Significance
Pyridoxal phosphate (PLP)-dependent enzymes rarely react with oxygen, but an emerging group of oxygen-, PLP-dependent enzymes oxidize l-arginine. Two types of oxidases are known: hydroxylases and desaturases. We demonstrate that arginine desaturases have a minor hydroxylase activity and then show through X-ray crystallographic, mutagenesis, spectroscopic, and computational studies that their mechanism involves two rounds of single-electron transfer to oxygen and superoxide rebound, ultimately giving a conjugated hydroperoxyl intermediate. Water can attack to give a hydroxylated product and release H2O2, but with water absent, the intermediate can be deprotonated, instead giving a desaturated product and H2O2. Our work outlines the unique mechanism and evolutionary history of these enzymes and sets the stage toward engineering these enzymes to catalyze oxidative reactions.
Keywords: pyridoxal phosphate, oxidase, X-ray crystallography, water, molecular dynamics
Abstract
The mechanism by which molecular oxygen is activated by the organic cofactor pyridoxal phosphate (PLP) for oxidation reactions remains poorly understood. Recent work has identified arginine oxidases that catalyze desaturation or hydroxylation reactions. Here, we investigate a desaturase from the Pseudoalteromonas luteoviolacea indolmycin pathway. Our work, combining X-ray crystallographic, biochemical, spectroscopic, and computational studies, supports a shared mechanism with arginine hydroxylases, involving two rounds of single-electron transfer to oxygen and superoxide rebound at the 4′ carbon of the PLP cofactor. The precise positioning of a water molecule in the active site is proposed to control the final reaction outcome. This proposed mechanism provides a unified framework to understand how oxygen can be activated by PLP-dependent enzymes for oxidation of arginine and elucidates a shared mechanistic pathway and intertwined evolutionary history for arginine desaturases and hydroxylases.
Enzymes that use molecular oxygen as a cosubstrate normally employ metal or redox-active cofactors to enable single-electron transfers. By contrast, cofactors like pyridoxal phosphate (PLP) are more rarely employed for O2-dependent reactions. Nonetheless, arginine oxidases have emerged as a group of PLP-, O2-dependent enzymes that are found in biosynthetic pathways to heterocyclic natural products indolmycin, enduracididine, and azomycin. All characterized arginine oxidases are members of the aspartate aminotransferase protein family (1). Among arginine oxidases, there are two groups: the desaturases and the hydroxylases. The desaturases are exemplified by Ind4, which converts l-arginine (1) to 4,5-dehydro-2-iminoarginine (2), a net four-electron oxidation reaction (2). The hydroxylases are exemplified by SwMppP/RohP, enzymes that convert 1 to (S)-4-hydroxy-2-ketoarginine (3) as demonstrated through crystallographic trapping experiments (Fig. 1) (3–5). Both the desaturases and hydroxylases also share a two-electron oxidized product, 2-ketoarginine (4), as well as similarities in their spectroscopic features, including two intermediates observed to have ultraviolet visible (UV-Vis) maxima at 510 to 515 and 560 to 567 nm, respectively (2, 3, 5). Recent work has demonstrated that the hydroxylase enzymes are oxidases, with the hydroxyl group in 3 coming from water rather than from molecular oxygen (4, 5). This result suggests that the desaturases and hydroxylases may traverse through similar unsaturated intermediates. However, the relationship between these two groups of enzymes remains underexplored, and mechanistic insight into how oxygen is used in either group is unknown.
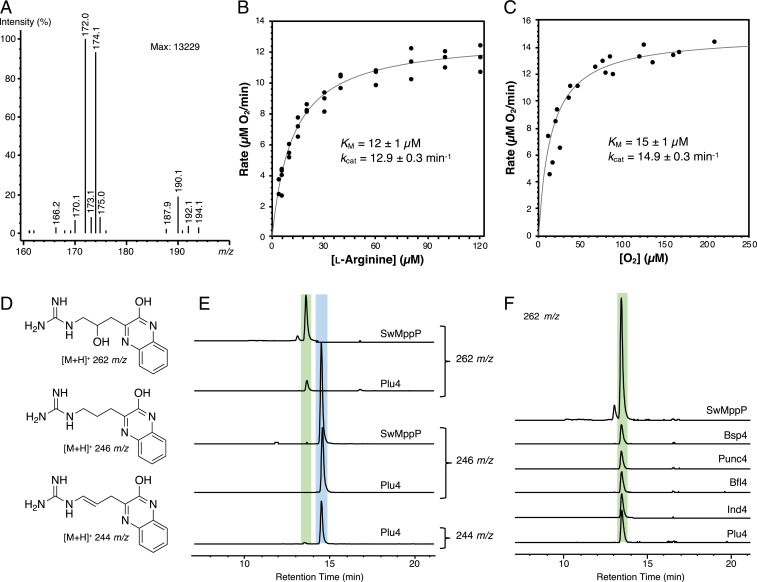
Fig. 1.
Scheme of PLP-, O2-dependent arginine oxidase reactivities. [M+H]+ masses are given below observed products. The Ind4-coupled reaction with Ind5 from the indolmycin biosynthetic pathway is also shown.
Here, we explore the relationship between these two types of arginine oxidases with the intention of unveiling the shared and distinctive mechanistic details for these two types of enzymes. Through phylogenetic, structural, and biochemical characterization, we demonstrate that these two groups of enzymes are more highly similar to one another than previously thought, despite catalyzing different major reactions. Importantly, molecular dynamics studies show that the presence or absence of an appropriately positioned water likely plays a critical role in delineating the product outcome. Furthermore, computations suggest that there may be a superoxide rebound onto the 4′ carbon of the PLP cofactor. This result raises the exciting possibility that this reaction has no theoretical requirement to employ l-arginine as a substrate and could be evolved to emerge in aminotransferases specific to other substrates.
Results
Biochemical Characterization of Plu4 Reveals that Arginine Desaturases Have a Minor Hydroxylase Activity.
To elucidate the determinants of catalysis in arginine desaturases, we attempted to solve the X-ray crystal structure of Ind4. When we were unable to crystallize Ind4, we focused on Plu4, encoded in the indolmycin gene cluster from Pseudoalteromonas luteoviolacea (6) and which has 32% sequence identity to Ind4. Using liquid chromatography–mass spectrometry (LC-MS) analysis, we found that purified Plu4 performs an identical reaction to that of Ind4, producing 174 and 172 m/z signals in the presence of catalase and 1 (Fig. 2A), corresponding to expected [M+H]+ ions for 4 and 5, respectively. We also found that this reaction likely traverses through 2 because we observed the corresponding [M+H]+ 171 m/z signal during shorter reaction times (SI Appendix, Fig. S1). In steady-state kinetic analyses using an oxygraph, Plu4 was most active at a pH of 8.5 to 9.0 (SI Appendix, Fig. S2). When assayed using [tris(hydroxymethyl)methylamino]propanesulfonic acid buffer (I = 0.1; pH 8.5) at 25 °C, Plu4 had KM and kcat values of 12 ± 1 µM and 12.9 ± 0.3 min−1, respectively, for l-arginine and KM and kcat values of 15 ± 1 µM and 14.9 ± 0.3 min−1, respectively, for O2 (Fig. 2 B and C). Altogether, these data support that Plu4 is a new member of the arginine desaturases, consistent with its predicted function in indolmycin biosynthesis in P. luteoviolacea.
Fig. 2.
Plu4 reaction products and kinetics. (A) LC-MS analysis of Plu4 reaction with 1. (B and C) Steady-state kinetic analyses of the Plu4-catalyzed reaction, with the lines representing fits of the Michaelis–Menten equation to the data: dependence of initial velocity on 1 (B) and O2 (C). Concentrations were measured by the rate of oxygen consumption. (D) Structures of OPD-derivatized products and their corresponding [M+H]+ signals. (E) Comparison of Plu4 and SwMppP products as extracted ion chromatograms from LC-MS after 20 min OPD incubations. The extracted ion chromatogram for the SwMppP reaction at m/z 244 is shown in SI Appendix, Fig. S6. (F) Comparison of OPD-derivatized products from several desaturases and SwMppP. Traces are extracted ion chromatograms at 262 m/z from LC-MS. Peaks corresponding to 3 are highlighted in green, while peaks corresponding to 4 and 5 are highlighted in blue in E and F.
In addition to this main reaction, we noticed a minor peak at 190 m/z, which matches the [M+H]+ signal for 3 (Fig. 2A and SI Appendix, Fig. S1). To determine the identity of this side product, we used o-phenylenediamine (OPD) to derivatize the products of the Plu4 reaction and separate them by LC-MS (Fig. 2D). An analysis of the derivatized products revealed a small peak with the same [M+H]+ signal of 262 m/z and retention time as derivatized 3 from SwMppP (Fig. 2E), suggesting that the structure could also be OPD-derivatized 3. The new peak we observed in the Plu4 reaction did not appear when the enzyme or 1 were omitted or when the enzyme had been boiled (SI Appendix, Fig. S3A), suggesting that it arises from an enzymatic process involving 1. To further validate that it arises from an enzymatic reaction, we demonstrated that the peak does not increase when the enzyme is removed (SI Appendix, Fig. S3B) and that the side product incorporates an 18O label from H218O, as in the SwMppP and RohP reactions (4, 5) (SI Appendix, Fig. S4). We then revisited whether this peak is seen in reactions with other homologs (Fig. 2F and SI Appendix, Fig. S5 and Table S1) and demonstrated the presence of this minor product under the same reaction conditions in the seven Ind4/Plu4 homologs available to us. We also tested whether the [M+H]+ signal of 244 m/z, consistent with OPD-derivatized 4,5-dehydro-2-ketoarginine (5), was present in SwMppP reactions. While this peak is present with SwMppP, we observed that it comes mainly from a nonenzymatic dehydration reaction of the hydroxylated product, with the m/z 244 peak appearing concurrently with the disappearance of the m/z 262 peak during longer incubation times (SI Appendix, Fig. S6). We quantified Plu4 products by high-performance liquid chromatography using a shorter incubation time of 3 min, determining that the amount of derivatized 3 is ∼35-fold less than the peak corresponding to derivatized 4 and 5. By contrast, in SwMppP, the production of derivatized 3 is ∼1.7-fold less than derivatized 4 and 5 (SI Appendix, Fig. S7). We also found that the amount of derivatized 3 detected from SwMppP is ∼29-fold more than the amount from Plu4 (SI Appendix, Fig. S7), suggesting a bona fide difference in reaction outcome for the two enzymes.
Sequence Similarity Networks Reveal Unexpected Evolutionary History.
The production of a minor hydroxylated product from Plu4 and other characterized desaturases suggests a close relationship between the desaturases and the hydroxylases. To probe the relationship between the two types of arginine oxidases, we made sequence similarity networks (SSNs) (7). Using Ind4 as a query, an initial SSN with a threshold of 10−30 was made with the top thousand sequences and showed that the known arginine oxidases are isolated from other top hits, which are uncharacterized bacterial aminotransferases (Fig. 3). An SSN with a threshold of 10−65 was then made using only the 296 sequences available from UniProt that clustered with the known arginine oxidases, revealing three distinct groups (Fig. 3). We also analyzed the genomic contexts of each of these nodes to assign probable function based on neighboring genes. Over 14 different gene organizations were observed (SI Appendix, Figs. S8–S11). Among these, four gene organizations—enduracididine, azomycin, mildiomycin, and indolmycin—have known functions and are therefore annotated in Fig. 3.
Fig. 3.
SSN analysis for the arginine oxidases. Each node represents a different enzyme from the UniProtKB database, and the edges represent sequence similarity relationships. Node shapes represent the characterized function of the enzymes, while node colors indicate information about surrounding genes in the genome. Previously characterized nodes and newly characterized Plu4 are labeled. Proximal indicates the plu5 gene homolog is adjacent to the ind4 homolog, whereas distal indicates a positioning of plu5 elsewhere in the gene cluster. Dashed boxes indicate nodes used for smaller networks at a higher threshold cutoff. These smaller networks at thresholds of 1 × 10−65 and 1 × 10−85 are shown as insets. Each predicted reactivity group is labeled as I to III.
The annotated SSN showed that hydroxylases and desaturases do not neatly separate into two groups. The main Group I, separated at a threshold of 10−65, consists of diverse gene clusters and previously characterized activities. Group I was further analyzed at a 10−85 threshold, revealing three isofunctional subgroups, Ia, Ib, and Ic (Fig. 3). Group Ia contains enzymes in both azomycin- and mildiomycin-like gene clusters (8, 9), including the previously characterized hydroxylase, RohP. Group Ib consists of enzymes in indolmycin-like gene clusters from Streptomyces, such as previously reported desaturases Ind4 and Pel4 (2). A genomic neighborhood analysis of nodes within Group Ib showed obligatory pairings of the arginine desaturase genes with an ind5-like gene and, in some cases, a nearby ind3-like gene (SI Appendix, Fig. S10), suggesting a bona fide role for an arginine desaturase. The function of Group Ic nodes and their associated gene clusters are unknown. The other major groups are Group II, which consists of enzymes in enduracididine-like gene clusters, including the characterized hydroxylase, SwMppP, and Group III, which contains Plu4 and other enzymes colocalized with plu5-like genes and sometimes ind3-like genes (SI Appendix, Fig. S11). These colocalized genes suggest that many members of Group III could also function as arginine desaturases. Sequence similarity patterns and phylogenetic trees of the arginine oxidases suggest that either the hydroxylase activity or the desaturase activity emerged on at least two different occasions during evolution (Fig. 3 and SI Appendix, Fig. S12). This phylogenetic relationship could make elucidating the basis of differentiation challenging because there could be more than one evolutionary pathway that each group took to reach the same reactivity.
Crystal Structure of Plu4 Shows Desaturases and Hydroxylases have Highly Similar Active Sites.
Our biochemical and bioinformatics analyses show a complex evolutionary relationship between the desaturase and hydroxylase activities. To better understand the relationship between the two types of enzymes, we crystallized Plu4 and collected datasets of Plu4 crystals soaked with PLP and/or l-arginine in aerobic conditions (SI Appendix, Table S4). During these soaking experiments, we observed crystals change from yellow to orange and back to yellow (SI Appendix, Fig. S13), and we froze crystals at multiple time points. The resulting crystal structures were solved by molecular replacement using RohP (Protein Data Bank [PDB] ID: 6C3D) as a search model.
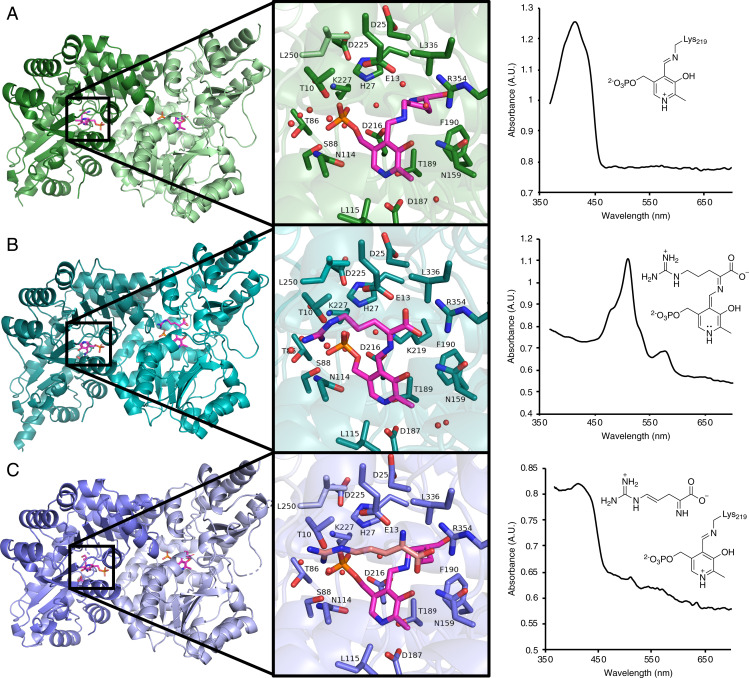
We obtained a holoenzyme structure of Plu4 (PDB: 7N79) at 2.0-Å resolution from soaking a crystal with 2 mM PLP for 2 h. This crystal shows the characteristic 415-nm peak for the Schiff base as observed by UV-Vis data on the crystal itself (Fig. 4A). This structure is a homodimer with an overall fold that matches SwMppP (5DJ1) (3), RohP (6C3B) (5), and other fold-type I PLP enzymes (SI Appendix, Table S5) (10, 11). Each monomer in the dimer contains a large domain (residues Asp31-Phe270) and small domain (residues Gly271-Lys381). The N-terminal helix (residues Met1-His30) is located at the dimer interface and helps enclose the active site. In contrast to holo-RohP and holo-SwMppP, in which the N-terminal region is disordered or pointed completely away from the active site (SI Appendix, Fig. S14), the N terminus of holo-Plu4 is fully ordered and closed over the active site. However, the ordering of the N-terminal helix in Plu4 may be the result of crystallographic contacts between asymmetric units and areas near the N terminus, creating a smaller cavity around the N-terminal helix for Plu4 than for RohP and SwMppP, which may result in restricted movement and reduce disordering of this helix (SI Appendix, Fig. S14).
Fig. 4.
X-ray crystal structures of Plu4. (A) Holoenzyme (PDB: 7N79). (B) Quinonoid I structure (PDB: 7RF9). (C) Product 2-bound structure (PDB: 7RGB). (Left) Overall fold. (Middle) Active-site view. (Right) Microspectrophotometer UV spectra obtained from the crystal used for X-ray crystallographic data collection and the corresponding molecules built into the active site. The PLP cofactor is shown with magenta carbons.
The holoenzyme contains the internal aldimine form of the PLP cofactor, attached to Lys219 via a Schiff base (SI Appendix, Fig. S15A). The PLP cofactor is also held in place by Asp187, which interacts with the nitrogen of the PLP pyridine ring (11) (SI Appendix, Fig. S15B) and by Asn159, which interacts with the phenolic oxygen (12). Furthermore, the phosphate group of PLP interacts with the Ala87 backbone nitrogen and the side chains of Ser88, Lys227, and His27 (SI Appendix, Fig. S15B). The majority of the active-site residues are identical between the holo-RohP, holo-SwMppP, and holo-Plu4 structures, but some residues, including His27, Asp25, Phe112, Asn114, Leu115, and Phe190, are observed in different conformations in holo-Plu4 compared to the hydroxylases (SI Appendix, Figs. S15 and S16).
We obtained a quinonoid-bound structure of Plu4 (PDB: 7RF9) to 1.9-Å resolution by soaking a Plu4 crystal with 3 mM l-Arg and 2 mM PLP for 8 min, during which the crystal changed from yellow to orange. By microspectrophotometer analysis, this crystal showed a major peak absorbance at λmax 510 nm and a minor peak at λmax 578 nm (Fig. 4B). In Ind4, the 511-nm species, which showed O2-dependent decay, was assigned as a quinonoid (Quinonoid I) via rapid-scanning, stopped-flow spectrophotometry experiments. These studies also suggest that the 567-nm species is a more conjugated quinonoid (Quinonoid II), which is also likely to show O2-dependent decay (2). When refinement was nearly complete, including the placement of waters, five possible intermediates were built into the active-site density (SI Appendix, Fig. S17). The Quinonoid I intermediate was chosen for the final structure based on the dominant UV microspectrophotomer signal observed at 510 nm; however, since oxygen was present during the crystallization process, it is likely that a mixture of intermediates was present, which is further evident from the additional signal at 578 nm. Although large structural changes can sometimes be observed in fold-type I PLP enzymes upon ligand binding (13, 14), there were no major changes between the holo structure and the intermediate-bound structure.
Comparing the internal aldimine of the holoenzyme with the Quinonoid I ligand of the intermediate-bound structure, the PLP cofactor rotates away from Lys219 by 23.8° (SI Appendix, Fig. S18), anchored by the heterocyclic nitrogen and phosphate group. Additionally, the bond between carbon and oxygen from the phosphate group rotates to accommodate this movement (SI Appendix, Fig. S18). In this structure, Lys219 is released from the imine bond and forms a polar interaction with His27, Asp216, and the PLP phosphate group, facilitated by a water molecule (SI Appendix, Fig. S19A). With the addition of substrate, new polar contacts are formed between the substrate and surrounding residues, including Thr10 and Glu13 from the N-terminal helix with the guanidine and carboxylate groups of 1, respectively, and Arg354, which forms a strong salt bridge interaction with the carboxylate group of 1 (SI Appendix, Fig. S19 B and C). Glu13 and His27 are within 4 Å of carbons 4 and 5 of the intermediate (SI Appendix, Fig. S19D). All other residues surrounding the active site appear to be unchanged in response to the formation of the Quinonoid I (SI Appendix, Fig. S18).
Finally, we obtained a 2.5-Å resolution product-bound structure (PDB: 7RGB) by soaking a Plu4 crystal with 2 mM PLP and 2 mM l-Arg for 16 h, during which the crystal changed from yellow to orange and back to yellow. The microspectrophotometer data showed a peak at 413 nm, indicative of an internal aldimine (Fig. 4C). Again, there were no large structural changes observed in this structure compared to the holo structure or the intermediate structure. However, segments of external loops (Asp302-Glu303 in Chain A and Gly293-Thr294 and Asp302-Glu303 in Chain B) became disordered in the product structure. Only one chain in each dimer in the asymmetric unit had electron density corresponding to a product in the active site. Before modeling any products in this density, we performed density functional theory (DFT) model calculations to determine which tautomeric form of the four-electron oxidized product was energetically more stable. Surprisingly, the imine form was calculated as more stable, despite the enamine form being more conjugated (SI Appendix, Fig. S20). Therefore, the isomer 2 was modeled into the active-site electron density corresponding to product formation (SI Appendix, Fig. S21A), although the active sites may contain a mixture of products, and at this resolution, it is not possible to provide crystallographic evidence for a specific tautomer. When modeled into the active site, the imine of 2 was pointed toward the carboxylate group of Glu13 (SI Appendix, Fig. S21B), in contrast to the interaction between the corresponding Glu and a product carboxylate oxygen in product-bound structures of RohP and SwMppP (SI Appendix, Fig. S21C). We determined that this unexpected binding conformation is energetically feasible through docking both products into their respective active sites (SI Appendix, Fig. S21D). These binding modes are triggered by the strong interaction between the product carboxylate group and Arg354. The active-site residues are, again, highly similar in conformation and identity to those in the previous holo and intermediate structures, with reformation of the internal aldimine making the position of the PLP cofactor comparable to the holoenzyme conformation (SI Appendix, Fig. S18).
Mutagenesis Reveals Residues Important for 4,5-dehydro-2-ketoarginine Formation.
Guided by our crystallographic studies of Plu4 and previous site-directed mutagenesis studies on the PLP-dependent arginine oxidases, we hypothesized that His27 and Asp225, two highly conserved active-site residues (SI Appendix, Fig. S22), could play a role in the second oxidative step. Previous site-directed mutagenesis of the His residue in RohP implicated it in catalysis during the second oxidation (5). We here made substitutions at residue His27 and nearby conserved residue Asp25, along with Asp225 to determine their effect on catalysis. In the H27A, D225A, D225N, and D25A variants, the first oxidation product 4 is still made, but the second oxidation product 5 is not (SI Appendix, Table S6). By contrast, the D25N variant had the same product profile as the wild-type enzyme (SI Appendix, Table S6), suggesting that Asp25 does not have an essential role in catalysis but may play a structural role during the second oxidation. While this result provides strong evidence that His27 and Asp225 are involved in catalysis during the second oxidative step, their specific role is still unclear. Glu15 in SwMppP, which corresponds to Glu13 in Plu4, was previously shown to be important in the formation of 3 through site-directed mutagenesis studies (4). While we did not test the corresponding variant of Plu4 (E13A), the Glu13 residue is positioned close enough to C4 (3.8 Å) and C5 (3.1 Å) in the intermediate-bound structure that it could also have an equivalent role in forming 5. Collectively, His27, Asp25, Asp225, and Glu13 may play important catalytic and/or structural roles in the second oxidation catalyzed by Plu4.
Next, we targeted residues within 5 Å of the active site that differ between the hydroxylases and the desaturases to try to find the cause of the different reactivities (SI Appendix, Fig. S23). Targeted residues include Thr189, Gly26, and Ala28, of which each has a different identity in both RohP and SwMppP. The corresponding substitutions were also made in Ind4 to match the RohP identity, since our SSN and phylogenetic tree analysis suggests that Group Ia, containing RohP, and Group Ib, containing Ind4, are the most closely related desaturases and hydroxylases. Variants G26A and A28T in Plu4 showed a complete loss of formation of both products, but T189C and T189S in Plu4 gave no change in the product profile (SI Appendix, Table S6). However, all substitutions at these positions in Ind4 resulted in loss of activity (SI Appendix, Table S6). Two nonconserved residues, Tyr29 and Cys251, near Asp225 were also targeted, but the substitutions in Plu4 either abolished production of both 4 and 5 or did not affect production (SI Appendix, Table S6). These residues seem likely involved in maintaining structure around the catalytic residues rather than determining hydroxylase and desaturase reactivity, similar to Asp25. We also targeted Leu117, which is in a hydrophobic tunnel leading to the active site in the Plu4 structure. This tunnel is far less hydrophobic in the RohP structure, which appears to be the result of an aspartate residue at the position of Leu117. However, we did not observe any products from the Plu4 L117D variant (SI Appendix, Table S6).
Presence and Reactivity of Superoxide.
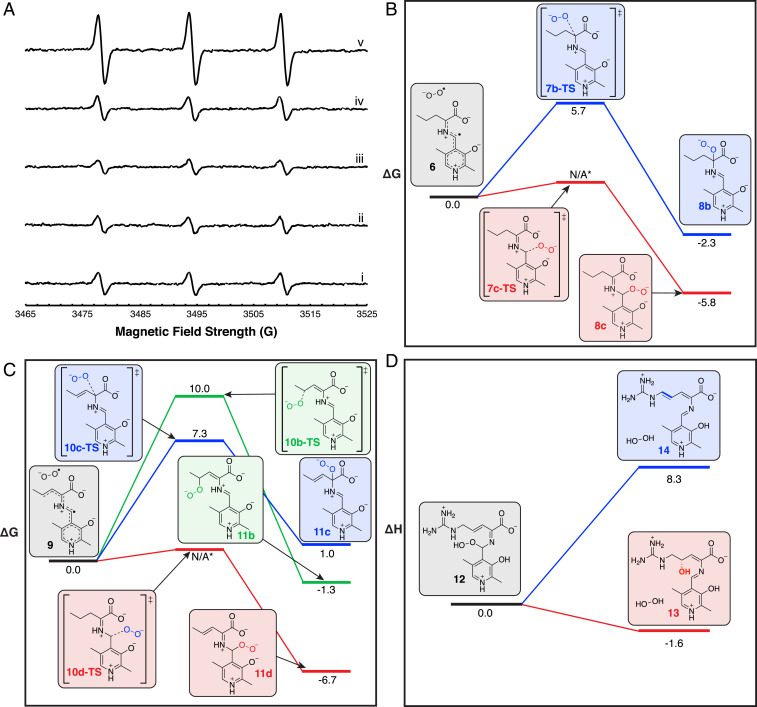
To better understand the role of oxygen in the arginine oxidase reactions, we used electron paramagnetic resonance (EPR) and a spin trap, cyclic hydroxylamine 1-hydroxy-3-methoxycarbonyl-2,2,5,5-tetramethylpyrrolidine (CMH), to look for the presence of superoxide during the course of the reaction (15, 16). We combined 0.5 mM ʟ-Arg and 1 mM Plu4 in the presence of 1 mM CMH to detect superoxide during the reaction. We observed a fourfold enhanced signal for CMH-trapped superoxide when both Plu4 and ʟ-Arg are present (Fig. 5A and SI Appendix, Fig. S24). When catalase was added to the reaction, the observed signal dropped to a twofold increase, and the signal increased slightly when only Plu4 and hydrogen peroxide were included. These controls indicate that the hydrogen peroxide produced during catalysis is able to react with CMH to some degree, but the remaining twofold signal from the catalase-containing reaction indicates that superoxide is produced from the Plu4 reaction. The presence of superoxide was further confirmed as an intermediate in the reaction by assaying Plu4 in the presence of cytochrome c, which uses superoxide to reduce Fe3+ to Fe2+, producing a UV-Vis signal at 550 nm (SI Appendix, Fig. S25). The rate of reduction in a reaction of Plu4, ʟ-Arg, and cytochrome c was 21.1 ± 0.5 μM ⋅ min−1, higher than the rate of 15.4 ± 0.5 μM ⋅ min−1 when superoxide dismutase—which consumes superoxide—was included in the assay. These results demonstrate that superoxide is produced in the Plu4 reaction.
Fig. 5.
EPR and DFT calculations used to determine the mechanism of arginine oxidases. (A) EPR spectra from CMH spin trap radical adduct. Traces show CMH (i); CMH and l-Arg (ii); CMH and Plu4 (iii); CMH, l-Arg, and boiled Plu4 (iv); and CMH, l-Arg, and Plu4 (v). For experimental parameters, see SI Appendix. (B) Free energy profile of possible peroxy adduct regioisomers during the first oxidation step (see SI Appendix, Fig. S26A). (C) Free energy profile of possible peroxy adduct regioisomers during the second oxidation step (see SI Appendix, Fig. S26B). (D) Change in enthalpy during deprotonation or hydroxylation of hydroperoxyl intermediate. Energies are given in kcal/mol. An asterisk indicates a transition state that could not be located.
To further investigate the role of superoxide, we explored the likelihood of possible mechanisms through DFT calculations using truncated computational models (see computational details in SI Appendix). We first calculated that a single-electron transfer to oxygen from Quinonoid I could generate intermediate 6 and superoxide (SI Appendix, Fig. S26A), consistent with our experimental studies demonstrating superoxide formation. We then explored the competing superoxide rebound at Cα or C4′ of PLP and Cβ hydrogen abstraction by superoxide (SI Appendix, Fig. S26A). The calculations showed that hydrogen abstraction at the β-carbon was less favorable than superoxide rebound, with ΔG‡ = 16.6 kcal/mol calculated for the C–H abstraction transition state (7a-TS) and ΔGR = 11.0 kcal/mol for the conjugated intermediate 8a. By comparison, superoxide rebound is more favorable in two scenarios. Rebound at the α-carbon was calculated to occur through the 7b-TS transition state (ΔG‡ = 5.7 kcal/mol, Fig. 5B and SI Appendix, Fig. S26A) to form the peroxy intermediate 8b (ΔGR = −2.3 kcal/mol). Furthermore, rebound at the C4′ position of PLP gives the thermodynamically more stable peroxy intermediate 8c, ΔGR = −5.8 kcal/mol. The putative transition state (7c-TS) could not be located for rebound of superoxide at the C4′ position of PLP, likely because of a very low energy barrier.
For the second oxidation, we calculated that a single-electron transfer to oxygen from Quinonoid II could form superoxide and intermediate 9 (SI Appendix, Fig. S26B). We then compared the possible superoxide hydrogen abstraction and superoxide rebound mechanisms to determine which was intrinsically more favorable. As in the first oxidation, superoxide C–H abstraction was energetically less favorable than superoxide rebound. The computed transition state for the hydrogen abstraction by superoxide at the δ-carbon (10a-TS) has a barrier of ΔG‡ = 18.8 kcal/mol, resulting in intermediate 11a formation (ΔGR = 8.4 kcal/mol). By contrast, the superoxide rebound mechanism was more favorable at the γ-carbon, α-carbon, and C4′ position of PLP. Calculated transition-state energies were ΔG‡ = 10.0 kcal/mol and ΔG‡ = 7.3 kcal/mol for rebound at the γ-carbon (10b-TS) and α-carbon (10c-TS), respectively, while the calculated intermediate formation reaction energies were ΔGR = −1.3 kcal/mol, ΔGR = 1.0 kcal/mol, and ΔGR = −6.7 kcal/mol for peroxy adducts formed at the γ-carbon (11b), α-carbon (11c), and C4′ position of PLP (11d), respectively (Fig. 5C and SI Appendix, Fig. S26B). Similar to the first oxidation, we could not locate the transition state to generate the peroxy adduct at the C4′ position of PLP (10d-TS), again likely due to a low-energy barrier, indicating the higher reactivity of this position. These results indicate that the proposed mechanism involving two single-electron transfer steps and the subsequent formation of two superoxide intermediates is energetically feasible (Fig. 6B, involving Quinonoid I and II).
Fig. 6.
Divergence of hydroxylase and desaturase mechanistic pathways. (A) Representative snapshots obtained from MD simulations with hydroperoxyl intermediate 15 bound in the RohP hydroxylase and Plu4 desaturase. The presence of a preorganized water molecule for nucleophilic addition at the substrate C4 position depends on the geometry adopted by intermediate 15 in the active site, which is influenced by the conformations explored by Phe119 (RohP) and Phe112 (Plu4) side chains. (B) Proposed mechanism of arginine desaturases and hydroxylases. Relevant atom labels are shown in blue. Products observed via LC-MS [M+H]+ signals are indicated with a box.
Origins of Divergence in Mechanistic Pathways.
We then interrogated how such an intermediate could proceed via a deprotonation or hydroxylation pathway by calculating the thermodynamics of these pathways on a more complete computational model containing the guanidino group. We calculated that the hydroperoxyl intermediate (12) can react with water to form the hydroxylated intermediate (13) and H2O2 (ΔGR = −1.5 kcal/mol), or it can directly produce the fully unsaturated intermediate (14) and H2O2 (ΔGR = −4.4 kcal/mol) (Fig. 5D and SI Appendix, Fig. S27). The hydroxylation pathway is enthalpically favored (ΔHR = −1.6 kcal/mol for hydroxylation versus ΔHR = +8.3 kcal/mol for deprotonation), which supports the fact that Plu4 has a minor hydroxylation activity despite being a desaturase. We note that while the hydroxylation pathway is enthalpically favored, it is not entropically favored, as combining a water molecule with intermediate 12 is entropically penalized (SI Appendix, Fig. S27). However, it is likely that in the context of the enzymatic reactions, water could be preorganized to facilitate the hydroxylation step to reduce the entropic cost. The enthalpic favorability of hydroxylation over deprotonation could thus suggest that the key distinction between hydroxylation and double bond formation is the position of water in the active site of the enzymes during catalysis (Fig. 6B). In order to test this hypothesis, we turned our attention to molecular dynamics (MD) simulations to model the hydroperoxyl intermediate (15, Fig. 6B) formed in Plu4 and RohP active sites.
MD simulations of Plu4 and RohP with 15 bound in the active sites (SI Appendix, Computational Methods) indicated that N-terminal regions are quite flexible in both Plu4 and RohP and that both systems behave similarly in solution (SI Appendix, Fig. S28). In both systems, the strong salt-bridge interaction between Arg357 (Arg367 in RohP) and the carboxylic group of 1 keeps the intermediate anchored in the same position in the active site (Fig. 6A). Simulations also show the presence of multiple water molecules in the active sites of both Plu4 and RohP. However, only one of these water molecules is ordered and preorganized for catalysis in RohP. When 15 is formed in RohP, simulations showed that an ordered water molecule hydrogen bonds with the protonated hydroperoxyl and phosphate groups in a bridging mode and appears persistently during the MD trajectories (Fig. 6A and SI Appendix, Figs. S29 and S30). This water molecule is preorganized to perform an enantioselective nucleophilic attack at the C4 position of 1 from the pro-S face to give the final hydroxylated product 3. The presence of this ordered water is possible thanks to the conformation adopted by 15 (SI Appendix, Fig. S29), which is maintained by Phe119 in RohP at the opposite face of the carbon–carbon double bond of 15, blocking the access of water molecules and preventing the displacement or rotation of the side chain of 1. By contrast, the protein backbone rms fluctuation values of residues ∼100 to 150 in Plu4 indicate higher flexibility than in RohP (SI Appendix, Fig. S28), and the Phe119 equivalent residue, Phe112, can explore alternative conformations in Plu4, permitting 15 to also adopt slightly different geometries (Fig. 6A and SI Appendix, Fig. S29). While in these alternative conformations, which are favored in Plu4, the hydroperoxyl group of 15 cannot preorganize a water molecule next to the C4 position of 1, thus disfavoring the hydroxylation pathway. However, MD simulations also showed that 15 in Plu4 can still explore minor conformations that allow the ordering of this water molecule (SI Appendix, Figs. S29 and S30), supporting the minor formation of the hydroxylated side product.
Discussion
Arginine desaturases and arginine hydroxylases both catalyze remarkable oxidation reactions using only molecular oxygen and PLP. Our work comparing the biochemical reactivity, structure, and sequence similarity patterns of arginine desaturases and hydroxylases revealed that the two types of enzymes are even more similar than has previously been thought. The highly similar active-site structures, including conserved catalytic residues, show the subtlety of the factors differentiating the two reactivities. Additionally, our mutagenesis study identified no active-site residues that could distinguish the reactivity. Besides the residues targeted in mutagenesis, only slight differences in the active-site conformations of residues Phe112, Asn114, Leu115, and Phe190 were observed when compared to the equivalent residues in RohP and SwMppP, possibly indicating the importance of dynamics in the catalytic mechanism (SI Appendix, Figs. S15, S16, S31, and S32). Our sequence similarity analysis furthermore reveals that there is a stronger evolutionary relationship between the Group Ia hydroxylases and the Group Ib desaturases than between the Group Ia hydroxylases and the Group II hydroxylases or the Group Ib desaturases and the Group III desaturases, an arrangement which is mirrored by a phylogenetic tree (SI Appendix, Fig. S12). These data suggest that either the hydroxylase or desaturase activity emerged separately, indicating a complex evolutionary history similar to other examples of nonisofunctional phylogenetic clustering reported in other protein families (SI Appendix, Fig. S33) (17–20). Furthermore, the consistent appearance of a minor hydroxylated side product in all characterized desaturases suggests that the hydroxylase and desaturase mechanisms are highly related and provide evidence for promiscuous enzyme evolution (21). The minor appearance of hydroxylated product 3 from the desaturases could have either served as a starting point for the evolution of hydroxylation reactions from a desaturase ancestor, or it could represent a vestigial feature of a hydroxylating ancestor.
EPR experiments and cytochrome c assays demonstrated the formation of superoxide, which establishes the ability of the quinonoid intermediates to transfer single electrons to oxygen, supporting the proposed mechanism (Fig. 6B). Both flavin-dependent enzymes and cofactor-independent enzymes form superoxide through a single-electron transfer to oxygen (16, 22–24), and the PLP-dependent enzyme, DOPA decarboxylase, is known to produce superoxide during a paracatalytic oxidative deamination (25). However, whether or not arginine oxidases also produced superoxide was unknown, and our results suggest that they may use oxygen similarly: a single-electron transfer to oxygen precedes formation of superoxide. Generally, positively charged species in the active site can contribute to oxygen activation by stabilizing the negatively charged peroxy intermediates (1, 22, 23). In the case of the arginine oxidases, Lys219, Lys227, or the arginine substrate itself could be responsible for providing the positive environment necessary to stabilize the formation of a peroxy intermediate in Plu4 (SI Appendix, Fig. S34).
DFT calculations showed that superoxide rebound, giving a peroxy intermediate, is most favorable at the C4′ position of PLP. This result can be rationalized because the arginine oxidases belong to the aspartate aminotransferase protein family. The mechanisms for this family typically include protonation at the C4′ position of PLP. By contrast, in the case of O2- and PLP-dependent enzymes from other protein families such as Cap15 (26) and CuaB (27), isotopic enhancement occurs in products hydroxylated at the Cα position when exposed to labeled oxygen. These results suggest that a peroxy intermediate is instead formed at the Cα position during the reaction, which reflects that they belong to the selenocysteine synthase and 8-amino-7-oxononanoate synthase families with enzymes that typically protonate at the Cα position. Additionally, we also compared the final deprotonation and hydroxylation steps proposed for the arginine oxidases by DFT calculations, which showed that the hydroxylation step is enthalpically more favorable than the deprotonation at C5. This result suggests that the eventual presence of an ordered water in the active site of a desaturase would form 3 as a side product. Calculations also support that the hydroxylation mechanism does not go through a fully oxidized intermediate with deprotonation at C5 because the hydroxylation step would likely precede deprotonation, since we predict hydroxylation is favorable in the enzyme active site.
Indeed, control of the positioning of water in the active site could be critical to the different reaction outcomes. While we could not identify any single active-site residue critical to controlling the reaction outcome, MD simulations showed that Plu4 exhibits higher flexibility than RohP in the ∼100 to 150 amino acid region, including at active-site residues like Phe112 (SI Appendix, Fig. S28). These differences may be due to distinct flexibilities and conformational dynamics inherent to these proteins, which might be caused by mutations introduced during evolution at distal positions but that are dynamically correlated with active-site residues (28–30). Our MD simulations showed that, when formed in RohP, intermediate 15 preferentially explores conformations in which an ordered water molecule is preorganized to perform the enantioselective hydroxylation at the C4 position (Fig. 6 and SI Appendix, Figs. S29 and S30). However, intermediate 15 in the Plu4 active site is more flexible, and it explores alternative geometries, of which most lack an ordered water molecule at the relevant site, thus favoring the desaturation pathway (SI Appendix, Figs. S29 and S30). An analysis of crystal structures for Plu4, RohP, and SwMppP revealed that each intermediate-bound structure has active-site water molecules that could become preorganized for reaction once intermediate 15 has formed and has adopted the appropriate geometry (SI Appendix, Fig. S35).
Overall, we have shown that the arginine desaturases and hydroxylases show a high degree of similarity despite catalyzing different reactions. Based on our presented data, we propose a mechanism that accounts for high active-site similarity, observed side products, and computational work (Fig. 6B). First, 1 binds and forms an external aldimine, similar to other PLP-dependent enzymes. Then, the α-carbon is deprotonated, forming the Quinonoid I intermediate. Quinonoid I is able to donate a single electron to oxygen, forming a semiquinone radical and superoxide, which was observed as a side product by EPR and cytochrome c reduction assays. The semiquinone radical and superoxide rebound at the C4′ position of PLP to form a hydroperoxyl intermediate, supported by DFT calculations. Then the C3 is deprotonated, eliminating the hydroperoxide group of this intermediate, to form an oxidized enamine intermediate. This enamine intermediate can either undergo reformation of the internal aldimine, releasing the product 4 from the cofactor, or it can proceed through a second round of oxidation in which C4 is deprotonated to form Quinonoid II.
Like Quinonoid I, Quinonoid II is also capable of donating a single electron to oxygen, forming a semiquinone radical and superoxide. Once again, as DFT calculations suggested, the rebound of these two species occurs at the C4′ position of PLP, forming intermediate 15. At this point, the reaction pathways diverge, in which the positioning of water, facilitated by intermediate 15 geometry, determines the fate of the reaction. When geometries conducive to water preorganization are adopted, as observed in MD simulations, the entropic cost of hydroxylation is overcome, and C4 could be hydroxylated. Alternatively, when other geometries are explored, water is not appropriately positioned, and deprotonation at C5 can preferentially occur. Both pathways result in elimination of the hydroperoxyl group from 15. The subtly of this geometric determinant from MD calculations is supported by the high similarity of the active-site structures between hydroxylases and desaturases, despite only ∼45% maximum sequence identity between the two types of enzymes. Additionally, the appearance of a preorganized water and a geometric configuration conducive to hydroxylation in desaturases, although infrequent, are supported by our observation of hydroxylated side products from desaturases. Finally, either product 2 or 3 is released by reformation of the internal aldimine. Our mechanism relies on only the C4′ position of PLP for peroxy adduct formation, which suggests that the reaction is not dependent on arginine as a substrate and could, theoretically, be engineered into other aminotransferases. Finally, our sequence similarity and phylogenetic analyses reveal that the desaturases and hydroxylases appear to each have two distinct phylogenetic groups, clustered nonisofunctionally, which will be essential to future studies aimed at understanding how the two activities evolved.
Methods
Detailed descriptions of network analysis, sequence alignment, phylogenetic tree, and homolog identification; cloning and mutagenesis; protein purification; in vitro characterization; crystallization; data collection, structure determination, and model refinement; EPR; and computational methods, including DFT calculations and MD simulations, are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Melanie Higgins for helping to solve the holo-Plu4 structure and Jason Hedges for helping plan EPR experiments and DFT calculations. Our work was supported by funding from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2016-03778 to K.S.R.), the NIH National Institute for General Medical Sciences (GM-124480 project to K.N.H.), a Natural Science and Engineering Research Council Alexander Graham Bell Canada Graduate Scholarship-Doctoral Award (CGSD2 – 504877 – 2017 to E.R.H.), the Spanish Ministerio de Ciencia e Innovación (PID2019-111300GA-I00 project to M.G.-B.), and the Generalitat de Catalunya Agency for Management of University and Research Grants (Beatriu de Pinós H2020 Marie Skłodowska-Curie Actions MSCA-Cofund 2018-BP-00204 project to M.G.-B.). The research described in this paper was performed using beamline 9-2 at the Stanford Synchrotron Radiation Light Source. Computations were performed on the Hoffman2 cluster at the University of California, Los Angeles and the Extreme Science and Engineering Discovery Environment, which is supported by the NSF (OCI-1053575).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012591118/-/DCSupplemental.
Data Availability
The atomic coordinates and structure factors have been deposited in the Protein Data Bank, (PDB ID codes: 7N79, 7RF9, and 7RGB). All other study data are included in the article and/or SI Appendix.
References
- 1.Hoffarth E. R., Rothchild K. W., Ryan K. S., Emergence of oxygen- and pyridoxal phosphate-dependent reactions. FEBS J. 287, 1403–1428 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Du Y.-L., et al., A pyridoxal phosphate-dependent enzyme that oxidizes an unactivated carbon-carbon bond. Nat. Chem. Biol. 12, 194–199 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Han L., Schwabacher A. W., Moran G. R., Silvaggi N. R., Streptomyces wadayamensis MppP is a PLP-dependent ʟ-arginine α-deaminase, γ-hydroxylase in the enduracididine biosynthetic pathway. Biochemistry 54, 7029–7040 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Han L., et al., Streptomyces wadayamensis MppP is a PLP-dependent oxidase, not an oxygenase. Biochemistry 57, 3252–3264 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Hedges J. B., Kuatsjah E., Du Y.-L., Eltis L. D., Ryan K. S., Snapshots of the catalytic cycle of an O2, pyridoxal phosphate-dependent hydroxylase. ACS Chem. Biol. 13, 965–974 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Du Y.-L., Higgins M. A., Zhao G., Ryan K. S., Convergent biosynthetic transformations to a bacterial specialized metabolite. Nat. Chem. Biol. 15, 1043–1048 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Copp J. N., Akiva E., Babbitt P. C., Tokuriki N., Revealing unexplored sequence-function space using sequence similarity networks. Biochemistry 57, 4651–4662 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Hedges J. B., Ryan K. S., In vitro reconstitution of the biosynthetic pathway to the nitroimidazole antibiotic azomycin. Angew. Chem. Int. Ed. Engl. 58, 11647–11651 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Wu J., Li L., Deng Z., Zabriskie T. M., He X., Analysis of the mildiomycin biosynthesis gene cluster in Streptoverticillum remofaciens ZJU5119 and characterization of MilC, a hydroxymethyl cytosyl-glucuronic acid synthase. ChemBioChem 13, 1613–1621 (2012). [DOI] [PubMed] [Google Scholar]
- 10.John R. A., Pyridoxal phosphate-dependent enzymes. Biochim. Biophys. Acta 1248, 81–96 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Schneider G., Käck H., Lindqvist Y., The manifold of vitamin B6 dependent enzymes. Structure 8, R1–R6 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Mehta P. K., Hale T. I., Christen P., Aminotransferases: Demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 214, 549–561 (1993). [DOI] [PubMed] [Google Scholar]
- 13.McPhalen C. A., Vincent M. G., Jansonius J. N., X-ray structure refinement and comparison of three forms of mitochondrial aspartate aminotransferase. J. Mol. Biol. 225, 495–517 (1992). [DOI] [PubMed] [Google Scholar]
- 14.McPhalen C. A., et al., Domain closure in mitochondrial aspartate aminotransferase. J. Mol. Biol. 227, 197–213 (1992). [DOI] [PubMed] [Google Scholar]
- 15.Kuzkaya N., Weissmann N., Harrison D. G., Dikalov S., Interactions of peroxynitrite, tetrahydrobiopterin, ascorbic acid, and thiols: Implications for uncoupling endothelial nitric-oxide synthase. J. Biol. Chem. 278, 22546–22554 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Thierbach S., et al., Substrate-assisted O2 activation in a cofactor-independent dioxygenase. Chem. Biol. 21, 217–225 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Glasner M. E., et al., Evolution of structure and function in the o-succinylbenzoate synthase/N-acylamino acid racemase family of the enolase superfamily. J. Mol. Biol. 360, 228–250 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Phillips G., et al., Functional promiscuity of the COG0720 family. ACS Chem. Biol. 7, 197–209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miles Z. D., Roberts S. A., McCarty R. M., Bandarian V., Biochemical and structural studies of 6-carboxy-5,6,7,8-tetrahydropterin synthase reveal the molecular basis of catalytic promiscuity within the tunnel-fold superfamily. J. Biol. Chem. 289, 23641–23652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez-Rodríguez S., Soriano-Maldonado P., Gavira J. A., N-succinylamino acid racemases: Enzymatic properties and biotechnological applications. Biochim. Biophys. Acta. Proteins Proteomics 1868, 140377 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Khersonsky O., Tawfik D. S., Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79, 471–505 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Mattevi A., To be or not to be an oxidase: Challenging the oxygen reactivity of flavoenzymes. Trends Biochem. Sci. 31, 276–283 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Chaiyen P., Fraaije M. W., Mattevi A., The enigmatic reaction of flavins with oxygen. Trends Biochem. Sci. 37, 373–380 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Fetzner S., Steiner R. A., Cofactor-independent oxidases and oxygenases. Appl. Microbiol. Biotechnol. 86, 791–804 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Bertoldi M., Cellini B., Montioli R., Borri Voltattorni C., Insights into the mechanism of oxidative deamination catalyzed by DOPA decarboxylase. Biochemistry 47, 7187–7195 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Huang Y., et al., Pyridoxal-5′-phosphate as an oxygenase cofactor: Discovery of a carboxamide-forming, α-amino acid monooxygenase-decarboxylase. Proc. Natl. Acad. Sci. U.S.A. 115, 974–979 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai G. Z., et al., Pyridoxal-5′-phosphate-dependent bifunctional enzyme catalyzed biosynthesis of indolizidine alkaloids in fungi. Proc. Natl. Acad. Sci. U.S.A. 117, 1174–1180 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang G., Miton C. M., Tokuriki N., A mechanistic view of enzyme evolution. Protein Sci. 29, 1724–1747 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrović D., Risso V. A., Kamerlin S. C. L., Sanchez-Ruiz J. M., Conformational dynamics and enzyme evolution. J. R. Soc. Interface 15, 20180330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gardner J. M., Biler M., Risso V. A., Sanchez-Ruiz J. M., Kamerlin S. C. L., Manipulating conformational dynamics to repurpose ancient proteins for modern catalytic functions. ACS Catal. 10, 4863–4870 (2020). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors have been deposited in the Protein Data Bank, (PDB ID codes: 7N79, 7RF9, and 7RGB). All other study data are included in the article and/or SI Appendix.