Significance
Increased tissue stiffness is a hallmark of several diseases such as organ fibrosis or cancer, and tissue fibroblasts play central roles in these pathologies. In tumors, stromal cells support the growth of cancer cells by supplying signaling molecules and nutrients, the latter also being provided by stromal autophagy. The mechanisms inducing these processes are, however, not well understood. Here, we demonstrate that an increased physical rigidity of a fibroblast’s environment is sufficient to evoke stromal autophagy. Mechanistically, matrix stiffness is transmitted via Integrin αV receptors and focal adhesion kinase, resulting in stabilization of AMPK at focal adhesion structures. Stiffness-mediated stromal autophagy supports the growth of neighboring tumor cells in vitro and in vivo, opening possibilities for anti-cancer therapies.
Keywords: AMPK, ITGAV, autophagy, tumor stroma, pancreatic stellate cells
Abstract
Increased stiffness of solid tissues has long been recognized as a diagnostic feature of several pathologies, most notably malignant diseases. In fact, it is now well established that elevated tissue rigidity enhances disease progression and aggressiveness and is associated with a poor prognosis in patients as documented, for instance, for lung fibrosis or the highly desmoplastic cancer of the pancreas. The underlying mechanisms of the interplay between physical properties and cellular behavior are, however, not very well understood. Here, we have found that switching culture conditions from soft to stiff substrates is sufficient to evoke (macro) autophagy in various fibroblast types. Mechanistically, this is brought about by stiffness-sensing through an Integrin αV–focal adhesion kinase module resulting in sequestration and posttranslational stabilization of the metabolic master regulator AMPKα at focal adhesions, leading to the subsequent induction of autophagy. Importantly, stiffness-induced autophagy in stromal cells such as fibroblasts and stellate cells critically supports growth of adjacent cancer cells in vitro and in vivo. This process is Integrin αV dependent, opening possibilities for targeting tumor-stroma crosstalk. Our data thus reveal that the mere change in mechanical tissue properties is sufficient to metabolically reprogram stromal cell populations, generating a tumor-supportive metabolic niche.
Changes in the physical properties of tissues, typically occurring during the course of malignant or fibrotic disease, have long been realized and utilized as a diagnostic feature by pathologists. Indeed, solid cancer progression is often associated with an increase in tissue stiffness or rigidity, driven mostly by activated stromal fibroblasts depositing large amounts of extracellular matrix (ECM) proteins such as collagen, fibronectin, or hyaluronic acid (1, 2). Tissue stiffness increases quite early during malignant progression and promotes the aggressiveness of tumors (3). Cancer cells can sense these physical alterations through Integrin receptors, which engage the actin cytoskeleton to relay extracellular information to mechanosensitive pathways such as MKL1/SRF or Hippo/YAP (4, 5). Therefore, drug-induced softening of cancer tissue has emerged as a novel therapeutic concept (6–10). Recently, cellular mechanics have been shown to regulate intracellular metabolism/glycolysis in an actin cytoskeleton–dependent manner, suggesting that physical features of a cell’s substrate can impinge on metabolic pathways (11–13). Two evolutionarily conserved master regulators of cellular metabolism exist: AMP-activated kinase (AMPK) and mTOR, driving catabolic and anabolic processes, respectively (14). As such, AMPK is induced by low energetic states and promotes increased glucose utilization, suppression of lipid and sterol biosynthesis, and the recycling of cellular material through autophagy (15). Functional AMPK is a heterotrimeric complex composed of the catalytic α-subunit and two regulatory subunits (β, γ). In mammalian cells, several isoforms of α, β, and γ subunits exist, which are encoded by distinct genes. AMPK activity is counteracted by mTOR, a multisubunit protein complex responsible for the promotion of cell growth, protein translation, and energy storage (16).
Here, we delineate a role for tissue/substrate stiffness in modulating stroma cell metabolism in an AMPK-dependent and actin-independent mechanism. Specifically, our data indicate that matrix stiffness alone is sufficient to evoke autophagy in stromal fibroblasts, enabling them to create a proproliferative niche supporting neighboring cancer cells. Thus, alterations in physical properties as encountered in malignancy, fibrosis, or scarring can, on their own, induce protumorigenic effects in noncancerous cells.
Results
Stiffness Increases AMPK Levels.
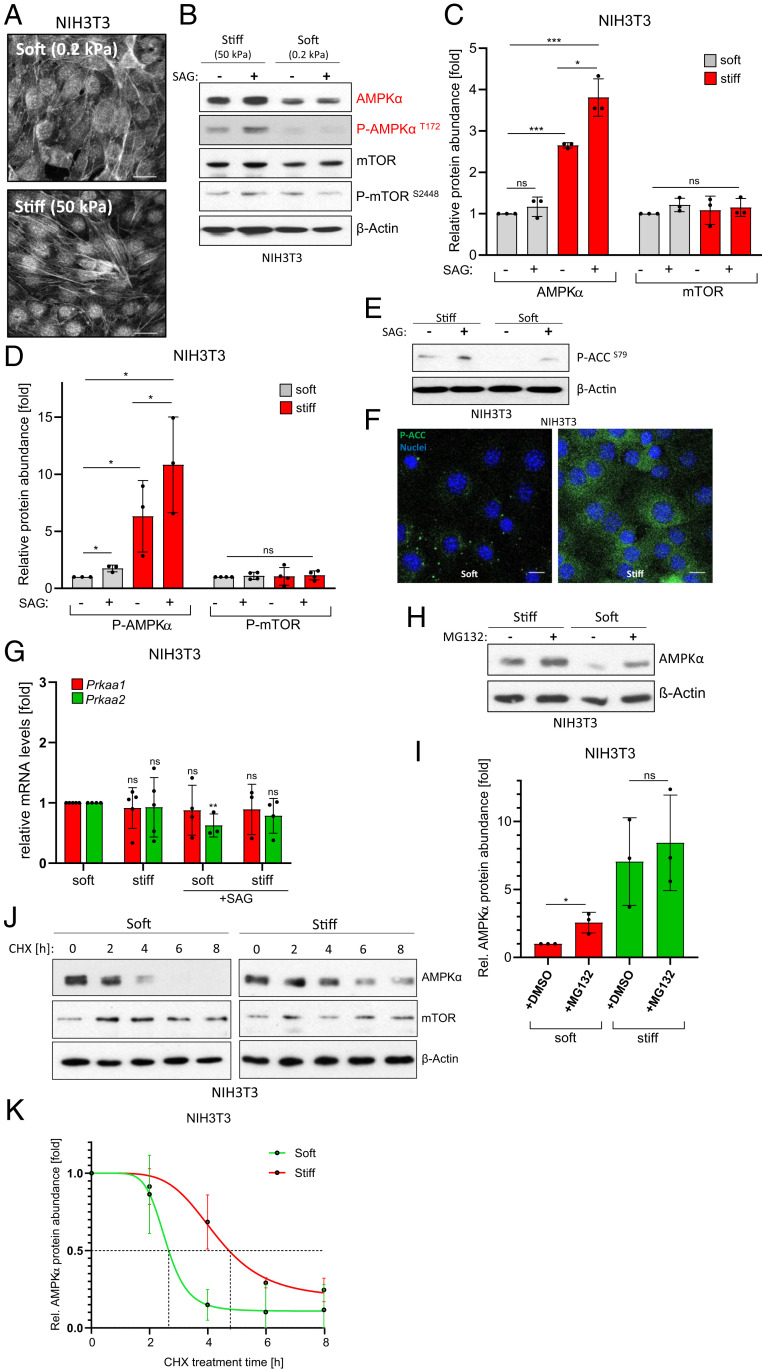
In order to investigate mechanotransduction in mesenchymal cells, we utilized an established in vitro model of substrate mechanics by culturing NIH 3T3 fibroblasts on collagen I–coated hydrogels of varying stiffness. Specifically, we grew cells on substrates possessing an elastic (Young’s) modulus of either 0.2 kPa (mimicking soft or healthy conditions) or 50 kPa (mimicking stiff or fibrotic conditions) (3, 7, 17). As expected, when compared with soft conditions, cells on stiff substrates adopted a more elongated morphology (SI Appendix, Fig. S1A), which was associated with the formation of numerous F-Actin fibers (Fig. 1A). In an attempt to investigate the impact of cell mechanics on metabolism, we analyzed the protein levels of the two master regulators of cellular metabolism, mTOR and AMPK, which are responsible for the general control of anabolic and catabolic processes, respectively. Intriguingly, we observed a significant increase in the amount of AMPKα (the catalytic subunit of the heterotrimeric AMPK complex) selectively under stiff conditions. Importantly, mTOR levels were unaffected (Fig. 1 B and C), demonstrating that stiffness does not unspecifically modulate global protein levels. These findings were not limited to NIH 3T3 fibroblasts and could also be observed in other cell types of mesenchymal origin such as mouse embryonic fibroblasts (MEFs) or human pancreatic stellate cells (hPSCs) (SI Appendix, Fig. S1B). Additionally, we investigated primary cancer-associated fibroblasts (CAFs) and detected a similar stiffness-dependent modulation of AMPKα levels, demonstrating that the observed effects were not caused by the previous use of immortalized cell lines (SI Appendix, Fig. S1C). In order to elucidate whether this increased pool of AMPK was functionally sensitive to physiological signaling cues, we activated endogenous Hedgehog (Hh) signaling by application of a Smoothened agonist (SAG). Hh signaling is an important regulator of CAFs in vivo and is known to trigger AMPK activation in an LKB1/CaMKK2-dependent manner (18). Indeed, Hh signaling induced by SAG promoted AMPKα activation as evidenced by an increase in phosphorylation of the activating Thr172 residue (Fig. 1 B and D). Similar findings were made using a structurally distinct partial SAG (Cyclopamine), which is known to activate AMPKα while blocking canonical Hh pathway transmission to the GLI transcription factors (18) (SI Appendix, Fig. S1D). These data show that stiff substrates increase the amounts of the catabolic master regulator AMPKα (but not mTOR) and that this pool of AMPK is sensitive to physiological stimuli. Next, we asked whether the increased pool of AMPK in cells with high mechanical tension was not only able to receive upstream signals but also to transmit them and evoke an established biochemical output. To this end, we analyzed the inhibitory phosphorylation of the well-established direct AMPK substrate Acetyl-CoA-carboxylase (ACC), a rate-limiting enzyme of fatty acid biosynthesis. As can be seen in Fig. 1 E and F, stiff conditions led to clearly up-regulated phosphorylation of the AMPK target site Ser79 in ACC, in line with stiff conditions favoring increased levels of a fully functional AMPK complex.
Fig. 1.
Substrate stiffness stabilizes AMPKα protein. (A) Black/white confocal images of Phalloidin (F-Actin)- and DAPI-stained NIH 3T3 cells on soft or stiff hydrogels. (Scale bar, 25 µm.) (B) Western blot depicting (phospho-)AMPKα and (phospho-)mTOR protein levels in NIH 3T3 cells. Actin was used as loading control. (C) Quantification of n = 3 independent experiments as shown in B. Shown is the mean ± SD. Significance by paired one-tailed t test. (D) Quantification of n = 3 independent experiments as shown in B. Shown is the mean ± SD. Significance by paired one-tailed t test. (E) Phospho-Acetyl-CoA-Carboxylase (P-ACCS79) levels in NIH 3T3. Shown is one representative experiment. (F) Confocal immunofluorescent image showing P-ACCS79 (green) in NIH 3T3 cells cultured on either soft or stiff hydrogels. Nuclei appear in blue. (Scale bar, 20 µm.) (G) Real-time PCR detecting the expression of Prkaa1 and Prkaa2 in NIH 3T3 cells. Shown is the mean ± SD of three to five independent experiments. (H) AMPKα protein levels in NIH 3T3 cells treated with DMSO or MG132 (10 µM, 8 h). Shown is one representative of n = 3 experiments. (I) Quantification of n = 3 independent experiments as shown in H. Shown is the mean ± SD. (J) AMPKα and mTOR stability as determined in immunoblotting experiments of NIH 3T3 cells cultured on soft/stiff matrices, each exposed to various times of CHX (100 µg/mL). (K) Quantification of AMPKα stability in NIH 3T3 cells as depicted in J. Shown is the mean ± SD of n = 3 experiments.
Interestingly, the observed rise in AMPKα abundance was not mirrored by a transcriptional up-regulation of the two genes encoding the AMPKα1 and AMPKα2 subunits (Prkaa1 and Prkaa2, respectively), implying a posttranscriptional mechanism of stabilization (Fig. 1G). In fact, proteasome inhibition rescued the low AMPKα levels on soft substrate (Fig. 1 H and I). Furthermore, Cycloheximide (CHX) blockade of de novo protein synthesis revealed that the half-life of AMPKα was considerably shorter on soft matrices (t1/2∼2.5 h) than on stiff ones (t1/2∼5 h); Fig. 1 J and K). Again, the protein stability of mTOR was not grossly affected by substrate rigidity, ruling out a general change in protein turnover kinetics (SI Appendix, Fig. S1E). Taken together, our data demonstrate that conditions of high substrate rigidity are sufficient to increase the levels of a functional AMPK pool by means of posttranscriptional protein stabilization.
Stiffness Information Is Conveyed by Integrin αV and FAK.
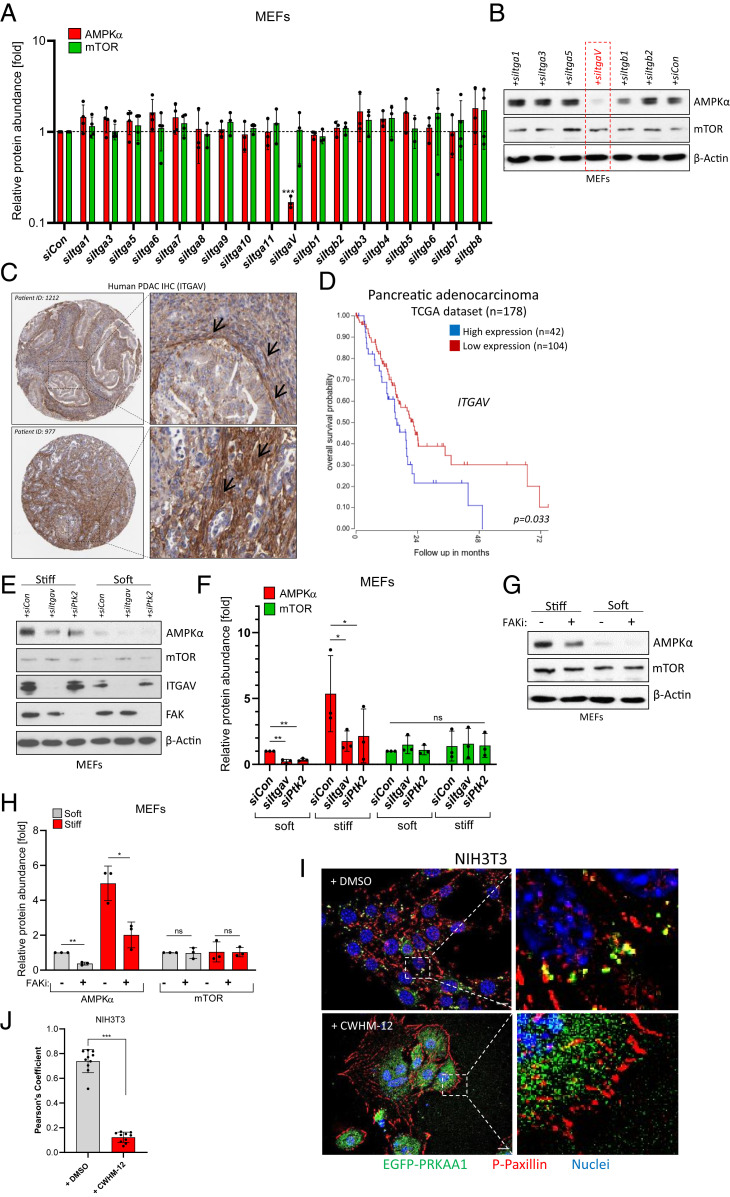
In order to find out more about the underlying mechanotransduction mechanism involved in AMPKα stabilization, we focused on Integrins as the main sensors of extracellular stiffness. To this end, we performed a small interfering RNA (siRNA) screen targeting various Integrin receptors on conventional plastic dishes (supraphysiological stiffness in the GPa range). Here, we used MEF cells as they behaved similarly to NIH 3T3 cells (SI Appendix, Fig. S1B) but were superior in transfection efficiency. Although each of the pools of four different siRNAs we used yielded a good knockdown efficiency (SI Appendix, Fig. S2A), only the knockdown of Integrin αV (Itgav) resulted in down-regulation of AMPKα protein (Fig. 2 A and B), despite the fact that many other Integrins are well-expressed in this cell type (SI Appendix, Fig. S2B). Supporting the specificity of the approach, the levels of mTOR were not affected by any siRNA (Fig. 2 A and B). Again, reduced messenger RNA (mRNA) levels of either Prkaa1 or Prkaa2 were not seen in siItgav-transfected cells (SI Appendix, Fig. S2C), underscoring the posttranscriptional regulation of AMPKα by biophysical substrate properties.
Fig. 2.
Integrin αV-FAK transmits stiffness information to AMPK. (A) Ribonucleic acid interference (RNAi) screen in MEFs cultured on plastic. Pools of four different siRNAs were transfected, and AMPKα or mTOR protein levels were subsequently determined by immunoblotting. Shown is an Actin-normalized quantification of n = 3 to 4 experiments (mean ± SD). Control siRNA = siCon. (B) Representative Western blot of MEF lysates after siRNA transfection on plastic dishes. (C) Immunohistochemical data (Human Protein Atlas) showing ITGAV protein expression in two PDAC patients. Black arrows denote ITGAV expression in stromal cells. (D) Kaplan–Meier curve (TCGA dataset) of PDAC patients depicting overall survival probability in relation to ITGAV gene expression (best cutoff by scan). (E) Levels of AMPKα, mTOR, ITGAV, and FAK in siRNA-transfected MEF cells cultured on soft/stiff substrates. (F) Quantification of n = 3 independent experiments as shown in E. Mean ± SD. Significance by paired one-tailed t test. (G) Levels of AMPKα and mTOR in soft/stiff-cultured MEF cells treated with the FAK inhibitor PF-573228 (FAKi, 10 µM, 16 h). (H) Quantification of n = 3 independent experiments as shown in G. Mean ± SD. (I) NIH 3T3 transiently expressing EGFP-AMPKα(PRKAA1) and stained for endogenous Phospho-Paxillin (P-PaxillinY118, red). Nuclei appear in blue. Images are three-dimensional (3D) deconvoluted images of cells cultured on glass slides. Cells were treated with DMSO or with CWHM-12 (500 nM, 24 h). (Scale bar, 10 µm.) (J) Pearson’s colocalization coefficient of EGFP-PRKAA1 and Phospho-Paxillin as depicted in I. Every data point represents the mean of two ROIs from one microscopic image.
Interestingly, considerable Integrin αV expression could readily be detected in stromal fibroblasts of human pancreatic ductal adenocarcinoma (PDAC) tissue, a disease characterized by an abundant desmoplastic stroma and high tissue stiffness (1, 2, 19) (Fig. 2C). Intriguingly, in some cases, it appeared that ITGAV expression was highest in the myofibroblast subpopulation of CAFs (Fig. 2C, Upper), which are located in close proximity to tumor cells and which, due to their contractile and ECM-synthesizing nature, would be expected to experience the highest mechanical tension (20). However, more detailed analyses revealed that in fact, both myCAFs and inflammatory iCAFs expressed ITGAV in human PDAC tissue (SI Appendix, Fig. S2D). Generally, stromal ITGAV expression was detected in the majority of human PDAC samples analyzed (61/75 cases; SI Appendix, Fig. S2 E and F).
Furthermore, in a dataset of 146 patients, high ITGAV gene expression was moderately but significantly associated with a poor survival in PDAC (Fig. 2D), suggesting that the mechanical ECM–fibroblast crosstalk might be of pathological importance (for survival impact of other Integrin/focal adhesion genes, refer to SI Appendix, Fig. S3 A–J). In order to shed more light on the necessary signaling elements downstream of ITGAV, we knocked down established Integrin mediators such as Integrin-linked kinase, SRC nonreceptor tyrosine kinase, focal adhesion kinase (FAK; encoded by Ptk2), or the actin regulators Cofilin (CFL1) and Profilin (PFN1). Despite good knockdown efficiencies for all siRNA pools (SI Appendix, Fig. S4A), only the knockdown of FAK (Ptk2) resulted in a reduction of AMPKα (but not mTOR) protein levels in MEF cells cultured on plastic dishes (SI Appendix, Fig. S4B), a finding verified also on physiologically soft (0.2 kPa) and pathologically stiff (50 kPa) hydrogels (Fig. 2 E and F). As expected, the levels of phospho-AMPK were also reduced by Itgav or Ptk2 knockdown (SI Appendix, Fig. S4C). These results were corroborated by using a small-molecule FAK inhibitor (PF-573228), which also lowered the abundance of AMPKα and phospho-AMPK protein in cells cultured under conditions of high mechanical tension (Fig. 2 G and H and SI Appendix, Fig. S4D). Surprisingly, the knockdown of actin-regulating molecules such as CFL1 and PFN1 had no significant impact on AMPK levels (SI Appendix, Fig. S4 A and B). Similar findings were obtained using small-molecule inhibitors of the actin cytoskeleton or the acto-myosin system such as Cytochalasin D, Blebbistatin, or the ROCK-inhibitor Y-27632, despite the fact that all of these drugs clearly impinged on the F-actin network within treated cells (SI Appendix, Fig. S4 E and F).
Our findings that ITGAV and FAK play a role in AMPK stability regulation pointed toward a potential involvement of focal adhesions (FAs) in this process. Indeed, we could detect EGFP-tagged AMPKα colocalizing with Phospho-Paxillin, which we used as a marker for FAs. However, this colocalization was only observed as long as the stiffness-sensing ITGAV was not blocked by the use of a small-molecule inhibitor (CWHM-12) (21) (Fig. 2I). In cells with inactive ITGAV, AMPKα-EGFP diffusely relocalized away from FAs toward the cytoplasm, which was reflected by a highly significant drop in Pearson’s colocalization coefficient between AMPKα and the FA marker Phospho-Paxillin (Fig. 2J). Comparable results were obtained when using a pharmacological FAK inhibitor (SI Appendix, Fig. S4G). In summary, the data provided suggest a mechanotransduction model in which extracellular stiffness is sensed by Integrin αV receptors and is subsequently relayed to FAK, resulting in the recruitment of AMPKα to FAs and the actin-independent stabilization/protection of the AMPKα protein.
Substrate Stiffness Induces Autophagy.
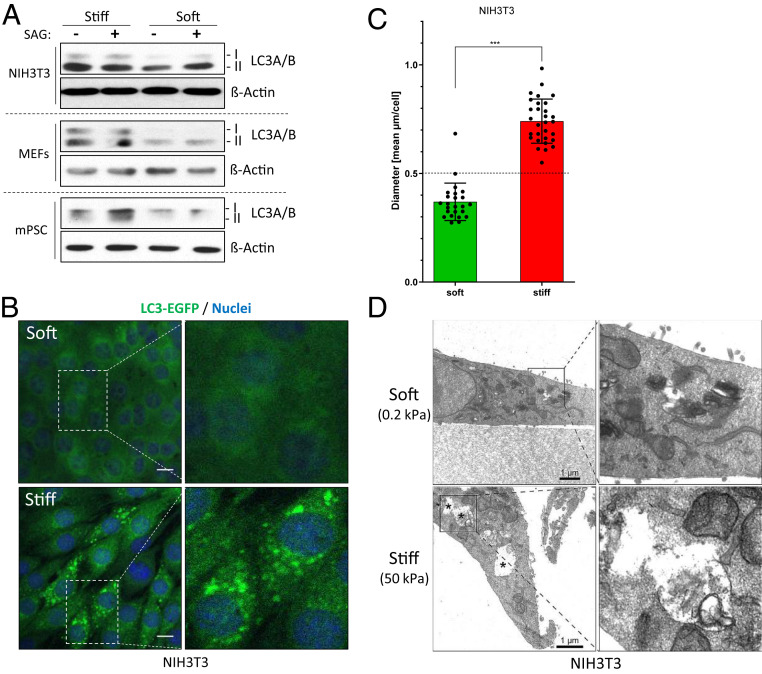
AMPK is a master regulator of catabolic processes, preventing energy storage and releasing metabolic building blocks. In this respect, one central feature of AMPK is its ability to induce macroautophagy (autophagy) to digest intracellular organelles or protein aggregates. In an attempt to investigate whether cell mechanics can regulate autophagy, we analyzed levels of the autophagy marker LC3A/B in cells cultured on soft and stiff substrates. Indeed, total LC3A/B levels (and in particular the autophagy-related conversion product LC3A/B-II form) were increased in stiff conditions (Fig. 3A and SI Appendix, Fig. S5A). As before, this AMPK-triggered event was sensitive to physiological stimuli such as Hh signaling (Fig. 3A and SI Appendix, Fig. S5A). In order to further verify the promotion of autophagy under conditions of high mechanical tension, we analyzed NIH 3T3 fibroblasts stably expressing an LC3B–EGFP fusion protein. To this end, we first determined a cutoff for the size of autophagic vesicles by serum-starving NIH 3T3 fibroblasts (SI Appendix, Fig. S5 B and C) and subsequently used this knowledge for cells cultured on varying stiffness. In agreement with our previous data and when compared to soft conditions, cells cultured on stiff substrate displayed significantly more and larger LC3-positive autophagic vesicles, indicative of a higher autophagic flux (Fig. 3 B and C and SI Appendix, Fig. S5D). In fact, stiffness was sufficient to significantly induce the formation of autophagic vesicles above the cutoff determined before (Fig. 3C). More-abundant and more-intense LC3-positive vesicular structures were also observed on stiff matrices when investigating endogenous LC3A/B (SI Appendix, Fig. S5 E and F). Moreover, we were readily able to document the presence of stiffness-induced autophagolysosomes (APLs) by means of transmission electron microscopy (TEM). As can be seen in Fig. 3D, cells cultured on a soft substrate possessed only a low number of small-sized APLs, whereas stiff-cultured cells displayed a high number of APLs which were considerably larger in size (SI Appendix, Fig. S5G) and contained digested material inside. Collectively, these results show that the mere change in substrate rigidity is sufficient to induce cellular autophagy.
Fig. 3.
Substrate stiffness is sufficient to induce autophagy. (A) Levels of the autophagy marker LC3A/B in various cell lines as determined by Western blotting. Shown is one representative of n = 1 to 3 independent experiments. (B) Confocal images of NIH 3T3 cells stably expressing LC3B-EGFP (green) and grown in soft or stiff conditions. Nuclei appear in blue (DAPI). (Scale bar, 10 µm.) (C) Mean diameter of LC3B-EGFP spots per cell (as shown in B). Each dot represents one cell. (D) TEM images of NIH 3T3 cells cultured on soft or stiff hydrogels. APLs are indicated by asterisks. (Scale bar, 1 µm.)
Stiffness Drives Autophagy-Mediated Stromal Support of Cancer Cell Growth.
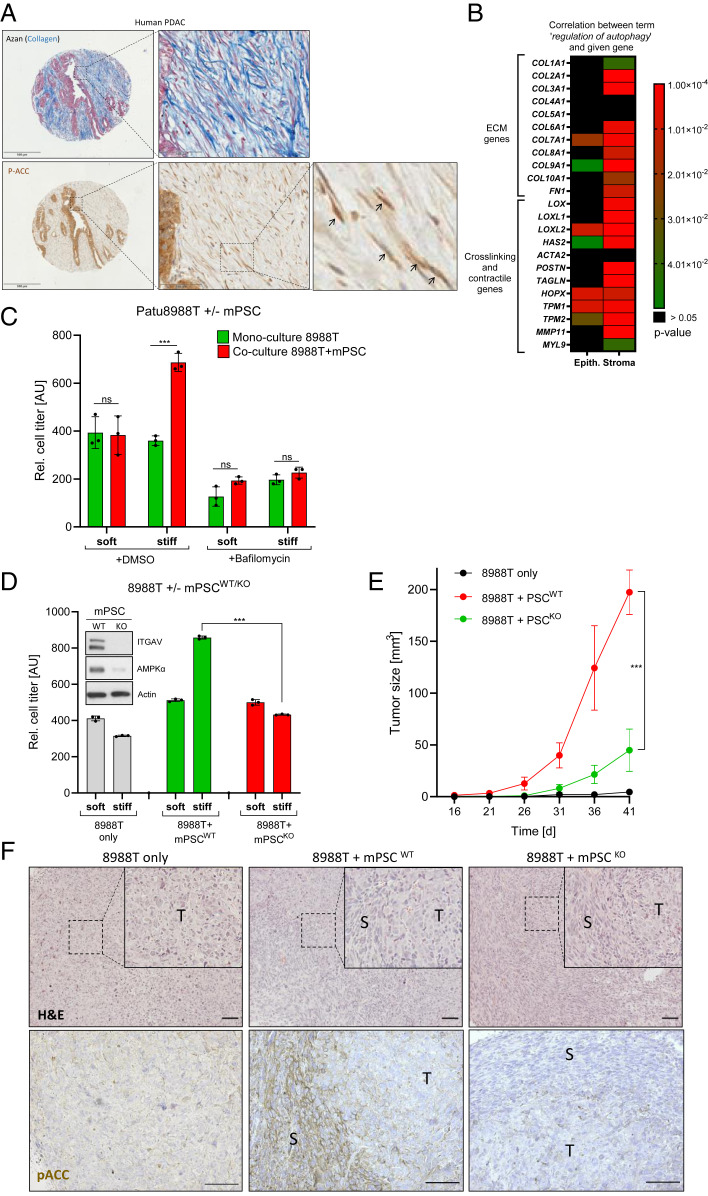
Finally, we wanted to investigate the functional consequences of AMPK-driven stromal autophagy in pancreatic cancer. To this end, we verified the presence of Ser79-phosphorylated ACC as a proxy for AMPK activity in stroma regions of human PDAC (Fig. 4A). Phospho-ACCS79 could readily be detected by immunohistochemistry in elongated mesenchymal cell populations surrounding tumor cell islands and cancer ducts, indicative of AMPK activity in CAFs. In a set of 75 human PDAC patient tissues, the levels of stromal phospho-ACC positively and significantly correlated with the expression of ITGAV in spindle-shaped cells (SI Appendix, Fig. S6 A and B).
Fig. 4.
Stiffness-induced stromal autophagy provides paracrine growth support for cancer cells. (A, Upper) Visualization of collagen fibers by means of Azan staining (blue) in human PDAC. (Lower) Phospho-ACCS79 expression in human PDAC (brown). Black arrows depict phospho-ACCS79–positive stromal cells. (Left scale bar is 500 µm, Middle scale bar is 100 µm.) Serial sections were used for Azan staining and immunohistochemistry (IHC). (B) Heatmap depicting the correlation between the expression of indicated stromal genes and the gene set “regulation of autophagy.” The data set (25) comprises microdissected epithelial and stromal material from n = 66 human PDAC patients each. (C) Cell viability assay using luciferase-labeled human PDAC cells (PaTu8988T). Tumor cells were cultured on soft/stiff substrates either alone or in coculture with mPSCs for 4 d. The autophagy inhibitor Bafilomycin A1 (100 nM, 4 d) was used as a control to demonstrate autophagy dependence. Shown is one representative experiment (mean ± SD of triplicate measurements) of n = 4 independent experiments. (D) Cell viability assay as in C, but with mPSCs either WT or KO for ITGAV. Shown is one representative experiment (mean ± SD of triplicate measurements) of n = 3 independent experiments. Inset depicts protein levels of ITGAV and AMPKα in WT and KO mPSCs grown on plastic. (E) Subcutaneous xenograft growth in athymic nude mice. Animals received either 8988T cells alone (n = 7), in combination with WT mPSCs (n = 7) or in combination with KO mPSCs (n = 6). (F) Representative histology of resulting tumors from E. Upper panel depicts hematoxylin–eosin staining (H&E) while Lower panel depicts phospho-ACCS79 stainings. T = tumor region; S = stroma region. (Scale bar, 100 µm in both panels.)
It is well-established that hepatic stellate cells (HSCs) and PSCs possess lipid droplets in quiescence, a feature which is lost upon activation and transdifferentiation into myofibroblasts, a phenomenon which can be recapitulated by transferring cells from soft matrigel to stiff plastic dishes (20, 22). We reasoned that the underlying mechanism behind this phenomenon may be stiffness-induced AMPK activity phosphorylating and inactivating ACC, thereby suppressing fatty acid biosynthesis and lipid droplet formation (SI Appendix, Fig. S6C). Indeed, treatment of quiescent PSCs with the pharmacological ACC inhibitor PF-05175157 reduced Oil Red O–stained lipid droplets by about 50% (SI Appendix, Fig. S6 D and E), strongly suggesting that stiffness-modulated ACC activity may indeed contribute to lipid droplet formation in PSCs. In line with previous data from HSCs (23), we could not find a functional role for lipid droplets (as assessed by ACC inhibitor treatment) in the polarization of quiescent PSCs toward either myofibroblastic or inflammatory subtypes (myCAFs or iCAFs, respectively) (20, 24) (SI Appendix, Fig. S6 F and G).
Next, we queried a public dataset comprising expression data from laser-microdissected human PDAC tumor and stroma compartments (n = 66 each) (25). Bioinformatics analysis revealed that the gene set Regulation of Autophagy was positively and highly significantly correlated with the mRNA expression levels of many stiffness-enhancing genes (Fig. 4B). For instance, numerous ECM genes (COL2A1, COL3A1, COL9A1, and FN1) as well as ECM-crosslinking (LOX, LOXL1, and LOXL2) and contractility genes (MYL9 and TAGLN [SM22α]) were all positively correlated with the autophagy gene set in the stromal but not in the epithelial compartment (Fig. 4B). Comparable findings were made for several Integrin and Focal adhesion genes (SI Appendix, Fig. S6 H and I). The results obtained from human patients support our in vitro data that stromal stiffness is associated with elevated AMPK activity and autophagy. In this respect, it is interesting to note that stromal autophagy has been linked to enhancement of cancer cell growth by providing critical metabolites and factors (26–28). With this knowledge about growth-promoting stromal effects in mind, we studied the proliferation of luciferase-labeled human PaTu8988T PDAC cells on soft and stiff matrices. When cultured alone, the substrate rigidity had only neglectable impact on their proliferation (Fig. 4C). However, when these cells were grown together with unlabeled murine PSCs, a significant promotion of cancer cell growth was noted (Fig. 4C). Most importantly, this growth-promoting effect of PSCs was only detectable in stiff, but not soft, culture conditions, strongly arguing that the physical properties of the substrate were responsible for this PSC-mediated effect (Fig. 4C). Similar findings were observed when human cancer cells were cocultured with human PSCs (SI Appendix, Fig. S6J). As a further control, we treated the different cell cultures with the autophagy inhibitor Bafilomycin A1 and observed a complete abrogation of the stromal and stiffness-mediated growth promotion (Fig. 4C).
Next, we generated PSCs lacking stiffness-sensing Integrin αV by means of CRISPR/Cas9. In agreement with our previous data, these cells displayed reduced levels of AMPKα and, most importantly, were completely unable to provide any paracrine growth support for cocultured cancer cells (Fig. 4D). Eventually, we wished to validate this concept also in an in vivo setting. To this end, we subcutaneously injected PaTu8988T cells alone or in combination with Itgav wild-type (WT) or knockout (KO) PSCs in nude mice (Fig. 4 E and F). Cancer cell–only tumors displayed a homogenous histology, whereas transplanted tumors containing PSCs possessed a more heterogeneous appearance with streams of cells having mesenchymal morphology (Fig. 4F, Upper). While cancer cells alone did not significantly grow under these experimental circumstances, tumor growth was significantly spurred when WT PSCs were coimplanted. In contrast, this PSC-mediated surplus in tumor growth was almost completely abrogated when KO PSCs were coinjected (Fig. 4E). Using the direct AMPK target Phospho-ACC as a readout for in vivo AMPK activity, we could detect high signals in WT PSCs but only very low signals in Itgav KO PSCs (Fig. 4F, Lower). Taken together, these data strengthen a model of stiffness-induced stromal autophagy supporting tumor cell growth by paracrine metabolic support.
Discussion
In summary, our data describe a function of cell mechanics in metabolic reprogramming through induction of AMPK-driven autophagy. The link between biophysical stimuli and metabolic regulation has recently become evident by focusing on glycolytic enzymes (11, 12) and on vascular cell functions (29). Our data on AMPK stabilization in fibroblasts fits nicely with these data suggesting globally enhanced glucose utilization under stiff conditions. Intriguingly, epithelial cells have been shown to activate AMPK through recruitment to E-Cadherin (30), a finding that is conceptually comparable to our result of Integrin-FAK–mediated AMPK regulation in mesenchymal cells, suggesting that mechano-sensitive membrane complexes (adherens junctions in epithelial cells and FAs in mesenchymal cells) might function as major metabolic regulators in different cell types. It can be hypothesized that these structures might provide protective reservoirs sheltering AMPK from cytoplasmic degradative processes. In cases in which FAs do not properly form (e.g., soft matrices) or under conditions of pharmacological Integrin αV blockade, AMPKα is no longer sequestered at FAs and becomes subject to cytoplasmic proteasomal degradation. Currently, it remains unclear why this process selectively requires Integrin αV (and not other α subunits) and why we could not observe a significant contribution of any β subunit. Future work will also be needed to exactly address the specific role of FAK in such a scenario. FAK could either stabilize AMPK through direct phosphorylation or through indirect effects enabling enhanced maturation of FAs. The presented FA-localized AMPK sequestration concept might even explain why, in our hands, the F-Actin network proved dispensable for AMPK stabilization.
In addition, AMPK-regulated metabolic effects such as fatty acid biosynthesis (ACC inhibition) contribute to well-known cellular processes in stromal tissue compartments, as evidenced by the mechano- and ACC inhibitor–sensitive loss of lipid droplets in HSCs and PSCs. These findings might contribute to the long-known observation that HSCs/PSCs lose their lipid droplets upon culture on stiff plastic dishes. From a functional point of view, it is interesting to note that Integrin αV has previously been identified as a driver of fibrosis in several organs (21), and it will therefore be important to delineate the contribution of ITGAV-regulated autophagy in organ fibrosis. In the cancerous setting, stromal autophagy has been established as a protumorigenic process (26, 27, 31), but the steps leading to its induction have remained obscure. Here, we provide evidence for an important contribution of stiffness-elicited and ITGAV-dependent stromal autophagy, highlighting ITGAV as an interesting drug target to disrupt certain aspects of the tumor-stroma crosstalk. In conclusion, we present evidence for a Stiffness-ITGAV-FAK-AMPK-Autophagy signaling axis providing a stromal metabolic niche in tumors (and potentially other fibrotic diseases), which might be worth considering in future therapeutic approaches.
Materials and Methods
Cell Lines.
NIH 3T3 and Panc1 cell lines were purchased from Cell Lines Service and Patu8988T cells from German Collection of Microorganisms and Cell Cultures (Deutsche Sammlung von Mikroorganismen und Zellkulturen [DSMZ]). Immortalized human (hPSC1) and mouse PSCs (mPSC4) were kindly obtained from Matthias Löhr (32) and Albrecht Neese (33), respectively. MEF cells were kindly provided by Wade Bushman (34). All cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM [high glucose plus glutamine and pyruvate], Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37 °C with 5% CO2 (35). If not otherwise stated, serum concentrations were reduced to 0.5% during experiments for all cell types. All cells were regularly checked for mycoplasma contamination.
Reagents.
Cells were cultured on Collagen I–coated polyacrylamide hydrogels of an elastic modulus of either 0.2 or 50 kPa (Matrigen Petrisoft/Softslip, Cell Guidance Systems). Inhibitors were purchased from the following companies: SelleckChem: PF-573228 (FAKi); Biomol/Cayman: PF-05175157 (ACCi), Y-27632, Bafilomycin A1, Cyclopamine, and CHX; Merck/Millipore: SAG; Adipogen: MG132; and Sigma-Aldrich: Cytochalasin D, Blebbistatin.
Generation of Stable Cell Lines.
PaTu8988T cells were transfected with PGK-Luc (firefly luciferase driven by PGK promoter) and cotransfected with pEF-MCS (= empty pEF6/V5-His [Invitrogen]) using TransIT-2020 [Mirus Bio]) according to the manufacturer’s protocol. Cells were subsequently selected with Blasticidin (Capricorn Scientific GmbH) at a concentration of 5 µg/mL.
NIH 3T3 cells were transfected with pMRX-IP-GFP-LC3-RFP (LC3B-EGFP; Addgene #84573) using TransIT-2020 (Mirus Bio) according to the manufacturer’s protocol. Cells were selected with Puromycin (PAA Laboratories GmbH) at a concentration of 2 µg/mL
Itgav KO Cell Line via CRISPR/Cas9.
For generating Itgav KO mPSC4 cells, the CRISPR/Cas9 gene-editing system was used (target sequence underlined): Primers mItgaV_Crispr1_s (5′-CACCGTGGAGTTTAAGTCCCACCAG) and mItgav_Crispr1_as (5′-AAACCTGGTGGGACTTAAACTCCAC) were annealed and cloned into the BbsI site of pU6-(BbsI)-CBh-Cas9-T2A-mCherry vector (Addgene: #64324). The KO cells were generated according to a strategy described in (36). To this end, the mPSC4 cell line was transfected with the cloned plasmid (pU6-mItgav_Crispr1), pTia-2A-Hygro (kindly provided by Till Adhikary), and empty pU6-(BbsI)-CBh-Cas9-T2A-mCherry (ratio 1:1:0.5) using Helix-In transfection reagent (Oz Biosciences, OZB-HX10500) for 24 h. Subsequently, cells were selected with Hygromycin (VWR International GmbH, J60681.MC) at a concentration of 400 µg/mL until resistant clones appeared. Single cell clones were picked, and the KO was confirmed via Western blot and RT-qPCR.
Western Blotting.
Separation of lysates by sodium dodecyl sulfate–polyacrylamide gel electrophoresis was followed by subsequent blotting on Immobilon–polyvinylidene difluoride membranes (Millipore) and incubation with the respective primary antibody, followed by incubation with a horseradish peroxidase (HRP)-coupled secondary antibody (Cell Signaling Technology). Detection of the HRP signal was performed using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) according to the manufacturer’s protocol (35).
qPCR Analyses.
Total RNA was extracted using NucleoSpin RNA II kit (Macherey–Nagel) according to the manufacturer’s protocol. Complementary DNA (cDNA) synthesis of 1 μg total RNA was performed using iScript cDNA Synthesis Kit (Biorad) following the manufacturer’s guidelines. qPCR reactions were performed using the Absolute QPCR SYBR Green Mix (ABGene). qPCR reactions were performed on 96-well qPCR plates (ABGene) using either the Mx3000P or Mx3005P qPCR systems (Agilent). Results were calculated as relative mRNA expression (2ΔΔCt). Data were obtained from at least three independent experiments and is shown as the mean ± StDev (35).
siRNA Transfections.
Cells were transfected with 35 nM siRNA (Dharmacon SMARTpools and Qiagen control siRNA [All-Stars siRNA; siCon] using RNAiMax (Invitrogen). For siRNA sequences, refer to the SI Appendix.
Microscopy.
Transfections with pEGFP-PRKAA1 (Addgene #30305) were done on plastic plates using TransIT-2020 (Mirus Bio) according to the manual. A total 24 h later, cells were transferred onto glass coverslips or hydrogels and incubated in 0.5% FBS-containing media for 1 to 2 d. Cells were subsequently fixed with 4% formaldehyde/phosphate buffered saline (PBS) for 10 min at room temperature (RT). After washing twice with PBS and permeabilization with 0.5% Triton-X100/PBS at RT for 5 min, hydrogels were blocked with 10% serum/PBS for 1 h at RT and washed once with PBS. Primary antibody was diluted in PBS containing 10% serum and 0.1% Saponin and incubated overnight at 4 °C. After washing twice with PBS at RT for 5 min, the hydrogels were incubated with fluorophore-coupled secondary antibodies diluted in PBS containing 10% serum and 0.1% Saponin at RT in the dark for 2 h. After washing twice with PBS for 5 min and rinsing with H2O, hydrogels were covered with mounting medium containing DAPI (Vectashield). Microscopy was performed on a Leica TCS SP8 confocal microscope (Leica Microsystems, Wetzlar, Germany).
In vitro PSC Polarization.
PSCs (4 × 105) were seeded in a 70-µL matrigel drop (Growth Factor Reduced Basement Membrane Matrix, Corning, 734 to 1,101) mixed at a 1:1 ratio with DMEM (10% FBS) on a 3.5-cm suspension plate (Sarstedt). The matrigel was covered with 0.5% FBS-containing DMEM, treated with ACCi (PF-05175157, 10 µM) and incubated for 48 h. Afterward, cells were treated with either recombinant human TGF-β1 (Peprotech, #100–21) or recombinant human IL-1α (Peprotech, #200–01A) at a concentration of 2 ng/mL or 1 ng/mL, respectively, followed by an incubation for 48 h. Warm medium was removed, and drops were collected in falcon tubes mixed with ice-cold DMEM. After centrifugation (400 rpm, 4 °C, 10 min) and removing of the medium, cells were resuspended in ice-cold DMEM and incubated for 30 min at 4 °C. Cells were centrifuged (400 rpm, 4 °C, 10 min), medium was removed, and pellets were frozen at −80 °C for later RNA preparation.
Oil Red O Staining and Quantification.
Oil Red O staining was performed as described in ref. 20. Briefly, stellate cells were fixed in 10% formaldehyde/Dulbecco's Balanced Salt Solution for 10 min at RT and incubated in 60% Isopropanol at RT for 5 min. The cells were air-dried, incubated with filtered Oil Red O solution (Sigma-Aldrich, O0625-25G), and diluted in water at a 3:2 ratio (Stock Solution: Water) for 10 min at RT. Afterward the cells were washed with water and microscopy was performed on a Leica DM3000 microscope (Leica Microsystems, Wetzlar, Germany). For quantification, cells were incubated in 100% isopropanol for 10 min on an Orbital Shaker. Supernatant was taken, and measurement was performed on a Spectra Max 340 microplate reader (Molecular Devices) at an absorbance wavelength of 510 nm.
Phalloidin Staining.
Cells were seeded on hydrogels and fixed with 4% formaldehyde/PBS for 10 min at RT. After washing twice with PBS, cells were permeabilized with 0.5% Triton-X100/PBS at RT for 5 min and washed twice with PBS. Phalloidin–California Red Conjugate (Biomol, #23103) was diluted 1:1,000 in PBS containing 1% Albumin Fraction V (Roth, 8076.1). Cells were incubated with Phalloidin for 20 min at RT. After washing twice with PBS and rinsing with water, hydrogels were covered with mounting medium containing DAPI (Vectashield). Microscopy was performed on a Leica TCS SP8 con focal microscope (Leica Microsystems, Wetzlar, Germany).
Immunohistochemistry.
For immunohistochemistry, heat-induced epitope retrieval was performed with ethylenediamine tetraacetic acid. Staining was performed on a DAKO Autostainer-Plus. After blocking endogenous peroxidase, sections were incubated for 45 min with rabbit polyclonal Anti-Phospho-Acetyl-CoA Carboxylase Antibody (1:100; Cell Signaling #3661). Sections were washed and incubated with Dako REAL EnVision HRP Rabbit/Mouse polymer, which reacts with 3,3′-diaminobenzidine Chromogen, according to the manufacturer’s protocol. The use of patient material (in form of tissue microarrays [TMAs]) was approved by the local ethics committee (Ethics Board University Hospital Marburg). Material on the TMA was from patients suffering from PDAC and eligible for surgery. Pathological diagnosis of patient from Fig. 4A is the following: G2 ductal adenocarcinoma of the pancreas, pT2, pN0, L0, V0, and Pn0.
TEM.
Cells were seeded on hydrogels and incubated for 48 h. TEM was done by the electron microscopy core facility at the University of Marburg.
Coculture Assays.
Cells were seeded on hydrogels in a cellular ratio of tumor cells (1/5) and stellate cells (4/5) or as tumor cell monoculture. Cells were treated with Bafilomyin A1 (100 nM) or DMSO and incubated for 4 d in 0% FBS DMEM. Afterward, cells were lysed in Passive Lysis Buffer (Promega, E1941) and Firefly Luciferase activity was measured using Beetle-Juice reagent (PJK, Kleinblittersdorf, Germany) on an Orion L microplate luminometer (Berthold Detection Systems).
Xenograft Experiment.
Female athymic nude mice were randomly divided into three groups. A total 0.5 × 106 PaTu8988T cells were suspended in a total volume of 150 μL of DMEM for group 1. 0.5 × 106 PaTu8988T cells + 1 × 106 mPSC4WT cells were suspended in a total volume of 200 μL of DMEM for group 2. 0.5 × 106 PaTu8988T cells + 1 × 106 mPSC4KO (Itgav KO) were suspended in a total volume of 200 μL of DMEM for group 3. The cell suspensions were injected subcutaneous at the posterior flank of the mice. Tumor volumes were calculated by the formula (length × width2)/2. The length represents the longer axis, and the width represents the shorter axis of the tumor. At the experimental endpoint mice were euthanized, and tumors were removed. The study was approved by the regional agency on animal experimentation (Regierungspräsidium Giessen).
Statistics.
Statistical comparisons were made of n ≥ 3 experiments using an unpaired two-tailed Student’s t test unless otherwise stated. Significances were indicated as ns (not significant) P > 0.05, *P < 0.05, **P < 0.01, and ***P < 0.001. Kaplan–Meier curves and gene correlations were done using the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl).
Supplementary Material
Acknowledgments
We thank Johanna Grass and Sina Wagner for help with animal handling, Viktoria Wischmann for immunohistochemistry, and Dr. Katrin Roth for help with microscopy (Center for Tumor and Immune Biology Microscopy Core Facility). We are indebted to Albrecht Neese, Matthias Löhr, Wade Bushman, Till Adhikary, and Thijn Brummelkamp for the kind provision of cell lines and plasmids and to the University of Marburg Biobanking Core Facility for provision of PDAC patient material. This work was supported by grants from the German Research Society (DFG-LA2829/9-1 and DFG-KFO325) and the German Cancer Aid (DKH, 70112599).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105367118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information. Previously published data were used for this work (25).
References
- 1.Neesse A., et al., Stromal biology and therapy in pancreatic cancer: Ready for clinical translation? Gut 68, 159–171 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Chu G. C., Kimmelman A. C., Hezel A. F., DePinho R. A., Stromal biology of pancreatic cancer. J. Cell. Biochem. 101, 887–907 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Rice A. J., et al., Matrix stiffness induces epithelial-mesenchymal transition and promotes chemoresistance in pancreatic cancer cells. Oncogenesis 6, e352 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iskratsch T., Wolfenson H., Sheetz M. P., Appreciating force and shape—The rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol. 15, 825–833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Dijk M., Göransson S. A., Strömblad S., Cell to extracellular matrix interactions and their reciprocal nature in cancer. Exp. Cell Res. 319, 1663–1670 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Provenzano P. P., et al., Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell 21, 418–429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang H., et al., Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat. Med. 22, 851–860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saatci O., et al., Targeting lysyl oxidase (LOX) overcomes chemotherapy resistance in triple negative breast cancer. Nat. Commun. 11, 2416 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neesse A., et al., CTGF antagonism with mAb FG-3019 enhances chemotherapy response without increasing drug delivery in murine ductal pancreas cancer. Proc. Natl. Acad. Sci. U.S.A. 110, 12325–12330 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hosein A. N., Brekken R. A., Maitra A., Pancreatic cancer stroma: An update on therapeutic targeting strategies. Nat. Rev. Gastroenterol. Hepatol. 17, 487–505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park J. S., et al., Mechanical regulation of glycolysis via cytoskeleton architecture. Nature 578, 621–626 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu H., et al., Phosphoinositide 3-kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton. Cell 164, 433–446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romani P., Valcarcel-Jimenez L., Frezza C., Dupont S., Crosstalk between mechanotransduction and metabolism. Nat. Rev. Mol. Cell Biol. 22, 22–38 (2021). [DOI] [PubMed] [Google Scholar]
- 14.González A., Hall M. N., Lin S.-C., Hardie D. G., AMPK and TOR: The Yin and Yang of cellular nutrient sensing and growth control. Cell Metab. 31, 472–492 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Herzig S., Shaw R. J., AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 19, 121–135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J., Guan K.-L., mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 21, 63–71 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Shi Y., et al., Pancreatic stiffness quantified with MR elastography: Relationship to postoperative pancreatic fistula after pancreaticoenteric anastomosis. Radiology 288, 476–484 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teperino R., et al., Hedgehog partial agonism drives Warburg-like metabolism in muscle and brown fat. Cell 151, 414–426 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Ying H., et al., Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 30, 355–385 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Öhlund D., et al., Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 214, 579–596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson N. C., et al., Targeting of αv integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat. Med. 19, 1617–1624 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calvo F., et al., Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 15, 637–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kluwe J., et al., Absence of hepatic stellate cell retinoid lipid droplets does not enhance hepatic fibrosis but decreases hepatic carcinogenesis. Gut 60, 1260–1268 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Biffi G., et al., IL1-induced JAK/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. 9, 282–301 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurer C., et al., Experimental microdissection enables functional harmonisation of pancreatic cancer subtypes. Gut 68, 1034–1043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sousa C. M., et al., Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker S. J., et al., Selective alanine transporter utilization creates a targetable metabolic niche in pancreatic cancer. Cancer Discov. 10, 1018–1037(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goruppi S., et al., Autophagy controls CSL/RBPJκ stability through a p62/SQSTM1-dependent mechanism. Cell Rep. 24, 3108–3114.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu M., et al., Substrate stiffness differentially impacts autophagy of endothelial cells and smooth muscle cells. Bioact. Mater. 6, 1413–1422 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bays J. L., Campbell H. K., Heidema C., Sebbagh M., DeMali K. A., Linking E-cadherin mechanotransduction to cell metabolism through force-mediated activation of AMPK. Nat. Cell Biol. 19, 724–731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferraresi A., et al., How autophagy shapes the tumor microenvironment in ovarian cancer. Front. Oncol. 10, 599915 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jesnowski R., et al., Immortalization of pancreatic stellate cells as an in vitro model of pancreatic fibrosis: Deactivation is induced by matrigel and N-acetylcysteine. Lab. Invest. 85, 1276–1291 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Neesse A., et al., Pancreatic stellate cells potentiate proinvasive effects of SERPINE2 expression in pancreatic cancer xenograft tumors. Pancreatology 7, 380–385 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Lipinski R. J., Bijlsma M. F., Gipp J. J., Podhaizer D. J., Bushman W., Establishment and characterization of immortalized Gli-null mouse embryonic fibroblast cell lines. BMC Cell Biol. 9, 49 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hupfer A., Brichkina A., Adhikary T., Lauth M., The mammalian Hedgehog pathway is modulated by ANP32 proteins. Biochem. Biophys. Res. Commun. 553, 78–84 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Blomen V. A., et al., Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. Previously published data were used for this work (25).