Significance
Bioinformatic analysis revealed that approximately 40% of human messenger RNAs contain upstream open reading frames (uORFs) in their 5′ untranslated regions. Some of these sequences are translated, but the function of the encoded peptides remains unknown. Our study revealed a uORF encoding for a peptide exhibiting kinase inhibitory activity. This uORF, upstream of a PKC family member, possess the typical pseudosubstrate motif, which autoinhibits the catalytic activity of all PKCs. Using mouse models and human cells, we show that this peptide inhibits cancer cell survival, tumor progression, invasion, and metastasis and synergizes with chemotherapy by interfering with DNA damage response. Together, we point to a previously unrecognized function of a uORF-encoded peptide as a kinase inhibitor, pertinent to cancer therapy.
Keywords: uORF, PKC, kinase inhibitor, pseudosubstrate
Abstract
Approximately 40% of human messenger RNAs (mRNAs) contain upstream open reading frames (uORFs) in their 5′ untranslated regions. Some of these uORF sequences, thought to attenuate scanning ribosomes or lead to mRNA degradation, were recently shown to be translated, although the function of the encoded peptides remains unknown. Here, we show a uORF-encoded peptide that exhibits kinase inhibitory functions. This uORF, upstream of the protein kinase C-eta (PKC-η) main ORF, encodes a peptide (uPEP2) containing the typical PKC pseudosubstrate motif present in all PKCs that autoinhibits their kinase activity. We show that uPEP2 directly binds to and selectively inhibits the catalytic activity of novel PKCs but not of classical or atypical PKCs. The endogenous deletion of uORF2 or its overexpression in MCF-7 cells revealed that the endogenously translated uPEP2 reduces the protein levels of PKC-η and other novel PKCs and restricts cell proliferation. Functionally, treatment of breast cancer cells with uPEP2 diminished cell survival and their migration and synergized with chemotherapy by interfering with the response to DNA damage. Furthermore, in a xenograft of MDA-MB-231 breast cancer tumor in mice models, uPEP2 suppressed tumor progression, invasion, and metastasis. Tumor histology showed reduced proliferation, enhanced cell death, and lower protein expression levels of novel PKCs along with diminished phosphorylation of PKC substrates. Hence, our study demonstrates that uORFs may encode biologically active peptides beyond their role as translation regulators of their downstream ORFs. Together, we point to a unique function of a uORF-encoded peptide as a kinase inhibitor, pertinent to cancer therapy.
Short open reading frames (ORF) upstream of the canonical ORF (uORFs) are found in about 40% of human protein-coding transcripts (1–3). uORF-containing genes were shown to participate in heavily regulated processes, including differentiation, cell cycle control, and stress responses (4–6). Ribosome profiling data revealed that many uORFs are translated (3, 7–11), of which ∼20% are also conserved (12). The current notion indicates that uORFs regulate the translation of their downstream canonical ORFs (13, 14), mostly by attenuating scanning ribosomes (15–17).
A role for uORF-encoded peptides beyond translational regulation was reported in few cases: uORF-encoded peptides were shown to selectively inhibit the non–G protein–coupled angiotensin receptor signaling (18), enhance mitochondrial fission (19), and suppress AP1-mediated transcription (20), suggesting that they operate in the same pathway as their main ORF. A recent large-scale analysis identified hundreds of uORF-encoded peptides, some of which formed complexes with the downstream canonical proteins encoded on the same transcript, suggesting they are functional (3). Here, we show a uORF-encoded peptide possessing a distinct biological function, acting as an inhibitor of the protein kinase activity of the downstream canonical ORF as well as of closely related kinases.
We previously identified two uORFs upstream of protein kinase C-eta (PKC-η) that regulate its translation under normal growth conditions and upon stress (21). PKC-η is a signaling and antiapoptotic stress kinase (22–27) that belongs to the PKC family of serine/threonine protein kinases and is involved in cell proliferation, differentiation, and apoptosis among other processes (28, 29). Members of the PKC family comprise ∼2% of the human kinome (30) and are included in three subfamilies which differ in their regulatory elements and cofactor requirements for activation (31, 32). These include the conventional (α, βΙΙ, βΙΙ,γ), novel (δ, ε, η, θ), and atypical (ζ, ι/λ) PKCs. All PKCs possess a short pseudosubstrate sequence (PS) (33) that resembles the sequence of generic PKC substrates, except for an alanine residue instead of the phosphorable serine/threonine (Fig. 1A). In resting cells, the pseudosubstrate region can associate with the substrate-binding pocket of PKC, autoinhibiting the kinase activity (34, 35). The activation of PKC is achieved by a conformational change due to the binding of phospholipids, diacylglycerol, or calcium ions to its regulatory domain, which relieves the autoinhibitory effect of the PS (36). Several studies characterized the impact of synthetic PS-derived peptides on PKC activity; for example, the PKC-ζ PS-derived peptide, ZIP, was shown to inhibit PKC-ζ activity (37, 38).
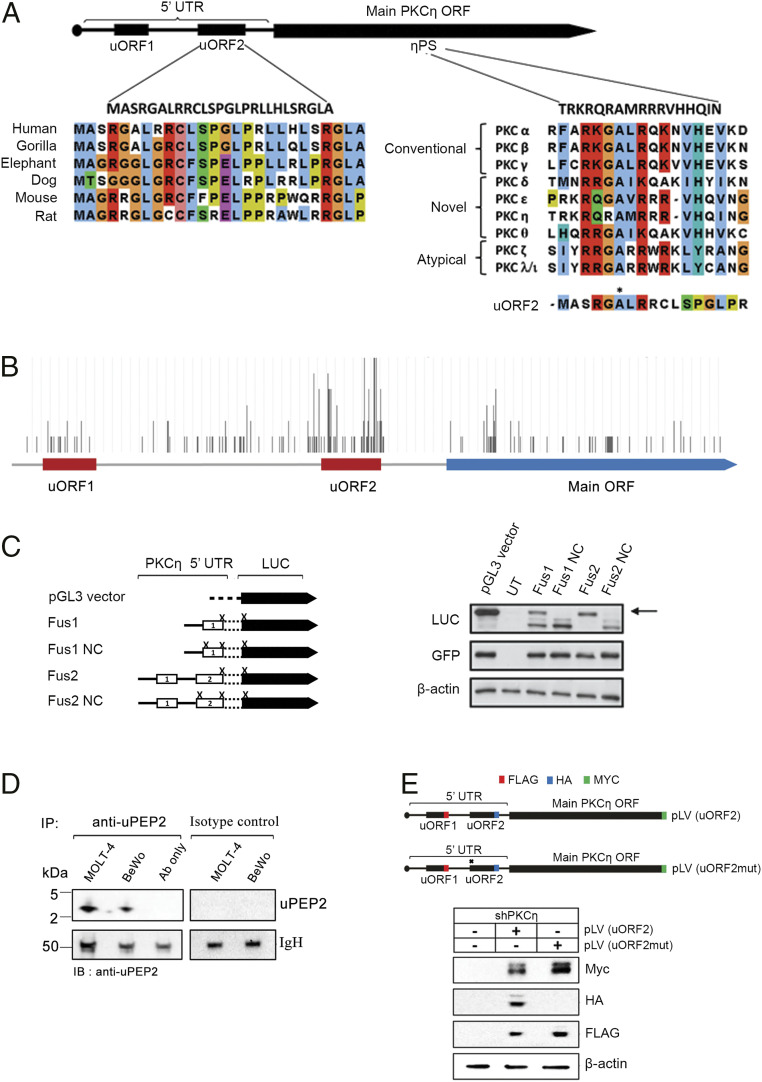
Fig. 1.
uORF2-encoded peptide contains a conserved PS-like motif. (A) The peptide encoded by uORF2 is conserved in other mammals (Left) and is similar to the PS domain of PKCs, including an alanine at position 6 (marked by *) instead of the phosphorable Ser/Thr of optimal PKC substrates (Right). (B) A representative example showing RP reads measured across PKC-η mRNA (40). Ribosomes accumulated mostly at the start codon and the stop codon of uORF2, also relative to uORF1 and PKC-η main ORF, supporting its translation (adapted from ref. 39; additional examples appear in SI Appendix, Fig. S1). (C) Experimental assessment of the translational initiation capacity of uORF1 and uORF2 shows it is more frequent at uORF2. We tested the following constructs: a pGL3 vector containing a firefly LUC reporter; uORF1 or uORF2 inserted without their stop codons upstream and in-frame with LUC that contained a mutated initiation codon (Fus1 and Fus2); and two similar constructs (negative controls) in which the initiation codons of uORF1 or uORF2 were mutated (Fus1NC and Fus2NC). Transfection efficiencies were examined by cotransfection with a GFP reporter plasmid. Cells transfected with the pGL3 vector, Fus1, and Fus2 yielded the expected 65-kDa LUC protein (marked by an arrow). NC, negative control; UT, untransfected. (D) The endogenous presence of uORF2-encoded peptide is depicted using immunoprecipitation experiments. Cell extracts of MOLT-4 and BeWo cells (1 mg/sample) were immunoprecipitated using uPEP2 specific antibody, separated on Tris-Tricine gel. Immunoblotting shows the presence of a band around 3 kDa of uPEP2. (E) Ablating the translation of uORF2 enhances the expression of PKC-η main ORF. The shPKC-η MCF-7 clone (sh3-3) was transduced with pLV(uORF2) or pLV(uORF2mut) lentiviral system in which uORF1, uORF2, and PKC-η were tagged with Flag, HA, and Myc, respectively. The pLV(uORF2mut) plasmid carries a point mutation in the start codon (AUG to ACG) of uORF2 to prevent its expression. Immunoblots were performed using anti-Flag/HA/Myc specific antibodies.
In the present study, we identified a PS-like motif in the second uORF (uORF2) of PKC-η. We show that the uORF2-encoded peptide (uPEP2) is an efficient inhibitor of the kinase activity of PKC-η as well as other members of the novel PKC subfamily. Importantly, uPEP2 suppresses the proliferation/survival and migration of cancer cells and acts in concert with the cell death–promoting effects of chemotherapy, apparently, by interfering with the cellular DNA damage response. We also demonstrate that uPEP2 inhibits tumor development in a mouse model of breast cancer xenograft. Altogether, our studies show that in addition to the uORF-mediated translational regulation, uORF-encoded peptides can also regulate biological functions of proteins, providing the basis for novel agents with potential therapeutic impact in cancer.
Results and Discussion
A Translatable uORF Upstream of PKC-η Possess a PS-like Motif.
The 5′ untranslated region (UTR) of PKC-η contains two uORFs, uORF1 and uORF2 (21). An analysis of a uORF2 sequence revealed the presence of a PS-like sequence (Fig. 1A), raising the possibility that this region encodes a peptide that could interact with the catalytic domain of PKC-η and regulates its activity. In support of its potential functionality, the genomic sequence of uORF2 upstream of PKC-η is conserved in several mammalian species (SI Appendix, Table S1). The conserved uORF2 sequences reside on the same transcripts as PKC-η homologs and encode closely related peptides of the same length (Fig. 1A and SI Appendix, Table S1).
To test whether uORF2 is translated, we viewed published ribosome profiling datasets on the genome browser of the GWIPS-viz database (39). Independent datasets collected from human embryonic kidney (HEK)-293, MCF10A, LCL 8664, and U2OS cells (12, 40–43) showed that ribosomes accumulated mostly at the start and stop codons of uORF2, supporting its translation (Fig. 1B and SI Appendix, Fig. S1). Moreover, ribosome profiles of harringtonine- or cycloheximide-treated cells, in which ribosomes are stalled at initiation sites (44), showed ribosomes accumulating at the start codon of uORF2 in MCF10A cells (12).
To further analyze whether uORF2 is translatable, we generated fusion constructs for each uORF and the luciferase (LUC) reporter gene. Specifically, we mutated the stop codon of each uORF1 and uORF2 as well as the AUG initiation codon of LUC, leading to the generation of the corresponding uORF-LUC fusion proteins (Fig. 1C, Fus1 and Fus2 plasmids). Following transfection into HEK-293T cells, protein expression was examined by Western blot analysis and an antibody against LUC protein. LUC products were not observed in cells transfected with constructs containing mutations at the initiation codons of the uORFs (Fig. 1C, Fus1 NC and Fus2 NC constructs). In contrast, LUC expression was observed in cells expressing uORF1 and uORF2 fusion proteins. The uORF2 fusion protein was expressed at higher levels compared to the uORF1 fusion protein (Fig. 1C), consistent with its stronger ribosome profiling signal (Fig. 1B and SI Appendix, Fig. S1) and its stronger match to the Kozak consensus sequence required for efficient translation initiation (21).
Furthermore, we show that the uORF2-encoded peptide is stably produced. Utilizing an uPEP2-specific antibody (Materials and Methods) and immunoprecipitation experiments on MOLT-4 and BeWo cell extracts, the presence of an ∼3-kDa peptide that was not immunoprecipitated by a control IgG antibody was depicted (Fig. 1D). Taken together, our studies revealed a translatable uORF upstream of a PKC gene and provided evidence that it is stably produced.
To determine the function of the uORF2-derived peptide on PKC-η expression, we generated an expression plasmid in which uORF1, uORF2, and PKC-η were tagged with Flag, HA, and Myc, respectively (Fig. 1E). A corresponding plasmid in which uORF2 expression was abolished by introducing a point mutation in its start AUG codon was also produced. Both plasmids were expressed in an MCF-7 clone in which PKC-η was down-regulated (shPKC-η) (23). Our results show that in the absence of uORF2, the expression of PKC-η is up-regulated (Fig. 1E) in accordance with our previous studies demonstrating that uORF2 acts as a cis-repressor element that suppresses expression of PKC-η and maintains its low basal levels (21). Taken together, we show that uORF2 is translated and suppresses the abundance of its downstream ORF. We further show (The uORF2-Encoded Peptide Inhibits the Kinase Activity of Novel PKCs) that beyond this role as a regulator of translation, the uORF2-encoded peptide also functions as an inhibitor of the kinase activity of novel PKC family members including PKC-η.
The uORF2-Encoded Peptide Inhibits the Kinase Activity of Novel PKCs.
The presence of a PS-like motif in the uORF2-encoded peptide (uPEP2) prompted us to test whether it can function as an inhibitor of the kinase activity of PKC-η or other PKCs (Fig. 2A). Computational modeling of the potential interaction between PKC-η and uPEP2 demonstrated that the N-terminal sequence of uPEP2 (residues 1 to 9) fits within the substrate-binding pocket of the kinase domain of PKC-η (Fig. 2B and SI Appendix, Fig. S2). Kinase activity assays were performed on different PKC isoforms that were expressed in HEK-293T cells, immunoprecipitated, and incubated in vitro in the presence of γ-[32P]ATP and myelin-binding protein (MBP) as a substrate (as described in ref. 23). Kinase activity was examined in the presence of either uPEP2, a control peptide derived from uPEP2 but lacking part of the N terminus PS motif (uPEP210–26), or a peptide derived from the internal autoinhibitory PS sequence of PKC-η (η-PS). We found that uPEP2 inhibited the kinase activity of PKCη with a half-maximal inhibitory concentration (IC50) of about 2 μM. In contrast, uPEP210–26 and η-PS exhibited weak inhibitory effects, even at a concentration of 10 μΜ (Fig. 2C and SI Appendix, Fig. S3A). uPEP2 also inhibited the kinase activity of other novel PKC isoforms, including PKCδ, PKCθ, and PKCε, with a similar IC50 of ∼2 μM. Notably, uPEP2 did not affect the activity of the classical PKC isoforms, PKC-α and PKC-γ, or the atypical isoform, PKC-ζ (Fig. 2D and SI Appendix, Fig. S3B). To test whether uPEP2 can physically interact with PKCs, we performed pulldown assays using bead-immobilized antibody–peptide complexes. uPEP2 was found to interact with the four novel PKC isoforms but not with PKC-γ or PKC-ζ (Fig. 2E and SI Appendix, Fig. S4), while the KS21 control peptide showed no binding to all isoforms tested. It appears, therefore, that uPEP2-mediated inhibition of the catalytic activity of novel PKCs is attributed to direct physical interaction between the peptide and the enzyme. Moreover, when the pivotal nonphosphorable alanine residue (at position 6) in uPEP2 was replaced by serine or threonine residues, uPEP2 transformed from an inhibitor into a favorable substrate for PKC-η and PKC-ε but not for PKC-α (Fig. 2F). Taken together, our results demonstrate that uPEP2 is a kinase inhibitor of novel PKCs and that the pseudosubstrate-like motif underlies this inhibition.
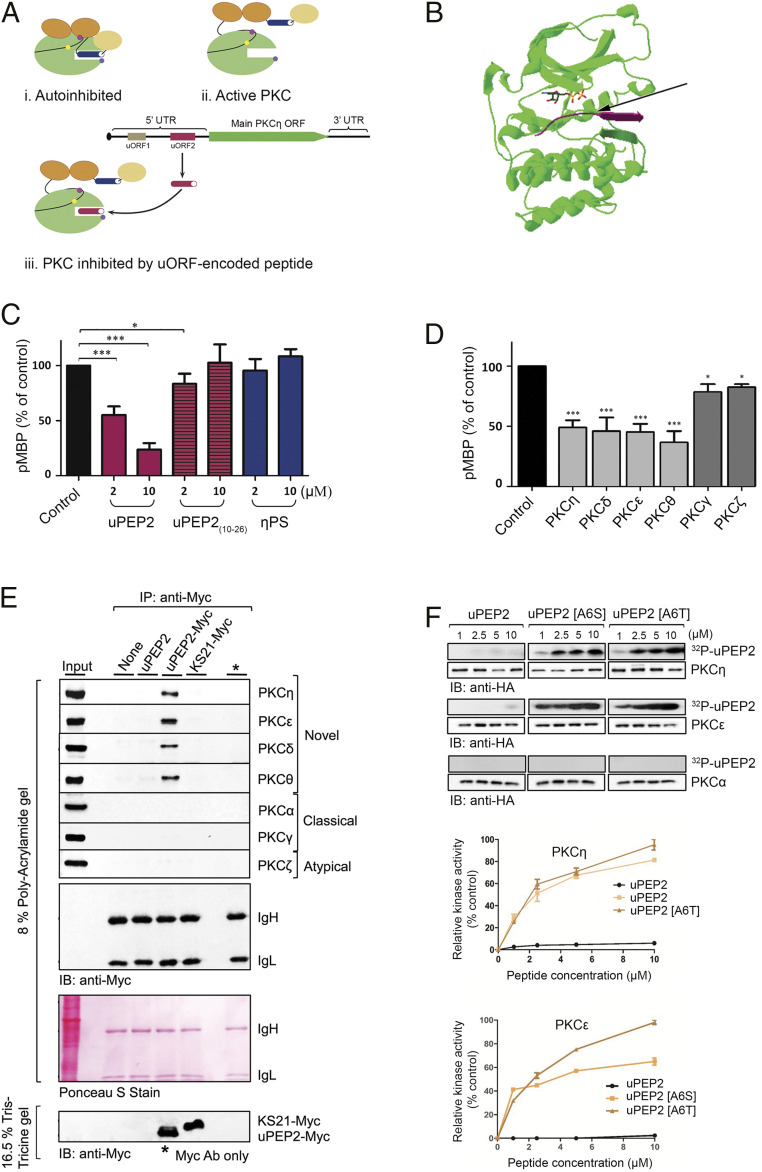
Fig. 2.
uORF2-encoded peptide inhibits the kinase activity of novel PKCs. (A) The active/inactive states of PKCs: (i) In the autoinhibited state, the PS domain (dark blue cylinder) binds to the kinase domain (green circle) leading to its inhibition. (ii) In the active state, the PS domain is detached from the kinase domain due to a conformation change. (iii) In the proposed state, the kinase domain is bound by the uORF2-encoded peptide (dark red cylinder) and thus inhibited. Oval shapes represent other PKC domains, and small circles represent phosphorylation sites. (B) A general view of the proposed PKC-η–uPEP2 complex. The model shows the catalytic domain of PKC-η (green) bound to ADP (stick model) and to residues 1 to 9 of uPEP2 (magenta). The nonphosphorable alanine 6 residue of uPEP2 (marked by an arrow) is positioned in the phosphorylation site by a small β-sheet formed by residues 7 to 9 of uPEP2 and residues 514 to 516 of PKC (magenta and green ribbons, respectively), which are unstructured in the unbound state. The model suggests several hydrophobic and electrostatic interactions (SI Appendix, Fig. S2) that may explain the stability and specificity of the complex. (C) uPEP2 inhibits the kinase activity of PKC-η. Kinase assays were performed using MBP as a substrate with/without tested peptides. uPEP2 was a more effective inhibitor than a peptide containing the internal PS motif of PKC-η (η-PS) and a peptide derived from uPEP2 but lacking its N-terminal part (uPEP2(10–26)). (D) uPEP2 inhibits the kinase activity of novel PKCs. Kinase activity assays depicting novel PKCs (light gray) and other PKC members (dark gray) were performed as in C. The data shown are means of three independent experiments. Details of kinase assays in C and D are presented in SI Appendix, Fig. S3. (E) uPEP2 selectively binds to novel PKC isoforms. HEK-293T lysates overexpressing different PKC isoforms were subjected to immunoprecipitation using protein A/G beads preadsorbed with anti-Myc antibody and Myc-tagged peptides (uPEP2, uPEP2-Myc, or the nonrelevant control peptide KS21-Myc). (F) Altering alanine at position 6 into S/T converts uPEP2 into an excellent substrate of novel PKC-η and PKC-ε but not PKC-α. Kinase assays were performed as in C using γ-ATP and uPEP2, uPEP2[A6S], or uPEP2[A6T] as substrates. P values calculated using one-way ANOVA. ***P ≤ 0.001 and *P ≤ 0.05.
The uORF2-Encoded Peptide Suppresses Proliferation and Migration of Cancer Cells.
PKC isoforms are known to be involved in the regulation of cancer cell proliferation, survival, invasion, migration, apoptosis, angiogenesis, and anticancer drug resistance (22, 45, 46). Since our experiments demonstrated that uPEP2 specifically inhibited the kinase activity of novel PKCs, we examined the effects of uPEP2 and its derivatives on the viability of breast cancer, leukemia, and other types of cancer cells (Fig. 3A and SI Appendix, Fig. S5A). All peptides tested contained an N-terminal myristoyl group to enable cell penetration. uPEP2 significantly reduced the viability of breast cancer and leukemia cells (Fig. 3A) but had no effect on the viability of the nontransformed MCF10A cells, suggesting it may have a therapeutic benefit in selected cancers. The nonmyristoylated peptide, uPEP2(-Myr), which cannot penetrate cells, did not affect cell viability. The control peptides, uPEP218–26 and η-PS, were less effective in inhibiting breast cancer and glioblastoma cell viability (Fig. 3B and SI Appendix, Fig. S5B). The deletion peptide uPEP21–17, containing the PS motif but lacking the peptide's C terminus, exhibited only partial inhibition of cell viability (SI Appendix, Fig. S5B), suggesting that the full-length peptide is required for efficient inhibition.
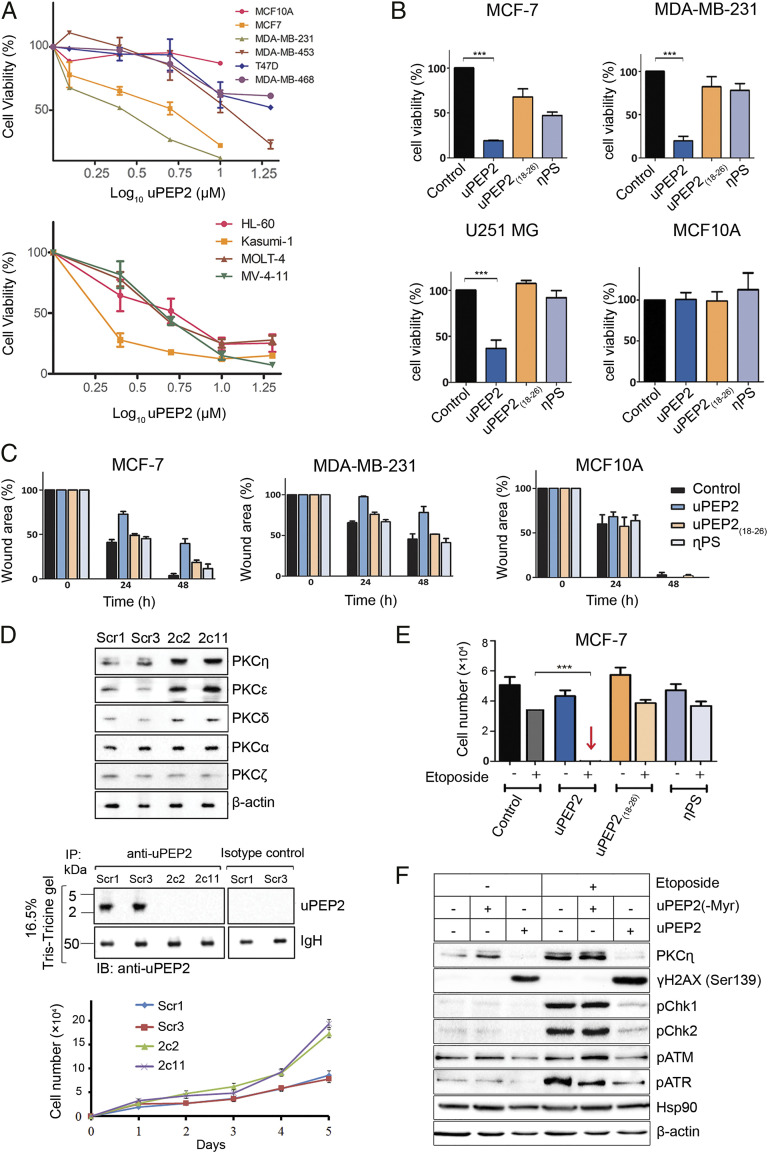
Fig. 3.
uORF2-encoded peptide inhibits cell proliferation and migration of cancer cells and is synergistic with chemotherapy by interfering with the response to DNA damage. (A) uPEP2 attenuates cell viability of breast cancer and leukemia-derived cells. Data shown represents three independent experiments carried with/without the presence of uPEP2 (1.25 to 10 μM). (B) uPEP2 reduces cell viability of MCF-7 and MDA-MB-231 breast cancer cells and U251 MG glioblastoma cancer cells but did not affect the viability of nontransformed MCF10A cells. uPEP2 was more effective compared to peptides containing the internal PS motif of PKC-η (η-PS) or lacking the PS-like sequence (uPEP2(18–26)). Data shown represents three independent experiments (peptide concentration, 10 μM). (C) uPEP2 inhibits migration of MDA-MB-231 and MCF-7 breast cancer cells but not of nontransformed MCF10A cells. Photographs were taken at indicated time points, and wound areas were normalized to time 0. Data shown represents at least three independent experiments (peptide concentration, 5 μM). uPEP2 is more potent in inhibiting migration compared to η-PS and uPEP2(18–26). All microscopy images appear in SI Appendix, Fig. S6. (D) Endogenous deletion of uORF2 in MCF-7 cells relieves suppression on novel PKCs expression. Using a CRISPR/Cas9 lentiviral system, uORF2 mutated (2c2 and 2c11) and control (Scr1 and Scr3) MCF-7 clones were generated. The elevation of novel PKC protein levels compared to scrambled controls were observed, while classical PKC-α and atypical PKC-ζ protein levels were unaffected. Immunoprecipitation experiments (as in Fig. 1D) verified the expression of uORF2-encoded peptide (uPEP2) in scrambled control (Scr1 and Scr3) clones but not in the deletion clones. Cell proliferation assay shows enhanced cell proliferation in uORF2-deleted MCF-7 clones. (E) uPEP2 is synergistic with etoposide in the induction of cell death. MCF-7 cells were treated with indicated peptides (2 μM) followed by the addition of etoposide (50 μM) for 48 h. Cell numbers were quantified by standard curves of absorbance versus cell numbers as described in Materials and Methods. Data shown are means of at least three separate experiments. (F) uPEP2 interferes with the cellular response to DNA damage. Experiments were carried out as in D followed by cell lysis (30 min after etoposide addition). A peptide without a myristoyl group at the N terminus of the peptide, unable to enter the cells, was used as a control. Phosphorylation on DNA damage response markers γ−H2AX, Chk1, Chk2, ATM, and ATR were detected using Western blot analysis and specific antibodies. Hsp90 and β-actin were used as markers for equal protein loading. The results shown are representative of three independent experiments. P values calculated using one-way ANOVA. ***P ≤ 0.001.
Malignant progression and cancer metastasis are strongly correlated with the ability of cancer cells to migrate. To further assess the impact of uPEP2 on cell migration, we tested its effect on wound closure in vitro using the scratch assay. We found that uPEP2 inhibited the migration of MCF-7 and MDA-MB-231 cells but had no effect on the migration of the nontransformed MCF10A cells (Fig. 3C and SI Appendix, Fig. S6). Taken together, the full-length uPEP2 was most effective in the inhibition of both cell viability and migration compared to the deletion variants or the PKC-η internal η-PS inhibitory peptide, in agreement with the effectiveness of these peptides as inhibitors of its kinase activity.
To determine the role of endogenously expressed uORF2 on cell survival, uORF2 was deleted in MCF-7 cells using CRISPR/Cas9 methodology (Fig. 3D). Knockout of uORF2 in 2c2 and 2c11 clones was verified by DNA sequencing, and its presence in Scr1 and Scr3 scrambled control cells (and absence in 2c2 and 2c11 clones) was depicted using immunoprecipitation experiments with anti-uPEP2–specific antibodies (Fig. 3D and SI Appendix, Fig. S7). A time-course analysis of cell proliferation of these clones showed increased cell numbers in uORF2-deleted cells compared to scrambled control lines (Fig. 3D). Notably, deletion of uORF2 was accompanied by increased expression of PKC-η in both uORF2-deleted clones. The protein abundance of novel PKC-ε and PKC-δ was also augmented in 2c2 and 2c11 cells, while that of classical PKC-α or atypical PKC-ζ was not altered compared to control cells (Fig. 3D). The effect of uORF2 on the expression of novel PKCs was further supported by the overexpression of uORF2 in MCF-7 cells (on the background of shPKC-η knockdown cells; see Fig. 1E), resulting in reduced endogenous expression of novel PKC-ε and PKC-δ compared to control nontransfected cells. The introduction of a mutation in the start codon of uORF2, ablating its expression, restored their expression (SI Appendix, Fig. S8).
In some reports, PKC-η expression was associated with cell proliferation and survival; overexpression of PKC-η in MCF-7 cells enhanced cell cycle progression and the expression of G1 cyclins, cyclin D and E, and p21Cip1 (47). In breast cancer patients’ biopsies, PKC-η expression was increased after chemotherapy, and its localization in cell membranes (activated) was a predictor for poor prognosis (27). PKC-η expression was augmented by estrogen, also enhancing proliferation induced by estrogen (48). In PKC-η−/− mice, PKC-η was required for T cell activation and proliferation (49). PKC-ε expression was also shown to promote cell proliferation, while PKC-δ effects on cell proliferation and survival were cell type dependent (29). Taken together, our studies show that the uORF2-encoded peptide affects the protein levels of novel PKCs; its deletion in MCF-7 cells increased protein levels of novel PKCs culminating in enhanced cell proliferation.
uORF2-Encoded Peptide Synergizes with Chemotherapy by Interfering with the Response to DNA Damage.
We and others previously published that PKC-η and other PKCs provide protection against DNA damage–induced cell death (22, 24–26, 50–54). Therefore, we examined if inhibition of PKCs by uPEP2 enhances cell death upon etoposide-induced DNA damage. Our data showed that under conditions in which death by either etoposide or uPEP2 was limited (10 to 30%), the presence of both agents highly increased cell death (95 to 100%), demonstrating synergism (Fig. 3E). Notably, the expression of PKC-η is increased by etoposide treatment (Fig. 3F), suggesting that its elevated expression is stress activated, needed to confer protection against DNA damage–induced stress (22). In the presence of uPEP2, PKC-η underwent enhanced degradation (Fig. 3F), most likely as a result of decreased stability of the protein because of diminished phosphorylation on its priming phosphorylation sites (36). We show that PKC-η is phosphorylated on the priming sites Ser675 and Thr655, the phosphorylation of which is diminished by uPEP2, accompanied by PKC-η degradation (SI Appendix, Fig. S9).
In response to DNA damage, the histone H2A variant H2AX undergoes phosphorylation on serine 139 (termed γ-H2AX). The phosphorylation occurs at sites of double-strand breaks (DSBs), thought to restructure the chromatin and assist in the recruitment of DNA repair and signaling factors (55, 56). DNA damage–induced phosphorylation of H2AX is mediated by ATM, ATR, and DNA-PK and is considered a marker for DNA DSBs initiating DNA damage response (55, 56). Upon induction of DSBs by etoposide, we show that phosphorylation of γ-H2AX markedly increased in uPEP2-treated MCF-7 cells (Fig. 3F). A similar effect was observed in X-ray or etoposide-treated PKC-η knocked-down MCF-7 cells or upon the expression of a kinase-dead PKC-η mutant (SI Appendix, Fig. S10 and see ref. 23). Thus, PKC-η indirectly interferes with γ-H2AX phosphorylation, and a catalytically active PKC-η is required for this inhibition of γ-H2AX phosphorylation. Moreover, while etoposide promoted the phosphorylation of ATM and ATR and their effector kinases Chk1 and Chk2, this phosphorylation was markedly decreased by uPEP2 (Fig. 3F). Hence, uPEP2 interferes with DNA repair processes in response to DNA damage, resulting in enhanced cell death.
The Antitumor Activity of uPEP2 in Breast Cancer Mice Models.
To further analyze the antitumor activity and therapeutic potential of uPEP2 in a preclinical model, we tested the in vivo effect of uPEP2 on breast cancer triple-negative MDA-MB-231 cells in tumor-bearing mice. In this model, MDA-MB-231 tumor cells were inoculated into the mammary fat pads of female mice, and when the tumors reached a volume of ∼100 mm3, the mice were treated with uPEP2 or the control peptide, uPEP2(-Myr), on alternate days. The results showed that uPEP2 inhibited the growth of MDA-MB-231 tumors in NOD-SCID gamma (NSG) and NOD-SCID mice as indicated by volume (Fig. 4 A and B and SI Appendix, Fig. S11). In contrast, parallel treatment of mice with uPEP2(-Myr) had no effect on tumor growth. Furthermore, treatment of NSG mice with uPEP2 inhibited MDA-MB-231 metastasis formation in the lung and liver (Fig. 4C). At the end of the experiments, histology analysis was performed, and tissue extracts were prepared from the collected tumors. Histology analysis of uPEP2-treated tumors showed fewer proliferating cells in these tumors compared to nontreated or uPEP2(-Myr)-treated tumors as indicated by Ki67 staining (Fig. 4D). Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays demonstrated increased apoptosis in uPEP2-treated tumors (Fig. 4E), supporting the exhibited reduced tumor progression. Immunoblotting studies demonstrated lower protein levels of novel PKC-η and PKC-ε in MDA-MB-231tumors treated by uPEP2 in NSG and NOD-SCID mice (Fig. 4F and SI Appendix, Fig. S11G). Notably, the expression levels of the classical PKC-α, PKC-γ, and atypical PKC-ζ were not affected by uPEP2 treatment, in correlation with the uPEP2-mediated inhibition of the catalytic activity of these kinases. Similar results were obtained in MDA-MB-231 cells grown in culture and treated by uPEP2 (SI Appendix, Fig. S12). Furthermore, phosphorylation of PKC substrates was also lower in uPEP2-treated MDA-MB-231 tumors grown in NSG or NOD-SCID mice (Fig. 4G and SI Appendix, Fig. S11H), supporting reduced PKC kinase activity in these tumors. Taken together, our studies demonstrate that treatment of breast cancer MDA-MB-231 xenografts in mice by uPEP2 suppresses kinase activity and protein abundance of novel PKCs accompanied by reduced tumor growth and metastasis, supporting the antitumor activity of uPEP2. However, the endogenous role of the uORF2-encoded peptide as a kinase inhibitor and suppressor of protein expression of novel PKCs should be studied further using genetic approaches to knockout or overexpress uORF2 in mice.
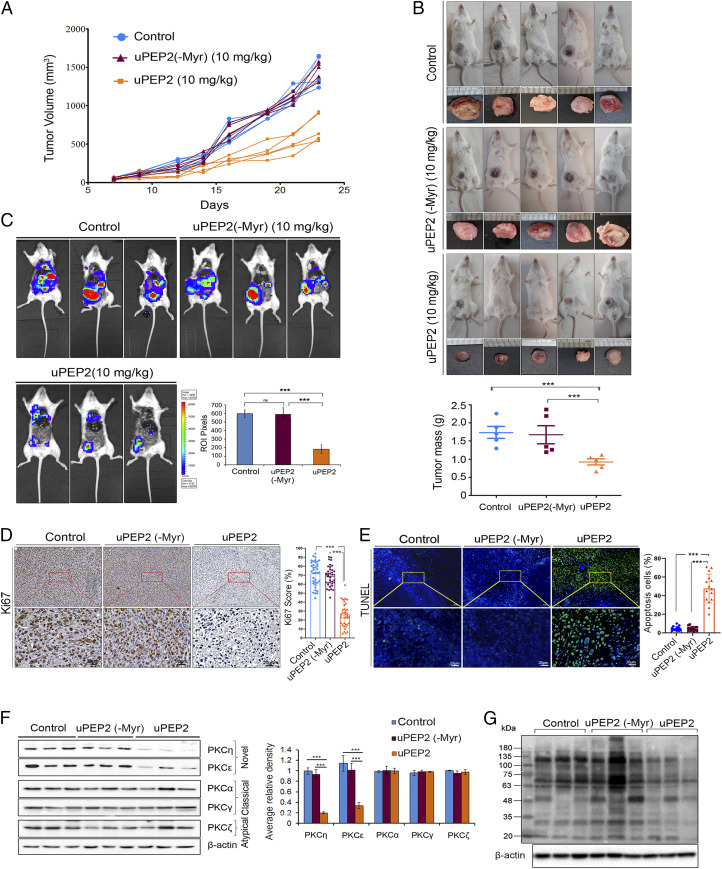
Fig. 4.
uPEP2 suppresses tumor development of MDA-MB-231 orthotopic xenograft in NSG mice. (A) Tumor growth curves depicting effects of uPEP2, uPEP2(-Myr), and control (PBS alone) treatments on tumor volume in NSG mice. (B) uPEP2 significantly attenuates breast tumor mass. Data shows tumor mass of uPEP2, uPEP2(-Myr), and PBS vehicle control– (mean ± SEM) treated mice (five animals/group). (C) Bioluminescence images depicting reduced tumor development and metastasis of MDA-MB-231-Luc cells in response to uPEP2 treatment. Breast tumor–bearing NSG mice were treated on alternate days with intratumor injections of uPEP2 and control peptide (10 mg/kg), and images were taken using the In Vivo Imaging System (Xenogen). The bar diagram shows quantification of mean bioluminescent intensity at regions of interest in Pixels. The images were captured from three independent mice (n = 3). (D) uPEP2-treated breast cancer xenografts from NSG mice show reduced proliferation. Tumor tissue sections obtained from MDA-MB-231 breast tumors treated with PBS (control), uPEP2(-Myr), and uPEP2, respectively, were immunostained with Ki67-specific antibodies. Shown are representative images and graphical representations of the quantitative analysis of tumor cells with positive Ki67 staining. (E) uPEP2-treated tumors showed increased apoptosis. Tissue slides of peptide-treated MDA-MB-231 breast tumors (as in D) were tested for apoptotic cells using TUNEL assays. Panels showing fluorescent images of cell nuclei (stained with DAPI; in blue) and fluorescent images of apoptotic cells (green). (F) uPEP2 down-regulates expression of novel PKCs in MDA-MB-231 xenograft tumors. Dissected tumors were lysed and subjected to immunoblot analysis as described in Materials and Methods. Comparative quantification of novel, classical, and atypical PKC protein band signals are represented in the bar diagram. (G) Phosphorylation on PKC substrates is reduced in uPEP2-treated tumor samples. Western blot analysis of tissue extracts prepared from PBS (control), uPEP2(-Myr), and uPEP2-treated tumors were immunoblotted with an antibody-detecting phospho-(Ser) of PKC substrates. P values calculated using one-way ANOVA. ***P ≤ 0.001 and ns ≥ 0.05.
The activation and stability of PKCs are dependent on their phosphorylation, predominantly on the activation loop, turn motif, and hydrophobic motif, by external kinases (e.g., PDK1) or autophosphorylation events (57). Moreover, phosphorylation of novel PKC isoforms appears to be functionally interdependent and cross-regulated (58, 59). Thus, the fact that uPEP2 specifically suppresses the kinase activities of novel PKCs and their protein levels was confirmed here by lower phosphorylation of PKC substrates, diminished cell proliferation, and enhanced apoptosis, further manifested by reduced tumor volume (Fig. 4 and SI Appendix, Fig. S11). As some PKCs might have contradictory effects on cell survival (29, 60), the outcome of the effect of uPEP2 is likely to be determined by the relative expression of the individual kinases in the specific tumors. Our studies show that endogenously expressed uORF2 has a restrictive effect on cell proliferation as demonstrated by its specific deletion (by CRISPR/Cas9) (Fig. 3D). When uORF2 is translated, it suppresses protein levels not only of its downstream ORF (PKC-η) but also of other novel PKCs (Fig. 3D). Our results suggest that binding of the uORF2-translated peptide to novel PKCs reduce their catalytic activity and protein phosphorylation, which may affect their protein stability. Although, it should be noted that in the case of PKC-η, uORF2 could also act as a translational repressor (21). It is of interest that in some tumors, the messenger RNA (mRNA) transcript of PKC-η is lower compared to their normal tissues (61), perhaps as a way to overcome limiting effects of uORF2 on tumor proliferation.
In summary, here we identified a uORF that encodes for a peptide exhibiting kinase inhibitory functions. This uORF2-encoded peptide, upstream of PKC-η, contains a PS-like motif, characteristic of all PKCs, inhibiting their kinase activity. By mimicking the internal inhibitory PS motif of PKCs, uORF2 possess the ability to directly bind and inhibit the catalytic activity of all members of the novel PKC subfamily. Moreover, we demonstrate that endogenous deletion of uORF2 relieves its suppressive effects on the protein abundance of PKC-η and novel PKCs, resulting in their elevated protein levels and increased cell proliferation. The uORF2-encoded peptide inhibited proliferation and migration of cancer cells and suppressed the growth of a xenograft of breast cancer tumor in mice models. The histology of these tumors showed reduced proliferation and enhanced cell death, exhibiting also lower protein abundance of novel PKCs and diminished phosphorylation on PKC substrates. Hence, our study suggests that uORFs may encode biologically active peptides beyond their role in regulating the translation of their downstream ORF. As protein kinase inhibitors, uORF-encoded peptides may play a role in signaling and stress, acting in cis and/or in trans, forming diverse regulatory networks, thereby opening new views into eukaryotic protein regulation and cancer biology.
Materials and Methods
Cell Lines.
MCF-7, MDA-MB-231, MDA-MB-231-Luc, HeLa, HaCat, Caco-2, DU145, and HEK- 293T cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (no. 01–055-1A, Biological Industries [BI]). BeWo cells were maintained in 1:1-DMEM-F12 (no. 01–170-1A, BI). T47D, Kasumi-4, MOLT-4, and U937 were maintained in Roswell Park Memorial Institute 1640 (no. 01–100-1A, BI). MDA-MB-453 and MDA-MB-468 were maintained in Leibovitz L-15 (no. 01–115-1A, BI). HL-60 and MV-4–11 cells were maintained in Iscove’s Modified Dulbecco’s Medium (no. 01–058-1A, BI). HCT-116 and U2OS cells were maintained in McCoy’s 5A (no. 01–075-1A, BI). Media were supplemented with penicillin (100 units/mL), streptomycin (0.1 mg/mL), and L-glutamine (2 mM) plus either 20% fetal bovine serum (FBS) for Caco-2, Kasumi-4, and HL-60 cells or 10% FBS for the rest of the cell lines. MCF10A cells were cultured in 1:1-DMEM-F12 supplemented with 5% horse serum, penicillin (100 units/mL), streptomycin (0.1 mg/mL), insulin (10 μg/mL), hydrocortisone (0.5 μg/mL), epidermal growth factor (10 ng/mL), and cholera toxin (0.1 μg/mL). shPKC-η (sh3-3) cells were maintained in DMEM complete media containing 0.2 mg/mL Geneticin (G418) (no. 509290, CalbioChem, MERCK). All cells were maintained in a 37 °C, 5% CO2 humidified incubator.
Peptides.
uPEP2(Myr-MASRGALRRCLSPGLPRLLHLSRGLA), uPEP2(10–26)(Myr-CLSPGLPRLLHLSRGLA), uPEP2(18–26)(Myr-MLLHLSRGLA), η-PS(Myr-TRKRQRAMRRVHQING), uPEP2(1–17)(Myr-MASRGALRRCLSPGLPR), uPEP2[A6S/T](Myr-MASRGS/TLRRCLSPGLPRLLHLSRGLA), uPEP2-Myc (MASRGALRRCLSPGLPRLLHLSRGLA-EQKLISEEDL), and KS21-Myc (KNEARPPCLPTPGKREPQGIS-EQKLISEEDL) were synthesized by GL-Biochem Ltd. All peptides contained N-terminal acetylation and C-terminal amidation. A myristoyl group was included at the N terminus to enable cell penetration.
Conservation Analysis.
We tested whether the nucleotide sequence of the human uORF2 is conserved at the genome level in other species by using the University of California, Santa Cruz browser track “Vertebrate Multiz Alignment & Conservation of 100 Species.” We focused on five mammalian species showing conservation, extracted the uORF2 sequence per organism, and used BLAST to identify the relevant transcript (SI Appendix, Table S1). RefSeq was used to confirm that the uORF is upstream of the main ORF of PKC-η. The nucleotide sequence of each uORF was translated to its corresponding amino acid sequence via ExPASy (https://web.expasy.org/translate/) with default parameters. We used Jalview to illustrate the multiple sequence alignment, highlighting amino acids with similar chemical features according to the Clustal X color scheme. Jalview was also used to view the alignment between the PS sequences of human PKCs.
Ribosome Profiling Analysis.
To test if PKC-η uORFs are translated, we used previously published ribosome profiling (RP) data from the GWIPS-viz that visualizes RP reads across the genome (39). We used the reference genomes hg38 for human and scanned datasets corresponding to initiating or elongating ribosomes, focusing on the 5′ UTR of PKC-η (SI Appendix, Fig. S1).
Modeling of the Proposed PKC-η–uPEP2 Complex.
A crystal structure of PKC-η was available only for the peptide-free enzyme (Protein Data Bank [PDB] code 3txo). To model the PKC-η–uPEP2 complex, we turned to homology-based modeling [Swiss-PdbViewer (62)]. A crystal structure of a peptide-bound PKC complex was available only for PKC-ι [PDB codes 5li1 and 5lih (63)]. The sequences of PKC-ι and PKC-η have over 50% identity overall, and identical peptide-binding segments (residues 412 to 417 of PKC-ι and 513 to 518 of PKC-η). The structures of the two proteins are also very similar (most residues are within 1Å RMSD), except for 1) phosphorylation of threonine 412 of PKC-ι and 2) PKC-η at residues 507 to 515 being unstructured. Modeling the structure of the peptide-binding segment of PKC-η was straightforward, as the alignment was unambiguous, and the neighboring segments were practically identical. The modeling of the bound peptide was more complex, as the template peptides had low sequence similarity among themselves and with uPEP2. Yet, starting from the phosphorylation-target serine, the known structures of the peptide-bound PKC-ι had four identical peptide residues. Alanine 6 of uPEP2 is analogous to the phosphorylation-target serine. Using it as an anchor for the alignment revealed two identical residues with the peptide of 5lih (arginine 8 and 9) within the structurally conserved segment, and thus, we used 5lih as the modeling template. The model included only the first nine residues of uPEP2, as none of the available templates could suggest the conformation of uPEP2 C terminus.
DNA Constructs.
The uORF-luciferase fusion plasmids were constructed based on sequences of the previously described wild-type (WT), mut1, and mut2 constructs (21). An intermediate plasmid harboring a mutation at the initiation codon of the main ORF (the Luc-mut plasmid) was generated using primers #1 and #2 (SI Appendix, Table S2) (synthesized at Sigma). A PKC-η mRNA segment containing uORF1 (not including the stop codon) was amplified from the WT plasmid using primer #3 (SI Appendix, Table S2). The resulted amplicon was then digested by SacI and AvrII (no. R0156S, no. R0174S, New England Biolabs, Inc. [NEB]) and cloned into the corresponding sites of the WT plasmid to generate the Fus1 plasmid. An identical amplicon containing a mutation in the initiation codon of uORF1 was amplified from the mut1 plasmid using these primers to generate the Fus1 NC plasmid. A PKC-η mRNA segment containing uORF1 and uORF2 (not including the stop codon of uORF2) was amplified from the WT plasmid using primer #4 (SI Appendix, Table S2). The resulting amplicon was digested by SacI and AvrII and cloned into the corresponding sites of the WT plasmid to generate the Fus2 plasmid. An identical amplicon containing a mutation in the initiation codon of uORF2 was amplified from the mut1 plasmid using these primers to generate the Fus2 NC plasmid.
Expression Plasmids pLV(uORF2) and pLV(uORF2mut) were designed (by us) and manufactured by VectorBuilder Inc. Vector details are available in SI Appendix, Table S3.
LENTICRISPRv2 Construction.
We engineered a CRISPR-Cas9 Lentiviral system based on the protocol perfected by the Zhang laboratory (63, 64). For PKC-η uORF2 guide RNA (gRNA) vector cloning, a custom single gRNA sequence that targets the uORF2 region of the human PRKCH gene (5′-CACCGTTAGGCGCTGCCTTTCCCCA-3′) and a nonspecific scrambled sequence (5′-CACCGATCGTTTCCGCTTAACGGCG-3′) were designed using Benchling software and synthesized from IDT-Syntezza. The LentiCRISPRv2Puro vector (no. 98290, Addgene) was digested using BsmBI (no. R0739, NEB) and dephosphorylated using shrimp Alkaline phosphatase (no. M0371S, NEB) for 30 mins at 37 °C. The digested plasmid was purified using a Universal DNA Purification Kit (no. DP214-02, TIANGEN BIOTECH). The gRNA and scramble oligos were phosphorylated and annealed using T4 PNK (no. M0201S, NEB) in T4 ligation buffer (no. M1801, Promega) in the following parameters: 37 °C, 30 min; 95 °C, 5 min; and ramp down to 25 °C at 5 °C/min. A total of 50 ng BsmBI digested plasmid and 1:100 diluted oligos were ligated using T4 ligase (no. M2200S, NEB) for 30 min at RT. The ligation mix was transformed into DH5-α (no. 18265017, Thermo Scientific Inc.), spread on Ampicillin agar plates, and incubated O/N at 37 °C. Positive clones were picked and cultured, and cloning was confirmed by sequencing.
Lentiviral Production and Transduction.
LentiCRISPRv2Puro carrying PKC-η uORF2 gRNA, scrambled constructs, or pLV(uORF2) plasmids (7 μg) were packaged into lentiviral particles using a mix of plasmids (7 μg) containing lentiviral packaging genes pCMV-VSV-G (no. 8454, Addgene) and psPAX2 (no. 12260, Addgene) in the ratio 1:9, respectively. The mix in combination with the specific plasmid was transfected into subconfluent HEK-293T using polyethylenimine reagent (no. 23966–1, Polysciences, Inc.). The 293T cells were then maintained in DMEM supplemented with 10% FBS and antibiotics and incubated overnight at 37 °C and 5% CO2. The medium was replaced 24 h after transfection, and 48 h later, the supernatants were collected, and the cell debris was removed by centrifugation and passed through a 0.45-µm filter. The viruses were concentrated using Amicon filter tubes (100 kDa MWCO) (no. ACS510024, Merck Millipore). A fresh culture of MCF-7 cells or the shPKC-η (sh3-3) clone was then infected with 100 μL concentrated virus suspension mix. Positive clones were isolated by Puromycin selection (5 μg/mL), and single-cell clones isolated were confirmed using PCR and sequencing. The uORF2 deletion in the single-cell clones established were confirmed by sequence analysis of PCR products obtained using primers (Forward: 5′-TTGGAAGGGACGGTCGG-3′; Reverse: 5′-GTTAGCGCAAAACTCCTCGT-3′) flanking the targeted DNA loci. Cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris HCl pH 8.0, 100 mM NaCl, 5 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), 1% Nonidet P-40, 45 mM β-mercaptoethanol, 10 mM ethylenediaminetetraacetic acid (EDTA) pH 8.0, and 50 mM NaF) including protease and phosphatase inhibitors (1 mM phenylmethylsulfonyl fluoride, 10 µg/mL Aprotinin, 10 µg/mL Leupeptin, 1 mM Na3VO4, 5 mM Na4P2O7, and 50 mM β-glycerol) and were separated on either 10% polyacrylamide or 16.5% Tris-Tricine gels and immunoblotted with the PKC-specific antibodies.
Immunoprecipitations and In Vitro Kinase Assays.
Immunoprecipitation and kinase assays were performed as described (23) using MBP (no. M1891, Sigma-Aldrich) (5 µg/100 μL reaction mix) as a substrate in the presence or absence of the indicated peptides. Briefly, HEK-293T lysates overexpressing HA-tagged PKCs (100 μg/sample) were immunoprecipitated using immobilized anti-HA Abs followed by the addition of indicated peptides and kinase reaction mixture (10 mM MgCl2, 20 mM Hepes, 0.1 mM EGTA, 50 µg/mL phosphatidylserine, 100 μM ATP, and 5 µCi γ-[32P]ATP [PerkinElmer]) plus either phorbol-12-myristate-13-acetate (PMA) (100 nM) and CaCl2 (0.3 mM) for classical PKCs or PMA (100 nM) for novel PKCs. Samples were incubated for 30 min at 32 °C with gentle shaking, and reactions were terminated by the addition of 5× sample buffer and boiling for 5 min. Samples were then subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis on 10% gels followed by a protein transfer to a nitrocellulose membrane (Sigma-Aldrich). After the detection of the phosphorylated substrate (32P-MBP) by autoradiography, the membranes were sequentially immunoblotted with anti-HA and anti-MBP Abs. 32P-MBP and MBP protein band signals were quantified using the Image Lab software (6.0.1).
Immunoprecipitation of the endogenous uORF2-encoded peptide was performed using MOLT4 and BeWo cells following treatment with 50 μM Bortezomib (PS-341) (no. S1013, SelleckChem) for 4 h in serum-free DMEM (to prevent protein degradation before lysis). Cells were washed with phosphate-buffered saline (PBS) and lysed as described before (23). Protein A beads (no. sc-2001, Santa Cruz Biotechnology) (20 μL) were coated with uORF2 antibody (anti-uPEP2; 30 μg/sample) for 1 h in 4 °C followed by two PBS washes. Cell lysates (1 to 1.5 mg/sample) were added to the bead antibody complex and incubated at 4 °C overnight. An anti-rabbit IgG isotype control was also used in immunoprecipitation simultaneously. Samples were prepared with Tricine sample buffer (no. 161–0739, Bio-Rad Laboratories, Inc.) and run on 16.5% Tris-Tricine gels.
To perform immunoprecipitation assays of Myc-tagged peptides with PKCs, HEK-293T cells were transfected with HA-tagged PKC constructs as described in ref. 23. Cell lysates were prepared using RIPA lysis buffer. Immunoprecipitation assays were performed using anti-Myc Abs. Anti-Myc mAbs were preadsorbed on protein A/G-agarose beads (no. sc-2003, Santa Cruz Biotechnology) for 1 h at 4 °C. Excess antibody was removed using wash buffer (25 mM Tris/HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100) followed by the addition of indicated control or Myc-tagged peptides and HEK-293T cell lysates overexpressing different PKC isoforms for 16 h at 4 °C. Immune complexes were washed extensively with wash buffer followed by the addition of SDS sample buffer or Tricine sample buffer, and the corresponding samples were subsequently run on either 10% polyacrylamide or 16.5% Tris-Tricine gels and immunoblotted with PKC-specific antibodies.
Antibodies.
Details of antibodies used in this study are given in SI Appendix, Table S4.
Cell Viability Assays.
Cells were seeded in 96-well plates (5 × 103 cells/well) and grown for 16 to 24 h. The indicated peptides were added in serum-free media for 4 h and replenished with serum for 24 h. XTT (2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide) assays were conducted according to the manufacturer’s instructions (no. 20–300-1000, BI). The plates were immediately read in a Multiskan Spectrum reader (Thermo Fisher Scientific). Cells plated at different densities in 96 wells at serial dilutions (1.25 to 20 × 103 cells) were assayed by XTT after 24 h to generate a standard curve of cell number versus absorbance for all cell lines. Each experiment was repeated at least three times in triplicates. For cell proliferation assays of uORF2-mutated/control (CRISPR/Cas9) MCF-7 clones, cells were seeded (5 × 103 cells/well) in 96-well plates. Cells were trypsinized, collected, and counted using an automated cell counter (Countess II FL, Thermo Scientific Inc.) at intervals of 24 h for a period of 5 d.
Wound Healing (Scratch) Assays.
MCF-7, MDA-MB-231, and MCF10A cells were seeded in 24-well plates and grown to confluence. A gap in the confluent monolayer (middle of the well) was created using a sterile pipette tip (200 μL) followed by two PBS washes. Subsequently, medium containing low serum (2% for MCF-7, 0.1% for MDA-MB-231, and 5% for MCF10A) and the indicated peptides (5 μM) were added to the wells for 4 h. The respective complete media (with peptides) were readded, and wells were photographed (three images for each well at the indicated time points) at 4× magnification with an IX70 Olympus Optical light microscope. The wound areas were measured and normalized to time 0 using ImageJ software (1.53i).
Orthotropic Breast Cancer Models in Mice.
All in vivo experiments were conducted using 6- 8-wk-old NSGS (The Jackson Laboratory, NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ), or NOD-SCID mice (Envigo, NOD.CB1 Prkdcscid/NCrHsd). MDA-MB-231-Luc cells (4 × 106) were suspended in PBS (100 μL) and injected subcutaneously in the mammary fat pad of NSG or NOD-SCID mice (65). When tumors reached an average size of ∼100 mm3, mice were randomized into three groups (five mice/group) and injected intratumor with PBS, uPEP2(-Myr), or uPEP2 (10 mg/kg) on alternate days.
Tumor volume was measured three times a week using a digital Vernier caliper. The tumor volumes (mm3) were calculated using the following formula V = (W2 × L)/2 (V, tumor volume; W, tumor width; L, tumor length). Mice were killed when tumors reached ∼10% of body weight (∼2,000 mm3). At the end of the experiment, all animals were weighed, euthanized, and liver, lung, spleen, kidney, brain, and tumors were harvested. The weights of tumors and organs were recorded. Part of the tumors were preserved in 4% paraformaldehyde for histology and immunohistochemistry analysis. For immunoblot analysis, the dissected tumors were snap frozen in liquid nitrogen. Tumor tissue samples (25 mg) were homogenized in RIPA buffer (500 μL) containing protease inhibitors followed by sonication for 3 to 5 min. Homogenized tissues were centrifuged at 16,000 × g for 20 min at 4 °C, and supernatants were collected and subjected to Western blot analysis.
Whole-body images of luciferase expression in NSG/NOD-SCID mice were monitored using the Xenogen In Vivo Imaging System (Xenogen). After anesthetizing the mice, 200 μL of D-luciferin (15 mg/mL) (no. LUCK-1G, Gold Biotechnology) was injected intraperitoneally, and in vivo imaging analysis was performed. Luciferase expression data were quantified using Living Image software (4.7.3) in a fixed region of interest in Pixels.
Immunohistochemistry and TUNEL Assay.
Formalin-fixed, paraffin-embedded blocks with tumor samples were sectioned at a thickness of 5 μm using a fully automated rotary microtome (no. RM2255, Leica), dried for 1 h at 65 °C, de-paraffinized, and rehydrated. The slides were incubated in 10 mM citric acid buffer, pH 6.0 at 100 °C for 20 min for antigen retrieval. The endogenous peroxidase activity was blocked with H2O2 (0.3%). Sections were then blocked for 1 h at room temperature with blocking solution (0.1% Tween, 5% bovine serum albumin) followed by incubation with primary antibody Ki67 (1:500). The VECTASTAIN ABC Kits (no. PK-6200, Vector Laboratories, Inc.) were used for detection according to the manufacturer’s protocol. Paraffin-embedded tissue slides were pretreated, and TUNEL assay (no. G3250, DeadEnd Fluorometric TUNEL System, Promega) was performed according to the manufacturer’s protocol to detect apoptosis. Images were captured by a PANNORAMIC MIDI scanner (3DHISTECH) and analyzed by QuPath software (0.2.1). The number of positive nuclei and the annotated area of each tissue of both Ki67 and TUNEL were calculated, and the results are denoted as the percentage of cells with positive nuclei using the software.
Statistical Analysis.
The statistical significance of differences between experimental groups was determined using the unpaired two-tailed Student's t test and the ANOVA test of variance in cases of multiple variables (using GraphPad Prism 5.2.1 software). P value < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by The Israel Science Foundation (Grant 2368/19 to E.L.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018899118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Johnstone T. G., Bazzini A. A., Giraldez A. J., Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J. 35, 706–723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young S. K., Wek R. C., Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J. Biol. Chem. 291, 16927–16935 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J., et al., Pervasive functional translation of noncanonical human open reading frames. Science 367, 1140–1146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu P. Y., Benfey P. N., Small but mighty: Functional peptides encoded by small ORFs in plants. Proteomics 18, e1700038 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Barbosa C., Peixeiro I., Romão L., Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 9, e1003529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spriggs K. A., Bushell M., Willis A. E., Translational regulation of gene expression during conditions of cell stress. Mol. Cell 40, 228–237 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Ingolia N. T., Ghaemmaghami S., Newman J. R., Weissman J. S., Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juntawong P., Girke T., Bazin J., Bailey-Serres J., Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, E203–E212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingolia N. T., Lareau L. F., Weissman J. S., Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fritsch C., et al., Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 22, 2208–2218 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingolia N. T., Hussmann J. A., Weissman J. S., Ribosome profiling: Global views of translation. Cold Spring Harb. Perspect. Biol. 11, a032698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji Z., Song R., Regev A., Struhl K., Many lncRNAs, 5'UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife 4, e08890 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S., et al., Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 109, E2424–E2432 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris D. R., Geballe A. P., Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20, 8635–8642 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Werner M., Feller A., Messenguy F., Piérard A., The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell 49, 805–813 (1987). [DOI] [PubMed] [Google Scholar]
- 16.Jorgensen R. A., Dorantes-Acosta A. E., Conserved peptide upstream open reading frames are associated with regulatory genes in angiosperms. Front Plant Sci 3, 191 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Law G. L., Raney A., Heusner C., Morris D. R., Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J. Biol. Chem. 276, 38036–38043 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Liu J., et al., Selective inhibition of angiotensin receptor signaling through Erk1/2 pathway by a novel peptide. Am. J. Physiol. Regul. Integr. Comp. Physiol. 306, R619–R626 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samandi S., et al., Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins. eLife 6, e27860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee C. F., Lai H. L., Lee Y. C., Chien C. L., Chern Y., The A2A adenosine receptor is a dual coding gene: A novel mechanism of gene usage and signal transduction. J. Biol. Chem. 289, 1257–1270 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raveh-Amit H., et al., Translational control of protein kinase Ceta by two upstream open reading frames. Mol. Cell. Biol. 29, 6140–6148 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zurgil U., et al., PKCη is an anti-apoptotic kinase that predicts poor prognosis in breast and lung cancer. Biochem. Soc. Trans. 42, 1519–1523 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Zurgil U., et al., PKCη promotes senescence induced by oxidative stress and chemotherapy. Cell Death Dis. 5, e1531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rotem-Dai N., Oberkovitz G., Abu-Ghanem S., Livneh E., PKCeta confers protection against apoptosis by inhibiting the pro-apoptotic JNK activity in MCF-7 cells. Exp. Cell Res. 315, 2616–2623 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Tamarkin A., et al., DNA damage targets PKCη to the nuclear membrane via its C1b domain. Exp. Cell Res. 317, 1465–1475 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Abu-Ghanem S., Oberkovitz G., Benharroch D., Gopas J., Livneh E., PKCeta expression contributes to the resistance of Hodgkin’s lymphoma cell lines to apoptosis. Cancer Biol. Ther. 6, 1375–1380 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Karp G., et al., Localization of PKCη in cell membranes as a predictor for breast cancer response to treatment. Onkologie 35, 260–266 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Newton A. C., Protein kinase C: Perfectly balanced. Crit. Rev. Biochem. Mol. Biol. 53, 208–230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isakov N., Protein kinase C (PKC) isoforms in cancer, tumor promotion and tumor suppression. Semin. Cancer Biol. 48, 36–52 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Rosse C., et al., PKC and the control of localized signal dynamics. Nat. Rev. Mol. Cell Biol. 11, 103–112 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Mellor H., Parker P. J., The extended protein kinase C superfamily. Biochem. J. 332, 281–292 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newton A. C., Regulation of protein kinase C. Curr. Opin. Cell Biol. 9, 161–167 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Newton A. C., Protein kinase C: Structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem. Rev. 101, 2353–2364 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Nishikawa K., Toker A., Johannes F. J., Songyang Z., Cantley L. C., Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol. Chem. 272, 952–960 (1997). [DOI] [PubMed] [Google Scholar]
- 35.House C., Kemp B. E., Protein kinase C contains a pseudosubstrate prototope in its regulatory domain. Science 238, 1726–1728 (1987). [DOI] [PubMed] [Google Scholar]
- 36.Newton A. C., Regulation of the ABC kinases by phosphorylation: Protein kinase C as a paradigm. Biochem. J. 370, 361–371 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laudanna C., Mochly-Rosen D., Liron T., Constantin G., Butcher E. C., Evidence of zeta protein kinase C involvement in polymorphonuclear neutrophil integrin-dependent adhesion and chemotaxis. J. Biol. Chem. 273, 30306–30315 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Volk L. J., Bachman J. L., Johnson R., Yu Y., Huganir R. L., PKM-ζ is not required for hippocampal synaptic plasticity, learning and memory. Nature 493, 420–423 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michel A. M., et al., GWIPS-viz: Development of a ribo-seq genome browser. Nucleic Acids Res. 42, D859–D864 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sidrauski C., McGeachy A. M., Ingolia N. T., Walter P., The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. eLife 4, e05033 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cenik C., et al., Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans. Genome Res. 25, 1610–1621 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elkon R., et al., Myc coordinates transcription and translation to enhance transformation and suppress invasiveness. EMBO Rep. 16, 1723–1736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jang C., Lahens N. F., Hogenesch J. B., Sehgal A., Ribosome profiling reveals an important role for translational control in circadian gene expression. Genome Res. 25, 1836–1847 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fields A. P., et al., A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol. Cell 60, 816–827 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mochly-Rosen D., Das K., Grimes K. V., Protein kinase C, an elusive therapeutic target? Nat. Rev. Drug Discov. 11, 937–957 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antal C. E., et al., Cancer-associated protein kinase C mutations reveal kinase’s role as tumor suppressor. Cell 160, 489–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fima E., et al., PKCeta enhances cell cycle progression, the expression of G1 cyclins and p21 in MCF-7 cells. Oncogene 20, 6794–6804 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Karp G., Maissel A., Livneh E., Hormonal regulation of PKC: Estrogen up-regulates PKCeta expression in estrogen-responsive breast cancer cells. Cancer Lett. 246, 173–181 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Fu G., et al., Protein kinase C η is required for T cell activation and homeostatic proliferation. Sci. Signal. 4, ra84 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basu A., The enigmatic protein kinase C-eta. Cancers (Basel) 11, 214 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akkaraju G. R., Basu A., Overexpression of protein kinase C-eta attenuates caspase activation and tumor necrosis factor-alpha-induced cell death. Biochem. Biophys. Res. Commun. 279, 103–107 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Raveh-Amit H., et al., Protein kinase Cη activates NF-κB in response to camptothecin-induced DNA damage. Biochem. Biophys. Res. Commun. 412, 313–317 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Hussaini I. M., et al., Protein kinase C-eta regulates resistance to UV- and gamma-irradiation-induced apoptosis in glioblastoma cells by preventing caspase-9 activation. Neuro-oncol. 4, 9–21 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonnemann J., et al., Down-regulation of protein kinase Ceta potentiates the cytotoxic effects of exogenous tumor necrosis factor-related apoptosis-inducing ligand in PC-3 prostate cancer cells. Mol. Cancer Ther. 3, 773–781 (2004). [PubMed] [Google Scholar]
- 55.Ward I. M., Chen J., Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276, 47759–47762 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Banáth J. P., Olive P. L., Expression of phosphorylated histone H2AX as a surrogate of cell killing by drugs that create DNA double-strand breaks. Cancer Res. 63, 4347–4350 (2003). [PubMed] [Google Scholar]
- 57.Newton A. C., Protein kinase C: Poised to signal. Am. J. Physiol. Endocrinol. Metab. 298, E395–E402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rybin V. O., et al., Cross-regulation of novel protein kinase C (PKC) isoform function in cardiomyocytes. Role of PKC epsilon in activation loop phosphorylations and PKC delta in hydrophobic motif phosphorylations. J. Biol. Chem. 278, 14555–14564 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Steinberg S. F., Structural basis of protein kinase C isoform function. Physiol. Rev. 88, 1341–1378 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duquesnes N., Lezoualc’h F., Crozatier B., PKC-delta and PKC-epsilon: Foes of the same family or strangers? J. Mol. Cell. Cardiol. 51, 665–673 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Doi S., Goldstein D., Hug H., Weinstein I. B., Expression of multiple isoforms of protein kinase C in normal human colon mucosa and colon tumors and decreased levels of protein kinase C beta and eta mRNAs in the tumors. Mol. Carcinog. 11, 197–203 (1994). [DOI] [PubMed] [Google Scholar]
- 62.Guex N., Peitsch M. C., SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997). [DOI] [PubMed] [Google Scholar]
- 63.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shalem O., et al., Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iorns E., et al., A new mouse model for the study of human breast cancer metastasis. PLoS One 7, e47995 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.