Abstract
Forkhead box (FOX) proteins are an evolutionarily conserved family of transcription factors that play numerous regulatory roles in eukaryotes during developmental and adult life. Dysfunction of FOX proteins has been implicated in a variety of human diseases, including cancer, neurodevelopment disorders and genetic diseases. The FOX family members share a highly conserved DNA-binding domain (DBD), which is essential for DNA recognition, binding and function. Since the first FOX structure was resolved in 1993, >30 FOX structures have been reported to date. It is clear now that the structure and DNA recognition mechanisms vary among FOX members; however, a systematic review on this aspect is lacking. In this manuscript, we present an overview of the mechanisms by which FOX transcription factors bind DNA, including protein structures, DNA binding properties and disease-causing mutations. This review should enable a better understanding of FOX family transcription factors for basic researchers and clinicians.
INTRODUCTION
Gene expression is a highly ordered and extremely complex process. This process is tightly controlled by transcription factors (TFs). TFs usually bind DNA regulatory elements upstream or downstream of the transcription start sites of target genes to modulate the level of gene transcription, thereby controlling protein synthesis and thus altering cellular function (1). To date, >1600 TFs have been annotated in the human genome (1). Among them, the evolutionarily conserved FOX proteins compose one of the largest TF families and play key regulatory roles in both embryonic and adult life (2–4).
The first member of the FOX protein family was identified in Drosophila in connection with the forkhead mutation phenotype (5,6). Subsequently, hundreds of FOX genes have been identified from various species. The FOX gene family is now known to be an evolutionarily ancient transcription family that is widely distributed in eukaryotes, from unicellular eukaryotes to mammals. However, FOX genes have not been discovered in the plant kingdom so far (7,8). The number of FOX genes varies largely among species. Fungi genomes encode only four FOX genes, whereas mammalian genomes typically encode dozens of members (9). All FOX proteins share an evolutionarily conserved DNA-binding core, while other parts of FOX proteins are poorly conserved. For example, the DBD of yeast forkhead protein 1 (FHL1) has 62% sequence similarity to human FOXP1-DBD. In the human genome, 50 members have been identified and can be classified into 19 subgroups (FOXA to FOXS) based on sequence conservation (10). These family members display large functional diversity in key biological processes, including development, proliferation, differentiation, apoptosis, and immune function (3,11,12). Many FOX TFs, especially FOXA, FOXO and FOXI subgroup members, are regarded as pioneer factors which open compacted chromatin and facilitate the binding of other transcription factors (13,14). Usually, each FOX TF possesses its own specific regulatory roles, and its expression is tissue- and cell type-specific (3). For example, FOXG1 is mainly expressed in forebrain and is essential for brain development (15); FOXO1 is highly expressed in B cells and dominates B-cell differentiation (16,17), whereas FOXO4 is highly expressed in muscle and FOXO6 is primarily expressed in the brain and liver (18). Meanwhile, FOX proteins also exhibit a degree of functional redundancy. For example, ovarian physiogenesis is regulated by three forkhead proteins, FOXL2, FOXC1 and FOXO3 (19,20). This functional redundancy may enhance the tolerance to decreased function of a specific FOX protein (21).
Genetic alterations and misregulation of FOX genes are closely associated with human diseases, especially cancers (22,23). The alterations, including deletion, point mutation, translocation and gene fusion, of FOX genes have been identified in a variety of cancers (24,25). Alterations and misregulation of almost all the 19 FOX subgroups have been shown to associate with tumorigenesis. Among them, the roles of FOXM1 (23,26), FOXO (22) and FOXA1 (27) in cancer have perhaps attracted the most attention from researchers. FOXM1 overexpression has been detected in a broad range of cancer types, such as glioblastoma (28) and chronic myelogenous leukemia (CML) (29), and it is critical for the initiation, progression and drug resistance of these cancers (26,30). Therefore, FOXM1 has been considered as a prognostic marker and an attractive drug target for some cancers. FOXOs are mostly considered as tumor suppressors, due to the involvement of FOXO gene activation in cell cycle arrest and apoptosis in both normal and cancer cells (31–33). However, increasing evidence suggests FOXO TFs also act as oncogenic regulators. The carcinogenic effects of FOXOs have been demonstrated in breast cancer, hepatocellular carcinoma and leukemia, among other cancers (31,34,35). More recently, the role of FOXA1 in prostate cancer has been intensively investigated. Mutations in the coding and non-coding sequences of the FOXA1 gene have been found to drive the development and progression of prostate cancer (36–40). The close relationship between dysfunction of FOX TFs and cancer suggests that targeting FOX proteins could be an effective anti-cancer therapeutic strategy.
The regulatory functions of FOX family TFs involve specific DNA sequence recognition. To accomplish this, all FOX proteins share a highly conserved DBD, also known as the forkhead domain (FKH) or winged-helix domain (41). Most pathogenic missense mutations reported in FOX TFs are located in the DBD region (37,39,42–46), emphasizing the importance of this domain in the function of FOX TFs. In this review, we aim to provide an overview of the biological features of the FOX DBD. The structural characteristics, DNA-binding mechanism, DNA-binding specificity and consequences of point mutations of FOX-DBD are summarized.
Overall domain organization of FOX proteins
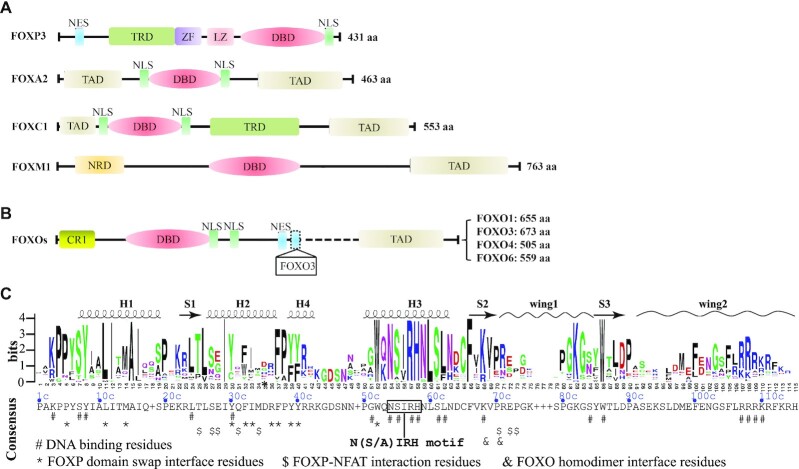
The FOX TF family proteins exhibit a wide range of functional diversity. This functional diversity is partially conferred by different spatio-temporal expression patterns. In addition, differences in protein size, structural domain composition and architecture also contribute significantly. The common functional domains or motifs present in FOX family TFs include, but are not limited to, nuclear export sequence (NES), transcriptional repressor domain (TRD), nuclear localization sequence (NLS), transactivation domain (TAD), negative-regulatory domain (NRD) and DBD (Figure 1A). The typical lengths of FOX TFs range from 300 to 800 residues, the differences in length often reflecting different domain compositions. For instance, FOXP3 lacks the transactivation domain (TAD), which is often linked with chromatin opening in FOX family TFs (Figure 1A). Nonetheless, FOXP3 possesses a transcription repressor domain, the presence of which is consistent with its well-known role as a transcription repressor (47,48). Furthermore, compared to FOXP3, FOXA1 has two TADs, one in its N-terminus and the other in the C-terminus. The two TADs confer FOXA1 a strong activity in promoting transcription. Indeed, the pioneering role of FOXA1 in the regulation of gene transcription has been well established (49,50).
Figure 1.
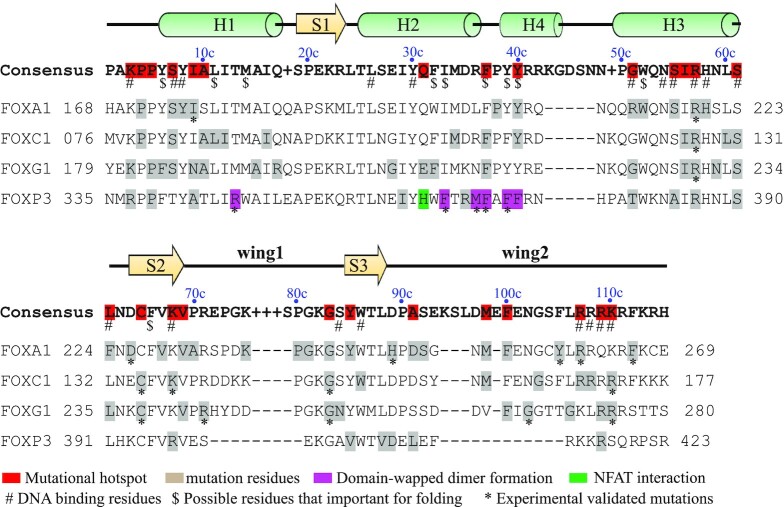
Domain organization of FOX TFs. (A) Major functional domains and motifs present in selected human FOX proteins; ZF, zinc finger; LZ, leucine zipper; CR1, conserved region 1. (B) Domain organization of human FOXO subgroup. (C) Conservation of human FOX-DBDs. The sequence logo was generated with WebLogo (http://weblogo.berkeley.edu/logo.cgi) by using a multiple sequence alignment profile of all the 50 human FOX-DBDs. The consensus sequence was provided by JalView based on the multiple sequences aligemnt file. Each residue in the consensus sequence represents the most frequent residue in each postion. If the highest frequency is shared by more than one residues, a ‘+’ symbol is used in the display. Resides that involved in DNA recognition and protein–protein interaction are indicated accordingly.
The organization of structural domains varies largely among FOX subfamilies, whereas they are similar within the same subfamily. For example, the four FOXO subgroup members (FOXO1, FOXO3, FOXO4 and FOXO6) have the same structural architecture, although they are of different lengths (Figure 1B). Interestingly, two NES have been reported for FOXO3, while the NES described in FOXO6 is under debate (51).
Although the domain arrangement of FOX TFs varies widely, all FOX members possess a DNA binding domain. As its name implies, the FOX-DBD is essential for DNA binding and is the core component for the regular function of FOX TFs. The position of DBD is different among family members; however, it is highly conserved in sequence and is why these proteins are grouped into the same family (52). The FOX-DBD is approximately 100 amino acids (aa) in length, with the residues in the N-terminal portion highly conserved, while the conservation of C-termini residues is limited (Supplementary Figure S1). According to the multiple sequence alignment, a consensus sequence for human FOX-DBD is proposed (Figure 1C). For descriptive convenience, the numbering schedule in the consensus sequence (position plus letter c) will be used throughout the text.
Structure of FOX-DBD
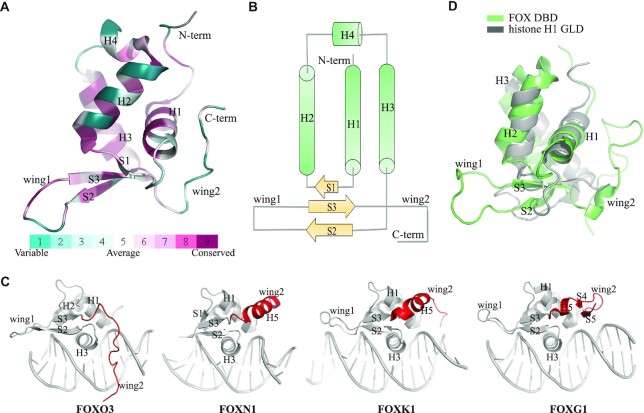
Since the first structure of FOX-DBD (FOXA3) determined in 1993 (41), the structure of forkhead domain has been extensively studied during the last 30 years. To date, >30 FOX-DBD structures have been deposited in the protein data bank (PDB, see table 1). These structures provide a thorough structural understanding of the core of FOX TFs. All FOX-DBDs adopt a similar overall compact folding, with three α-helices (H1, H2 and H3) forming a core that packs against an antiparallel three-stranded β-sheet (S1, S2 and S3); there are two flanking wing-like loops (wing 1 and wing 2) (Figure 2A). This folding pattern has been named as winged-helix fold, which is often found in DNA-binding proteins (41,53,54). In addition to the three typical helices, several structures show that the residues between H2 and H3 helix can also fold into a 310-helix (H4) (Figure 2A) (55–59). Topologically, the FOX-DBD is arranged as H1-S1-H2-H4-H3-S2-wing1-S3-wing2 (Figure 2B). Interestingly, the S1 strand is not observed in most of the reported FOX-DBD structures; instead, this region has been shown to be a flexible loop in these structures.
Table 1.
Determined FOX-DBD structures
| PDB entry | Protein | DNA Core motif | Method | Resolution (Å) | Assembly | Reference |
|---|---|---|---|---|---|---|
| 5X07 | FOXA2 | GTAAACA | X-ray | 2.80 | Monomer | (61) |
| 1VTN | FOXA3 | TAAGTCA | X-ray | 2.50 | Monomer | (41) |
| 1D5V | FOXC2 | Protein alone | NMR | n/a | Monomer | (81) |
| 6AKO | FOXC2 | GTAAACA | X-ray | 2.40 | Monomer | (59) |
| 6AKP | FOXC2 | GTACACA | X-ray | 2.32 | Monomer | (59) |
| 6O3T | FOXC2 | TGTTTATAAACA | X-ray | 3.06 | Dimer, lack PPI | (82) |
| 6LBM | FOXC2 | GTAAACA | X-ray | 2.84 | Monomer | (83) |
| 2HFH | Foxd3 (mouse) | Protein alone | NMR | n/a | Monomer | (84) |
| 2HDC | Foxd3 (mouse) | GCTTAAAATAACAATAC | NMR | n/a | Monomer | (58) |
| 7CBY | FOXG1 | GTAAACA | X-ray | 1.65 | Monomer | (44) |
| 2A3S | Foxk1 (mouse) | Protein alone | NMR | n/a | Monomer | n/a |
| 2C6Y | FOXK1 | GTAAACA | X-ray | 2.40 | Dimer, lack PPI | (65) |
| 3G73 | FOXM1 | TGTTTATAAACA | X-ray | 2.21 | Dimer, weak PPIs | (56) |
| 6EL8 | FOXN1 | GACGC | X-ray | 1.61 | Monomer | (66) |
| 5OCN | FOXN1 | Protein alone | X-ray | 2.70 | Monomer | (66) |
| 6NCE | FOXN3 | AGTAAACA | X-ray | 2.60 | Monomer | (67) |
| 6NCM | FOXN3 | GACGC | X-ray | 2.70 | Monomer | (67) |
| 3CO7 | FOXO1 | GTAAACA | X-ray | 2.91 | Monomer | (60) |
| 3CO6 | FOXO1 | GTAAACA | X-ray | 2.10 | Monomer | (60) |
| 3COA | FOXO1 | CAAAACAA | X-ray | 2.20 | Monomer | (60) |
| 5DUI | FOXO1 | ATGATTTACGTAAAATAGAAA | X-ray | 2.31 | Dimer, lack PPI | (85) |
| 4LG0 | FOXO1/ETS1 | AAACAATAACAGGAAACCGTG | X-ray | 2.19 | Heterodimer, no PPI | (5) |
| 6QVW | FOXO1 | Protein alone | NMR | n/a | Monomer | (86) |
| 6LBI | FOXO1 | GTAAACATGTTTAC | X-ray | 3.07 | Dimer, with PPIs | (83) |
| 2UZK | FOXO3 | GTAAACA | X-ray | 2.70 | Monomer | (62) |
| 2K86 | FOXO3 | Protein alone | NMR | n/a | Monomer | (87) |
| 1E17 | FOXO4 | Protein alone | NMR | n/a | Monomer | (88) |
| 3L2C | FOXO4 | CTATGTAAACAAC | X-ray | 1.87 | Monomer | (55) |
| 2KIU | FOXP1 | Protein alone | NMR | n/a | Monomer | (89) |
| 2A07 | FOXP2 | AACTATGAAACAAATTTTCCT | X-ray | 1.90 | Domain-swapped dimer | (63) |
| 2AS5 | FOXP2/NFAT | AACTATGAAACAAATTTTCCT | X-ray | 2.70 | Heterodimer, with PPIs | (90) |
| 3QRF | FOXP3/NFAT | AACTATGAAACAAATTTTCCT | X-ray | 2.80 | Heterotrimer, With PPIs | (64) |
| 4WK8 | FOXP3 | AACTATGAAACAAATTTTCCT | X-ray | 3.40 | Domain-swapped dimer | (91) |
Figure 2.
Structure of FOX-DBD. (A) Overall structure of FOX-DBD. The figure shows the structure of FOXM1 (PDB code: 3G73). Structural cartoons are colored by conservation. (B) Topology of FOX-DBD. Both the S1 and H4 are only presented in certain FOX-DBDs. (C) Different wing2 conformations of FOX-DBDs. The PDB codes for FOXO3, FOXN1, FOXK1 and FOXG1 are 2UZK, 6EL8, 2C6Y and 7CBY, respectively. (D) Structural comparison between human FOXA3-DBD (PDB code: 1VTN) and human histone H1 globular domain (PDB code: 6HQ1).
The conformation of wing2 is the most variable region in the FOX-DBD, which is consistent with the fact that wing2 shows only limited sequence conservation (Figure 1C). In some structures, wing2 is not observable due to the flexibility of this region (60,61). However, biochemical assays indicate that the wing2 is important for optimal DNA binding by enhancing protein–DNA interaction (60,61). In some structures, such as FOXC2 (59), FOXO3 (62) and FOXD3, the wing2 presented as a loop (58). In contrast, in the structure of FOXP2 (63), FOXP3 (64), FOXK1 (65), FOXN1 (66) and FOXN3 (67), the wing2 folds into an α-helix. More recently, our reported FOXG1-DBD structure shows that FOXG1-DBD wing2 folds into two antiparallel β-strands (44) (Figure 2C). Thus, wing2 of FOX TFs can adopt various conformations and perhaps is a source of DNA binding specificity and functional diversity of FOX TFs.
As mentioned, many FOX TFs exhibit a pioneering role in transcription regulation. The structural comparison reveals a highly similar architecture between histone H1 globular domain and FOX-DBD (Figure 2D). The linker histone H1 is an important component of chromatin. Histone H1 is essential for maintaining chromatin in a compact and transcription-repressed conformation (68). The DNA binding region (globular domain) of histone H1 also adopts a typical winged-helix fold (69,70). The structural similarity may help FOX protein displace histone H1 to open chromatin and function as a pioneer transcription factor.
DNA recognition by FOX-DBD
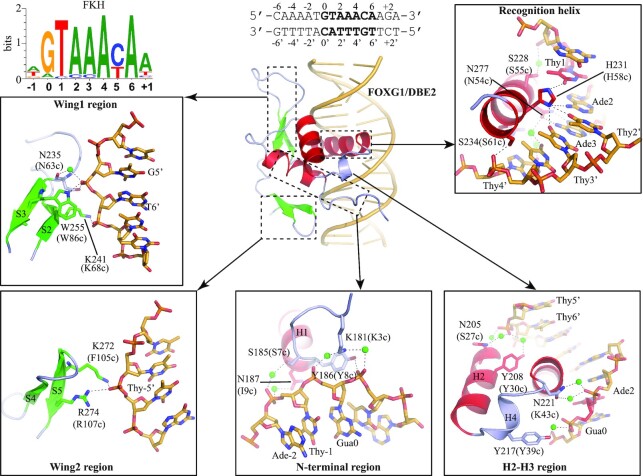
Through its DBD, FOX TF binds to specific DNA motifs in the genome and regulates gene expression. Most FOX TFs recognize the canonical forkhead (FKH) sequence, 0RYAAAYA6 (R = purine, Y = pyrimidine) (71,72). A FKH PWM (Positional weight matrix) is shown in Figure 3. The binding of FOX-DBD to FKH DNA site has been well established. Upon binding to DNA, the H3 helix of FOX-DBD inserts into the DNA major groove and mediates most of the base-specific recognition (Figure 3). Therefore, the H3 helix has been considered as the recognition helix (41). Usually, only the conserved N54c and H58c from the so-called ‘54cN(S/A)IRH58c’ motif (see Figure 1C) of recognition helix form direct H-bonds with DNA bases. In addition to direct H-bonds, water-mediated interactions and van der Waals contacts are also involved in H3-DNA interactions (44,60). Functional assays have shown that all direct and indirect contacts are important for DNA binding (44,59,62).
Figure 3.
DNA recognition by FOX-DBD. The hydrogen bonds are indicated by dashed lines. Residues at the corresponding positions in the FOX-DBD consensus sequence of Figure 1C are shown in parentheses (and hereafter). Water molecules are represented by green balls. Van der Waals contacts are not shown. The figures were prepared from the FOXG1/DBE2 complex structure (PDB code: 7CBY, resolution at 1.6 Å).
Other regions of FOX-DBD also participate in DNA binding outside the core-site. The N-terminal, H4 helix, wing1 and wing2 are involved in DNA binding by forming direct H-bonds, water-mediated H-bonds and van de Waals contacts with DNA. Interestingly, though wing2 is important for DBD–DNA contacts, it is invisible in some of FOX/DNA complex structures. Furthermore, in the structures where wing2 is well ordered, such as FOXM1 (56), FOXP2 (63) and FOXN1 (66), no direct wing2–DNA interaction is observed. These observations may indicate the binding between wing2 and DNA is highly dynamic and difficult to be captured during crystallizing. Interestingly, in addition to DBD, the LZ motif immediately adjacent to the N-term of FOXP3-DBD may also take part in DNA binding. The presence of the LZ motif largely enhances the DNA binding ability of FOXP3 in vitro (73,74).
The DNA-binding potential of FOX-DBD can be modified by various post-translational modifications (PTMs), including phosphorylation, acetylation and methylation (27,75). For example, the FOXO-DBD is frequently phosphorylated by various kinases. Kinase MST1 mediates phosphorylation of FOXO3-DBD at S207 (S55c: the corresponding residue in the consensus sequence), S213 (S61c), S229 (G83c: the amino acid sequence may be different from the consensus sequence in some cases because the consensus sequence only shows the residue with highest frequency) and S230 (S84c) to promote apoptosis (76). In vitro assay showed phosphorylation of the four corresponding serine residues in FOXO1-DBD blocks its binding to DNA because these residues are involved in DNA backbone interactions (direct or water-mediated) (60). Interestingly, the phosphorylation of FOXO1-DBD at S249 by CDK1/CDK2 (77,78) can impact its subcellular localization without disturbing its DNA-binding ability (60).
In addition to phosphorylation, two acetylation sites, K245 (K94c) and K248 (S104c) have been identified within the wing2 of FOXO1-DBD. These two sites can be specifically acetylated by CBP/p300 (60). Since acetyl groups are negatively charged, acetylation introduces charge repulsion between wing2 and DNA, thus reducing the DNA-binding affinity. Several acetylation sites in the wing1 or wing2 regions of FOXA1 and FOXP3 have also been reported. Acetylation of FOXA1-DBD decreases its DNA-binding and ability to remodel chromatin (79). Similarly, acetylation of FOXP3 K382 (Q53c) and K393 (D64c) impairs the DNA binding and inhibits Treg suppressive function (80). Collectively, FOX-DBD is often subjected to PTM and the phosphorylation sites in FOX-DBD are highly conserved, which may be a regulatory mechanism adopted by FOX family TFs. However, the acetylation sites of FOX proteins are primarily localized in the variable wing1 or wing2 region, thus these sites are less conserved compared to phosphorylation sites. The variable acetylation sites may indicate different regulatory mechanisms of individual FOX proteins.
In summary, the conserved recognition helix of FOX-DBD dominates base-specific DNA recognition, while less conserved regions, including the N-terminus, H2-H3 region, wing1 and wing2, make adjustable but essential connections with DNA. In addition, the DNA-binding capability of FOX-DBD is frequently modified by various PTMs, such as phosphorylation and acetylation.
Mechanisms for DNA binding diversity
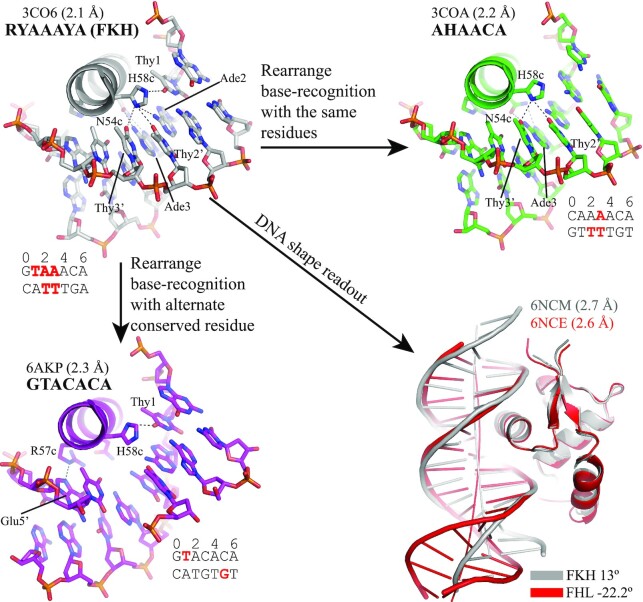
The functional diversity of FOX TFs is usually associated with the diversity of their DNA binding. In addition to the canonical FKH site, FOX TFs have been shown to bind other DNA motifs, including AHAACA (60,92,93), GTACACA (59) and GACGC (66,67,72). The AHAACA (H = A, C, T) motif was first reported to be a lower affinity binding site of FOXO1, compared to the FKH site (94,95). Crystal structures of FOXO1 in complex with FKH and AHAACA sites, respectively, reveal that FOXO1 rearranges histidine (H58c) and asparagine (N54c) in the N(S/A)IRH motif to bind different DNA bases (60) (Figure 4).
Figure 4.
Binding of FOX-DBD to different DNA motifs. Direct H-bonds are indicated by dash lines. The recognized bases are highlighted in red.
Interestingly, in vitro DNA-binding analysis demonstrates that FOXC2 prefers to bind the GTACACA DNA sites rather than a canonical RYAAAYA site, ACAAATA (59). When bound to the GTACACA site, FOXC2 engages R57c other than N54c in the 54cN(S/A)IRH58c motif to carry out base recognition (Figure 4). Furthermore, this mechanism also applies to the cases where the A3 in FKH motif is substituted by other nucleotides (59,65). These observations suggest the FOX TFs may have specific preferences within the core motif. Moreover, flanking bases outside the core motif are reported to be important for optimal DNA-binding bases (61,62,96).
Both AHAACA and GTACACA are variants of the FKH site, and the binding of FOX TFs to these variants typically involves the rearrangement of contacts in the recognition helix while maintaining similar overall DNA deformation (17,44,59). However, compared to the RYAAAYA site, the GACGC (FHL) site is different in length and base composition. The FHL site was first reported as a binding site of yeast Fhl1 (97). Later studies revealed that the FHL site was an alternative binding site for FOXN subgroup members, and thus, FOXNs have been regarded as bispecific FOX TFs (72,98,99). Rogers et al. showed FOXN3 adopted an almost identical structure to contact the FKH and FHL sites and utilized the same amino acid residues for base-specific recognition (67). However, the binding-induced DNA shape deformation is strikingly different, suggesting an indirect DNA-shape readout mechanism (Figure 4). A similar DNA conformation was observed in the FOXN1-FHL structure, indicating the unusual FHL conformation is unlikely to be induced by crystal packing (66).
Subdomain swap experiments suggest the two wings (wing1 and wing2) are necessary but not enough to determine the bispecificity of FOXN3 (67). The two-wing switch between FOXN1 and monospecific FOXJ3 abolished the FHL site binding ability of FOXN1, but did not confer FOXJ3 the ability to bind the FHL site (67). Further, it has been shown that the H2-H3 region has a dominant role in determining the FHL site binding ability of FOXN1 (67). Interestingly, another study of the DNA-binding properties of a series of FKH protein chimeras also suggests that the less conserved region between H2 and H3 helices plays an important role in regulating the binding specificity of the forkhead domain (92). The H2-H3 region, where H4 helix is located, does not participate in direct DNA binding, showing only water-mediated interaction with DNA in some structures. These observations suggest water-mediated interactions may be important for DNA binding specificity.
Furthermore, even when bound to the same DNA site, FOX-DBDs exhibit considerable variations in DNA recognition. The differences occurred both in the recognition helix and other less conserved regions (44,59–61) (and the references hereafter). In base-specific recognition, FOXA2 engages H54c to bind a single base of the DBE2 (A FKH DNA) site, while the numbers of base contacts mediated by the same residue in FOXC2, FOXO1 and FOXG1 are two, three and four, respectively (Supplementary Figure S2). In addition, FOXA2 also employs R208 (G51c) and S212 (S55c) of the H3 helix to bind the DBE2 phosphate backbone; however, these interactions are not observed in FOXC2, FOXO1 and FOXG1. DNA recognition by the N-terminal segment is different among the four structures. FOXC2 binds Thy1 with its K72 (K3c), while FOXO1 utilizes N158 (P1c) to contact Gua0. Nevertheless, no direct interaction is observed in the other two structures. Although the wing2 has been included in all four proteins for crystallizing, it has been observed only in FOXG1 and FOXC2 structures. Again, the observed wing2–DNA interactions are different in the two structures. FOXC2 wing2 contacts the DBE2 site with R163 (R107c) and R164 (R108c), while FOXG1 wing2 binds the DNA with K272 (R107c) and R274 (R109c). The mentioned interactions do not include water-mediated contacts, the observation of which depends largely on the diffraction resolution. Notably, this does not mean that water-mediated interactions are not important. On the contrary, water-mediated hydrogen bonds may play a key role in FOX–DNA interaction (44,60). In addition, the reported differences may arise from the fact that different groups used different protein constructs (with different lengths of N- and C- terminal segments) in their DNA-binding studies.
Altogether, the DNA-binding specificity of FOX-DBDs is determined by rearranging base-specific contacts of the H3 helix, DNA shape recognition and the regulation of the less conserved regions, especially wing1, wing2 and the H2-H3 region. In addition, flanking bases outside the core sequence also affect the binding of FOX-DBD. Due to the plasticity and complexity of DNA recognition by FOX-DBDs (different FOX-DBDs bind to the same DNA with varied interactions and the same FOX-DBD often behaves differently when binding to different DNA sites), it is difficult to come up with an overarching model to explain the different binding specificities of FOX-DBDs. Compared to most sequence-specific transcription factors, an unusual feature of forkhead protein binding to DNA is its extensive use of van der Waals contacts and relatively few hydrogen bonds to bases within the major groove. This shape recognition may allow forkhead proteins to bind a wide range of sequences if the DNA maintains a few hydrogen bonding determinants in the core region of the binding site and has a shape that is complementary to the DNA binding surface of the forkhead protein.
DNA-binding induced conformational change of FOX-DBD
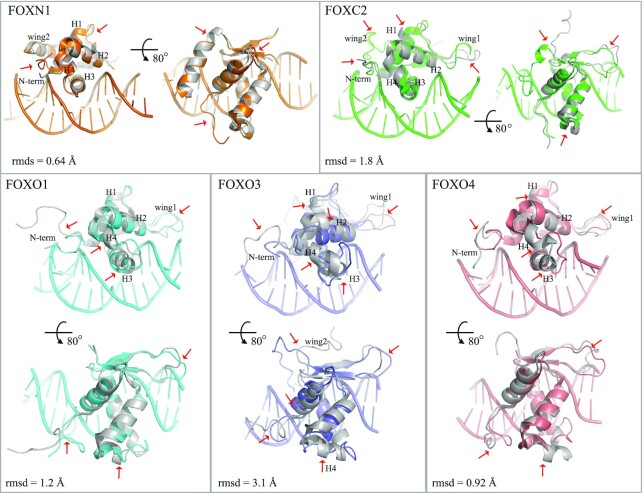
Some of the FOX-DBD structures are determined both in the presence and absence (apo) of DNA, including FOXC2, FOXN1, FOXO1, FOXO3 and FOXO4 (Table 1). These structure pairs allow a close inspection of the conformational changes induced by the binding of DNA (Figure 5). Superimposing the DNA-bound DBD to unbound DBD of the five structure pairs, the root mean square deviation (rmsd) values range from 0.64 to 3.1 Å, indicating no significant rearrangements (Figure 5). However, mild structural changes are presented, which mainly occurred in regions around the DNA-binding surface (H3 helix, H4 helix, N-terminal segment, wing1 and wing2). These regions may function through structural plasticity, adopting ordered conformations when bound to DNA.
Figure 5.
DNA-binding-induced conformational change of FOX-DBD. DNA-bound structures are shown in color and apo FOX-DBD structures are shown in gray. Major conformational differences are indicated by red arrows. The PDB codes are FOXN1, 6EL8 (DNA-bound) and 5OCN (apo); FOXC2, 6AKO (DNA-bound) and 1D5V (apo); FOXO1, 3CO7 (DNA-bound) and 6QVW (apo); FOXO3, 2UZK (DNA-bound) and 2K86 (apo); FOXO4, 3L2C (DNA-bound) and 1E17 (apo).
DNA-binding-induced conformational changes are observed at the N-terminal segment, H1-H2 loop and wing2 for FOXN1-DBD (rmsd, 0.64 Å). The orientations of FOXC2 N-term, H1 helix, H4 helix, wing1 and wing2 are rearranged when bound to DNA. The C-terminus of H1 helix shifts by 2 Å toward the S3 strand when bound to DNA. Similar H1 orientations have also been observed in the DNA-bound FOXO3 and FOXO4 structures, but not for FOXO1. The interactions between wing2 and DNA induce a ∼180º flip of wing2 compared to apo FOXC2 structure. FOXO3 wing2 also interacts with DNA and adopts a similar orientation as FOXC2 wing2 when compared to its DNA-unbound conformation. The N-terminal part of H3 helix shifts by about 2 Å toward the H1 helix in the DNA-bound forms of all four FOX-DBDs, except FOXN1. Accordingly, the H2-H3 region (H4 helix) undergoes some deformations upon DNA binding in these four structures. Therefore, we speculate that the H2-H3 region may act as a regulatory element that allows the H3 helix to rearrange upon binding to DNA. Of course, it cannot be excluded that some of the observed differences may arise due to crystal packing, but we can expect most of them are induced by DNA binding.
These observations show that the DNA binding-induced conformational changes are varied in different FOX members, which may partially explain the observed differences in binding affinity to similar or identical DNA sites of individual FOX proteins (44,59,60,83). Furthermore, the plasticity of FOX-DBD conformation in DNA recognition may confer FOX TFs the ability to bind DNA sites of vastly different sequences.
Various DNA-binding modes of FOX-DBD
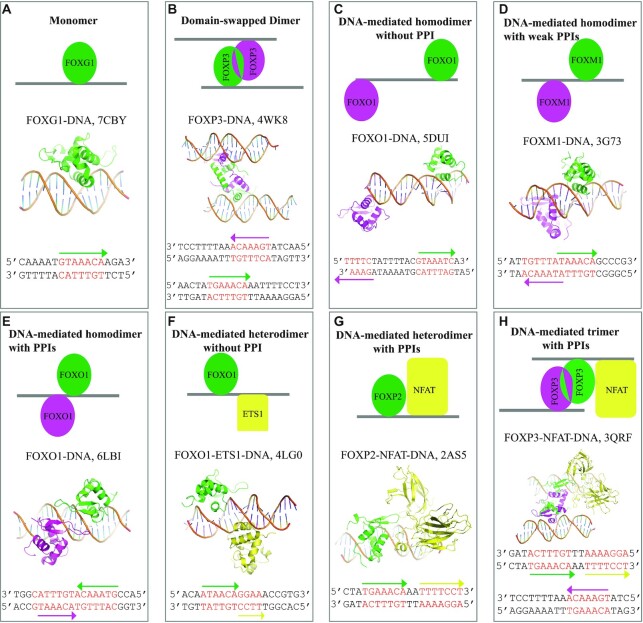
In vertebrate genomes, clustering of multiple binding sites for the same transcription factors is widely distributed (100–102). Increasing data also suggests that FOX TF binding sites are often arranged in a tandem fashion in the genome, which allows two or more FOX members to bind (72,103). For example, recent studies suggest that both FOXA1 and FOXO1 are capable of binding the compact DIV0 (termed ‘DIV’ for its diverging half-sites) site as a homodimer in a highly cooperative manner, where the first monomer binding facilities the binding of the second one (83,103,104). In addition, FOX TFs also recruit other partners to cooperatively recognize DNA. For instance, FOXOs interact with transcription factors like p53, STAT3 and retinoic acid receptors to cooperatively regulate gene transcription (105,106). The homotypic or heterotypic complex can enhance the specificity of DNA recognition as they increase the criteria for the binding (107). By analyzing the current available FOX–DNA complex structures, the DNA-binding modes of FOX TFs can generally be classified into four types: Monomer, homodimer, DNA-mediated homodimer and DNA-mediated heterodimer (Figure 6).
Figure 6.
DNA-binding modes of FOX TFs. (A) DNA recognition by the FOXG1 monomer. (B) DNA recognition by the FOXP2 domain-swapped dimer. FOXP2 forms domain swapped-dimer to bind two separate DNA sites simultaneously. (C) DNA recognition by the FOXO1 homodimer. No PPI is present. (D) DNA recognition by the FOXM1 homodimer. Weak PPIs are present. (E) DNA recognition by the FOXO1 homodimer. Extensive PPIs are present in the dimer interface. (F) DNA recognition by the FOXO1-ETS1 heterodimer. No PPI is present. (G) DNA recognition by the FOXP2-NFAT heterodimer. Extensive PPIs are present. (H) DNA recognition by the FOXP3-FOXP3-NFAT trimer. NFAT directly interacts with the domain-swapped FOXP3 dimer. FOX-DBDs are colored in green or magenta. Ets1 and NFAT are colored in yellow. DNA core sites are highlighted in red. DNA reading orientations are indicated by arrows.
Almost all FOX-DBD bind the consensus forkhead recognition site as monomer (Figure 6A), except FOXPs. The FOXP subgroup members can bind two separate DNA sites and form domain-swapped dimer (Figure 6B). The domain-swap dimer interface is mediated by residues from H1, H2, H3 and H4 helixes (Figure 1C). Moreover, the FOXP homodimer is formed prior to DNA binding (64,91,73); thus, the dimer is formed on the protein level independent of DNA. Autoimmune disease (IPEX)-associated mutations have been identified in the domain-swap interface. These mutations have been shown to eliminate T cell-suppressive activity conferred by FOXP3, both in vitro and in a murine model of autoimmune diabetes in vivo (63,64). The special DNA-binding pattern of the FOXP subgroup is believed to have relevance in chromatin bridging and long-range transcriptional regulation. In addition, it has been found that Saccharomyces cerevisiae FOX proteins Fhl1 and Fhl2 may adopt a similar domain-swapped binding mechanism as FOXPs to loop DNA (108). With the high conservation of the DBD among FOXP subgroup members, FOXPs may form hetero-domain-swapped dimer to bind DNA.
Unlike the FOXP homodimer, other observed FOX homodimers are mediated by DNA. In these binding modes, intramolecular protein–protein interactions (PPIs) may or may not be presented. For instance, FOXO1 binds the glucose-6-phosphatase catalytic subunit 1 (G6PC1) gene promoter as a homodimer in a cooperative manner (109). However, crystal structure shows the two binding sites of FOXO1 in G6PC1 promoter DNA are apart from each other and no direct PPI is presented (Figure 6C) (85). In another case, FOXM1 forms a dimer on a tandem sequence, where limited PPIs are observed (Figure 6D). However, functional and bioinformatical research suggests that the binding of FOXM1 on such DNA sites has no biological significance (56). Our recent work shows that FOXO1 binds to DIV2 sites (termed ‘DIV’ for its diverging half-sites) as a dimer (Figure 6E) (83). The structure of FOXO1/DIV2 complex revealed that the two FOXO1 monomers bind DIV2 from the same side and form PPIs mediated by the two wing1s. The wing1 residues R225 (K68c) and Q227 (P70c) play key roles at the dimer interface, and the PPIs greatly enhanced the FOXO1-DIV2 binding affinity and thermal stability (83). The further bioinformatical analysis found that the DIV2 motif is widespread in FOXO1 target genes, indicating this motif is an important regulatory element of FOXOs (83). However, the positions 68c and 70c in FOX family DBD are not well conserved, suggesting that the binding modes on DIV2 sites may vary in the FOX TF family (Figure 1C). Indeed, FOXC2 has been shown only able to bind DIV2 as a monomer (83).
The currently known interactions of FOX TFs with other TF family proteins are mediated by DNA. It is known that the FOX:Ets cis-acting element is synergistically activated by FOX-Ets TF pairs (110). The FOXO1-Ets1-DNA structure shows the two TFs binding the DNA from opposite sides without direct PPI exhibited (Figure 6F). Another study revealed that the DNA shape is the driver for the FOX-Ets pair’s cooperativity, as the binding of FOXO1 to the FOX:Ets element requires the presence of Ets1 (111). FOXP3 and nuclear factor of activated T cells (NFAT) form a cooperative complex on DNA to control regulatory T-cell function (112,113). Structural study shows that FOXP2 binds the DNA from the same side as NFAT and makes interactions with NFAT through its wing1. Mutagenesis experiments reveal that these interactions are essential for the normal function of regulatory T cells (Figure 6G) (90). Further, the FOXP3-NFAT-DNA structure suggests a complex with more subunits, where FOXP3 forms a homodimer and binds simultaneously with NFAT at the interleukin-2 promoter (Figure 6H) (64). In both FOXP2- and FOXP3-NFAT-DNA structures, the intermolecular PPIs are predominately mediated by H2 and wing1 residues at 25c, 27c, 28c 31c, 34c, 70c, 72c and 73c (Figure 1C). However, these residues are not well conserved in the FOX family, suggesting that this binding mode may be unique for the FOXP subgroup. Furthermore, it has been reported that interactions between FOX TF and other TF can occur at regions besides the DBD (114). Therefore, it cannot be excluded that the observed heterodimers may form prior to DNA binding.
Collectively, cooperative DNA recognition is widely adopted by FOX TFs. The cooperativity may arise by three mechanisms. (i) The cooperative recognition is mediated by PPIs, in which case, the TFs bind to each other in the absence of DNA. For example, the domain-swapped FOXP homodimer is formed prior to DNA binding; (ii) the cooperativity is partially mediated by DNA, where the presence of PPIs requires specific orientation- and spacer-arrangements between the tandem binding sites. In turn, DNA-mediated PPIs enhance the binding affinity and stability of the protein to DNA, for instance, the FOXO1 homodimer formed on a DIV2 site and the FOXP-NFAT regulatory pairs in regulatory T cells; (iii) cooperativity is mediated entirely by DNA, where no PPI is presented. One possible mechanism for DNA-mediated cooperativity may be attributed to the shape of the DNA. DNA shape has been shown to be a driving factor for Forkhead-Ets pairs (111).
Pathogenic mutations of FOX-DBD
Mutations of FOX proteins underlie many human diseases. For example, mutation of FOXG1 is linked to FOXG1 syndrome, a serious neurodevelopmental disorder (115,116); FOXA mutations are frequently identified in prostate and breast cancers (42,117–120); FOXC mutations are associated with Axenfeld–Rieger syndrome (ARS) and Lymphoedema distichiasis syndrome (LDS) (43,121,122); and the inactivating mutations in FOXP3 result in IPEX (immune dysregulation, polyendocrinopathy, enteropathy, X-linked) autoimmune syndrome (123,124).
The high conservation of the DBD emphasizes its critical role in the regulatory activity of FOX TFs. Therefore, it is not surprising that missense mutations in FOX-DBD are poorly tolerated and lead to dysfunctional FOX TFs. Many of the disease-causing point mutations have been found to occur in this domain (Figure 7 and Supplementary Table S1). Early functional studies characterized that the mutations in FOXC1-DBD can affect almost every aspect of its function, including reduced DNA binding, blocking nuclear localization, impairing transactivation, and so on (125–128). Recent biochemical studies demonstrate that the mutations in FOX-DBD usually affect its DNA binding affinity, protein thermal stability, or both (44,59). Furthermore, the mutations in the DBD may directly affect the interaction with other co-factors. For example, some mutations in FOXP3 do not affect its DNA binding but disrupt the interaction with NFAT, thereby eliminating its T cell-suppressive activity (64). Interestingly, biochemical studies revealed that some point mutations that reduce DNA binding are not involved in direct DNA interactions, and circular dichroism spectra showed that these mutations do not affect the folding of the DBD (44). The underlying mechanism of such dysfunction-inducing mutations of FOX TFs is elusive.
Figure 7.
Mutations identified among selected FOX-DBDs and possible mutational hotspots in FOX-DBD. Mutation data are sourced from COSMIC database or literatures (see Supplementary Table S1 for details).
Some of the pathogenic mutation residues have been found in the same positions in different FOX members and could be potentially FOX family mutational hotspots (Figure 7), including K3c, P4c, P5c, S7c, I9c, A10c, F37c, Y39c, Y40c, I56c, R57c, S61c, C65c, K68c, V69c, G83c, M98c, F100c, R107c and R109c. Specifically, K3c, S7c, R57c, S61c, K68c, R107c and R109c are shown to participate in DNA backbone interaction, and mutation in these sites may impact the DNA binding; R57c is involved in base-specific DNA recognition; F37c, Y39c and Y40c are located at the end of H2. These residues form extensive hydrophobic contacts with other buried non-polar resides; hence, mutation in these positions may impact the overall folding of FOX-DBD. The other residues (P4c, P5c, I9c, A10c, C65c, I56c, V69c, G83c and M98c) are involved with neither binding to DNA nor contact with partner TFs. We have shown that the mutations in some of these sites impact the DNA binding or protein thermal stability of FOXC2 and FOXG1 (44,82). The consequences may be caused by impairing the structural dynamics of FOX-DBD when mutated.
SUMMARY AND FUTURE PERSPECTIVES
FOX family TFs are central regulatory players in eukaryotic cells. The functional diversity of FOX TFs relies on the binding of FOX members to specific genomic loci. Structural studies of FOX–DNA complexes have provided an informative insight into the DNA recognition by FOX TFs in vitro. DNA binding outside the recognition helix of FOX-DBD is highly variable, which leads to the preferences on DNA sequences in and outside the core motif of FOX TFs. However, the recognized motifs are too short to define unique genomic binding sites. To compensate, FOX TFs usually collaboratively bind DNA with other families of TFs (129). Therefore, the cooperative DNA recognition by FOX TFs is still poorly studied and should be given more attention in the future. Accumulating evidence highlights the important role of FOX TFs in tumorigenesis and progression, making this family of proteins an attractive drug target. Designing modifiers that can modulate FOX function through its DBD is common sense in developing FOX TF-targeted drugs. However, the high sequence conservation and structural similarity of FOX-DBD makes targeting DBD challenging, as unpredictable off-target effects may be raised. Since other regions outside the DBD can impact the DNA binding properties of FOX TFs (87,130,131), targeting less conserved regions to regulate FOX-DBD function may be an alternative option. As current structural studies focus on the isolated DBD, the structure of other regions outside the DBD remains largely unknown, and the study of the DNA recognition of FOX-DBD in the context of other FOX domains is urgently needed.
Many FOX proteins have been reported to function as pioneer transcription factors, especially FOXA1, which binds to DNA sites on nucleosomes and induces an open chromatin conformation to allow the binding of other TFs (49,50). The high structural similarity between FOXA1-DBD and linker histone H1 globular domain suggests FOXA1 may displace histone H1, open the local chromatin and allow other transcription factors to bind and regulate transcription. However, it is still a mystery how FOXA1 binds the tightly compact chromatin and which DNA sequence is engaged by FOXA1 to initiate the binding. Recently, nucleosome-bound pioneer transcription factors SOX2, SOX11 and OCT4-SOX2 structures have been solved using cryogenic electron microscopy (cryo-EM) (132,133). It will be exciting to have a structure of an FKH domain bound to a nucleosome, which will reveal the mechanism by which Forkhead proteins function as pioneer transcription factors.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Michael R. Stallcup for proofreading.
Contributor Information
Shuyan Dai, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
Linzhi Qu, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
Jun Li, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
Yongheng Chen, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [81570537 to Y.C.; 81974074 to Y.C.]; China Postdoctoral Science Foundation [2021M693574 to S.D.]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T.. The human transcription factors. Cell. 2018; 172:650–665. [DOI] [PubMed] [Google Scholar]
- 2. Laissue P. The forkhead-box family of transcription factors: key molecular players in colorectal cancer pathogenesis. Mol. Cancer. 2019; 18:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Herman L., Todeschini A.L., Veitia R.A.. Forkhead transcription factors in health and disease. Trend. Genet. 2020; 37:460–475. [DOI] [PubMed] [Google Scholar]
- 4. Zhu H. Forkhead box transcription factors in embryonic heart development and congenital heart disease. Life Sci. 2016; 144:194–201. [DOI] [PubMed] [Google Scholar]
- 5. Weigel D., Jürgens G., Küttner F., Seifert E., Jäckle H.. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989; 57:645–658. [DOI] [PubMed] [Google Scholar]
- 6. Weigel D., Jäckle H.. The fork head domain: a novel DNA binding motif of eukaryotic transcription factors?. Cell. 1990; 63:455–456. [DOI] [PubMed] [Google Scholar]
- 7. Wang D.Y., Kumar S., Hedges S.B.. Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proc. Biol. Sci. 1999; 266:163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hannenhalli S., Kaestner K.H.. The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 2009; 10:233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mazet F., Yu J.K., Liberles D.A., Holland L.Z., Shimeld S.M.. Phylogenetic relationships of the Fox (Forkhead) gene family in the Bilateria. Gene. 2003; 316:79–89. [DOI] [PubMed] [Google Scholar]
- 10. Jackson B.C., Carpenter C., Nebert D.W., Vasiliou V.. Update of human and mouse forkhead box (FOX) gene families. Hum. Genomics. 2010; 4:345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown A.K., Webb A.E.. Regulation of FOXO factors in mammalian cells. Curr. Top. Dev. Biol. 2018; 127:165–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bacchetta R., Barzaghi F., Roncarolo M.G.. From IPEX syndrome to FOXP3 mutation: a lesson on immune dysregulation. Annal. N. Y. Acad. Sci. 2018; 1417:5–22. [DOI] [PubMed] [Google Scholar]
- 13. Yan J., Xu L., Crawford G., Wang Z., Burgess S.M.. The forkhead transcription factor FoxI1 remains bound to condensed mitotic chromosomes and stably remodels chromatin structure. Mol. Cell. Biol. 2006; 26:155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lalmansingh A.S., Karmakar S., Jin Y., Nagaich A.K.. Multiple modes of chromatin remodeling by Forkhead box proteins. Biochim. Biophys. Acta. 2012; 1819:707–715. [DOI] [PubMed] [Google Scholar]
- 15. Hettige N.C., Ernst C.. FOXG1 dose in brain development. Front. Pediatr. 2019; 7:482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dengler H.S., Baracho G.V., Omori S.A., Bruckner S., Arden K.C., Castrillon D.H., DePinho R.A., Rickert R.C.. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat. Immunol. 2008; 9:1388–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Amin R.H., Schlissel M.S.. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat. Immunol. 2008; 9:613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Y., Zhou Y., Graves D.T.. FOXO transcription factors: their clinical significance and regulation. Biomed. Res. Int. 2014; 2014:925350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uhlenhaut N.H., Treier M.. Forkhead transcription factors in ovarian function. Reproduction. 2011; 142:489–495. [DOI] [PubMed] [Google Scholar]
- 20. Uhlenhaut N.H., Treier M.. Foxl2 function in ovarian development. Mol. Genet. Metab. 2006; 88:225–234. [DOI] [PubMed] [Google Scholar]
- 21. Schmitt-Ney M. The FOXO’s advantages of being a family: considerations on function and evolution. Cells. 2020; 9:e787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiramongkol Y., Lam E.W.. FOXO transcription factor family in cancer and metastasis. Cancer Metastasis Rev. 2020; 39:681–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Raychaudhuri P., Park H.J.. FoxM1: a master regulator of tumor metastasis. Cancer Res. 2011; 71:4329–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Katoh M., Igarashi M., Fukuda H., Nakagama H., Katoh M.. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013; 328:198–206. [DOI] [PubMed] [Google Scholar]
- 25. Bach D.H., Long N.P., Luu T.T., Anh N.H., Kwon S.W., Lee S.K.. The dominant role of forkhead box proteins in cancer. Int. J. Mol. Sci. 2018; 19:e3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gartel A.L. FOXM1 in cancer: interactions and vulnerabilities. Cancer Res. 2017; 77:3135–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Teng M., Zhou S., Cai C., Lupien M., He H.H.. Pioneer of prostate cancer: past, present and the future of FOXA1. Protein Cell. 2021; 12:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kruiswijk F., Hasenfuss S.C., Sivapatham R., Baar M.P., Putavet D., Naipal K.A., van den Broek N.J., Kruit W., van der Spek P.J., van Gent D.C.et al.. Targeted inhibition of metastatic melanoma through interference with Pin1-FOXM1 signaling. Oncogene. 2016; 35:2166–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jin B., Wang C., Li J., Du X., Ding K., Pan J.. Anthelmintic niclosamide disrupts the interplay of p65 and FOXM1/β-catenin and eradicates leukemia stem cells in chronic myelogenous leukemia. Clin. Cancer Res. 2017; 23:789–803. [DOI] [PubMed] [Google Scholar]
- 30. Koo C.Y., Muir K.W., Lam E.W.. FOXM1: From cancer initiation to progression and treatment. Biochim. Biophys. Acta. 2012; 1819:28–37. [DOI] [PubMed] [Google Scholar]
- 31. Hornsveld M., Smits L.M.M., Meerlo M., van Amersfoort M., Groot Koerkamp M.J.A., van Leenen D., Kloet D.E.A., Holstege F.C.P., Derksen P.W.B., Burgering B.M.T.et al.. FOXO transcription factors both suppress and support breast cancer progression. Cancer Res. 2018; 78:2356–2369. [DOI] [PubMed] [Google Scholar]
- 32. Paik J.H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R.et al.. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007; 128:309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Coomans de Brachène A., Demoulin J.B.. FOXO transcription factors in cancer development and therapy. Cell. Mol. Life Sci. 2016; 73:1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yamaguchi F., Hirata Y., Akram H., Kamitori K., Dong Y., Sui L., Tokuda M.. FOXO/TXNIP pathway is involved in the suppression of hepatocellular carcinoma growth by glutamate antagonist MK-801. BMC Cancer. 2013; 13:468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu H. Targeting forkhead box transcription factors FOXM1 and FOXO in leukemia (review). Oncol. Rep. 2014; 32:1327–1334. [DOI] [PubMed] [Google Scholar]
- 36. Parolia A., Cieslik M., Chu S.C., Xiao L., Ouchi T., Zhang Y., Wang X., Vats P., Cao X., Pitchiaya S.et al.. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature. 2019; 571:413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gao S., Chen S., Han D., Barrett D., Han W., Ahmed M., Patalano S., Macoska J.A., He H.H., Cai C.. Forkhead domain mutations in FOXA1 drive prostate cancer progression. Cell Res. 2019; 29:770–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu B., Song B., Lu X., Kim J., Hu M., Zhao J.C., Yu J.. Altered chromatin recruitment by FOXA1 mutations promotes androgen independence and prostate cancer progression. Cell Res. 2019; 29:773–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adams E.J., Karthaus W.R., Hoover E., Liu D., Gruet A., Zhang Z., Cho H., DiLoreto R., Chhangawala S., Liu Y.et al.. FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature. 2019; 571:408–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim J., Jin H., Zhao J.C., Yang Y.A., Li Y., Yang X., Dong X., Yu J.. FOXA1 inhibits prostate cancer neuroendocrine differentiation. Oncogene. 2017; 36:4072–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Clark K.L., Halay E.D., Lai E., Burley S.K.. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993; 364:412–420. [DOI] [PubMed] [Google Scholar]
- 42. Fu X., Pereira R., De Angelis C., Veeraraghavan J., Nanda S., Qin L., Cataldo M.L., Sethunath V., Mehravaran S., Gutierrez C.et al.. FOXA1 upregulation promotes enhancer and transcriptional reprogramming in endocrine-resistant breast cancer. Proc. Natl. Acad. Sci. USA. 2019; 116:26823–26834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Elian F.A., Yan E., Walter M.A.. FOXC1, the new player in the cancer sandbox. Oncotarget. 2018; 9:8165–8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dai S., Li J., Zhang H., Chen X., Guo M., Chen Z., Chen Y.. Structural basis for DNA recognition by FOXG1 and the characterization of disease-causing FOXG1 mutations. J. Mol. Biol. 2020; 432:6146–6156. [DOI] [PubMed] [Google Scholar]
- 45. Torgerson T.R., Ochs H.D.. Immune dysregulation, polyendocrinopathy, enteropathy, X-linked: forkhead box protein 3 mutations and lack of regulatory T cells. J. Allergy Clin. Immunol. 2007; 120:744–750. [DOI] [PubMed] [Google Scholar]
- 46. Ochs H.D., Gambineri E., Torgerson T.R.. IPEX, FOXP3 and regulatory T-cells: a model for autoimmunity. Immunol. Res. 2007; 38:112–121. [DOI] [PubMed] [Google Scholar]
- 47. Schubert L.A., Jeffery E., Zhang Y., Ramsdell F., Ziegler S.F.. Scurfin (FOXP3) acts as a repressor of transcription and regulates T cell activation. J. Biol. Chem. 2001; 276:37672–37679. [DOI] [PubMed] [Google Scholar]
- 48. Kwon H.K., Chen H.M., Mathis D., Benoist C.. Different molecular complexes that mediate transcriptional induction and repression by FoxP3. Nat. Immunol. 2017; 18:1238–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sérandour A.A., Avner S., Percevault F., Demay F., Bizot M., Lucchetti-Miganeh C., Barloy-Hubler F., Brown M., Lupien M., Métivier R.et al.. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 2011; 21:555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lupien M., Eeckhoute J., Meyer C.A., Wang Q., Zhang Y., Li W., Carroll J.S., Liu X.S., Brown M.. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008; 132:958–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim D.H., Perdomo G., Zhang T., Slusher S., Lee S., Phillips B.E., Fan Y., Giannoukakis N., Gramignoli R., Strom S.et al.. FoxO6 integrates insulin signaling with gluconeogenesis in the liver. Diabetes. 2011; 60:2763–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Katoh M., Katoh M.. Human FOX gene family (Review). Int. J. Oncol. 2004; 25:1495–1500. [PubMed] [Google Scholar]
- 53. Gajiwala K.S., Burley S.K.. Winged helix proteins. Curr. Opin. Struct. Biol. 2000; 10:110–116. [DOI] [PubMed] [Google Scholar]
- 54. Benayoun B.A., Caburet S., Veitia R.A.. Forkhead transcription factors: key players in health and disease. Trends in genetics : TIG. 2011; 27:224–232. [DOI] [PubMed] [Google Scholar]
- 55. Boura E., Rezabkova L., Brynda J., Obsilova V., Obsil T.. Structure of the human FOXO4-DBD-DNA complex at 1.9 Å resolution reveals new details of FOXO binding to the DNA. Acta Crystallogr. Sect. D, Biol. Crystallogr. 2010; 66:1351–1357. [DOI] [PubMed] [Google Scholar]
- 56. Littler D.R., Alvarez-Fernández M., Stein A., Hibbert R.G., Heidebrecht T., Aloy P., Medema R.H., Perrakis A.. Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence. Nucleic Acids Res. 2010; 38:4527–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sheng W., Rance M., Liao X.. Structure comparison of two conserved HNF-3/fkh proteins HFH-1 and genesis indicates the existence of folding differences in their complexes with a DNA binding sequence. Biochemistry. 2002; 41:3286–3293. [DOI] [PubMed] [Google Scholar]
- 58. Jin C., Marsden I., Chen X., Liao X.. Dynamic DNA contacts observed in the NMR structure of winged helix protein-DNA complex. J. Mol. Biol. 1999; 289:683–690. [DOI] [PubMed] [Google Scholar]
- 59. Chen X., Wei H., Li J., Liang X., Dai S., Jiang L., Guo M., Qu L., Chen Z., Chen L.et al.. Structural basis for DNA recognition by FOXC2. Nucleic Acids Res. 2019; 47:3752–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brent M.M., Anand R., Marmorstein R.. Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure. 2008; 16:1407–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li J., Dantas Machado A.C., Guo M., Sagendorf J.M., Zhou Z., Jiang L., Chen X., Wu D., Qu L., Chen Z.et al.. Structure of the forkhead domain of FOXA2 bound to a complete DNA consensus site. Biochemistry. 2017; 56:3745–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tsai K.L., Sun Y.J., Huang C.Y., Yang J.Y., Hung M.C., Hsiao C.D.. Crystal structure of the human FOXO3a-DBD/DNA complex suggests the effects of post-translational modification. Nucleic Acids Res. 2007; 35:6984–6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stroud J.C., Wu Y., Bates D.L., Han A., Nowick K., Paabo S., Tong H., Chen L.. Structure of the forkhead domain of FOXP2 bound to DNA. Structure. 2006; 14:159–166. [DOI] [PubMed] [Google Scholar]
- 64. Bandukwala H.S., Wu Y., Feuerer M., Chen Y., Barboza B., Ghosh S., Stroud J.C., Benoist C., Mathis D., Rao A.et al.. Structure of a domain-swapped FOXP3 dimer on DNA and its function in regulatory T cells. Immunity. 2011; 34:479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tsai K.L., Huang C.Y., Chang C.H., Sun Y.J., Chuang W.J., Hsiao C.D.. Crystal structure of the human FOXK1a-DNA complex and its implications on the diverse binding specificity of winged helix/forkhead proteins. J. Biol. Chem. 2006; 281:17400–17409. [DOI] [PubMed] [Google Scholar]
- 66. Newman J.A., Aitkenhead H., Gavard A.E., Rota I.A., Handel A.E., Hollander G.A., Gileadi O.. The crystal structure of human forkhead box N1 in complex with DNA reveals the structural basis for forkhead box family specificity. J. Biol. Chem. 2020; 295:2948–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rogers J.M., Waters C.T., Seegar T.C.M., Jarrett S.M., Hallworth A.N., Blacklow S.C., Bulyk M.L.. Bispecific forkhead transcription factor FoxN3 recognizes two distinct motifs with different DNA shapes. Mol. Cell. 2019; 74:245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fyodorov D.V., Zhou B.R., Skoultchi A.I., Bai Y.. Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 2018; 19:192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ramakrishnan V., Finch J.T., Graziano V., Lee P.L., Sweet R.M.. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993; 362:219–223. [DOI] [PubMed] [Google Scholar]
- 70. Ono K., Kusano O., Shimotakahara S., Shimizu M., Yamazaki T., Shindo H.. The linker histone homolog Hho1p from Saccharomyces cerevisiae represents a winged helix-turn-helix fold as determined by NMR spectroscopy. Nucleic Acids Res. 2003; 31:7199–7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G.et al.. DNA-binding specificities of human transcription factors. Cell. 2013; 152:327–339. [DOI] [PubMed] [Google Scholar]
- 72. Nakagawa S., Gisselbrecht S.S., Rogers J.M., Hartl D.L., Bulyk M.L.. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc. Natl. Acad. Sci. USA. 2013; 110:12349–12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li J., Jiang L., Liang X., Qu L., Wu D., Chen X., Guo M., Chen Z., Chen L., Chen Y.. DNA-binding properties of FOXP3 transcription factor. Acta Biochim. Biophys. Sin. (Shanghai). 2017; 49:792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Koh K.P., Sundrud M.S., Rao A.. Domain requirements and sequence specificity of DNA binding for the forkhead transcription factor FOXP3. PLoS One. 2009; 4:e8109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Obsil T., Obsilova V.. Structure/function relationships underlying regulation of FOXO transcription factors. Oncogene. 2008; 27:2263–2275. [DOI] [PubMed] [Google Scholar]
- 76. Lehtinen M.K., Yuan Z., Boag P.R., Yang Y., Villén J., Becker E.B., DiBacco S., de la Iglesia N., Gygi S., Blackwell T.K.et al.. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006; 125:987–1001. [DOI] [PubMed] [Google Scholar]
- 77. Yuan Z., Becker E.B., Merlo P., Yamada T., DiBacco S., Konishi Y., Schaefer E.M., Bonni A.. Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science. 2008; 319:1665–1668. [DOI] [PubMed] [Google Scholar]
- 78. Huang H., Regan K.M., Lou Z., Chen J., Tindall D.J.. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006; 314:294–297. [DOI] [PubMed] [Google Scholar]
- 79. Kohler S., Cirillo L.A.. Stable chromatin binding prevents FoxA acetylation, preserving FoxA chromatin remodeling. J. Biol. Chem. 2010; 285:464–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Liu Y., Wang L., Han R., Beier U.H., Hancock W.W.. Two lysines in the forkhead domain of foxp3 are key to T regulatory cell function. PLoS One. 2012; 7:e29035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. van Dongen M.J., Cederberg A., Carlsson P., Enerbäck S., Wikström M.. Solution structure and dynamics of the DNA-binding domain of the adipocyte-transcription factor FREAC-11. J. Mol. Biol. 2000; 296:351–359. [DOI] [PubMed] [Google Scholar]
- 82. Li S., Pradhan L., Ashur S., Joshi A., Nam H.J.. Crystal structure of FOXC2 in complex with DNA target. ACS Omega. 2019; 4:10906–10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li J., Dai S., Chen X., Liang X., Qu L., Jiang L., Guo M., Zhou Z., Wei H., Zhang H.et al.. Mechanism of forkhead transcription factors binding to a novel palindromic DNA site. Nucleic Acids Res. 2021; 49:3573–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Marsden I., Jin C., Liao X.. Structural changes in the region directly adjacent to the DNA-binding helix highlight a possible mechanism to explain the observed changes in the sequence-specific binding of winged helix proteins. J. Mol. Biol. 1998; 278:293–299. [DOI] [PubMed] [Google Scholar]
- 85. Singh P., Han E.H., Endrizzi J.A., O’Brien R.M., Chi Y.I.. Crystal structures reveal a new and novel FoxO1 binding site within the human glucose-6-phosphatase catalytic subunit 1 gene promoter. J. Struct. Biol. 2017; 198:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Psenakova K., Kohoutova K., Obsilova V., Ausserlechner M.J., Veverka V., Obsil T.. Forkhead domains of FOXO transcription factors differ in both overall conformation and dynamics. Cells. 2019; 8:e966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wang F., Marshall C.B., Yamamoto K., Li G.Y., Plevin M.J., You H., Mak T.W., Ikura M.. Biochemical and structural characterization of an intramolecular interaction in FOXO3a and its binding with p53. J. Mol. Biol. 2008; 384:590–603. [DOI] [PubMed] [Google Scholar]
- 88. Weigelt J., Climent I., Dahlman-Wright K., Wikström M.. 1H, 13C and 15N resonance assignments of the DNA binding domain of the human forkhead transcription factor AFX. J. Biomol. NMR. 2000; 17:181–182. [DOI] [PubMed] [Google Scholar]
- 89. Chu Y.P., Chang C.H., Shiu J.H., Chang Y.T., Chen C.Y., Chuang W.J.. Solution structure and backbone dynamics of the DNA-binding domain of FOXP1: insight into its domain swapping and DNA binding. Protein Sci. 2011; 20:908–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wu Y., Borde M., Heissmeyer V., Feuerer M., Lapan A.D., Stroud J.C., Bates D.L., Guo L., Han A., Ziegler S.F.et al.. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006; 126:375–387. [DOI] [PubMed] [Google Scholar]
- 91. Chen Y., Chen C., Zhang Z., Liu C.C., Johnson M.E., Espinoza C.A., Edsall L.E., Ren B., Zhou X.J., Grant S.F.et al.. DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions. Nucleic Acids Res. 2015; 43:1268–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Overdier D.G., Porcella A., Costa R.H.. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol. Cell. Biol. 1994; 14:2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Badis G., Berger M.F., Philippakis A.A., Talukder S., Gehrke A.R., Jaeger S.A., Chan E.T., Metzler G., Vedenko A., Chen X.et al.. Diversity and complexity in DNA recognition by transcription factors. Science. 2009; 324:1720–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Brunet A., Bonni A., Zigmond M.J., Lin M.Z., Juo P., Hu L.S., Anderson M.J., Arden K.C., Blenis J., Greenberg M.E.. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999; 96:857–868. [DOI] [PubMed] [Google Scholar]
- 95. Guo S., Rena G., Cichy S., He X., Cohen P., Unterman T.. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J. Biol. Chem. 1999; 274:17184–17192. [DOI] [PubMed] [Google Scholar]
- 96. Pierrou S., Hellqvist M., Samuelsson L., Enerbäck S., Carlsson P.. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 1994; 13:5002–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhu C., Byers K.J., McCord R.P., Shi Z., Berger M.F., Newburger D.E., Saulrieta K., Smith Z., Shah M.V., Radhakrishnan M.et al.. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009; 19:556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Schlake T., Schorpp M., Nehls M., Boehm T.. The nude gene encodes a sequence-specific DNA binding protein with homologs in organisms that lack an anticipatory immune system. Proc. Natl. Acad. Sci. USA. 1997; 94:3842–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Luo H., Jin K., Xie Z., Qiu F., Li S., Zou M., Cai L., Hozumi K., Shima D.T., Xiang M.. Forkhead box N4 (Foxn4) activates Dll4-Notch signaling to suppress photoreceptor cell fates of early retinal progenitors. Proc. Natl. Acad. Sci. USA. 2012; 109:E553–E562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gotea V., Visel A., Westlund J.M., Nobrega M.A., Pennacchio L.A., Ovcharenko I.. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 2010; 20:565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. He X., Duque T.S., Sinha S.. Evolutionary origins of transcription factor binding site clusters. Mol. Biol. Evol. 2012; 29:1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kazemian M., Pham H., Wolfe S.A., Brodsky M.H., Sinha S.. Widespread evidence of cooperative DNA binding by transcription factors in Drosophila development. Nucleic Acids Res. 2013; 41:8237–8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wang X., Srivastava Y., Jankowski A., Malik V., Wei Y., Del Rosario R.C., Cojocaru V., Prabhakar S., Jauch R.. DNA-mediated dimerization on a compact sequence signature controls enhancer engagement and regulation by FOXA1. Nucleic Acids Res. 2018; 46:5470–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Jankowski A., Szczurek E., Jauch R., Tiuryn J., Prabhakar S.. Comprehensive prediction in 78 human cell lines reveals rigidity and compactness of transcription factor dimers. Genome Res. 2013; 23:1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. van der Vos K.E., Coffer P.J.. FOXO-binding partners: it takes two to tango. Oncogene. 2008; 27:2289–2299. [DOI] [PubMed] [Google Scholar]
- 106. Baar M.P., Brandt R.M.C., Putavet D.A., Klein J.D.D., Derks K.W.J., Bourgeois B.R.M., Stryeck S., Rijksen Y., van Willigenburg H., Feijtel D.A.et al.. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell. 2017; 169:132–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Smith N.C., Matthews J.M.. Mechanisms of DNA-binding specificity and functional gene regulation by transcription factors. Curr. Opin. Struct. Biol. 2016; 38:68–74. [DOI] [PubMed] [Google Scholar]
- 108. Ostrow A.Z., Kalhor R., Gan Y., Villwock S.K., Linke C., Barberis M., Chen L., Aparicio O.M.. Conserved forkhead dimerization motif controls DNA replication timing and spatial organization of chromosomes in S. cerevisiae. Proc. Natl. Acad. Sci. USA. 2017; 114:E2411–E2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Onuma H., Vander Kooi B.T., Boustead J.N., Oeser J.K., O’Brien R.M.. Correlation between FOXO1a (FKHR) and FOXO3a (FKHRL1) binding and the inhibition of basal glucose-6-phosphatase catalytic subunit gene transcription by insulin. Mol. Endocrinol. 2006; 20:2831–2847. [DOI] [PubMed] [Google Scholar]
- 110. De Val S., Chi N.C., Meadows S.M., Minovitsky S., Anderson J.P., Harris I.S., Ehlers M.L., Agarwal P., Visel A., Xu S.M.et al.. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008; 135:1053–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ibarra I.L., Hollmann N.M., Klaus B., Augsten S., Velten B., Hennig J., Zaugg J.B.. Mechanistic insights into transcription factor cooperativity and its impact on protein-phenotype interactions. Nat. Commun. 2020; 11:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Josefowicz S.Z., Rudensky A.. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009; 30:616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Shevach E.M. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009; 30:636–645. [DOI] [PubMed] [Google Scholar]
- 114. Penrad-Mobayed M., Perrin C., Herman L., Todeschini A.L., Nigon F., Cosson B., Caburet S., Veitia R.A.. Conventional and unconventional interactions of the transcription factor FOXL2 uncovered by a proteome-wide analysis. FASEB J. 2020; 34:571–587. [DOI] [PubMed] [Google Scholar]
- 115. Wong L.C., Wu Y.T., Hsu C.J., Weng W.C., Tsai W.C., Lee W.T.. Cognition and evolution of movement disorders of FOXG1-Related syndrome. Front. Neurol. 2019; 10:641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Vegas N., Cavallin M., Maillard C., Boddaert N., Toulouse J., Schaefer E., Lerman-Sagie T., Lev D., Magalie B., Moutton S.et al.. Delineating FOXG1 syndrome: From congenital microcephaly to hyperkinetic encephalopathy. Neurol. Genet. 2018; 4:e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Grasso C.S., Wu Y.M., Robinson D.R., Cao X., Dhanasekaran S.M., Khan A.P., Quist M.J., Jing X., Lonigro R.J., Brenner J.C.et al.. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012; 487:239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Sahu B., Laakso M., Ovaska K., Mirtti T., Lundin J., Rannikko A., Sankila A., Turunen J.P., Lundin M., Konsti J.et al.. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011; 30:3962–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Barbieri C.E., Baca S.C., Lawrence M.S., Demichelis F., Blattner M., Theurillat J.P., White T.A., Stojanov P., Van Allen E., Stransky N.et al.. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012; 44:685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Arruabarrena-Aristorena A., Maag J.L.V., Kittane S., Cai Y., Karthaus W.R., Ladewig E., Park J., Kannan S., Ferrando L., Cocco E.et al.. FOXA1 mutations reveal distinct chromatin profiles and influence therapeutic response in breast cancer. Cancer Cell. 2020; 38:534–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Seifi M., Walter M.A.. Axenfeld-Rieger syndrome. Clin. Genet. 2018; 93:1123–1130. [DOI] [PubMed] [Google Scholar]
- 122. Bell R., Brice G., Child A.H., Murday V.A., Mansour S., Sandy C.J., Collin J.R., Brady A.F., Callen D.F., Burnand K.et al.. Analysis of lymphoedema-distichiasis families for FOXC2 mutations reveals small insertions and deletions throughout the gene. Hum. Genet. 2001; 108:546–551. [DOI] [PubMed] [Google Scholar]
- 123. Bennett C.L., Christie J., Ramsdell F., Brunkow M.E., Ferguson P.J., Whitesell L., Kelly T.E., Saulsbury F.T., Chance P.F., Ochs H.D.. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 2001; 27:20–21. [DOI] [PubMed] [Google Scholar]
- 124. Wildin R.S., Ramsdell F., Peake J., Faravelli F., Casanova J.L., Buist N., Levy-Lahad E., Mazzella M., Goulet O., Perroni L.et al.. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 2001; 27:18–20. [DOI] [PubMed] [Google Scholar]
- 125. Saleem R.A., Banerjee-Basu S., Berry F.B., Baxevanis A.D., Walter M.A.. Analyses of the effects that disease-causing missense mutations have on the structure and function of the winged-helix protein FOXC1. Am. J. Hum. Genet. 2001; 68:627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Saleem R.A., Banerjee-Basu S., Berry F.B., Baxevanis A.D., Walter M.A.. Structural and functional analyses of disease-causing missense mutations in the forkhead domain of FOXC1. Hum. Mol. Genet. 2003; 12:2993–3005. [DOI] [PubMed] [Google Scholar]
- 127. Saleem R.A., Murphy T.C., Liebmann J.M., Walter M.A.. Identification and analysis of a novel mutation in the FOXC1 forkhead domain. Invest. Ophthalmol. Vis. Sci. 2003; 44:4608–4612. [DOI] [PubMed] [Google Scholar]
- 128. Murphy T.C., Saleem R.A., Footz T., Ritch R., McGillivray B., Walter M.A.. The wing 2 region of the FOXC1 forkhead domain is necessary for normal DNA-binding and transactivation functions. Invest. Ophthalmol. Vis. Sci. 2004; 45:2531–2538. [DOI] [PubMed] [Google Scholar]
- 129. Morgunova E., Taipale J.. Structural perspective of cooperative transcription factor binding. Curr. Opin. Struct. Biol. 2017; 47:1–8. [DOI] [PubMed] [Google Scholar]
- 130. Kim J., Ahn D., Park C.J.. FOXO4 transactivation domain interaction with forkhead DNA binding domain and effect on selective DNA recognition for transcription initiation. J. Mol. Biol. 2021; 433:166808. [DOI] [PubMed] [Google Scholar]
- 131. Marceau A.H., Brison C.M., Nerli S., Arsenault H.E., McShan A.C., Chen E., Lee H.W., Benanti J.A., Sgourakis N.G., Rubin S.M.. An order-to-disorder structural switch activates the FoxM1 transcription factor. eLife. 2019; 8:e46131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Michael A.K., Grand R.S., Isbel L., Cavadini S., Kozicka Z., Kempf G., Bunker R.D., Schenk A.D., Graff-Meyer A., Pathare G.R.et al.. Mechanisms of OCT4-SOX2 motif readout on nucleosomes. Science. 2020; 368:1460–1465. [DOI] [PubMed] [Google Scholar]
- 133. Dodonova S.O., Zhu F., Dienemann C., Taipale J., Cramer P.. Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function. Nature. 2020; 580:669–672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.