Abstract
Translation of eukaryotic mRNAs begins with binding of their m7G cap to eIF4E, followed by recruitment of other translation initiation factor proteins. We describe capCLIP, a novel method to comprehensively capture and quantify the eIF4E (eukaryotic initiation factor 4E) ‘cap-ome’ and apply it to examine the biological consequences of eIF4E–cap binding in distinct cellular contexts. First, we use capCLIP to identify the eIF4E cap-omes in human cells with/without the mTORC1 (mechanistic target of rapamycin, complex 1) inhibitor rapamycin, there being an emerging consensus that rapamycin inhibits translation of TOP (terminal oligopyrimidine) mRNAs by displacing eIF4E from their caps. capCLIP reveals that the representation of TOP mRNAs in the cap-ome is indeed systematically reduced by rapamycin, thus validating our new methodology. capCLIP also refines the requirements for a functional TOP sequence. Second, we apply capCLIP to probe the consequences of phosphorylation of eIF4E. We show eIF4E phosphorylation reduces overall eIF4E–mRNA association and, strikingly, causes preferential dissociation of mRNAs with short 5′-UTRs. capCLIP is a valuable new tool to probe the function of eIF4E and of other cap-binding proteins such as eIF4E2/eIF4E3.
INTRODUCTION
Control of protein synthesis (mRNA translation) is crucial in regulating the composition of the cellular proteome (1). Mammalian cells employ diverse control mechanisms to regulate translation of specific mRNAs or subsets of mRNAs. Such mechanisms act primarily at translation initiation, where ribosomes are recruited to mRNAs (1). eIF4E (eukaryotic initiation factor 4E) is critical for both translation initiation and its regulation (2). eIF4E binds the 7-methyl-GTP (m7GTP) moiety of the 5′-cap structure of cellular cytoplasmic mRNAs. This interaction is crucial for the efficient translation of almost all mRNAs (3). eIF4E interacts with the scaffold protein eIF4G, and indirectly with other eIFs, including eIF3. The eIF4E–eIF4G–eIF3 complex then recruits 40S ribosomal subunits to the 5′ end of the mRNA, permitting scanning of 40S-associated initiation complexes to the start codon, 60S subunit joining and translation of the message (4). While eIF4E+/− mice are viable, eIF4E+/− embryonic fibroblasts resist oncogenic transformation (5), indicating that cellular translational activity is acutely sensitive to levels of eIF4E. Other cap-binding proteins also exist, including eIF4E2 (4EHP) and eIF4E3, although their cellular roles remain poorly understood (6).
Here, we describe a novel technique that identifies the set of mRNAs associated with eIF4E (the ‘cap-ome’). We term this method ‘capCLIP’. It is based on cross-linking immunoprecipitation (CLIP)-based methodology (7,8) and utilizes the ability of UV irradiation of living cells to generate photo-cross-links between eIF4E and the m7G cap, thereby covalently linking eIF4E to bound, capped mRNAs (9). Immunoprecipitation (IP) and subsequent purification of eIF4E selectively co-purify these cross-linked mRNAs; cloning and sequencing of this material identifies the cellular eIF4E ‘cap-ome’.
To validate capCLIP and explore the effects of specific regulatory inputs into the binding of eIF4E to capped mRNAs, we first use this method to examine eIF4E–cap binding behaviour in cells treated with rapamycin. This drug inhibits mTORC1 (mechanistic target of rapamycin, complex 1), a multi-subunit protein kinase that is activated by amino acids, growth factors and other trophic stimuli. mTORC1 inhibition markedly impairs the synthesis of ribosomal proteins (10), which are encoded by mRNAs that possess a 5′-terminal tract of oligopyrimidines (TOP). Growing evidence suggests that control of TOP mRNA translation by mTORC1 is mediated by LARP1, which binds the cap and the adjacent pyrimidines of TOP motifs (11,12). Thus, in rapamycin-treated cells, LARP1 likely displaces eIF4E specifically from TOP mRNAs and capCLIP should reveal this loss of TOP mRNAs from the eIF4E cap-ome.
We show, using HeLa cells, that capCLIP identifies an eIF4E cap-ome of ∼3300 unique mRNAs and rapamycin causes significant depletion of 86 mRNAs from the eIF4E cap-ome. Of these mRNAs, 62 (72%) are known or newly discovered TOP mRNAs. Overall, our data indicate that the capCLIP method successfully identifies the cellular eIF4E cap-ome, and that, in the context of mTORC1 inhibition, it also effectively indicates an mRNA’s translational activity.
Second, we use capCLIP to probe the poorly understood consequences of phosphorylation of eIF4E, which occurs at a single site (Ser209), catalysed by the MNKs (MAP kinase-interacting kinases: MNK1 and MNK2), which are themselves directly activated by MAP kinases (13). There is a long-recognized association between elevated P-eIF4E levels and cancer (14). While phosphorylation of eIF4E reportedly increases the polysomal association of some mRNAs, both the mechanistic basis for this and its consequences for their translation remain unclear (15).
In sum, capCLIP provides a novel and robust method to capture and identify cellular eIF4E cap-omes. As capCLIP relies on a covalent link between eIF4E and the cap, which withstands very rigorous purification strategies, it can be adapted to work in complex samples such as tissues. Furthermore, capCLIP could readily be used to examine the regulation and roles of other cap-binding proteins, including eIF4E2 and eIF4E3.
MATERIALS AND METHODS
Cell line information
Human HeLa cells were obtained from ATCC. Cat# CCL-2 (RRID:CVCL_0030) authentication: Original stock of CCL-2 was obtained directly from the ATCC and the laboratory stock of cells was re-verified in 2016 by Garvin Molecular Genetics (Sydney, Australia), the Garvin Institute’s cell line identification service.
CRISPR/Cas9-based gene editing
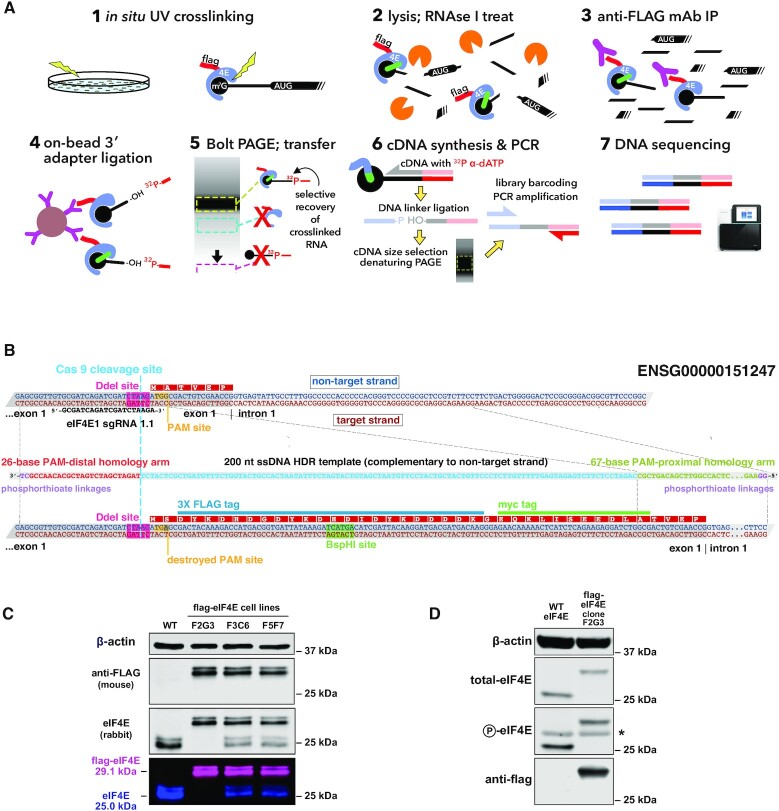
To introduce a 3X-flag epitope tag at the N-terminus of the endogenous eIF4E gene in HeLa cells, an sgRNA guide sequence targeting the first exon of human eIF4E was designed, with the Cas9 endonuclease cut site lying three bases upstream of the AUG of the eIF4E open-reading frame (ORF) (scheme is shown in Figure 1B). A 200-nt asymmetric, single-stranded homology-directed repair (HDR) DNA template (16) containing two modified phosphorothioate linkages at each end (17) was synthesized to permit the introduction of a 3X-flag/1X-myc epitope tag at the extreme N-terminus of the eIF4E protein. The HDR template, along with a GeneArt CRISPR Nuclease Vector (Life Technologies) expressing the eIF4E guide RNA and a Cas9 nuclease-human CD4 pre-protein, was introduced into HeLa cells using nucleofection (Lonza). Forty-eight hours post-nucleofection, Cas9-expressing cells were isolated using anti-CD4 Dynabeads, and the purified cells were plated at 1 cell/well in four 96-well TC dishes. Following ∼7 days of growth, wells containing clonal cell colonies were replica-plated, to allow further growth of each colony and provide cells for genomic DNA analysis. To check for insertion of the epitope tag, PCR products spanning the editing region of eIF4E were screened for length and for digestion with BspHI, a restriction site provided by the HDR template. Colonies positive in these screens were cultured further, and genomic DNA was again isolated for additional PCR amplification of the 5′-region on the eIF4E coding sequence, followed by forward and reverse Sanger sequencing of the entire edited region of eIF4E. Clones passing sequencing verification were further analysed by western blot, using anti-flag, anti-eIF4E and anti-P-eIF4E antibodies (Figure 1C and D).
Figure 1.
capCLIP method and flag-eIF4E expressing cell line. (A) Individual steps of the capCLIP method: (1) short-wavelength (∼254 nm) UV irradiation of living HeLa cells (clone F2G3; see below) expressing flag-eIF4E creates a ‘zero-order’ photo-cross-link between an mRNA’s m7G cap and an associated protein, e.g. eIF4E; (2) cells are then lysed and the mRNA fragmented using a limiting concentration of RNAse I; (3) flag-eIF4E (and cross-linked m7G-capped mRNA fragments) are captured by anti-FLAG mAb complexed to protein G-coated magnetic beads; (4) an RNA adapter is ligated ‘on bead’ to the 3′ end of the capped RNA fragment; (5) IP material is then subjected to neutral pH PAGE and then transferred to a membrane to size-separate cross-linked flag-eIF4E:mRNA:adapter complexes from unwanted IP products; (6) flag-eIF4E is proteolytically removed from capped mRNA:adapter complexes and the purified material used for cDNA synthesis, followed by ligation of a 3′-end DNA adapter and then PCR amplification; and (7) finally, barcoded capCLIP libraries are combined for high-throughput sequencing. (B) Outline of the CRISPR/Cas9-mediated addition of a 3X-flag/1X-myc epitope tag to the N-terminus of human eIF4E (Ensembl gene ENSG00000151247) in HeLa cells (henceforth identified as ‘flag-eIF4E’). Experimental details of this eIF4E gene editing strategy are provided in the ‘Materials and Methods’ section. (C) Anti-eIF4E/anti-flag westerns of wild-type (WT) HeLa cells and three CRISPR-edited flag-eIF4E HeLa clonal cell lines. The 29-kDa flag-eIF4E and the 25-kDa WT eIF4E bands detected by the rabbit anti-eIF4E antibody, and the 29-kDa flag-eIF4E band detected solely by the anti-flag antibody, are shown both as separate and as merged LiCor image channels. (D) Westerns using antibodies for total eIF4E, P-eIF4E or flag of lysates from WT and the 3X-flag-eIF4E-expressing HeLa cell clonal line F2G3. The asterisk denotes a non-specific band consistently seen with this anti-P-eIF4E antibody.
capCLIP
A detailed account of the pilot capCLIP experiments and ‘preparative’ capCLIP experiments is provided here. A comprehensive, step-by-step capCLIP protocol (including a reagent list), along with detailed commentary on specific novel aspects of the method, has been included as a Supplementary Protocol.
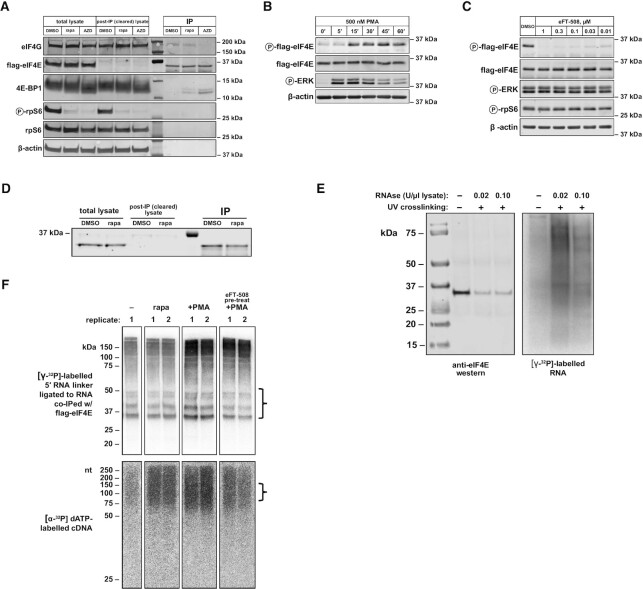
capCLIP was initially piloted on untreated HeLa cells endogenously expressing flag-eIF4E. Cells were exposed to 350 mJ/cm2 of short-wavelength (∼254 nm) UV irradiation, followed by cell lysis in 1× PXL (1× PBS supplemented with 0.1% SDS, 0.5% deoxycholate and 0.5% NP-40). Both a ‘high’ (0.10 U/μl lysate) and a ‘low’ (0.02 U/μl lysate) concentration of Ambion RNase I were tested to determine the optimal RNase I amount for generation of RNA tags of length ∼30–80 nt. Anti-flag mAb bound to protein G Dynabeads was used to immunoprecipitate flag-eIF4E and cross-linked mRNAs, followed by three washes in 1× PXL and one wash using 5× PXL (1× PXL with 5× PBS). [γ-32P]ATP was used to 5′ end-label the RNA molecules, followed by a final wash, elution and Bis–Tris PAGE size separation of the labelled eIF4E:RNA complexes (Figure 2E). Based on the initial RNase concentration experiments, an RNase I concentration of 0.02 U/μl of cell lysate was used for all subsequent capCLIP experiments.
Figure 2.
(A) Western blot of IPs of flag-eIF4E from HeLa cells using indicated antibodies. Anti-P-rpS6 and rpS6 antibodies were used to evaluate the ability of rapamycin and AZD to inhibit mTORC1 signalling. Molecular weight markers appear between the depleted and IP samples. The relative loading volume for total lysate, cleared lysate and IP lanes is 1:1:1 (thus, the amount of protein used as input for each IP reaction is the same as the amount of protein loaded into each total lysate lane). (B) Western analysis of P-flag-eIF4E stimulation in flag-eIF4E HeLa cells. Overnight serum-starved cells were treated with 500 nM phorbol myristate acetate (PMA) for the indicated times. (C) eFT-508 dose response of P-eIF4E levels in flag-eIF4E HeLa cells. Continued activation of MAPK/ERK signalling was confirmed with anti-P-ERK; mTORC1 activity assayed with anti-P-rpS6. (D) Western analysis of flag-eIF4E IPs from untreated (DMSO) and rapamycin-treated (rapa) HeLa cells. The relative loading volume for total lysate, cleared lysate and IP lanes is 1:1:2.5. (E) Left: anti-eIF4E western of flag-eIF4E (29.1 kDa) immunoprecipitated from HeLa cells in a pilot capCLIP experiment to test optimal RNase concentration. Right: phosphorimage of 32P-labelled RNA present on the same anti-flag-eIF4E co-IP western membrane shown on left. (F) Top: phosphorimage of 32P-labelled RNA after anti-flag-eIF4E co-IP of eIF4E protein and cross-linked mRNA, and subsequent PAGE separation and transfer to nitrocellulose for all capCLIP samples referenced in this study. Bracket at right indicates the portion of the membrane taken for subsequent isolation of RNA. Bottom: phosphorimage of 32P-labelled cDNA separated by 7 M urea denaturing PAGE for the same capCLIP samples. Bracket at right indicates the size range of cDNA products used for subsequent PCR library amplification.
To verify the effects of mTORC1 inhibition on translation initiation factors that affect eIF4E function and to validate the correct behaviour of our flag-eIF4E edited HeLa cell line under mTORC1 inhibition, flag-eIF4E HeLa cells were treated with 100 nM AZD8055 (an mTOR-KI), 100 nM rapamycin or DMSO alone for 2 h, followed by IP of flag-eIF4E from the resulting cell lysates (Figure 2A). Rapamycin treatment does not appear to affect the co-IP of eIF4G with eIF4E, though we do observe a modest increase in 4E-BP binding to eIF4E with this treatment.
For the preparative capCLIP experiments ± rapamycin, HeLa cells endogenously expressing flag-eIF4E were treated for 2 h with 100 nM rapamycin, or with the equivalent amount of DMSO vehicle, and then UV irradiated as above. For the capCLIP experiment to study the effects of phosphorylation of eIF4E on mRNA binding, we needed to compare cells with high or very low levels of p-eIF4E. To do so, flag-eIF4E HeLa cells were serum-starved overnight. They were then pre-treated for 1 h with either the potent and specific MNK inhibitor 0.1 μM eFT-508 (18) or vehicle (DMSO). Cells were then stimulated with 500 nM PMA to activate MAP kinase signalling and thus MNK activity for 30 min immediately prior to UV irradiation.
Cell pellets for all samples were lysed in 1× PXL supplemented with 0.02 U of RNase I per μl of cell lysate and DNase I. Cell lysates were subject to anti-flag IP and washed, as above. A 5′-32P-labelled RNA adapter was ligated to the 3′ end of the eIF4E-cross-linked RNA ‘on bead’, followed by a final wash, elution and Bis–Tris PAGE size separation of the 32P-labelled eIF4E–RNA complexes. The material resolved by Bis–Tris PAGE was transferred electrophoretically to a nitrocellulose membrane and the radioactive RNA detected using phosphor imaging (see Figure 2F, top). The ∼37–60-kDa region of the membrane, corresponding to eIF4E:RNA complexes with RNAs ∼30–80 nt in length, was excised, protease treated and the liberated RNA precipitated.
cDNA synthesis, followed by ligation of a UMI (unique molecular identifier)-containing DNA adapter to the 3′ end of the cDNA, was conducted, and the resulting material was purified and size-selected on denaturing PAGE (Figure 2F, bottom). Gel slices corresponding to ssDNAs of ∼80–130 nt were cut from the gel, and the cDNA was eluted and precipitated. This DNA was subsequently amplified by PCR, and individual capCLIP samples were barcoded prior to sequencing. Following a final size purification of the amplified PCR product, and QC checks of library concentration and length, all sample libraries were subject to high-throughput single-end sequencing on the NextSeq 500 with a read length of 80.
Bioinformatic analysis
Raw reads were adaptor trimmed and filtered for short sequences during base calling with bcl2fastq2 (--find-adapters-with-sliding-window --adapter-stringency 0.9 --mask-short-adapter-reads 35 --minimum-trimmed-read-length 35). The resulting FASTQ files, averaging 45 million reads per sample, were analysed and quality checked using FastQC and Picard. The filtered reads were mapped against the human reference genome (hg19) using the STAR alignment algorithm (19) (version 2.5.3a with default parameters and --chimSegmentMin 20, --quantMode GeneCounts) returning an average unique alignment rate of 30%. UMI-tools (20) was used to deduplicate reads in each sample using UMIs (default settings with --method adjacency --edit-distance-threshold 1). Enriched regions of the genome were identified separately for each strand with the MACS2 peak caller (21) (version 2.1.1.20160309) using default parameters and reporting only peaks with an FDR cut-off (−q) <0.05. The resulting peak files from each strand were merged. Differential binding analysis was performed using R (version 3.4.3) and the DiffBind package (22). Alignments were visualized and interrogated using the Integrative Genomics Viewer (IGV) v2.3.80 (23). For peak calling and differential binding analyses of the rapamycin capCLIP data, two pseudo-replicate datasets were generated from the single DMSO-only control dataset; these are termed controls 1A and 1B in Figure 3.
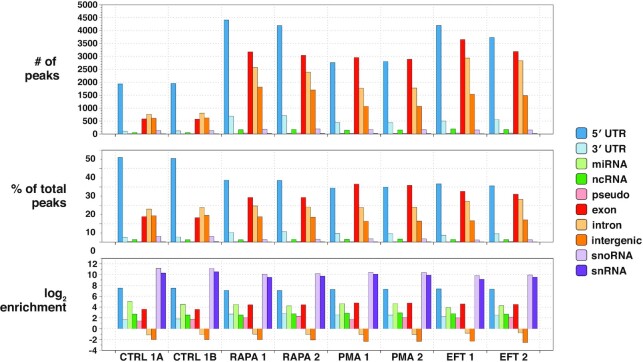
Figure 3.
capCLIP bioinformatic data. Peak calls from each of the six individual capCLIP sequencing datasets (x-axis) are categorized into 10 separate transcript/genome features indicated in the figure’s legend and plotted across three separate bar graphs. The top graph displays raw peak numbers for each of the 10 transcript/genome features. In the middle graph, the percentage of peaks in each dataset assigned to each of the 10 transcript/genome features is plotted. The bottom graph displays peak data according to the log2 enrichment per transcript/genome feature (the number of actual peaks per feature versus the number of peaks expected based on the relative fraction of the specific feature occupies in the human genome).
CAGE data
The CAGE (cap analysis of gene expression) dataset used for analysis of mRNA transcription start sites (TSSs) is ENCSR000CJJ (HeLa S3 cells) from the ENCODE portal (www.encodeproject.org) (24).
Validation of the TOP consensus motif identified by capCLIP
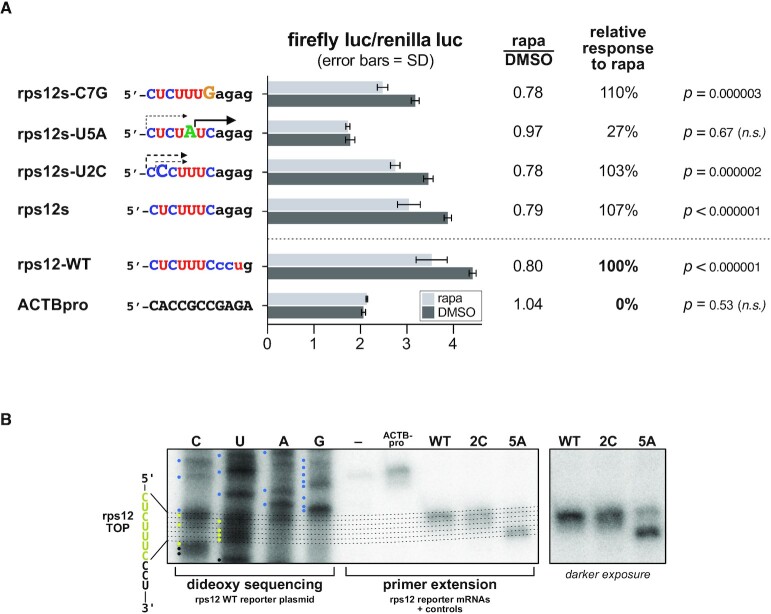
To test our TOP motif consensus motif analysis data, a TOP reporter system using a 588-bp DNA fragment encoding the human rpS12 promoter and adjacent 5′-UTR of rps12 was constructed. This DNA was incorporated into a dual luciferase plasmid to drive the transcription of an mRNA that begins with the first 11 nt of the rps12 mRNA, followed by a PEST-destabilized firefly ORF (the plasmid also harbours a second mRNA encoding Renilla luciferase driven by an SV40 promoter). Transfection of this reporter plasmid into HeLa cells (rps12-WT) or a control plasmid (ACTBpro), where transcription of a non-TOP firefly mRNA is driven by a 477-bp fragment of the human β-actin (ACTB) promoter, shows that firefly luciferase translation is specifically inhibited by the rps12 TOP sequence in the presence of 100 nM rapamycin (Figure 6A). To verify that the intended TSS of the rps12-WT firefly mRNA was indeed correct, we conducted RNA primer extension analysis of the WT reporter (and two single-nucleotide substitution reporters) and mapped the extension products to dideoxy sequencing lanes of the reporter DNA (Figure 6B).
Figure 6.
(A) Mutational analysis of the rps12 TOP motif using dual luciferase reporter assays. Firefly luciferase reporter mRNAs incorporating either the first 11 nt of the WT rps12 mRNA 5′-UTR or the indicated mutations were expressed in HeLa cells ± rapamycin. Firefly activity is normalized to the activity of a co-expressed non-TOP Renilla luciferase mRNA. (B) Primer extension assay to verify reporter mRNA TSSs. A common, 24-nt reverse complement primer to the 5′ end of the firefly luciferase ORF was used for both dideoxy sequencing of the WT rps12 reporter plasmid DNA and individual primer extension reactions of the indicated rps12 TOP reporter mRNAs and non-TOP control mRNA (ACTBpro).
To test whether the first seven pyrimidines of the rps12 mRNA are indeed sufficient for TOP functionality, the CCU at positions 8–10 of the rps12-WT reporter mRNA were substituted with AGA to produce the reporter rps12s. Firefly luciferase translation was repressed as potently from this reporter as for rps12-WT (all reporter data in Figure 6A). The seventh pyrimidine (7C) of the rps12s reporter is also dispensable for TOP activity, as its substitution with G in the reporter rps12s-C7G still confers TOP behaviour on the mRNA. Two additional single-nucleotide substitutions to the rps12s reporter were tested, rps12-U2C and rps12-U5A. Strikingly, while only 1 out of 52 TOPs in our analysis had a C at position 2, the rps12s-U2C reporter still functions as a TOP. Unfortunately, it was not possible to evaluate the effect of the U5A substitution, as most of transcription products for this reporter begin at +6 rather than at +1 (see Figure 6B).
Analysis of 5′-UTR length and nucleotide composition for eFT-508 capCLIP data
In total, 90 mRNAs were selected: the 45 mRNAs with the greatest reduction in eIF4E binding upon eIF4E phosphorylation, and correspondingly, the 45 mRNAs with the greatest relative increase in eIF4E binding upon eIF4E phosphorylation. All selected mRNAs were subject to a minimum mRNA abundance cut-off of 2.5 to remove low-confidence mRNAs (low tag coverage) from the analysis. Ensembl Biomart and the NCBI RefSeq collection were used to source the 5′-UTR sequence of all 90 mRNAs selected. For mRNAs where the database queries returned multiple mRNA isoforms (often due to the presence of multiple TSSs and/or alternative splicing in the 5′-UTR regions of these mRNAs), eFT-508 capCLIP tag data and HeLa CAGE data were viewed in IGV to determine which sequence best fits the observed capCLIP and CAGE data. When capCLIP data did not identify the precise pattern of 5′-UTR alternative exon usage in HeLa cells, all potential 5′-UTR isoforms were included, and the average 5′-UTR length used for subsequent calculations.
Dual luciferase reporter assays
Dual luciferase plasmids have been constructed from the vector backbone of Promega firefly luciferase vector pGL4.11, which provides a firefly luciferase ORF (plus PEST element). A fragment encoding the Renilla luciferase ORF (driven by an SV40 promoter) was subcloned from Promega vector pGL4.73 into the pGL4.11 backbone to allow the simultaneous expression of both luciferase proteins in mammalian cells.
To construct the TOP mRNA reporter vectors, a 588-bp DNA fragment encoding the human rps12 promoter and adjacent 5′-UTR of rps12 were cloned immediately upstream of the firefly luciferase ORF. Individual nucleotides of the rps12 TOP motif and adjacent 5′-UTR sequence were changed using PCR methods. To test how eIF4E phosphorylation affects the translational activity of mRNAs with specific short or long 5′-UTR sequences, a dual luciferase vector was constructed as above but using a fragment of the human ACTB promoter to drive transcription of the firefly luciferase ORF. MluI and BglII restriction sites were introduced to allow the insertion of specific sequences immediately downstream of the firefly mRNA TSS and immediately upstream of a common Kozak element preceding the firefly CDS. Individual short 5′-UTR sequences were synthesized as ssDNA oligonucleotides and converted into dsDNA using a common reverse primer and Klenow exo-polymerase. Long 5′-UTR sequences were PCR amplified from human genomic DNA.
Individual 5′-UTR reporter plasmids were transfected into HeLa cells using Lipofectamine 3000. For TOP mRNA analysis, transfected cells were treated with 100 nM rapamycin (in DMSO) for 2 h, or with DMSO alone. For the analysis of 5′-UTR reporters, after overnight serum starvation, cells were pre-treated for 1 h with eFT-508 (or DMSO only) and then stimulated with PMA for additional 1 or 8 h. All transfections were done in triplicate. Firefly and Renilla luciferase activities were measured in duplicate, and the data plotted as a normalized firefly/Renilla luciferase ratio.
Quantification and statistical analysis
Statistical parameters used in the bioinformatic analysis of capCLIP data are listed above in the ‘Bioinformatic analysis’ section; in general, statistically significant differences are defined by an FDR cut-off <0.05 and/or a P-value <0.05.
For the mRNA reporter assays of both TOP motif function and 5′-UTR length on translational activity, transfections were performed in triplicate for each unique TOP mRNA or 5′-UTR reporter plasmid. Firefly and Renilla luciferase protein activity for each transfection was assayed in duplicate; mean firefly and Renilla luciferase activities were background corrected using luciferase measurements from non-transfected cells. A firefly/Renilla luciferase activity ratio was then calculated for each transfection, and the mean value of the three independent transfections was plotted; error bars represent standard deviation.
In general, for all other plotted data, each individual figure indicates value of n, and all error bars represent the standard deviation of data. Except for specific bioinformatic analyses specified above, statistical analysis was conducted using GraphPad Prism, with a P-value <0.05 considered statistically significant. Statistical tests are indicated in individual methods where appropriate.
Resource availability
Plasmids generated in this study have been deposited to Addgene: pKJ-500 (#165025) and pKJ-501 (#165027).
RESULTS
Capture and identification of the eIF4E–mRNA interactome
Individual steps of the capCLIP method are depicted in Figure 1A and briefly summarized in the legend. Full details of these steps are found in the ‘Materials and Methods’ section, and a step-by-step protocol suitable for reproduction of the capCLIP method in the laboratory is provided in the Supplementary Data. In essence, capCLIP uses short-wavelength (∼254 nm) UV irradiation of living cells to create a ‘zero-order’ photo-cross-link between an mRNA’s m7G cap (9) and an adjacent residue in bound eIF4E. The cross-link stably captures the in vivo protein:RNA interaction and permits a rigorous biochemical co-purification of eIF4E with only those mRNA fragments that were initially photo-cross-linked to it.
capCLIP requires efficient and specific IP of eIF4E. As commercially available anti-eIF4E antibodies do not work in IP, we applied a CRISPR/Cas9 genome editing strategy (Figure 1B) to introduce a 3X-flag epitope tag at the ‘N-termini’ of the chromosomal EIF4E genes in HeLa cells. Clone F2G3, which exhibits homozygous expression of the 3X-flag-eIF4E (Figure 1C), was used for subsequent experiments; levels of eIF4E expression and phosphorylation were similar in WT and flag-eIF4E cells (Figure 1D).
Using capCLIP to probe the effects of mTORC1 inhibition on translation initiation
The mechanisms for mTORC1 regulation of TOP mRNA translation initiation are controversial, as several translation-related proteins serve as direct or indirect substrates of mTORC1, including the eIF4E-binding proteins [4E-BPs, reviewed in (1)] and LARP1 (25,26). mTORC1 phosphorylates 4E-BPs at multiple residues and hyperphosphorylated 4E-BPs cannot bind to eIF4E, allowing eIF4E to bind eIF4G and mediate translation initiation (27). While mTOR kinase inhibitors (mTOR-KIs) inhibit 4E-BP phosphorylation and consequently disrupt eIF4E–eIF4G binding (28), rapamycin, which indirectly impairs mTORC1’s function (29), generally has little effect on 4E-BP1 phosphorylation or eIF4E:eIF4G binding.
Inhibition of TOP mRNA translation by mTOR-KIs (and presumably rapamycin) was proposed to be mediated through binding of 4E-BPs to eIF4E (30), but it remained unclear how TOP mRNA specificity was achieved (10). Growing evidence suggests that LARP1 regulates TOP mRNA translation; inhibition of mTORC1 by mTOR-KIs or rapamycin leads to hypophosphorylation of LARP1, which then competes with eIF4E for binding specifically to the 5′ ends of TOP mRNAs (11,12). Recently, ribosome profiling (RP) revealed that LARP1 is essential for translational repression of TOP mRNAs during mTORC1 inhibition (31).
We first evaluated the consequences of mTORC1 inhibition on binding of flag-eIF4E to its protein partners in our engineered HeLa cells (see Figure 2A). Importantly, eIF4E–eIF4G binding was not affected by rapamycin, but was strongly decreased by AZD8055 (an mTOR-KI that completely inhibits mTORC1 activity). These data agree with our earlier findings for WT HeLa cells (10). We conclude that normal levels and regulation of eIF4E are preserved in our gene-edited cells.
We next verified that we could quantitatively immunoprecipitate flag-eIF4E from cell lysates under the high-detergent and high-salt conditions used in CLIP and that UV irradiation permitted the co-capture of RNA fragments with flag-eIF4E (Figure 2D and E). For preparative capCLIP, flag-eIF4E HeLa cells were treated for 2 h with rapamycin (or DMSO), UV cross-linked on ice and then processed through the remainder of the capCLIP methodology (see the ‘Materials and Methods’ section, capCLIP experimental data in Figure 2F and method summary in Figure 1A). A UMI linker was added to the cross-linked co-IP flag-eIF4E RNA fragments, which were converted to cDNA and used to prepare indexed PCR libraries that were combined for high-throughput sequencing (see the ‘Materials and Methods’ section).
capCLIP specifically captures mRNA 5′ ends
Filtered, deduplicated capCLIP tag reads were mapped to the hg19 reference genome. Peak calling software was then used to create peak data for each replicate (see the ‘Materials and Methods’ section for bioinformatic analyses). Figure 3 displays key peak statistics for the experimental replicates, showing that the majority of peak calls in all replicates align within spliced mRNA sequences. The largest peak fraction in each sample (representing 34–47% of all peak calls) maps to the first exon of the mRNA (the 5′-UTR), consistent with our expectation that UV irradiation generates cross-links between eIF4E and the m7GTP moiety of each mRNA. Critically, these 5′-UTR peak sets both identify the mRNAs bound by eIF4E and provide quantitative data on how binding of eIF4E to each mRNA species is altered under each experimental condition. While the peak data generally conform to our expectations, we were somewhat puzzled by the (small) number of peaks in ncRNAs, particularly in snoRNAs and snRNAs (which are highly enriched with respect to their small fraction of the genome). Investigation of these peaks revealed that all had a common 5′-AGAU-3′ at their 5′ ends, which has been identified as a common mis-priming artefact of CLIP methods (32).
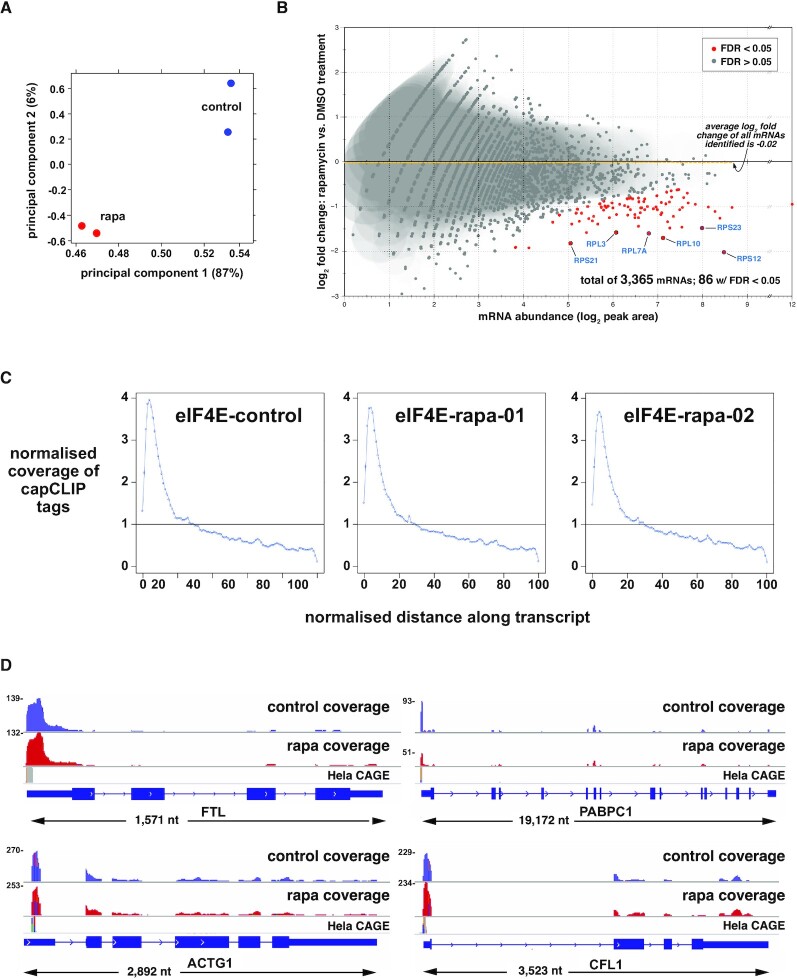
Principal component analysis (PCA) of the four dataset replicates (Figure 4A) indicates that rapamycin leads to a significant and consistent change in the composition of the peak data. DiffBind (22) was used to identify those mRNAs whose association with eIF4E is significantly altered by rapamycin. 5′-UTR peak data for the replicates specify the eIF4E cap-ome as comprising 3372 individual mRNAs. Figure 4B plots each mRNA as a function of its abundance (log2 of peak area; x-axis) and the log2 fold change of its binding to eIF4E in response to rapamycin (y-axis). As a population (dashed yellow line; log2 fold change = 0.02), the eIF4E cap-ome is essentially unchanged upon rapamycin treatment and this is also true for the abundance of the vast majority of individual mRNAs (depicted as grey spots). However, rapamycin significantly decreases the association with eIF4E of 86 mRNAs (red spots, log2 fold-change ≶0.50; P-value <0.01; FDR ≤ 0.05). About two-thirds of these were previously identified (30,33) as ones whose translation is impaired by mTOR-KIs (or rapamycin). Additional details are available in Supplementary Table S1.
Figure 4.
capCLIP of rapamycin-treated and control HeLa cells. (A) PCA of the control and rapamycin sequencing datasets. (B) Log2 fold change in eIF4E affinity for 3365 individual mRNAs versus mRNA abundance (log2 of peak area). Eighty-six mRNAs show a statistically significant log2 fold change in eIF4E binding (P > 0.01; FDR > 0.05). Average log2 fold change in eIF4E binding of the total mRNA population is −0.02. (C) Distribution of capCLIP tags as a function of the normalized distance along the transcript for the single control (DMSO) replicate and two rapamycin (rapa) replicates. (D) IGV gene-level views, reformatted to aid visualization (all genes displayed 5′ to 3′, left to right). Displayed above each gene are histogram plots of control (in blue) and rapamycin (in red) capCLIP tag alignments, along with HeLa CAGE data (in grey). Numbers to the left of each tag histogram indicate raw tag counts at the highest point of each 5′-UTR tag peak.
In Figure 4C, capCLIP tag coverage from the control and two rapamycin datasets for all tags aligning to processed (spliced) mRNA is plotted over a normalized mRNA ‘length space’. All three plots show robust enrichment of tags at the extreme 5′ end of the mRNAs, consistent with the mapping data in Figure 3, and demonstrating that capCLIP functions as intended. Figure 4D displays capCLIP tag coverage on gene-level IGV (23) representations for FTL, PABPC1, CFL1 and ACTG1 mRNAs, and histograms of the control and rapamycin-treated capCLIP tag data mapping to them. Figure 4D also shows HeLa CAGE (34,35) data for each mRNA, revealing that the 5′-boundary of each mRNA’s capCLIP tag data aligns with the corresponding CAGE-identified TSS.
Rapamycin specifically depletes TOP mRNAs from the eIF4E cap-ome
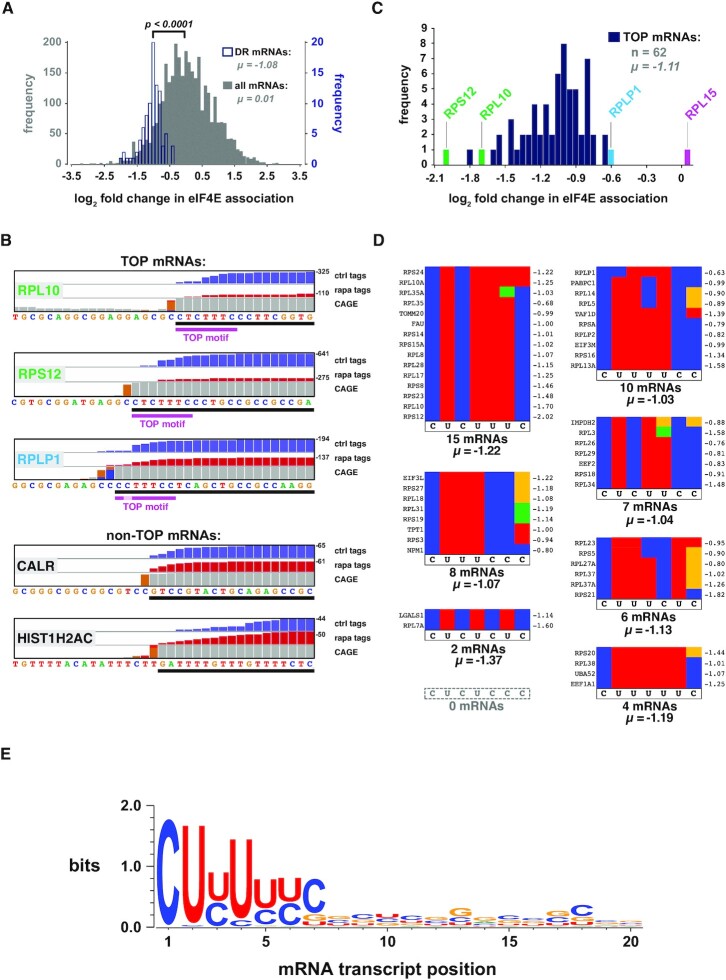
Figure 5A shows a frequency plot depicting both the 86 mRNAs whose eIF4E association is significantly altered by rapamycin [‘differentially regulated (DR) mRNAs’, in blue] and all mRNAs identified by capCLIP (in grey), arranged according to the fold change in eIF4E association caused by rapamycin. While the overall shift in eIF4E association of the total population of mRNAs is essentially 0 (μ = 0.01), there is an average 2-fold decrease among the DR mRNAs (μ = −1.08; P < 0.0001). Using CAGE data to verify the precise TSS for each of the DR mRNAs reveals that 61 of them (72%) possess functional TOPs (initial C, followed by 5–14 pyrimidines) (36) (Supplementary Table S1). An additional 13 appear to have a TOP TSS and at least one non-TOP TSS. The remaining 11 DR mRNAs seem to lack any identifiable TOP motif. capCLIP also identifies two novel TOP mRNAs in HeLa cells, TAF1D and LGALS1. TAF1D is an essential component of the SL1 complex that regulates transcription of the major ribosomal RNAs (37), which is driven by mTORC1. This may provide a further way in which mTORC1 signalling promotes ribosome biogenesis (38). We are not aware of any link between LGALS1 (galectin-1; a secreted lectin) and mTORC1 signalling.
Figure 5.
Rapamycin specifically depletes TOP mRNAs from the eIF4E cap-ome. (A) Frequency plot of the log2 fold change in eIF4E association upon treatment with rapamycin. All peak-called mRNAs (grey) and the 86 DR mRNAs (blue) that show a statistically significant change with drug (fold change ← 0.5 or >0.5; FDR < 0.05; P < 0.01). (B) IGV plots of the extreme mRNA 5′ ends of five mRNAs; views are reformatted to run 5′ to 3′, left to right, to aid visualization. The mRNA (black bar) is depicted below the sequence (the 5′-most nucleotide is the TSS). Histogram plots of control (blue) and rapamycin-treated (red) raw capCLIP tag data, along with HeLa CAGE data (grey and orange) are shown above the sequence. In general, the first grey bar 5′ of an orange bar in the CAGE histogram denotes the TSS (34). The 7-nt TOP consensus motif is indicated with a magenta bar. (C) Log2 fold change in eIF4E association for the 62 TOP mRNAs identified by capCLIP. (D) Sorting of the 52 TOP mRNAs into seven of eight possible sequence permutations of the 7-nt TOP consensus motif. Individual sequences are depicted as coloured strips (red = U, blue = C, green = A and yellow = G). Gene name is shown left of each strip; log2 fold change value (μ) upon rapamycin treatment is shown at right. (E) WebLogo analysis of the first 20 nucleotides of 52 capCLIP-identified TOP mRNAs.
Rapamycin-sensitive mRNAs possess a strikingly well-defined TOP motif
Nucleotide-level views of three TOP and two non-TOP mRNA 5′ ends (Figure 5B), displaying normalized control and rapamycin capCLIP tag histograms (along with HeLa CAGE data), clearly indicate the specific loss of TOP mRNAs from the eIF4E cap-ome following rapamycin treatment and show the consistent tag coverage provided by capCLIP at each mRNA’s extreme 5′ end. To assess whether the capCLIP log2 fold change data for the 62 TOP mRNAs (Figure 5C) provide deeper insight into features that define a functional TOP motif, TOP mRNAs were allocated into seven unique permutation groups (Figure 5D) (excluding 10 mRNAs, as we could not use CAGE and capCLIP data to unambiguously identify their TSSs). While there is no significant difference in each group’s mean-fold rapamycin-induced change in eIF4E binding, suggesting that all represented permutations are equally functional, we note that all 52 sequences are devoid of purines at positions 1–4 of the TOP and only 3/52 mRNAs have a purine at position 5/66 [IMPDH2 (G5), RPL3 (A5) and RPL35A (A6); Figure 5D]. In contrast, a purine at position 7 of the motif (seen in 13/52 mRNAs) clearly does not abrogate TOP function. Our findings are in close agreement with those of Philippe et al. (31) who, by analysing which mRNAs are targets for control by LARP1 by RP, also concluded that a C at position +1 and a stretch of six pyrimidines suffice to confer control by mTORC1 on the translation of specific mRNAs in a LARP1-dependent manner.
capCLIP data should allow us to derive a ‘functional’ TOP mRNA consensus motif. The first 20 nucleotides of the 52 TOP mRNAs were therefore inputted into WebLogo 3 (39) (Figure 5E). The results show that functional TOP mRNAs invariably have a C base at position 1 (C1) consistent with earlier work. They also suggest optimal TOP motifs possess other conserved features. First, TOP motifs are at most seven pyrimidines in length, positioned directly at the 5′ end, with absolutely no preference for additional pyrimidines (or indeed any specific base) at positions 8–20. Second, there are strong-to-mild preferences for a pyrimidine at three other positions: U2, U4 and C7 (Figure 5E). Thus, our analysis reveals the 7-nt sequence 5′-CUYUYYC-3′ as the consensus TOP motif and that functional TOPs are significantly shorter than previously suggested (36). Luciferase reporter assays validated the functionality of both 7- and 6-nt TOP motifs (Figure 6A and B), further supporting the effectiveness of capCLIP as a novel method to probe mRNA translational behaviour.
Together, these data indicate that capCLIP successfully identifies the cellular eIF4E cap-ome. Bioinformatic analysis of capCLIP tag data indicates that capCLIP replicates the finding that rapamycin specifically inhibits TOP mRNA translation and demonstrates that this is associated with decreased binding of such mRNAs to eIF4E. Our results are consistent with the recent model whereby, when hypophosphorylated (due to mTORC1 inhibition), LARP1 regulates TOP mRNA translation by competing with eIF4E for binding to their caps.
Furthermore, it is also clear that the nature and magnitude of the effects of rapamycin on the eIF4E cap-ome are very similar to those obtained from RP experiments measuring the impact of mTOR-KIs on translational efficiency (30,33). While the mTOR-KIs are much more potent inhibitors of mTORC1 activity than rapamycin, the ∼3400 mRNAs identified by rapamycin capCLIP compare favourably to the ∼4800 mRNAs captured by RP with the inhibitor Torin-1 (30). Furthermore, to better understand whether eIF4E capCLIP and RP have similar sensitivity for detecting the translational effects of rapamycin on individual mRNA species, we plotted (in Supplementary Figure S1) the fold change in eIF4E–cap binding versus the fold change in TE for the 45 TOP mRNAs common to both the DR capCLIP target list (P < 0.01; FDR < 0.05) and the RP TE data of Hsieh et al. [(33); FDR and P-values not published]. Spearman r-value analysis indicates that there is a significant correlation (r = 0.3216; P = 0.031) between eIF4E binding and TE. As translational repression of TOP mRNAs is likely driven by the specific displacement of eIF4E from TOP mRNA caps by LARP1 (see below), this similarity is unsurprising. However, it is critical to note that capCLIP and RP may not yield similar results in other biological contexts where translation is regulated independently of eIF4E–mRNA binding. This is because numerous additional control mechanisms regulate translation initiation, including stem–loop structures and upstream ORFs, cap-independent internal ribosome entry sites and regulatory processes that affect the elongation phase of translation.
Using capCLIP to probe the effect of phosphorylation of eIF4E on its cap-ome
We next used capCLIP to study a long-standing translational ‘mystery’: the effect of phosphorylation of eIF4E on its association with (specific) mRNAs. The flag-eIF4E HeLa cells exhibit essentially identical levels of eIF4E phosphorylation to WT HeLa cells (Figure 1D). Treating serum-starved cells with PMA, a potent activator of ERK signalling and thus MNK activity (40), rapidly increased eIF4E phosphorylation (lanes 2–6, Figure 2B). To obtain cells with undetectable P-eIF4E as controls, we used the highly specific MNK inhibitor eFT-508 (18) at 0.1 μM, the lowest effective dose.
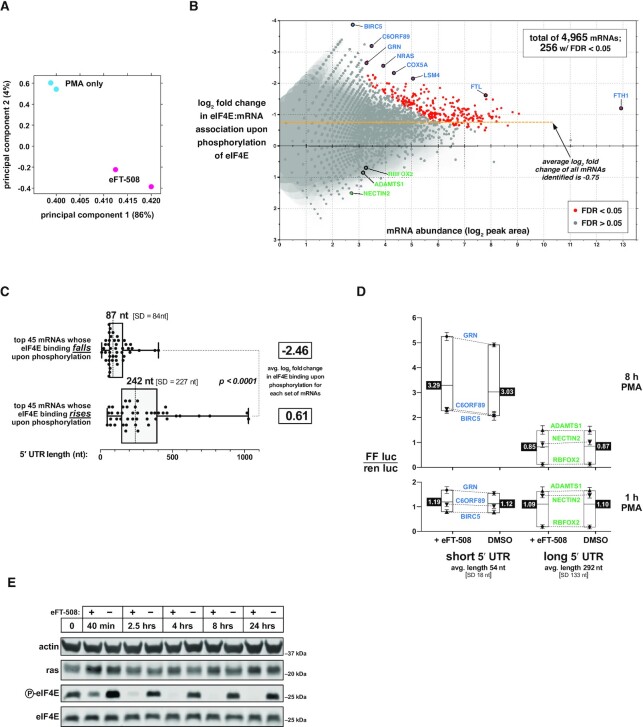
For capCLIP, serum-starved flag-eIF4E HeLa cells were treated with eFT-508 or DMSO for 1 h, and then with PMA for 30 min, followed by an essentially identical work-up to that for the rapamycin capCLIP study (see Figure 2B and C for determination of optimal PMA stimulation time and eFT-508 concentration, and Figure 2F and ‘Materials and Methods’ section for capCLIP itself). Raw sequencing reads were aligned and used to define peak regions with FDR < 0.05. PCA of the four dataset replicates (Figure 7A) indicates that eFT-508 pre-treatment causes a significant and consistent change in the composition of the peak data. In total, capCLIP identified 5′-UTR peaks for 4965 unique mRNAs, 256 of which exhibited statistically significant decreases in eIF4E binding upon eFT-508 treatment (FDR < 0.05) (full list in Supplementary Table S2); Figure 7B plots each mRNA’s abundance versus the change in eIF4E binding with eFT-508 (left y-axis). Thus, there is an extensive subset of mRNAs that is particularly sensitive to phosphorylation of eIF4E. Interestingly, the entire population of 4965 mRNAs exhibits a modest reduction in eIF4E binding, with a log2 fold change in eIF4E binding of −0.75 (Figure 7B, dashed yellow line). This finding is consistent with in vitro biophysical studies demonstrating that phosphorylation of eIF4E lowers its affinity for the cap (41–43).
Figure 7.
mRNAs with short 5′-UTR are depleted from the cap-ome upon phosphorylation of eIF4E. (A) PCA of the two control (PMA-only) DNA sequencing datasets and two eFT-508 pre-treated (eFT-508) DNA sequencing datasets. (B) Plot of mRNA abundance (x-axis; log2 of the capCLIP peak area) versus the log2 fold change in eIF4E binding upon eIF4E phosphorylation (left y-axis). Two hundred fifty-six mRNAs (out of a total of 4965 mRNAs) exhibit a significant change in log2 fold change binding to eIF4E upon phosphorylation (P > 0.01; FDR > 0.05). (C) 5′-UTR length distribution for the 45 mRNAs with the greatest reduction (top) or the greatest apparent increase (bottom) in eIF4E binding upon eIF4E phosphorylation. (D) Dual luciferase assays to measure the effects of reporter mRNA 5′-UTR length on translational output ± eFT-508. Individual reporter mRNAs and treatments are described in the text and ‘Materials and Methods’ section. (E) Western blots of ras protein levels in WT HeLa cells treated with DMSO only or 100 nM eFT-508 for indicated times. Inhibition of eIF4E phosphorylation by eFT-508 was monitored with anti-P-eIF4E antibody.
To determine whether the alteration in an mRNA’s binding to eIF4E upon its phosphorylation (either up or down) correlates with specific sequence features within its 5′-UTR, we focused our subsequent analyses on the top 45 mRNAs with the greatest fall in eIF4E binding upon phosphorylation, along with a second set of 45 mRNAs that exhibited the greatest rise in eIF4E binding upon phosphorylation (Figure 7C). The mean log2 fold change in eIF4E binding of the ‘rising’ group is just 0.61, while that of the ‘falling’ group is −2.46; accordingly, we term them, respectively, ‘P-eIF4E-insensitive’ and ‘P-eIF4E-sensitive’ mRNAs. The median 5′-UTR length of 242 nt (SD = 227) and GC content of 58% of the former group are indistinguishable from the average for human 5′-UTRs [either 293 nt (SD = 27) (UTRdb) or 218 nt (NCBI RefSeq), and 58% GC using UTRdb] (44,45). In contrast, 5′-UTRs of P-eIF4E-sensitive mRNAs are significantly shorter than average [median length 87 nt (SD = 84)] and more GC-rich (68% GC). Interestingly, ∼25% of the P-eIF4E-sensitive mRNAs are TOP mRNAs, consistent with their shorter than average 5′-UTR length.
These data suggest that phosphorylation of eIF4E binding particularly decreases its binding to mRNAs defined by short, GC-rich 5′-UTRs, which would be expected to impair their translation. To test this, we made a series of six reporters in which the firefly luciferase ORF is preceded by 5′-UTR sequences chosen from either P-eIF4E-sensitive and P-eIF4E-insensitive mRNAs; these reporters also contain a Renilla luciferase ORF, for normalization, and were transfected into WT HeLa cells. Serum-starved cells were pre-treated for 1 h with eFT-508 or DMSO, and then stimulated with PMA for 1 or 8 h, followed by lysis and measurement of luciferase activities. Data were plotted as normalized firefly/Renilla activity ratios (Figure 7D). The sets of P-eIF4E-sensitive and P-eIF4E-insensitive 5′-UTR reporters show a striking difference in firefly protein output after 8 h PMA treatment, which suggests that short 5′-UTR mRNAs have a significantly higher intrinsic translational efficiency. As the Kozak and coding sequences of all reporters are identical, the data imply that the short 5′-UTR reporters complete translation initiation faster than long 5′-UTR reporters. Indeed, such a finding was recently made using TCP-seq in human cells, including HeLa cells (46–48). Our data also indicate that inhibiting eIF4E phosphorylation specifically reduces the translational output from short 5′-UTR reporters, but this effect was not statistically significant.
The mRNA for NRAS (a Ras family member mutated in some cancers) is a P-eIF4E-sensitive mRNA. We also therefore assessed the effect of eIF4E phosphorylation on levels of NRAS (Figure 7E). We observed a small (∼1.8-fold) increase in steady-state NRAS protein levels after 4 or 8 h of MNK inhibition. Thus, the luciferase reporter and NRAS western experiments provide a modest indication that translation of short 5′-UTR mRNAs is repressed by eIF4E phosphorylation; this merits further exploration.
Ours is the first study to examine the effects of phosphorylation of eIF4E on its binding to specific mRNAs in a cellular context. Further validation of our data and model requires methods that provide direct quantification of de novo rates of synthesis of specific polypeptides such as pulsed stable-isotope labelling technology (49,50).
DISCUSSION
We envision that capCLIP will be developed into a general technique to acquire deep, quantitative information on a cell’s or tissue’s eIF4E cap-ome, and may be particularly attractive where the introduction of a flag tag on eIF4E through genome editing can also serve to distinguish a cell population of interest from other cells or tissues; for example, the effects of tumorigenesis or anti-tumour agents on a cancer cell’s cap-ome could be specifically measured in a mouse tumorigenesis model using human patient-derived cancer cell lines that have been edited to express flag-eIF4E. An especially promising feature of capCLIP in this context is its ability to report on the translational status of mRNAs specifically in metastatic cancer cells without needing to remove cells from the surrounding tumour stroma. As such an application is not amenable to RP or direct proteomic approaches (unless they are specifically altered to differentiate tumour from stroma), capCLIP offers a novel way to expand our still limited understanding of translational regulation in cancer cells in situ, as well as in other disease settings where dysregulation of translation is implicated.
Similarly, capCLIP also provides a unique means to elucidate the specific biological role(s) and control of other cytoplasmic cap-binding proteins, particularly eIF4E2 and eIF4E3, whose roles in translational control remain poorly understood. Here again, the addition by genome editing of a flag tag to a specific alternative cap-binding protein such as eIF4E2 provides the means to specifically identify that protein’s (likely much more limited) cap-ome in a ‘wild-type’ cellular context where the major eIF4E cap-binding protein remains fully functional (as there is no need to knock out/down other cap-binding proteins). As eIF4E2 is generally thought to function in translational repression [(51,52); although it may promote mRNA translation in certain settings (53)], an eIF4E2 capCLIP experiment may therefore primarily identify a translationally repressed cap-ome. Likewise, capCLIP of LARP1, or of the nuclear cap-binding protein NCBP2, will also likely identify groups of repressed mRNAs given the known biology of these proteins.
Finally, it is important to reiterate that while our eIF4E capCLIP experiments primarily identified translationally active mRNAs (given eIF4E’s essential role in most all cap-dependent translation), the capCLIP methodology does not intrinsically measure translational activity but merely the binding between a specific cap-binding protein and specific cellular mRNAs. Hence, while RP and capCLIP approaches yield similar data for investigations of the effects of mTOR inhibition on eIF4E’s cap-ome, this similarity is likely limited to this specific case. As we believe capCLIP’s greatest potential lies in revealing the full biological roles of alternative cap-binding proteins, capCLIP occupies a separate, but equally powerful, methodological niche from existing high-throughput translational profiling techniques.
DATA AVAILABILITY
The raw sequencing files for the rapamycin capCLIP and eFT-508 capCLIP samples, along with processed peak, .bed and .tsv files for each sample, have been uploaded to the Gene Expression Omnibus repository at the NCBI under the accession number GSE138473.
Supplementary Material
Contributor Information
Kirk B Jensen, Lifelong Health, South Australian Health and Medical Research Institute, North Terrace, Adelaide, SA 5000, Australia; School of Biological Sciences, Faculty of Sciences, University of Adelaide, Adelaide, SA 5005, Australia.
B Kate Dredge, Centre for Cancer Biology, SA Pathology and University of South Australia, Adelaide, SA 5000, Australia.
John Toubia, Centre for Cancer Biology, SA Pathology and University of South Australia, Adelaide, SA 5000, Australia; ACRF Cancer Genomics Facility, Centre for Cancer Biology, SA Pathology and University of South Australia, Frome Road, Adelaide, SA 5000, Australia.
Xin Jin, Lifelong Health, South Australian Health and Medical Research Institute, North Terrace, Adelaide, SA 5000, Australia; School of Medicine and Pharmacy, Ocean University of China, 5 Yushan Road, Qingdao 266003, China.
Valentina Iadevaia, School of Biosciences and Medicine, University of Surrey, Guildford, Surrey GU2 7XH, UK.
Gregory J Goodall, School of Biological Sciences, Faculty of Sciences, University of Adelaide, Adelaide, SA 5005, Australia; Centre for Cancer Biology, SA Pathology and University of South Australia, Adelaide, SA 5000, Australia; Adelaide Medical School, Faculty of Health and Medical Sciences, University of Adelaide, Adelaide, SA 5005, Australia.
Christopher G Proud, Lifelong Health, South Australian Health and Medical Research Institute, North Terrace, Adelaide, SA 5000, Australia; School of Biological Sciences, Faculty of Sciences, University of Adelaide, Adelaide, SA 5005, Australia.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council [GNT1089167, GNT1118170 and GTN1126711 to G.J.G.]; South Australian Health and Medical Research Institute (SAHMRI) [to C.G.P.]. Funding for open access charge: SAHMRI.
Conflict of interest statement. None declared.
REFERENCES
- 1. Proud C.G. Phosphorylation and signal transduction pathways in translational control. Cold Spring Harb. Perspect. Biol. 2019; 11:a033050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Merrick W.C., Pavitt G.D.. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb. Perspect. Biol. 2018; 10:a033092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rhoads R.E. eIF4E: new family members, new binding partners, new roles. J. Biol. Chem. 2009; 284:16711–16715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Merrick W.C. eIF4F: a retrospective. J. Biol. Chem. 2015; 290:24091–24099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Truitt M.L., Conn C.S., Shi Z., Pang X., Tokuyasu T., Coady A.M., Seo Y., Barna M., Ruggero D.. Differential requirements for eIF4E dose in normal development and cancer. Cell. 2015; 162:59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pelletier J., Schmeing T.M., Sonenberg N.. The multifaceted eukaryotic cap structure. Wiley Interdiscip. Rev. RNA. 2021; 12:e1636. [DOI] [PubMed] [Google Scholar]
- 7. Ule J., Jensen K.B., Ruggiu M., Mele A., Ule A., Darnell R.B.. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003; 302:1212–1215. [DOI] [PubMed] [Google Scholar]
- 8. Ule J., Jensen K., Mele A., Darnell R.B.. CLIP: a method for identifying protein–RNA interaction sites in living cells. Methods. 2005; 37:376–386. [DOI] [PubMed] [Google Scholar]
- 9. Pelletier J., Sonenberg N.. Photochemical cross-linking of cap binding proteins to eucaryotic mRNAs: effect of mRNA 5′ secondary structure. Mol. Cell. Biol. 1985; 5:3222–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huo Y., Iadevaia V., Yao Z., Kelly I., Cosulich S., Guichard S., Foster L.J., Proud C.G.. Stable isotope-labelling analysis of the impact of inhibition of the mammalian target of rapamycin on protein synthesis. Biochem. J. 2012; 444:141–151. [DOI] [PubMed] [Google Scholar]
- 11. Philippe L., Vasseur J.J., Debart F., Thoreen C.C.. La-related protein 1 (LARP1) repression of TOP mRNA translation is mediated through its cap-binding domain and controlled by an adjacent regulatory region. Nucleic Acids Res. 2018; 46:1457–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lahr R.M., Fonseca B.D., Ciotti G.E., Al-Ashtal H.A., Jia J.J., Niklaus M.R., Blagden S.P., Alain T., Berman A.J.. La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. eLife. 2017; 6:e24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Proud C.G. Mnks, eIF4E phosphorylation and cancer. Biochim. Biophys. Acta. 2015; 1849:766–773. [DOI] [PubMed] [Google Scholar]
- 14. Xie J., Merrett J.E., Jensen K.B., Proud C.G.. The MAP kinase-interacting kinases (MNKs) as targets in oncology. Expert Opin. Ther. Targets. 2019; 23:187–199. [DOI] [PubMed] [Google Scholar]
- 15. Furic L., Rong L., Larsson O., Koumakpayi I.H., Yoshida K., Brueschke A., Petroulakis E., Robichaud N., Pollak M., Gaboury L.A.et al.. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl Acad. Sci. U.S.A. 2010; 107:14134–14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E.. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR–Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016; 34:339–344. [DOI] [PubMed] [Google Scholar]
- 17. Renaud J.B., Boix C., Charpentier M., De Cian A., Cochennec J., Duvernois-Berthet E., Perrouault L., Tesson L., Edouard J., Thinard R.et al.. Improved genome editing efficiency and flexibility using modified oligonucleotides with TALEN and CRISPR–Cas9 nucleases. Cell Rep. 2016; 14:2263–2272. [DOI] [PubMed] [Google Scholar]
- 18. Reich S.H., Sprengeler P.A., Chiang G.G., Appleman J.R., Chen J., Clarine J., Eam B., Ernst J.T., Han Q., Goel V.K.et al.. Structure-based design of pyridone–aminal eFT508 targeting dysregulated translation by selective mitogen-activated protein kinase interacting kinases 1 and 2 (MNK1/2) inhibition. J. Med. Chem. 2018; 61:3516–3540. [DOI] [PubMed] [Google Scholar]
- 19. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith T., Heger A., Sudbery I.. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 2017; 27:491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B.. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008; 456:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R.et al.. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012; 481:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., Hilton J.A., Jain K., Baymuradov U.K., Narayanan A.K.et al.. The Encyclopedia of DNA Elements (ENCODE): data portal update. Nucleic Acids Res. 2018; 46:D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fonseca B.D., Zakaria C., Jia J.J., Graber T.E., Svitkin Y., Tahmasebi S., Healy D., Hoang H.D., Jensen J.M., Diao I.T.et al.. La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1). J. Biol. Chem. 2015; 290:15996–16020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hong S., Freeberg M.A., Han T., Kamath A., Yao Y., Fukuda T., Suzuki T., Kim J.K., Inoki K.. LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. eLife. 2017; 6:e25237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gingras A.C., Raught B., Sonenberg N.. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001; 15:807–826. [DOI] [PubMed] [Google Scholar]
- 28. Choo A.Y., Yoon S.O., Kim S.G., Roux P.P., Blenis J.. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc. Natl Acad. Sci. U.S.A. 2008; 105:17414–17419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aylett C.H., Sauer E., Imseng S., Boehringer D., Hall M.N., Ban N., Maier T.. Architecture of human mTOR complex 1. Science. 2016; 351:48–52. [DOI] [PubMed] [Google Scholar]
- 30. Thoreen C.C., Chantranupong L., Keys H.R., Wang T., Gray N.S., Sabatini D.M.. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012; 485:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Philippe L., van den Elzen A.M.G., Watson M.J., Thoreen C.C.. Global analysis of LARP1 translation targets reveals tunable and dynamic features of 5′ TOP motifs. Proc. Natl Acad. Sci. U.S.A. 2020; 117:5319–5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gillen A.E., Yamamoto T.M., Kline E., Hesselberth J.R., Kabos P.. Improvements to the HITS-CLIP protocol eliminate widespread mispriming artifacts. BMC Genomics. 2016; 17:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hsieh A.C., Liu Y., Edlind M.P., Ingolia N.T., Janes M.R., Sher A., Shi E.Y., Stumpf C.R., Christensen C., Bonham M.J.et al.. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012; 485:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kodzius R., Kojima M., Nishiyori H., Nakamura M., Fukuda S., Tagami M., Sasaki D., Imamura K., Kai C., Harbers M.et al.. CAGE: cap analysis of gene expression. Nat. Methods. 2006; 3:211–222. [DOI] [PubMed] [Google Scholar]
- 35. Takahashi H., Lassmann T., Murata M., Carninci P.. 5′ end-centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat. Protoc. 2012; 7:542–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meyuhas O., Kahan T.. The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Acta. 2015; 1849:801–811. [DOI] [PubMed] [Google Scholar]
- 37. Gorski J.J., Pathak S., Panov K., Kasciukovic T., Panova T., Russell J., Zomerdijk J.C.. A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. EMBO J. 2007; 26:1560–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iadevaia V., Liu R., Proud C.G.. mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin. Cell Dev. Biol. 2014; 36:113–120. [DOI] [PubMed] [Google Scholar]
- 39. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang X., Flynn A., Waskiewicz A.J., Webb B.L., Vries R.G., Baines I.A., Cooper J.A., Proud C.G.. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses, and cytokines is mediated by distinct MAP kinase pathways. J. Biol. Chem. 1998; 273:9373–9377. [DOI] [PubMed] [Google Scholar]
- 41. Scheper G.C., van Kollenburg B., Hu J., Luo Y., Goss D.J., Proud C.G.. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J. Biol. Chem. 2002; 277:3303–3309. [DOI] [PubMed] [Google Scholar]
- 42. Slepenkov S.V., Darzynkiewicz E., Rhoads R.E.. Stopped-flow kinetic analysis of eIF4E and phosphorylated eIF4E binding to cap analogs and capped oligoribonucleotides: evidence for a one-step binding mechanism. J. Biol. Chem. 2006; 281:14927–14938. [DOI] [PubMed] [Google Scholar]
- 43. Zuberek J., Wyslouch-Cieszynska A., Niedzwiecka A., Dadlez M., Stepinski J., Augustyniak W., Gingras A.C., Zhang Z., Burley S.K., Sonenberg N.et al.. Phosphorylation of eIF4E attenuates its interaction with mRNA 5′ cap analogs by electrostatic repulsion: intein-mediated protein ligation strategy to obtain phosphorylated protein. RNA. 2003; 9:52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Leppek K., Das R., Barna M.. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018; 19:158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grillo G., Turi A., Licciulli F., Mignone F., Liuni S., Banfi S., Gennarino V.A., Horner D.S., Pavesi G., Picardi E.et al.. UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2010; 38:D75–D80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Archer S.K., Shirokikh N.E., Beilharz T.H., Preiss T.. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature. 2016; 535:570–574. [DOI] [PubMed] [Google Scholar]
- 47. Shirokikh N.E., Archer S.K., Beilharz T.H., Powell D., Preiss T.. Translation complex profile sequencing to study the in vivo dynamics of mRNA–ribosome interactions during translation initiation, elongation and termination. Nat. Protoc. 2017; 12:697–731. [DOI] [PubMed] [Google Scholar]
- 48. Bohlen J., Fenzl K., Kramer G., Bukau B., Teleman A.A.. Selective 40S footprinting reveals cap-tethered ribosome scanning in human cells. Mol. Cell. 2020; 79:561–574. [DOI] [PubMed] [Google Scholar]
- 49. Kenney J.W., Genheden M., Moon K.M., Wang X., Foster L.J., Proud C.G.. Eukaryotic elongation factor 2 kinase regulates the synthesis of microtubule-related proteins in neurons. J. Neurochem. 2016; 136:276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu R., Kenney J.W., Manousopoulou A., Johnston H.E., Kamei M., Woelk C.H., Xie J., Schwarzer M., Garbis S.D., Proud C.G.. Quantitative non-canonical amino acid tagging (QuaNCAT) proteomics identifies distinct patterns of protein synthesis rapidly induced by hypertrophic agents in cardiomyocytes, revealing new aspects of metabolic remodeling. Mol. Cell. Proteomics. 2016; 15:3170–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jafarnejad S.M., Chapat C., Matta-Camacho E., Gelbart I.A., Hesketh G.G., Arguello M., Garzia A., Kim S.H., Attig J., Shapiro M.et al.. Translational control of ERK signaling through miRNA/4EHP-directed silencing. eLife. 2018; 7:e35034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Peter D., Weber R., Sandmeir F., Wohlbold L., Helms S., Bawankar P., Valkov E., Igreja C., Izaurralde E.. GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression. Genes Dev. 2017; 31:1147–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Uniacke J., Holterman C.E., Lachance G., Franovic A., Jacob M.D., Fabian M.R., Payette J., Holcik M., Pause A., Lee S.. An oxygen-regulated switch in the protein synthesis machinery. Nature. 2012; 486:126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing files for the rapamycin capCLIP and eFT-508 capCLIP samples, along with processed peak, .bed and .tsv files for each sample, have been uploaded to the Gene Expression Omnibus repository at the NCBI under the accession number GSE138473.