Abstract
In order to achieve efficient therapeutic post-transcriptional gene-silencing mediated by the RNA interference (RNAi) pathway, small interfering RNAs (siRNAs) must be chemically modified. Several supra-RNA structures, with the potential to stabilize siRNAs metabolically have been evaluated for their ability to induce gene silencing, but all have limitations or have not been explored in therapeutically relevant contexts. Covalently closed circular RNA transcripts are prevalent in eukaryotes and have potential as biomarkers and disease targets, and circular RNA mimics are being explored for use as therapies. Here we report the synthesis and evaluation of small circular interfering RNAs (sciRNAs). To synthesize sciRNAs, a sense strand functionalized with the trivalent N-acetylgalactosamine (GalNAc) ligand and cyclized using ‘click’ chemistry was annealed to an antisense strand. This strategy was used for synthesis of small circles, but could also be used for synthesis of larger circular RNA mimics. We evaluated various sciRNA designs in vitro and in vivo. We observed improved metabolic stability of the sense strand upon circularization and off-target effects were eliminated. The 5′-(E)-vinylphosphonate modification of the antisense strand resulted in GalNAc-sciRNAs that are potent in vivo at therapeutically relevant doses. Physicochemical studies and NMR-based structural analysis, together with molecular modeling studies, shed light on the interactions of this novel class of siRNAs, which have a partial duplex character, with the RNAi machinery.
INTRODUCTION
Circular RNAs (circRNAs) are covalently closed transcripts that we now know are highly prevalent and conserved in eukaryotes (1–3). Viruses also encode circRNAs that appear to be involved in disease pathogenesis (4). In eukaryotes, circRNAs are formed by backsplicing of linear RNA transcripts. Although these circular molecules were discovered >40 years ago, they were largely ignored until recently. There is now convincing evidence that circRNAs are not just byproducts of the splicing process. Certain circRNAs clearly have regulatory functions. For example, circRNAs initiate a cascade of reactions leading to macrophage activation (5). Some circRNAs bind to mRNA or microRNA, some participate in intracellular transport of RNA binding proteins, and some act to sequester proteins (6). circRNA dysregulation has been implicated in cancer development, and their stability in bodily fluids makes these circRNAs promising biomarkers (7). An example is circAGO2. This circRNA is detected at high levels in various tumors, promotes tumor growth in animal models, and is associated with poor prognosis of patients (8). Moreover, circRNAs are highly abundant in brain and have been implicated in neurodegenerative disorders (9,10).
Because of these functions, there is considerable interest in synthetic circRNAs as potential drugs, as biomarkers of disease, as therapeutic targets, and as research tools (11). Supporting the use of circRNAs in therapy, a synthetic circRNA with multiple regions complementary to miR-21 suppresses proliferation of cancer cells (12). circRNAs can be prepared by in vitro transcription followed by splint ligation to cyclize the molecule as was done to prepare an miRNA sponge (13). The potential use of synthetic circRNAs is limited by difficulties with generating a large amount of circRNAs.
Another potential application of RNA circles is in the field of RNA interference (RNAi). In 2021, 15 years after Andrew Fire and Craig Mello were awarded the 2006 Nobel Prize in Physiology or Medicine for their discovery of the RNAi pathway (14), four small interfering RNAs (siRNAs) are now approved for clinical use, and numerous investigational RNAi therapeutics are advancing through human clinical trials (15). The current clinical development pipeline is dominated by siRNAs conjugated to the trivalent N-acetylgalactosamine (GalNAc) ligand (16,17), which results in targeted delivery to hepatocytes (18–23). The three approved GalNAc-siRNA conjugates are fully modified with 2′-deoxy-2′-fluoro (2′-F) and 2′-O-methyl (2′-OMe) and strategically modified with phosphorothioate (PS) to confer stability.
Chemical modifications of the nucleobases, ribose sugar, and phosphate backbone of siRNAs conjugated to GalNAc are necessary to impart favorable pharmacological properties (24–31), and a large number of chemical modifications have been evaluated (32). Alterations at the level of the three-dimensional structure of siRNAs have also been explored (33). Supra-RNAi structures evaluated include hairpin siRNAs (34), dumbbell-shaped nanocircular siRNAs (35), siRNA nanosheets (36), branched siRNAs (37), caged circular siRNAs for photomodulation of gene expression (38), circular single-strand RNAs that serve as siRNA precursors (39), and circular siRNAs with reduced off-target effects (40,41). However, the experiments evaluating these structures often did not utilize therapeutically relevant siRNA chemical modifications or experimental systems or, in the case of covalent cyclization, employed inefficient strategies such as peptide coupling or T4 ligation techniques (38,41). The benefits of circular siRNA design, as previously described by others, are reduced off-target effects of the sense strand, prolonged duration due to enhanced nuclease resistance, and the preservation of potency (41). In the early reports describing in vivo analysis of circular siRNAs, they were administered locally at high doses and endogenous gene silencing was not evaluated (38,41). In a recent evaluation of a prodrug-type covalently closed siRNA in mice, systemic administration resulted in prolonged circulation and improved gene silencing activity in the liver, kidney, and muscle relative to the non-circularized siRNA (42); however, the doses used (20 mg/kg, q 3 d × 3) were not therapeutically realistic.
Here, we describe a straightforward synthetic strategy for the preparation of fully chemically modified small circular interfering RNAs (sciRNAs) utilizing a ‘click’-cyclized sense strand carrying the trivalent GalNAc ligand. Both strands were strategically modified with 2′-OMe, 2′-F, and PS. The antisense strand was also modified with a 5′ (E) vinylphosphonate (26,43) (Figure 1). We evaluated sciRNA potency in vitro and in vivo. In addition, we characterized the metabolic stability of sciRNAs in the liver and liver extracts and determined the ability of antisense strands to associate with Argonaute 2 (Ago2) after subcutaneous administration in mice. Physicochemical and NMR-based structural analyses and molecular modeling studies shed light on the interactions of the sciRNAs, which have a partial duplex character, with the RNAi machinery. Our study established that sciRNAs induce silencing in vitro and in vivo with efficacy comparable to siRNAs used clinically. What we have learned regarding synthesis, chemical modifications, and physical characterization of sciRNAs will be applicable to other novel circRNAs.
Figure 1.
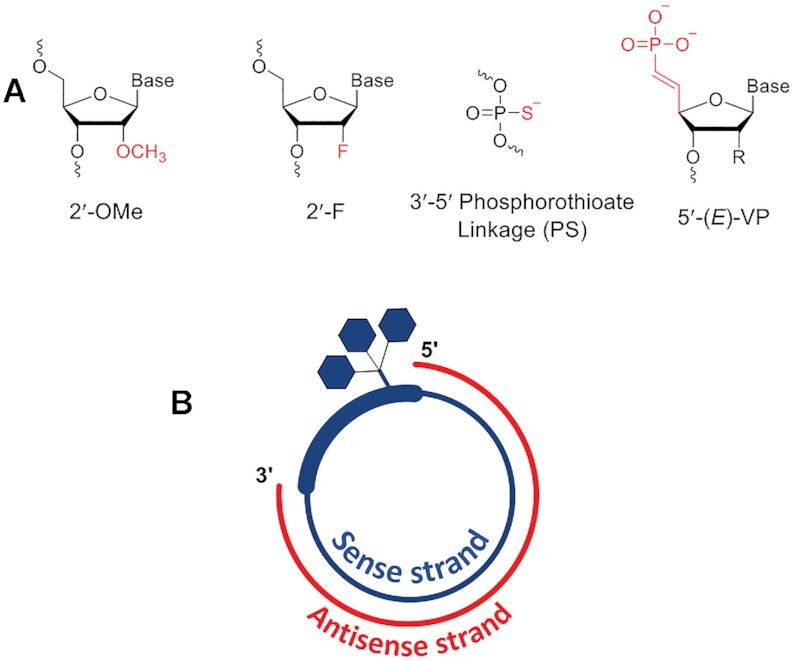
(A) Chemical modifications used in sciRNA. (B) Schematic representation of GalNAc-conjugated sciRNA.
MATERIALS AND METHODS
Synthesis of oligonucleotides
Oligonucleotides were synthesized on a MerMade-12 DNA/RNA synthesizer. Sterling solvents/reagents from Glen Research, 500-Å controlled pore glass (CPG) solid supports from Prime Synthesis, 2′-deoxy 3′-phosphoramidites from Thermo, and 2′-OMe and 2′-F nucleoside 3′-phosphoramidites from Hongene were all used as received. The 2′-OMe-uridine-5′-bis-POM-(E) vinylphosphonate (VP) 3′-phosphoramidite was synthesized according to previously published procedures (44), dissolved to 0.15 M in 85% acetonitrile 15% dimethylformamide (DMF), and coupled using standard conditions on the synthesizer. GalNAc CPG support (L, Scheme 1) was prepared and used as previously described (16). 5-Bromohexyl phosphoramidite (Glen Research, Cat# 10-1946) was dissolved to 0.15 M in acetonitrile and coupled using standard conditions on the synthesizer. Alkyne CPG support and alkyne hydroxyprolinol phosphoramidite (Y) were prepared and used as previously described (45). Low-water content acetonitrile was purchased from EMD Chemicals. A solution of 0.6 M 5-(S-ethylthio)-1H-tetrazole in acetonitrile was used as the activator. The phosphoramidite solutions were 0.15 M in anhydrous acetonitrile with 15% DMF as a co-solvent for 2′-OMe uridine and cytidine. The oxidizing reagent was 0.02 M I2 in THF/pyridine/water. N,N-dimethyl-N′-(3-thioxo-3H-1,2,4-dithiazol-5-yl)methanimidamide (DDTT), 0.09 M in pyridine, was used as the sulfurizing reagent. The detritylation reagent was 3% dichloroacetic acid (DCA) in dichloromethane (DCM).
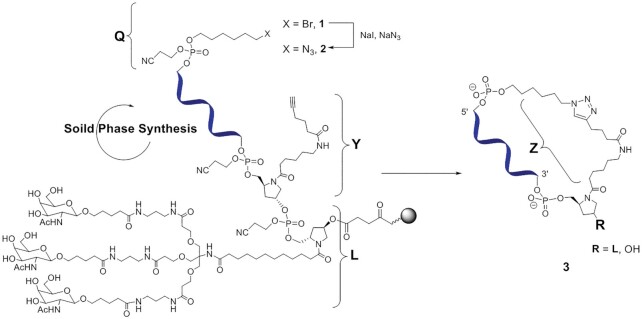
Scheme 1.
Synthesis of click-cyclized sense strand of the sciRNA-GalNAc conjugate. Abbreviations: Q, 6-azidohexyl handle; Y, alkyne hydroxyprolinol phosphoramidite; L, GalNAc ligand; and Z, linker.
After completion of the solid-phase synthesis, the CPG solid support was washed with 5% (v/v) piperidine in anhydrous acetonitrile three times with 5-min holds after each flow. The support was then washed with anhydrous acetonitrile and dried with argon. The oligonucleotides were then incubated with 28–30% (w/v) NH4OH, at 35°C for 20 h. For, VP-containing oligonucleotides, the CPG solid support was incubated with 28–30% (w/v) NH4OH containing 5% (v/v) of diethylamine at 35°C for 20 h (46). The solvent was collected by filtration, and the support was rinsed with water prior to analysis. Oligonucleotide solutions of ∼1 OD260 units/mL were used for analysis of the crudes, and 30 – 50 μl of solution were injected. LC/ESI-MS was performed on an Agilent 6130 single quadrupole LC/MS system using an XBridge C8 column (2.1 × 50 mm, 2.5 μm) at 60°C. Buffer A consisted of 200 mM 1,1,1,3,3,3-hexafluoro-2-propanol and 16.3 mM triethylamine in water, and buffer B was 100% methanol. A gradient from 0% to 40% of buffer B over 10 min followed by washing and recalibration at a flow rate of 0.70 mL/min. The column temperature was 75°C. All oligonucleotides were purified and desalted, and further annealed to form GalNAc-siRNAs as previously described. (16). Complete characterization of all oligonucleotides, conjugates and duplexes are given in Supplementary Tables S1 and S2 and Supplementary Figure S1.
Circularization of sense-strand oligonucleotides using click chemistry and formation of sciRNA-GalNAc conjugates
To functionalize the sense strand, commercially available 5-bromohexyl phosphoramidite was coupled to the 5′ end during solid-phase synthesis. To replace the bromide group with an azido group to provide the 6-azidohexyl click chemistry ‘handle’, a suspension of 10 μmol of CPG support loaded with 5′-(5-bromohexyl) modified oligonucleotides (47) in 15 ml of an anhydrous DMF solution containing 130 mg of sodium azide and 300 mg of sodium iodide was vigorously shaken at 65°C for 75 min. After cooling to room temperature, the solution was removed by filtration, and the CPG beads with the resulting 5′-(5-azidohexyl) solid-supported oligonucleotides were washed with DMF (2 × 10 ml) and dried under a stream of argon. The oligonucleotides were released from the solid support and purified and desalted as described above. The oligonucleotides were then dissolved in water to a concentration of approximately 10 OD260 units/ml. For a typical copper(I)-catalyzed alkyne-azide cycloaddition click cyclization reaction, we used a modified version of the protocol published by Lietard et al. (48): 2 mL of the oligonucleotide solution (∼200 OD260 units) were mixed with 2 ml of methanol, 1.1 ml of 0.1 mM sodium l-ascorbate and 1.1 ml of 20 mM copper sulfate. The reaction mixture was placed into a microwave tube container, equipped with a stirring bar, and placed in a microwave reactor for 40 min at 60°C (power ∼ 8 W, stirring, cooling, P = 0).
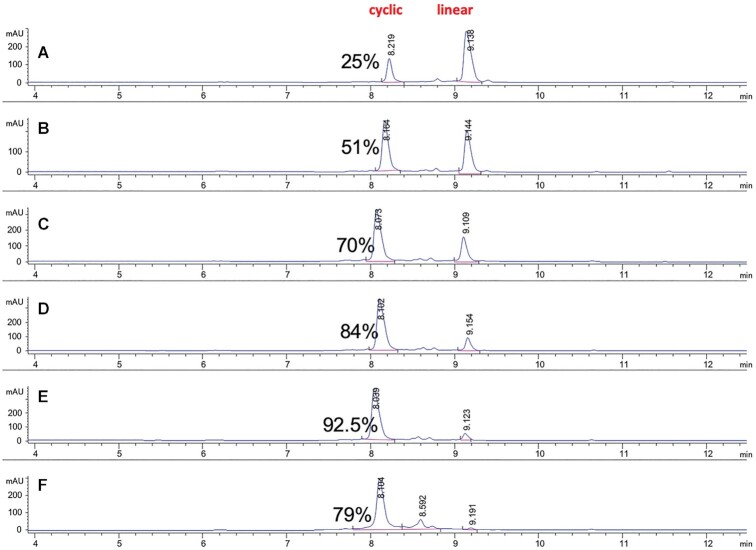
The click cyclization reaction was optimized to proceed at room temperature. It provided almost quantitative (>92.5%) conversion of the starting linear oligonucleotides within 4 h at room temperature. A time-course HPLC plot is presented in the Supplementary Figure S2. A representative room-temperature reaction was performed as follows: Linear oligonucleotide (9 mg, 0.9 mL, 10 mg/ml) was suspended in 1.8 ml of methanol and 0.4 ml of water. Solvents were degassed with argon. A freshly prepared mixture of 0.25 ml of 0.1 mM sodium l-ascorbate and 0.25 ml of 20 mM copper sulfate in argon-degassed water was added. The reaction was placed in a vial and closed under argon and then shaken at room temperature for up to 17 h. Optimal yield was achieved at about 4 h with high conversion and minimal byproduct formation (Supplementary Figure S2). Cyclic oligonucleotides were HPLC purified quickly in order to minimize complexation of copper to the phosphate backbone. After ion-exchange HPLC purification and size-exclusion HPLC desalting, sterile filtration, and lyophilization, about 3 mg (30% isolated yield, comparable to regular oligonucleotide synthesis and purification) of cyclic oligonucleotide was obtained with the required high purity needed for in vivo experiments. Cyclic oligonucleotides were then annealed with an antisense strand to form the GalNAc-sciRNAs.
Enzymatic stability assays
Modified oligonucleotide was added at 0.1 mg/ml to a solution of 50 mM Tris–HCl (pH 7.2) and 10 mM MgCl2. Stability was evaluated in the presence of snake venom phosphodiesterase (SVPD) (Worthington, Cat# LS003926) and of phosphodiesterase II (PDII) from bovine spleen (Worthington, Cat# LS003 602). SVPD was added to the oligonucleotide at 750 mU/ml. Enzyme was prepared as a stock of 1000 mU/mL aliquoted into 1 ml tubes and stored at −20°C. A new aliquot was used each week. PDII was added at 500 mU/ml. Enzyme was prepared as a stock of 2000 mU/ml, aliquoted into 1 ml tubes and stored at −20°C. A new aliquot was used each week.
Immediately after addition of the enzyme, the sample was injected onto a Dionex DNAPac PA200 column (4 mm × 250 mm) at 30°C and run at a flow rate of 1 ml/min with a gradient of 40–55% Buffer B over 7.5 min. Buffer A was 20 mM sodium phosphate, 15% acetonitrile, pH 11; Buffer B was Buffer A containing 1 M sodium bromide (pH 11). Aliquots were analyzed every hour for 24 h. The area under the peak corresponding to full-length oligonucleotide was normalized to the area from the 0 h time point (first injection). First order decay kinetics were assumed in calculation of half-lives. For experiments with SVPD, the control oligonucleotide was dT19•dT, where dT19 is 2′-deoxythymidine and ‘•’ indicates a single 3′-terminal PS linkage. For experiments with PDII, the control oligonucleotide was dT•dT19 with a single 5′-terminal PS linkage. Half-lives are reported relative to the half-life of the control sequence. Experiments were performed in triplicate.
Analysis of oligonucleotide stability in plasma and liver homogenates
Rat plasma (BioIVT, Cat# RAT00PL38NCXNN) and liver homogenate (BioIVT, custom order) were diluted with a 10× cofactor solution to achieve a final concentration of 1 mM MgCl2, 1 mM MnCl2 and 2 mM CaCl2. The sciRNA was added to 50 μl of the plasma or the liver homogenate to achieve the final concertation of 20 μg/ml. The reaction mixture was incubated with gently shaking at 37°C. At each predetermined time point (0, 1, 4, 8 and 24 h), the reaction was stopped by adding 450 μl of Clarity OTX lysis-loading buffer (Phenomenex, Cat# AL0-8579) containing internal standard (oligonucleotide U21 at 1 μg/ml final concentration) and frozen at −80°C until analysis. Experiments were performed in triplicate.
Oligonucleotide enrichment for LC–MS analysis was performed using Clarity OTX 96-well solid-phase extraction plates as described by Liu et al. (49). The SPE columns were conditioned initially with 1 ml of methanol followed by equilibration with 2 ml of 50 mM ammonium acetate with 2 mM sodium azide in HPLC-grade water. The samples were loaded on to SPE column by applying positive pressure. The columns were then washed five times with 1 ml of 50 mM ammonium acetate in 50/50 (v/v) water and acetonitrile (pH 5.5). Finally, the oligonucleotides are eluted using elution buffer containing 10 mM EDTA, 100 mM ammonium bicarbonate in 40/10/50 (v/v/v) acetonitrile/tetrahydrofuran/water (pH 8.8). The eluant was dried under nitrogen and resuspended in 120 μl of LC–MS grade water for LC–MS analysis.
Relative quantitation and metabolite identification of modified oligonucleotides was performed using high-resolution mass spectrometry on a Thermo Scientific Q Exactive coupled to ion-pairing reverse-phase liquid chromatography (Dionex Ultimate 3000) (LC-HRMS). A Waters X-Bridge BEH C8 XP Column (Cat# 176002554,130 Å, 2.5 μm, 2.1 mm × 30 mm, 80°C) was used for the chromatographic separation. The injection volume and flow rates were 30 μl and 1 ml/min, respectively. Mobile phase A consisted of 16 mM triethylamine (Sigma, Cat# 471283), 200 mM 1,1,1,3,3,3-hexafluoro-2-propanol (Fisher, Cat# 67-56-1) in LC–MS grade water (Fisher, Cat# 7732-18-5); mobile phase B was 100% methanol (Fisher, Cat# 67-56-1). The gradient started with 1% mobile phase B and progressed to 35% B over 4.3 min, then the column was equilibrated with 1% mobile phase B for 1 min. The mass spectrometer data acquisition was performed in full scan mode with a scan range of 500–3000 m/z at a resolution setting of 35 000. Spray voltage was 2.8 kV. The auxiliary gas temperature and the capillary temperature were set to 300°C.
The Thermo Quan browser was used calculate the area ratio of extracted ion chromatograms (XIC) of test oligonucleotide to internal standard with 10 ppm mass accuracy. The m/z ions for used for XIC of the test oligonucleotide were 1018.7752, 1018.8868, 1019.1101, 1019.2220, 1019.3325, 1019.4436, 1019.5559 and 1019.6671, and m/z ions used for XIC of the internal standard were 1258.5073, 1258.8408, 1259.1740, 1259.8411, 1289.5061. After LC-HRMS analysis, data were processed using ProMass HR Deconvolution software (Novatia, LLC) to identify linearization and major metabolism of the modified oligonucleotides as described in Liu et al. (49). Representative chromatograms are shown in the Supplementary Figures S3 and S4. Complete characterization of all metabolites of oligonucleotides, conjugates and duplexes are given in Supplementary Tables S3–S5.
The half-lives were calculated as described by Chan et al. by monitoring loss of full-length test oligonucleotide for 24 h (50). The amounts of test oligonucleotide and internal standard were normalized to time 0 h for each time-point for respective oligonucleotides. The natural log of percentage full length remaining and the slope were calculated using linear regression. The half-life was calculated using equation (1) as described previously (50,51):
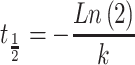 |
(1) |
Thermal melting studies
Melting studies were performed in 1-cm path length quartz cells on a Beckman DU800 spectrophotometer equipped with a thermoprogrammer. Samples were diluted to obtain a final concentration of oligonucleotide strand of approximately 1 μM in 0.1× PBS buffer (pH 7.4). Melting curves were monitored at 260 nm with a heating rate of 1°C/min from 10 to 90°C. Melting temperatures (Tm) were calculated from the first derivatives of the heating curves, and the reported values are the result of two independent measurements.
NMR studies
Lyophilized RNA was dissolved in a mixture of 10% 2H2O/90% H2O with 20 mM NaCl and 10 mM sodium phosphate buffer (pH 7). Equimolar ratios of sense and antisense strands were mixed to yield linear or circular duplexes. Final concentrations of duplexes in 600 μl were in the range from 20 to 60 μM. All spectra were acquired at 25°C on an Agilent VNMRS 800 MHz NMR spectrometer equipped with a cold probe. The full-size 1H NMR spectra (0–14 ppm) overlay is provided as Supplementary Figure S5.
Circular dichroism spectroscopy
The circular dichroism (CD) spectra were obtained on a Jasco J-815 spectropolarimeter equipped with a Julaba F25 circulating bath. The sample was allowed to equilibrate for 5 min at 10 ºC in 1× PBS at a final duplex concentration of 1.57 μM. The spectrum was an average of 5 scans. Spectra were collected at a rate of 50 nm/min, with a bandwidth of 1 nm and sampling wavelength of 0.2 nm using fused quartz cells (Starna 29-Q-10). The CD spectra were recorded from 350 to 200 nm at 10 ºC. The molar ellipticity was calculated from the equation [0] = 0/10Cl, where 0 is the ellipticity (mdeg), C is the molar concentration of oligonucleotide (M), and l is the path length of the cell (cm). Representative CD spectra are provided in Supplementary Figure S6.
Analysis of silencing activity in vitro
Silencing was evaluated in primary mouse hepatocytes by transfection and by free uptake of siRNAs. For transfection, siRNA (5 μl) at the indicated concentration was mixed with 4.9 μl of Opti-MEM and 0.1 μl of Lipofectamine RNAiMAX (Invitrogen, Cat# 13778-150) per well of a 384-well plate and incubated at room temperature. After 15 min, 40 μl of William's E medium or EMEM medium containing approximately 5 × 103 primary mouse hepatocytes were added to the wells. Cells were incubated for 24 h prior to RNA purification. For free uptake experiments, siRNA (5 μl at the indicated concentration) was mixed with 5 μl of Opti-MEM per well of a 384-well plate. After 15 min, 45 μl of William's E medium or EMEM medium containing ∼5 × 103 cells were added to the wells. Cells were incubated for 48 h prior to RNA purification.
RNA was isolated using an automated protocol on a BioTek-EL406 platform using Dynabeads (Invitrogen, Cat# 61012). To each well was added 50 μl of lysis/binding buffer (Tris–HCl pH 7.5, LiCl, EDTA pH 8.0, DTT) and 25 μl of lysis/binding buffer containing 3 μl of magnetic beads. The plates were incubated on an electromagnetic shaker for 10 min at room temperature, and then the magnetic beads were captured, and the supernatant removed. The bead-bound RNA was then washed twice with 150 μl/well of Buffer A (Tris–HCl pH 7.5, LiCl, EDTA pH 8.0, DTT) and once with 150 μl/well of Buffer B (Tris–HCl pH 7.5, LiCl, EDTA pH 8.0). The beads were then washed with 150 μl of Elution Buffer, re-captured, and the supernatant was collected.
cDNA synthesis was performed using an ABI kit (Cat# 4368813). To the wells of a 384-well plate containing the RNA isolated using Dynabeads was added 10 μl of a master mix containing 1 μl 10× Buffer, 0.4 μl 25 × dNTPs, 1 μl 10× random primers, 0.5 μl reverse transcriptase, 0.5 μl RNase inhibitor and 6.4 μl of nuclease free water. The plates were sealed, mixed, and incubated on an electromagnetic shaker for 10 min at room temperature, followed by 2 h at 37°C.
RNA quantification was performed using the Life Technologies Taqman gene expression system with dual labeled probes. Target gene expression was normalized to endogenous Gapdh. Ct values were measured using a Light Cycler 480 (Roche). To calculate relative fold change real time data were analyzed using the ΔΔCt method and normalized to assays performed with cells treated with a non-targeting siRNA control. Taqman probes used were Mouse C5 (Mm00439275_m1), Mouse Gapdh 4352339E and Mouse Ttr (Mm00443267_m1).
Analysis of silencing activity in mice
All procedures involving mice were conducted by certified laboratory personnel using protocols consistent with local, state, and federal regulations. Experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC), the Association for Assessment and Accreditation of Laboratory Animal Care International (accreditation number: 001345), and the office of Laboratory Animal Welfare (accreditation number: A4517-01). When deciding on sample numbers for animal studies, we determined the final number required to ensure confidence in the resulting data while utilizing the least number of animals, as required by IACUC guidelines. Female C57BL/6 mice approximately 8 weeks of age were obtained from Charles River Laboratories and randomly assigned to each group. Mice were acclimated in-house for 48 h prior to study start.
Animals were dosed subcutaneously at 10 μl/g with sciRNA, siRNA, or with PBS saline control. Doses used in this study were 3 mg/kg. The test compounds were diluted into phosphate buffered saline (PBS, pH 7.4). All solutions were stored at 4°C until time of injection. Animals were sacrificed at either 5 or 7 days post dose. Livers were harvested and snap frozen for analysis. Blood was collected utilizing the retro-orbital eye bleed procedure 24 h post the final dose in accordance with the IACUC approved protocol. The sample was collected in Becton Dickinson serum separator tubes (Fisher Scientific, Cat# BD365967).
For analysis of TTR, serum samples were kept at room temperature for 1 h and then spun in a microcentrifuge at 21 000 × g at room temperature for 10 min. Serum was transferred into 1.5 ml microcentrifuge tubes for storage at −80°C until the time of assay. Serum samples were diluted 1:4000 and assayed using a commercially available kit from ALPCO specific for detection of mouse prealbumin (Cat# 41-PALMS-E01). Protein concentrations (μg/ml) were determined by comparison to a purified TTR standard and the manufacturer's instructions were followed.
Serum collected for the analysis of circulating C5 was kept at room temperature for 15 min and then immediately transferred to 4°C prior to spinning in a microcentrifuge at 21 000 × g at room temperature for 10 min. Serum was transferred into 1.5 ml microcentrifuge tubes for storage at −80°C until the time of assay. The serum samples were diluted 1:5000 for analysis by ELISA. The primary antibody was goat-anti-human C5 (Complement Technologies, Cat# A220), and the secondary antibody was bovine anti-goat IgG-HRP (Jackson ImmunoResearch, Cat# 805-035-180), which had minimal cross-reactivity to other species. Antibodies were used at 0.8 mg/ml. The assay was developed using a TMB substrate kit (R&D Systems, Cat# DY999), and the reaction was stopped using sulfuric acid prior to measurement.
Analysis of in vivo liver exposure and Ago2 loading
Mice were sacrificed on day 7 post-dose, and livers were snap frozen in liquid nitrogen and ground into powder for further analysis. Total siRNA liver levels were measured by reconstituting liver powder at 10 mg/mL in PBS containing 0.25% Triton-X 100. The tissue suspension was further ground with 5-mm steel grinding balls at 50 cycles/s for 5 min in a tissue homogenizer (Qiagen TissueLyser LT) at 4°C. Homogenized samples were then heated at 95°C for 5 min, briefly vortexed, and allowed to rest on ice for 5 min. Samples were then centrifuged at 21 000 × g for 15 min at 4°C. The siRNA-containing supernatants were transferred to new tubes. The siRNA sense and antisense strand levels were quantified by stem loop reverse transcription followed by Taqman PCR (SL-RT QPCR) based on a previously published method (52,53) and adapted to chemically modified siRNAs (24,27). Ago2-bound siRNA from mouse liver was quantified by preparing liver powder lysates at 100 mg/mL in lysis buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.5% Triton-X 100) supplemented with freshly added protease inhibitors (Sigma-Aldrich, Cat# P8340) at 1:100 dilution and 1 mM PMSF (Sigma-Aldrich, Cat# P7626). Total liver lysate (10 mg) was used for each Ago2 immunoprecipitation and control immunoprecipitation. The siRNA not bound to Ago2 was purified with pre-saturated QAE Resin (GE Healthcare, Cat# 17-0200-01) in lysis buffer (16 mg/ml) supplemented with protease inhibitors at 1:100 dilution and 1 mM PMSF. Samples were filtered through cellulose acetate filter (Fisher, Cat# P169702) to remove the resin before proceeding with the Ago2 immunoprecipitation. Anti-Ago2 antibody was purchased from Wako Chemicals (Clone No. 2D4). Control mouse IgG was from Santa Cruz Biotechnology (Cat# sc-2025). Protein G Dynabeads (Life Technologies, Cat# 10003D) were used to precipitate antibodies. Ago2-associated antisense strands were eluted by heating (50 μl PBS, 0.25% Triton X-100; 95°C, 5 min) and quantified by SL-RT QPCR as described previously (52,53) and adapted to chemically modified siRNAs (24,27). The Ttr siRNA RT-qPCR primers and probes sequences were as follows: RT: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAACAGTGT; Forward qPCR: GCCGCGCTTATAGAGCAAG; TaqMan Probe: CTGGATACGACAAAACAGT; Universal Reverse qPCR: GTGCAGGGTCCGAGGT.
Molecular modeling
Coordinates of the complex between human Ago2 and an siRNA with seed region pairing were retrieved from the Protein Data Bank (www.rcsb.org; ID code 4W5T) (54). In the complex, the sense strand is comprised of residues S1 to S9 (5′ to 3′ direction). The Z linker was built with the program UCSF Chimera (55) using the structure-editing ‘build structure’ and ‘modify structure’ tools and starting from the 3′-terminus of the sense strand (S9). After adding a 3′-phosphate group, the linker was constructed using ideal bond lengths and angles and an extended conformation in the direction of the 5′ end of the sense strand wherever possible, while avoiding short contacts with the antisense strand and side chains of the Ago2 PIWI and MID domains and L2 linker region. The 5′-terminal S1 residue was looped around in order to bring the 5′-hydroxyl group toward the Z linker. After addition of a phosphate to the last methylene carbon of the linker, a distance of some 5 Å remained between the 5′-OH of S1 and phosphorous. This distance was systematically shortened by altering S1 and linker torsion angles until a final distance of 1.6 Å allowed cyclization of the sense strand with acceptable bond and torsion angles.
RESULTS
Chemical synthesis of GalNAc-sciRNA conjugates
It has been reported previously that cyclizing the antisense strands in an siRNA results in the loss of silencing activity (38). A circular strand may not be loaded onto Ago2 protein, which would preclude the formation of an active RNA-induced silencing complex (RISC), the functional component of RNAi-mediated mRNA silencing. In contrast, siRNAs with cyclized sense strands reportedly have potency nearly identical to that of linear siRNA controls (41). Here we designed and synthesized sciRNA constructs in which both strands are constructed with 2′-OMe, 2′-F and PS backbone modifications (Figure 1A). The sense strand was conjugated at the 3′ end to the trivalent GalNAc ligand as in typical linear GalNAc–siRNA conjugates (16). The sense strand was cyclized and annealed to a linear antisense strand (Figure 1B).
The sense strand was synthesized on solid support functionalized with the GalNAc ligand (L) (16). As a click chemistry handle, we introduced an N-alkyne linker using a hydroxyprolinol phosphoramidite building block (Y) at the 3′ end of the sense strand (45) as the first step in solid-phase synthesis of the oligonucleotide (Scheme 1). Both the hydroxyprolinol linker and the trivalent GalNAc ligand are clinically validated moieties in RNAi therapeutics. After completion of the solid-phase synthesis of the sense stand, the commercially available 6-bromohexyl phosphoramidite was coupled to the 5′ end of the sense strand. Subsequently, the bromide (1, Scheme 1) was substituted with an azido group (2, Scheme 1), providing the 6-azidohexyl handle (Q) (47). After cleavage from the solid support, deprotection and purification of the full-length 5′-azido oligonucleotide, the copper(I)-catalyzed alkyne-azide cycloaddition click reaction resulted in cyclization (48,56).
For a typical copper(I)-catalyzed alkyne-azide cycloaddition click cyclization reaction, we used a modified version of the protocol published by Lietard et al. (48). We first performed the cyclization reaction in a microwave reactor for 40 min at 60°C. The click oligonucleotide cyclization proceeded smoothly with almost quantitative conversion of the linear material to the cyclic product, with some hydrolysis byproducts observed (<10%). No linearization or cross-polymerization products were observed. With the aim of further expanding the scope of the reaction and to minimize the formation of hydrolysis byproducts, the click cyclization reaction was optimized at room temperature, providing clean and almost quantitative (>92.5%) conversion of the starting linear oligonucleotides to cyclic products within about 4 h (Supplementary Figure S2). After 17 h, when the starting material was fully consumed, the crude HPLC purity of the cyclic product decreased from 92.5% to 79%, suggestive of decomposition (Supplementary Figure S2). We found that it is important to purify the cyclic oligonucleotides immediately after the click reaction to minimize copper-mediated degradation of the phosphate backbone. For solution-phase click reactions, this can potentially be avoided with the use of immobilized-copper supports as we have previously reported (57). After ion-exchange HPLC purification and size-exclusion HPLC desalting, sterile filtration, endotoxin removal, and lyophilization, we obtained cyclic RNAs in about 30% isolated yield, comparable to yields for synthesis of linear oligonucleotides. The reaction profiles of α-azido-ω-alkyne linear oligonucleotide and cyclic oligonucleotide are shown in Figure 2. Annealing with the corresponding complementary antisense strand yielded the sciRNA. The oligonucleotides were characterized as detailed in the Supplementary Table S1.
Figure 2.
Ion-exchange HPLC time-course of the cyclization ‘click’ reaction profiles (AU at 260 nm) of cyclic ON-6 versus α-azido-ω-alkyne linear ON-5 Aliquots were taken off from the reaction at each timepoint and analyzed. Timepoints are shown as follows: (A) 0 h (analysis performed immediately after start of the reaction); (B) 1 h; (C) 2 h; (D) 3 h; (E) 4 h and (F) 17 h. Formation of the cyclic oligonucleotide product (tR ∼8.1 min) is observed with increased amounts as starting material linear oligonucleotide (tR ∼9.1 min) is being consumed. Integration and area values are shown. Percentage of cyclic oligonucleotide product is also shown for each timepoint. See also the Supplementary Figure S2 (pages 23-24) and experimental section for method details.
Enzymatic stability of cyclic oligonucleotides
Circularization significantly stabilizes phosphodiester oligonucleotides since their degradation in biological media is primarily driven by the action of 5′ and 3′ exonucleases (58). This strategy has been employed to stabilize siRNAs composed of only natural ribonucleotides and phosphodiester linkages (41). In order to assess the stabilization of our cyclic sense strands, we analyzed half-lives of full-length strands in the presence of either a 3′-exonuclease, SVPD, or a 5′-exonuclease, PDII.
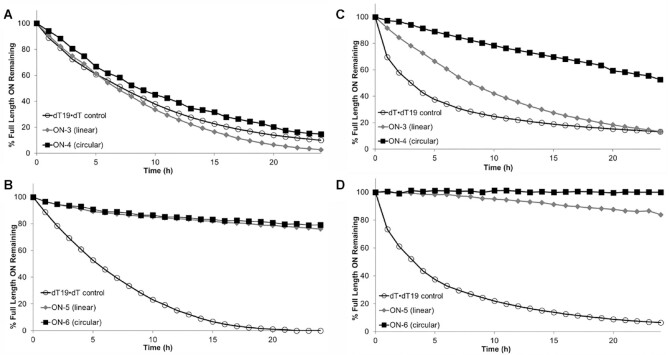
We prepared single-stranded 23-mer phosphodiester oligonucleotides composed of 2′-deoxythymidine or of fully chemically 2′-modified nucleotides. We analyzed the circularized oligonucleotide strands as well as the linear oligonucleotides containing the ‘unclicked’ precursor moieties Q and Y (Table 1). The results from the exonuclease assays (Table 1 and Figure 3) revealed that the cyclized oligonucleotides, ON-4 (deoxythymidine) and ON-6 (chemically modified), were more stable than the linear counterparts (ON-3 and ON-5, respectively) and the control oligonucleotides (ON-1 and ON-2). The fully chemically 2′-modified and cyclized oligonucleotide ON-6 was not degraded to a significant extent by either nuclease over the time course of the experiment. Interestingly, the 3′-exonuclease showed less differentiation between linear and circular oligonucleotides than did the 5′-exonuclease.
Table 1.
Enzymatic stability of linear and circular single-stranded oligonucleotides
| Oligonucleotide | Sequences (5′-3′)a | 3′-Exonuclease half-lifeb | Intact after 24 h (%) | 5′-Exonuclease half-lifec | Intact after 24 h (%) |
|---|---|---|---|---|---|
| ON-1 | (dT)19•dT | 1 | 10.0 | ||
| ON-2 | dT•(dT)19 | 1 | 6.5 | ||
| ON-3 | Q(dT)23Y | 0.7 | 2.7 | 8.2 | 13.3 |
| ON-4 | Z(dT)23 | 1.2 | 14.8 | 26.8 | 52.5 |
| ON-5 (linear) | QaaAaCaGuGuUCUuGcUcUaUaAY | 12.8 | 76.1 | 14.1 | 83.8 |
| ON-6 (cyclic) | ZaaAaCaGuGuUCUuGcUcUaUaA | 14.5 | 79.1 | 423.0 | 100.0 |
aChemistry modifications legend: •, terminal PS linkage; dT, 2′-deoxythimidine; lower case nucleotides, 2′-OMe; upper case nucleotides in italics, 2′-F; Q, 6-azidohexyl-phosphate. For structures of Y and Z, see Scheme 1. Z denotes a cyclic oligonucleotide.
bHalf-life value determined in hours and reported relative to that of the (dT)19•dT control.
cHalf-life value was determined in hours and reported relative to that of the dT•(dT)19 control. First-order decay kinetics were assumed in calculation of half-lives. Data are representative of two independent experiments with similar results.
Figure 3.
Percent full-length oligonucleotide remaining as a function of time in the presence of (A and B) 3′-exonuclease or (C and D) 5′-exonuclease.
To further understand the metabolism of the circular siRNA designs, we extended the stability studies from the individual single strands to the annealed sciRNA duplexes. The in vitro stabilities of two sciRNAs (si-4 and si-5, Table 2, Supplementary Table S2) and the linear siRNA containing the same nucleotide sequence and modifications (si-3) were assessed in rat plasma and rat liver homogenates. Proposed metabolite identification of si-3 from rat liver homogenate or plasma is shown in Supplementary Table S3.
Table 2.
Sequences, thermal stabilities, and in vitro potencies of linear GalNAc-siRNAs and circular GalNAc-sciRNAs.
| siRNA | Target | Sequences (5′-3′)a | T m (°C)b (ΔTm)c | Transfection IC50 (pM)d | Free uptake IC50 (pM)e |
|---|---|---|---|---|---|
| si-1 | Ttr | A•a•CaGuGuUCUuGcUcUaUaAL | 68.2 | 151.1 ± 21.8 | 53.1 ± 15.6 |
| u•U•aUaGaGcAagaAcAcUgUu•u•u | |||||
| si-2 | Ttr | A•a•CaGuGuUCUuGcUcUaUaAL | 67.8 (−0.4) | 41.9 ± 13.8 | 21.6 ± 3.3 |
| VPu•U•aUaGaGcAagaAcAcUgUu•u•u | |||||
| si-3 (linear sense) | Ttr | Q A•a•CaGuGuUCUuGcUcUaUaAYL | 67.3 (−0.9) | 81.3 ± 55.5 | 58.5 ± 1.9 |
| u•U•aUaGaGcAagaAcAcUgUu•u•u | |||||
| si-4 (cyclic sense) | Ttr | Z A•a•CaGuGuUCUuGcUcUaUaAL | 52.4 (−15.7) | 836.8 ± 140.4 | 197.9 ± 31.2 |
| u•U•aUaGaGcAagaAcAcUgUu•u•u | |||||
| si-5 (cyclic sense) | Ttr | Z A•a•CaGuGuUCUuGcUcUaUaAL | 52.5 (−15.7) | 124.7 ± 64.1 | 63.9 ± 9.1 |
| VPu•U•aUaGaGcAagaAcAcUgUu•u•u | |||||
| si-6 | C5 | G•a•CaAaAuAACuCaCuAuAaUL | 62.3 | 60.5 ± 30.1 | 109.2 ± 53.8 |
| a•U•uAuAgUgAguuAuUuUgUc•a•a | |||||
| si-7 (cyclic sense) | C5 | Z G•a•CaAaAuAACuCaCuAuAaUL | 41.8 (−20.5) | 830.2 ± 195.0 | 824.4 ± 72.7 |
| a•U•uAuAgUgAguuAuUuUgUc•a•a |
aSense strand sequences are the top rows; antisense strand sequences are the bottom rows. Z denotes a cyclic oligonucleotide. Chemistry modifications are indicated as follows: •, PS linkage; lower case nucleotides, 2′-OMe; upper case nucleotides in italics, 2′-F; Q, 6-azidohexyl-phosphate; VP, 5′-(E)-vinylphosphonate. For structures of L, Y, and Z, see Scheme 1.
b T m refers to melting temperatures obtained from the maxima of the first derivatives of the melting curves (A260 vs. temperature) recorded in 0.1 × PBS buffer (pH 7.4) using 1.0 μM concentrations of each strand.
cΔTm refers to the change in melting temperature compared to the unmodified duplex (for si-2 through si-5, reference duplex is si-1, and for entry si-7, reference duplex is si-6). Average of two independently annealed sample preparations are reported.
dsiRNAs were transfected into primary mouse hepatocytes with Lipofectamine RNAiMAX, and mRNA levels were quantified after 24 h and normalized to Gapdh. Mean IC50 values are reported with SD (n = 4).
eCompounds were added to media over primary mouse hepatocytes, and mRNA levels were quantified after 48 h. mRNA levels were normalized to Gapdh. Mean IC50 values are reported with SD (n = 4).
Rat plasma and liver homogenates have been used to characterize the metabolism of GalNAc-conjugated oligonucleotides (59,60). No degradation of either the circular sciRNA strands or the linear siRNA strands was detected in plasma for up to 8 h (Figure 4A). However, in our metabolite analysis by LC–MS, we observed that after 24 h of incubation in rat plasma, an addition of a water molecule to the circular strand was observed (Supplementary Figure S3, Supplementary Tables S4 and S5). Proposed metabolite identification of si-4 from rat liver homogenate or plasma is shown in Supplementary Table S4 and proposed metabolite identification of si-5 from rat liver homogenate or plasma is shown in Supplementary Table S5. We theorize that this species resulted from opening of the cyclic structure yielding a linearized oligonucleotide. In rat liver homogenates, the circular sciRNAs (si-4 and si-5) were more stable than the linear counterpart (si-3). There was no degradation of si-4 or si-5 at 24 h, whereas for si-3, only ∼55% of the full-length sense strand was detected at 24 h (Figure 4B). The sciRNAs have significantly longer half-lives of 29 and 30 h compared to the linear siRNA, which has a 4-h half-life. This correlates with the results of the analysis of the stabilities of the single strands, where circularization enhanced stability in the presence of exonucleases. In the liver homogenates, we did not observe opening of the cyclic structure of the sense strands of either si-4 or si-5 (Supplementary Figure S4, Supplementary Tables S4 and S5).
Figure 4.
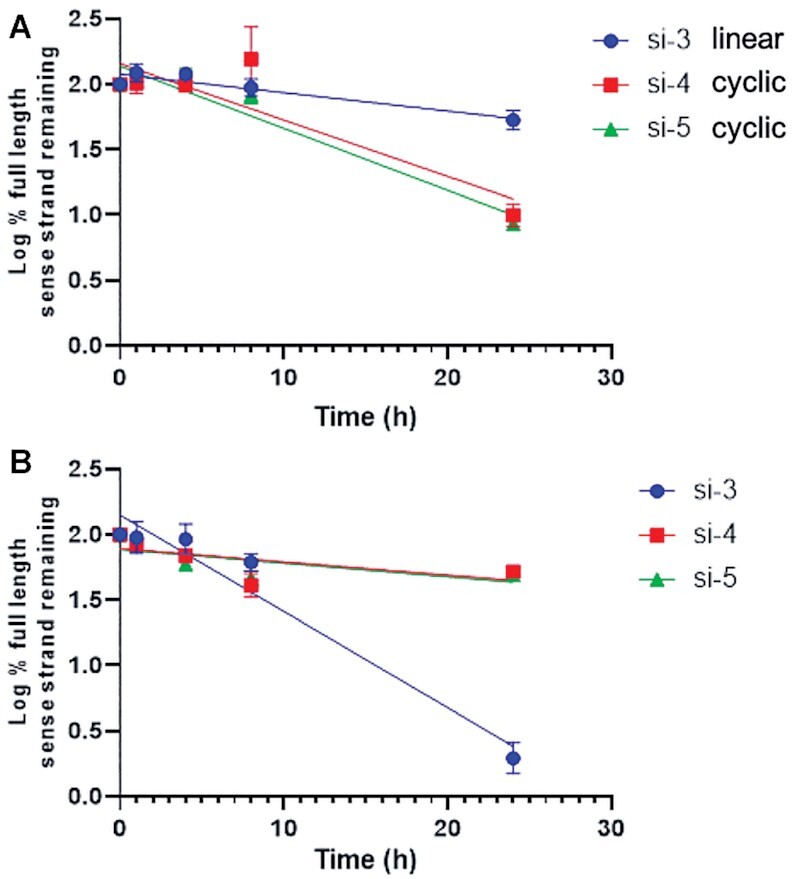
Stability of full-length sense strands after incubation of linear GalNAc-siRNA si-3 and GalNAc-sciRNAs with cyclic sense strands si-4 and si-5 in plasma and liver homogenate measured using LC–MS. Percent of full-length sense strand remaining (log10) in (A) rat plasma or (B) rat liver homogenate. Plotted are means with error bars as standard deviation of three replicates per time point.
Thermal stability and structures of GalNAc-sciRNA conjugates
The melting temperatures (Tm) of each sciRNA (si-4, si-5 and si-7) were determined and compared to siRNAs formed from linear sense strands (si-1, si-2 and si-6) and an siRNA composed of a linear sense strand with Q and Y modifications (si-3). The sciRNA duplexes were considerably destabilized, by 15–20°C, compared to si-3 and to the standard siRNA duplexes (Table 2). This thermal destabilization results from the strain of the cyclic sense strand. It is likely that only a subset of the nucleobases form Watson–Crick base pairs in the sciRNA duplexes.
This destabilized nature of the cyclic duplex structures was corroborated by solution-state 1H NMR analysis of the siRNA duplexes. The spectra of siRNA duplexes formed from linear sense strands (si-1, si-2, si-3 and si-6) exhibit well-resolved resonances due to imino proton in the region from δ 12 to 14 ppm (Figure 5 and Supplementary Figure S5), which are indicative of Watson-Crick base pairing. The numbers and intensities of peaks suggest that fully complementary duplexes are formed. In contrast, in all regions of the spectra of circular duplexes, mostly broad signals are observed with only a few low intensity, broad signals in the imino regions (Figure 5). These data support our hypothesis that only a subset of the possible 21 base pairs form in the si-4 and si-5 duplexes.
Figure 5.
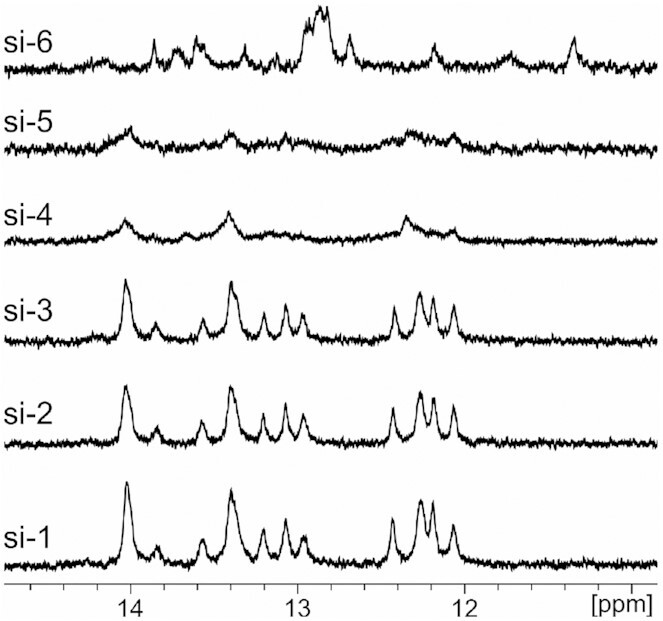
Base pairing is disrupted by cyclization of the sense strand. Imino base-pair regions of 1D 1H NMR spectra of linear GalNAc-siRNAs (si-1, si-2, si-3 and si-6) and GalNAc-sciRNA duplexes with cyclic sense strands (si-4 and si-5).
In addition, in CD spectroscopy experiments (Supplementary Figure S6), the linear siRNAs (si-1 and si-3) as well as the sciRNAs (si-4 and si-5) showed identical profiles. The spectra of the linear and cyclic siRNAs had A-type duplex signatures with negative peaks at about 211 nm diagnostic of a global A-type helix.
In vitro and in vivo activity of GalNAc–sciRNA conjugates
We evaluated sciRNAs designed to inhibit expression of rodent Ttr and C5 (Table 2). The sequences and chemical modifications of the linear siRNAs were previously described (16,61). The potency of the sciRNAs was determined in primary mouse hepatocytes, after transfection into cells using Lipofectamine RNAiMAX or after simple free uptake mediated by interaction between the GalNAc ligand and its receptor, the asialoglycoprotein receptor (ASGPR) (16), which is ubiquitously expressed on the surface of hepatocytes. We tested sciRNAs and linear ‘unclicked’ siRNA controls.
In the Ttr series, the cyclic sciRNA compound si-4 had a 5-fold (transfection) and 4-fold (free uptake) loss of in vitro potency relative to the linear controls si-1 and si-3, correlating with the strong thermal destabilization observed in the Tm studies (Table 2). We also evaluated the use of the 5′-(E)-vinylphosphonate VP modification of the antisense strands, a modification that enhances the metabolic stability and Ago2 loading of siRNAs (24,26). There was an increase of about 3-fold in potency upon the introduction of the VP modification as shown by comparison of si-1 and si-2 (Table 2). The sciRNA with a VP-modified antisense strand (si-5) was only 3-fold less potent than the VP-modified control siRNA with a linear sense strand si-2 and had similar potency to the control non-VP siRNA si-1. For the C5-targeting compounds, the cyclic sciRNA si-7 was 14-fold less potent by transfection and 8-fold less potent by free uptake than si-6 (Table 2).
We next evaluated GalNAc-sciRNAs activities in vivo. For this experiment, all sciRNAs and linear counterparts were subcutaneously administered as a single dose (3 mg/kg on Day 0), and the serum levels of circulating TTR or C5 proteins were determined over time. For the Ttr siRNA series, the data in mice mirrored the in vitro results: The sciRNAs si-4 and si-5 were considerably less potent than si-1 and si-2 with linear sense strands (Figure 6A). However, for the series targeting C5, the siRNAs with linear (si-6) and circular (si-7) sense strands had similar potency (Figure 6B), whereas at least an ∼8-fold potency loss was observed in vitro upon sense strand cyclization (Table 2).
Figure 6.
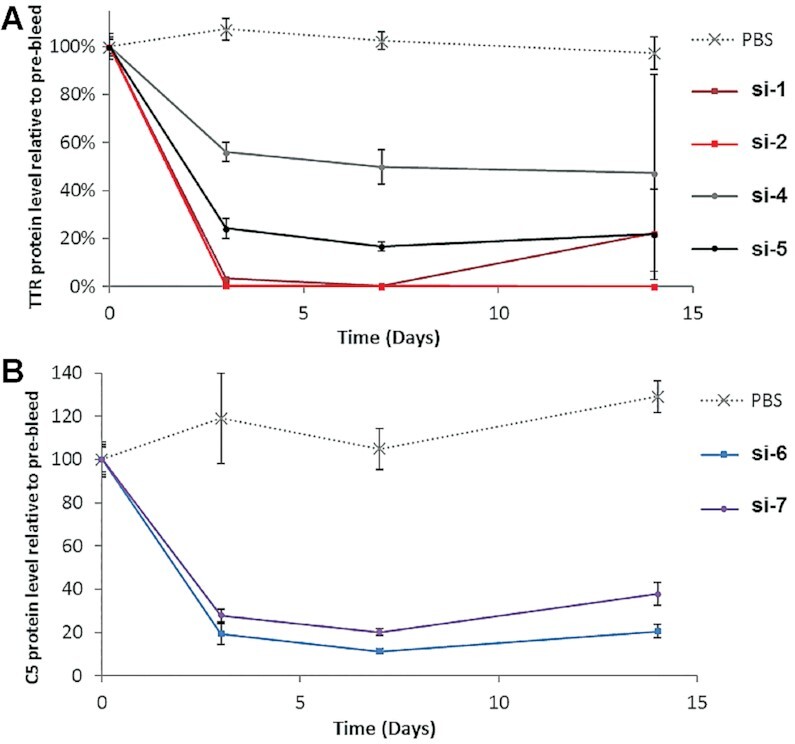
Gene silencing effects of sciRNAs vs. siRNAs in mice. A single dose of siRNA or sciRNA (3 mg/kg) was administered to mice on Day 0, and serum was collected on Days 0 (pre-dose), 3, 7 and 14. Circulating serum protein levels for (A) TTR and (B) C5 were determined relative to PBS-treated groups. Error bars are SD (n = 3). GalNAc-siRNAs with linear sense strands (si-1, si-2 and si-6); GalNAc-sciRNAs with cyclic sense strands, si-4, si-5 and si-7).
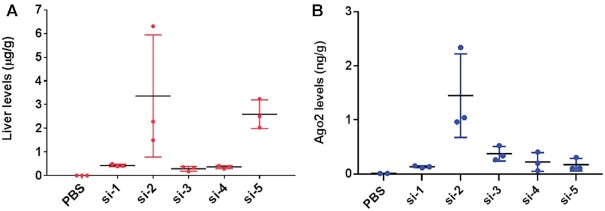
We next evaluated in vivo metabolic stability of sciRNA and linear siRNA controls by measuring the total liver levels and levels of antisense strand loaded in Ago2 following subcutaneous administration of a 3 mg/kg dose in mice. Mouse livers were harvested 7 days after the dose and treated to isolate and quantify the antisense strands by SL-RT qPCR based on a previously published method (52,53) adapted to chemically modified siRNAs (24,27). Modification of the antisense strand with the metabolically stable VP increased the liver levels as both the linear siRNA si-2 and sciRNA si-5, which are modified with VP, were about 5-fold more potent than respective controls with a linear sense strand si-1 and si-4, respectively. Interestingly, whole liver levels of si-2 and si-5 were comparable (Figure 7A), although their pharmacodynamic profiles in mice differed significantly (Figure 7A).
Figure 7.
Antisense strands of sciRNAs are less efficiently loaded onto Ago2 than those of siRNAs and the VP modification enhances metabolic stability of the antisense strands. (A) Liver levels of antisense strands of indicated siRNAs and sciRNAs after subcutaneous dosing in mice. (B) Levels of antisense strand associated with Ago2 in livers treated subcutaneously with indicated siRNAs and sciRNAs. A single dose of each conjugate (3 mg/kg) was administered on Day 0, and livers were collected on Day 7. Levels were determined using SL-RT QPCR relative to PBS groups. Error bars are SD (n = 3). GalNAc-siRNAs with linear sense strands (si-1, si-2, and si-3); GalNAc-sciRNAs with cyclic sense strands, si-4 and si-5).
To evaluate the efficiency of Ago2 loading, we immunoprecipitated Ago2 from mouse whole liver lysate and extracted and quantified antisense strand levels. Consistent with previous reports (24,26), the VP modification enhanced Ago2 loading as shown by comparison of the amount of si-2 antisense strand relative to the non-VP-modified antisense strand of si-1 (Figure 7B). Interestingly, the VP modification did not enhance loaded of the sciRNA to the same extent as shown by comparison of si-5 to si-3 (Figure 7B). VP-modified si-2 with a linear sense strand and VP-modified si-5 with a circular sense strand had similar antisense strand liver levels (Figure 7A) but had different levels of Ago2-loaded antisense strand (Figure 7B). Thus, the antisense strands of sciRNAs had similar metabolic stability as those hybridized to linear sense strands, but their loading efficiency into Ago2 was lower.
DISCUSSION
Straightforward synthesis and characterization of sciRNAs
We describe here a straightforward strategy for the solid-phase synthesis of a new class of fully chemically modified, GalNAc-conjugated siRNAs with a click-cyclized sense strand; we refer to these RNAi-mediating agents as small circular interfering RNAs (sciRNAs) (16). After completion of the synthesis of the linear sense strand carrying a trivalent GalNAc ligand, an alkyne-functionalized linker was coupled at the 5′-end, and the bromide group of the linker was then substituted with an azido group. After cleavage from the solid support, deprotection, and purification, standard click reaction conditions allowed cyclization of the sense strand (48,56). By adapting previously reported methods of click cyclization of azido-alkyne oligonucleotides (48), we demonstrated that the cyclization click reaction can proceed either under microwave activation or at room temperature with almost quantitative conversion of the starting linear oligonucleotide. The isolated yields after stringent purification of the circular strands of about 30% were similar to those of linear oligonucleotides. No linearization or cross-polymerization by-products were observed.
Annealing of the circular sense strands with the corresponding complementary antisense strands yielded the sciRNA duplexes. The sciRNAs evaluated in this study were modified with 2′-OMe, 2′-F and PS as are the GalNAc-conjugated siRNAs used clinically. Our analyses of these sciRNAs are the first to evaluate the effect of sense strand cyclization in therapeutically relevant model systems.
We confirmed that cyclization of oligonucleotides significantly reduced the Tm and thereby the thermal stability of the resulting RNA duplexes. Cyclic DNA and RNA duplex structures have been previously studied by Kool et al. (58,62,63) and Morvan et al. (48). When annealed to short oligonucleotide targets (6-nt to 10-nt), a 20-nt cyclic oligonucleotide formed duplexes with higher Tm values than a 20-nt linear oligonucleotide, demonstrating that a short linear target can be structurally well accommodated with a cyclic DNA strand. However, the complex of a 20-nt cyclic oligonucleotide with a 20-nt target had the same Tm as that of the complex with the 10-nt target, suggesting that only about 10 nucleotides of the target were able to hybridize with the cyclic oligonucleotide. The Tm of the duplex formed by the 20-nt cyclic oligonucleotide with a 20-nt target was about 20°C lower than that of the 20-nt linear duplex (48). Indeed, for DNA, longer nucleotide cycles have been reported to be required for stable circular duplexes (64).
Our thermal melting and NMR analysis of the sciRNAs designed to form 21 bp indicate that when the sense strand has a rigid circular structure only a subset of the nucleobases form Watson–Crick base pairs. We observed fewer and broadened NMR signals in spectra of sciRNAs than linear siRNAs suggesting that in the sciRNAs a heterogeneous mixture of partially base paired duplexes coexists in solution. A recent publication on circular siRNA indicated that a 27-nt circle size appears to be optimal for binding to a 21-nt antisense strand to maintain the biological properties of these molecules (42). The CD spectroscopy analysis, on the other hand, revealed no difference in the duplex structure between linear and circular duplexes, both exhibiting the overall global structure of the typical A-type family.
sciRNAs interact with Ago2
We modeled the interaction of an sciRNA with Ago2 based on the previously described crystal structure of human Ago2 with antisense (AS) and sense (S) strands of an siRNA (54). We built a model in which the sense strand functionalized with the Z linker was cyclized (Figure 8) and in which base pairing was maintained in the entire seed region (AS2:S20–AS8:S14). Pairing across the entire seed region seen in the model is consistent with the stability of the duplex formed by short oligonucleotides and a 20-nt cyclic strand. In our model, the Z linker connects the 3′-terminal sense strand nucleotide (labeled S21 in Figure 8) and the first sense strand residue visible in the crystal structure (labeled S13 in Figure 8). The latter residue was flipped relative to the conformation in the crystal. The Z linker crosses the major groove of the duplex. The sciRNAs tested experimentally have 21-nt sense strand circles rather than the 12-nt circle of the model. However, the Z linker can presumably bridge no more than 8 pairs as per our model, so a sense strand circle of 21 nucleotides must have internal structure such as a hairpin loop. The prolinol moiety adjacent to the 3′-phosphate of residue S21 in our model is oriented such that the hydroxyl group juts outwards, thus placing the GalNAc ligand on the surface of the complex and precluding potential clashes with Ago2 (Figure 8).
Figure 8.
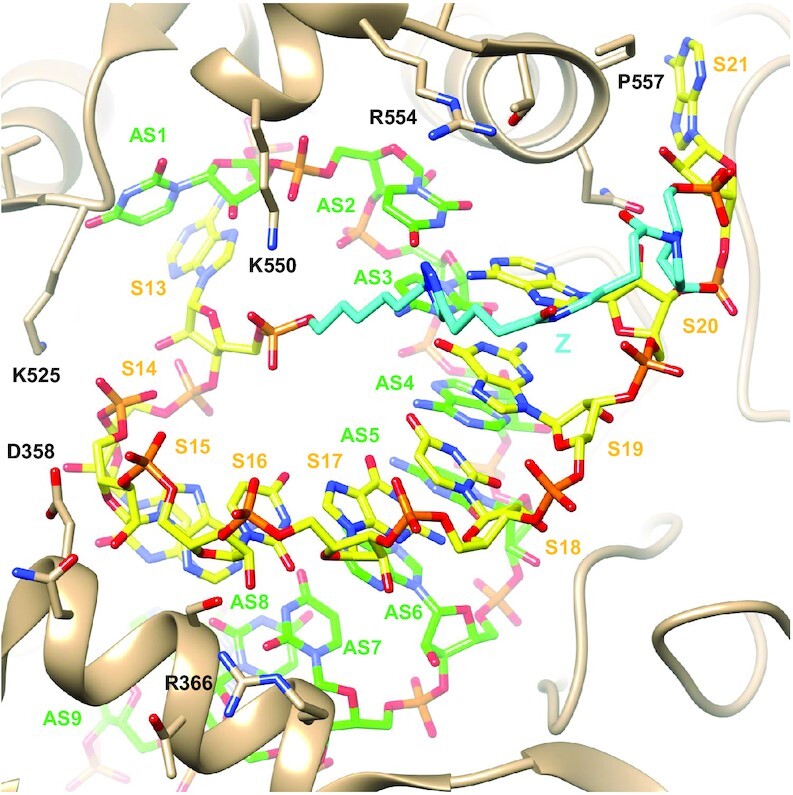
Model of an sciRNA:Ago2 complex suggests that a circular sense strand can be accommodated. The model is based on the crystal structure of Ago2 bound to a duplex siRNA with seed region pairing (54). Carbon atoms of the sense strand (nucleotides S13–S21) are colored in yellow, and carbon atoms of the antisense strand (nucleotides AS1-AS9) are in green. The Z linker carbons are highlighted in cyan. Selected side chains of the Ago2 PIWI and MID domains and the L2 linker region (tan ribbon) are labeled. The view is across the major (top) and minor grooves (bottom) of the seed region duplex.
Sense strand cyclization slightly impairs RNAi activity in vivo but recuperated by chemical modifications
The thermal destabilization induced by the cyclization of the sense strand in sciRNAs adversely impacted potency of RNAi-mediated silencing in vitro, especially when using transfection reagents for intracellular uptake. The potency differences between linear GalNAc–siRNAs and circular GalNAc–sciRNAs was generally less pronounced in mice than in cell culture. The reason for the discrepancy between in vitro and in vivo potency is at this point unclear, but we speculate that it results from the greater impact of metabolic stability on efficacy in vivo. For example, in the Ttr series, addition of the stabilizing VP group in the sciRNA (si-5) had a greater fold impact on in vivo potency than in vitro potency. In mice at Day 14 post dose, the sciRNA with an antisense strand modified with VP had potency equivalent to that of the linear non-VP-modified siRNA (si-1). Analyses of the levels of antisense strands loaded in Ago2, which is a predictive measure of pharmacological effect (13), revealed that levels of antisense strands of sciRNAs are significantly lower than those of the linear siRNA counterparts. This observation supports the hypothesis that Ago2 loading is negatively impacted by the circular nature of sense strand. In addition, despite similar levels of antisense strand associated with Ago2 for si-1, with a linear sense strand, and sciRNAs si-4 and si-5, without and with the VP modification respectively, si-4 was less potent. The reason for this is unclear to us, although additional factors, such as impaired target recognition, slower kinetics of strand cleavage, or decreased removal of the circular sense strand could negatively impact efficacy.
In rat plasma, neither sense nor antisense strands of sciRNAs or siRNAs were degraded at 8 h. However, we observed considerable linearization of the circular sense strand of the sciRNA after 24 h of incubation. This indicates that the circular structure can act as a pro-drug, with hydrolysis (presumably of a phosphate group), resulting in a linear oligonucleotide. Interestingly, this hydrolysis did not occur when the compounds were incubated in rat liver homogenates. GalNAc-conjugated siRNA is rapidly absorbed into liver through efficient ASGPR-mediated uptake (27,65). Thus, linearization probably does not have an impact on the potency in vivo due to rapid uptake of GalNAc–siRNA by hepatocytes.
The in vitro and in vivo potencies we measured and the partial duplex structure observed in our molecular models of the sciRNA with Ago2 positions the sciRNA constructs somewhere between canonical siRNA duplexes and single-stranded siRNAs (ss-siRNAs). The ss-siRNAs are between 10- and 100-fold less potent than linear, duplexed siRNAs (66,67). It is also worth noting that in the case of ss-siRNA the use of a VP group is strictly required for loading onto Ago2 and RNAi activity (66), whereas this was not the case for sciRNAs, although there was a potency benefit when sciRNAs were modified with VP. Our data suggest that the sense strand plays a critical role in loading of the siRNA onto Ago2 (56). In these studies, the sense strand is also important as the carrier for the GalNAc ligand (16). The efficacy of GalNAc-conjugated sciRNAs into hepatocytes demonstrates that the cyclic sense strand is complexed with the antisense strand despite the relative instability. Additionally, this confirms that the GalNAc ligand can effectively deliver cargo when placed internally, as well as terminally, which is in line with previous results from our group (68). Thus, VP in the antisense strand and the internal GalNAc ligand placement can compensate for the slight loss in potency.
Potential benefits of sense strand cyclization
As RNAi therapeutics, sciRNAs could provide beneficial features including metabolic stabilization, resulting in extended duration of action, and reduced off-target binding. Further circular RNA constructs could be engineered to exhibit tailored three-dimensional structures that could influence tissue distribution and tissue retention, minimize undesired protein binding, and optimize ligand-receptor interactions due to changes in physical properties. In the current landscape of RNAi therapies, a number of approved medicines and advanced drug candidates in human clinical trials have been optimized for targeted delivery to the liver. It remains to be seen whether similar rules of drug design will universally apply to the successful delivery of siRNAs to extrahepatic tissues and various associated cell types as well. Work along these challenging lines warrants exploration of new classes of chemical and structural modifications. An understanding of the impact of structural motifs on potency and safety will pave the paths for future development for cell- and tissue-specific delivery of RNAi therapeutics. siRNAs with modified structures will also expand the toolbox of RNAi therapeutics and off-target reduction and enhanced metabolic stabilization will provide additional benefits when incorporated into the initial RNAi screening process.
The present study shed light on the architecture of the sense strand of the siRNA duplex, which has not been as well studied as the antisense strand, despite its importance in the RNAi process. In our hands, single-stranded siRNAs do not function well, but sciRNAs have potency only slightly less than an siRNA in vivo. To the best of our knowledge, this work is the first example of partial-duplex siRNA constructs that are functionally effective at pharmacologically and therapeutically relevant doses in vivo. This work sets the stage for development of innovative designs for therapeutic siRNA molecules likely to have enhanced potency resulting from increased metabolic stability and decreased off-target properties. The linker length of the circle could be optimized to increase the base-pairing interactions between sense and antisense strands, which may lead to further improvement in potency. Evaluating the impact of changes in the three-dimensional siRNA duplex structure and study of interactions of the resulting structures with enzymes involved in metabolic pathways and RISC machinery will also improve our mechanistic understanding of our RNAi therapeutic platform.
Broader implications and conclusions
With the current explosion in use of RNA-based molecules in therapeutic applications, circular RNAs, both natural and unnatural have been the focus of considerable attention (69). Such circular RNAs can have regulatory functions (70) by, for example, serving as sponges for miRNAs (71). Unmodified exogenous circular RNAs have been shown to bypass cellular RNA sensors such as RIG-I and Toll-like receptors, and thus do not provoke an immune response (72). With the revolution of mRNA vaccines, novel cyclic RNA therapeutic modalities such as endless RNAs and O RNAs have been proposed. Effective methods for large-scale synthesis will be required for preparation of circRNAs used clinically. In this context, it is provocative to think about how we can apply the methods and chemistry described here for sciRNAs to larger circRNAs. We expect that longer RNAs can be circularized using the click-type chemistry strategy, perhaps using an oligonucleotide to bring ends into proximity. Furthermore, our strategy will allow introduction of chemical modifications into circRNAs that will affect stability, localization, immune response, and therapeutic activities. Interestingly, a circAgo2 RNA has been identified (8) and sciRNAs may be useful tools to study the function of this molecule.
A number of questions remain: The circular sense strand binding to the guide strand is considerably less stable thermodynamically than binding of a linear strand as proven by the NMR experiment. The level of tolerance for thermodynamic stability remains unclear. Although we have demonstrated circle formation by click chemistry, approaches more pharmacologically friendlier than copper-mediated click chemistry are desired. Although circularization improves nuclease mediated biostability, metabolism is a species-dependent phenomenon, and this creates uncertainty in clinical translation. Finally, the safety features of such novel constructs need to be evaluated.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor François Morvan (Université de Montpellier) and Dr Derek O’Flaherty (Alnylam) for critical review of the manuscript and Dr Kevin Fitzgerald (Alnylam) for suggestions and support for this work. We thank Dr Anne Noronha and Professor Christopher J. Wilds (Concordia University) for performing the CD experiments.
Contributor Information
Hartmut Jahns, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Rohan Degaonkar, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Peter Podbevsek, Slovenian NMR Center, National Institute of Chemistry, Hajdrihova 19, SI-1001 Ljubljana, Slovenia, EU.
Swati Gupta, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Anna Bisbe, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Krishna Aluri, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
John Szeto, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Pawan Kumar, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Sarah LeBlanc, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Tim Racie, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Christopher R Brown, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Adam Castoreno, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Dale C Guenther, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Vasant Jadhav, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Martin A Maier, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Janez Plavec, Slovenian NMR Center, National Institute of Chemistry, Hajdrihova 19, SI-1001 Ljubljana, Slovenia, EU.
Martin Egli, Department of Biochemistry, School of Medicine, Vanderbilt University, Nashville, TN 37232, USA.
Muthiah Manoharan, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Ivan Zlatev, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Alnylam Pharmaceuticals. Funding for open access charge: Alnylam Pharmaceuticals.
Conflict of interest statement. H.J., R.D., S.G., A.B., P.K., K.A., J.S., S.LB., T.R., C.R.B., D.C.G., A.C., V.J., M.A.M., M.M. and I.Z. are currently, or were during the time this work was conducted, employees of Alnylam Pharmaceuticals.
REFERENCES
- 1. Danan M., Schwartz S., Edelheit S., Sorek R.. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2011; 40:3131–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O.. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012; 7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lukiw W. Circular RNA (circRNA) in Alzheimer's disease (AD). Front. Genet. 2013; 4:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tan K.-E., Lim Y.-Y.. Viruses join the circular RNA world. The FEBS J. 2020; 288:4488–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu M., Xie F., Tang X., Wang T., Wang S.. Insights into the role of circular RNA in macrophage activation and fibrosis disease. Pharmacol. Res. 2020; 156:104777. [DOI] [PubMed] [Google Scholar]
- 6. Patop I.L., Kadener S.. circRNAs in cancer. Curr. Opin. Genet. Dev. 2018; 48:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sarkar D., Diermeier S.D.. Circular RNAs: potential applications as therapeutic targets and biomarkers in breast cancer. Non-Coding RNA. 2021; 7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Y., Yang F., Fang E., Xiao W., Mei H., Li H., Li D., Song H., Wang J., Hong M.et al.. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019; 26:1346–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Errichelli L., Dini Modigliani S., Laneve P., Colantoni A., Legnini I., Capauto D., Rosa A., De Santis R., Scarfò R., Peruzzi G.et al.. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017; 8:14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hanan M., Simchovitz A., Yayon N., Vaknine S., Cohen-Fultheim R., Karmon M., Madrer N., Rohrlich T.M., Maman M., Bennett E.R.et al.. A Parkinson's disease CircRNAs resource reveals a link between circSLC8A1 and oxidative stress. EMBO Mol. Med. 2020; 12:e11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang Q., Li F., He A.T., Yang B.B.. Circular RNAs: expression, localization, and therapeutic potentials. Mol. Ther. 2021; 29:1683–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu X., Abraham J.M., Cheng Y., Wang Z., Wang Z., Zhang G., Ashktorab H., Smoot D.T., Cole R.N., Boronina T.N.et al.. Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol. Ther. - Nucleic Acids. 2018; 13:312–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jost I., Shalamova L.A., Gerresheim G.K., Niepmann M., Bindereif A., Rossbach O.. Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol. 2018; 15:1032–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C.. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998; 391:806–811. [DOI] [PubMed] [Google Scholar]
- 15. Setten R.L., Rossi J.J., Han S.-P.. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019; 18:421–446. [DOI] [PubMed] [Google Scholar]
- 16. Nair J.K., Willoughby J.L., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel’in A.V., Milstein S.et al.. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014; 136:16958–16961. [DOI] [PubMed] [Google Scholar]
- 17. Springer A.D., Dowdy S.F.. GalNAc-siRNA conjugates: leading the way for delivery of RNAi therapeutics. Nucleic Acid Ther. 2018; 28:109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fitzgerald K., Kallend D., Simon A.. A highly durable RNAi therapeutic inhibitor of PCSK9. N. Engl. J. Med. 2017; 376:e38. [DOI] [PubMed] [Google Scholar]
- 19. Pasi K.J., Rangarajan S., Georgiev P., Mant T., Creagh M.D., Lissitchkov T., Bevan D., Austin S., Hay C.R., Hegemann I.et al.. Targeting of antithrombin in hemophilia A or B with RNAi therapy. N. Engl. J. Med. 2017; 377:819–828. [DOI] [PubMed] [Google Scholar]
- 20. Zimmermann T.S., Karsten V., Chan A., Chiesa J., Boyce M., Bettencourt B.R., Hutabarat R., Nochur S., Vaishnaw A., Gollob J.. Clinical proof of concept for a novel hepatocyte-targeting GalNAc-siRNA conjugate. Mol. Ther. 2017; 25:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sardh E., Harper P., Balwani M., Stein P., Rees D., Bissell D.M., Desnick R., Parker C., Phillips J., Bonkovsky H.L.et al.. Phase 1 trial of an RNA interference therapy for acute intermittent porphyria. N. Engl. J. Med. 2019; 380:549–558. [DOI] [PubMed] [Google Scholar]
- 22. Raal F.J., Kallend D., Ray K.K., Turner T., Koenig W., Wright R.S., Wijngaard P.L.J., Curcio D., Jaros M.J., Leiter L.A.et al.. Inclisiran for the treatment of heterozygous familial hypercholesterolemia. N. Engl. J. Med. 2020; 382:1520–1530. [DOI] [PubMed] [Google Scholar]
- 23. Ray K.K., Wright R.S., Kallend D., Koenig W., Leiter L.A., Raal F.J., Bisch J.A., Richardson T., Jaros M., Wijngaard P.L.J.et al.. Two Phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N. Engl. J. Med. 2020; 382:1507–1519. [DOI] [PubMed] [Google Scholar]
- 24. Elkayam E., Joshua-Tor L., Brown C.R., Theile C.S., Willoughby J.L., Parmar R., Manoharan M.. siRNA carrying an (E)-vinylphosphonate moiety at the 5′ end of the guide strand augments gene silencing by enhanced binding to human Argonaute-2. Nucleic Acids Res. 2016; 45:3528–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Janas M.M., Jiang Y., Duncan R.G., Hayes A.N., Liu J., Kasperkovitz P.V., Placke M.E., Barros S.A.. Exposure to siRNA-GalNAc conjugates in systems of the standard test battery for genotoxicity. Nucleic Acid Ther. 2016; 26:363–371. [DOI] [PubMed] [Google Scholar]
- 26. Parmar R., Willoughby J.L., Liu J., Foster D.J., Brigham B., Theile C.S., Charisse K., Akinc A., Guidry E., Pei Y.et al.. 5′-(E)-Vinylphosphonate: a stable phosphate mimic can improve the RNAi activity of siRNA-GalNAc conjugates. ChemBioChem. 2016; 17:985–989. [DOI] [PubMed] [Google Scholar]
- 27. Nair J.K., Attarwala H., Sehgal A., Wang Q., Aluri K., Zhang X., Gao M., Liu J., Indrakanti R., Schofield S.et al.. Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc-siRNA conjugates. Nucleic Acids Res. 2017; 45:10969–10977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Foster D.J., Brown C.R., Shaikh S., Trapp C., Schlegel M.K., Qian K., Sehgal A., Rajeev K.G., Jadhav V., Manoharan M.et al.. Advanced siRNA designs further improve in vivo performance of GalNAc-siRNA conjugates. Mol. Ther. 2018; 26:708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Janas M.M., Harbison C.E., Perry V.K., Carito B., Sutherland J.E., Vaishnaw A.K., Keirstead N.D., Warner G.. The nonclinical safety profile of GalNAc-conjugated RNAi therapeutics in subacute studies. Toxicol. Pathol. 2018; 46:735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janas M.M., Schlegel M.K., Harbison C.E., Yilmaz V.O., Jiang Y., Parmar R., Zlatev I., Castoreno A., Xu H., Shulga-Morskaya S.et al.. Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat. Commun. 2018; 9:723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Janas M.M., Zlatev I., Liu J., Jiang Y., Barros S.A., Sutherland J.E., Davis W.P., Liu J., Brown C.R., Liu X.et al.. Safety evaluation of 2′-deoxy-2′-fluoro nucleotides in GalNAc-siRNA conjugates. Nucleic Acids Res. 2019; 47:3306–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khvorova A., Watts J.K.. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotech. 2017; 35:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Egli M., Manoharan M.. Re-engineering RNA molecules into therapeutic agents. Acc. Chem. Res. 2019; 52:1036–1047. [DOI] [PubMed] [Google Scholar]
- 34. Yu J.-Y., DeRuiter S.L., Turner D.L.. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abe N., Abe H., Ito Y.. Dumbbell-shaped nanocircular RNAs for RNA interference. J. Am. Chem. Soc. 2007; 129:15108–15109. [DOI] [PubMed] [Google Scholar]
- 36. Kim H., Lee J.S., Lee J.B.. Generation of siRNA nanosheets for efficient RNA interference. Sci. Rep. 2016; 6:25146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Avino A., Ocampo S.M., Perales J.C., Eritja R.. Branched RNA: a new architecture for RNA interference. J. Nucleic Acids. 2011; 2011:586935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang L., Liang D., Wang Y., Li D., Zhang J., Wu L., Feng M., Yi F., Xu L., Lei L.et al.. Caged circular siRNAs for photomodulation of gene expression in cells and mice. Chem. Sci. 2018; 9:44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kimura Y., Shu Z., Ito M., Abe N., Nakamoto K., Tomoike F., Shuto S., Ito Y., Abe H.. Intracellular build-up RNAi with single-strand circular RNAs as siRNA precursors. Chem. Commun. 2020; 56:466–469. [DOI] [PubMed] [Google Scholar]
- 40. Abe N., Abe H., Ohshiro T., Nakashima Y., Maeda M., Ito Y.. Synthesis and characterization of small circular double-stranded RNAs. Chem. Commun. 2011; 47:2125–2127. [DOI] [PubMed] [Google Scholar]
- 41. Zhang L., Liang D., Chen C., Wang Y., Amu G., Yang J., Yu L., Dmochowski I.J., Tang X.. Circular siRNAs for reducing off-target effects and enhancing long-term gene silencing in cells and mice. Mol. Ther. - Nucleic Acids. 2018; 10:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hagiwara K., Honma M., Harumoto T., Harada K., Sawada T., Yamamoto J., Shinohara F.. Development of prodrug type circular siRNA for in vivo knockdown by systemic administration. Nucleic Acid Ther. 2020; 30:346–364. [DOI] [PubMed] [Google Scholar]
- 43. Prakash T.P., Kinberger G.A., Murray H.M., Chappell A., Riney S., Graham M.J., Lima W.F., Swayze E.E., Seth P.P.. Synergistic effect of phosphorothioate, 5′-vinylphosphonate and GalNAc modifications for enhancing activity of synthetic siRNA. Bioorg. Med. Chem. Lett. 2016; 26:2817–2820. [DOI] [PubMed] [Google Scholar]
- 44. Parmar R.G., Brown C.R., Matsuda S., Willoughby J.L.S., Theile C.S., Charissé K., Foster D.J., Zlatev I., Jadhav V., Maier M.A.et al.. Facile synthesis, geometry, and 2′-substituent-dependent in vivo activity of 5′-(E)- and 5′-(Z)-vinylphosphonate-modified siRNA conjugates. J. Med. Chem. 2018; 61:734–744. [DOI] [PubMed] [Google Scholar]
- 45. Jayaprakash K.N., Peng C.G., Butler D., Varghese J.P., Maier M.A., Rajeev K.G., Manoharan M.. Non-nucleoside building blocks for copper-assisted and copper-free click chemistry for the efficient synthesis of RNA conjugates. Org. Lett. 2010; 12:5410–5413. [DOI] [PubMed] [Google Scholar]
- 46. O'Shea J., Theile C.S., Das R., Babu I.R., Charisse K., Manoharan M., Maier M.A., Zlatev I.. An efficient deprotection method for 5′-[O,O-bis(pivaloyloxymethyl)]-(E)-vinylphosphonate containing oligonucleotides. Tetrahedron. 2018; 74:6182–6186. [Google Scholar]
- 47. Lietard J., Meyer A., Vasseur J.-J., Morvan F.. An efficient reagent for 5′-azido oligonucleotide synthesis. Tetrahedron Lett. 2007; 48:8795–8798. [Google Scholar]
- 48. Lietard J., Meyer A., Vasseur J.-J., Morvan F.. New strategies for cyclization and bicyclization of oligonucleotides by click chemistry assisted by microwaves. J. Org. Chem. 2008; 73:191–200. [DOI] [PubMed] [Google Scholar]
- 49. Liu J., Li J., Tran C., Aluri K., Zhang X., Clausen V., Zlatev I., Guan L., Chong S., Charisse K.et al.. Oligonucleotide quantification and metabolite profiling by high-resolution and accurate mass spectrometry. Bioanalysis. 2019; 11:1967–1980. [DOI] [PubMed] [Google Scholar]
- 50. Chan T.S., Yu H., Moore A., Khetani S.R., Tweedie D. Meeting the challenge of predicting hepatic clearance of compounds slowly metabolized by cytochrome P450 using a novel hepatocyte model, HepatoPac. Drug Metab. Dispos. 2013; 41:2024–2032. [DOI] [PubMed] [Google Scholar]
- 51. Ramsden D., Zhou J., Tweedie D.J.. Determination of a degradation constant for CYP3A4 by direct suppression of mRNA in a novel human hepatocyte model, HepatoPac. Drug Metab. Dispos. 2015; 43:1307–1315. [DOI] [PubMed] [Google Scholar]
- 52. Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R.et al.. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res. 2005; 33:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pei Y., Hancock P.J., Zhang H., Bartz R., Cherrin C., Innocent N., Pomerantz C.J., Seitzer J., Koser M.L., Abrams M.T.et al.. Quantitative evaluation of siRNA delivery in vivo. RNA. 2010; 16:2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schirle N.T., Sheu-Gruttadauria J., MacRae I.J.. Structural basis for microRNA targeting. Science. 2014; 346:608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 56. Huisgen R. Pure Appl. Chem. 1989; 61:613. [Google Scholar]
- 57. Eltepu L., Jayaraman M., Rajeev K.G., Manoharan M.. An immobilized and reusable Cu(I) catalyst for metal ion-free conjugation of ligands to fully deprotected oligonucleotides through click reaction. Chem. Commun. 2013; 49:184–186. [DOI] [PubMed] [Google Scholar]
- 58. Kool E.T., Design Topological modification of oligonucleotides for potential inhibition of gene expression. Persp. Drug Discov. 1996; 4:61–75. [Google Scholar]
- 59. Crooke R.M., Graham M.J., Martin M.J., Lemonidis K.M., Wyrzykiewiecz T., Cummins L.L.. Metabolism of antisense oligonucleotides in rat liver homogenates. J. Pharma. Exp. Ther. 2000; 292:140–149. [PubMed] [Google Scholar]
- 60. Baek M.-S., Yu R.Z., Gaus H., Grundy J.S., Geary R.S.. In vitro metabolic stabilities and metabolism of 2′-O-(methoxyethyl) partially modified phosphorothioate antisense oligonucleotides in preincubated rat or human whole liver homogenates. Oligonucleotides. 2010; 20:309–316. [DOI] [PubMed] [Google Scholar]
- 61. Kusner L.L., Yucius K., Sengupta M., Sprague A.G., Desai D., Nguyen T., Charisse K., Kuchimanchi S., Kallanthottathil R., Fitzgerald K.et al.. Investigational RNAi therapeutic targeting C5 is efficacious in pre-clinical models of myasthenia gravis. Mol. Ther. - Meth. Clin. Dev. 2019; 13:484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Prakash G., Kool E.T.. Structural effects in the recognition of DNA by circular oligonucleotides. J. Am. Chem. Soc. 1992; 114:3523–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kool E.T. Recognition of DNA, RNA, and proteins by circular oligonucleotides. Acc. Chem. Res. 1998; 31:502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kumar R., El-Sagheer A., Tumpane J., Lincoln P., Wilhelmsson L.M., Brown T.. Template-directed oligonucleotide strand ligation, covalent intramolecular DNA circularization and catenation using click chemistry. J. Am. Chem. Soc. 2007; 129:6859–6864. [DOI] [PubMed] [Google Scholar]
- 65. Ramsden D., Wu J.-T., Zerler B., Iqbal S., Jiang J., Clausen V., Aluri K., Gu Y., Dennin S., Kim J.et al.. In vitro drug-drug interaction evaluation of GalNAc conjugated siRNAs against CYP450 enzymes and transporters. Drug Metab. Dispos. 47. 2019; 1183–1194. [DOI] [PubMed] [Google Scholar]
- 66. Lima Walt F., Prakash Thazha P., Murray Heather M., Kinberger Garth A., Li W., Chappell Alfred E., Li Cheryl S., Murray Susan F., Gaus H., Seth Punit Pet al.. Single-stranded siRNAs Activate RNAi in animals. Cell. 2012; 150:883–894. [DOI] [PubMed] [Google Scholar]
- 67. Prakash T.P., Lima W.F., Murray H.M., Elbashir S., Cantley W., Foster D., Jayaraman M., Chappell A.E., Manoharan M., Swayze E.E.et al.. Lipid nanoparticles improve activity of single-stranded siRNA and gapmer antisense oligonucleotides in animals. ACS Chem. Biol. 2013; 8:1402–1406. [DOI] [PubMed] [Google Scholar]
- 68. Rajeev K.G., Nair J.K., Jayaraman M., Charisse K., Taneja N., O'Shea J., Willoughby J.L.S., Yucius K., Nguyen T., Shulga-Morskaya S.et al.. Hepatocyte-specific delivery of siRNAs conjugated to novel non-nucleosidic trivalent N-acetylgalactosamine elicits robust gene silencing in vivo. ChemBioChem. 2015; 16:903–908. [DOI] [PubMed] [Google Scholar]
- 69. Kosik K.S. Circles reshape the RNA world. Nature. 2013; 495:322–324. [DOI] [PubMed] [Google Scholar]
- 70. Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M.et al.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013; 495:333–338. [DOI] [PubMed] [Google Scholar]
- 71. Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J.. Natural RNA circles function as efficient microRNA sponges. Nature. 2013; 495:384–388. [DOI] [PubMed] [Google Scholar]
- 72. Wesselhoeft R.A., Kowalski P.S., Parker-Hale F.C., Huang Y., Bisaria N., Anderson D.G.. RNA circularization diminishes immunogenicity and can extend translation duration. Mol. Cell. 2019; 74:508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.