Significance
Microvilli are used by immune cells to sense the surface features of pathogens and antigen presenting cells. However, microvilli’s contribution in T cell signaling and activation is largely unknown. Here, we introduce a material-based platform for induction of microvilli formation in T cells, in which the dimensions of the microvilli can be controlled by tuning the dimensions of the nanotopographical features, such as pore depth, pore size, and interpore distance. We demonstrate the direct causality between microvilli formation and altered gene expression in T cells. We discover that the size of the microvilli critically influences T cell receptor agonistic independent signaling in T cells. The results provide a physical strategy for T cell activation and expansion for immunotherapy applications.
Keywords: T cell microvilli, nanoconfinement, immunoengineering, mechanobiology, cell-surface interactions
Abstract
T cells sense and respond to their local environment at the nanoscale by forming small actin-rich protrusions, called microvilli, which play critical roles in signaling and antigen recognition, particularly at the interface with the antigen presenting cells. However, the mechanism by which microvilli contribute to cell signaling and activation is largely unknown. Here, we present a tunable engineered system that promotes microvilli formation and T cell signaling via physical stimuli. We discovered that nanoporous surfaces favored microvilli formation and markedly altered gene expression in T cells and promoted their activation. Mechanistically, confinement of microvilli inside of nanopores leads to size-dependent sorting of membrane-anchored proteins, specifically segregating CD45 phosphatases and T cell receptors (TCR) from the tip of the protrusions when microvilli are confined in 200-nm pores but not in 400-nm pores. Consequently, formation of TCR nanoclustered hotspots within 200-nm pores allows sustained and augmented signaling that prompts T cell activation even in the absence of TCR agonists. The synergistic combination of mechanical and biochemical signals on porous surfaces presents a straightforward strategy to investigate the role of microvilli in T cell signaling as well as to boost T cell activation and expansion for application in the growing field of adoptive immunotherapy.
Microvilli are small actin-rich protrusions that are used by cells to explore and respond to their local environment, and they are used by immune system cells to sense the surface features of pathogens and antigen presenting cells (1–3). Besides their exploratory role in maximizing surface contact to detect cognate antigens (4), microvilli can regulate T cell signaling by forming areas with high membrane curvature (5). Formation of high membrane curvature is thought to be sensed by triggering biological processes, such as the assembly of signaling microclusters. Formation of high membrane curvatures (200 to 1,000 nm in diameter) supports antigen recognition in T cells, through amplified and sustained signaling at the tip of the protrusions (1, 6). Recent studies suggest that sporadic nonspecific T cell receptor (TCR) phosphorylation can occur at the tip of microvilli, where CD45 is sterically excluded through the formation of “tight contacts” (7, 8). However, the underlying mechanism and the biological processes triggered by microvilli formation in the process of T cell activation remain largely unknown.
Understanding signaling processes induced by microvilli formation is important both from a physiological and technological point of view. This becomes obvious in Whiskott–Aldrich Syndrome and Leukemia, where disrupted formation of microvilli leads to compromised function of various immune cells (9). Importantly, microvilli formation can be regulated by common anticancer drugs such as cytokines, chemokines, or cytoskeletal inhibitors, potentially impairing T cells in being able to efficiently recognize their cognate antigens, thus limiting the efficiency of the therapy (10, 11). Beside chemical signals, the physical environment of the cell can also regulate microvilli formation, particularly at the nanoscale (12). The consequences of T cell microvilli regulation by nanomaterial implants is largely unexplored, which is particularly significant considering the rise of nanotechnology-based immunotherapy approaches and a large pipeline of associated treatments in clinical trials (13, 14). From an engineering perspective, one may exploit the possibility of engineering T cell functions by regulating microvilli formation ex vivo, which can have an impact in the growing field of adoptive T cell therapy.
Despite the importance of microvilli as a structural unit of the cell and their role in T cell signaling, there is a limited amount of studies that investigated microvilli function in T cell activation (4, 5, 9). This is mainly due to the fact that microvilli are nanoscale projections from the cell membrane displaying high dynamics of formation and retraction, therefore difficult to investigate by standard optical microscopy (4, 15, 16). While electron microscopy enables their visualization, their dynamic behavior still remains largely unresolved (1). The fast and transient nature of microvilli protrusions makes it challenging to detect downstream signaling events. Using lattice light-sheet microscopy and real-time tracing of dynamic microvilli on two-dimensional (2D) surfaces, the lifetime of the microvilli protrusions is estimated to be roughly 1 min (16).
To investigate how microvilli interact with nanoscale surface features, we studied the natural behavior of T cells in exploring their environment when in contact with engineered nanoporous materials. Surprisingly, microvilli formation was triggered by presenting a physical cue. The dimensions of the T cell projections are defined by the pore size, and it matches the dimensions of membrane protrusions used by T cells to explore antigen presenting cells. Particularly, confinement of the microvilli into pores with 200 nm in diameter markedly alters cell signaling cascades and subsequent gene expression, even enabling physical activation of T cells without TCR engagement. We explain this phenomenon by nanoconfinement enforced spatiotemporal segregation of the CD45 phosphatase from the TCR. Physically separated from the inhibitory function of CD45, TCR downstream signaling is enhanced even in the absence of specific TCR agonistic stimuli. Scientifically, this discovery that size-matched nanopores promote microvilli formation provides a platform to study microvilli formation processes and their role in T cell activation. Technologically, this opens unforeseen opportunities to boost T cell activation and expansion ex vivo by inducing microvilli formation with specific dimensions, which has potential applications in cell-based immunotherapies.
Results and Discussion
T Cell Microvilli Induction and Stabilization by Nanoporous Substrates.
While microstructured surfaces were shown to induce formation of membrane protrusions in T cells (17–19), T cells reside in nanostructured environments, where they explore the nanoscale features of antigen presenting cells. To match with the membrane protrusions used by T cells, we studied the effect of nanotopographical constraints on microvilli formation in nonactivated T cells. Anodic aluminum oxide (AAO) was used as a substrate to create nanotopographical constraints for T cell microvilli induction. AAO is produced by anodization of aluminum films, which results in formation of quasi-ordered arrays of nanopores with a narrow distribution of pore size and interpore distance (SI Appendix, Fig. S1). Nonporous films, as control samples, were prepared by atomic layer deposition (ALD) of aluminum oxide on flat glass coverslips.
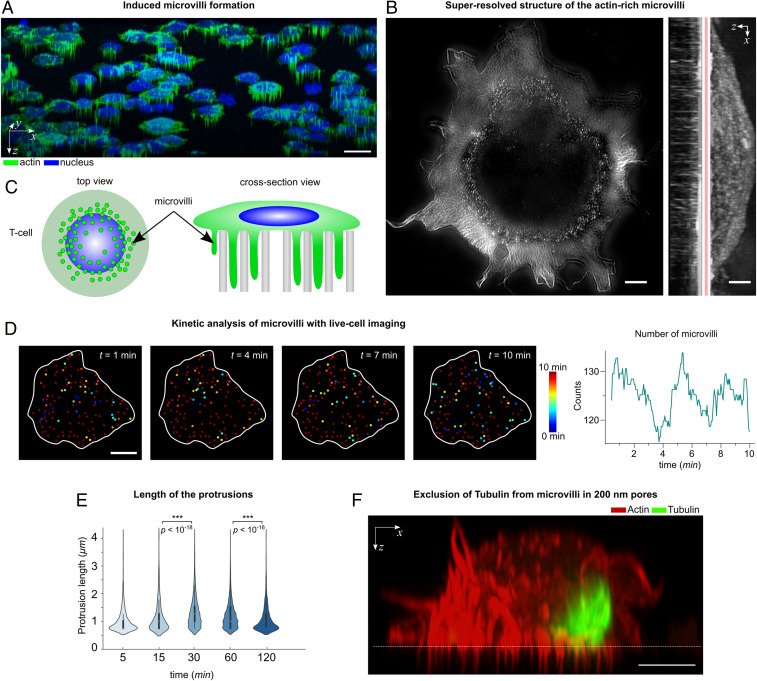
The attachment of the T cells to the nanoporous surface (without agonistic TCR stimuli) was first visualized by confocal microscopy (Fig. 1), showing how the cells extend their actin-rich microvilli into the nanopores (Fig. 1A, also see the z-stack Movie S1). The actin-rich protrusions appeared as dotted structures under the microscope (schematically shown in Fig. 1C). By obtaining a three-dimensional profile of the cells using actin staining, the length and depth of the protrusions were estimated. A more detailed picture of the actin cytoskeleton was obtained by superresolution microscopy (structured illumination microscopy), revealing the protrusions being mostly concentrated around the periphery of the cell (Fig. 1B).
Fig. 1.
Microvilli protrusions in T cells on porous surfaces. (A) The confocal fluorescence microscopy images of Jurkat T cells on a porous AAO with 200-nm pore size (three-dimensional isometric projection). (Scale bar, 10 μm.) (B) A superresolution image of the actin cytoskeleton in a T cell using structured illumination microscopy. The protrusions are seen as a dotted structure in the top-view intensity projection and mostly concentrated around the cells’ periphery. (Scale bar, 1 μm.) (C) A schematic presentation of the actin-rich protrusions in T cells (not to scale). (D, Left) Snapshots of the actin protrusions in a live cell, obtained by fluorescent microscopy. The color corresponds to the lifetime of each protrusion. (Scale bar, 5 μm.) (Right) The number of protrusions in each frame, for the duration of 10 min. (E) The protrusions’ lengths were measured on fixed cells at the indicated time points. Violin plots show the distribution of the measured lengths (n = 20 cells). The P values were determined by two-sided Mann–Whitney U tests using R. (F) A cross-section view of a Jurkat T cell on a porous AAO with 200-nm pore size, captured by Airyscan confocal microscopy. The cells were stained with phalloidin (actin cytoskeleton) and antitubulin (microtubules), indicating that the microtubules were mostly excluded from entering the 200-nm pores. (Scale bar, 5 μm.)
The microvilli formation occurred very fast and within the first minute after contact with the porous surface (without agonistic TCR stimuli), as seen in Movie S2, which was recorded by live cell imaging of Jurkat T cells transfected with Lifeact-GFP, for fluorescently tagging the actin cytoskeleton. The appearance of the bright dotted structure in 2D images is an indication of microvilli formation inside the pores, therefore allowing for the estimation of formation/retraction kinetics. As an example, we collected snapshots (every 10 s) of the microvilli just below the surface, showing that the formation or retraction of the microvilli is a highly dynamic process with an estimated average rate of ∼14 microvilli being formed (and ∼13 being retracted) per minute (Fig. 1D and Movie S3). The highly dynamic infiltration of microvilli into the pores enables the cells to maintain their mobility on the surface and to retract from the surface. Retraction of microvilli from the pores was captured by live cell imaging, where we observed, for example, detachment of some protrusions at the trailing edge of a migrating cell (Movie S4) or gradual yet complete detachment of protrusions upon the cell’s retraction from the surface (Movie S5). Moreover, cells can spread their lamellipodia at the upper interface and initiate actin treadmilling (Movie S6).
By looking into the kinetics of the microvilli length over time (t = 5, 15, 30, 60, and 120 min), it was observed that the average protrusion length increased with time until t = 30 min, reaching to an average length of Lave ∼1.3 μm, and then decreased after 120 min (Lave ∼0.9 μm), suggesting that the cells can loosen their contacts to retract from the surface (Fig. 1E). Notably, protrusions within 200-nm pores are both longer and more stable than those observed on flat surfaces (16, 20), therefore nanotopography encourages and stabilizes formation of microvilli.
The advantage of using nanoengineered surfaces is that the length, thickness, and number of the microvilli can be controlled by tuning the dimensions of the nanotopographical constraints, such as pore depth, pore size, and interpore distance. For example, the pore size (and spacing) in AAO films can be finely tuned by anodizing conditions ranging from 20 to 400 nm (21), or even smaller via ion irradiation (22), whereas the pore depth can be tuned by anodization time or by filling the pores with other polymeric materials (SI Appendix, Fig. S3). Surface functionalization can also play a role in microvilli formation, for example, hydrophobic pores were created via silane modification, inhibiting penetration of protrusions into the porous membrane (SI Appendix, Fig. S3).
In order to establish the link between nanotopographical constraints and microvilli dimensions, Jurkat T cells were cultured on AAO membranes with different pore sizes, i.e. 20, 100, 200, and 400 nm, as well as on PDMS membranes with 1-μm pores. Notably, T cells were not able to form protrusions on smaller pores, i.e. 20 and 100 nm pores, most probably due to the fact that the pores could not physically accommodate microvilli and their protrusive actin-based machinery (SI Appendix, Fig. S4). On the other hand, 1-μm pores can be penetrated but are not sufficient to induce microvilli formation. This is particularly evident from the microscope images, where a micrometer-size pore is only partially infiltrated by a cell protrusion (SI Appendix, Fig. S4), suggesting nanoscale topographical constraints are required to stabilize microvilli. The 200- and 400-nm pores were particularly potent to induce microvilli formation. On the mechanistic side, nanopores create a physical barrier—either by size of the pore or by the high membrane curvature at the pore edge—that may exclude some cellular components from entering the pores. Notably, microtubules could not enter the 200-nm pores (Fig. 2E), whereas they were found in 400-nm pores (SI Appendix, Fig. S4). Microtubules are one of the main tracks for organelle positioning and protein transport (23, 24).
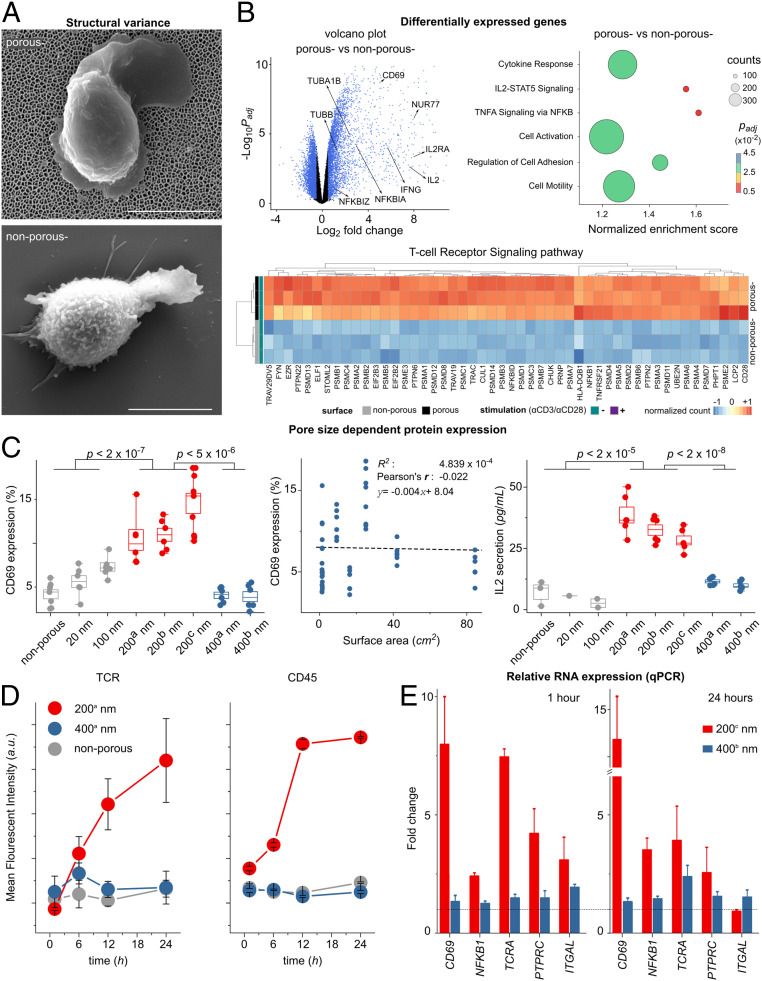
Fig. 2.
Altered T cell gene expression in 200-nm porous substrates. (A) Scanning electron microscope images of nonstimulated primary human T cell on top of the porous (−) and nonporous (−) surfaces (without αCD3/CD28 coatings). (Scale bar, 4 μm.) (B) Nanoconfinement-induced alteration in the gene program of the primary human T cells after 4 h of seeding on the surfaces. Volcano plot shows differences in RNA expression of nonstimulated T cells on porous (−) versus nonporous (−) surfaces. Blue color indicates significance with adjusted P value <0.05 and log2 (fold changes) >0.5. Dot plot of gene set enrichment analysis shows differences in differentially enriched pathways of nonstimulated T cells on porous (−) versus nonporous (−) surfaces. The size of the circle corresponds to the gene counts (from the reference pathway), the color corresponds to the adjusted P value. (Bottom) Heatmap of the top 50 significantly up-regulated genes (adjusted P values <0.05) in the TCR signaling pathway, where genes in porous (−) cells were compared to nonporous (−) cells (n = 3 replicates, human primary T cells). The complete set of the significantly differentially expressed genes in TCR signaling pathway can be found in SI Appendix. (C) Jurkat T cells were seeded on porous and nonporous surfaces (without activating antibodies αCD3/CD28) with the indicated pore size (Table 1). CD69 and IL-2 were measured 24 h after activation by flow cytometry and ELISA, respectively. Box diagrams show data pooled from two or three independent experiments performed in triplicates. The scatter plot represents the pulled data from all experiments, plotted as surface area versus CD69 expression. The dashed line is the best linear fit to the data, which shows correlation between CD69 expression and increased surface area (Pearson’s r = −0.022). (D) The scatter plot shows the kinetics of the mean fluorescent intensity of TCR and CD45 of primary human T cells cultured on the indicated surfaces at different time points, measured by flow cytometry. Data represent one of two independent experiments performed in triplicates. Error bars are mean ± SD. (E) Relative RNA expression of selected genes measured by RT-qPCR. Primary human T cells were cultured on the indicated surfaces for 1 and 24 h. Fold expression is measured against the control sample (nonporous) and normalized by the reference gene RNA18S. The dashed line indicates fold expression =1, the expression of the genes in the control sample. The P values were determined by two-sided Mann–Whitney U tests in R.
Similar to the signaling effect of receptor segregation imposed by 2D micropatterns (25), we might expect that the exclusion of cellular components from microvilli via nanoscale confinement in pores may have significant biological consequences. For example, vesicles that are normally transported via microtubules to the tip of the protrusions may not be transported to the confined regions, potentially altering cellular output. Moreover, size exclusion of membrane proteins can occur, which is very critical for T cell signaling (see Membrane Protein Sorting by Size Exclusion).
Nanoscale Microvilli Formation Alters Gene Expression and T Cell Signaling.
In order to evaluate whether microvilli nanoconfinement has an effect on signaling of primary human T cells, early gene expression analysis was conducted 4 h after T cell seeding. Naive T cells were incubated on nonporous and porous membranes (200 nm), without the presence of TCR activating antibodies (αCD3/CD28) on the surface, referred to as nonporous (−) and porous (−), respectively (Fig. 2A). The expression of ∼6,300 genes distinguished T cells being exposed to porous (−) or nonporous (−) conditions, with ∼3,300 genes having higher expression in cells being exposed to porous (−) versus nonporous (−) conditions. Gene ontology reveals that the set of genes up-regulated in T cells being exposed to porous (−) conditions exhibited enrichment for those encoding proteins involved in diverse molecular functions and cellular processes involving transcription, protein transport, ribosome activity, and cell division, among others (reference Dataset S1 for a complete list). Moreover, T cells being exposed to porous conditions had higher expression of genes encoding for transcription factors, cytokine receptors, and signaling molecules associated with T cell activation. In particular, the TCR signaling pathway was significantly enriched in T cells being exposed to porous surfaces (Fig. 2B and SI Appendix, Fig. S7).
Among the most significant genes that were up-regulated on the porous membrane, CD69 and IL-2 genes stood out as activation markers for T cells. Protein expression (CD69 and IL-2) was measured for both primary human T cells as well as immortalized human T cell line (Jurkat). Primary human T cells (SI Appendix, Fig. S5) and Jurkat T cells (Fig. 2C) exhibited a significant increase in expression of the membrane protein CD69 as well as secretion of IL-2 after 24 h of culture on porous compared to nonporous surfaces. Strikingly, up to 20% of the primary human cells exhibited a significant increase in CD69 expression after 24 h, and ∼10% of the cells expressed CD25 and proliferated after 4 d in conditions promoting formation of microvilli, and remarkably, this is happening in the absence of any TCR agonistic triggers (SI Appendix, Fig. S5).
T Cell Activation in the Absence of TCR Agonistic Antibodies Is Independent of the Surface Area.
Other studies have shown that in the presence of TCR agonistic antibodies, lymphocyte activation has a higher yield on micropatterned substrates due to the greater exposure of the cells to a higher number of the activating antibody owing to the larger surface area (26, 27). To confirm that TCR signaling in the absence of agonistic TCR triggers is indeed induced by nanoconfinement of microvilli, one needs to first assess the possibility of induced stochastic TCR signaling by the greater contact area of the cells with the porous surfaces.
To perform a complete assessment of the relationship between protrusion dimensions and TCR signaling, we used nanoporous membranes with different pore sizes, interpore distances, pore densities, and of different materials in the absence of TCR agonistic antibodies (Table 1). Interestingly, the expression of CD69 and IL-2 secretion by T cells was only enhanced on the porous surface with 200-nm pore size without TCR agonistic antibodies (Fig. 2C). Moreover, we plotted CD69 expression versus the surface area of the porous membrane, which clearly indicates that there is no correlation between the up-regulation of activation markers and the surface area. For example, AAO with 400-nm pores has a four times larger surface area compared to AAO with 200-nm pores (200b nm versus 400b nm in Table 1), but CD69 and IL-2 were not up-regulated on the 400-nm substrates.
Table 1.
List of the surfaces used for microvilli induction
| Sample name | Material | Average pore diameter (nm) | Pore density (cm−2) | Surface area (cm2)* |
| 20 | AAO | 20 | 1011 | 84.5 |
| 100 | AAO | 100 | 1010 | 42.1 |
| 200a | Polycarbonate | 200 | 107 | 1.4 |
| 200b | AAO | 200 | 109 | 9.3 |
| 200c | AAO | 200 | 3 × 109 | 25.1 |
| 400a | Polystyrene | 400 | 107 | 1.5 |
| 400b | AAO | 400 | 109 | 16.4 |
| Reference | Aluminum Oxide or glass | Nonporous | 0 | 1.3 |
In surface area calculations, it is assumed that 1-μm pore depth is an accessible surface for the microvilli, according to the data in Fig. 1E.
Gene transcription analysis revealed that several genes associated with T cell activation—such as CD69, NFKB1, TCRA, PTPRC, and ITGAL—were up-regulated on 200-nm pore surfaces. To ask how the pore size tunes the gene expression in T cells, we performed qPCR analysis on primary human T cells exposed to 200- and 400-nm AAO membranes without TCR agonistic antibodies, 1 h (early regulation) and 24 h (late regulation) after seeding (Fig. 2D). The expression of the target genes was calculated relative to the expression of the genes on the nonporous surface as control. qPCR analysis revealed that the target genes were significantly up-regulated on 200-nm pores, but not on 400-nm pores. To verify the results, we tested the protein expression of TCR and CD45 at different time points (30 min to 24 h), which indicated that cells cultured on 200-nm pores exhibited higher expression of TCR and CD45 (Fig. 2D).
In another attempt, by controlling the depth of the 200-nm pores, we showed that despite an eightfold increase in the surface area (depth from 0.5 to 4 μm), CD69 expression did not change on 200-nm pore membranes (SI Appendix, Fig. S5). These results suggest that 0.5-μm-long protrusions can equally induce enhanced signaling, compared to longer protrusions with an average length of ∼1.3 μm.
To further establish the correlation between nanoconfined microvilli protrusions and T cell signaling, we perturbed the protrusions with the cytoskeleton-interfering agent Latrunculin B (Lat B), which blocks actin polymerization, and observed that CD69 expression was reduced with increasing Lat B concentrations (SI Appendix, Fig. S5). The influence of actin polymerization inhibition on CD69 expression was much more significant on 200-nm pores compared to the other substrates, indicating that the enhanced activation on 200-nm pores is due to microvilli protrusions.
Given the above observations, increased surface area by the pores is not necessarily an improving effect for T cell activation, unless T cells exploit it via microvilli formation. Furthermore, microvilli formation does not necessarily promote T cell signaling, unless its dimensions are in the right range, most pronounced with pores ∼200 nm in diameter. We conclude that nanoconfinement of microvilli via infiltration in the porous substrates with 200-nm pore size induces altered cell signaling and potentially T cell activation. But how does imposing a nanoconfinement lead to T cell signaling, particularly in a TCR-triggering independent manner?
Membrane Protein Sorting by Size Exclusion.
We tested “kinetic segregation” as a model for antibody-independent T cell activation (28). In this model, when the inhibitory phosphatase CD45 and TCR are spatiotemporally segregated, a signaling cascade occurs, leading to activation of T cells (29–32). When CD45 is absent, sporadic nonspecific interactions lead to temporary phosphorylation of TCR associated signaling hubs; in its presence, CD45 shields and inhibits nonspecific TCR signals in its proximity. In case of CD45 spatiotemporal exclusion from TCR regions [e.g., in “close contacts” (7, 8)], sustained downstream TCR signaling and thus T cell activation can take place (8). Spatiotemporal segregation is observed to occur both in vitro and in vivo, even without specific TCR-ligand engagement; however, fast transient triggering does not lead to T cell activation (8, 33, 34).
With the above observations, we hypothesized that T cell microvilli nanoconfinement in 200-nm pores might enforce the spatiotemporal segregation of CD45 and TCR, due to size exclusion. CD45 [∼40 nm (35)] is much larger compared to TCR [∼15 nm (35)]; therefore, it is plausible that segregation could occur in nanoconfined spaces. In fact, it has been shown in previous studies that physical barriers can hinder diffusion of CD45 (12, 25). More recent studies have shown that in areas of high membrane curvature, such as at the tip of a microvilli, segregation of TCR and CD45 occurs, establishing a phosphatase-free zone, prone for TCR triggering (5, 36).
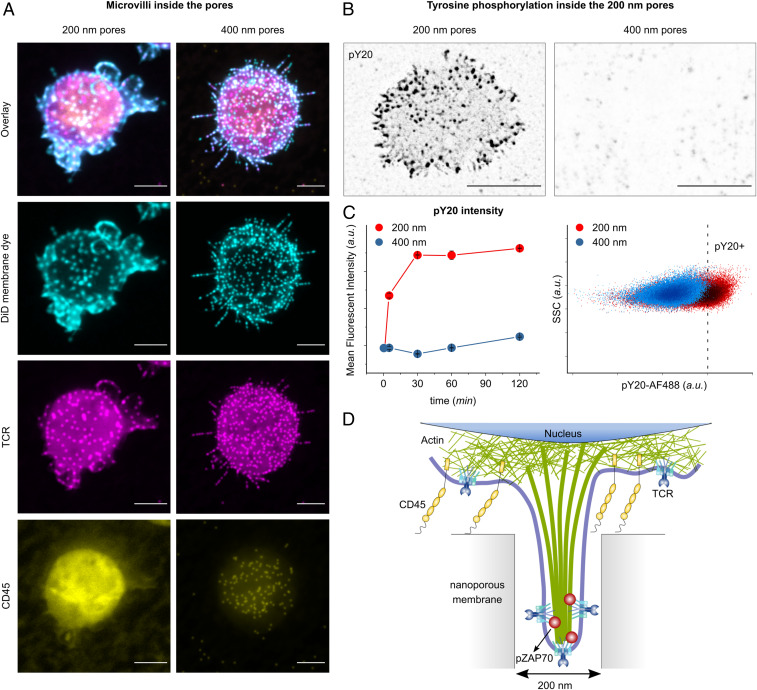
In our experiments, fluorescence images obtained from Jurkat T cells on porous surfaces demonstrated that the nanoconfinement inhibited or reduced the penetration of large molecules such as CD45 in 200-nm pores (Fig. 3A). Smaller molecules such as TCR could be found abundantly inside of the microvilli protrusions—recognizable by the dotted structure in the fluorescence images. The induced exclusion of CD45 was not significant in 400-nm pores. The cross-sectional profile of individual microvilli in 200- and 400-nm pores is shown in SI Appendix, Fig. S4. Exclusion of CD45 from 200-nm pores could be caused by several likely synergistic mechanisms. First, the plasma membrane is highly curved around the pore edges, and it is well known that high membrane curvature can act as a size-selective diffusion barrier (37). Either the high curvature of the plasma membrane around the pore edge creates a size-selective diffusion barrier, as correlations between membrane curvature and proteins are well established (12, 35, 38–40), or the accessible pore volume is largely occupied by the cell membrane and the actin cytoskeleton, leaving little space for the diffusion of proteins with large extracellular domains, such as CD45. Our observations suggest that the highly confined space of the nanopore enforces spatiotemporal segregation of the signaling complex and that this phenomenon is completely lost on 1-µm porous membranes (SI Appendix, Fig. S4).
Fig. 3.
Membrane-bound protein sorting in confined spaces. (A) The projection of confocal fluorescence microscopy images of the cell membrane (stained with DiD), TCR, and CD45, from fixed T cells on porous surfaces with (Left) 200-nm and (Right) 400-nm pore size, 30 min after seeding. (Scale bar, 5 μm.) (B) The projection of confocal fluorescence microscopy images of pY20, from fixed T cells on 200- and 400-nm pores, fixed 5 min after seeding. The fluorescent signal from inside of the pores is indicated by black dots. (Scale bar, 5 μm.) (C, Left) The scatter plot shows the kinetics of the mean fluorescent intensity of pY20 of primary human T cells cultured on the indicated surfaces at different time points, measured by flow cytometry. Data represent one of two independent experiments performed in triplicates. Error bars are mean ± SD. (Right) Representative dot plots of pY20 measured by flow cytometry on primary human T cells seeded on 200-nm (red) and 400-nm (blue) porous substrates, 30 min after seeding. (D) Schematics of the spatiotemporal segregation model induced by nanoconfinement in 200-nm pores.
Furthermore, to verify that the exclusion of CD45 is due to the large extracellular domain of the protein, we tested antibody-independent activation of CD45 chimera cells with large (∼45 nm) and small (∼7 nm) ectodomains (41) on 200-nm pores (SI Appendix, Fig. S5). T cells containing small ectodomain of CD45 were not stimulated by exposure to the 200-nm pores, unlike the cells with full CD45 (with large ectodomain). Additionally, microscope images indicate that CD45 with small ectodomain were able to enter the 200-nm pores, pointing to the fact that large ectodomain is required for exclusion of CD45 from 200-nm pores (SI Appendix, Fig. S5).
To test whether the spatiotemporal segregation is sustained or transient, cells were stained with anti-phosphotyrosine (pY20) as an indicator of TCR signaling (Fig. 3 B and C), as well as pZAP70 (SI Appendix, Fig. S6). Most significantly, fluorescence microscopy revealed the presence of (phosphotyrosine) pY signal inside of the 200-nm pores in a dotted pattern, whereas no fluorescent signal for pY was observed inside of the 400-nm pores (Fig. 3B). In another attempt, the cells were removed from the surface at different time points (5 to 120 min) and stained with pY20 for flow cytometry analysis (Fig. 3B). Notably, the cells cultured on 200-nm pores exhibited a strong pY signal. This is rather surprising because studies on T cell activation on flat surfaces shows that pY signaling is transient and decays within several minutes after activation. The prolonged pY signaling on the porous surface indicates that the nanoconfinement-induced segregation leaves the TCRs accessible to kinases while protecting them from phosphatases that could potentially reverse the phosphorylation, allowing for sustained signaling during their residence inside of the pores (Fig. 3D).
Augmented and Sustained ERK Phosphorylation Induced by Nanoconfinement.
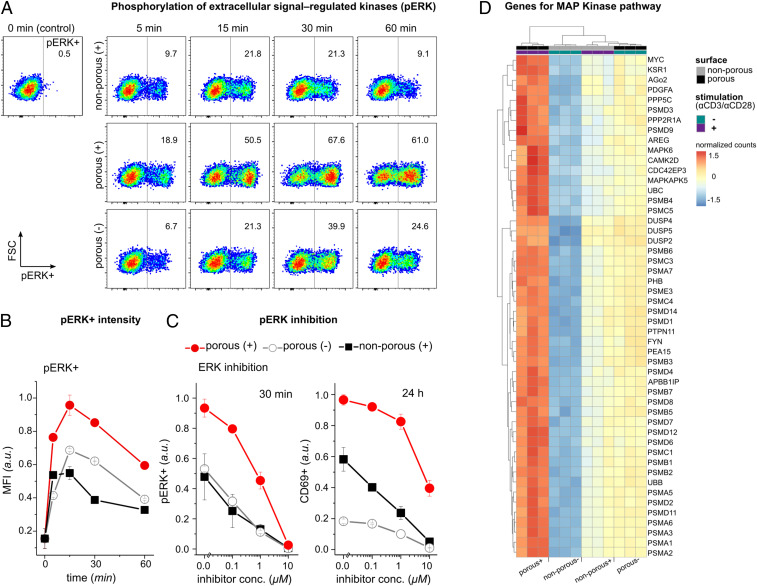
Next, we asked whether TCR phosphorylation inside of the 200-nm pores can initiate downstream signaling to activate T cells with/without TCR agonistic antibodies. Therefore, we looked into the extracellular-signal-regulated kinase (ERK) pathway in T cells exposed to 200-nm porous surfaces and compared it with agonistic TCR antibody-activated T cells as a reference. The ERK pathway is part of the mitogen-activated protein kinase (MAP Kinase) pathway, a major signaling cascade in regulation of T cell activation, differentiation, and proliferation (42). ERK signaling is activated during antigen recognition by T cells, where the strength of the signal and its duration depend on the affinity and avidity of the TCR–antigen interactions (33, 43). Additionally, recent studies propose a correlation between ERK phosphorylation and podosome maturation (i.e., nanoscale actin-rich protrusions) (44–46).
The cells were cultured on either TCR agonistic antibody (i.e., αCD3/αCD28) coated (+) and noncoated (−) porous (200 nm) or nonporous surfaces. T cells activated on AAO with 200-nm pores with TCR agonistic antibody [shown as porous (+)] exhibited a significantly higher phosphorylation of ERK, which was sustained much longer than in cells activated on the nonporous (+) surface (Fig. 4 A and B). ERK signaling in nonporous (+) peaked at 15 min and ceased in most cells after 60 min. On the other hand, porous (+) had a higher amount of ERK phosphorylation (pERK) per cell (as measured by mean fluorescent intensity), which was sustained for longer than in nonporous (+). Interestingly, cells cultured on AAO with 200-nm pores with TCR agonistic antibody [shown as porous (−)] also exhibited amounts of pERK comparable to those in nonporous (+) cells. ERK was not activated in resting cells on the nonporous (−) surfaces (Fig. 4A, the control experiment).
Fig. 4.
ERK phosphorylation is augmented and sustained on the porous membranes. (A) Representative dot plots of phosphorylated ERK measured by flow cytometry on Jurkat T cells under the indicated conditions. Data are representative from two independent experiments performed in duplicates or triplicates. (B) The bar diagram shows the kinetics of the mean fluorescent intensity (MFI) of ERK-phosphorylated cells (pERK+) with the indicated treatment. Data represent one of two independent experiments performed in duplicates or triplicates. Error bars are mean ± SD. (C) Jurkat T cells were incubated with ERK inhibitor U0126 in different concentrations for 30 min before being activated on the indicated surfaces. Bar diagrams show percentages of pERK+ cells after 30 min and CD69+ cells after 24 h measured by flow cytometry (n = 3 replicates). Error bars are mean ± SD. (D) Heatmap of the top 50 significantly up-regulated genes in the MAP Kinase pathway of primary human T cells activated on the indicated surfaces for 4 h, adjusted P values <0.05. Genes were selected based on the comparison between porous (+) and nonporous (−) (n = 3 replicates). The complete set of the significantly differentially expressed genes in MAP Kinase pathway can be found in SI Appendix.
To further elucidate the correlation between nanoconfined microvilli protrusions and ERK signaling, we used ERK signaling inhibitor U0126 before culturing T cells on the surfaces. As expected, we observed reduced ERK phosphorylation in a concentration-dependent fashion (Fig. 4C). ERK inhibition correlated with reduced percentages of CD69+ cells 24 h after activation in all conditions. Moreover, pathway enrichment analysis for differentially expressed genes indicated that the MAP Kinase pathway was enriched in all of the three groups [i.e., porous (+), porous (−), and nonporous (+) compared to nonporous (−), as shown in Fig. 4D and SI Appendix, Fig. S8]. There was a large overlap in the corresponding gene expression patterns between porous (−) and nonporous (+), whereas expression levels were much higher on porous (+). These results suggest that nanotopographical constraints have a central role in the enhanced and sustained MAP Kinase signaling.
T Cell Activation Is Boosted by Nanotopographical Constraints.
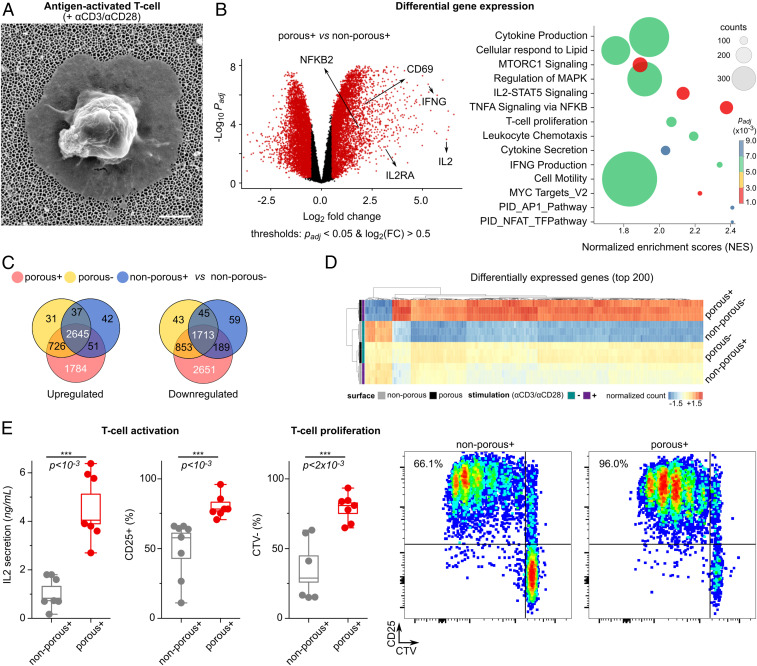
The process of T cell activation and signaling in response to the physical nanoconstraints can be exploited for many applications, including the topographic design of biomaterials for enhanced activation and proliferation of the cells (47–51). We examined here the suitability of 200-nm pore AAO substrates for T cell activation, to validate its potential for application in immunotherapy, by enhancing activation of T cells via imposing a physical constraint (Fig. 5). For polyclonal T cell activation and expansion, the surfaces of porous and nonporous aluminum oxide films were coated using activating antibodies against CD3 (αCD3; TCR stimulus) and CD28 (αCD28; costimulatory cue).
Fig. 5.
Boosted T cell activation and proliferation by combining nanotopographical and biochemical cues. (A) Scanning electron microscope image of an activated primary human T cell on top of the nanoporous membrane with 200-nm pore size. (Scale bar, 1 μm.) (B, Left) Volcano plot showing differences in RNA expression of T cells activated with antibodies on porous (+) versus nonporous (+) surfaces. Red color indicates significance with adjusted P value <0.05, and log2 (fold changes) >0.5. (Right) Pathway enrichment of differentially expressed genes in porous (+) cells versus nonporous (+) T cells shows increased stimulation of porous (+) T cells. The size of the circle corresponds to the gene counts (from the reference pathway), the color corresponds to the adjusted P value. (C) Venn diagrams of the differentially expressed genes (up-regulated and down-regulated) in three conditions [porous (+), porous (−), and nonporous (+) compared to the nonporous (−)]. (D) The top 200 significantly up-regulated genes in porous (+) compared to the nonporous (−), based on adjusted P values <0.05 (n = 3 replicates). +/− signs indicate the presence of activation antibodies (αCD3/CD28) on the surface. The complete set of the significantly differentially expressed genes can be found in SI Appendix. (E) Box diagrams show activation of human primary T cells on porous surfaces. IL-2 secretion was measured after 24 h, and CD25 expression was measured after 4 d. Three independent experiments were performed in duplicates or triplicates. The P values were determined by two-sided Mann–Whitney U tests in R. The proliferation assay (CTV, CellTrace Violet) was used to assess the expansion of the cells after 4 d. The graphs represent examples of the measurements by flow cytometry.
Differential gene expression analysis revealed that more than 2,700 genes were up-regulated in T cells cultured on porous (+) (i.e., AAO 200 nm now in the presence of TCR agonistic antibodies), compared to cells cultured on nonporous (+) surfaces, whereas more than 2,300 genes were down-regulated. Venn diagrams of the differentially expressed genes (compared to nonstimulated cells) indicate the significance of the nanotopography-induced gene regulation. The list of the differentially expressed genes is provided in Dataset S1. Gene ontology comparison showed that the set of genes up-regulated by porous (+) compared to nonporous (+) surfaces are enriched for biological processes such as regulation of cellular amino acid metabolic process, NIK/NF-kappaB signaling, TCR signaling pathway, and cell division (SI Appendix, Fig. S7).
After activation with TCR agonistic antibodies, T cells exhibited a significant increase in expression and secretion of IL-2 after 24 h of culture on porous compared to nonporous surfaces (Fig. 5E). Higher IL-2 levels correlated with more proliferating primary T cells with high CD25 expression, as measured 4 d after activation via a dye dilution assay (Fig. 5E). The observed boosted T cell response on 200-nm pores is therefore due to the concurring biochemical and biomechanical signals that are induced by TCR activation and nanotopographical constraints, respectively. Moreover, it is worth pointing out that cells activated on porous and nonporous share similar phenotypic characteristics despite enhanced activation and proliferation (SI Appendix, Fig. S9).
Conclusions
In summary, we show here that nanoporous membranes with 200-nm pores promote the creation of TCR nanoclusters at the tips of the microvilli protrusions, thereby boosting T cell signaling, activation, and proliferation. TCR-independent T cell signaling is facilitated via pore size–dependent segregation of membrane proteins, amplifying and sustaining the necessary signaling events that lead to T cell activation in the absence of cognate antigen-mediated T cell activation. We discovered that kinetic segregation can be locally induced when T cells protrude into nanoporous substrates. Notably, cell signaling cascades and gene regulation are maximally altered in cells cultured on nanoporous membranes with 200-nm pores, including ERK phosphorylation as a component of MAP Kinase pathway and T cell activation. Future studies will need to address what other proteins/molecules might be involved in the membrane-curvature–induced segregation of the TCR and CD45 phosphatase signaling complex and to what extent they resemble the previously described microsynapses/podosynapses that are enriched in TCR signaling molecules (1, 52). Our discovery is of fundamental importance in immunology and cell biology because we established a direct causality between size-dependent microvilli formation and markedly altered gene expression in T cells, which supports T cell activation. Also, the synergistic combination of protrusion formation and antibody stimulation presents a simple strategy to optimize in vitro T cell activation and may be of value for protocols for adoptive immune cell therapy.
Methods
Materials.
Porous AAO samples were round 13-mm diameter Whatman Anodisc Circles (Sigma-Aldrich), with 20-nm (WHA68097003), 100-nm (WHA68097013), and 200-nm (WHA68097023) pore size. Nonporous aluminum oxide samples were prepared by deposition of 40 nm of Al2O3 on a 13-mm round coverslip (1.5 H, 0117530, Marienfeld) using ALD (Picosun Sunale R-150B). In order to assess the effect of surface area, different materials (Table 1) were used with different pore size and pore density: AAO 200 nm (A2041011, ACS Material), AAO 400 nm (A2061011, ACS Material); ion-track etched Polycarbonate membrane filters 200 nm, 13 mm (PCT0213100, Sterlitech); and ion-track etched Polystere membrane filters 400 nm, 13 mm (PET0413100, Sterlitech). More information on the materials in SI Appendix.
Cleaning the Surfaces.
The samples were cleaned in an air plasma (3 min at 18 W, using a PDC-32G; Harrick Plasma). For glass coverslips, the samples were sonicated in acetone and isopropanol solutions (3 min each), rinsed with MilliQ water, and dried with a nitrogen flow prior to the plasma cleaning. All samples were autoclaved at 120 °C for 2 h for experiments with primary human T cells.
Antibody Coating of the Surfaces.
To coat the surface of the samples with activating antibodies (αCD3 and αCD28), a streptavidin intermediate was used. The samples were placed on a stretched parafilm over a plate. Streptavidin (Thermo Fisher Scientific, 434301) was diluted in phosphate-buffered saline (PBS, ROTI Cell, Carl Roth) to the concentration of 10 μg/mL. Then, 100 μL diluted streptavidin was added directly on top of the samples and was incubated for 30 min at room temperature. The samples were washed with 100 μL PBS (three times) before adding the activating antibodies. The quality of the coating was checked with fluorescent microscopy (SI Appendix, Fig. S2). Monoclonal CD3 and CD28 antibodies were used as activating antibodies. In total, 100 μL of 5 μg/mL biotinylated CD3 (Thermo Fisher Scientific, 13-0037-82) and CD28 (Thermo Fisher Scientific, 13-0289-82) antibodies were added on top of each sample and were incubated for 20 min at room temperature. The samples were then washed with PBS three times and were transferred to 24-well plates for cell seeding.
Cell Seeding.
T cells were freshly plated at a density of 2.5 × 105 cells per well in a 24-well plate containing the 13-mm samples at the bottom. The medium consisted of Roswell Park Memorial Institute medium (RPMI 1640, Invitrogen) with 10% fetal bovine serum (FBS) (ATCC-LGC Standards). Cells were incubated for the planned duration at 37 °C and 5% CO2, without any further supplements.
Human Primary T Cells.
Collection of plasma and PBMCs (peripheral blood mononuclear cell) was approved by the Kantonale Ethikkommission Zürich (KEK-ZH-Nr. 2012-0111), and written consent was obtained from all subjects. A total of 10 separate healthy donors participated in this study. All experiments were approved by the Kantonale Ethikkommission Zürich (KEK-ZH-Nr. 2012-0111).
Cell Culture.
Isolated primary T cells were cultured in RPMI 1640 supplemented with 10% FBS (ATCC-LGC Standards), 1× penicillin/streptomycin, 2-ME (50 mM), nonessential amino acids (ThermoFischer Scientific), sodium pyruvate (ThermoFischer Scientific), Hepes (ThermoFischer Scientific), and glutamate (ThermoFischer Scientific). Isolation protocol can be found in SI Appendix. Jurkat T cells (an immortalized line of human T cell) were cultured in RPMI 1640 (Invitrogen) supplemented with 10% FBS and 1× penicillin–streptomycin (Invitrogen) and were kept in an incubator at 37 °C and 5% CO2 for at least 3 d before measurements. To maintain the concentration of less than 106 cells/mL, cells were split 1:4 with fresh media every 3 to 4 d. Prior to the experiments, cells were counted, centrifuged at 300 × g for 3 min at room temperature, and then resuspended in the fresh medium to concentration of 5 × 105 cells/mL. The amount of 0.5 mL of the cell suspension was then seeded into 24-well plates, which contained the 13-mm samples in the bottom. The cells were incubated at 37 °C and 5% CO2 before further analysis.
High-Throughput RNA-Seq (RNA sequencing).
RNeasy Mini Kit (Qiagen) was used for RNA isolation (reference SI Appendix for detailed protocols). llumina TruSeq mRNA (messenger RNA) protocol was used for library preparation, and sequencing was performed in an Illumina NovaSeq6000 (200 million reads and 100 cycles). The workflow for data analysis is shown in SI Appendix.
Real-Time qPCR.
Real-time qPCR (RT-qPCR) was performed on primary human T cells cultured on 200- and 400-nm AAO surfaces, 1 and 24 h after seeding. Total RNA extraction was performed using RNeasy Mini Kit (Qiagen). The purity of the isolated RNA was assessed with 260-/280-nm absorption ratio (>2.0) using Nanodrop. iScript Advanced cDNA (complementary DNA) Synthesis Kit for RT-qPCR (172-5038, Bio-Rad) was used to prepare cDNA. RT-qPCR was performed using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) in a CFX96 Connect RT PCR Detection System (Bio-Rad). The reference gene 18S RNA (RNA18S) was chosen for normalization of the RT-qPCR data. Primers were synthesized by Microsynth (100 nmol/mL). The sequences used for the amplification of target gene mRNAs are shown in SI Appendix, Table S1.
Measurements.
Details of the analytical measurements for flow cytometry, ELISA (enzyme-linked immunosorbent assay), immunostaining, optical microscopy, and electron microscopy are shown in SI Appendix, Experimental.
Supplementary Material
Acknowledgments
This project was funded by Swiss National Science Foundation Spark Grant CRSK-3_190394 to M.A. and D.S. This work was supported by the Human Frontiers Science Program RGY0065/2017 to E.K. M.A. is thankful to The Holcim Foundation for the Promotion of Scientific Training for partially funding the project. We are grateful to members of our laboratories for assistance during the project, in particular Lina Aires, Johanna Mehl, Arianna Menghini, Cameron Moshfegh, Lion Raaz, Norma De Giuseppe, Chantel Spencer-Hänsch, and Isabel Gerber. We acknowledge the valuable and insightful discussions with János Vörös, Jonas Ries, Michael Dustin, and Gerhard Schütz. We thank Daniel Hoces and Leonhard Sigel for the valuable clinical assistance. We are thankful to Tobias Wolf and Federica Sallusto for the discussions as well as materials. We thank Simon J. Davis for the kind donation of cell lines. We acknowledge support of the Scientific Center for Optical and Electron Microscopy ScopeM of the Swiss Federal Institute of Technology, Eidgenössche Technische Hochschule Zürich (ETH), and Tobias Schwarz. We thank the Functional Genomics Center Zürich at ETH Zürich and the University of Zürich, as well as Emilio Yángüez and Ge Tan for conducting the high-throughput RNA-seq.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107535118/-/DCSupplemental.
Data Availability
The RNA-seq data reported in this paper and all codes for RNA-seq analysis and image analysis are available on github (https://github.com/mortimus-p/T-cell-microvilli) (53). All other study data are included in the article and/or supporting information.
References
- 1.Sage P. T., et al., Antigen recognition is facilitated by invadosome-like protrusions formed by memory/effector T cells. J. Immunol. 188, 3686–3699 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ueda H., Morphew M. K., McIntosh J. R., Davis M. M., CD4+ T-cell synapses involve multiple distinct stages. Proc. Natl. Acad. Sci. U.S.A. 108, 17099–17104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leithner A., et al., Dendritic cell actin dynamics controls T cell priming efficiency at the immunological synapse. J Cell Biol 220, e202006081 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aramesh M., et al., Functionalized bead assay to measure three-dimensional traction forces during T-cell activation. Nano Lett. 21, 507–514 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Jung Y., Wen L., Altman A., Ley K., CD45 pre-exclusion from the tips of microvilli establishes a phosphatase-free zone for early TCR triggering. Nat. Commun 12, 3872 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung Y., Wen L., Altman A., Ley K., CD45 pre-exclusion from the tips of T cell microvilli prior to antigen recognition. Nat. Commun. 12, 3872 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Razvag Y., et al., T cell activation through isolated tight contacts. Cell Rep. 29, 3506–3521.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Chang V. T., et al., Initiation of T cell signaling by CD45 segregation at ‘close contacts’. Nat. Immunol. 17, 574–582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orbach R., Su X., Surfing on membrane waves: Microvilli, curved membranes, and immune signaling. Front. Immunol. 11, 2187 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang F.-K., et al., Targeted inhibition of fascin function blocks tumour invasion and metastatic colonization. Nat. Commun. 6, 7465 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Kumari S., et al., Actin foci facilitate activation of the phospholipase C-γ in primary T lymphocytes via the WASP pathway. eLife 4, e04953 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai H., et al., Full control of ligand positioning reveals spatial thresholds for T cell receptor triggering. Nat. Nanotechnol. 13, 610–617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong N., Sheppard N. C., Billingsley M. M., June C. H., Mitchell M. J., Nanomaterials for T-cell cancer immunotherapy. Nat. Nanotechnol. 16, 25–36 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Fadel T. R., et al., A carbon nanotube-polymer composite for T-cell therapy. Nat. Nanotechnol. 9, 639–647 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Fritzsche M., et al., Cytoskeletal actin dynamics shape a ramifying actin network underpinning immunological synapse formation. Sci. Adv. 3, e1603032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai E., et al., Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science 356, eaal3118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamzalit F., et al., Interfacial actin protrusions mechanically enhance killing by cytotoxic T cells. Sci. Immunol. 4, eaav5445 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin W., et al., T cell activation and immune synapse organization respond to the microscale mechanics of structured surfaces. Proc. Natl. Acad. Sci. U.S.A. 116, 19835–19840 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaudhuri P. K., Wang M. S., Black C. T., Huse M., Kam L. C., Modulating T cell activation using depth sensing topographic cues. Adv. Biosyst. 4, e2000143 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fritz-Laylin L. K., et al., Actin-based protrusions of migrating neutrophils are intrinsically lamellar and facilitate direction changes. eLife 6, e26990 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee W., Park S.-J., Porous anodic aluminum oxide: Anodization and templated synthesis of functional nanostructures. Chem. Rev. 114, 7487–7556 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Aramesh M., Mayamei Y., Wolff A., Ostrikov K. K., Superplastic nanoscale pore shaping by ion irradiation. Nat. Commun. 9, 835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanchard N., Di Bartolo V., Hivroz C., In the immune synapse, ZAP-70 controls T cell polarization and recruitment of signaling proteins but not formation of the synaptic pattern. Immunity 17, 389–399 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Fourriere L., Jimenez A. J., Perez F., Boncompain G., The role of microtubules in secretory protein transport. J. Cell Sci. 133, jcs237016 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Mossman K. D., Campi G., Groves J. T., Dustin M. L., Altered TCR signaling from geometrically repatterned immunological synapses. Science 310, 1191–1193 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Le Saux G., et al., Nanoscale mechanosensing of natural killer cells is revealed by antigen-functionalized nanowires. Adv. Mater. 31, e1805954 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Dang A., et al., Enhanced activation and expansion of T cells using mechanically soft elastomer fibers. Adv. Biosyst. 2, 1700167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis S. J., van der Merwe P. A., The kinetic-segregation model: TCR triggering and beyond. Nat. Immunol. 7, 803–809 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Campi G., Varma R., Dustin M. L., Actin and agonist MHC-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. J. Exp. Med. 202, 1031–1036 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varma R., Campi G., Yokosuka T., Saito T., Dustin M. L., T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity 25, 117–127 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choudhuri K., Wiseman D., Brown M. H., Gould K., van der Merwe P. A., T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature 436, 578–582 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Grakoui A., et al., The immunological synapse: A molecular machine controlling T cell activation. Science 285, 221–227 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Stefanová I., et al., TCR ligand discrimination is enforced by competing ERK positive and SHP-1 negative feedback pathways. Nat. Immunol. 4, 248–254 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Santos A. M., et al., Capturing resting T cells: The perils of PLL. Nat. Immunol. 19, 203–205 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Junghans V., Santos A. M., Lui Y., Davis S. J., Jönsson P., Dimensions and interactions of large T-cell surface proteins. Front. Immunol. 9, 2215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franke C., et al., Unraveling nanotopography of cell surface receptors. bioRxiv [Preprint] (2020). 10.1101/2019.12.23.884460. Accessed 28 September 2021. [DOI]
- 37.McMahon H. T., Boucrot E., Membrane curvature at a glance. J. Cell Sci. 128, 1065–1070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quemeneur F., et al., Shape matters in protein mobility within membranes. Proc. Natl. Acad. Sci. U.S.A. 111, 5083–5087 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryu Y.-S., et al., Reconstituting ring-rafts in bud-mimicking topography of model membranes. Nat. Commun. 5, 4507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Derganc J., Čopič A., Membrane bending by protein crowding is affected by protein lateral confinement. Biochim. Biophys. Acta 1858, 1152–1159 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Irles C., et al., CD45 ectodomain controls interaction with GEMs and Lck activity for optimal TCR signaling. Nat. Immunol. 4, 189–197 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Smith-Garvin J. E., Koretzky G. A., Jordan M. S., T cell activation. Annu. Rev. Immunol. 27, 591–619 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McNeil L. K., Starr T. K., Hogquist K. A., A requirement for sustained ERK signaling during thymocyte positive selection in vivo. Proc. Natl. Acad. Sci. U.S.A. 102, 13574–13579 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelley L. C., Hayes K. E., Ammer A. G., Martin K. H., Weed S. A., Cortactin phosphorylated by ERK1/2 localizes to sites of dynamic actin regulation and is required for carcinoma lamellipodia persistence. PLoS One 5, e13847 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murphy D. A., Courtneidge S. A., The ‘ins’ and ‘outs’ of podosomes and invadopodia: Characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12, 413–426 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buccione R., Orth J. D., McNiven M. A., Foot and mouth: Podosomes, invadopodia and circular dorsal ruffles. Nat. Rev. Mol. Cell Biol. 5, 647–657 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Shah N. J., et al., An injectable bone marrow-like scaffold enhances T cell immunity after hematopoietic stem cell transplantation. Nat. Biotechnol. 37, 293–302 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H., Mooney D. J., Biomaterial-assisted targeted modulation of immune cells in cancer treatment. Nat. Mater. 17, 761–772 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Wang C., Ye Y., Hu Q., Bellotti A., Gu Z., Tailoring biomaterials for cancer immunotherapy: Emerging trends and future outlook. Adv. Mater. 29, 1606036 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Dunn Z. S., Mac J., Wang P., T cell immunotherapy enhanced by designer biomaterials. Biomaterials 217, 119265 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aramesh M., Stoycheva D., Raaz L., Klotzsch E., Engineering T-cell activation for immunotherapy by mechanical forces. Curr. Opin. Biomed. Eng. 10, 134–141 (2019). [Google Scholar]
- 52.Hashimoto-Tane A., Saito T., Dynamic regulation of TCR-microclusters and the microsynapse for T cell activation. Front. Immunol. 7, 255 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aramesh M., Data for Nanoconfinement of Microvilli Alters Gene Expression and Boosts T cell Activation. GitHub. https://github.com/mortimus-p/T-cell-microvilli. Deposited 20 August 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data reported in this paper and all codes for RNA-seq analysis and image analysis are available on github (https://github.com/mortimus-p/T-cell-microvilli) (53). All other study data are included in the article and/or supporting information.