Abstract
Controlling proper RNA pool for nuclear export is important for accurate gene expression. ZFC3H1 is a key controller that not only facilitates nuclear exosomal degradation, but also retains its bound polyadenylated RNAs in the nucleus upon exosome inactivation. However, how ZFC3H1 retains RNAs and how its roles in RNA retention and degradation are related remain largely unclear. Here, we found that upon degradation inhibition, ZFC3H1 forms nuclear condensates to prevent RNA trafficking to nuclear speckles (NSs) where many RNAs gain export competence. Systematic mapping of ZFC3H1 revealed that it utilizes distinct domains for condensation and RNA degradation. Interestingly, ZFC3H1 condensation activity is required for preventing RNA trafficking to NSs, but not for RNA degradation. Considering that no apparent ZFC3H1 condensates are formed in normal cells, our study suggests that nuclear RNA degradation and retention are two independent mechanisms with different preference for controlling proper export RNA pool—degradation is preferred in normal cells, and condensation retention is activated upon degradation inhibition.
INTRODUCTION
In eukaryotes, genetic information needs to be transferred from the nucleus to the cytoplasm through mRNA export. Except for mRNAs, many other kinds of RNAs are produced by RNA polymerase II (RNAPII). Some of these RNAs are processed and transported to the cytoplasm, while others, like RNAs produced by pervasive transcription, are rapidly degraded shortly after transcription (1,2). To ensure proper gene expression, nascent transcripts need to be rapidly and accurately sorted into the export and the degradation pathway.
Transcripts produced by RNAPII are capped co-transcriptionally. The cap structure is bound by the cap-binding complex (CBC), consisting of the canonical components CBP80 and CBP20, and non-canonical component ARS2 (3,4). CBC facilitates mRNA export via directly interacting with and recruiting the mRNA export adaptor ALYREF to the 5′ region of mRNAs (5–7). In addition to CBC, the nuclear polyadenylate (polyA)-binding protein PABPN1 that binds mRNAs post-transcriptionally also facilitates ALYREF recruitment, mainly to the 3′ region of the mRNA (5). It was recently reported that ALYREF is co-transcriptionally recruited (8), and eIF4A3, a component of the exon junction complex that is loaded on mRNAs during co-transcriptional splicing, associates with ALYREF and promotes ALYREF recruitment (8,9). Thus, ALYREF is likely recruited both co-transcriptionally and post-transcriptionally, with post-transcriptional recruitment mainly occurring in nuclear speckles (NSs), where mRNA export factors are enriched and many mRNAs gain export competence (10–14).
The evolutionarily conserved exosome complex (exosome) is the central RNA degradation machine (15–17). To recognize its diverse types of substrates and to achieve its full activity, exosome needs to associate with many co-factors. A central co-factor is the conserved RNA helicase MTR4, which facilitates exosome functions alone or through forming multiple complexes with other proteins (1,2,18–20). These complexes include the nuclear exosome targeting complex (MTR4–RBM7–ZCCHC8), the polyA tail exosome targeting (PAXT) connection (MTR4–ZFC3H1–PABPN1) and the TRAMP complex (MTR4–PAPD5–ZCCHC7) (1,2,18). Among these, PAXT is localized in the nucleoplasm and is involved in the degradation of polyA transcripts, such as snoRNA host gene (SNHG) transcripts, prematurely terminated RNAs (ptRNAs) and polyA + promoter upstream transcripts (PROMPTs) (1,18,21–24). Different from the above-mentioned complexes that all facilitate exosome functions, MTR4 also forms a complex with NRDE2 that is enriched in NSs and negatively impacts exosomal degradation. In this way, NRDE2 ensures NSs as perfect sites for mRNAs gaining export competence (10,25).
Accurate sorting of nascent RNAs into the nuclear export and the nuclear degradation pathway is a fundamental issue that every eukaryotic cell must deal with. Recent studies revealed that the competition between the export and the degradation machinery provides an important mechanism for RNA sorting (20,24,26,27). For most polyA RNAs, including mRNAs and lncRNAs, MTR4 competes with ALYREF for CBC bound at the 5′ region before RNA traffics to NSs (20,27). As a consequence, MTR4 targets RNAs, which cannot rapidly and/or efficiently recruit ALYREF, to degradation in the nucleoplasm (20,27). At the 3′ region, the PAXT component ZFC3H1 functionally competes with ALYREF for PABPN1 and serves as a nuclear retention factor for its bound RNAs upon exosome inactivation (24). Recent studies demonstrated that exosome inactivation triggers the formation of specific nucleoplasmic polyA foci, in which exosome target RNAs as well as PAXT components are accumulated (24,27). However, it is still an open question whether these polyA foci are functionally important for retaining exosome target RNAs in the nucleus. Interestingly, ZFC3H1 knockdown (KD) resulted in diminished polyA foci (24). However, it is unclear whether ZFC3H1 directly functions in foci formation, or just promotes their formation indirectly by retaining polyA RNAs in the nucleus.
In this study, we report that ZFC3H1 forms nuclear condensates to prevent its target RNAs trafficking to NSs, from where they could be exported to the cytoplasm. Nuclear RNA retention and degradation are independent mechanisms for controlling proper nuclear RNA pool for export, as condensation and degradation activities are attributed to different ZFC3H1 domains, and the condensation activity is required for retention, but not degradation. Together with previous findings, our data suggest that in normal cells, ZFC3H1 preferentially triggers exosomal degradation to prevent the accumulation of unwanted RNAs; upon nuclear RNA degradation inactivation, ZFC3H1 forms nuclear condensates to trap accumulated RNAs in the nucleus.
MATERIALS AND METHODS
Plasmids and antibodies
To construct the expression plasmids of Flag-ZFC3H1 full length (FL) or fragments, the corresponding sequences were cloned into Flag-Phage using ClonExpress® Ultra One Step Cloning Kit (Vazyme Biotech Co., Ltd). The deletion or point mutation ZFC3H1 expression plasmids were constructed by mutagenesis using the KOD-Plus-Mutagenesis Kit (Takara). ZFC3H1-FL-ZsGreen and ZFC3H1-NP-ZsGreen were constructed by deletion of the sequence between ZFC3H1 and ZsGreen in Flag-ZFC3H1-FL and Flag-ZFC3H1-NP, respectively. Plasmids encoding Flag-eIF4A3 and Flag-DDX3 were described previously (20,25). To generate construct for gene editing, sgRNA targeting ZFC3H1 was synthesized, annealed and ligated to the pX330-mCherry plasmid. The sgRNA sequence is shown in Supplementary Table S1. The Smad and β-globin constructs were described previously (28).
The CBP80, MTR4, UAP56 and ARS2 antibodies were described previously (20). Antibodies against PABPN1 (Abcam), ZFC3H1 (Novus), Flag (Sigma-Aldrich), GAPDH (Proteintech), Tubulin (Proteintech), SC35 (Sigma-Aldrich), digoxin (Roche) and SON (Thermo Fisher) were purchased. Alexa Fluor 546, Alexa Fluor 488 or Alexa Fluor 647 conjugated secondary antibodies were purchased from Life Technologies.
Cell culture, transfections and RNAi
HeLa cells were cultured in DMEM supplemented with 10% FBS (Biochrom) and penicillin/streptomycin. Lipofectamine 2000 (Invitrogen) was used for DNA transfection. For RNAi, siRNA transfection was carried out with Lipofectamine RNAiMax (Invitrogen) following manufacturer’s protocol. The siRNA targeting sequences are shown in Supplementary Table S1. It is of note that both UAP56 and its homolog URH49 must be knocked down to observe a robust export block (29).
RNA isolation, reverse transcription and PCR analysis
Total RNAs were extracted with TRIzol (Invitrogen) and treated with the RNase-free RQ DNase I (Promega) to remove genomic DNA. cDNAs were synthesized from 1 μg of RNAs with random primer using M-MLV reverse transcriptase (Promega). Quantitative PCR was carried out using SYBR qPCR SuperMix (Novoprotein) according to the manufacturer’s instruction. Primer sequences are listed in Supplementary Table S2.
FISH, immunofluorescence and DNA microinjections
To detect polyA RNAs, HeLa cells were fixed with 4% formaldehyde/acetic acid in 1× PBS. Cells were washed with 1× PBS three times and permeabilized with 0.1% Triton in 1× PBS for 15 min, followed by washes with 2× saline–sodium citrate buffer (SSC) twice and incubation at 37°C with a high-performance liquid chromatography (HPLC)-purified Alexa 546-conjugated oligo dT (50-nt) probe for 12–16 h. Cells were then washed with 2× SSC twice and 0.5× SSC once, followed by DAPI staining. For immunofluorescence, SC35 (1:1000), SON (1:200) or Flag (1:1000) antibody were used. After primary antibody incubation for 1 h, cells were washed three times with 1× PBS and incubated with Alexa 488- or 647-labeled anti-mouse or anti-rabbit secondary antibody (1:1000) in blocking buffer (1× PBS, 0.1% Triton X-100 and 2 mg/ml BSA) at room temperature for 1 h. Then cells were washed with 1× PBS three times for 10 min each. Confocal imaging was performed on a Leica TCS SP8 WLL. All structured illumination microscopy (SIM) experiments were performed on a DeltaVision OMX SR system (GE Healthcare) equipped with a 60×/1.42 NA Plan Apo oil-immersion objective (Olympus) and three laser beams (405, 488 and 546 nm). FISH quantitation was carried out using the ImageJ software (National Institutes of Health). After stacking the FISH and IF images, FISH signals in NSs and nucleus were measured for NS/N ratios. Measurements were obtained for fluorescence in NSs for FISH probe (F1, F2, …, Fn), fluorescence in the nucleus for FISH probe (F′), area of NSs (A1, A2, …, An) and area of the nucleus (A′). NS/N ratios were calculated as NS/N = (A1F1 + A2F2 + ··· + AnFn)/A′F′. N/T and N/C ratios were calculated as described (30).
To analyze the distribution of β-globin and Smad spliced mRNAs, HeLa cells transfected with β-globin and Smad constructs were fixed with 4% PFA in 1× PBS for 15 min, followed by washes with 1× PBS three times and permeabilization with 1× PBS/0.1% Triton for 15 min. Cells were then washed with 1× PBS three times and 1× SSC/50% formamide twice, and were incubated with an HPLC-purified Alexa 546-conjugated 70-nt probe (vector probe) that hybridizes to the pcDNA3 vector sequence at 37°C for 12–16 h. The cells were then washed with 1× SSC/50% formamide four times, followed by DAPI staining. The vector probe targeting sequence is shown in Supplementary Table S3. Images were captured with a DP72-CCD camera (Olympus) on an inverted microscope using the DP-BSW software (Olympus). FISH quantitation was carried out using the ImageJ software (National Institutes of Health). N/C ratios were calculated as described (30).
To detect endogenous SNHG19 RNA and NS protein SON simultaneously, HeLa cells were fixed with 4% formaldehyde/acetic acid in 1× PBS for 20 min, followed by three washes with 1× PBS and permeabilization with 1× PBS/0.1% Triton X-100/2 mM ribonucleoside vanadyl complex for 15 min. The cells were incubated with specific probes that were labeled with digoxin at 50°C for 16 h. The targeting sequence of the SNHG19 probe is shown in Supplementary Table S3. After extensive wash, cells were incubated with the digoxin antibody diluted in blocking buffer for 1 h. After three washes with 1× PBS, cells were incubated with the Alexa Flour 488-labeled anti-sheep antibody (1:1000) for 1 h and washed with 1× PBS three times. For SON IF, cells were incubated with SON antibody (1:5000) for 40 min and the Alexa Fluor 546-labeled anti-rabbit antibody (1:5000) for another 40 min, followed by DAPI staining and three washes with 1× PBS. Confocal imaging was performed on a Leica TCS SP8 WLL. FISH quantitation was carried out using the ImageJ software (National Institutes of Health).
For DNA microinjection, HeLa cells were plated on 20-mm coverslips at the bottom of 35-mm dishes, and 100 ng/μl plasmid DNA was injected into cells. For each experiment, ∼200 cells were microinjected, followed by incubation at 37°C. Two hours post-microinjection, cells were fixed with 4% PFA and followed by DAPI staining.
Protein immunoprecipitations
For each assay, 106 cells were suspended in the lysis buffer (20 mM Tris–HCl, pH 7.4, 100 mM NaCl, 2 mM EDTA, pH 8.0, 0.1% Triton, 1 mM DTT, 1 mM PMSF). After sonication and centrifugation, the lysate was treated with RNase A for 20 min at 30°C, followed by incubation with antibody-crosslinked beads at 4°C overnight. The beads were washed three times with the lysis buffer, and proteins were eluted for western blot analysis.
RNA immunoprecipitations
Cells treated with different siRNAs for 72 h were washed with cold 1× PBS and then suspended in 1 ml NET-2 buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% NP-40, 0.2 mM PMSF), followed by sonication on ice. The lysates were incubated with the ZFC3H1 or Myc antibody at 4°C for 2 h, followed by rotation with nProtein A Sepharose (GE) for another 2 h at 4°C. The beads were washed three times with NET-2 buffer. One fifth of the immunoprecipitate was analyzed by western blotting. The rest of the immunoprecipitate was treated with proteinase K, and RNAs were recovered by phenol/chloroform extraction and ethanol precipitation. RNAs were then treated with the RNase-free RQ DNase I (Promega) for 2 h at 37°C. RT-qPCR was carried out as described above.
UV-crosslinking RNA immunoprecipitations
Cells transfected with different plasmids for 24 h were washed with cold 1× PBS and then UV crosslinked at 150 mJ/cm2. Cells were resuspended with 1 ml RNA immunoprecipitation (RIP) buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.2% NP-40, 0.1% sodium deoxycholate, 0.2 mM PMSF), followed by sonication on ice. Cell lysates were precleared with 20 μl nProtein A Sepharose (GE) for 1 h to get rid of non-specific binding. Then, the precleared lysates were incubated with the Flag or Myc antibody at 4°C for 2 h, followed by rotation with nProtein A Sepharose (GE) for another 2 h at 4°C. The beads were washed twice with buffer 1 (50 mM Tris, pH 7.4, 300 mM NaCl, 0.2% NP-40, 0.2% sodium deoxycholate, 2 mM VRC, 0.2 mM PMSF) and once with buffer 2 (50 mM Tris, pH 7.4, 300 mM NaCl, 0.2% NP-40, 0.2% sodium deoxycholate, 2 mM VRC, 0.2 M urea, 0.2 mM PMSF). One fifth of the immunoprecipitate was analyzed by western blotting. The rest of the immunoprecipitate was treated with proteinase K, and RNAs were recovered by phenol/chloroform extraction and ethanol precipitation. RNAs were then treated with the RNase-free RQ DNase I (Promega) for 2 h at 37°C. RT-qPCR was carried out as described above.
Fluorescence recovery after photobleaching
Cells were transfected using 1 μg of plasmid DNA per dish. Twenty-four hours post-transfection, cells were imaged using a Nikon A1 confocal microscope operated by Nikon Elements, a 60× objective lens, and a heating chamber with CO2 pre-warmed to 37°C. The regions of interest (ROIs) were outlined with Elements and photobleached using a 488 nm laser set at 70% power, 1 pulse/s × 7 s. Fluorescence recovery was monitored up to 10 min after photobleaching. Images were acquired every 10 s. The fluorescence intensity in the obtained images was measured in ROIs using the ImageJ software. The recovery ratio is defined as (F∞ − F0)/(Fi − F0), where F∞ is the fluorescence in the bleached region after full recovery, Fi is the fluorescence before bleaching and F0 is the fluorescence just after bleaching (31).
Live cell imaging
For time-lapse microscopy of droplet fusion, HeLa cells were transfected with ZFC3H1-FL-ZsGreen or ZFC3H1-NP-ZsGreen plasmid. Twenty-four hours post-transfection, live cell imaging was performed on a Zeiss Celldiscoverer 7. Images were acquired every 5 min. All images were acquired using ZEN software.
RESULTS
ZFC3H1 prevents RNA trafficking to NSs
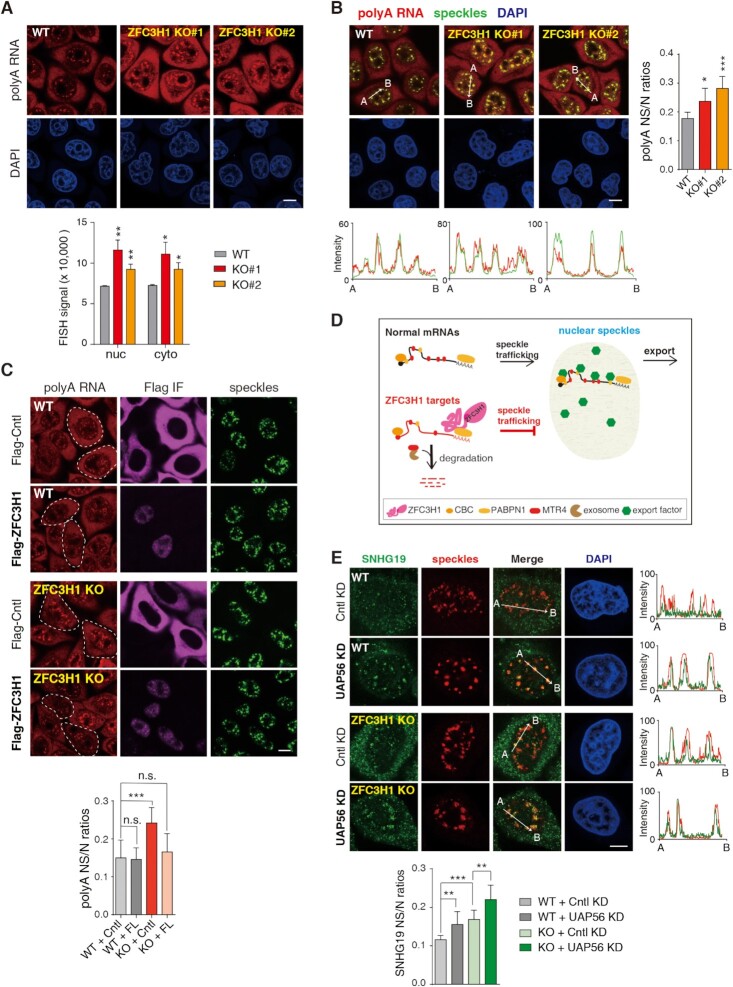
In a previous study, we found that ZFC3H1 KD resulted in elevated polyA signals in NSs (27). At first glance, this observation seemed to conflict with the reported roles of ZFC3H1 in nuclear RNA retention (24), as increased polyA RNA signals in NSs are usually associated with inhibited mRNA nuclear export. To study how ZFC3H1 affects polyA RNA distribution, we knocked it out in HeLa cells using CRISPR–Cas9 and obtained two independent knockout (KO) cell lines. Western blot and DNA sequencing confirmed successful KO (Supplementary Figure S1A and B). As expected from previous studies (18,21,23,24), in these KO cells, SNHG transcripts, well-known PAXT targets, were significantly upregulated (Supplementary Figure S1C), and cytoplasmic polyA RNA signals were elevated (Figure 1A). Interestingly, same as previous observations with ZFC3H1 KD (27), nuclear polyA signals apparently increased in ZFC3H1 KO cells (Figure 1A), and these increased polyA signals were colocalized with SC35 immunofluorescence signals used to indicate NSs (Figure 1B). To specifically examine polyA signal change in NSs, we quantified the fluorescence intensities of polyA RNAs at NSs and normalized them to that in the nucleus (NS/N). The polyA RNA NS/N ratios increased in both KO lines in comparison to WT cells (Figure 1B). Importantly, exogenous expression of ZFC3H1 with proper level apparently rescued the increased polyA signals in NSs (Figure 1C). Note that in cells with very high ZFC3H1 expression, nuclear polyA signals were not reduced but even enhanced (Supplementary Figure S1D) (see results and discussion later).
Figure 1.
ZFC3H1 depletion results in polyA RNA accumulation in NSs. (A) (Top) FISH with an oligo(dT) probe to detect polyA RNAs in WT and ZFC3H1 KO cells. Scale bar, 10 μm. (Bottom) Quantification of nuclear and cytoplasmic FISH signals of polyA RNAs in WT and ZFC3H1 KO cells. FISH signals of 30 cells were calculated in each experiment. Error bars, standard deviations (n = 3). (B) (Left) Confocal microscopic imaging to examine the colocalization of polyA RNAs with NSs in WT and ZFC3H1 KO cells. FISH with an oligo(dT) probe and IF using the SC35 (as an NS marker) antibody were carried out. The red and green lines in the graphs show the intensities of FISH and SC35 IF signals along the freely positioned arrow indicated from A to B, respectively. Scale bar, 10 μm. (Right) Quantification of NS/N ratios of polyA RNA signals in WT and ZFC3H1 KO cells. Error bars, standard deviations (n = 15). (C) (Top) Confocal microscopy analysis to examine the effect of Flag-DDX3 (Cntl) and Flag-ZFC3H1 on polyA RNA signals in WT and ZFC3H1 KO cells. FISH with an oligo(dT) probe and IF using Flag and SON (as an NS marker) antibodies were carried out. Exemplified cells with proper expression of Flag-ZFC3H1 or Flag-Cntl are indicated by white dashed lines. Scale bar, 10 μm. (Bottom) Quantification of polyA RNA NS/N ratios in cells transfected with corresponding constructs. Error bars, standard deviations (n = 10). (D) The illustration of possible fate of normal mRNAs and ZFC3H1 targets. Normal mRNAs traffic into NSs to gain export competence and are consequently exported to the cytoplasm. ZFC3H1 prevents its target RNAs trafficking into NSs and facilitates their degradation. (E) (Top) Confocal microscopic imaging to examine the colocalization of endogenous SNHG19 RNA with NSs in WT and ZFC3H1 KO cells treated with Cntl or UAP56/URH49 siRNAs. FISH with an SNHG19-specific probe and IF using a SON (as an NS marker) antibody were carried out. The green and red lines in the graphs show the intensities of FISH and SON IF signals along the freely positioned arrow indicated from A to B, respectively. Scale bar, 10 μm. (Bottom) Quantification of NS/N ratios of SNHG19 RNA. Error bars, standard deviations (n = 10). Statistical analysis was performed using Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001, n.s., not significant.
Why did polyA signals in NSs increase upon ZFC3H1 downregulation? Considering that many mRNAs gain export competence in NSs (10) and ZFC3H1 functions in nuclear RNA retention (24), we reasoned that ZFC3H1 might prevent its bound polyA RNAs trafficking to NSs (Figure 1D), and thus in the absence of ZFC3H1, they traffic to NSs from where a part of them are released to the cytoplasm. If true, one would expect that in ZFC3H1 KO cells, cytoplasmic accumulated polyA RNAs had trafficked through NSs before nuclear export. Indeed, KD of UAP56, a key export factor required for polyA RNA release from NSs (32), abolished cytoplasmic polyA signals in ZFC3H1 KO, and led to even more apparently enhanced NS polyA signals compared to UAP56-depleted WT cells (Supplementary Figure S2A–C). Importantly, when a specific ZFC3H1 target RNA, SNHG19, was examined in ZFC3H1 KO cells, similar observations were obtained. SNHG19 was largely detected in both NSs and the cytoplasm in Cntl KD cells, and was mostly accumulated in NSs upon UAP56 depletion (Figure 1E, KO; Supplementary Figure S2D). Together, these data are consistent with the view that ZFC3H1 prevents its bound RNAs trafficking into NSs. Note that in WT cells, UAP56 depletion also resulted in SNHG19 accumulation in NSs (Figure 1E, WT; Supplementary Figure S2D), suggesting that in normal condition, a small part of SNHG19 escaped from ZFC3H1 binding and trafficked through NSs prior to transport to cytoplasm.
Without ZFC3H1, why are some accumulated polyA RNAs retained in NSs, rather than all released to the cytoplasm? This retention could be due to low abilities of ZFC3H1 target RNAs in recruiting export factors in NSs. Consistent with this possibility, ZFC3H1 target levels were elevated not only in the cytoplasm, but also in the nucleus upon its downregulation (1,24). However, it was also possible that accumulated ZFC3H1 targets occupied export factors, resulting in impaired nuclear export of bulk mRNAs. To examine this possibility, we compared export efficiencies of two different reporters, spliced β-globin and Smad mRNAs, which are not exosome targets (20), in WT and KO cells. For both reporters, no apparent difference in nucleocytoplasmic distribution was detected between WT and KO cells (Supplementary Figure S2E). Thus, it is likely that many ZFC3H1 target RNAs have limited export activities. In the absence of ZFC3H1, although they were free to traffic to NSs, a part of them were still retained there.
PABPN1 plays a primary role in ZFC3H1 recruitment to many targets
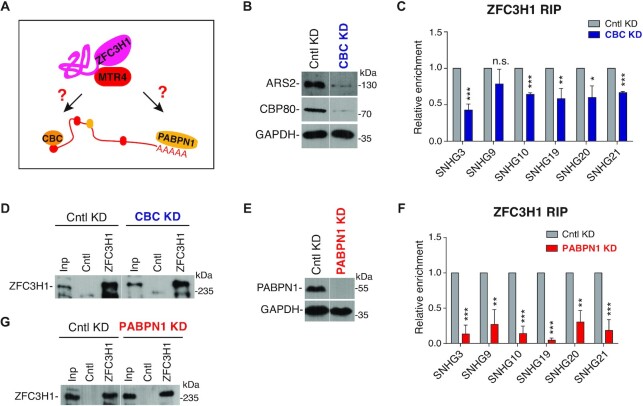
We next asked how ZFC3H1 is recruited to its target RNAs, including SNHG RNAs, polyA + PROMPTs and ptRNAs. As expected (1,18,22), the levels of these RNAs were apparently elevated in ZFC3H1 KO cells (Supplementary Figures S1C and S3A). ZFC3H1 was found to associate with nuclear CBC and nuclear polyA-binding protein PABPN1, both of which are important platforms for recruiting export and degradation machineries (Figure 2A) (1,5,7,18,20). We examined how CBC and PABPN1 function in ZFC3H1 recruitment by carrying out RIPs with a ZFC3H1 antibody followed by RT-qPCRs. For all SNHG RNAs we tested, co-KD of CBP80 and ARS2 demonstrated moderate or no inhibitory effect on ZFC3H1 associations (Figure 2B–D). Compared to CBC KD, PABPN1 KD showed more dramatic effects, with ZFC3H1 associations with all tested SNHG RNAs apparently weakened (Figure 2E–G). Similar to SNHG RNAs, ZFC3H1 recruitment to three polyA + PROMPTs was more sensitive to PABPN1 KD in comparison to CBC KD (Supplementary Figure S3B). For some unclear reasons, no apparent binding of ZFC3H1 with ptRNAs was detected in normal cells (Supplementary Figure S3C). One possibility is that ptRNAs might be rapidly degraded upon binding by ZFC3H1. Together, these data suggest that PABPN1 plays a primary role in facilitating ZFC3H1 recruitment, at least to SNHGs and polyA + PROMPTs. In agreement with this, PABPN1 KD led to apparent accumulation of SNHG19 RNA in NSs (Supplementary Figure S3D and E). Note that this accumulation was even more pronounced compared to that in ZFC3H1 KO, probably due to the requirement for PABPN1 in ALYREF recruitment for release from NSs (5,32).
Figure 2.
PABPN1 facilitates ZFC3H1 to its targets. (A) The illustration of possible roles of CBC and PABPN1 in ZFC3H1 recruitment. (B) Western blot to examine the KD efficiency of ARS2 and CBP80. GAPDH was used as a loading control. The white line delineates the boundary where irrelevant lanes have been removed from the same blot. (C, D) RNA-IP analysis to examine the association of ZFC3H1 with SNHG RNAs upon CBC KD. IPs were carried out with a ZFC3H1 antibody in cells treated with Cntl or CBC siRNAs, followed by RT-qPCRs to detect RNAs (C) and western blot to detect ZFC3H1 (D). Error bars, standard deviations (n = 3). The white line delineates the boundary where irrelevant lanes have been removed from the same blot. (E) Western blot to examine the KD efficiency of PABPN1. GAPDH was used as a loading control. The white line delineates the boundary where irrelevant lanes have been removed from the same blot. (F, G) Same as (C) and (D), except that PABPN1 KD cells were used instead of CBC KD cells. Error bars, standard deviations (n = 3). Statistical analysis was performed using Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001, n.s., not significant.
ZFC3H1 interacts with CBC, PABPN1 and MTR4 through distinct domains
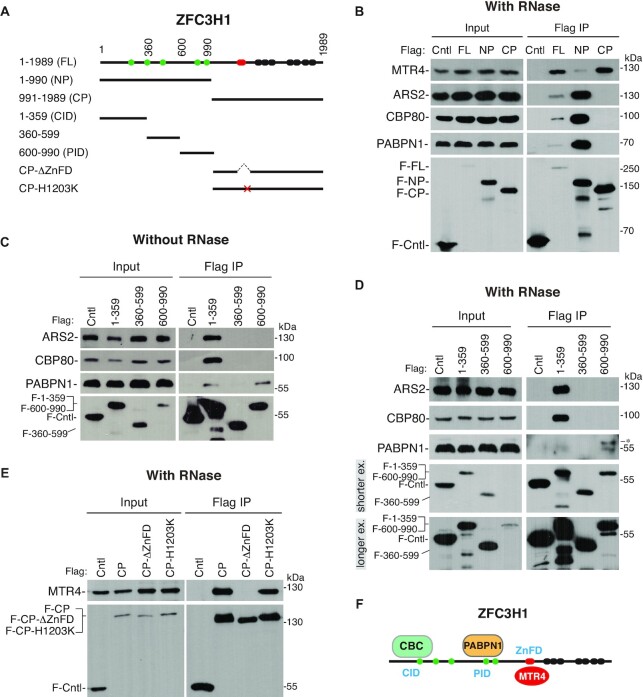
We next sought to determine how ZFC3H1 interacted with CBC, PABPN1 and MTR4. ZFC3H1 contains five predicted coiled-coil domains in the N terminal part (1–990 aa, NP), and a highly conserved zinc finger domain (ZnFD) and seven TPRs in the C terminal part (991–1989 aa, CP) (Figure 3A). We first separated it into the NP and the CP fragment and Flag-tagged. Flag immunoprecipitations (IPs) with or without RNase treatment showed that CBC and PABPN1 interacted with NP, whereas MTR4 mainly associated with CP (Figure 3B; Supplementary Figure S4). Although the expression of the FL protein was quite low, it efficiently associated with CBC, PABPN1 and MTR4 (Figure 3B; Supplementary Figure S4). Further mapping revealed that two N-terminal domains, ZFC3H11–359aa (CID, CBC-interacting domain) and ZFC3H1600–990aa (PID, PABPN1-interacting domain), mainly associated with CBC and PABPN1, respectively (Figure 3C and D). Although CID also weakly associated with PABPN1, this association became undetectable upon RNase treatment (Figure 3D).
Figure 3.
ZFC3H1 interacts with CBC, PABPN1 and MTR4 via distinct domains. (A) Domain schematic representation of ZFC3H1. The green, red and black bars indicate coiled-coil, zinc finger and TPR repeat domains, respectively. (B) Flag IP-WB to examine the associations of ZFC3H1 fragments with CBC, PABPN1 and MTR4 in the presence of RNase A. (C) Flag IP-WB to examine the associations of NP sub-fragments with CBC and PABPN1 in the absence of RNase A. (D) Same as (C), except that RNase A-treated cell lysates were used for IPs. Short and long exposures of the same blot probed with Flag antibody were shown. An asterisk indicates a non-specific band. (E) IP-WB to examine the role of ZnFD in the ZFC3H1–MTR4 interaction in the presence of RNase A. (F) Schematic illustration of ZFC3H1-binding domains with CBC, PABPN1 and MTR4.
Considering that the MTR4–ZFC3H1 interaction is conserved in fission yeast (33,34), and the ZnF domain (ZnFD, 1185–1206 aa) is the most conserved part between ZFC3H1 and its fission yeast counterpart Red1, we examined whether ZnFD is required for interacting with MTR4. Truncation of ZnFD in FL led to very low expression; we thus made the deletion on the basis of CP instead. ZnFD deletion diminished CP interaction with MTR4, indicating that ZnFD is required for the ZFC3H1–MTR4 interaction (Figure 3E and F). Mutation of H1203 supposed to directly bind the zinc ion to lysine did not show a similarly strong effect to ZnFD deletion (Figure 3E).
PID and ZnFD, but not CID, are required for ZFC3H1 function in target RNA degradation
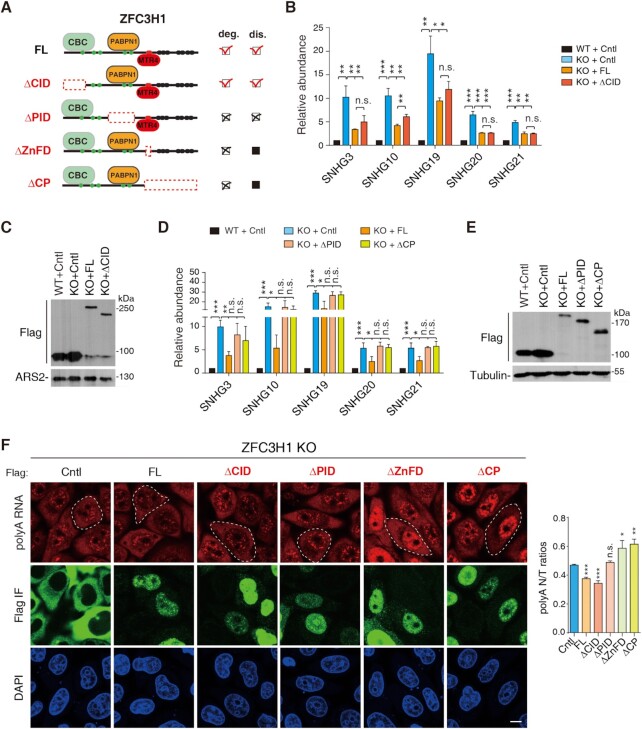
We next asked how the interactions with CBC, PABPN1 and MTR4 contribute to ZFC3H1 functions in RNA degradation and distribution. To answer this question, we first examined whether expression of ZFC3H1 mutants lacking CID, PID or ZnFD could rescue increased target RNA levels in KO cells (Figure 4A). As mentioned earlier, deletion of ZnFD in FL resulted in very low expression; we thus used ΔCP here instead. When these mutants as well as FL were expressed in ZFC3H1 KO cells, the increased SNHG RNA levels were inhibited by expression of FL and the ΔCID mutant, indicating that CID is not important for ZFC3H1 function in RNA degradation (Figure 4B and C). In agreement with the primary role of PABPN1 in ZFC3H1 recruitment, the ΔPID mutant did not apparently affect SNHG levels in KO cells. As expected from the loss of MTR4 interaction, the ΔCP mutant did not repress increased SNHG RNA levels either (Figure 4D and E).
Figure 4.
Interactions with PABPN1 and MTR4 are important for ZFC3H1 functions in RNA degradation. (A) Domain schematic representation of ZFC3H1 mutants. Functions of these mutants in target polyA RNA degradation and distribution are summarized and showed on the right. (B) RT-qPCRs to detect SNHG RNA levels in ZFC3H1 KO cells expressing with Flag-tagged DDX3 (Cntl), FL and ΔCID, respectively. The bars show RNA levels relative to 18S rRNA. Error bars, standard deviations (n = 3). (C) Western blots to examine the levels of ZFC3H1 mutants shown in (B). ARS2 was used as a loading control. (D) Same as (B), except that ΔPID and ΔCP were used instead of ΔCID in ZFC3H1 KO cells. (E) Western blots to examine the levels of ZFC3H1 mutants in (D). Tubulin was used as a loading control. (F) (Left) Confocal microscopy analysis to detect polyA RNAs in ZFC3H1 KO cells expressing indicated ZFC3H1 mutants. FISH with an oligo(dT) probe and IF using the Flag antibody were carried out. The white dashed lines indicate the cells with similarly low expression of ZFC3H1 FL and mutants. Scale bar, 10 μm. (Right) Quantification of nuclear (N) and total (T) FISH signals of polyA RNAs in cells transfected with corresponding constructs. N/T ratios of 30 cells were calculated in each experiment. Error bars, standard deviations (n = 3). Statistical analysis was performed using Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001, n.s., not significant.
Next, we examined how these mutants affected polyA signal intensities and distribution patterns in ZFC3H1 KO cells. Considering that highly overexpressed FL caused even more apparently increased nuclear polyA signals (Supplementary Figure S1D), here we examined cells with similarly low expression of FL and mutants. Consistent with RT-qPCR data, similar to that of FL, expression of the ΔCID mutant led to the reversal of elevated polyA RNA signals in both the nucleus and the cytoplasm (Figure 4F). In contrast, expression of the ΔPID mutant did not display an apparent effect (Figure 4F). Interestingly, expression of the ΔZnFD or the ΔCP mutant led to further increased nuclear polyA signals (Figure 4F). This additionally reinforced nuclear polyA RNA signals might be due to distribution change of these RNAs (see results later). Consistent with this possibility, when the same set of proteins were expressed in WT cells, the ΔCP mutant and in some cases ΔZnFD, but not others, caused increased nuclear polyA signals (Supplementary Figure S5). Together, the interactions of ZFC3H1 with PABPN1 and MTR4 are important for its functions in both degradation and distribution of RNAs.
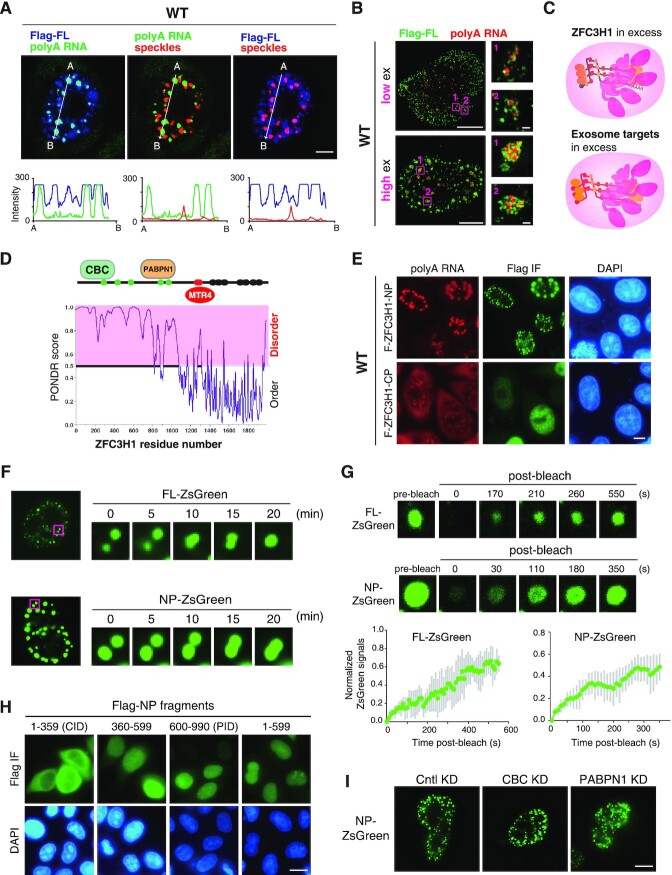
ZFC3H1 forms condensates via its N terminal IDR
How does ZFC3H1 prevent RNAs trafficking into NSs? We noticed that overexpressed FL formed nuclear condensates that did not colocalize with NSs in WT cells (Figure 5A). Super-resolution 3D SIM analysis revealed that polyA RNAs were intertwined with ZFC3H1 in these condensates (Figure 5B). These data, together with previous studies showing polyA foci formation upon exosome inactivation (24,27), suggested that high concentration of either exosome target polyA RNAs or the protein itself triggers ZFC3H1 condensation. ZFC3H1 condensates trap polyA RNAs, resulting in the formation of nucleoplasmic polyA foci (Figure 5C). This explains why high overexpression of ZFC3H1 in KO cells led to even more apparently increased polyA signals in the nucleus (Supplementary Figure S1D).
Figure 5.
The N-terminal IDR is sufficient and necessary for ZFC3H1 condensation. (A) Confocal microscopic imaging to examine the colocalization of polyA RNAs, NSs and overexpressed Flag-ZFC3H1-FL in WT cells. FISH with an oligo(dT) probe and IF using the SON (as an NS marker) and Flag antibodies were carried out. The green, blue and red lines in the graphs show the intensity of FISH, Flag IF and SON IF signals along the freely positioned arrow indicated from A to B, respectively. Scale bar, 5 μm. (B) SIM imaging to detect the distribution of polyA RNAs and overexpressed Flag-ZFC3H1-FL at different expression levels in WT cells. FISH with an oligo(dT) probe and IF using the Flag antibody were carried out. Higher magnification of the boxed regions is shown. Scale bar is 5 μm in the left panel and 0.2 μm in the right panel. (C) ZFC3H1 forms nuclear condensates when itself or its target polyA RNAs are present in excess. (D) Disorder score was calculated for ZFC3H1 using the PONDR program. (E) FISH and Flag IF to detect the distribution of polyA RNAs and Flag-ZFC3H1-NP or Flag-ZFC3H1-CP in WT cells. Scale bar, 5 μm. (F) Living cell images to show mobility and fusion of FL-ZsGreen or NP-ZsGreen granules. (G) Fluorescence recovery after photobleaching (FRAP) of FL-ZsGreen or NP-ZsGreen granules. One granule of a cell at each time point is shown. FRAP recovery curves are shown at the bottom. Error bars, standard deviations (n = 4). (H) Flag IF to detect the localization of Flag-NP sub-fragments. Scale bar, 10 μm. (I) Fluorescence microscopic imaging to examine the effect of CBC and PABPN1 KD on granule formation of NP-ZsGreen. NP-ZsGreen construct was microinjected into cells treated with Cntl, CBC or PABPN1 siRNAs for 72 h, and microscopy was carried out 2 h after microinjection. Scale bar, 5 μm.
We next asked how ZFC3H1 forms condensates. Based on sequence analysis, ZFC3H1-NP is an intrinsic disordered region (IDR) (Figure 5D). Indeed, NP apparently formed droplet-like granules in the cells, and the sizes of these granules increased along with their elevated expression level (Figure 5E). In contrast, CP showed a diffused pattern (Figure 5E). Note that like nuclear condensates formed by FL, these granules also colocalized with polyA RNAs and were distinct from NSs (Supplementary Figure S6A). Although like FL condensates, NP granules were also colocalized with polyA RNAs and distinct from NSs, polyA RNAs were mostly attached to the surfaces of these granules (Supplementary Figure S6B). Interestingly, nuclear condensates formed by both FL and NP exhibited dynamic properties, moving freely in the nucleus and fusing when approached one another, indicative of liquid-like behavior (Figure 5F). We next examined whether these condensates showed internal rearrangement by photobleaching condensates, and tracking the recovery of fluorescence intensity by laser scanning confocal microscopy. Condensates formed by FL and NP both displayed internal rearrangement over the course of minutes consistent with liquid-like properties (Figure 5G).
When we tried to further narrow down the condensation domain by separating NP into 1–359 aa (CID), 360–599 aa and 600–990 aa (PID) fragments, none of them formed condensates (Figure 5H). Because CID was mainly cytoplasmic, we also examined the distribution pattern of 1–599 aa and found it diffused in the nucleus (Figure 5H). Bearing in mind that NP contains CBC- and PABPN1-binding domains, it was possible that NP sub-fragments cannot form condensates due to their reduced interactions with CBC or PABPN1. However, in cells depleted of CBC or PABPN1, the granule formation activity of NP was largely unchanged (Figure 5I). Together, these data indicate that NP is required and sufficient for ZFC3H1 forming condensates with liquid-like properties.
Condensation of many proteins also involves nucleic acid binding (35–37). This could be also true for ZFC3H1 as its condensation is triggered upon target RNA accumulation. Further, UV-crosslinking RIPs showed that NP that possesses condensation activity also exhibited direct RNA binding ability, while CP that lacks condensation activity did not (Supplementary Figure S6C–E). These data are in agreement with the view that RNA binding is involved in ZFC3H1 condensation.
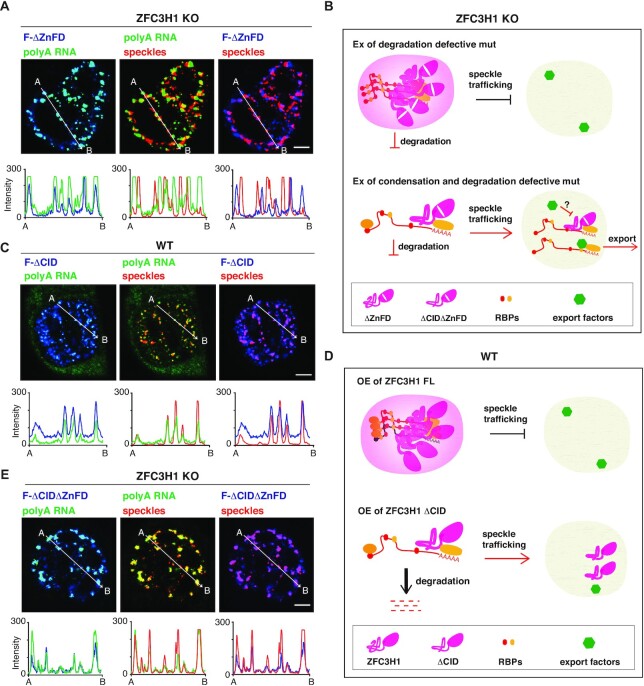
ZFC3H1 condensation activity is important for nuclear RNA retention, but not degradation
The next important question is whether ZFC3H1 condensation activity is required for preventing RNAs trafficking into NSs. To answer this question, we first sought to disrupt its degradation function but keep its condensation activity, and detect where the accumulated RNAs distributed. The ΔZnFD mutant, which contained the intact IDR but lost MTR4 interaction, perfectly satisfied this requisite. Indeed, in KO cells overexpressing ΔZnFD, consistent with degradation inhibition, the overall polyA signals increased (Figure 4F). Importantly, these increased nuclear polyA RNAs did not overlap with NSs, but were mostly colocalized with nuclear condensates formed by the mutant protein (Figure 6A and B; Supplementary Figure S7A). We next sought to inhibit ZFC3H1 condensation without affecting its degradation activity. Considering the whole NP is required for condensation, we reasoned that the ΔCID mutant, which showed similar RNA degradation functions to FL, might have reduced condensation activity. Indeed, different from FL (Figure 5A), overexpressed ZFC3H1ΔCID did not apparently form nucleoplasmic foci, but was enriched in NSs as well as diffused in the nucleoplasm in WT cells (Figure 6C and D; Supplementary Figure S7B). An important indication from these data is that the condensation activity is not required for ZFC3H1-mediated exosomal degradation. We next made CID and ZnFD double deletion mutant (ZFC3H1ΔCIDΔZnFD) to repress ZFC3H1 degradation and condensation activities simultaneously. Significantly, ZFC3H1ΔCIDΔZnFD did not form nuclear condensates in KO cells like ZFC3H1ΔZnFD, and polyA RNAs trafficked into NSs again (Figure 6B and E; Supplementary Figure S7C). These data together indicate that ZFC3H1 condensation activity is required for preventing RNA trafficking into NSs. We noted that overexpression of the ΔCID ΔZnFD mutant in KO cells caused enhanced polyA RNA retention in NSs and concomitantly reduced accumulation in the cytoplasm (Supplementary Figure S7D). This might be due to precluded ALYREF recruitment by binding of the mutant protein on ZFC3H1 target polyA RNAs.
Figure 6.
The condensation activity of ZFC3H1 is required for retaining target RNAs in the nucleoplasm. (A) Confocal microscopic imaging to examine the colocalization of polyA RNAs, the ΔZnFD mutant and NSs in ZFC3H1 KO cells. FISH with an oligo(dT) probe and IF using the SON (as an NS marker) and Flag antibodies were carried out. The green, blue and red lines in the graphs show the intensity of the FISH, Flag IF and SON IF signals along the freely positioned arrow indicated from A to B, respectively. Scale bar, 5 μm. (B) Illustration of the roles of the ΔZnFD and ΔCIDΔZnFD mutants in degradation and retention of ZFC3H1 target polyA RNAs in ZFC3H1 KO cells. (Top) The ΔZnFD mutant that loses degradation function but possesses condensation activity forms nucleoplasmic condensates that prevent RNA trafficking into NSs in ZFC3H1 KO cells. (Bottom) The ΔCIDΔZnFD mutant lacking both condensation and degradation functions does not form nucleoplasmic granules, but co-traffic with its target polyA RNAs to NSs, from where some were further transported to the cytoplasm while the others were retained there possibly due to occupation by the mutant protein. (C) Confocal microscopic imaging to examine the colocalization of polyA RNAs, the overexpressed ΔCID mutant and NSs in WT cells. FISH with an oligo(dT) probe and IF using the SON (as an NS marker) and Flag antibodies were carried out. The green, blue and red lines in the graphs show the intensity of the FISH, Flag IF and SON IF signals along the freely positioned arrow indicated from A to B, respectively. Scale bar, 5 μm. (D) Illustration of the roles of FL and the ΔCID mutant in degradation and retention of ZFC3H1 bound polyA RNAs in WT cells. (Top) Overexpressed FL forms nucleoplasmic condensates that prevent its bound polyA RNA trafficking into NSs. (Bottom) The ΔCID mutant loses its condensation activity but keeps degradation function. Overexpressed ΔCID mutant does not form nucleoplasmic granules, but traffics to NSs. (E) Same as (A), except that the ΔCIDΔZnFD is used instead of ΔZnFD in ZFC3H1 KO cells.
DISCUSSION
To ensure accurate gene expression, nascent RNAs need to be rapidly sorted into the export and the degradation pathway. ZFC3H1 is a key RNA sorter that mediates exosomal degradation and nuclear retention of exosome target RNAs (1,18,24). To date, how ZFC3H1 executes these two functions remains largely unclear. For example, whether its retention activity is required for facilitating degradation, and how does ZFC3H1 retain its bound RNAs in the nucleus? We found that forming condensates and consequently preventing trafficking of its bound RNAs into NSs is an important mechanism for ZFC3H1 retaining RNAs. Further, ZFC3H1 condensation-retention activity is not important for its RNA degradation functions. Together with previous work, our data suggest that ZFC3H1 plays two-layer surveillance roles in keeping unwanted RNAs from the cytoplasm, removing these RNAs rapidly as the first choice in normal condition and preventing their entry into NSs for gaining export competence upon degradation inhibition.
Taking degradation as the first choice makes sense, as accumulated target RNAs occupy functional proteins, i.e. PABPN1, ARS2 and MTR4, in the nucleus (24). However, when unwanted RNAs accumulate anyway, cells then activate the second-choice surveillance mechanism, forming nuclear condensates that trap these RNAs to prevent their transport to the cytoplasm. When ZFC3H1 lost its condensation activity, no polyA foci are formed even in the presence of accumulated target polyA RNAs. This suggests that ZFC3H1 directly triggers the formation of nucleoplasmic polyA foci through condensation. Our data also imply that polyA foci formed upon exosome inactivation are important for retaining exosome targets in the nucleus, as disruption of ZFC3H1 condensation led to the release of its bound polyA RNAs into NSs as well as the cytoplasm. Several exosome subunits are downregulated during epidermal and erythroid cell maturation (38,39). ZFC3H1-mediated condensation retention might have a more dominant role under such circumstances. In addition, mutations in exosome components have been associated with neurodevelopmental delay and intellectual disability (40,41). It would be interesting to examine whether polyA foci are formed in neurons of patients.
In normal cells, no apparent nuclear condensates were formed by ZFC3H1, probably due to rapid target RNA degradation. From this point of view, RNA degradation seems not very active in condensates resulted from ZFC3H1 overexpression, as polyA RNAs were clearly detected in them. In Schizosaccharomyces pombe, Red1 is the counterpart of ZFC3H1 that promotes selective removal of meiotic mRNAs containing determinant of selective removal (DSR) sequences in mitotic cells (42). Different from ZFC3H1, in normal condition, Red1 localizes in nuclear dots in which meiotic RNA degradation is thought to occur (42,43). Thus, not like mammalian cells that have an ordered choice for degradation and retention, Red1 might execute these functions simultaneously. Further, Mmi1, a Red1-interacting protein, forms nuclear dots through self-interaction and tethers DSR-containing meiotic transcripts to prevent the mistimed expression (44). Thus, it is possible that the condensation-retention mechanism we found here is evolutionarily conserved.
NSs are subnuclear structures in which mRNA export factors are enriched and many mRNAs are packaged into export-competent mRNPs (9–13). Previously, we found that MTR4 competes with ALYREF to sort nascent mRNAs into the degradation and export pathways prior to trafficking into NSs (20,27). Upon MTR4 depletion, its target mRNAs passed through NSs before being released to the cytoplasm (27). Here, we showed that in the absence of nuclear RNA decay, ZFC3H1 condenses and retains its bound RNAs in the nucleus through preventing their entry into NSs. In the absence of the ZFC3H1 protein or its condensation activity, its target RNAs trafficked through NSs from where they were exported to the cytoplasm. Thus, preventing NS entry seems to be a common and effective way for precluding unwanted RNAs from transporting to the cytoplasm.
Data shown here suggest that PABPN1 is the primary determinant for ZFC3H1 recruitment. However, PABPN1 ubiquitously binds to RNAs with a polyA tail; how is ZFC3H1 selectively recruited to its target RNAs, i.e. SNHG RNAs? It is possible that ZFC3H1, like Red1 in yeast, directly or indirectly recognizes certain RNA sequences or structures. In this case, although PABPN1 provides an important platform for ZFC3H1 recruitment, ZFC3H1 itself determines the binding specificity. Alternatively, PABPN1 might determine ZFC3H1 binding specificity through other unclear mechanisms. In future, these possibilities need to be investigated.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Cheng Lab members for useful discussion. The authors gratefully acknowledge the support of SA-SIBS scholarship program.
Contributor Information
Yimin Wang, State Key Laboratory of Molecular Biology, Shanghai Key Laboratory of Molecular Andrology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200031, China.
Jing Fan, State Key Laboratory of Molecular Biology, Shanghai Key Laboratory of Molecular Andrology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200031, China.
Jianshu Wang, State Key Laboratory of Molecular Biology, Shanghai Key Laboratory of Molecular Andrology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200031, China.
Yi Zhu, State Key Laboratory of Molecular Biology, Shanghai Key Laboratory of Molecular Andrology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200031, China.
Lin Xu, State Key Laboratory of Molecular Biology, Shanghai Key Laboratory of Molecular Andrology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200031, China.
Deng Tong, State Key Laboratory of Molecular Biology, Shanghai Key Laboratory of Molecular Andrology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200031, China.
Hong Cheng, State Key Laboratory of Molecular Biology, Shanghai Key Laboratory of Molecular Andrology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200031, China; School of Life Science, Hangzhou Institute for Advanced Study, University of Chinese Academy of Sciences, Hangzhou 310024, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31925008, 31770880, 32071287, 31800686 and 91540104]; National Key Research and Development Program of China [2017YFA0504400]; ‘Strategic Priority Research Program’ of Chinese Academy of Sciences [XDB19000000]; Shanghai Municipal Science and Technology Commission [20JC1410300]; Youth Innovation Promotion Association of the Chinese Academy of Sciences [2021269]. Funding for open access charge: National Natural Science Foundation of China [31925008].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ogami K., Richard P., Chen Y., Hoque M., Li W., Moresco J.J., Yates J.R. 3rd, Tian B., Manley J.L.. An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev. 2017; 31:1257–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lubas M., Christensen M.S., Kristiansen M.S., Domanski M., Falkenby L.G., Lykke-Andersen S., Andersen J.S., Dziembowski A., Jensen T.H.. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell. 2011; 43:624–637. [DOI] [PubMed] [Google Scholar]
- 3. Izaurralde E., Lewis J., McGuigan C., Jankowska M., Darzynkiewicz E., Mattaj I.W.. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994; 78:657–668. [DOI] [PubMed] [Google Scholar]
- 4. Gruber J.J., Zatechka D.S., Sabin L.R., Yong J., Lum J.J., Kong M., Zong W.X., Zhang Z., Lau C.K., Rawlings J.et al.. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell. 2009; 138:328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shi M., Zhang H., Wu X., He Z., Wang L., Yin S., Tian B., Li G., Cheng H.. ALYREF mainly binds to the 5′ and the 3′ regions of the mRNA in vivo. Nucleic Acids Res. 2017; 45:9640–9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chi B., Wang Q., Wu G., Tan M., Wang L., Shi M., Chang X., Cheng H.. Aly and THO are required for assembly of the human TREX complex and association of TREX components with the spliced mRNA. Nucleic Acids Res. 2013; 41:1294–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng H., Dufu K., Lee C.S., Hsu J.L., Dias A., Reed R.. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006; 127:1389–1400. [DOI] [PubMed] [Google Scholar]
- 8. Viphakone N., Sudbery I., Griffith L., Heath C.G., Sims D., Wilson S.A.. Co-transcriptional loading of RNA export factors shapes the human transcriptome. Mol. Cell. 2019; 75:310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gromadzka A.M., Steckelberg A.L., Singh K.K., Hofmann K., Gehring N.H.. A short conserved motif in ALYREF directs cap- and EJC-dependent assembly of export complexes on spliced mRNAs. Nucleic Acids Res. 2016; 44:2348–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang K., Wang L., Wang J., Chen S., Shi M., Cheng H.. Intronless mRNAs transit through nuclear speckles to gain export competence. J. Cell Biol. 2018; 217:3912–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Masuda S., Das R., Cheng H., Hurt E., Dorman N., Reed R.. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005; 19:1512–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gatfield D., Le Hir H., Schmitt C., Braun I.C., Kocher T., Wilm M., Izaurralde E.. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 2001; 11:1716–1721. [DOI] [PubMed] [Google Scholar]
- 13. Zhou Z., Luo M.J., Straesser K., Katahira J., Hurt E., Reed R.. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000; 407:401–405. [DOI] [PubMed] [Google Scholar]
- 14. Kataoka N., Yong J., Kim V.N., Velazquez F., Perkinson R.A., Wang F., Dreyfuss G.. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell. 2000; 6:673–682. [DOI] [PubMed] [Google Scholar]
- 15. Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D.. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997; 91:457–466. [DOI] [PubMed] [Google Scholar]
- 16. Houseley J., LaCava J., Tollervey D.. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006; 7:529–539. [DOI] [PubMed] [Google Scholar]
- 17. Lebreton A., Seraphin B.. Exosome-mediated quality control: substrate recruitment and molecular activity. Biochim. Biophys. Acta. 2008; 1779:558–565. [DOI] [PubMed] [Google Scholar]
- 18. Meola N., Domanski M., Karadoulama E., Chen Y., Gentil C., Pultz D., Vitting-Seerup K., Lykke-Andersen S., Andersen J.S., Sandelin A.et al.. Identification of a nuclear exosome decay pathway for processed transcripts. Mol. Cell. 2016; 64:520–533. [DOI] [PubMed] [Google Scholar]
- 19. LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D.. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005; 121:713–724. [DOI] [PubMed] [Google Scholar]
- 20. Fan J., Kuai B., Wu G., Wu X., Chi B., Wang L., Wang K., Shi Z., Zhang H., Chen S.et al.. Exosome cofactor hMTR4 competes with export adaptor ALYREF to ensure balanced nuclear RNA pools for degradation and export. EMBO J. 2017; 36:2870–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beaulieu Y.B., Kleinman C.L., Landry-Voyer A.M., Majewski J., Bachand F.. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet. 2012; 8:e1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu G., Schmid M., Rib L., Polak P., Meola N., Sandelin A., Jensen T.H.. A two-layered targeting mechanism underlies nuclear RNA sorting by the human exosome. Cell Rep. 2020; 30:2387–2401. [DOI] [PubMed] [Google Scholar]
- 23. Bresson S.M., Hunter O.V., Hunter A.C., Conrad N.K.. Canonical poly(A) polymerase activity promotes the decay of a wide variety of mammalian nuclear RNAs. PLoS Genet. 2015; 11:e1005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silla T., Karadoulama E., Makosa D., Lubas M., Jensen T.H.. The RNA exosome adaptor ZFC3H1 functionally competes with nuclear export activity to retain target transcripts. Cell Rep. 2018; 23:2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang J., Chen J., Wu G., Zhang H., Du X., Chen S., Zhang L., Wang K., Fan J., Gao S.et al.. NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction. Genes Dev. 2019; 33:536–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Giacometti S., Benbahouche N.E.H., Domanski M., Robert M.C., Meola N., Lubas M., Bukenborg J., Andersen J.S., Schulze W.M., Verheggen C.et al.. Mutually exclusive CBC-containing complexes contribute to RNA fate. Cell Rep. 2017; 18:2635–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fan J., Kuai B., Wang K., Wang L., Wang Y., Wu X., Chi B., Li G., Cheng H.. mRNAs are sorted for export or degradation before passing through nuclear speckles. Nucleic Acids Res. 2018; 46:8404–8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shi M., Zhang H., Wang L., Zhu C., Sheng K., Du Y., Wang K., Dias A., Chen S., Whitman M.et al.. Premature termination codons are recognized in the nucleus in a reading-frame dependent manner. Cell Discov. 2015; 1:15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kapadia F., Pryor A., Chang T.H., Johnson L.F.. Nuclear localization of poly(A)+ mRNA following siRNA reduction of expression of the mammalian RNA helicases UAP56 and URH49. Gene. 2006; 384:37–44. [DOI] [PubMed] [Google Scholar]
- 30. Valencia P., Dias A.P., Reed R.. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl Acad. Sci. U.S.A. 2008; 105:3386–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reits E.A., Neefjes J.J.. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat. Cell Biol. 2001; 3:E145–E147. [DOI] [PubMed] [Google Scholar]
- 32. Dias A.P., Dufu K., Lei H., Reed R.. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat. Commun. 2010; 1:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Egan E.D., Braun C.R., Gygi S.P., Moazed D.. Post-transcriptional regulation of meiotic genes by a nuclear RNA silencing complex. RNA. 2014; 20:867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee N.N., Chalamcharla V.R., Reyes-Turcu F., Mehta S., Zofall M., Balachandran V., Dhakshnamoorthy J., Taneja N., Yamanaka S., Zhou M.et al.. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell. 2013; 155:1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang H., Elbaum-Garfinkle S., Langdon E.M., Taylor N., Occhipinti P., Bridges A.A., Brangwynne C.P., Gladfelter A.S.. RNA controls polyQ protein phase transitions. Mol. Cell. 2015; 60:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Van Treeck B., Parker R.. Emerging roles for intermolecular RNA–RNA interactions in RNP assemblies. Cell. 2018; 174:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gotor N.L., Armaos A., Calloni G., Torrent Burgas M., Vabulas R.M., De Groot N.S., Tartaglia G.G.. RNA-binding and prion domains: the Yin and Yang of phase separation. Nucleic Acids Res. 2020; 48:9491–9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McIver S.C., Kang Y.A., DeVilbiss A.W., O’Driscoll C.A., Ouellette J.N., Pope N.J., Camprecios G., Chang C.J., Yang D., Bouhassira E.E.et al.. The exosome complex establishes a barricade to erythroid maturation. Blood. 2014; 124:2285–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mistry D.S., Chen Y., Sen G.L.. Progenitor function in self-renewing human epidermis is maintained by the exosome. Cell Stem Cell. 2012; 11:127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fasken M.B., Morton D.J., Kuiper E.G., Jones S.K., Leung S.W., Corbett A.H.. The RNA exosome and human disease. Methods Mol. Biol. 2020; 2062:3–33. [DOI] [PubMed] [Google Scholar]
- 41. Towler B.P., Pashler A.L., Haime H.J., Przybyl K.M., Viegas S.C., Matos R.G., Morley S.J., Arraiano C.M., Newbury S.F.. Dis3L2 regulates cell proliferation and tissue growth through a conserved mechanism. PLoS Genet. 2020; 16:e1009297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sugiyama T., Sugioka-Sugiyama R.. Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J. 2011; 30:1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamashita A., Takayama T., Iwata R., Yamamoto M.. A novel factor Iss10 regulates Mmi1-mediated selective elimination of meiotic transcripts. Nucleic Acids Res. 2013; 41:9680–9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shichino Y., Otsubo Y., Kimori Y., Yamamoto M., Yamashita A.. YTH-RNA-binding protein prevents deleterious expression of meiotic proteins by tethering their mRNAs to nuclear foci. eLife. 2018; 7:e32155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.