Abstract
The twister ribozyme is widely distributed over numerous organisms and is especially abundant in Schistosoma mansoni, but has no confirmed biological function. Of the 17 non-LTR retrotransposons known in S. mansoni, none have thus far been associated with ribozymes. Here we report the identification of novel twister variant (T-variant) ribozymes and their function in S. mansoni non-LTR retrotransposition. We show that T-variant ribozymes are located at the 5′ end of Perere-3 non-LTR retrotransposons in the S. mansoni genome. T-variant ribozymes were demonstrated to be catalytically active in vitro. In reporter constructs, T-variants were shown to cleave in vivo, and cleavage of T-variants was sufficient for the translation of downstream reporter genes. Our analysis shows that the T-variants and Perere-3 are transcribed together. Target site duplications (TSDs); markers of target-primed reverse transcription (TPRT) and footmarks of retrotransposition, are located adjacent to the T-variant cleavage site and suggest that T-variant cleavage has taken place inS. mansoni. Sequence heterogeneity in the TSDs indicates that Perere-3 retrotransposition is not site-specific. The TSD sequences contribute to the 5′ end of the terminal ribozyme helix (P1 stem). Based on these results we conclude that T-variants have a functional role in Perere-3 retrotransposition.
INTRODUCTION
The twister ribozyme was originally identified by bioinformatics. Twister RNA sequences are remarkably widespread, with close to 2700 twister ribozyme RNA sequences present in bacteria and diverse eukaryotic genomes; including yeasts, plants, insects and worms. The twister RNA is composed of highly conserved structural domains that have self-cleavage ribozyme activity in vitro (1). Crystal structures of the RNA, supported by biochemical data, confirm that the four helical stems (P1 to P4), two internal loops (L1 and L2) and hairpin loop (L4) adopt a compact fold stabilized by two pseudoknots (T1 and T2) with the U–A cleavage site buried in the center (2–6). Within the RNA sequence, ten nucleotides are >97% conserved. A highly conserved Guanosine plays a key catalytic role in cleavage of the scissile U–A bond. A function for the twister ribozyme has yet to be shown.
Retrotransposons are transposable genetic elements that require an RNA intermediate for transposition (7,8). They are abundant in the genomes of organisms across all kingdoms of life, for example, 45% of the human genome and at least 50% of the maize genome are made up of retrotransposon sequences (9,10). Retrotransposon insertion contributes to genomic diversity and complexity (11,12). In contrast to LTR retrotransposons (13) non-LTR retrotransposons, long interspersed nuclear elements (LINEs) and non-autonomous short interspersed nuclear elements (SINEs) and SVA (SINE/VNTR/Alu) elements lack long terminal repeats at each end (14–18). In general, the non-LTR retrotransposons may contain an internal promoter and open reading frames (ORFs) that encode reverse transcriptase (RT) and/or endonuclease domains and short sequence repeats at their 3′ boundary (19,20). The promoter sequences of functional non-LTR retrotransposons are not conserved across species (21,22) and some elements lack internal promoters and are transcribed as introns of larger host transcripts (23). Some elements may be transcribed by a nearby upstream cellular promoter, while some elements specifically insert into genes and may be expressed as precise cotranscripts (24). The features and regulation of the transcription of non-LTR retrotransposons are likely to vary from species to species and within particular retrotransposon clades (25). The main feature of the non-LTR retrotransposons is the presence of a reverse transcriptase (RT)/endonuclease domain (8,26,27), which generates DNA copies from the retrotransposon RNA transcripts for insertion of a transposon DNA copy into the new genomic target (25,28,29). For transposition, the non-LTR retrotransposons undergo a replicative cycle, the broad features of which are outlined in Supplementary Figure S1 (25,30,31). The mRNA is exported from the nucleus and the RT/endonuclease domains translated in the cytoplasm, mRNA and proteins are subsequently assembled into ribonucleoprotein particles (RNP) (32). Translation of the ORFs may be cap dependent (33) or through internal ribosomal entry (23). Ribonucleoprotein particles are then transported into the nucleus, for retrotransposon insertion at a new site in the host genome (34,35). Non-LTR retrotransposon integration into the host genome is thought to take place by a multi-step process termed target-primed reverse transcription (TPRT) (15,36).
A simplified TPRT model has the following steps:
Firstly, a free 3′ hydroxyl group is generated by an initial endonucleolytic cleavage at the target site on the bottom strand, by a retrotransposon encoded endonuclease (7,15). Non-LTR retrotransposons can be grouped into 2 functional classes; either encoding restriction enzyme-like endonucleases (RLE), or apurinic/apyrimidic endonucleases (APE). Non-LTR retrotransposition can be either site-specific or non-specific (26–28). The 3′-hydroxyl (3′-OH) product of the endonucleolytic cleavage serves as a priming site for the reverse transcriptase at the target site (7,8,30). RT initiates reverse transcription using the exposed 3′ end as a primer and the mRNA of the non-LTR retrotransposon as a template (25,28–30,37–40).
The subsequent integration of the freshly synthesised LINE DNA is not fully understood (41,42). A second cleavage on the top strand is then introduced for the synthesis of the second cDNA (30). This cleavage may generate blunt, 5′ or 3′ overhangs, and insertion at 3′ overhangs leads to target site duplication (TSD), and at 5′ overhangs to target site truncation (TST) (25,43,44). For either TSDs or TSTs endogenous repair enzymes are believed to contribute to the final transposon integration (45). The presence of a TSD in an integrated transposon is therefore a consequence of target-primed reverse transcription and also a footprint characteristic of TPRT (46). The asymmetry and sequence differences between the initial target endonucleolytic cleavage site and the second cleavage site, support a role for additional factors or changes to the DNA tertiary structure in the selection and cleavage of the second site (as discussed in (42)). Synthesis of the second strand, has not yet been efficiently verified in vitro (38,42). Second strand synthesis by the LINE reverse transcriptase ‘template jumping’ has been proposed to take place through priming at the 3′-OH of the second endonuclease cleavage site, the biochemical complexities and specificities of this reaction have been discussed (38,42). In some cases host polymerase activities may account for second strand synthesis as with the analogous group II intron retrohoming reverse splicing reaction, or by strand invasion through the host repair/recombination machinery (47,48).
The human parasite, Schistosoma mansoni, causes Schistosomiasis, a disease that affects ∼250M people worldwide in more than 70 countries (49). The parasite has a complex life cycle with snail and human hosts mediating the six stages of its life-cycle: egg, miracidia, sporocysts, cercaria, schistosomula and adult. The S. mansoni genome sequence is available (50,51) and transcriptome profiles and EST of S. mansoni have been reported (52,53). More than 20% of S. mansoni genome is considered to be composed of retrotransposons and reverse transcriptase activity has been detected in S. mansoni extracts (54,55). Studies have identified 28 different S. mansoni retrotransposon elements including members of LTR and non-LTR retrotransposon. The members of the S. mansoni non-LTR retrotransposon elements belong to the RTE (Perere-3), the CR1 (Perere, Perere-2, Perere-4, Perere-5, Perere-6 and Perere-7) clade, the R2 (Perere-9) and the Jockey clade (56,57). Perere-3 is a member of the RTE family of non-LTR retrotransposons elements and has a single ORF coding for a protein with endonuclease and reverse transcriptase domains (58). Perere-3 has an estimated genomic copy number of 2400–24 000 and is transcriptionally active (56). All the S. mansoni non-LTR retrotransposon elements are archived in the Repbase (59). Although the twister ribozyme is abundantly present in S. mansoni (1), no association of non-LTR retrotransposon elements and the twister ribozyme has been reported so far.
Historically, self-cleaving ribozymes were identified through their association with biological functions (60). Analysis of the well characterized R2 LINE retrotransposon that inserts into the 28S rRNA of Drosophila melanogaster showed that the 5′ junction of the retrotransposon contained an embedded self-cleaving ribozyme that was similar to the previously characterized hepatitis delta virus (HDV) ribozyme and was proposed to have a role in 5′ processing of the R2 RNA for insertion (61–65).
Here, we have investigated the function of novel twister ribozyme variants in non-LTR retrotransposon RNA processing. We show biochemically that the twister ribozyme variants are active in vitro and in reporter constructs and present evidence that twister ribozyme variants process the RNA of non-LTR retrotransposons in schistosoma mansoni by specific ribozyme cleavage.
MATERIALS AND METHODS
The materials used in this study were obtained from the following sources. 5′ 6-FAM labeled RNA were synthesized by Takara. DNA primers for T-variant in vitro transcription template amplification is purchased from Sangon Biotech (Shanghai, China). Phanta max DNA polymerase Mix was purchased from Vazyme (Nanjing, China). dNTP and NTP were purchased from Sangon Biotech. T7 RNA polymerase was produced in our lab. Plasmid insertion fragments for reporter assay and real-time PCR were synthesized by GenScript (Nanjing, China). Yeast extract, glucose, leucine, tryptone, agar and thiamine for strain culture were purchased from Sigma. Phenol (pH 4.3 ± 0.2) and EDTA for RNA extraction were purchased from Sigma. Acetic acid for RNA extraction were purchased from Sinopharm (China). DNase I for genome DNA digestion was purchased from Thermo Fisher.
T-variant search and sequence function prediction
The RNABOB program (66) was used to search genome sequence data from the NCBI Refseq database (release 90, https://ftp.ncbi.nlm.nih.gov/genomes/refseq) using the descriptor detailed in Figure 1C for the T-variant searching; the sequences are listed Supplementary Document 1. The secondary structure was built using information from the twister ribozyme covariance model (1). Downstream and upstream 10kb sequences were extracted from Refseq database and coding sequences were identified by GENSCAN (67) and ExPASy translate tool (68). Predicted amino acids sequence identities were further compared with known functional proteins by BLAST searching the UniProt protein database (69). Conserved protein domains were identified by SMART (Simple Modular Architecture Research Tool) (70).
Figure 1.
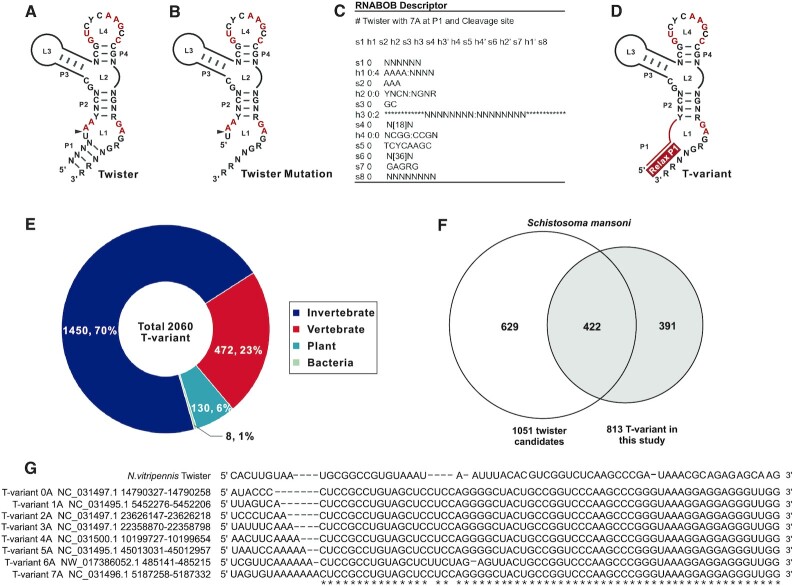
Identification of T-variants in Schistosoma mansoni. (A) Covariance model of twister ribozyme. (B) Twister ribozyme lacking the P1 helix. (C) RNABOB descriptor of twister with seven ‘A’s neighbouring the cleavage site. (D) Covariance model of twister ribozyme variants with altered helix P1. (E) Distribution of T-variants by organism. (F) Overlap between published twister sequences (1) and T-variants in S. mansoni. (G) Primary sequence alignment of typical T-variant-0A∼7A in S.mansoni, compared to published N. vitripennis twister ribozyme.
To obtain the 10 kb sequences downstream of the T-variants, sequences were extracted from the NCBI nucleotide database using an in-house script. The extracted T-variant (10 kb downstream) sequences, accession numbers and locations were assembled into a FASTA format database. The in-house script is available at https://github.com/threadtag/SPSA/tree/main/snippet. A common endonuclease-reverse transcriptase nucleic acid sequence was obtained by alignment of six endonuclease-reverse transcriptase DNA sequences downstream of T-variants 3–8. The Alignment of T-variant and amino acid sequences were performed by UniProt Align (https://www.uniprot.org/align). The Alignment parameters are as follows: Sequence Type (DNA), Dealign Input Sequences (no), Output Alignment Format (clustal_num), mBed-like Clustering Guide-tree (true), mBed-like Clustering Iteration (true), Number of Combined Iterations (Values 0), Max Guide Tree Iterations (Values -1), Max HMM Iterations (Values –1), Order (Aligned). The common endonuclease-reverse transcriptase nucleic acid sequence was then used to BLAST against the 10Kb downstream sequence database to predict bulk sequence function. Promoter prediction of upstream sequences was implemented on the neural network promoter prediction server (71): (https://www.fruitfly.org/seq_tools/promoter.html).
Determination of TSD
TSDs were determined individually by searching for identical nucleotide sequences at the 5′ and 3′ end of the sequences that were located 5′ to the cleavage site.
Synthesis and purification of oligoribonucleotides
RNA was prepared by in vitro transcription using T7 RNA Polymerase. The reaction contained 0.4 μM dsDNA template, 40 mM Tris–HCl, 40 mM KCl, 10 mM MgCl2, 2.5 mM DTT, 1 mM rNTP, and 3000 U/ml T7 RNA polymerase at pH 8. After incubating the mixture at 42°C for 3 h, the DNA template was digested by DNase I at 37°C for 1 h. RNA transcripts were purified on 8%, 8M urea denaturing polyacrylamide gel and eluted with 0.3 M sodium acetate at pH 5.2 with 1 mM EDTA. It was precipitated with ethanol and dissolved finally in sterile water.
T-variant in vitro cleavage in presence of divalent metal ions
10 μM ribozyme and 200nM 6-FAM-labeled substrate strands were annealed separately with 30 mM HEPES, pH7.5, 100 mM KCl, the mixture was heated at 95°C for 1.5 min, and cooled to room temperature for over 2 h. MgCl2 or other metal ions were then added to a final concentration of 10 mM. After incubation at 25°C for 15 min the cleavage reaction was initiated by mixing the two solutions. After incubation at 37°C for 15 min or 2 h as indicated, the cleavage reactions were stopped by adding 1 volume of stop buffer (80% v/v deionized formamide, 50 mM EDTA at pH 8.0, 0.025% w/v bromophenol blue, 0.025% w/v xylene cyanol). Substrate and cleavage products were separated on 20% polyacrylamide/8 M urea gels, and the fraction of substrate cleaved was quantitated by using ImageJ 1.51j8. The observed rate constant for the cleavage reaction was obtained using GraphPad Prism 6.01.
T-variant single-turnover kinetics
For twister ribozyme and T-variant kinetics under single-turnover conditions, 10 μM ribozyme and 200 nM 6-FAM-labeled substrate strands were annealed separately as previously described (72). The cleavage reaction was initiated by mixing the two solutions. At each time point, the cleavage reactions were stopped by adding 1 volume of stop buffer (80% deionized formamide, 50 mM EDTA at pH 8.0, 0.025% bromophenol blue, 0.025% xylene cyanol). Substrate and cleavage products were separated on 20% polyacrylamide/8 M urea gels, and the fraction of substrate cleaved was quantitated by using ImageJ 1.51j8 software. The first order rate constants (kobs) with and without antibiotic were calculated by plotting the fraction of substrate cleaved (ft) versus time (t) and fitting to the equation ft = 1 – exp(kobst) with GraphPad Prism 6.01.
T-variant in vitro cleavage site mapping
For each T-variant transcription product, 500 ng was annealed with 1 μM T-variant-RT-primer, and reverse transcribed using the SuperScript III Reverse Transcriptase Kit (Invitrogen). Sequence markers were generated by reverse transcription of the RNA in the presence of ddNTPs. cDNA sequences were analyzed by capillary electrophoresis (TsingKe, Beijing).
Reporter plasmid constructs
For the wild type T-variant 3A reporter plasmid, T-variant 3A, T-variant Δ3A with the original 5′-UTR of the predicted endonuclease-reverse transcriptase and T-variant sequences 1-N were synthesized with Xho I complementary ends and cloned into Xho I-digested REP41X-lacZ (73,74). For plasmids without the T-variant 3A, the 5′-UTR of the predicted endonuclease-reverse transcriptase lacking the T-variant 3A was cloned into Xho I-digested REP41X-lacZ. An HCV-IRES was cloned into the Xho I-digested REP41X-lacZ as an additional control and a further five genomic T-variant sequences were also cloned as T-variant controls (all sequences are given in Supplementary Table S4).
5′RACE detection of in vivo cleavage site
The wild type T-variant 3A-REP41X-lacZ plasmid was transformed into fission yeast hleu1-32 competent cells by electroporation, and cultured on an EMM plate at 30°C for 3–5 days. Positive clones were transferred into fresh EMM, and cells were grown to OD600 = 0.5, 10ml of culture was used for total RNA extraction. DNA was removed by DNase I (Thermo Fisher Scientific) from the RNA sample. Reverse transcription and PCR was carried out using SMARTer RACE 5′/3′ kit (Clotech). Genespecific primer P1 and P2 were respectively used for T-variant-3A cleavage site and transcription start site identification. The T-variant 3A cleavage site and transcription start sites were determined from the DNA sequence.
Real-time PCR analysis
The wild type T-variant 3A-REP41X-lacZ plasmid and three control plasmids were transformed into fission yeast hleu1-32 competent cells by electroporation and cultured on EMM plates. Total RNA was extracted by the hot phenol protocol and DNase I digested. cDNA was synthesized using PrimeScript RT Regent Kit (Takara, RR037A) according to the manufacturer's instructions. Messenger RNA abundance of lacZ (β-galactosidase reporter) from the reporter plasmid was detected by real-time PCR (oligonucleotide PCR primer sequences are detailed in Supporting data using SYBR Premix Ex Mix II (Takara, RR820A) with Amp as an internal reference. Error bars are the mean ± SD of three biological replicates.
Reporter assays
Fission yeast hleu1-32 competent cells transformed with the wild type T-variant 3A REP41X-lacZ plasmid and two control plasmids containing no T-variant and HCV-IRES were initially grown on EMM plates for 3∼5 days, followed by transfer to EMM liquid medium. Cells were diluted to OD600 = 0.1 in 3 × 10 ml of EMM. Cells were harvested and resuspended in 1 ml of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM 2-mercaptoethanol, pH 7.0). Cells were diluted thrice with Z buffer, and 600 μl of cell suspension was mixed with 70 μl of chloroform and 60 μl of 0.1% SDS, followed by mixing for 10 s and incubated at 30°C for 15 min, after adding 120 μl of 4 mg/ml o-nitrophenyl β-d-galactopyranoside (ONPG), and further incubated for 15–20 min (30°C). The reaction was quenched by the addition of 400 μl of 1 M sodium carbonate. The OD420 and OD600 were measured, and Miller units were calculated from the formula: U = 1000 × OD420/(time) × (volume) × OD600 (75). Error bars are the mean ± SD from three individual replicates.
S. mansoni transcriptome data analysis
The RNA-seq data of the six developmental stages of S. mansoni was obtained from the NCBI SRA database, with the following accession numbers; Egg (SRR2245469), Miracidia (SRR922067), Sporocyst (SRR922068), Cercaria (SRR5860351), Schistosomula (SRR5054493) and Adult (SRR2245496) (50–52). The RNABOB descriptor was built to search T-variants with different numbers of ‘A’s around the cleavage site. The T-variant candidate sequences were mapped to the S.mansoni genome (NCBI Genome Accession number: Assembly ASM23792v2) by GMAP (76), then base quality control implemented using Trimmomatic (Parameter:LEADING: 3TRAILING:3 SLIDINGWINDOW:4:15 AVGQUAL:20) (77), the positions of T-variant sample sequences were mapped onto the genome using hisat2 (78). Counts were based on htseq-count, and calculated as FPKM (fragments per kb per million reads) by the following formula:
 |
The distance between the 345 T-variants and the AUG of the downstream RT domains were each analysed manually. The AUGs of the downstream RT domains are divided in three main groups: reported AUGAGGCCGAUGCACCUUCUU (56), predicted AUGACGUCUCAUGAUGAA and predicted II AUGCACCUUCU by ExPASy translate tool (68).
RESULTS
Identification of twister ribozyme variant sequences
Twister ribozymes self-cleave at the U–A position within the (UAA) L1 loop of the ribozyme; one nucleotide 3′ to the P1 helical stem (Figure 1A) (1,4). The P1 stem typically contains at least two base pairs, although mutational analysis of the ribozyme has shown that inefficient ribozyme cleavage can take place in the absence of the P1 stem (Figure 1B) (79). The P1 stem is immediately adjacent to the cleavage site in the (UAA) L1 loop. The majority of the sequences contain 2A’s in L1 at the cleavage site and a P1 stem (Figure 1A), on closer examination of the published natural twister ribozyme sequences (1), a small number of the sequences contain fewer or more than 2 adenines (0,1, 3–7A) in the L1 loop that overlap the position of the cleavage site and impinge upon the stem P1 (Table 1).
Table 1.
Published twister ribozyme candidate sequences with different As neighbouring the cleavage site
| Twister ID | P1 | L1 | P2 | L2 | P3 | L3 | P3 | P4 | L4 | P4 | L2 | P2 | L1 | P1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sma-1–680 | UUUA | UCA | CUCC | GC | CUGUAGCUC | UUCUA | GAGUUACUG | CCG | GUCCCAAGC | CCGG | GUAAA | GGAG | GAGGG | UUGG |
| Sma-1–15 | UGCU | UAA | CUCC | GC | GUCUGUAGCUCC | UCUG | GGGGUUACUG | CCG | GUCCCAAGC | CCGG | GUAAA | GGAG | GAGGG | UUGG |
| Sma-1–119 | AGAU | AAA | CUCC | GC | CUGUAGCU | CUUCUAA | AGUUACUUG | CCG | GUUCCAAGC | CCGG | GUAAA | GGAG | GAGGG | UUGG |
| Sma-1–146 | CCUA | AAA | CUCC | GC | CUGUAGCUCC | UCUG | GGGGUUACUG | CCG | GUCCCAAGC | CCGG | GUAAA | GGAG | GAGGG | UUGG |
| Sma-1–94 | UGAA | AAA | CUCC | GC | GUAGCUCC | UCCG | GGGGCUACUG | CCG | GUCCCAAGC | CCGG | GUAAA | GGAG | GAGGG | UUAU |
| Sma-1–102 | UAAA | AAA | CUCC | GC | CUGUAGCUCC | UCCG | GGGGCUACUC | CCG | GUCCCAAGC | CCGG | GUGAA | GGAG | GAGGG | UUGA |
| Sma-1–73 | AAAA | AAA | CUCC | GC | CUGUAGCUC | CUCCUA | GGGCCACUG | CCG | GUCCCAAGC | CCGG | AUAAA | GGAG | GAGGG | UUGG |
The variation in the number of A’s in L1, neighbouring the cleavage site was intriguing to us and, based on the known twister ribozyme sequence domains, a further search was initiated using RNABOB (http://eddylab.org/ software.html) (80) (the exemplar syntax for the T-variant containing seven As at the cleavage site is shown in Figure 1C) to search for sequences that retained the conserved twister ribozyme sequence domains, but had 0–7A’s in the L1 loop next to the cleavage site with an allowance of up to four mismatches in the stem P1 (Figure 1D). A total of 2060 twister-like variant sequences were identified in vertebrate, invertebrate, plant and bacterial genomes and their distribution is displayed (Supplementary Figure S2 and Supplementary Document 1). To distinguish these sequences from the characterized twister ribozyme and to avoid confusion, these twister-like variant sequences were designated as twister-variant (T-variant (n)A where n = 0–7), in this study. Examples of T-variant 0–7A sequences are listed in Supplementary Table S1. The distribution of T-variants by organism is shown in Figure 1E. In invertebrates, the majority of T-variant sequences are found to be in Schistosoma mansoni. There are 813 S. mansoni T-variant sequences, most of which contain 2As at the cleavage site and the distribution of numbers of A are shown in Supplementary Document 2. Out of the 813 S. mansoni T-variant sequences, 422 sequences had been previously identified in the published twister ribozyme sequences (1), a further 391 novel T-variant sequences were identified in this study (Figure 1F, Supplementary Document 3). Examples of published S. mansoni twister sequences that have T-variant sequences (0–7A) at the cleavage site are displayed as Figure 1G.
T-variant ribozymes are associated with Perere-3 non-LTR retrotransposon elements in the S. mansoni genome
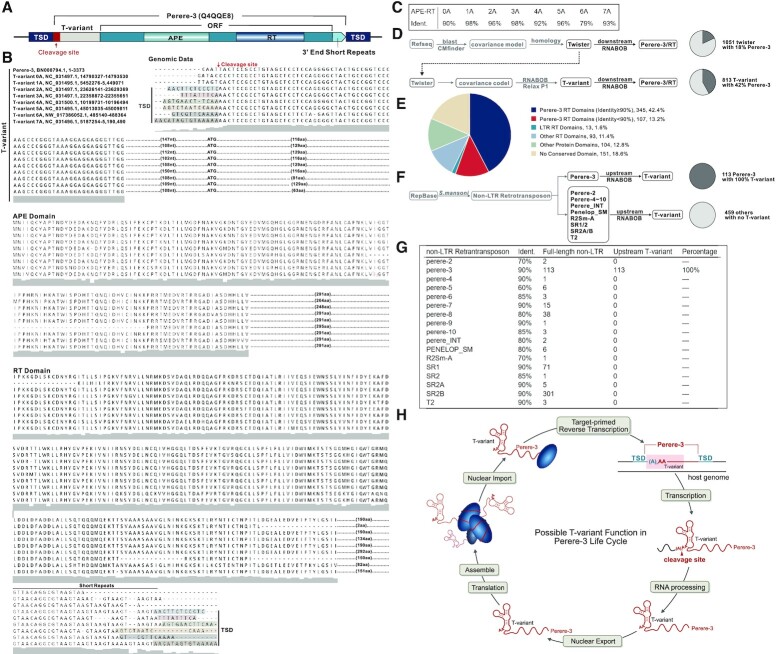
Among the 813 T-variants, there are T-variant sequences that lack an A at the cleavage site. The T-variant 0A consists of only the highly conserved region lacks both a P1 stem and an A adjacent to the scissile bond (Figure 1G), which had not been previously reported. These observations led us to investigate the origin of such sequences. We randomly selected fifty T-variant sequences (0–7A). Ten kilobase of the downstream sequences of these T-variants (0–7A) were searched for proximal protein coding sequences. Because the majority of the sequences had not been annotated, two independent peptide prediction programs (GENSCAN (67) and ExPASy translate tool (68) were used to predict peptide sequences. The potential protein coding sequences were further blasted against the UniProt protein database (69). For one subset of the T-variant downstream sequences, we identified potential protein domains that shared high identities with known apurinic/apyrimidinic endonucleases and reverse transcriptases (APE and RT Domain) (UniProtKB Code: Q4QQE8), that are key components of S. mansoni Perere-3 non-LTR retrotransposon elements (56,81). Since this subset of the T-variant downstream sequences are enriched with the RT domain of Perere-3, we subsequently choose 8 examples sequences to analyse the association between T-variants (0–7) A and the RT domain of Perere-3. A schematic representation of genomic organization of T-variants and Perere-3 is shown in Figure 2A (Supplementary Figure S3) and the sequence alignments in Figure 2B. The high similarity of protein domains downstream of T-variants (0–7A) to known APE and RT domains are listed in Figure 2C. The Perere-3 APE and RT domains, which play central roles in TPRT during retrotransposition into the genome, are found downstream of T-variant sequences (Figure 2A, B and Supplementary Figure S3). Target Site Duplications (TSDs) are the end product of non-LTR retrotransposon replication in the genome and are evidence that retrotransposition has taken place. TSDs are found flanking the Perere-3 and the T-variant sequences, confirming that these sequences are the product of retrotransposition. Note that TSD is immediately adjacent to the AA at cleavage site of the T-variant (Figure 2A, B and Supplementary Figure S3). In the case of the T-variants 0A and 1A where there is no evidence of TSD, it may be that for these sequences, retrotransposition has taken place with deletion of the target site (25). In addition, short repeats of the sequence GTAA are found at the 3′ boundary of non-LTR retrotransposons (Figure 2B), which may be an additional feature of Perere-3 retrotransposons, and may be analogous to the tandem UAA repeats at the 3′-end of the transcripts of non-LTR retrotransposons in Drosophila melanogaster (82).
Figure 2.
Genomic location of T-variant and Perere-3. (A) Schematic representation of the Perere-3 non-LTR retrotransposable element (UniProtKB Code: Q4QQE8) containing T-variant. T-variant sequences at the retrotransposon 5′ ends are marked as light grey boxes, with the different numbers of As at the cleavage site highlighted in the red box. The single open reading frame (ORF) of perere-3 is indicated as a turquoise box, with the embedded gradient boxes denoting the APE (pea green) and RT domains (sky blue). The light green arrow after the ORF represents the short repeats at the 3′ end. The TSDs flanking the whole retrotransposon element are marked as navy-blue boxes. (B) Alignment of representative (0–7A) T-variant sequences with accession numbers and genomic locations. The site of ribozyme self-cleavage is marked with the red arrow. TSDs are shown in shaded boxes at the 5′ and 3′ ends and the short 3′ sequence repeats indicated. The predicted amino acid sequences of the APE and RT domains downstream of the T-variants in Perere-3 retrotransposable elements are aligned. Similarities between the two domains are indicated as a grey shadow below the sequences. (C) Identity of T-variant downstream endonuclease-reverse transcriptases compared to the reported perere-3 non-LTR retrotransposon (56). (D) Pipeline for identification of twister (upper path) and T-variant sequences (lower branch-point). T-variants were identified by retaining the conserved structural components of Twister and relaxing the constraints on the P1 stem as an additional search criterion. The pie charts indicate the percentage of the published twister (top) and the enrichment of T-variant (bottom) sequences in S. mansoni that possess RT domains within 10kb downstream of the ribozyme sequence (marked as charcoal grey). (E) Analysis of the 813 T-variant downstream sequences in S. mansoni by domain identity: Perere-3 RT domains ≥ 90% (Blue segments), Perere-3 RT domains 60–90% (Red segments), LTR RT domains (green), other RT domains (light blue), other protein domains (light green) and no conserved domain (sand). Chromosomal locations and accession numbers are listed in Supplementary Table S2. (F) Pipeline for the reciprocal searching of all 17 non-LTR retrotransposons classes in S.mansoni. The full-length published non-LTR retrotransposon sequences were obtained from Repbase (https://www.girinst.org/repbase) and searched against the S.mansoni genome. The upstream sequences (1 kb) of these non-LTR retrotransposons were searched for T-variants with RNABOB. The pie charts indicate the percentage of the full-length Perere-3 (100%) and other non-LTR retrotransposons in S. mansoni that possess T-variants up to 1kb upstream (marked as charcoal gray). (G) Counts of each full-length non-LTR retrotransposons with respective identities and their upstream T-variants. (H) The Possible function of T-variants in the Perere-3 non-LTR retrotransposon replication cycle.
The analysis of the downstream sequences of the eight exemplar T-variant sequences revealed characteristic Perere-3 APE/RT domains. The numbers of the RT domains downstream of the total 813 S. mansoni T-variant sequences were next investigated. All of the 813 T-variant downstream 10 kb sequences were collated using an in-house script (Materials and Methods). Although protein domain prediction is feasible on a gene-by-gene basis, it is challenging to predict protein domains on bulk sequences due to the absence of prediction tools that can directly annotate functional protein domains from a large number of DNA sequences. However, we found that at the DNA level the reverse transcriptase domain sequences downstream of the T-variants share high sequence identities. The downstream DNA sequences of the 813 T-variants were then searched for the presence of RT domains by 90% similarity. In Schistosoma mansoni, of the 813 T-variants 42% (345) contain RT domains downstream (Figure 2D). In contrast, no RT domains were identified in the sequences up to 10 kb upstream of the T-variants (Supplementary Figure S4). In addition, only 18% of published Twister sequences (1) contain RT domains downstream (Figure 2D and Supplementary Document 4). The downstream T-variant sequences that have RT domains include the majority of the known Twister sequences with RT-domains (180 of 190) (Supplementary Figure S5). Twister was initially identified by a bioinformatics pipeline based on sequence homology and the T-variants were found by adding additional searching criteria based on the conserved structural components of Twister (both search strategies for Twister/T-variant and downstream protein domain in this study are displayed in Figure 2D). By using search criteria that focus on allowing up to four mismatches in the P1 stem, the downstream sequences of the T-variants were found to be enriched in RT domains, suggesting an association between T-variant ribozyme and the RT domains, a key component of Perere-3 non-LTR retrotransposons. Although 42% (>90% identity) of T-variants contain downstream RT domains this was probably an underestimate of the true RT content. Further sequence analysis (83,84) of the downstream sequences of the remaining 58% of T-variants revealed a further 13.2% Perere-3 (90%-60% identity) encoded RT domains, 11.4% other, 1.6% LTR RT domains, 12.8% known protein domains and 18.6% contained no conserved domain (Figure 2E). RT domain, known protein domains, chromosomal locations and accession numbers are listed in Supplementary Table S2.
Here we have identified RT domains that belong to Perere-3 non-LTR retrotransposons by searching downstream sequences of T-variants. Alternatively, a reciprocal approach is to search the upstream sequences of all S. mansoni non-LTR retrotransposon elements for T-variant sequences. In Repbase, there are 17 S. mansoni non-LTR retrotransposon elements based on RT domain similarity (59) (Figure 2F, G). The numbers of the full-length S. mansoni non-LTR retrotransposons and their relative RT sequence identities are listed in Figure 2F, G. There are 113 full-length Perere-3, all of which contain T-variant sequences upstream (Supplementary Document 5). Complete conservation of T-variant sequences upstream of Perere-3 implies a functional role for T-variants in Perere-3 retrotransposition. However, no T-variant sequence was found upstream of the other 16 non-LTR retrotransposons elements, for example 301 full-length SR2B non-LTR retrotransposons were found but no T-variant sequences can be detected upstream (Figure 2G). The analysis in Figure 2F was performed on full-length non-LTR retrotransposon elements that contain the whole protein including RT and Endonuclease domains. This excludes the possibility that elements containing only the RT domains can associate with T-variants. When the sequences of all of the other 16 non-LTR retrotransposons that contain only RT domains were collected and used to search for T-variants, no T-variant sequences were found upstream of the RT domains (Supplementary Table S3). Therefore, there appears to be a specific association between the T-variants and Perere-3 that is unlikely to have occurred at random in the genome.
Taken together, the bidirectional searching results confirms the genomic association of the T-variant and Perere-3 non-LTR retrotransposons element. T-variants are potential self-cleaving ribozymes. The presence of TSDs are footprints and evidence of Perere-3 non-LTR retrotransposition. In our analysis, the TSDs are positioned right next to the potential T-variant cleavage sites (Figure 2B). We speculate that T-variants may function during the life cycle of Perere-3 non-LTR retrotransposon elements (Figure 2H). T-variant ribozyme cleavage of RNA transcript would generate a 5′AA at the cleavage site for TPRT genome insertion with TSD. The location of TSD in the genomic sequence directly correlates to the nucleotides of the T-variant P1 stem in the RNA, which is ultimately related to the self-cleavage activity of the T-variant. The efficiency of Perere-3 non-LTR retrotransposition may be affected by the sequence at the genomic insertion site (TSD) which forms P1 of the T-variant. There may be a close relationship between the activity of T-variant and efficiency of the Perere-3 non-LTR retrotransposition.
T-variant ribozyme activity in vitro
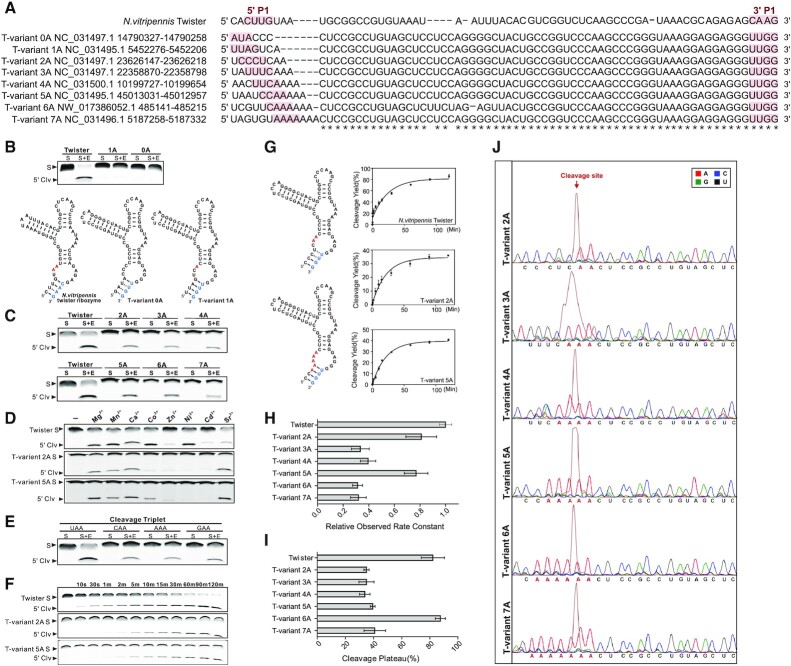
The T-variants identified here have not previously been shown to have ribozyme activity and differ, compared to previously characterized twister ribozymes, in the sequences neighbouring the scissile position in the P1 stem. For the T-variants to have a function in Perere-3 non-LTR retrotransposition their ribozyme activity must be established. The potential ribozyme activity of the representative T-variants (0–7A) (Figure 3A) was investigated and compared to previously characterised twister ribozymes in vitro in Figure 3. T-variants (0–7A) were separated into substrate and enzyme strands based on twister. The FAM labeled substrate strand was mixed with the enzyme strand and the ribozyme cleavage was measured by gel electrophoresis. No cleavage was detected for either T-variant 0A or T-variant 1A under standard twister ribozyme cleavage conditions compared to the control (Figure 3B). However, for the T-variants (2–7A), enzyme strand dependent cleavage of the substrate RNA was observed under the same conditions, confirming ribozyme activity (Figure 3C).
Figure 3.
T-variant catalytic activity in vitro. (A) Sequences of typical T-variants in S.mansoni for in vitro cleavage activity investigation, compared to published N. vitripennis twister ribozyme. Sequence accession numbers and locations are shown. The 5′ end and 3′ ends of the P1 stem are marked as red shadow. (B) Test of in vitro cleavage activity of T-variants 1A and 0A, compared to the N. vitripennis twister ribozyme, based on the structure of the N. vitripennis ribozyme, T-variants 1A and 0A RNAs were divided into substrate (S) and enzyme (E) strands. Purified strands were mixed in the combinations shown in the figure and incubated at 37°C for 2 h in 30 mM HEPES, pH 7.5, 100 mM KCl and 10 mM Mg2+, the cleavage products (5′ Clv) of 5′ 6-FAM-labeled substrate RNA samples were resolved on 8% denaturing polyacrylamide gels. (C) T-variant-2A-7A and N. vitripennis ribozyme cleavage activities in vitro, T-variants 2A-7A RNAs were divided into substrate (S) and enzyme (E) strands and cleavage products identified as before. (D) T-variant activity in the presence of divalent metal ions. The 5′ 6-FAM-labeled substrate was incubated with excess enzyme RNA for 15 min in the absence (–) or presence of 10 mM divalent metal ion as indicated. (E) Comparison of in vitro cleavage activity of T-variant sequences 5′ to the cleavage site for the cleavage triplet NAA where N = A, G, C or U. (F) T-variant-2A and 5A and N. vitripennis ribozyme cleavage kinetics in vitro. Time courses were performed; E + S strands were mixed and incubated at 37°C in 30 mM HEPES, pH 7.5, 100 mM KCl and 10 mM Mg2+, and samples removed after incubation at the given times (t). (G) Plots of ribozyme cleavage for the N. vitripennis twister, T-variant-2A and 5A ribozymes taken from Supplementary Figure S7, the first order rate constants (kobs) of T-variant were calculated by plotting the fraction of substrate cleaved (ft) versus time (t) and fitting to the equation ft = 1 – exp(kobst) with GraphPad Prism 6.01. Error bars are the standard deviation of three independent experiments. (H) Comparison of kobs for each T-variant (2A–7A) and N. vitripennis twister ribozyme as measured in (E), relative kobs, krel = kT-variant/k N. vitripennis twister. (I) Maximal cleavage (cleavage plateau) for the N. vitripennis and T-variant-2A to 7A and ribozymes. (J) Cleavage site mapping of T-variants 2A–7A by capillary electrophoresis. The purified transcription products of self-cleaved T-variant 2A–7A RNAs were reverse transcribed and subjected to capillary electrophoresis, relative to dideoxy markers. In each panel the peak corresponding to self-cleavage is shown in russet and the location of the cleavage site marked with a red arrow above the marker peaks.
For the T-variants (2A–7A), when compared to the twister control, broadly similar divalent cation dependent ribozyme activity was observed for Mg2+, Mn2+, Ca2+ and Sr2+, but different specificities were observed for Co2+, Zn2+, Ni2+and Cd2+ (1) (Figure 3D and Supplementary Figure S6). For the T-variants (2A–7A) time courses were used to measure ribozyme kinetics, in comparison with a twister ribozyme control (Figure 3F and Supplementary Figure S7). Plots of cleavage versus time yield ribozyme cleavage rates, showing that all of the T-variants catalyze RNA self-cleavage on a similar time-frame to known ribozymes (Figure 3F, G, H, I and Supplementary Figure S7). T-variants 2A and 5A, have similar activities to twister. Although the T-variants 3A, 4A and 7A have lower efficiencies, they show typical ribozyme activity (Figure 3H and I). To investigate and map the potential cleavage sites of the T-variants, 6-FAM labeled substrate strands were also analyzed by capillary gel electrophoresis. The positions of cleavage (red arrows) were resolved by capillary electrophoresis (russet trace) and mapped relative to sequence markers (Figure 3J). The cleavage positions of the T-variants (2A–7A) are the same as for the established twister ribozyme, such that cleavage of the RNA generates a 5′-AA end. Structural, modelling and mechanistic studies have shown that the product of phosphodiester bond scission; the free 5′ HO-AA, is generated through acid-base catalysis utilising the N3 of the terminal A as a proton donor, and the conserved catalytic G of loop 1 as a general base (2–6,85). Analysis of the T-variant (2A) sequences identified T-variant substrate sequences composed of C*AA, G*AA and A*AA (as T-variant (3A)) (where * indicates the position of the scissile bond) as potential T-variant ribozymes, in addition to the well characterised (U*AA). For these RNAs, enzyme strand dependent cleavage of the substrate RNA also took place under ribozyme cleavage conditions, confirming ribozyme activity (Figure 3E) and suggesting that ribozyme activity is not contingent on the identity of the nucleobase 5′ to the scissile bond. Thus, the T-variant sequences are catalytically active ribozymes.
T-variant ribozyme activity and function in reporter constructs
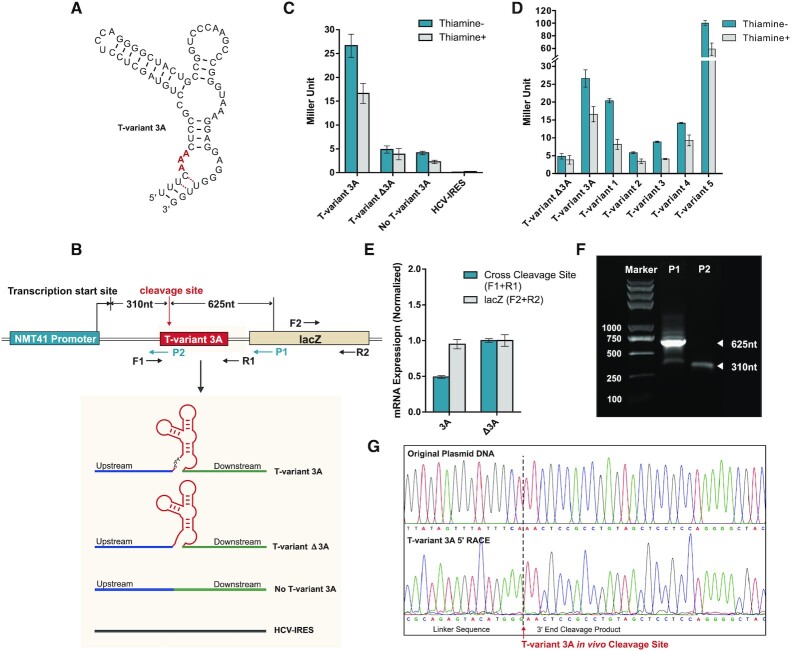
To investigate T-variant ribozyme function and its effect on downstream gene translation, a reporter plasmid was constructed using the plasmid REP41X-LacZ in fission yeast. The plasmid REP41X-LacZ contains the thiamine repressive NMT41 promoter, the polylinker sites for insertion and the LacZ protein coding sequence. The transcription of downstream sequences is dependent on the NMT41 promoter in the absence of thiamine. In the presence of thiamine, transcription from the NMT41 promoter should be significantly repressed although incomplete repression with reduced levels of transcription has been reported (86–88). Thiamine repression can be used as a control for reporter assays. The DNA fragment corresponding to the active T-variant 3A sequence and control sequences was inserted downstream of the NMT41 promoter and upstream of LacZ (Figure 4A and B). The effect of T-variant on downstream gene expression can be measured through expression of the LacZ reporter. Control constructs, consisting of T-variant 3A that lacks a cleavage site (T-variant Δ3A), a deletion of the T-variant 3A which has only the upstream and downstream sequences (No T-variant 3A) were constructed in parallel (Supplementary Table S4). The HCV internal ribosomal entry site (HCV-IRES) RNA is a highly structured RNA (65) that is unrelated to the T-variant sequences in S. mansoni was also used as a control (Supplementary Table S4).
Figure 4.
Reporter constructs and T-variant in vivo catalytic activity. (A) Sequence and secondary structure of T-variant 3A. (B) The reporter plasmid constructs. The wild type T-variant 3A for 5′ RACE is located behind an NMT41 thiamine repressible promoter and in front of a lacZ reporter gene. The gene specific primers P1 and P2 were used to map the T-variant 3A in vivo cleavage site and transcription start sites respectively. The T-variant 3A cleavage site as determined in vitro is marked by the red arrow and the predicted P1 and P2 primer fragment sizes of cleaved and whole transcript shown. The control plasmid constructs for real-time PCR and Miller assay are shown in the grey box: In parallel with T-variant 3A (red box), a sequence containing only the peripheral sequence of the Perere-3 5′-UTR without the ribozyme (No T-variant 3A) or with an HCV-IRES were substituted into the position of the red box in the lacZ reporter. (C) Miller assay of lacZ reporter activity ± thiamine for T-variant 3A, no T-variant and HCV IRES plasmid constructs. Note the high levels of reporter gene expression for T-variant 3A relative to the controls when the ribozyme is active. (D) Miller assay of lacZ reporter activity ± thiamine for T-variant sequences 1 to 5, relative to the control constructs T-variants Δ3A and 3A. (E) Real-time PCR analysis of mRNA abundance of the lacZ RNA (F2 and R2 primers) relative to the Ampicillin internal reference, showing the level of mRNA abundance of lacZ in the wild type T-variant 3A remains stable after T-variant cleavage, compared to the controls. Error bars represent the standard deviation of three independent experiments. (F) Agarose gel of PCR product of 5′RACE; the P1 primer generates a 625 nt T-variant cleavage product. (G) Capillary electrophoresis sequencing of original plasmid DNA compared to the 5′ RACE of cleaved T-variant 3A in vivo, the T-variant 3A cleavage site is marked by the red arrow.
The plasmids were transformed into the host strain hleu1-32 (a gift from Jurg Bahler) and grown in the presence or absence of thiamine. In the absence of thiamine (NMT41 promoter is active), the expression of Lac Z was detected in the construct containing active T-variant 3A, while only very low levels of Lac Z expression were detected in the construct with the inactive T-variant Δ3A that lacks cleavage site or in the construct with only the upstream and downstream sequences (No T-variant 3A) in which the T-variant 3A has been deleted (Figure 4C), suggesting that LacZ expression is associated with the activity of the T-variant 3A. However, in the presence of thiamine when the NMT41 promoter is repressed, reduced levels of Lac Z expression were observed in the constructs containing active T-variant 3A, inactive T-variantΔ3A or No T-variant 3A, compared to the samples in the absence of thiamine (Figure 4C). These control results suggest that the expression of Lac Z is dependent on both the transcription and the activity of T-variant 3A. No Lac Z expression was observed in the control construct containing the unrelated HCV-IRES sequence in the presence or absence of thiamine (Figure 4C). To further investigate if T-variants have effects on downstream gene expression in general, constructs containing 5 genomic Perere-3 T-variant sequences were made and the LacZ expression measured. In each case LacZ expression of the additional sequences was observed. The LacZ expression in the constructs containing T-variants 1–5 and is comparable with that of T-variant 3A (Supplementary Table S4, Figure 4D).
These results indicate a relationship between the activity of T-variant 3A RNA and the LacZ expression in vivo. To further investigate the in vivo cleavage activity of the T-variant 3A, RNA was extracted from the strains expressing the active T-variant 3A and cleavage site deletion T-variant Δ3A RNAs. Real-time PCR experiment was performed to compare the amount of T-variant 3A mRNA and T-variant Δ3A mRNA by using cross cleavage site primer pairs (F1 + R1) (Figure 4B). The amount of LacZ mRNA was measured in each of the two constructs by primer pairs (F2 + R2) (Figure 4B). The results show that the amount of T-variant 3A mRNA is less than half (49%) of T-variant Δ3A mRNA, due to T-variant 3A mRNA in vivo cleavage activity that is not present in the T-variant Δ3A (Figure 4E). In contrast, similar amounts of LacZ mRNA were detected in cells with T-variant 3A mRNA or T-variant Δ3A mRNA using (F2 + R2) primer pairs (Figure 4E). Although similar amounts of LacZ mRNA were observed, the LacZ protein expression was still much higher in the T-variant 3A constructs in the reporter assay (Figure 4C). These results suggest that the expression of LacZ is dependent on whether the T-variant is cleaved. 5′RACE was further used to map the T-variant 3A cleavage site in vivo. The reverse primer P2 in the 5′RACE generated a fragment of 310 nt up to the transcription start site and the reverse primer P1 in the 5′RACE generates a fragment of 625 nt from the cleavage site for T-variant 3A in vivo (Figure 4B, F). The 5′RACE sequence (Figure 4G) revealed the cleavage site of the T-variant 3A in vivo to be the same as observed in vitro (Figure 3J). T-variant cleavage removes the 5′ end of the RNA including the 5′-cap but leaves the residual structured ribozyme. These results suggest that T-variant cleaves in vivo and that the cleaved T-variant is sufficient for the translation of its downstream genes, implying that cleavage of T-variants in S. mansoni is required for translation of APE/RT the key protein for Perere-3 retrotranspostion.
T-variant ribozyme activity and Perere-3 non-LTR retrotransposon element replication in S. mansoni
If T-variants function as ribozymes to process RNA in Perere-3 non-LTR retrotransposon elements in S. mansoni, as proposed in Figure 2H, two primary requirements have to be met. The T-variants and Perere-3 elements must be transcribed together into RNA, and the T-variants must subsequently cleave the transcribed RNA. To identify RNA transcripts for T-variants and Perere-3, we carried out a search in the Ensembl EST database that contains the assembled RNA transcripts in S. mansoni (ftp://ftp.ensemblgenomes.org/pub/metazoa/current/fasta/schistosoma_mansoni/cdna). The S. mansoni genome is 65% AT rich (50), promoter prediction identified a number of possible promotor sequences upstream of the T-variant sequences (71) (Supplementary table S5). The cDNA data in the Ensembl EST database confirms the transcription of active promoters in vivo. The results for representative transcript RNA sequences are shown in Figure 5A. The genomic locations of these RNA transcripts and the transcript IDs are shown in Figure 5B and C. These results reveal that there are indeed RNA transcripts for T-variants with Perere-3, suggesting that there is an active promoter upstream of the T-variants. In Perere-3 the APE/RT domain is a single ORF in S. mansoni (58).
Figure 5.
T-variant and Perere-3 retrotransposition. (A) cDNA sequences of four T-variants and downstream ORFs containing APE domain and RT domain from Ensembl cDNA database. The T-variant sequences are highlighted in pink, with secondary structural stems and loops marked. The arrow marks the T-variant cleavage site. The 5′ TSD next to the cleavage site and the 3′ TSDs next to the short repeats are marked with red boxes. The APE and RT domains are highlighted in turquoise and blue. (B) The location of the sequences (from A) on S. mansoni chromosomes. (C) Genomic accession number, location number and transcript location number of the sequences in (A). (D) Schematic of the possible process by which T-variant cleavage and retrotransposon insertion in the Perere-3 replication cycle leads to TSD and the formation of a new ribozyme P1 stem. The formation of an active ribozyme generates a viable retrotransposon that is available for further retrotransposition, conversely the formation of a lower efficiency ribozyme would be predicted to lead to a reduction in retrotransposon activity.
For each sequence, TSDs are observed flanking the APE/RT/Perere-3 element, and the TSDs are directly adjacent to the T-variant cleavage site AA (Figure 5A, D). TSDs are a footprint for TPRT and evidence that retrotransposition of the Perere-3 element has taken place. Each TSD is generated by the insertion of double-stranded DNA that was synthesized from the cleaved T-variant RNA template. The observation that the TSDs are immediately next to the T-variant cleavage sites provides evidence that T-variant cleavage has taken place in vivo.
The TSD for each of the sequences is different, suggesting that insertion of the cleaved T-variant associated with Perere-3 is not site-specific (Figure 5A, D). Since the S. mansoni genome is AT rich, the insertion would be predicted to be more likely next to sequences with an A or T (50). Because the TSDs are located 5′ to the genomic position corresponding to the T-variant cleavage site in the RNA, they also form the P1 stem of the T-variant and the sequence composition of the P1 stem also affects the activity of the T-variants (Figure 3F, G, H, I and Supplementary Figure S7). Thus, the TSD sequence generated by T-variant/Perere-3 insertion is closely linked to the activity of the T-variant through the P1 stem (Figure 5D). TSD generation in the replication cycle is located in the position that corresponds to the 5′ end of P1 stem of T-variant. The P1 stem is formed by base paring of the 5′ and 3′ ends of the T-variant. Since the 3′ end of the P1 stem is imbedded within Perere-3, it remains unchanged during retrotransposition. However, 5′ TSD can generate variable sequences in the 5′ strand of the P1 stem. Changes in base-pairing between the variable 5′ strand and the unchanged 3′ strand of P1 may stabilise or destabilise P1, potentially enhancing or inhibiting T-variant cleavage (Figure 5D), which is consistent the T-variant activities observed in vitro (Figure 3F, G, H, I and Supplementary Figure S7). A T-variant/Perere-3 retrotransposition event that makes a TSD sequence and P1 stem that generates an active T-variant would enable Perere-3 to remain active during the replication cycle (Figure 5D). In contrast, a T-variant/Perere-3 retrotransposition that generates a TSD that leads to a loss of T-variant activity would potentially impact Perere-3 replication through effects on downstream gene expression as seen in the reporter assays (Figure 4). Therefore, the dependence of T-variant activity on the insertion site (by TSD) through retrotransposition is linked to the activity of Perere-3 during its replication cycle.
The proposed model in Figure 5D explains the function of the T-variant in the Perere-3 replication based on these results. If the proposed model is reasonable, the distances between the T-variant and the AUG of the Perere-3 elements are expected to be similar. The distances between the T-variant and the AUG of all 345 Perere-3 elements was analysed manually. For the majority of the Perere-3 elements (∼306 out of 345 with RT domain), the distance between the T-variant and the reported AUG is ∼147 nt (211 sequences) and there are 95 sequences with distances ∼112 nt (T-variant to predicted AUG I). There are small numbers of other distances (Supplementary Table S6). The average and mean distances between T-variant and the AUG are shown in (Supplementary Table S6). The distances between the T-variants and the AUG naturally falls into two main groups (Supplementary Table S6), supporting the notion that within each group the proposed model explains the function of the T-variant in Perere-3 replication.
DISCUSSION
Here, we have identified over 800 T-variant sequences (Figure 1) and investigated their potential function in S. Mansoni. Several lines of evidence point to an important functional role for T-variant ribozymes in the non-LTR retrotransposon replication cycle: (1) The genomic location of T-variant ribozyme is upstream of the Perere-3 retrotransposon element containing APE/RT domains (Figures 2A-G and 5). (2) T-variant ribozymes were shown to be active in vitro and in vivo (Figures 3 and 4). (3) Reporter assays show that T-variant cleavage is required for translation of the downstream gene (Figure 4). (4) T-variants and Perere-3 are cotranscribed in S. Mansoni (Figure 5A). TSDs are generated by the repair of the intermediates of retrotransposon DNA insertion in the final integration step and are therefore evidence of retrotransposon insertion (53,89). TSDs flank the inserted retrotransposons of T-variant sequences (Figures 5A, 2A, B) and are positioned right next to the T-variant cleavage sites, suggesting T-variant cleavage in vivo and active ribozyme sequences were involved in successful Perere-3 retrotransposition. Differences in TSDs suggest that Perere-3 transposition is not site-specific. The TSDs contribute to the 5′ P1 stem of the T-variants and may effect T-variant structure and function which can in turn impact Perere-3 transposition. Together this evidence suggests a function for the T-variants in Perere-3 retrotransposon replication (Figure 5D).
An understanding of the RNA template that is involved in reverse transcription is required for dissection of retrotransposon integration reaction. There are similarities and differences between the Perere-3 LINE retrotransposon in S. Mansoni and the well characterized R2 LINEs from Drosophila melanogaster and Bombyx mori retrotransposon. The R2 LINEs encode a restriction enzyme-like endonuclease, that directs LINE insertion to a precise position in 28S rRNA genes (90,91). Analysis of 28S rRNA/R2 co-transcripts identified that the 5′ junction of the inserted R2 element contained small deletions suggesting that R2 element insertion was dependent on 5′ processing of the R2 RNA (62,63). In vitro transcription experiments showed that the exact 5′ junction between the 28S rRNA and the R2 RNA mapped to the cleavage site of an embedded self-cleaving ribozyme that was similar to the previously characterized hepatitis delta virus (HDV) ribozyme (61,64,65). For R2 LINES the ribozyme cleavage contributes to processing of the 5′ end of the inserted transcript (65). A clear parallel between the Perere-3 and R2 classes of retroelements is that they each incorporate an efficient 5′-ribozyme; the Twister ribozyme variants in Perere-3 (S. mansoni) and the HDV-like ribozyme in the R2 elements. In contrast to the R2 elements, that have a specific target site in the 28SrRNA gene (62,92), the Perere-3 LINEs encode an APE endonuclease which leads to non-specific retrotransposon insertion and consequent TSD. The inserted Perere-3 retrotransposon retains the active 5′-AA ribozyme and generates a new P1 substrate strand in the TSD, the presence of the inserted T-variants adjacent to the TSDs are indicative of the insertion of a cleaved T-variant. Compared to the limited number of R2 element target sites, there are a greater variety of Perere-3 target sites (38,56). Like the R2 elements, Perere-3 appears to be transcribed from host promoters, RNA Pol I for the R2 elements and RNA Pol II for Perere-3 (23). In reporter strains both ribozymes appear to effect downstream reporter gene expression in vivo (65) (Figure 4). Interestingly, for Perere-3, although T-variant cleavage would be predicted to excise the 5′ methyl-guanosine cap, the expression of downstream genes, is comparable to previously characterized structured UTRs in similar constructs (73,74), suggesting that in reporter strains, the residual cleaved twister ribozyme is sufficient for effective ribosome recruitment and translation of the downstream gene. The cleaved ribozyme would be predicted to retain its tertiary fold (79). Although both Perere-3 and R2 retrotransposons encode characteristic endonuclease and reverse transcriptases, the positions of the endonuclease domains relative to the reverse transcriptase are inverted (38,56).
It is noteworthy that the inactive T-variants (0–1A) (Figure 3A) lack TSDs and such retrotransposon RNAs would be predicted to be deficient in ribozyme dependent RNA maturation. The non-self-cleaving T-variant (0–1A) retrotransposon RNA’s may use a different mechanism for RNA maturation, and/or, retrotransposon integration may incorporate upstream host RNA sequences. Alternatively, the loss of ribozyme activity may reduce the efficiency of transposition, rendering them inactive. Such a loss of activity would represent the end-point of the retrotransposon replication cycle and we note that, to preserve genomic integrity, the majority of genomic transposons are no longer active (93). Inactive T-variant (0–1A) ribozymes may be generated by inaccurate reverse transcription of the 5′ end of the self-cleaved retrotransposon RNA or by mis-repair of the inserted top-strand intermediate.
For each T-variant the local environment at each scissile bond varies, and these differences are reflected in the contrasting cleavage rates observed for each T-variant in comparison with the well characterized and efficient N. vitripennis twister ribozyme in Figure 4. Due to these differences, each individual T-variant ribozyme can be regarded as a novel sub-class of ribozyme that will require further analysis and optimization. It may well be the case that in vivo cellular conditions further modulate the activity of these novel ribozymes (72,94).
The transcriptome profile of the six developmental stages (egg, miracidia, sporocysts, cercaria, schistosomula and adult) of S. mansoni from RNA-seq data is available (50–52,95,96). The transcriptome data for each stage comes from different sources, and cannot be quantitatively compared. However, the data can indicate if a transcript is present or not. The transcription levels of four examples of T-variant/Perere-3 in all the development stages of S. mansoni are shown in Supplementary Table S7. Transcription of T-variant/Perere-3 between the different developmental stages can be seen to be discontinuous. The function of T-variant/Perere-3 in the S. mansoni developmental stages requires further investigation.
Here, we have shown that embedded T-variant ribozymes are an integral component of Perere-3, an abundant retrotransposon in S. mansoni, and that the T-variants also associate with other reverse transcriptase domains. Suggesting that T-variants may have a wider role in retrotransposition in S. mansoni.
DATA AVAILABILITY
The supporting data for this manuscript are available as supplementary data.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Murchie lab for discussion.
Contributor Information
Getong Liu, Fudan University Pudong Medical Center, and Institutes of Biomedical Sciences, Shanghai Medical College, Key Laboratory of Medical Epigenetics and Metabolism, Fudan University, Shanghai 200032, China; Key Laboratory of Metabolism and Molecular Medicine, Ministry of Education, School of Basic Medical Sciences, Fudan University, Shanghai 200032, China.
Hengyi Jiang, Fudan University Pudong Medical Center, and Institutes of Biomedical Sciences, Shanghai Medical College, Key Laboratory of Medical Epigenetics and Metabolism, Fudan University, Shanghai 200032, China; Key Laboratory of Metabolism and Molecular Medicine, Ministry of Education, School of Basic Medical Sciences, Fudan University, Shanghai 200032, China.
Wenxia Sun, Fudan University Pudong Medical Center, and Institutes of Biomedical Sciences, Shanghai Medical College, Key Laboratory of Medical Epigenetics and Metabolism, Fudan University, Shanghai 200032, China; Key Laboratory of Metabolism and Molecular Medicine, Ministry of Education, School of Basic Medical Sciences, Fudan University, Shanghai 200032, China.
Jun Zhang, Fudan University Pudong Medical Center, and Institutes of Biomedical Sciences, Shanghai Medical College, Key Laboratory of Medical Epigenetics and Metabolism, Fudan University, Shanghai 200032, China; Key Laboratory of Metabolism and Molecular Medicine, Ministry of Education, School of Basic Medical Sciences, Fudan University, Shanghai 200032, China.
Dongrong Chen, Fudan University Pudong Medical Center, and Institutes of Biomedical Sciences, Shanghai Medical College, Key Laboratory of Medical Epigenetics and Metabolism, Fudan University, Shanghai 200032, China; Key Laboratory of Metabolism and Molecular Medicine, Ministry of Education, School of Basic Medical Sciences, Fudan University, Shanghai 200032, China.
Alastair I H Murchie, Fudan University Pudong Medical Center, and Institutes of Biomedical Sciences, Shanghai Medical College, Key Laboratory of Medical Epigenetics and Metabolism, Fudan University, Shanghai 200032, China; Key Laboratory of Metabolism and Molecular Medicine, Ministry of Education, School of Basic Medical Sciences, Fudan University, Shanghai 200032, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2016YFA0500604] and Natural Science Foundation [31420103907, 31770873, 31330022], to A.M.; Natural Science Foundation [31370107 to D.C., 31470777]. Funding for open access charge: Laboratory publication fund.
Conflict of interest statement. None declared.
REFERENCES
- 1. Roth A., Weinberg Z., Chen A.G.Y., Kim P.B., Ames T.D., Breaker R.R.. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat. Chem. Biol. 2014; 10:56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eiler D., Wang J., Steitz T.A.. Structural basis for the fast self-cleavage reaction catalyzed by the twister ribozyme. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:13028–13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gebetsberger J., Micura R.. Unwinding the twister ribozyme: from structure to mechanism. Wiley Interdiscip. Rev. RNA. 2017; 8:e1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu Y., Wilson T.J., McPhee S.A., Lilley D.M.J.. Crystal structure and mechanistic investigation of the twister ribozyme. Nat. Chem. Biol. 2014; 10:739–744. [DOI] [PubMed] [Google Scholar]
- 5. Ren A., Košutić M., Rajashankar K.R., Frener M., Santner T., Westhof E., Micura R., Patel D.J.. In-line alignment and Mg2+ coordination at the cleavage site of the env22 twister ribozyme. Nat. Commun. 2014; 5:5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wilson T.J., Liu Y., Domnick C., Kath-Schorr S., Lilley D.M.J.. The novel chemical mechanism of the twister ribozyme. J. Am. Chem. Soc. 2016; 138:6151–6162. [DOI] [PubMed] [Google Scholar]
- 7. Kapitonov V.V., Tempel S., Jurka J.. Simple and fast classification of non-LTR retrotransposons based on phylogeny of their RT domain protein sequences. Gene. 2009; 448:207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Malik H.S., Burke W.D., Eickbush T.H.. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 1999; 16:793–805. [DOI] [PubMed] [Google Scholar]
- 9. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W.et al.. Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 10. Schnable P.S., Ware D., Fulton R.S., Stein J.C., Wei F., Pasternak S., Liang C., Zhang J., Fulton L., Graves T.A.et al.. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009; 326:1112–1115. [DOI] [PubMed] [Google Scholar]
- 11. Mustafin R.N., Khusnutdinova E.K.. The role of reverse transcriptase in the origin of life. Biochemistry (Mosc). 2019; 84:870–883. [DOI] [PubMed] [Google Scholar]
- 12. Göke J., Ng H.H.. CTRL+INSERT: retrotransposons and their contribution to regulation and innovation of the transcriptome. EMBO Rep. 2016; 17:1131–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boeke J.D., Stoye J.P.. Coffin J.M., Hughes S.H., Varmus H.E.. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. Retroviruses. 1997; NY: Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- 14. Dewannieux M., Heidmann T.. LINEs, SINEs and processed pseudogenes: parasitic strategies for genome modeling. Cytogenet. Genome Res. 2005; 110:35–48. [DOI] [PubMed] [Google Scholar]
- 15. Eickbush T.H., Malik H.S.. Origins and evolution of retrotransposons. Mobile DNA II. 2002; 2:1111–1144. [Google Scholar]
- 16. Kajikawa M., Okada N.. LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell. 2002; 111:433–444. [DOI] [PubMed] [Google Scholar]
- 17. Konkel M.K., Batzer M.A.. A mobile threat to genome stability: The impact of non-LTR retrotransposons upon the human genome. Semin. Cancer Biol. 2010; 20:211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roy-Engel A.M. A tale of an A-tail: the lifeline of a SINE. Mob Genet Elements. 2012; 2:282–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McLean C., Bucheton A., Finnegan D.J.. The 5′ untranslated region of the I factor, a long interspersed nuclear element-like retrotransposon of Drosophila melanogaster, contains an internal promoter and sequences that regulate expression. Mol. Cell. Biol. 1993; 13:1042–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mizrokhi L.J., Georgieva S.G., Ilyin Y.V.. jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell. 1988; 54:685–691. [DOI] [PubMed] [Google Scholar]
- 21. Khan H. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2005; 16:78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haas N.B., Grabowski J.M., North J., Moran J.V., Kazazian H.H., Burch J.B.. Subfamilies of CR1 non-LTR retrotransposons have different 5′UTR sequences but are otherwise conserved. Gene. 2001; 265:175–183. [DOI] [PubMed] [Google Scholar]
- 23. George J.A., Eickbush T.H.. Conserved features at the 5 end of Drosophila R2 retrotransposable elements: implications for transcription and translation. Insect Mol. Biol. 1999; 8:3–10. [DOI] [PubMed] [Google Scholar]
- 24. Eickbush T.H. Transposing without ends: the non-LTR retrotransposable elements. New Biol. 1992; 4:430–440. [PubMed] [Google Scholar]
- 25. Han J.S. Non-long terminal repeat (non-LTR) retrotransposons: mechanisms, recent developments, and unanswered questions. Mob DNA. 2010; 1:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang J., Malik H.S., Eickbush T.H.. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:7847–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Feng Q., Moran J.V., Kazazian H.H., Boeke J.D.. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996; 87:905–916. [DOI] [PubMed] [Google Scholar]
- 28. Fujiwara H. Site-specific non-LTR retrotransposons. Mobile DNA III. 2015; John Wiley & Sons, Ltd; 1147–1163. [Google Scholar]
- 29. Eickbush T.H. R2 and related site-specific non-long terminal repeat retrotransposons. Mobile DNA II. 2002; 2:813–835. [Google Scholar]
- 30. Luan D.D., Korman M.H., Jakubczak J.L., Eickbush T.H.. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993; 72:595–605. [DOI] [PubMed] [Google Scholar]
- 31. Beauregard A., Curcio M.J., Belfort M.. The take and give between retrotransposable elements and their hosts. Annu. Rev. Genet. 2008; 42:587–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hohjoh H., Singer M.F.. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996; 15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 33. Dmitriev S.E., Andreev D.E., Terenin I.M., Olovnikov I.A., Prassolov V.S., Merrick W.C., Shatsky I.N.. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5′ untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated. Mol. Cell. Biol. 2007; 27:4685–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kubo S., Seleme M.D.C., Soifer H.S., Perez J.L.G., Moran J.V., Kazazian H.H., Kasahara N.. L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl. Acad. Sci. USA. 2006; 103:8036–8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kinsey J.A. Tad, a LINE-like transposable element of Neurospora, can transpose between nuclei in heterokaryons. Genetics. 1990; 126:317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shapiro J.A. How chaotic is genome chaos. Cancers (Basel). 2021; 13:1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cost G.J., Feng Q., Jacquier A., Boeke J.D.. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002; 21:5899–5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eickbush T.H., Eickbush D.G.. Integration, regulation, and long-term stability of R2 retrotransposons. Microbiol. Spectrum. 2015; 3:MDNA3–0011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kojima K.K. Structural and sequence diversity of eukaryotic transposable elements. Genes Genet. Syst. 2020; 94:233–252. [DOI] [PubMed] [Google Scholar]
- 40. Moran J.V., Gilbert N.. Mammalian LINE-1 retrotransposons and related elements. Mobile DNA II. 2002; 2:836–869. [Google Scholar]
- 41. Kajikawa M., Yamaguchi K., Okada N.. A new mechanism to ensure integration during LINE retrotransposition: a suggestion from analyses of the 5′ extra nucleotides. Gene. 2012; 505:345–351. [DOI] [PubMed] [Google Scholar]
- 42. Khadgi B.B., Govindaraju A., Christensen S.M.. Completion of LINE integration involves an open ‘4-way’ branched DNA intermediate. Nucleic Acids Res. 2019; 47:8708–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gilbert N., Lutz-Prigge S., Moran J.V.. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002; 110:315–325. [DOI] [PubMed] [Google Scholar]
- 44. Stage D.E., Eickbush T.H.. Origin of nascent lineages and the mechanisms used to prime second-strand DNA synthesis in the R1 and R2 retrotransposons of Drosophila. Genome Biol. 2009; 10:R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee W., Mun S., Kang K., Hennighausen L., Han K.. Genome-wide target site triplication of Alu elements in the human genome. Gene. 2015; 561:283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Plasterk R.H. The origin of footprints of the Tc1 transposon of Caenorhabditis elegans. EMBO J. 1991; 10:1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yao J., Truong D.M., Lambowitz A.M.. Genetic and biochemical assays reveal a key role for replication restart proteins in group II intron retrohoming. PLoS Genet. 2013; 9:e1003469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fujimoto H., Hirukawa Y., Tani H., Matsuura Y., Hashido K., Tsuchida K., Takada N., Kobayashi M., Maekawa H.. Integration of the 5′ end of the retrotransposon, R2Bm, can be complemented by homologous recombination. Nucleic Acids Res. 2004; 32:1555–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gryseels B., Polman K., Clerinx J., Kestens L.. Human schistosomiasis. Lancet. 2006; 368:1106–1118. [DOI] [PubMed] [Google Scholar]
- 50. Berriman M., Haas B.J., LoVerde P.T., Wilson R.A., Dillon G.P., Cerqueira G.C., Mashiyama S.T., Al-Lazikani B., Andrade L.F., Ashton P.D.et al.. The genome of the blood fluke Schistosoma mansoni. Nature. 2009; 460:352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Protasio A.V., Tsai I.J., Babbage A., Nichol S., Hunt M., Aslett M.A., De Silva N., Velarde G.S., Anderson T.J.C., Clark R.C.et al.. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl. Trop. Dis. 2012; 6:e1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Anderson L., Amaral M.S., Beckedorff F., Silva L.F., Dazzani B., Oliveira K.C., Almeida G.T., Gomes M.R., Pires D.S., Setubal J.C.et al.. Schistosoma mansoni egg, adult male and female comparative gene expression analysis and identification of novel genes by RNA-Seq. PLoS Negl Trop Dis. 2015; 9:e0004334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Verjovski-Almeida S., DeMarco R., Martins E.A.L., Guimarães P.E.M., Ojopi E.P.B., Paquola A.C.M., Piazza J.P., Nishiyama M.Y., Kitajima J.P., Adamson R.E.et al.. Transcriptome analysis of the acoelomate human parasite Schistosoma mansoni. Nat. Genet. 2003; 35:148–157. [DOI] [PubMed] [Google Scholar]
- 54. Laha T., Brindley P.J., Verity C.K., McManus D.P., Loukas A.. pido, a non-long terminal repeat retrotransposon of the chicken repeat 1 family from the genome of the Oriental blood fluke, Schistosoma japonicum. Gene. 2002; 284:149–159. [DOI] [PubMed] [Google Scholar]
- 55. Ivanchenko M.G., Lerner J.P., McCormick R.S., Toumadje A., Allen B., Fischer K., Hedstrom O., Helmrich A., Barnes D.W., Bayne C.J.. Continuous in vitro propagation and differentiation of cultures of the intramolluscan stages of the human parasite Schistosoma mansoni. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:4965–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. DeMarco R., Machado A.A., Bisson-Filho A.W., Verjovski-Almeida S.. Identification of 18 new transcribed retrotransposons in Schistosoma mansoni. Biochem. Biophys. Res. Commun. 2005; 333:230–240. [DOI] [PubMed] [Google Scholar]
- 57. DeMarco R., Kowaltowski A.T., Machado A.A., Soares M.B., Gargioni C., Kawano T., Rodrigues V., Madeira A.M.B.N., Wilson R.A., Menck C.F.M.et al.. Saci-1, -2, and -3 and Perere, four novel retrotransposons with high transcriptional activities from the human parasite Schistosoma mansoni. J. Virol. 2004; 78:2967–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Valentim C.L.L., Gomes M.S., Jeremias W.J., Cunha J.C., Oliveira G.C., Botelho A.C.C., Pimenta P.F.P., Janotti-Passos L.K., Guerra-Sá R., Babá E.H.. Physical localization of the retrotransposons Boudicca and Perere 03 in Schistosoma mansoni. J. Parasitol. 2008; 94:993–995. [DOI] [PubMed] [Google Scholar]
- 59. Bao W., Kojima K.K., Kohany O.. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015; 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Weinberg C.E., Weinberg Z., Hammann C.. Novel ribozymes: discovery, catalytic mechanisms, and the quest to understand biological function. Nucleic Acids Res. 2019; 47:9480–9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Been M.D., Wickham G.S.. Self-cleaving ribozymes of hepatitis delta virus RNA. Eur. J. Biochem. 1997; 247:741–753. [DOI] [PubMed] [Google Scholar]
- 62. Eickbush D.G., Eickbush T.H.. R2 retrotransposons encode a self-cleaving ribozyme for processing from an rRNA cotranscript. Mol. Cell. Biol. 2010; 30:3142–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Eickbush D.G., Ye J., Zhang X., Burke W.D., Eickbush T.H.. Epigenetic regulation of retrotransposons within the nucleolus of Drosophila. Mol. Cell. Biol. 2008; 28:6452–6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ferré-D’Amaré A.R., Zhou K., Doudna J.A.. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998; 395:567–574. [DOI] [PubMed] [Google Scholar]
- 65. Ruminski D.J., Webb C.-H.T., Riccitelli N.J., Lupták A.. Processing and translation initiation of non-long terminal repeat retrotransposons by hepatitis delta virus (HDV)-like self-cleaving ribozymes. J. Biol. Chem. 2011; 286:41286–41295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gautheret D., Major F., Cedergren R.. Pattern searching/alignment with RNA primary and secondary structures: an effective descriptor for tRNA. Comput. Appl. Biosci. 1990; 6:325–331. [DOI] [PubMed] [Google Scholar]
- 67. Burge C., Karlin S.. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997; 268:78–94. [DOI] [PubMed] [Google Scholar]
- 68. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A.. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003; 31:3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schultz J., Milpetz F., Bork P., Ponting C.P.. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Reese M.G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2001; 26:51–56. [DOI] [PubMed] [Google Scholar]
- 72. Zhang J., Liu G., Sun W., Chen D., Murchie A.I.H.. The effects of aminoglycoside antibiotics on twister ribozyme cleavage. FEBS J. 2020; 288:1586–1598. [DOI] [PubMed] [Google Scholar]
- 73. Sun W., Zhang X., Chen D., Murchie A.I.H.. Interactions between the 5′ UTR mRNA of the spe2 gene and spermidine regulate translation in S. pombe. RNA. 2020; 26:137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang X., Sun W., Chen D., Murchie A.I.H.. Interactions between SAM and the 5′ UTR mRNA of the sam1 gene regulate translation in S. pombe. RNA. 2020; 26:150–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang X., Bremer H.. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J. Biol. Chem. 1995; 270:11181–11189. [DOI] [PubMed] [Google Scholar]
- 76. Raghava G.P., Sahni G.. GMAP: a multi-purpose computer program to aid synthetic gene design, cassette mutagenesis and the introduction of potential restriction sites into DNA sequences. BioTechniques. 1994; 16:1116–1123. [PubMed] [Google Scholar]
- 77. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Goldstein L.D., Cao Y., Pau G., Lawrence M., Wu T.D., Seshagiri S., Gentleman R.. Prediction and quantification of splice events from RNA-Seq data. PLoS One. 2016; 11:e0156132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Košutić M., Neuner S., Ren A., Flür S., Wunderlich C., Mairhofer E., Vušurović N., Seikowski J., Breuker K., Höbartner C.et al.. A mini-twister variant and impact of residues/cations on the phosphodiester cleavage of this ribozyme class. Angew. Chem. Int. Ed. Engl. 2015; 54:15128–15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Riccitelli N.J., Lupták A.. Computational discovery of folded RNA domains in genomes and in vitro selected libraries. Methods. 2010; 52:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. DeMarco R., Kowaltowski A.T., Machado A.A., Soares M.B., Gargioni C., Kawano T., Rodrigues V., Madeira A.M.B.N., Wilson R.A., Menck C.F.M.et al.. Saci-1, -2, and -3 and Perere, four novel retrotransposons with high transcriptional activities from the human parasite Schistosoma mansoni. J. Virol. 2004; 78:2967–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chambeyron S., Bucheton A., Busseau I.. Tandem UAA repeats at the 3′-end of the transcript are essential for the precise initiation of reverse transcription of the I factor in Drosophila melanogaster. J. Biol. Chem. 2002; 277:17877–17882. [DOI] [PubMed] [Google Scholar]
- 83. Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Marchler G.H., Song J.S.et al.. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020; 48:D265–D268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C.J., Lu S., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R.et al.. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017; 45:D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gaines C.S., York D.M.. Ribozyme catalysis with a twist: active state of the twister ribozyme in solution predicted from molecular simulation. J. Am. Chem. Soc. 2016; 138:3058–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Forsburg S.L. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 1993; 21:2955–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Moreno M.B., Durán A., Ribas J.C.. A family of multifunctional thiamine-repressible expression vectors for fission yeast. Yeast. 2000; 16:861–872. [DOI] [PubMed] [Google Scholar]
- 88. Tamm T. A thiamine-regulatable epitope-tagged protein expression system in fission yeast. Methods Mol. Biol. 2012; 824:417–432. [DOI] [PubMed] [Google Scholar]
- 89. Kryatova M.S., Steranka J.P., Burns K.H., Payer L.M.. Insertion and deletion polymorphisms of the ancient AluS family in the human genome. Mob DNA. 2017; 8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dawid I.B., Rebbert M.L.. Nucleotide sequences at the boundaries between gene and insertion regions in the rDNA of Drosophilia melanogaster. Nucleic Acids Res. 1981; 9:5011–5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Roiha H., Miller J.R., Woods L.C., Glover D.M.. Arrangements and rearrangements of sequences flanking the two types of rDNA insertion in D. melanogaster. Nature. 1981; 290:749–753. [DOI] [PubMed] [Google Scholar]
- 92. Christensen S.M., Ye J., Eickbush T.H.. RNA from the 5′ end of the R2 retrotransposon controls R2 protein binding to and cleavage of its DNA target site. Proc. Natl Acad. Sci. 2006; 103:17602–17607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Venancio T.M., Wilson R.A., Verjovski-Almeida S., DeMarco R.. Bursts of transposition from non-long terminal repeat retrotransposon families of the RTE clade in Schistosoma mansoni. Int. J. Parasitol. 2010; 40:743–749. [DOI] [PubMed] [Google Scholar]
- 94. Messina K.J., Bevilacqua P.C.. Cellular small molecules contribute to twister ribozyme catalysis. J. Am. Chem. Soc. 2018; 140:10578–10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wang B., Collins J.J., Newmark P.A.. Functional genomic characterization of neoblast-like stem cells in larval Schistosoma mansoni. Elife. 2013; 2:e00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wang B., Lee J., Li P., Saberi A., Yang H., Liu C., Zhao M., Newmark P.A.. Stem cell heterogeneity drives the parasitic life cycle of Schistosoma mansoni. Elife. 2018; 7:e35449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The supporting data for this manuscript are available as supplementary data.