabolic pathways were methionine metabolism, betaine metabolism, glycine and serine metabolism, and purine metabolism (Fig. 2E). Betaine metabolism, glycine and serine metabolism, and purine metabolism are tightly linked to methionine metabolism. In the methionine cycle, SAM is used for methylation reactions and generates SAH, which is further hydrolyzed into homocysteine. Homocysteine can be remethylated back to methionine using betaine (betaine metabolism pathway) or 5-methyl-THF as methyl donors and can thus be retained in the methionine cycle (Fig. 1C). Glycine and serine (glycine and serine metabolism pathway) serve as precursors for one-carbon metabolism, which is intimately linked with purine biosynthesis (purine metabolism pathway) and serves as an additional source of a methyl group (5-methyl-THF) for the methionine cycle. Altogether, our results suggest that Methioninase is an efficient tool for in vivo methionine depletion.
Comparison of Methioninase (Genetic MetR) and Food-Deprived MetR.
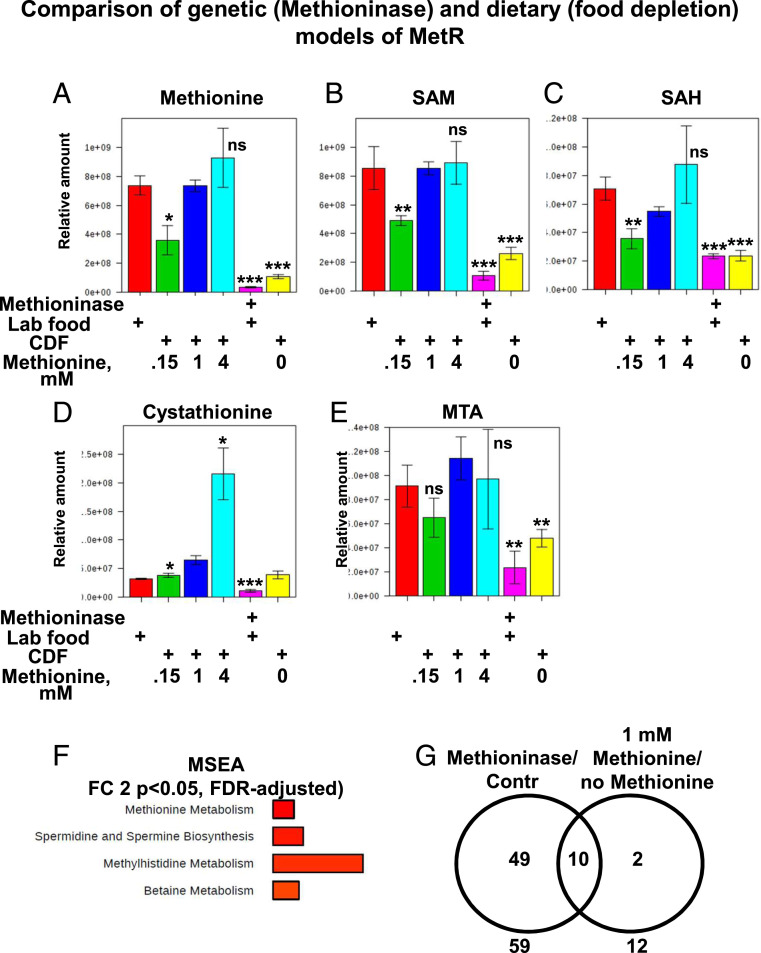
Several groups have developed semi- or fully defined fly diets for testing the effects of specific food components on lifespan. The basal level of methionine significantly varies in these diets, ranging from 0.34 to 1.65 g/L (38). To compare the effects of Methioninase expression to MetR caused by methionine deprivation from fly food, we performed a metabolomics analysis of flies ubiquitously expressing Methioninase and maintained on standard laboratory food (same as Fig. 2E) and control flies maintained either on laboratory food or on chemically defined food supplemented with a range of concentrations of methionine (no methionine, 0.15 mM, 1 mM, or 4 mM). Flies maintained on chemically defined food supplemented with 1 mM of methionine had methionine pathway metabolite levels similar to those of control flies maintained on standard laboratory food (Fig. 3 A–E). Increasing the concentration of methionine from 1 mM to 4 mM led to dramatically increased levels of cystathionine, an intermediary metabolite in the transsulfuration pathway that irreversibly removes methionine from the methionine cycle toward the production of cysteine (Fig. 3 A–E). Decreasing the concentration of methionine from 1 mM to 0.15 mM or no methionine led to a concentration-dependent decrease in methionine, SAM, SAH, and MTA, and the effects were very similar to those observed during the expression of Methioninase (Fig. 3 A–E). In addition, the expression of Methioninase decreased levels of cystathionine, implying that there was a highly decreased flux of methionine into the transsulfuration pathway, while complete depletion of methionine from the food did not affect cystathionine levels (Fig. 3D). These differences can be explained by the fact that enzymatic depletion of methionine is much faster and degrades both food-derived methionine and methionine derived from internal protein degradation, whereas upon depletion of methionine from food, cells still have accumulated methionine in the body and it only eliminates food-derived methionine.
Fig. 3.
Comparison of genetic (Methioninase) and dietary (food depletion) models of MetR. Box plots of relative levels of methionine (A), SAM (B), SAH (C), Cystathionine (D), MTA (E) in tubulin-Gal4,tubulin-Gal80ts > control flies or flies expressing Methioninase maintained on either standard laboratory food (marked as Lab food) or on the chemically defined food (CDF) supplemented with a range of concentrations of methionine (no methionine, 0.15 mM, 1 mM, or 4 mM). Four biological replicates per condition. Means ± SD, *P < 0.05, **P < 0.01, ***P < 0.001. (F) MSEA of the metabolites that changed significantly in tubulin-Gal4,tubulin-Gal80ts > control flies maintained on chemically defined food supplemented with either no methionine or 1 mM methionine. (G) Overlap of metabolites with more than a twofold change in the two models of methionine deprivation: 1) tubulin-Gal80ts tubulin-Gal4 flies > control flies (marked as Contr) or flies expressing Methioninase and 2) tubulin-Gal4,tubulin-Gal80ts > control flies maintained on chemically defined food supplemented with either no methionine (marked as no Met) or 1 mM methionine (marked as 1 mM Met).
We next applied MSEA to metabolites that were significantly changed between flies fed with 1 mM methionine (analogous to control flies) and those fed with no methionine in the food (analogous to Methioninase expression). Similar to Methioninase expression, methionine metabolism and betaine metabolism were among the most significantly changed metabolic pathways (Fig. 3F). Among 12 metabolites significantly changed by complete depletion of methionine from food, 10 were in common with the set of metabolites changed by Methioninase expression (Fig. 3G). It should be noted that the metabolomes of control flies maintained on standard laboratory food and control flies maintained on chemically defined food were dramatically different (SI Appendix, Fig. S3 B and C), which could explain the differences in basal lifespan, developmental timing, and other phenotypic traits. Altogether, we demonstrated that the genetic model of MetR that we created is biochemically similar to dietary MetR.
Methionine Requirements for Normal Maintenance of Lifespan.
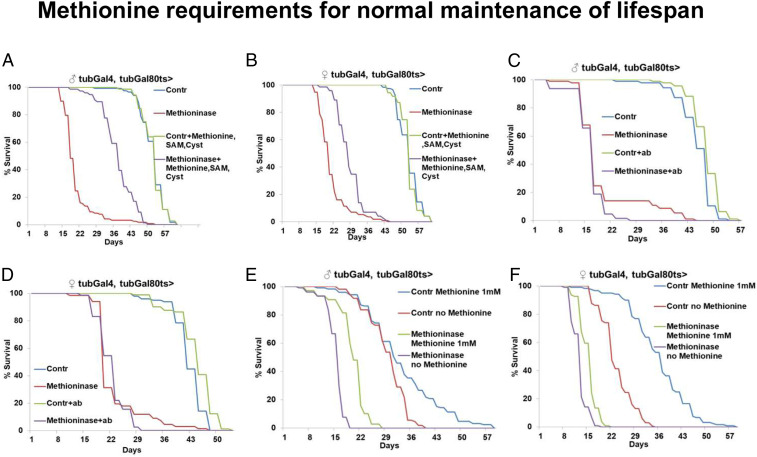
Since we established a genetic system for efficient methionine degradation, we further tested how long flies could survive without methionine. Specifically, we compared the lifespans of control flies and flies that ubiquitously expressed Methioninase at 29 °C. Methioninase expression dramatically decreased lifespan in both males (mean lifespan for controls, 51 d, and for Methioninase, 20 d; P < 0.0001, log-rank test) and females (mean lifespan for controls, 52 d, and for Methioninase, 20 d; P < 0.0001, log-rank test) (Fig. 4 A and B). Because Methioninase almost completely depleted levels of methionine, SAM, SAH, and MTA (Fig. 3 A–C and E), we tested whether supplementing the food with different intermediates from the methionine metabolism pathway could at least partially rescue the lifespan. We supplemented adult flies with one or a combination of metabolites belonging to the methionine metabolism in concentrations ranging from 1 to 3 mM. Interestingly, combinations of SAM, MTA, and cysteine or SAM, methionine, and cysteine effectively rescued the lifespan of flies expressing Methioninase (Fig. 4 A and B). In males, a combination of SAM, methionine, and cysteine (3 mM each) rescued mean lifespan from 20 d to 37 d, while it did not affect the lifespan of control flies (both groups had a mean lifespan of 51 d). In females, the combination of SAM, methionine, and cysteine (3 mM each) had much weaker effects, rescuing mean lifespan from 20 d to 27 d; it also did not affect the lifespan of control flies (both groups had a mean lifespan of 52 d). The incomplete rescue of lifespan in flies exposed to a mix of metabolites may be linked to factors such as the inefficient spread of metabolites to all the cells in the fly body or an insufficient amount of methionine for protein synthesis. Although Methioninase products are volatile substances and it is believed that they are easily excreted upon methionine degradation, we cannot exclude the possibility that they underlie different effects associated with Methioninase expression. We also noticed that although Methioninase expression abruptly decreased lifespan, a small fraction of flies survived much longer than others. Because methionine can also be obtained from gastrointestinal microbes in addition to food, we hypothesized that microbiota-derived methionine could promote survival under Methioninase expression. To test this, we treated control and ubiquitously expressed Methioninase flies with a mix of antibiotics (chloramphenicol, kanamycin, and rifamycin). Interestingly, while the antibiotic mix did not suppress the survival of the main group of flies expressing Methioninase or the survival of control flies, it completely abrogated the fraction of “super survivors” under Methioninase expression (Fig. 4 C and D).
Fig. 4.
Methionine requirements for normal maintenance of lifespan. Ubiquitous adult-onset expression of Methioninase suppresses lifespan in both male (A) and female (B) flies, which can be rescued by supplementation with a mixture of 3 mM of methionine, SAM, and cysteine. In males, mean lifespan for controls 51 d, and for Methioninase 20 d; P < 0.0001, log-rank test. In females, mean lifespan for controls 52 d, and for Methioninase 20 d; P < 0.0001, log-rank test. Survival of super survivor flies under ubiquitous adult-onset expression of Methioninase can be suppressed by treatment with a mixture of antibiotics in both male (C) and female (D) flies. Survival of male (E) and female (F) flies with ubiquitous adult-onset expression of control or Methioninase maintained on chemically defined food with either 1 mM methionine or with no methionine.
These data suggest that under certain circumstances, microbiota can support survival under severe MetR and potentially abrogate the effects associated with MetR. We also tested whether food-derived methionine can interfere with Methioninase expression and promote fly survival. We used chemically defined food with a normal amount of methionine (1 mM) or with no methionine and compared the lifespans of controls and flies ubiquitously expressing Methioninase. Although Methioninase expression decreased lifespan more strongly than depletion of methionine from food (potentially due to methionine already present in fly cells in the food-depleted group, which would be eliminated in the Methioninase-depleted group), the combination of Methioninase expression and methionine depletion from the food was associated with no or very moderate lifespan suppression (Fig. 4 E and F). Altogether, our data demonstrate that the complete depletion of methionine (either enzymatically or via nutrient intervention) significantly suppressed lifespan, but that this effect could be modified by intestinal microbiota.
Methioninase Expression Extends Lifespan Independently of Amino Acid Status.
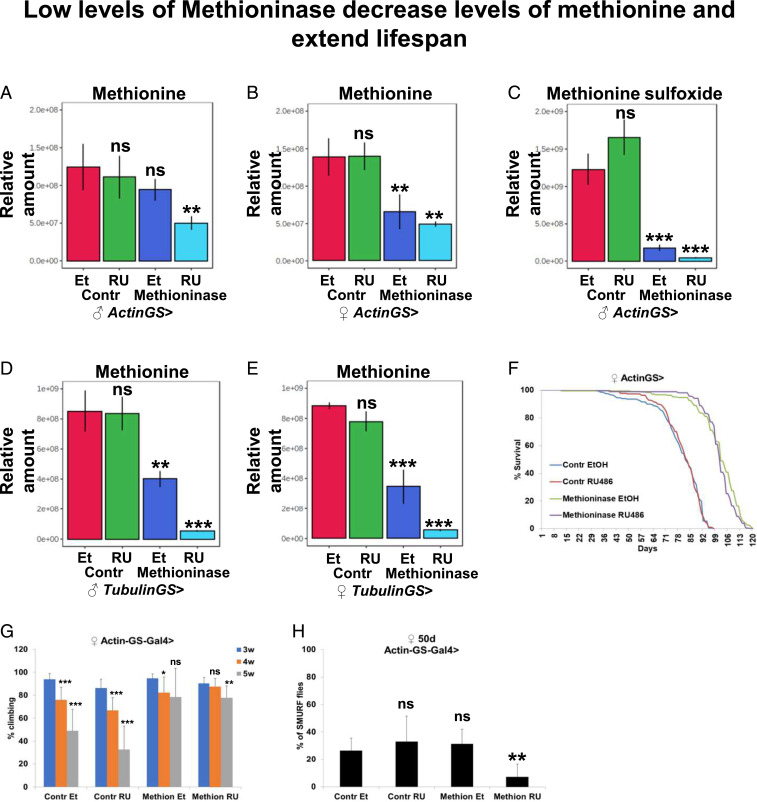
We further tested whether some moderate level of Methioninase expression could phenocopy MetR, reset age-dependent changes in methionine metabolism, and extend lifespan. Because the tubulin-Gal4,tubulin-Gal80ts temperature-inducible system requires a switch to 29 °C (nonoptimal temperature for studying lifespan) and does not easily allow for switching between “on” and “off” states of Methioninase expression, for the next set of studies we used the GeneSwitch (GS) system. Actin Gene-Switch (ActinGS) is a ubiquitous inducible Gal4/UAS expression system (39, 40), whereby the UAS-GeneX or RNAi expression is driven by Gal4 when flies are fed mifepristone (RU486). We first performed a metabolomics analysis of control and Methioninase-expressing male and female flies crossed to the ActinGS driver (ActinGS-Gal4/+; control/+ or ActinGS-Gal4/+; UAS-Methioninase/+), which were fed with solvent or RU486 for 10 d. As a control line, we used a line that was injected with UAS-Methioninase to produce transgenic UAS-Methioninase flies and that had the same genetic background. In male flies, the level of methionine was not significantly different in ActinGS > control flies fed with solvent or RU486, while in ActinGS > Methioninase flies, the level of methionine was significantly reduced upon feeding with RU486 (Fig. 5A). It should be noted that the level of methionine in ActinGS > Methioninase flies fed with solvent was lower than in ActinGS > control flies fed with solvent, although this difference was not statistically significant, potentially indicating a leaky expression of Methioninase in ActinGS > Methioninase flies without addition of RU486 (Fig. 5A). In female flies, the level of methionine was significantly lower in both ActinGS > Methioninase flies fed with solvent or RU486, indicating a stronger leaky expression of Methioninase in female flies (Fig. 5B).
Fig. 5.
Low levels of Methioninase decrease levels of methionine and extend lifespan independently of the amino acid status in the food. Relative levels of methionine in male (A) and female (B) ActinGS > control or Methioninase flies fed with either EtOH or RU486. (C) Relative levels of methionine sulfoxide in male ActinGS > control or Methioninase flies fed with either EtOH or RU486. Relative levels of methionine in male (D) and female (E) TubulinGS > control or Methioninase flies fed with either EtOH or RU486. (F) Survival of female ActinGS > control or Methioninase flies fed with either EtOH or RU486. (G) Climbing of female ActinGS > control or Methioninase flies fed with either EtOH or RU486. (H) Proportion of smurf females at 50 d of age in female ActinGS > control or Methioninase flies fed with either EtOH or RU486. **P < 0.01, ***P < 0.001.
In addition, we compared levels of methionine sulfoxide, the oxidized form of methionine that indirectly reflects the concentration of methionine. The level of methionine sulfoxide was significantly reduced in male ActinGS > Methioninase flies fed with solvent compared with ActinGS > control flies fed with solvent, further confirming the reduced levels of methionine in uninduced flies (Fig. 5C). To estimate the level of leaky expression, we performed qRT-PCR for control and Methioninase male and female flies crossed to the ActinGS driver, which were fed with solvent or RU486 for 10 d. Because Methioninase is absent from control flies, it was undetectable by qRT-PCR in the controls, but its expression was detected at low levels in female ActinGS > UAS-Methioninase flies without the addition of RU486. As expected, the addition of RU486 caused strong expression of Methioninase in both male and female flies (SI Appendix, Fig. S4 A and B) that was further confirmed by immunoblotting (SI Appendix, Fig. S4 C and D).
We further tested whether an alternative ubiquitous tubulin-GS-Gal4 driver can be used to avoid leaky expression of Methioninase in noninduced flies. We performed a metabolomics analysis of control and Methioninase-expressing male and female flies crossed to the Tubulin-GS driver (Tubulin-GS-Gal4/+; control/+ or Tubulin-GS-Gal4/+; UAS-Methioninase/+), which were fed with solvent or RU486 for 10 d. We observed strongly reduced methionine levels in both male (Fig. 5D) and female (Fig. 5E) flies fed with solvent, while the addition of RU486 further decreased methionine levels (Fig. 5 D and E). Similarly, the levels of methionine sulfoxide were also dramatically reduced (SI Appendix, Fig. S4 E and F). Because we could not either confirm or exclude the possibility of leaky Methioninase expression in male ActinGS-Gal4 flies, we further tested lifespans of control and Methioninase-expressing female flies crossed to the ActinGS driver (ActinGS-Gal4/+; control/+ or ActinGS-Gal4/+; UAS-Methioninase/+). Interestingly, we found that female flies (Fig. 5F) without RU486 addition had much longer lifespans than control flies (mean lifespan for controls, 78 d, and for Methioninase, 100 d, P < 0.0001). In female flies, the lifespan was not changed after the addition of RU486 (Fig. 5F), which was consistent with the observation that methionine levels also were not changed after the addition of RU486 and sex-specific differences in the response to MetR. In addition to its leakage, GS drivers may exhibit mosaic patterns of expression. Mosaic expression precludes degradation of methionine in cells where ActinGS driver is not expressed, providing an explanation why an increase in Methioninase expression in ActinGS > Methioninase + RU female flies does not further decrease the amount of methionine (Methioninase is simply not expressed in all cells). Note that using the tubulinGal4;tubulinGal80ts system to drive Methioninase expression completely depletes methionine from flies (Fig. 3A).
It should be noted that we performed our lifespan experiments on regular laboratory food, which contains a normal amount of all amino acids. Previously, Lee et al. (15) demonstrated that MetR extends lifespan in flies only in low amino acid conditions. Using the genetic model of MetR, we found that the lifespan effect can be increased on regular food without lowering the amino acid levels. To characterize how Methioninase expression affects healthspan, we tested its ability to delay age-dependent tissue functional decline as measured by maintained climbing activity (indicative of neuromuscular function), intestine barrier, and egg laying. Normally, skeletal muscle function declines during aging in flies (41). However, Methioninase expression suppressed age-dependent decreases in climbing activity, as determined by negative geotaxis assays (Fig. 5G). Aging is also characterized by decreased reproductive function, which by itself can extend lifespan (42). Although methionine levels are known to regulate egg production (15, 43), we did not observe differences in egg laying between control and Methioninase-expressing flies (SI Appendix, Fig. S4G). In addition, we measured the intestinal integrity of flies fed with a nonabsorbable blue food dye (the “Smurf” assay) (44). Strikingly, Methioninase expression retarded the age-related onset of the Smurf phenotype, suggesting improved intestinal integrity in old flies (Fig. 5H). In addition, we compared food uptake between control and Methioninase-expressing flies and we did not find any differences (SI Appendix, Fig. S4H). Altogether, our data suggest that Methioninase can be used to manipulate methionine levels, creating a MetR state and extending Drosophila health- and lifespan without reducing the levels of other amino acids in fly food.
Tissue-Specific Methioninase Expression Decreases Levels of Methionine and Extends Lifespan Independently of Amino Acid Status.
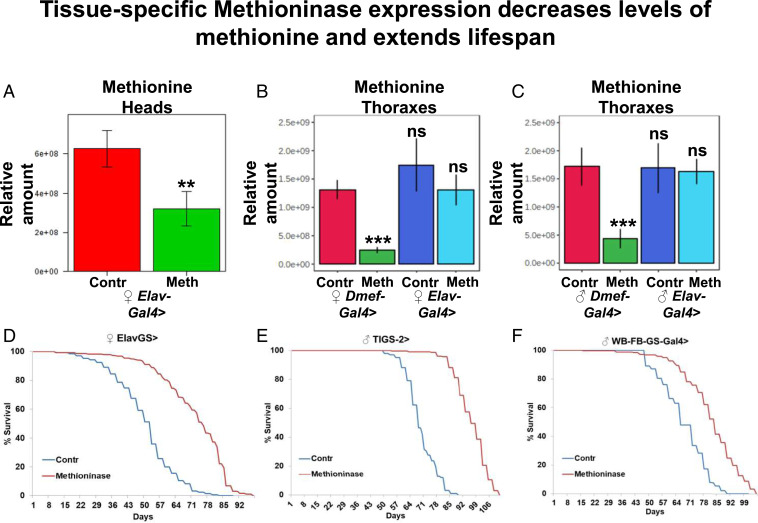
Manipulations of enzymes that either belong to methionine metabolism pathways or those that affect the levels of methionine metabolism metabolites can extend lifespan when manipulated in just one tissue (8, 21, 22). Thus, we tested if Methioninase expression in one tissue would decrease levels of methionine in that tissue, or if transport of methionine from other organs would compensate for the deficiency. To do this, we expressed Methioninase specifically in pan-neuronal tissues in adult flies using the elav-Gal4,tubulin-Gal80ts temperature-inducible system. Flies were grown at 18 °C and switched to 29 °C after eclosion to induce the neuronal-specific expression of Methioninase for 10 d. Expression of Methioninase caused a significant reduction of methionine in female fly heads (Fig. 6A). It should be noted that because fly heads contain a mix of fat, glial, and neuronal cells and Methioninase was expressed only in neuronal cells, a drop in methionine levels in neuronal tissues may be even more significant than what we observed in whole heads. We intentionally used the Gal4 system instead of the GS-Gal4 system because GS-Gal4 may be mosaic and leaky in other tissues that would complicate interpretation of the experiment. We further tested whether degrading methionine in one tissue may function as a “sink” and decrease the level of methionine in another organ. To address this question, we used neuronal-specific (elav-Gal4,tubulin-Gal80ts) and muscle-specific (Dmef-Gal4,tubulin-Gal80ts) temperature-inducible systems to express Methioninase in neuronal- or muscle-specific cells, respectively.
Fig. 6.
Tissue-specific Methioninase expression decreases levels of methionine and extends lifespan independently of the amino acid status in the food. (A) Relative levels of methionine in heads of tubulin-Gal80ts, elav-Gal4 > control or Methioninase female flies. Means ± SD. Relative levels of methionine in thoraxes of female (B) and male (C) tubulin-Gal80ts, Dmef-Gal4 > or tubulin-Gal80ts, elav-Gal4 > control or Methioninase flies. (D) Neuronal-specific leaky expression of Methioninase in ElavGS > Methioninase female flies extends lifespan. (E) Intestine-specific leaky expression of Methioninase in TIGS-2 > Methioninase male flies extends lifespan. (F) Fat body-specific leaky expression of Methioninase in WB-FB-GS > Methioninase male flies extends lifespan. **P < 0.01, ***P < 0.001.
We then compared methionine levels in both male and female thoraxes (consisting mostly of muscle cells). As expected, muscle-specific Methioninase expression significantly decreased the levels of methionine in thoraxes, while neuronal-specific expression of Methioninase did not affect levels of methionine in thoraxes (Fig. 6 B and C). This potentially means that tissue-specific expression of Methioninase does not function as a sink for methionine from other organs. We then tested whether similar to a whole-body ActinGS system, potentially leaky Methioninase expression would extend the lifespan using nervous system-specific (ElavGS or nSybGS) (40, 45), intestine-specific (TIGS-2) (44, 46), or fat body-specific (WB-FB-GS) (45, 47) GS systems. In both neuronal-specific GS lines ElavGS and nSybGS, potentially leaky levels of Methioninase expression (without addition of RU486) extended the lifespan of female flies (ElavGS: mean lifespan for controls, 49 d, and for Methioninase, 71 d, P < 0.0001; nSybGS: mean lifespan for controls, 60 d, and for Methioninase, 68 d, P < 0.0001) (Fig. 6D and SI Appendix, Fig. S5A) and male flies (ElavGS: mean lifespan for controls, 66 d, and for Methioninase, 75 d, P < 0.0001; nSybGS: mean lifespan for controls, 66 d, and for Methioninase, 76 d, P < 0.0001) (SI Appendix, Fig. S5 B and C).
In addition, potentially leaky levels of Methioninase expression in the intestine and fat body dramatically extended lifespan of male flies (TIGS-2: mean lifespan for controls, 68 d, and for Methioninase, 95 d, P < 0.0001; WB-FB-GS: mean lifespan for controls, 67 d, and for Methioninase, 81 d, P < 0.0001) (Fig. 6 E and F) but had a very moderate effect (SI Appendix, Fig. S5D) or no effect (SI Appendix, Fig. S5E) in female flies. We note that we did not test leaky expression in specific organs and the differences in the extension of lifespan may simply depend on the levels of leakiness in the GS lines and are not necessarily linked to sex-specific differences. It is also possible that the extension of lifespan happens due to leaky expression of Methioninase in nonspecific tissues.
We further measured whether Methioninase expression in addition to lifespan extension also extends healthspan. Interestingly, we found that intestine-specific Methioninase expression was associated with a significantly delayed decline in climbing activity with age (SI Appendix, Fig. S5 F and G) but did not significantly change the number of flies with the SUMRF phenotype (SI Appendix, Fig. S5H); while neuronal-specific Methioninase expression significantly suppressed the number of flies with the SMURF phenotype (SI Appendix, Fig. S5I), it was not associated with improved climbing (SI Appendix, Fig. S5 J and K). It should be noted that in both GS systems, Methioninase expression was not associated with decreased egg laying capacity (SI Appendix, Fig. S5 L and M). In addition, the lifespan extension in TIGS-2 male flies was not associated with altered food uptake (SI Appendix, Fig. S5N). Altogether, we demonstrated that expression of Methioninase in a specific tissue can effectively decrease levels of methionine in a targeted tissue without changing the levels of methionine in other tissues, and that even potentially a very low (leaky) level of Methioninase expression in different organs can be sufficient to result in a dramatic extension of health- and lifespan.
Reprogramming of Methionine Metabolism Flux in Aged and Tau-Expressing Drosophila Heads.
Increased levels of plasma homocysteine (one of the metabolites in the methionine metabolism pathway) have been recognized as a risk factor for dementia and Alzheimer’s disease (AD) (48, 49). Thus, we wanted to compare whether the fate of methionine and its distribution via different branches of the methionine metabolism pathway is affected with age in wild-type Drosophila heads (which predominantly consist of neuronal cells) in a similar way as we observed in whole flies and whether overexpression of human Tau phenocopies the aging phenotype in young heads. To address this, we fed either wild-type OreR flies of different ages (1 wk and 4 wk) or 1-wk-old flies with neuronal-specific control or Tau overexpression with 1 mM labeled 13C5-methionine tracer using chemically defined food and analyzed the distribution of labeled methionine to methionine metabolism metabolites in fly heads. Interestingly, expression of human Tau had a much stronger effect on the reprogramming of methionine metabolism in fly heads compared to aging in fly heads. We found that aging only significantly decreased the level of unlabeled methionine (M0) (SI Appendix, Fig. S6A), while it did not affect the enrichment ratio (M1/M4+M5+M1) between methionine (M1) produced from the salvage pathway and total labeled methionine (SI Appendix, Fig. S6C), the enrichment ratio (M4/M4+M5+M1) between methionine (M4) produced from the methionine cycle and total labeled methionine (SI Appendix, Fig. S6E), as well as the labeling in downstream metabolites: MTA (SI Appendix, Fig. S6G) and SAH (SI Appendix, Fig. S6I). Accordingly, with the decreased amount of unlabeled methionine (M0), the level of its downstream product, unlabeled SAM (M0), was also decreased (SI Appendix, Fig. S6K).
In contrast to aging, overexpression of human Tau causes strong changes in the fate of labeled methionine (Fig. 6B), ratio of methionine (M1/M4+M5+M1) produced from the salvage pathway (SI Appendix, Fig. S6D) and via the methionine cycle (M4/M4+M5+M1) (SI Appendix, Fig. S6F), and labeling of its downstream metabolites: MTA, SAH, and SAM (SI Appendix, Fig. S6 H, J and L). Like in the results of our analysis of aging in whole flies, this suggests that the fate of the labeled methionine is altered in the heads of flies with an overexpression of human Tau. It would be interesting to test the effect of neuronal-specific genetic MetR on the phenotype of flies with Tau overexpression. However, as AD-relevant phenotypes require very high level of Tau overexpression and we observe health- and lifespan benefits only with leaky Methioninase expression, generating such flies is problematic. Similarly, we cannot combine Methioninase expression and methionine labeling because Methioninase would destroy labeled methionine. Moreover, AD is characterized by extensive reprogramming of metabolism (50) and targeting of multiple metabolic pathways may be required to prevent neurodegeneration. Altogether, we demonstrated that impairment of methionine metabolism flux is common between aging and in Tau-expressing Drosophila heads.
Discussion
By feeding 13C5-Methionine to Drosophila and tracing its fate in different branches of the methionine metabolism, we were able to identify impairments of methionine flux associated with aging and AD. By creating transgenic flies that express Methioninase, which is capable of degrading methionine, we were able to create a genetic tool for the efficient depletion of methionine and its downstream metabolites, thus “resetting” methionine metabolism. Moreover, whole-body or tissue-specific low-level expression of Methioninase could significantly extend health- and lifespan irrespective of amino acid levels in the food. Altogether, our study highlights Methioninase, which has been tested in several clinical trials, as a potential agent for health- and lifespan extension.
Methionine Metabolism and Aging.
MetR extends the lifespan across different species and exerts beneficial effects on metabolic health and inflammatory responses (51). To test the effect of MetR in flies and its interaction with dietary restriction, Lee et al. (15) compared the lifespans of flies using diets with different levels of amino acids and different concentrations of methionine. Restriction of amino acids increased lifespan at any concentration of methionine, whereas MetR extended lifespan only in conditions in which the levels of amino acids were reduced. The caveat of studying MetR in flies is the necessity of using a chemically defined diet that requires defining the basal (1×) levels of each component. One of the advantages of using Methioninase to mimic MetR is that it can be expressed in flies cultured on a standard sugar/yeast diet. In contrast to Lee et al. (15), we found that low levels of Methioninase expression in flies cultured on a standard diet containing normal levels of amino acids extended lifespan. Moreover, metabolomics analysis of flies with control or Methioninase expression did not reveal a global decrease in amino acids, while the level of methionine was significantly decreased.
Methionine Is an Essential Amino Acid.
Because methionine is an essential amino acid, one question is how long flies can survive under complete depletion of methionine. Ubiquitous expression of Methioninase is an ideal tool for testing this since it is expressed in all cells where it efficiently degrades methionine. Metabolomics analysis of whole flies expressing Methioninase verifies the manipulations cause a complete lack of methionine and its downstream metabolites, SAM and SAH. However, half of the flies with depleted methionine can still survive up to 15 d and a small fraction can survive much longer (super survivors), which might be explained by the activation of compensatory mechanisms. We demonstrated that microbiota affects the survival of flies under MetR. Treatment of flies with a mix of antibiotics known to kill all Drosophila flora did not affect the survival of wild-type flies or the survival of the main fraction of flies expressing Methioninase, but treatment with antibiotics did remove the fraction of super survivors, suggesting that microbiota reprogramming can be an important factor interacting with methionine metabolism and aging. Studies in mice demonstrated that MetR or methionine supplementation affect the mouse gut microbiome (52). Moreover, rearing flies with Escherichia coli mutants for distinct genes relevant to methionine metabolism alters Drosophila starvation response and longevity (53, 54). We also tested whether dietary methionine can at least partially compensate for methionine degradation by Methioninase. By combining Methioninase expression and methionine depletion from the food, we demonstrated that dietary methionine cannot substantially compensate for methionine degradation by Methioninase. It is possible that protein degradation is also involved in maintaining methionine levels. In summary, we demonstrated that bacterial enzymes that are not present in the Drosophila genome can be useful tools for dissecting the role of specific metabolites, such as methionine.
Mechanisms of Lifespan Extension.
Lifespan extension is often accompanied and dependent on target of rapamycin (TOR) inhibition and the activation of autophagy (55, 56). In yeast, MetR is accompanied by activation of autophagy flux, and the deletion of autophagy essential genes abolished MetR-induced lifespan extension (57). Moreover, MetR-induced lifespan extension could not be further extended by TOR inhibition (57). Similar to yeast, knockdown of sams-1 failed to extend the lifespan of eat-2 mutant worms (a genetic model for studying dietary restriction) and exhibited phenotypes reminiscent of dietary restriction, such as slender worms with reduced brood size and delayed reproduction. In mammalian cells, it has been shown that SAM (58) and homocysteine (59) can directly activate mTORC1 via its relocalization to the lysosomal surface. Moreover, by regulating the production of the methyl donor SAM, alterations in methionine metabolism can affect the function of 30 methyltransferases that have been previously shown to regulate lifespan (51). The precise mechanisms of lifespan extension by MetR could be clarified through experiments that use two independent binary expression systems—such as UAS/GS-GAL4/RU486 and QUAS/QF/QS—for the inducible expression of Methioninase and potential downstream mediators of lifespan extension.
Methioninase in Human Clinical Trials.
Certain cancer cells exhibit methionine auxotrophy. Thus, MetR represents an intervention that can extend lifespan with a complementary effect of delaying tumor growth (60). In addition to suppressing tumor growth and extending lifespan, MetR in rodents also prevents visceral fat mass accumulation and from the negative effects of a high-fat diet (16, 61–63). Based on these results, MetR has also been tested in a clinical trial of obese adults with metabolic syndrome (64, 65). Dietary MetR can be compensated by methionine production through protein degradation, homocysteine remethylation, or by intestinal flora, which may limit the clinical benefits exerted by MetR. As an alternative method for lowering methionine levels in humans, the enzyme Methioninase has been isolated from Clostridium sporogene (66). rMetase has been used in a human clinical trial (67). It would be interesting to test whether rMetase could extend lifespan in rodents and humans as in flies. However, the extent of MetR should be precisely controlled. As we show in flies and, as previously demonstrated in mice (68), decreasing the concentration of methionine outside of the beneficial range results in the switch from MetR to methionine starvation, which is detrimental for lifespan.
Limitations of This Study.
We note that because of experimental constraints there may be some limitations of our study. We compared the lifespans of GS > Control + EtOH vs. GS > Methioninase + EtOH flies rather than GS > Methioninase + EtOH vs. GS > Methioninase + RU flies due to the leakage of the GS system, which we demonstrated by qRT-PCR, immunoblotting, measuring methionine, and methionine sulfoxide levels, and because MetR extends lifespan only in the narrow range of the dietary concentration of methionine, as previously demonstrated both in mice (68) and in flies (15). Decreasing the concentration of methionine outside of this range results in the switch from MetR to methionine starvation, which is detrimental for lifespan. It has been also previously shown that different GS drivers may exhibit leaky expression both during development and adult stage (46, 69). While it may be negligible for most of the lifespan studies with other transgenes, it is problematic with our experiments as even a low amount of Methioninase can efficiently degrade a significant amount of methionine. One of the potential problems with comparing GS > Methioninase + EtOH vs. GS > control + EtOH is a potential difference in genetic background or other factors, which is why we decided to use as a control the line that was used for the injection of Methioninase.
Additional evidence that lifespan differences cannot simply be explained by differences between control and Methioninase lines comes from the fact that female WB-FB-GS flies crossed to control or Methioninase flies show no difference in their lifespan. Additionally, comparing the absolute lifespan of female ActinGS > Methioninase + EtOH flies to previously published studies from different laboratories (70–72) indicates that these flies are exceptionally long-lived (surviving up to 120 d). Due to the low level of leakage even in ubiquitous ActinGS-Gal4 flies, and the mixture of different cell types, we could not use similar assays to detect leakage in tissue-specific GS lines. We interpret our results with tissue-specific GS lines by extrapolating from our results with ActinGS flies and the observed differences in lifespans can be caused by other factors.
Materials and Methods
For survival analysis, flies were collected within 24 h from eclosion, sorted by sex, and reared at standard density (20 to 25 flies per vial) on cornmeal/soy flour/yeast fly food at 25 °C, as previously described (8, 10). RU486 dissolved in ethanol was administered in the media at a final concentration of 150 μg/mL. Flies were transferred to fresh vials every 2 d, and the dead flies were counted. Detailed methods and materials are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank John Tower for the GeneSwitch fly stocks, Scott Pletcher for the TIGS-2 GeneSwitch driver line, and Stephanie Mohr for critical manuscript reading. This work was supported by National Institute of Aging Grants R00AG057792 (to A.A.P.), 5P01CA120964 (to N.P.), and American Federation for Aging Research (N.P.). N.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110387118/-/DCSupplemental.
Data Availability
All study data are included in the main text and supporting information.
References
- 1.López-Otín C., Blasco M. A., Partridge L., Serrano M., Kroemer G., The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkhitko A. A., Filine E., Mohr S. E., Moskalev A., Perrimon N., Targeting metabolic pathways for extension of lifespan and healthspan across multiple species. Ageing Res. Rev. 64, 101188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs S., et al., A metabolic signature of long life in Caenorhabditis elegans. BMC Biol. 8, 14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Houtkooper R. H., et al., The metabolic footprint of aging in mice. Sci. Rep. 1, 134 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darst B. F., Koscik R. L., Hogan K. J., Johnson S. C., Engelman C. D., Longitudinal plasma metabolomics of aging and sex. Aging (Albany NY) 11, 1262–1282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menni C., et al., Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int. J. Epidemiol. 42, 1111–1119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu Z., et al., Human serum metabolic profiles are age dependent. Aging Cell 11, 960–967 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parkhitko A. A., et al., Tissue-specific down-regulation of S-adenosyl-homocysteine via suppression of dAhcyL1/dAhcyL2 extends health span and life span in Drosophila. Genes Dev. 30, 1409–1422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma S., et al., Organization of the mammalian Metabolome according to organ function, lineage specialization, and longevity. Cell Metab. 22, 332–343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkhitko A. A., et al., Downregulation of the tyrosine degradation pathway extends Drosophila lifespan. eLife 9, e58053 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uthus E. O., Brown-Borg H. M., Methionine flux to transsulfuration is enhanced in the long living Ames dwarf mouse. Mech. Ageing Dev. 127, 444–450 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mudd S. H., Hypermethioninemias of genetic and non-genetic origin: A review. Am. J. Med. Genet. C. Semin. Med. Genet. 157C, 3–32 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Ostrakhovitch E. A., Tabibzadeh S., Homocysteine and age-associated disorders. Ageing Res. Rev. 49, 144–164 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Kozieł R., et al., Methionine restriction slows down senescence in human diploid fibroblasts. Aging Cell 13, 1038–1048 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee B. C., et al., Methionine restriction extends lifespan of Drosophila melanogaster under conditions of low amino-acid status. Nat. Commun. 5, 3592 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orentreich N., Matias J. R., DeFelice A., Zimmerman J. A., Low methionine ingestion by rats extends life span. J. Nutr. 123, 269–274 (1993). [DOI] [PubMed] [Google Scholar]
- 17.Miller R. A., et al., Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell 4, 119–125 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ables G. P., Hens J. R., Nichenametla S. N., Methionine restriction beyond life-span extension. Ann. N. Y. Acad. Sci. 1363, 68–79 (2016). [DOI] [PubMed] [Google Scholar]
- 19.McIsaac R. S., Lewis K. N., Gibney P. A., Buffenstein R., From yeast to human: Exploring the comparative biology of methionine restriction in extending eukaryotic life span. Ann. N. Y. Acad. Sci. 1363, 155–170 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Sanchez-Roman I., Barja G., Regulation of longevity and oxidative stress by nutritional interventions: Role of methionine restriction. Exp. Gerontol. 48, 1030–1042 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Kabil H., Kabil O., Banerjee R., Harshman L. G., Pletcher S. D., Increased transsulfuration mediates longevity and dietary restriction in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 108, 16831–16836 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tain L. S., et al., Longevity in response to lowered insulin signaling requires glycine N-methyltransferase-dependent spermidine production. Aging Cell 19, e13043 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obata F., et al., Necrosis-driven systemic immune response alters SAM metabolism through the FOXO-GNMT axis. Cell Rep. 7, 821–833 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Hansen M., Hsu A. L., Dillin A., Kenyon C., New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 1, 119–128 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takakura T., et al., Physicochemical and pharmacokinetic characterization of highly potent recombinant L-methionine gamma-lyase conjugated with polyethylene glycol as an antitumor agent. Cancer Res. 66, 2807–2814 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Hoffman R. M., Development of recombinant methioninase to target the general cancer-specific metabolic defect of methionine dependence: A 40-year odyssey. Expert Opin. Biol. Ther. 15, 21–31 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Agrawal V., Alpini S. E., Stone E. M., Frenkel E. P., Frankel A. E., Targeting methionine auxotrophy in cancer: Discovery & exploration. Expert Opin. Biol. Ther. 12, 53–61 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Tan Y., et al., Recombinant methioninase infusion reduces the biochemical endpoint of serum methionine with minimal toxicity in high-stage cancer patients. Anticancer Res. 17 (5B), 3857–3860 (1997). [PubMed] [Google Scholar]
- 29.Perła-Kaján J., Twardowski T., Jakubowski H., Mechanisms of homocysteine toxicity in humans. Amino Acids 32, 561–572 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Sauer U., Metabolic networks in motion: 13C-based flux analysis. Mol. Syst. Biol. 2, 62 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buescher J. M., et al., A roadmap for interpreting (13)C metabolite labeling patterns from cells. Curr. Opin. Biotechnol. 34, 189–201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jang C., Chen L., Rabinowitz J. D., Metabolomics and isotope tracing. Cell 173, 822–837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zamboni N., 13C metabolic flux analysis in complex systems. Curr. Opin. Biotechnol. 22, 103–108 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Park A., Tran T., Atkinson N. S., Monitoring food preference in Drosophila by oligonucleotide tagging. Proc. Natl. Acad. Sci. U.S.A. 115, 9020–9025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pegg A. E., Functions of polyamines in mammals. J. Biol. Chem. 291, 14904–14912 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minois N., Carmona-Gutierrez D., Madeo F., Polyamines in aging and disease. Aging (Albany NY) 3, 716–732 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGuire S. E., Le P. T., Osborn A. J., Matsumoto K., Davis R. L., Spatiotemporal rescue of memory dysfunction in Drosophila. Science (New York, NY) 302, 1765–1768 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Piper M. D., Using artificial diets to understand the nutritional physiology of Drosophila melanogaster. Curr. Opin. Insect Sci. 23, 104–111 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Roman G., Endo K., Zong L., Davis R. L., P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 98, 12602–12607 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osterwalder T., Yoon K. S., White B. H., Keshishian H., A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. U.S.A. 98, 12596–12601 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demontis F., Piccirillo R., Goldberg A. L., Perrimon N., Mechanisms of skeletal muscle aging: Insights from Drosophila and mammalian models. Dis. Model. Mech. 6, 1339–1352 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sgró C. M., Partridge L., A delayed wave of death from reproduction in Drosophila. Science 286, 2521–2524 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Grandison R. C., Piper M. D., Partridge L., Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 462, 1061–1064 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rera M., et al., Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 14, 623–634 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen J., Curtis C., Tavaré S., Tower J., A screen of apoptosis and senescence regulatory genes for life span effects when over-expressed in Drosophila. Aging (Albany NY) 1, 191–211 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poirier L., Shane A., Zheng J., Seroude L., Characterization of the Drosophila gene-switch system in aging studies: A cautionary tale. Aging Cell 7, 758–770 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Giannakou M. E., et al., Dynamics of the action of dFOXO on adult mortality in Drosophila. Aging Cell 6, 429–438 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Ravaglia G., et al., Homocysteine and folate as risk factors for dementia and Alzheimer disease. Am. J. Clin. Nutr. 82, 636–643 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Seshadri S., et al., Plasma homocysteine as a risk factor for dementia and Alzheimer’s disease. N. Engl. J. Med. 346, 476–483 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Yan X., Hu Y., Wang B., Wang S., Zhang X., Metabolic dysregulation contributes to the progression of Alzheimer’s disease. Front. Neurosci. 14, 530219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parkhitko A. A., Jouandin P., Mohr S. E., Perrimon N., Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species. Aging Cell 18, e13034 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallis K. F., Melnyk S. B., Miousse I. R., Sex-specific effects of dietary methionine restriction on the intestinal microbiome. Nutrients 12, 781 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Judd A. M., et al., Bacterial methionine metabolism genes influence Drosophila melanogaster starvation resistance. Appl. Environ. Microbiol. 84, e00662-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matthews M. K., et al., Genetic influences of the microbiota on the life span of Drosophila melanogaster. Appl. Environ. Microbiol. 86, e00305-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parkhitko A. A., Favorova O. O., Henske E. P., Autophagy: Mechanisms, regulation, and its role in tumorigenesis. Biochemistry (Mosc.) 78, 355–367 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Parkhitko A. A., Favorova O. O., Khabibullin D. I., Anisimov V. N., Henske E. P., Kinase mTOR: Regulation and role in maintenance of cellular homeostasis, tumor development, and aging. Biochemistry (Mosc.) 79, 88–101 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Ruckenstuhl C., et al., Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification. PLoS Genet. 10, e1004347 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu X., et al., SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 358, 813–818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khayati K., et al., The amino acid metabolite homocysteine activates mTORC1 to inhibit autophagy and form abnormal proteins in human neurons and mice. FASEB J. 31, 598–609 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Cavuoto P., Fenech M. F., A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat. Rev. 38, 726–736 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Wanders D., et al., The components of age-dependent effects of dietary methionine restriction on energy balance in rats. Obesity (Silver Spring) 26, 740–746 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ables G. P., Perrone C. E., Orentreich D., Orentreich N., Methionine-restricted C57BL/6J mice are resistant to diet-induced obesity and insulin resistance but have low bone density. PLoS One 7, e51357 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malloy V. L., et al., Methionine restriction decreases visceral fat mass and preserves insulin action in aging male Fischer 344 rats independent of energy restriction. Aging Cell 5, 305–314 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Olsen T., et al., Effects of dietary methionine and cysteine restriction on plasma biomarkers, serum fibroblast growth factor 21, and adipose tissue gene expression in women with overweight or obesity: A double-blind randomized controlled pilot study. J. Transl. Med. 18, 122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Plaisance E. P., et al., Dietary methionine restriction increases fat oxidation in obese adults with metabolic syndrome. J. Clin. Endocrinol. Metab. 96, E836–E840 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kreis W., Hession C., Isolation and purification of L-methionine-alpha-deamino-gamma-mercaptomethane-lyase (L-methioninase) from Clostridium sporogenes. Cancer Res. 33, 1862–1865 (1973). [PubMed] [Google Scholar]
- 67.Hoffman R. M., et al., Pilot phase I clinical trial of methioninase on high-stage cancer patients: Rapid depletion of circulating methionine. Methods Mol. Biol. 1866, 231–242 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Forney L. A., Wanders D., Stone K. P., Pierse A., Gettys T. W., Concentration-dependent linkage of dietary methionine restriction to the components of its metabolic phenotype. Obesity (Silver Spring) 25, 730–738 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scialo F., Sriram A., Stefanatos R., Sanz A., Practical recommendations for the use of the GeneSwitch Gal4 system to knock-down genes in Drosophila melanogaster. PLoS One 11, e0161817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slack C., et al., The Ras-Erk-ETS-signaling pathway is a drug target for longevity. Cell 162, 72–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alic N., Hoddinott M. P., Vinti G., Partridge L., Lifespan extension by increased expression of the Drosophila homologue of the IGFBP7 tumour suppressor. Aging Cell 10, 137–147 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tian X., et al., SIRT6 is responsible for more efficient DNA double-strand break repair in long-lived species. Cell 177, 622–638.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and supporting information.