Abstract
DNA is intrinsically dynamic and folds transiently into alternative higher-order structures such as G-quadruplexes (G4s) and three-way DNA junctions (TWJs). G4s and TWJs can be stabilised by small molecules (ligands) that have high chemotherapeutic potential, either as standalone DNA damaging agents or combined in synthetic lethality strategies. While previous approaches have claimed to use ligands that specifically target either G4s or TWJs, we report here on a new approach in which ligands targeting both TWJs and G4s in vitro demonstrate cellular effects distinct from that of G4 ligands, and attributable to TWJ targeting. The DNA binding modes of these new, dual TWJ-/G4-ligands were studied by a panel of in vitro methods and theoretical simulations, and their cellular properties by extensive cell-based assays. We show here that cytotoxic activity of TWJ-/G4-ligands is mitigated by the DNA damage response (DDR) and DNA topoisomerase 2 (TOP2), making them different from typical G4-ligands, and implying a pivotal role of TWJs in cells. We designed and used a clickable ligand, TrisNP-α, to provide unique insights into the TWJ landscape in cells and its modulation upon co-treatments. This wealth of data was exploited to design an efficient synthetic lethality strategy combining dual ligands with clinically relevant DDR inhibitors.
Graphical Abstract
Graphical Abstract.
Small molecules that concomitantly target three-way DNA junctions (TWJs) and G-quadruplexes (G4s) are used here to study the landscape of alternative DNA folds and their potential as therapeutic targets to trigger DNA structure-dependent DNA damage in human cells.
INTRODUCTION
DNA is constantly damaged by exogeneous and endogenous processes. Lesions, such as modifications and loss of bases or replication errors, interfer with DNA transactions (i.e. replication and transcription) and induce DNA damage signalling and single- (SSB) and double-strand breaks (DSB) (1–3). A complex mechanism, known as the DNA damage response (DDR), exists in higher organisms to sense damage, block the cell cycle and repair DNA lesions via a large variety of mechanisms. DSBs are the most dangerous and deleterious of all lesions, and two major pathways exist to repair them: homologous recombination (HR) and non-homologous end joining (NHEJ) (4–5). HR relies on a homologous template for repair, is mediated by a large number of proteins, including RAD51, and is stimulated by the activation of two kinases, namely ATM (ataxia telangiectasia mutated) kinase at DSBs, and ATR (ATM and Rad3-related) kinase at stalled replication forks. NHEJ consists in the direct ligation of the two DNA ends, and is mediated by the DNA-dependent protein kinase (DNA-PK) and DNA ligase IV-XRCC4 complexes. Prolonged DNA damage signalling induces senescence or programmed cell death (4–6).
Cancer cells are commonly deficient in DDR pathways, which can be seen as a therapeutic advantage to exploit in the fight against cancer cell proliferation (1,7). Classic chemotherapy and radiotherapy function by inducing DNA damage (3,8), which is particularly lethal in fast dividing cells with impaired DDR machinery. More recently developed treatments have improved specificity by exploiting synthetic lethal interactions between the inactivation of different DNA repair mechanisms: while the inactivation of a single DNA repair mechanism is tolerated via use of a second compensatory mechanism, the concomitant inactivation of the two mechanisms is synthetic lethal. Based on cancer's biomarkers, specific DDR deficiencies can be identified and treated by inhibitors targeting the compensatory mechanisms which are necessary for cancer cell survival (9–10).
An alternative to creating DNA damage with broad-spectrum agents, such as DNA-alkylating chemicals, ionising radiation and radiomimetics, is to create structure-specific DNA damage. Indeed, DNA is no longer seen as a rigid double helix: mounting evidence indicates that it behaves as a highly dynamic biopolymer (11–15), folding into alternative (i.e. non-B helical) DNA structures, including left-handed Z-DNA (16–17), triplex DNA (18), R-loops (19–21) and point structures such as G-quadruplexes (G4s) (22–25), i-motifs (26), three-way junctions (TWJ, or slipped loops or hairpins) (13) and four-way junctions (FWJ, or cruciform DNA or Holliday junctions) (13,27). Secondary structures are most likely transient and have a higher tendency to form during DNA transactions when the chromatin structure is open, the two strands are separated and torsional stress twists the DNA. G4s are the most studied higher-order structures for their possible regulatory roles in gene expression and replication origins (28–29). Besides these useful roles, excessive or prolonged formation of alternative structures is also correlated to replication stress (30–32), and non-B structure-forming sequences are enriched at mutation sites in cancer genomes (33). For example, TWJ- and FWJ-forming sequences are more concentrated at replication fork stall sites in various cell types in independent studies (34–36). DNA structures are known to impede DNA transactions in vitro, acting as roadblocks that cannot be bypassed efficiently by DNA-associated enzymes (27,37) and represent a window of opportunity to create DNA damage by using ad hoc ligands (3).
Numerous G4-interacting small molecules (G4 ligands (38), Figure 1A) have been shown to induce DNA damage and replicative stress signalling (39–40), in several cases at telomeres (41–44), and have shown promising therapeutic properties in synthetic lethality strategies when used in DDR-deficient systems (40,42,45–47) and in combination with DNA-damaging or DDR-inhibiting drugs (48–52). In contrast, DNA junction-targeting agents are far less developed in a cellular context (3,53). Hannon and co-workers first characterized a TWJ-binding supramolecular iron(II) helicate ligand (Figure 1B) (54), demonstrated its ability to impede DNA transactions in vitro (55) and its modest cytotoxic activity in cells (56). More recently, Vazquez and co-workers used a fluorescently labelled iron(II) helicate to label TWJs in cells (57), and Nitschke, Keyser and co-workers used a tetrahedral cage to detect mismatched DNA architectures in vitro via fluorescence quenching (58). We also reported on the ability of metallacages to interact with TWJs (59–60), but have chosen to focus on the pharmacologically more relevant small organic molecules, notably the azacryptand TrisPOB (vide infra), to study the antitumoral and DNA damage-inducing properties of TWJ ligands and their synergistic combination with DNA repair inhibitors (61).
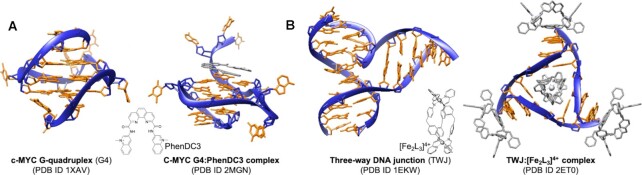
Figure 1.
Crystal or NMR structures of G4-/TWJ-forming oligonucleotides without or with ligand: (A) G4 (62) with PhenDC3; (63) (B) TWJ (64) with the iron supramolecular helicate [Fe2L3]4+(54).
Mergny et al. pioneered the study of azacryptands as ligands of higher-order DNA structures by demonstrating the ability of tris-acridine TrisA to bind to imperfect DNA hairpins that fold from trinucleotide repeats (65). Tris-naphthalene TrisNP (Figure 2A) was the first azacryptand described to bind TWJs in vitro (66); however, this ligand had initially been discarded from our search for highly specific TWJ ligands because of unwarranted interactions with G4s (67). In this regard, we studied TrisPOB, another azacryptand (Figure 2A), for its promising cellular properties (61). We now show that the potential concomitant targeting of two secondary DNA structures can yield real therapeutic dividends, echoing the ‘multitargeting concept’ by S. Neidle (38) that describes the targeting of mutiple G4 sites, here extended to multiple folded DNA structures. We thus investigated the genotoxic potential of a novel class of dual TWJ-/G4-ligands and report herein on the powerful antitumoral and synergistic properties of TrisNP upon co-treatment with DDR inhibitors in cancer models.
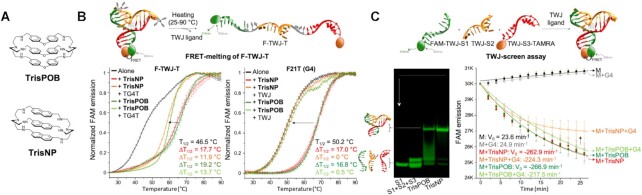
Figure 2.
(A) Chemical structures of TrisPOB and TrisNP. (B) Competitive FRET-melting assays using labelled intramolecular TWJ- and G4-forming oligonucleotides: experiments performed from 25 to 90°C with F-TWJ-T (0.2 μM) in the presence of TrisNP or TrisPOB (1.0 μM, 5 mol. eq.) and competitive non-fluorescent G4 TG4T (50 mol. eq.) (left panel) and labelled G4 F21T (0.2 μM) in the presence of TrisNP or TrisPOB (1.0 μM, 5 mol. eq.) and competitive non-fluorescent TWJ (15 mol. eq.) (right panel). (C) TWJ-folding monitored by either PAGE (TWJ-S1 (5.0 μM), TWJ-S1 + TWJ-S2 + TWJ-S3 (M, 5.0 μM), M + TrisPOB and M + TrisNP, (5 mol. eq., 1 h; gels post-stained with SybrGold)) (left) or TWJ-Screen assays performed with a mixture of FAM-TWJ-S1, TWJ-S2 and TWJ-S3-TAMRA (M, 0.2 μM) alone or in presence of ligand (5 mol. eq.), with or without competitive TG4T (5 mol. eq.). Control experiments are provided in Supplementary Figure S1.
MATERIALS AND METHODS
FRET-melting assay
Experiments were performed in a 96-well plate (Agilent) using an Agilent Mx3005P equipped with FAM filters (λex = 492 nm; λem = 516 nm). Experiments were performed in CacoK buffer (10 mM lithium cacodylate buffer with 10 mM KCl and 90 mM LiCl, pH 7.2) (final volume: 100 μl per well) with 0.2 μM DNA (labelled sequence F-TWJ-T and F21T) and 1 μM ligand. The microplate was centrifuged quickly (10 s) and then placed into the Mx3005P. After an initial equilibration step (25°C, 30 s), a rapid increase to 90°C is followed by a stepwise decrease (1°C every 30 s, 65 cycles) to 25°C and then a stepwise increase (1°C every 30 s, 65 cycles) to reach 90°C again, and measurements were made after each cycle. Final data were analyzed with Excel and OriginPro 9. The emission of FAM was normalized (0 to 1), and T1/2 was defined as the temperature for which the normalized emission was 0.5; the reported ΔT1/2 values were means of 3 experiments. Competitive experiments were performed similarly, with labelled DNA (F-TWJ-T and F21T, 0.2 μM) in the presence of ligand (1.0 μM, 5 molar equiv.) and increasing amounts of the unlabelled competitor TWJ and TG4T (3.0 and 10.0 μM, 15 and 50 molar equiv.). F-TWJ-T: FAM-d[5′A(CT)2(TC)2G-T6-C-(GA)2GCGAC-T6-GTCGC(AG)2T3′]-TAMRA); F21T: FAM-d[5′G3(T2AG3)33′]-TAMRA; TG4T: d[5′(TG4T)3′]4; TWJ: TWJ-S1 (d[5′CG2A2CG2CACTCG3′]) + TWJ-S2 (d-[5′CGAGTGCAGCGTG23′]) + TWJ-S3 (d[5′C2ACGCTCGT2C2G3′]).
Polyacrylamide gel electrophoresis
Nondenaturing PAGE was performed in 15% polyacrylamide gel. Samples were prepared in 15 μl solutions of DNA or DNA/ligand mixtures. Each solution was prepared separately: TWJ-S1 alone (5 μM), [TWJ-S1 + TWJ-S2 + TWJ-S3] (or M) (5 μM), [M (5 μM) + 5 molar equiv. ligand (25 μM)]. The solutions were stirred for 1 h at 25°C during which time the gel was stacked at 7 W (5 min, 150−180 V, 43−38 mA) in TBE buffer enriched with 100 mM NaCl, pH 8.3 (68). DNA loading dye (6×, 3 μl) was added to each 15 μl solution of DNA/ligand, mixed briefly and loaded onto the gel (10 μl per well), and electrophoresis migration was performed at 7 W (<1 h). After migration, gels were stained (SYBR Gold solution, 1:10 000, 15 min, 25°C under gentle agitation) and visualized with a UVP MultiDoc-It imaging system (λex = 302 nm).
TWJ-Screen
Experiments were performed in a 96-well format using a BMGLabtech CLARIOStar machine equipped with FAM filters (λex = 492 nm; λem = 516 nm) at 25°C (59). Each experiment is performed in CacoK buffer (final volume: 100 μl per well), with FAM-TWJ-S1 alone (0.2 μM) and in presence of ligands (5 molar equiv.), or with mixture of FAM-TWJ-S1, TWJ-S2 and TWJ-S3-TAMRA (M, 0.2 μM), alone or in presence of ligand (5 molar equiv.), with or without a competitive G4 (TG4T, 5 molar equiv.). Fluorescence intensity measurements were taken every 2 min over 30 min. Initial velocity (V0) of ligand-mediated TWJ assembly is calculated over the first 10 min of incubation.
Cell culture and cell proliferation assay
MCF7 or MDA-MB-231 cells were routinely cultured in 75 or 175 cm2 tissue culture flasks (Nunc) at 37°C in a humidified, 5% CO2 atmosphere in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% foetal bovine serum (FBS, Gibco) and 1% Penicillin-Streptomycin mixture (10,000 units/mL penicillin, 10,000 μg/mL streptomycin, Gibco). Cells were subcultured twice a week using standard protocols. For the cell viability assays: cells were seeded in a 96-well plate (6000 cells/well) in 160 μl of growth medium for 24 h at 37°C. Then, 40 μl of ligand solution was added to reach the final concentration of the ligands between 50 and 0.005 μM and incubated for 72 h at 37°C. The media was removed and the cells fixed with trichloroacetic acid 10% (120 μl, 1 h at 4°C). The supernatant was removed, the fixed cells were washed 5 times with water and then dried. A solution of sulforhodamine B (SRB, 100 μl, 0.057% w/v in 1% acetic acid) was added to each well. After 30 min, the supernatant was removed and the wells were washed 3 times with 150 μl of acetic acid (1%) and dried. Tris base (150 μl, 10 mM) was added to each well and the microplate was gently shaken for 10 min at 25°C. Optical density (OD) values were determined at 530 nm. Final data were analyzed with GraphPad Prism: transforming to log concentration values, normalising (from 0 to 100; 0 for ligand-treated wells where absolute cell death was observed and 100 for ligand-untreated SRB-stained cells). LD50 (defined as the concentration at which 50% of the cell growth inhibition is reached) was determined by non-linear regression function for inhibition dose response. Reported LD50 values were means of 3 experiments. Statistics were calculated with GraphPad Prism.
Immunodetection and optical imaging protocols
MCF7 cells were seeded on glass coverslips in a 24-well plate for 24 h at 37°C. For γH2AX quantification, cells were either untreated (control) or incubated with TrisPOB and TrisNP at toxic concentrations (10× the LD50 determined for 72 h treatment) but for shorter times (4 h), in order to maximize the investigated effect and avoid downstream signalling such as apoptosis. For co-treatments, cells were incubated for 1 h with BNS-22 (50 μM), aphidicolin (10 μM) or DRB & BMH-21 (50 and 0.5 μM respectively), to which TrisNP (22 μM for γH2AX quantification) was added without removing the inhibitor, and incubated for a further 4 h. Cells were fixed with ice-cold PFA (2%, 15 min) and blocked with blocking buffer (1% BSA, 0.1% Triton X-100 in PBS) for 10 min at room temperature (rt). Coverslips were incubated with γH2AX antibody (1/1000 dilution in blocking buffer, EMD Millipore Corp.) for 2 h at rt in a humid dish, then cells were washed with PBS + 0.1% Triton X-100 (3 × 5 min), incubated with Alexa Fluor 647-conjugated secondary antibody (1/500 in blocking buffer, EMD Millipore Corp.) for 45 min at rt in the dark. Cells were then washed with PBS + 0.1% Triton X-100 (3 × 5 min), incubated with DAPI (10 min, 1 μg/ml in PBS) and mounted with Fluoromount. Confocal imaging was performed using a confocal laser-scanning microscope (Leica TCS SP8) with a 63× objective lens and LASX software (Leica Microsystems CMS GmbH). The samples were excited at 405 nm (DAPI) and 638 nm (Alexa Fluor 647) and the fluorescence collected at 409−499 nm (DAPI) and 649−775 nm (Alexa Fluor 647). Image processing was carried out using ImageJ and the 3D Object Counter plugin.
Click imaging
MCF7 cells were seeded on glass coverslips in a 24-well plate for 24 h at 37°C. In the case of post-fixation labelling, cells were fixed with ice cold PFA (2%, 15 min), with or without treatment with CSK buffer containing 0.7% Triton X-100 and RNase A (Sigma-Aldrich, 0.3 mg/ml) (henceforth CSK + RNAse) (2 × 3 min, rt) and treated with clickable ligand (10 μM). In the case of live cell ligand treatment, cells were either untreated (control) or incubated with TrisNP-α (3 μM, 4 h). In the case of RNAse pre-extraction, cells were treated with or without ligand, then with CSK + RNAse (5 min, rt) and fixed. For co-treatments, cells were incubated for 1 h with BNS-22 (50 μM), aphidicolin (10 μM) or DRB & BMH-21 (100/1 μM), to which TrisNP-α was added without removing the inhibitor, and incubated for a further 4 h. For AAV pre-treatment, cells were seeded with AAV1-mCherry (Vector Biolabs, #7103) (multiplicity of infection (MOI): 75000) and allowed to attach overnight, washed with DMEM, and incubated with TrisNP-α (3 μM, 4 h). Cells were then fixed with ice cold PFA (2%, 15 min), washed 3 times with PBS, and click staining was carried out with Alexa Fluor 594-azide (or Alexa Fluor 488-azide in experiments with AAV1-mCherry) (1 μM) in PBS enriched with 0.05% IGEPAL CA-630, 1 mM CuSO4 and 10 mM sodium ascorbate for 30 min, followed by washing with PBS + 0.1% Triton (3 × 5 min), incubated with DAPI (10 min, 1.5 μg/ml in PBS) and mounted with Fluoromount. Confocal imaging was performed as described above. Image processing was carried out using ImageJ. Staining pattern was identical in MDA-MB-231 cells.
TrisNP-α and BG4 co-staining
MCF7 cells were treated with or without TrisNP-α (3 μM, 3 h), washed with PBS and pre-extracted by a 5 min incubation with CSK + RNAse at rt. Cells were washed 3 times on ice with cold PBS, fixed on ice (PFA 2% in PBS, 15 min) and washed 3 times with PBS. For TrisNP-α post-fixation labelling, cells were fixed then incubated with or without TrisNP-α (10 μM, 3 h) at rt. After PBS washing, click reaction was performed in PBS containing 0.05% IGEPAL CA-630, 4 mM CuSO4, 10 mM sodium ascorbate and 1 μM Alexa Fluor 488-azide for 30 min at rt. Finally, cells were washed with PBS and blocked (1 h in 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 2% BSA, 0.2% fish gelatine, 0.1% Triton-X 100), incubated overnight at 4°C with BG4 antibody (0.25 μg/ml in blocking buffer, mouse monoclonal, Absolute antibody, Ab00174-1.1), washed with PBS + 0.1% Tween-20 and incubated with secondary goat antibody Alexa Fluor 594 in blocking buffer (45 min, rt). DNA was stained with DAPI (1 μg/ml in PBS, 15 min) and mounted with Vectashield (Vector Laboratories). Images were acquired on a ZEISS Elyra 7 3D Lattice SIM super-resolution microscope with a 63× objective (PLANAPO NA 1.4, Zeiss) and dual sCMOS cameras (pco.edge). 3D-SIM reconstructions were performed using Zen Black (2.3). Imaging intensity parameters were adjusted differently between live and fixed treated cells using Zen Blue (3.3).
Flow cytometry
MCF7 cells were seeded (3 × 106 per T75 flask or 6 × 106 per T175 flask) for 24 h at 37°C. Cells were either untreated (control) or treated with 10 × LD50 of ligands for 4 h. Cells were trypsinised and fixed in suspension with PFA 1% for 15 min on ice. The PFA was removed by centrifugation, cells were resuspended and washed with PBS, and ice-cold EtOH was added for overnight storage at –20°C until analysis (max. three nights). 1 × 106 fixed cells per condition were labelled as in optical imaging protocols: incubation with γH2AX antibody (1/1000) for 2 h at 25°C, rinsed twice with blocking buffer, then incubated with Alexa Fluor 647-conjugated secondary antibody (1/500) for 45 min before counterstaining with DAPI (2 μg/ml) for 30 min. Stained samples were analyzed by flow cytometry with a 3-laser LSRII (Becton Dickinson) using 633 nm excitation for Alexa Fluor 647 (670/30 BP filter) and 355 nm excitation for DAPI (450/50 BP filter). Debris were excluded from the analysis by gating a forward scatter versus side scatter plot (gating strategy in Supplementary Figure S3B). Integrated fluorescence measurements for Alexa Fluor 647 and DAPI were recorded for 104 single non-debris events. Data were plotted using FlowJo software, and cell aggregates and false positives were excluded.
Synthetic lethality cytotoxicity matrices
The antiproliferative properties of combinations of ligands and DNA repair inhibitors (DNA-PKi, ATMi, and RAD51i) were assessed via the SRB assay. Cells were seeded in a 96-well plate (6000 cells per well) in 160 μl of growth medium for 24 h at 37°C prior to addition of ligand/inhibitor solution prepared in DMEM in a fresh 96-well plate. Seven serial dilutions of the inhibitor were performed: 12–0.18 μM for NU7441 (DNA-PKi), 30–0.47 μM for KU55933 (ATMi) and 15–0.23 μM for B02 (RAD51i), before being distributed into a fresh 96-well plate to be mixed with ligand, which is diluted within the plate (40–0.04 μM for ligands TrisNP and TrisPOB), with the final column containing no ligand and the final row containing no inhibitor. Cell viability was measured after 72 h according to the SRB protocol described above. The IC50 (or LD50) values (called Dm, for median-effect dose) were calculated for each inhibitor:ligand ratio (from 24:1 to 0.1875:1) according to the Chou and Talalay method (69–70). The IC50 values of inhibitors and ligands alone (IC50 of inhibitor = Dm1, IC50 of ligand = Dm2) were determined in control wells (single agent only). The contribution to Dm of each drug in the mixture (Dinhibitor or D1, Dligand or D2) was calculated for each ratio, as follows: at 24:1 inhibitor:ligand ratio, D1 = [Dm/(24 + 1)] × 24 and D2 = [Dm/(24 + 1)] × 1; at 12:1 inhibitor:ligand ratio, D1 = [Dm/(12 + 1)] × 12 and D2 = [Dm/(12 + 1)] × 1, etc. Then, isobolograms were constructed by plotting [D2/Dm2] versus [D1/Dm1] for each ratio. The combination index (CI, with CI < 1, = 1 and > 1 for synergistic, additive, and antagonistic effects, respectively) was also calculated for each ratio as follows: CI = (D1/Dm1) + (D2/Dm2).
RESULTS
Azacryptands interact with both TWJ and G4 in vitro, with a preference for TWJs
A battery of five in vitro techniques were employed to gain insights into how TrisNP and TrisPOB interact with both TWJ and G4: FRET-melting, PAGE gel, TWJ-Screen, ESI-MS and equilibrium dialysis. FRET-melting investigations were performed with both labelled F-TWJ-T (66) and G4-forming labelled human telomere sequence F21T (71) (Figure 2B and Supplementary Figure S1). This assay conveniently quantifies apparent TWJ- or G4-affinity by the change in the mid-transition temperature (ΔT1/2, °C) of fluorophore-labelled TWJ- and G4-forming oligonucleotides. TrisNP and TrisPOB induce a large and comparable stabilisation effect on both TWJ- and G4-forming oligonucleotides, with ΔT1/2 = 17.7 and 19.2°C for F-TWJ-T, and 17.0 and 16.8°C for F21T, for TrisNP and TrisPOB, respectively. Next, competition experiments with an excess of non-labelled competitor oligonucleotide (15 and 50 molar equiv. with respect to the labelled oligonucleotide) were performed to assess the structural preferences of azacryptands. When a G4 competitor (TG4T, selected as a highly thermally stable G4 with T1/2 = 85°C) (72) was added to a F-TWJ-T/azacryptand system, the ligand-induced stabilization of F-TWJ-T was maintained to a large extent (ΔT1/2 = 11.9 and 13.7°C for TrisNP and TrisPOB in the presence of 50 molar equiv. TG4T). Conversely, when non-labelled TWJ (15 mol. equiv.) was added as a competitor to the F21T/azacryptand system, the stabilisation of the G4 structure was totally lost (ΔT1/2 = 0 and 0.5°C for TrisNP and TrisPOB, respectively). This shows that azacryptands interact with both G4 and TWJs, with a strong preference for TWJs.
This was further assessed by complementary isothermal assays. First, polyacrylamide gel electrophoresis (PAGE) (Figure 2C) (60–61,66), which provides a qualitative demonstration of the ability of azacryptands to readily assemble a trimolecular TWJ from the three separate strands (TWJ-S1, TWJ-S2 and TWJ-S3). Second, the TWJ-Screen assay (59–61), which quantifies the TWJ-folding ability of each ligand at a constant temperature (25°C), relies on a trimolecular system of fluorophore-labelled oligonucleotides capable of forming a TWJ structure (Figure 2C). In initial conditions, the three strands remain separate, thus a maximum fluorescence signal is observed, and upon addition of a TWJ-stabilising ligand, the TWJ structure assembles and FRET quenching reduces the fluorescent signal. The speed of TWJ-assembly promoted by the ligand is quantified by the initial velocity (V0, min−1) of the TWJ-folding-mediated FAM quenching. Both TrisNP and TrisPOB accelerate TWJ folding (V0 = −262.9 and − 266.9 min−1, respectively) compared to ligand-free conditions in which there is no spontaneous folding of the three-strand mixture (FAM-TWJ-S1, TWJ-S2 and TWJ-S3-TAMRA, or M (mixture), V0 = 23.6 min−1, Figure 2C). Controls were performed to show that no non-specific interactions are observed between the ligand and the FAM label (ligand + TWJ-S1, V0 = 70.0 and 17.4 min−1, for TrisNP and TrisPOB, respectively, Supplementary Figure S1G). When performed in a competitive manner, that is, in the presence of the unlabelled TG4T, this ligand-induced TWJ assembly is marginally affected for both ligands (V0 = −224.3 versus −262.9 min−1 for TrisNP, V0 = −217.5 versus −266.9 min−1 for TrisPOB (Figure 2C); control: 24.9 versus 23.6 min−1 for M in absence and presence of TG4T, Supplementary Figure S1G). These results are in line with those obtained in competitive FRET-melting, demonstrating again that azacryptands do interact with both G4 and TWJs, but with a preferential affinity for TWJs.
The high TWJ affinity of TrisNP for TWJ was also shown by electrospray mass spectrometry (ESI-MS) performed with equimolar (10 μM) mixtures of TrisNP with either TWJ, G4 (TG5T) or dsDNA (ds17) (Supplementary Figure S2A–C). The spectra collected demonstrated the exclusive formation of the 1:1 TrisNP/TWJ complex, and no 2:1 TrisNP/TWJ complex is observed even when 20 μM of ligand is used. The peak of unbound TWJ–DNA is negligible, whilst the bound TrisNP/dsDNA complex is undetectable, and partial formation of the TrisNP/G4 complex is observable in identical experimental conditions. Thus for TWJ and dsDNA, the apparent equilibrium constant (Kapp) could not be reliably calculated; only the Kapp of the TrisNP/G4 complex could be determined (1.0 × 104 M–1). To address this issue, we performed competitive equilibrium dialysis assay according to previously described protocols (61,73). This isothermal, solution-phase assay compares the affinity of a ligand for several oligonucleotide structures, each isolated within a separate dialysis chamber suspended in a solution of diffusible ligand. After 24 h of equilibration, the quantity of ligand bound to each DNA structure was quantified by exploiting the fluorescence of the azacryptands, and relative concentrations of free and bound ligands are used to determine the apparent affinity constant (Supplementary Figure S2D). Kapp values for TrisNP and TrisPOB were high for TWJ (5.1 × 106 and 9.6 × 106 M−1), at least one order of magnitude lower for G4-DNA (human telomeric sequence 22AG, 7.6 × 104 and 1.2 × 105 M−1), and two orders of magnitude lower for duplex-DNA (ds17, 2.1 × 104 and 2.8 × 104 M−1 respectively). These results further confirm that both azacryptands bind strongly to TWJs and G4s in vitro with a marked preference for TWJs.
Insights into how TrisNP and TrisPOB interact with TWJs
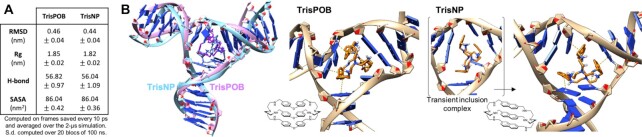
While structural data are now widely available for a number of small molecules interacting with G4s (notably via NMR and X-ray crystallography studies) (74–75), little is known about how ligands bind to TWJ, with the notable exception of the solid-state investigations performed by Hannon et al. (vide supra) (54). We thus investigated how TrisPOB and TrisNP interact with TWJs via molecular dynamics (MD) simulations. After optimisation of TrisPOB and TrisNP conformations (Gypsum-DL, see Supplementary Data for details) (76), the ligands were docked in a TWJ structure (built from PDB: 2ET0) (54) using AutoDock 4.2 (77) and 2 μs molecular dynamics simulations were performed (Gromacs 2019.4 with the Amber99-BSC1 force field for DNA, see SI) to analyze their binding modes (78–81). The ligand-TWJ interactions were quantified by four metrics: the root-mean square deviation (RMSD), which measures the deviation of the structure throughout the simulation from the starting structure of the 2 μs simulation (i.e. the end of the equilibration); the radius of gyration (Rg); the number of hydrogen bonds between the DNA strands (H-bonds); and the solvent-accessible surface area (SASA) (Figure 3A and Supplementary Figure SI). These metrics were found to be similar for TrisNP and TrisPOB.
Figure 3.
(A) Metrics quantifying the interaction between TrisPOB and TrisNP with TWJs. (B) Representative conformations of TrisNP and TrisPOB bound in a TWJ, obtained via MD simulations; π-stacking and polar–π interactions are shown as dotted lines; the transient inclusion complex in which a nucleobase (dT) is sandwiched inside TrisNP is shown in brackets.
The second half of the simulations was clustered, and representative structures (populated at 84 and 91% of the time for TrisPOB and TrisNP, respectively) are shown in Figure 3B. The steric hindrance of both ligands induces a nucleotide base pair disruption at the cavity site, an energy penalty that is compensated by the creation of multiple π-stacking interactions. However, the way the two ligands interact with TWJ was different due to the inherent difference in structure and flexibility of the ligands: TrisPOB was found in an oblong conformation which nestles within the TWJ cavity, while TrisNP was found in a globular conformation which can even transiently form an inclusion complex (ca. 1% of the time) in which one isolated dT residue is sandwiched in between its naphthalene units (Figure 3B and Supplementary movies 1–4). This transient binding mode correlates with the known ability of azacyclophanes, which contain only two aromatic units, to sandwich aromatic hosts (as demonstrated by both NMR and X-ray crystallography studies) (82–84) but is more surprising for azacryptands, known to act as hosts for small anions and metal ions so far (85–87). Altogether, these findings comply with the nature of the ligands, since the diphenyl ether units give TrisPOB more flexibility due to the rotation about the C–O bonds, allowing it to adapt to, and to efficiently π-stack with the surrounding nucleobases, while the rigid naphthalene units make TrisNP prone to transiently catch isolated nucleobases in addition to π-stacking with the nucleobases that form the cavity walls.
Azacryptands are cytotoxic and induce DNA damage
The cytotoxicity of both TrisPOB and TrisNP was tested in hormone-responsive adenocarcinoma cells MCF7 (88–89), expressing oestrogen (ER), progesterone (PR) and glucocorticoid receptors (GR), and triple-negative MDA-MB-231 (90), which are ER−, PR− and HER2− (human epidermal growth factor receptor 2) adenocarcinoma cells, typically more aggressive. These two cell lines are routinely used to validate experimental therapeutics as they correspond to two different types of breast cancers. The median lethal dose (LD50) was calculated using the Sulforhodamine B (SRB) assay (91–92) and both compounds were found quite active, with average LD50 values of 2.7 and 0.6 μM for TrisPOB against MCF7 and MDA-MB-231, respectively, and 5.8 and 1.6 μM for TrisNP against MCF7 and MDA-MB-231, respectively, averaged over three to six biological replicates (Supplementary Figure S3).
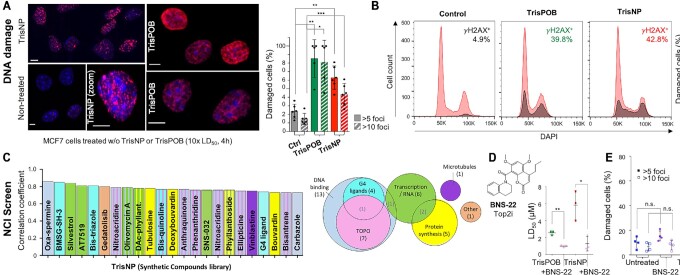
To prove that this cellular activity originates in the ligands’ ability to trigger DNA damage, as previously described for TrisPOB (61), we assessed the induction of DNA damage through immunostaining experiments with an anti-γH2AX antibody, which reveal the phosphorylation of histone H2AX (Figure 4A, B), an established marker of DSBs (93–94). Confocal microscopy showed a significant increase in DSBs in treated cells compared to non-treated (Ctrl) cells (Figure 4A), quantified by the percentage of nuclei with >5 and >10 γH2AX foci. 75% and 71% of TrisPOB-treated and 63% and 43% of TrisNP-treated cells showed >5 and >10 foci respectively, compared to 24% and 15% in non-treated cells as an average of five or more experiments. Flow cytometry quantification also showed significant DNA damage by γH2AX immunostaining, similar to the levels previously observed for TrisPOB (Figure 4B). Results are shown as a percentage of γH2AX-stained cells (an average of 45% (TrisPOB) and 49% (TrisNP) of cells when gating is performed to assign 5% of non-treated cells as γH2AX-positive (n = 3). No significant difference in cell cycle was observed (Supplementary Figure S3C).
Figure 4.
(A,B) Immunodetection of DNA damage in MCF7 cells non-treated or treated with 10 × LD50 of TrisPOB and TrisNP (9 and 22 μM) for 4 h at 37°C prior to immunolabeling with antibodies raised against γH2AX (secondary antibody labelled with Alexa Fluor 647, λem = 670 nm) and DAPI nuclear staining (λem = 450 nm); scale bars = 5 μm. γH2AX foci are quantified by fluorescence imaging (% of cells with > 5 and > 10 γH2AX foci) with ImageJ plugin 3D Object Counter (80 to 200 cells from >8 different images per condition (A), or quantified by flow cytometry and data treatment using FlowJo software (B), repeated in at least three separate experiments. (C) NCI databases of Synthetic Compounds (left panel and Venn diagram) tested in the NCI-60 cytotoxicity assay show the compounds with the highest correlations of cytotoxicity fingerprint with TrisNP and TrisPOB. Colour-coding shows the mechanism of action described for the compounds. (D) Antiproliferative activity of TrisPOB and TrisNP is markedly increased in the presence of a subtoxic concentration of BNS-22 (12.5 μM) in MCF7 cells over 72 h. (E) Immunodetection of DNA damage in MCF7 cells treated with or without BNS-22 (50 μM, 5 h) and/or 10 × LD50 of TrisNP (4 h) prior to immunolabeling with γH2AX-specific antibody. Quantified as previously. P values were calculated using a paired, two-tailed t test. ns: P> 0.05; *P< 0.05; **P< 0.005; **P < 0.0005 (A, B, D, E).
To further investigate the mechanism of the antiproliferative activity of TrisNP and TrisPOB, the compounds were submitted to the USA National Cancer Institute's (NCI) NCI-60 Human Tumor Cell Lines Screen. In this screen, the cytotoxicity fingerprint of novel compounds obtained in 60 well-established tumour cell lines is compared with that of known, previously tested compounds using the COMPARE algorithm, in order to put forward mechanistic hypotheses (95–96). The cytotoxicity patterns of TrisNP and TrisPOB (NSC 818603 and NSC 818604, respectively) were compared with the molecules in two chemical libraries: the Synthetic Compounds library, the largest and most varied (400 compounds) and the Marketed Drugs library, which contains the drugs with the best studied mechanisms (90 compounds, Figure 4C and Supplementary Figure S3). In both libraries, the highest correlation coefficients were obtained with DNA-binding compounds, including several G4 ligands (e.g. the naphthalene diimide BMSG-SH-3 developed as a pancreatic cancer drug (97) and bis-triazole G4 ligands) (98). Topoisomerase (TOP) inhibitors, e.g. phenanthridinium derivatives (99), were also found to rank highly. A full list of correlated drugs and proposed mechanisms is available in the Supplementary Data. These findings are in line with recent genetic screens indicating that ligands targeting higher-order DNA structures, such as pyridostatin (PDS) and CX-5461, exert at least a part of their toxicity via G4-mediated TOP2 trapping (100–102), which ultimately triggers DNA damage (3,39). This series of results thus indicates that the toxicity signatures of both TrisNP and TrisPOB resemble those of strong antiproliferative agents, which mostly act through DNA binding, with a particular ability to interact with higher-order DNA structures, and might induce topoisomerase-mediated DNA damage.
Mechanistic studies of dual ligands with topoisomerase inhibition
Following the indications from the NCI60 data that topoisomerases may be involved in cytotoxicity, we tested the effect of co-treatment with a catalytic TOP2 inhibitor BNS-22 (103). TOP2 inhibitors are categorized into two families: the most used family is the TOP2 poisons, which stabilize the covalently bound TOP2 enzyme-DNA cleaving complex (TOP2cc), and lead to DSBs; the second family is the TOP2 catalytic inhibitors, which are able to rescue the DNA damaging effects of TOP2 poisons (104). BNS-22 efficiently suppressed cytotoxicity and γH2AX induction of the TOP2 poison etoposide (Supplementary Figure S4A) in MCF7 cells, indicating that it efficiently functions as a catalytic TOP2 inhibitor in MCF7 cells. While inhibition of TOP2 has been described to block the cytotoxic activity of G4 ligands (102), it had the opposite effect on TrisNP by repeatably reducing its LD50 by a factor of ca. 4.5, from an average of 5.8 to 1.3 μM (Figure 4D) when BNS-22 is used at a single non-toxic concentration. Similar effects were observed with TrisPOB (from 2.54 to 0.98, a 2.6-fold decrease). DNA damage induction was quantified via γH2AX immunostaining, proving to be significantly increased when cells were co-treated with TrisNP and BNS-22, compared to TrisNP alone (Figure 4E), whilst BNS-22 alone at this concentration induces a negligible amount of DNA damage. No effect was observed on co-treatment with etoposide, but the latter had little cytotoxicity in MCF7 cells at all (Supplementary Figure S4A). The synergic relationship between TrisNP and BNS-22 is thus opposite to what was reported for PDS (102), indicating that azacryptands act through a distinct mechanism of action than a known G4 ligand, which reinforces our hypothesis regarding preferential TWJ targeting in cells. This effect could originate in the higher prevalence of TWJ-forming motifs compared to G4-forming motifs, as there are approximately 3-fold more direct repeat sequences in the human genome than G4-forming sequences (33), and ca. 50% of the genome is made up of repetitive sequences (105).
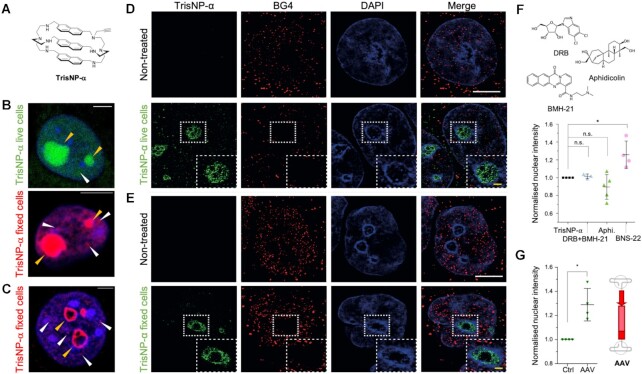
Traceable TrisNP analogue localizes in nucleoli and nucleoplasm
To further investigate this, an analogue of TrisNP with a small terminal alkyne modification (TrisNP-α, Figure 5A, see Supplementary Data for synthetic procedure) was synthesized as a chemical biology tool to quantify the accumulation of TrisNP in cells by in situ click imaging, and to assess the modulation of the TWJ landscape upon co-treatments (106). TrisNP-α presented in vitro TWJ-binding properties similar to TrisNP, albeit with a slightly lower affinity (Supplementary Figure S1E), presumably due to the steric hindrance introduced by the alkyne appendage. MCF7 cells were incubated with TrisNP-α, prior to fixation and bioorthogonal copper-catalysed alkyne-azide click reaction with an azide-tagged fluorophore, in order to localize TrisNP-α in cells (Supplementary Figure S5A). TrisNP-α localized primarily in the nucleus (nucleoplasm), and particularly in the nucleoli (yellow arrows, Figure 5B and Supplementary Figure S5B), when incubated both in live cells and in fixed cells. Nucleoplasmic foci were observable (white arrows) in weakly DAPI-labeled zones (euchromatin marker). The staining obtained when incubating TrisNP-α with living cells or after fixation was very similar, indicating that TWJ structures are present in cells even without being stabilised by TWJ ligands. Pre-extraction with CSK + RNase A before fixation (108) was also performed to focus on the DNA-associated staining (Figure 5C). This revealed that the inner part of the nucleolar TrisNP-α staining is lost upon RNase A treatment, indicating that this treatment efficiently removed RNA and that part of the staining observed without CSK + RNase pre-extraction is likely to correspond to RNA secondary structures. This also revealed that TrisNP-α specifically labels an RNase-resistant perinucleolar compartment partly colocalizing with the perinucleolar heterochromatin. This region around the nucleoli is likely to represent nucleolus-associated DNA domains (NADs) (109–111), which contain weakly transcribed heterochromatic regions of low gene density including centromeric and pericentromeric satellite repeats and subtelomeric regions, all of which containing palindromic and repetitive sequences, prone to form secondary structures including TWJs (112).
Figure 5.
(A) Structure of TrisNP-α. (B,C) In situ click imaging obtained with TrisNP-α (3 μM, 4 h) incubated in live or fixed cells, then illuminated with Alexa Fluor 488-azide (green, live cell incubation) or Alexa Fluor 594-azide (red, post-fixation incubation). Ligand is present in the nucleoli (yellow arrows) and the nucleoplasm (white arrows). Cells pre-extracted (C) with CSK + RNAse show perinucleolar staining (yellow arrows). Nuclei are counterstained with DAPI (blue). Scale bars = 5 μm. (D, E) Super-resolution images of MCF7 cells are treated with TrisNP-α (3 h, 3 μM), pre-extracted with CSK + RNase A, fixed, clicked, and co-stained with BG4 antibody (D) or pre-extracted with CSK + RNase A, fixed, incubated with TrisNP-α (3 h, 10 μM), clicked and co-stained with BG4 antibody (E). Nuclei are counterstained with DAPI. White scale bar = 5 μm, yellow scale bar = 1 μm. (F) Change in nuclear intensity of live TrisNP-α click staining after pre-treatment with inhibitors DRB + BMH-21 (100/1 μM), aphidicolin (10 μM) and BNS-22 (50 μM). (G) TrisNP-α nuclear intensity increases with AAV1-mCherry treatment, stained as previously described and labelled after fixation via click reactions with AF488-azide. Nuclear intensity after inhibitor or AAV treatment was quantified in FIJI ImageJ and normalised to TrisNP-α staining. P values were calculated using a paired, two-tailed t test. ns: P> 0.05; *P< 0.05 (F, G).
We then used super-resolution 3D structured illumination microscopy (3D-SIM) to evaluate the relative patterns obtained with the TrisNP-α staining and with the G4-specific antibody BG4, which is here a recombinant IgG1 antibody in which the variable parts have been replaced by the single-chain variable fragment (scFv) of the initially described BG4 (107) (Figure 5D, E and Supplementary Figure S5C, D). CSK + RNAse A treatment was also performed to improve BG4 staining. Aside from the perinucleolar regions, small and weak TrisNP-α foci were observable dispersed in the nucleoplasm, which did not colocalize with the denser nucleoplasmic foci of BG4. Importantly, no nucleolar or perinucleolar staining was observed with BG4. Altogether these data support that TrisNP binds principally to non-BG4 labelled structures, associated to or in proximity with NADs. The lack of colocalization between the BG4 and TrisNP-α staining together with the fact that the intensity of the BG4 signal is not modified by the incubation of living cells with TrisNP-α, support the notion that TrisNP-α preferentially targets DNA junctions in cells.
TrisNP-α staining is modulated by inhibitors of DNA transactions and exogenous TWJs
BNS-22 pre-treatment was found to increase the intensity of TrisNP-α staining, indicating that TWJs become more prevalent upon TOP2 inhibition (Figure 5F). This presents a novel tool to study dynamic DNA mechanisms, and corresponds to the role of topoisomerases in reducing helical strain in DNA (113), thus reducing the formation of alternative DNA structures (114–115). More specifically, TOP2 is described not only to release DNA supercoiling, but also to recognise DNA structures (116–117) and induce DSB formation at these sites (118–119). TOP2 poisons are frequently included in first and second line anticancer therapies (120–121), and cancer cells can develop resistance by multiple mechanisms including through reduced TOP2 activity (122). Altogether, our results imply that azacryptands could prove useful to treat cancers that have developed resistance to TOP2 poisons, since azacryptand treatment could confer a greater sensitivity to these molecules through increased levels of stabilized TWJs.
We also investigated if certain DNA transactions are more favourable to TWJ formation. Cells were pre-treated with either transcription inhibitors (BMH-21 (123) and 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) (124), which inhibit RNA polymerases I and II, respectively) or replication inhibitor aphidicolin (Aphi, Figure 5F) (125). Inhibitor concentrations were optimized for efficient inhibition of transcription/replication using clickable nucleotides EdU and EU (Supplementary Figure S6A, B). Using these conditions, we found that transcription inhibition with DRB/BMH-21 had no effect on TrisNP-α click staining intensity, whilst replication inhibition with Aphi reduced TrisNP-α staining, albeit in a variable and non-statistically significant manner, indicating that TWJ-formation might be preferentially dependent on replication rather than transcription. This could not be confirmed by monitoring TrisNP-induced γH2AX signalling, since no significant change in γH2AX staining by transcription/replication inhibition was observed, whilst transcription inhibition alone induced important amounts of damage signalling (Supplementary Figure S6A). Again, these results are in contrast with what was described for PDS, where Aphi and DRB pre-treatments were found to reduce PDS-induced γH2AX signalling (39). This indicates that G4 formation is linked to these two major DNA transactions, whereas TWJ formation may be more specific to replication, and more difficult to disrupt once stabilized by ad hoc ligands. Direct comparison with PDS was hindered by the non-toxicity of PDS and other G4-ligands in MCF7 cells (LD50 > 100 μM, Supplementary Figure S6C).
To further demonstrate that TrisNP-α targets TWJs in a cellular context, we employed a common viral gene vector, the adeno-associated virus (AAV) (126), which contains a TWJ in each of its two inverted terminal repeats (ITRs, the only part of the genome required for AAV uptake) (Figure 5G) (127). AAV is quickly taken up into cells and transcribed. We first verified that MCF7 cells could take up AAV1-mCherry (henceforth AAV1) and express mCherry by fluorescence microscopy. Next, we incubated MCF7 cells with TrisNP-α and monitored the ligand/ITRs interaction by click chemistry. As seen in Figure 5G, TrisNP-α staining increased significantly on AAV1 incorporation (1.3-fold averaged over 4 biological repeats), thus providing an additional strong argument in favour of the interaction of TrisNP-α with TWJ structures in cells.
DNA junction ligands are synthetically lethal with DDR inhibitors
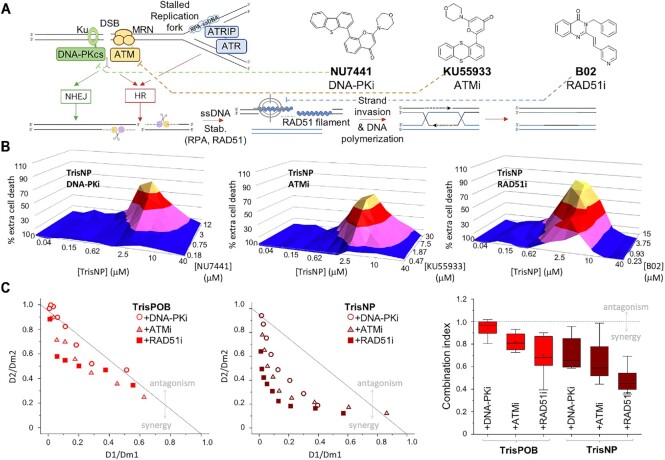
A strategy that has received growing interest is the development of drug combinations to provoke chemically-induced synthetic lethality, notably by co-treating cancer cells with DNA damaging agents and DDR inhibitors (3,10,46,128). DDR inhibitors targeting clinically relevant cellular processes were used (Figure 6A): NU7441, or DNA-PKi (inhibitor of DNA-PK, involved in NHEJ) (129), KU55933, or ATMi (inhibitor of ATM, involved in HR and checkpoint activation) (130) and B02, or RAD51i (inhibitor of RAD51, involved in HR) (131). Synergic effects between two cytotoxic drugs were tested by measuring the survival of cells in an 8 × 12 matrix of drug concentrations. TrisNP showed strong synergy with DDR inhibitors, as represented by the pyramid graphs (Figure 6B) where a peak, indicative of a synergic effect, shows extra cell death compared to the simple addition of cell death induced by the two drugs alone, according to the Loewe additivity model (132). Another reliable representation of the cytotoxic effects of drug combinations is provided by the combination index (CI), indicating synergistic, additive and antagonistic effects respectively when CI < 1, = 1, and > 1 (70). The synergy of TrisNP with DDR inhibitors was even stronger than what was previously reported for TrisPOB (61). TrisNP shows low, strongly synergic CI values with the three DDR inhibitors (average CI = 0.71, 0.64 and 0.49 with DNA-PKi, ATMi and RAD51i, respectively, and 0.94, 0.82 and 0.70 for TrisPOB) (Figure 6C). This implies that both NHEJ and HR pathways are implicated in the repair of azacryptand-induced damage, and strongly supports the conclusion that TrisNP induces genotoxic DSBs.
Figure 6.
(A) Double-strand breaks (DSB) and stalled replication forks are repaired by HR and NHEJ, mediated by DNA-PK and ATM kinases and RAD51. Created with BioRender. Chemical structure of DDR inhibitors NU7441 (DNA-PKi), KU55933 (ATMi) and B02 (RAD51i). (B) 3D pyramid plots showing the percentage of extra cell death for the combination of TrisNP (lethal concentration range: 0–40 μM), with DNA-PKi, ATMi, or RAD51i (non-lethal concentration range, see Supplementary Data). Results are the average of three separate experiments each containing technical duplicates. (C) Normalised isobolograms for combination of TrisPOB or TrisNP with DNA-PKi (circles), ATMi (triangles), and RAD51i (squares). Points above the grey horizontal line show antagonism between agents, below the line show synergism. Combination index graph (right) of the same co-treatments. Derived from the same raw data as the pyramid plots in Figure 5B.
DISCUSSION
In a continuation of our studies of the therapeutic potential of small molecules that target alternative DNA structures, we describe here a novel family of highly promising candidates, able to interact with two non-B DNA structures: three-stranded three-way DNA junctions and four-stranded G-quadruplex-DNA. These two ligands, TrisPOB and TrisNP, that belong to the family of azacryptands, display high toxicity in two cancer cell lines (MCF7 and MDA-MB-231) via induction of DNA damage, most notably DSBs. TrisNP and its traceable analogue TrisNP-α, the first ‘clickable’ probe belonging to this novel family of molecules, were used as chemical biology tools to study the formation and toxicity of ligand-stabilized DNA structures in cells, and provide mechanistic insights into the origins of their antiproliferative activity. Although further validation is required, this molecule could be seen as the first cellular probe for TWJ formation. Due to the elusive and transient nature of TWJs, currently available molecular tools are not fully suitable for their studies; however we continue searching for methods that could prove that TrisNP(-α) genuinely binds to TWJs in cells.
Interestingly, the cellular activity of these dual ligands is rather different from that of typical G4-ligands, for which the opposite response was reported for co-treatments with both TOP2 and transcription inhibitors. Azacryptands also distinguish themselves from G4-ligands by their high toxicity in MCF-7 cells, whilst benchmark G4-ligands such as PDS (39,133), PhenDC3 (134–135) and 360A (135–137) are non-toxic (LD50 > 100 μM, Supplementary Figure S6E). The chemotherapeutic potential of these molecules can be strongly potentiated when included in synthetic lethality combinations, demonstrated here by their highly synergic cytotoxicity profiles in combination with DDR inhibitors. Thus, azacryptands act via a distinct mechanism to that of G4-ligands and topoisomerase poisons, thus representing new and promising weapons in the chemical arms race with cancers, especially to treat cancers resistant to TOP2 poisons.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Anne Cucchiarini (Institut Curie) for HPLC analyses, Marc Pirrotta (ICMUB) for FRET-melting and TWJ-Screen investigations, and both the flow cytometry (Plateforme de Cytométrie de l’UMR1231) and optical imaging platforms (Plateforme Dimacell, INRA, Dijon) of the Université de Bourgogne Franche-Comté (supported by Conseil Régional de Bourgogne) and the TRI-IPBS Imaging Core Facility, member of TRI-Genotoul. Data of the NCI60 screening assay were obtained thanks to the National Cancer Institute Developmental Therapeutics Program (NCI/DTP, https://dtp.cancer.gov).
Contributor Information
Joanna Zell, Institut de Chimie Moléculaire de l’Université de Bourgogne (ICMUB), CNRS UMR 6302, UBFC Dijon, 21078 Dijon, France.
Katerina Duskova, Institut de Chimie Moléculaire de l’Université de Bourgogne (ICMUB), CNRS UMR 6302, UBFC Dijon, 21078 Dijon, France.
Leïla Chouh, Institut Curie, CNRS UMR 9187, INSERM U1196, PSL Research University, 91405 Orsay, France; Université Paris Saclay, CNRS UMR 9187, INSERM U1196, 91405 Orsay, France.
Madeleine Bossaert, Institut de Pharmacologie et de Biologie Structurale (IPBS), CNRS UMR 5089, Université de Toulouse, UPS, Équipe labellisée la Ligue Contre le Cancer, 31077 Toulouse, France.
Nicolas Chéron, Pasteur, Département de chimie, École Normale Supérieure (ENS), CNRS UMR8640, PSL Research University, Sorbonne Université, 75005 Paris, France.
Anton Granzhan, Institut Curie, CNRS UMR 9187, INSERM U1196, PSL Research University, 91405 Orsay, France; Université Paris Saclay, CNRS UMR 9187, INSERM U1196, 91405 Orsay, France.
Sébastien Britton, Institut de Pharmacologie et de Biologie Structurale (IPBS), CNRS UMR 5089, Université de Toulouse, UPS, Équipe labellisée la Ligue Contre le Cancer, 31077 Toulouse, France.
David Monchaud, Institut de Chimie Moléculaire de l’Université de Bourgogne (ICMUB), CNRS UMR 6302, UBFC Dijon, 21078 Dijon, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CNRS (to N.C., A.G., S.B., D.M.); INSERM Plan Cancer 2014–2019 [19CP117-00 for S.B. and D.M.]; Agence Nationale de la Recherche [ANR-17-CE17-0010-01 to A.G. and D.M., ANR-17-CE18-0002-01 for S.B.]; European Union [PO FEDER-FSE Bourgogne 2014/2020 programs, grant BG0021532]; HPC resources of IDRIS under the allocation 2020–077156 made by GENCI. Funding for open access charge: INSERM Plan Cancer 2014-2019 [19CP117-00].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zell J., Rota Sperti F., Britton S., Monchaud D.. DNA folds threaten genetic stability and can be leveraged for chemotherapy. RSC Chem. Biol. 2021; 2:47–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hustedt N., Durocher D.. The control of DNA repair by the cell cycle. Nat. Cell Biol. 2017; 19:1–9. [DOI] [PubMed] [Google Scholar]
- 5. Scully R., Panday A., Elango R., Willis N.A.. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019; 20:698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Verma N., Franchitto M., Zonfrilli A., Cialfi S., Palermo R., Talora C.. DNA damage stress: cui prodest?. Int. J. Mol. Sci. 2019; 20:1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. O’Connor M.J. Targeting the DNA damage response in cancer. Mol. Cell. 2015; 60:547–560. [DOI] [PubMed] [Google Scholar]
- 8. Swift L.H., Golsteyn R.M.. Genotoxic anti-cancer agents and their relationship to DNA damage, mitosis, and checkpoint adaptation in proliferating cancer cells. Int. J. Mol. Sci. 2014; 15:3403–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaelin W.G. The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer. 2005; 5:689–698. [DOI] [PubMed] [Google Scholar]
- 10. O’Neil N.J., Bailey M.L., Hieter P.. Synthetic lethality and cancer. Nat. Rev. Genet. 2017; 18:613–623. [DOI] [PubMed] [Google Scholar]
- 11. Belotserkovskii B.P., Mirkin S.M., Hanawalt P.C.. DNA sequences that interfere with transcription: implications for genome function and stability. Chem. Rev. 2013; 113:8620–8637. [DOI] [PubMed] [Google Scholar]
- 12. Técher H., Koundrioukoff S., Nicolas A., Debatisse M.. The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat. Rev. Genet. 2017; 18:535. [DOI] [PubMed] [Google Scholar]
- 13. Khristich A.N., Mirkin S.M.. On the wrong DNA track: Molecular mechanisms of repeat-mediated genome instability. J. Biol. Chem. 2020; 295:4134–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao J., Bacolla A., Wang G., Vasquez K.M.. Non-B DNA structure-induced genetic instability and evolution. Cell. Mol. Life Sci. 2010; 67:43–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rich A., Zhang S.. Z-DNA: the long road to biological function. Nat. Rev. Genet. 2003; 4:566. [DOI] [PubMed] [Google Scholar]
- 17. Ravichandran S., Subramani V.K., Kim K.K.. Z-DNA in the genome: from structure to disease. Biophys. Rev. 2019; 11:383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Duca M., Vekhoff P., Oussedik K., Halby L., Arimondo P.B.. The triple helix: 50 years later, the outcome. Nucleic Acids Res. 2008; 36:5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aguilera A., Gómez-González B.. DNA–RNA hybrids: the risks of DNA breakage during transcription. Nat. Struct. Mol. Biol. 2017; 24:439. [DOI] [PubMed] [Google Scholar]
- 20. Santos-Pereira J.M., Aguilera A.. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. [DOI] [PubMed] [Google Scholar]
- 21. Crossley M.P., Bocek M., Cimprich K.A.. R-loops as cellular regulators and genomic threats. Mol. Cell. 2019; 73:398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spiegel J., Adhikari S., Balasubramanian S.. The structure and function of DNA G-quadruplexes. Trends Chem. 2020; 2:123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Varshney D., Spiegel J., Zyner K., Tannahill D., Balasubramanian S.. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020; 21:459–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tian T., Chen Y.-Q., Wang S.-R., Zhou X.. G-quadruplex: a regulator of gene expression and its chemical targeting. Chem. 2018; 4:1314–1344. [Google Scholar]
- 26. Abou Assi H., Garavís M., González C., Damha M.J.. i-Motif DNA: structural features and significance to cell biology. Nucleic Acids Res. 2018; 46:8038–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mirkin E.V., Mirkin S.M.. Replication fork stalling at natural impediments. Microbiol Mol. Biol. Rev. 2007; 71:13–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Valton A.-L., Prioleau M.-N.. G-quadruplexes in DNA replication: a problem or a necessity?. Trends Genet. 2016; 32:697–706. [DOI] [PubMed] [Google Scholar]
- 29. Kim N. The interplay between G-quadruplex and transcription. Curr. Med. Chem. 2019; 26:2898–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zeman M.K., Cimprich K.A.. Causes and consequences of replication stress. Nat. Cell Biol. 2014; 16:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dobbelstein M., Sørensen C.S.. Exploiting replicative stress to treat cancer. Nat. Rev. Drug Discov. 2015; 14:405–423. [DOI] [PubMed] [Google Scholar]
- 32. Gaillard H., García-Muse T., Aguilera A.. Replication stress and cancer. Nat. Rev. Cancer. 2015; 15:276. [DOI] [PubMed] [Google Scholar]
- 33. Georgakopoulos-Soares I., Morganella S., Jain N., Hemberg M., Nik-Zainal S.. Noncanonical secondary structures arising from non-B DNA motifs are determinants of mutagenesis. Genome Res. 2018; 28:1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Voineagu I., Narayanan V., Lobachev K.S., Mirkin S.M.. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:9936–9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Voineagu I., Surka C.F., Shishkin A.A., Krasilnikova M.M., Mirkin S.M.. Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nat. Struct. Mol. Biol. 2009; 16:226–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shastri N., Tsai Y.-C., Hile S., Jordan D., Powell B., Chen J., Maloney D., Dose M., Lo Y., Anastassiadis T.. Genome-wide identification of structure-forming repeats as principal sites of fork collapse upon ATR inhibition. Mol. Cell. 2018; 72:222–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lerner L.K., Sale J.E.. Replication of G quadruplex DNA. Genes. 2019; 10:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neidle S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017; 1:0041. [Google Scholar]
- 39. Rodriguez R., Miller K.M., Forment J.V., Bradshaw C.R., Nikan M., Britton S., Oelschlaegel T., Xhemalce B., Balasubramanian S., Jackson S.P.. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012; 8:301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu H., Di Antonio M., McKinney S., Mathew V., Ho B., O’Neil N.J., Santos N.D., Silvester J., Wei V., Garcia J.et al.. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017; 8:14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rizzo A., Salvati E., Porru M., D’Angelo C., Stevens M.F., D’Incalci M., Leonetti C., Gilson E., Zupi G., Biroccio A.. Stabilization of quadruplex DNA perturbs telomere replication leading to the activation of an ATR-dependent ATM signaling pathway. Nucleic. Acids. Res. 2009; 37:5353–5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Salvati E., Leonetti C., Rizzo A., Scarsella M., Mottolese M., Galati R., Sperduti I., Stevens M.F.G., D’Incalci M., Blasco M.et al.. Telomere damage induced by the G-quadruplex ligand RHPS4 has an antitumor effect. J. Clin. Invest. 2007; 117:3236–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pennarun G., Granotier C., Hoffschir F., Mandine E., Biard D., Gauthier L.R., Boussin F.D.. Role of ATM in the telomere response to the G-quadruplex ligand 360A. Nucleic Acids Res. 2008; 36:1741–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gauthier L.R., Granotier C., Hoffschir F., Etienne O., Ayouaz A., Desmaze C., Mailliet P., Biard D.S., Boussin F.D.. Rad51 and DNA-PKcs are involved in the generation of specific telomere aberrations induced by the quadruplex ligand 360A that impair mitotic cell progression and lead to cell death. Cell. Mol. Life Sci. 2012; 69:629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zimmer J., Tacconi E.M.C., Folio C., Badie S., Porru M., Klare K., Tumiati M., Markkanen E., Halder S., Ryan A.et al.. Targeting BRCA1 and BRCA2 deficiencies with G-quadruplex-interacting compounds. Mol. Cell. 2016; 61:449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McLuckie K.I.E., Di Antonio M., Zecchini H., Xian J., Caldas C., Krippendorff B.F., Tannahill D., Lowe C., Balasubramanian S.. G-quadruplex DNA as a molecular target for induced synthetic lethality in cancer cells. J. Am. Chem. Soc. 2013; 135:9640–9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y., Yang J., Wild A.T., Wu W.H., Shah R., Danussi C., Riggins G.J., Kannan K., Sulman E.P., Chan T.A.et al.. G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat. Commun. 2019; 10:943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tauchi T., Shin-ya K., Sashida G., Sumi M., Nakajima A., Shimamoto T., Ohyashiki J.H., Ohyashiki K.. Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: involvement of ATM-dependent DNA damage response pathways. Oncogene. 2003; 22:5338–5347. [DOI] [PubMed] [Google Scholar]
- 49. Biroccio A., Porru M., Rizzo A., Salvati E., D’Angelo C., Orlandi A., Passeri D., Franceschin M., Stevens M.F., Gilson E.. DNA damage persistence as determinant of tumor sensitivity to the combination of Topo I inhibitors and telomere-targeting agents. Clin. Cancer Res. 2011; 17:2227–2236. [DOI] [PubMed] [Google Scholar]
- 50. Leonetti C., Scarsella M., Riggio G., Rizzo A., Salvati E., D’Incalci M., Staszewsky L., Frapolli R., Stevens M.F., Stoppacciaro A.et al.. G-quadruplex ligand RHPS4 potentiates the antitumor activity of camptothecins in preclinical models of solid tumors. Clin. Cancer Res. 2008; 14:7284–7291. [DOI] [PubMed] [Google Scholar]
- 51. Zyner K.G., Mulhearn D.S., Adhikari S., Cuesta S.M., Di Antonio M., Erard N., Hannon G.J., Tannahill D., Balasubramanian S.. Genetic interactions of G-quadruplexes in humans. eLife. 2019; 8:e46793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Day T.A., Layer J.V., Cleary J.P., Guha S., Stevenson K.E., Tivey T., Kim S., Schinzel A.C., Izzo F., Doench J.et al.. PARP3 is a promoter of chromosomal rearrangements and limits G4 DNA. Nat. Commun. 2017; 8:15110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. del Mundo I.M., Vasquez K.M., Wang G.. Modulation of DNA structure formation using small molecules. Biochim. Biophys. Acta - Mol. Cell Res. 2019; 1866:118539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oleksi A., Blanco A.G., Boer R., Usón I., Aymamí J., Rodger A., Hannon M.J., Coll M.. Molecular recognition of a three-way DNA junction by a metallosupramolecular helicate. Angew. Chem. Int. Ed. 2006; 45:1227–1231. [DOI] [PubMed] [Google Scholar]
- 55. Ducani C., Leczkowska A., Hodges N.J., Hannon M.J.. Noncovalent DNA-binding metallo-supramolecular cylinders prevent DNA transactions in vitro. Angew. Chem. Int. Ed. 2010; 49:8942–8945. [DOI] [PubMed] [Google Scholar]
- 56. Hotze A.C., Hodges N.J., Hayden R.E., Sanchez-Cano C., Paines C., Male N., Tse M.-K., Bunce C.M., Chipman J.K., Hannon M.J.. Supramolecular iron cylinder with unprecedented DNA binding is a potent cytostatic and apoptotic agent without exhibiting genotoxicity. Chem. Biol. 2008; 15:1258–1267. [DOI] [PubMed] [Google Scholar]
- 57. Gómez-González J., Pérez Y., Sciortino G., Roldan-Martín L., Martínez-Costas J., Maréchal J.-D., Alfonso I., Vázquez López M., Vázquez M.E.. Dynamic stereoselection of peptide helicates and their selective labeling of DNA replication foci in cells. Angew. Chem. Int. Ed. 2021; 60:8859–8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhu J., Haynes C.J.E., Kieffer M., Greenfield J.L., Greenhalgh R.D., Nitschke J.R., Keyser U.F.. FeII4L4 tetrahedron binds to nonpaired DNA bases. J. Am. Chem. Soc. 2019; 141:11358–11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Guyon L., Pirrotta M., Duskova K., Granzhan A., Teulade-Fichou M.-P., Monchaud D.. TWJ-Screen: an isothermal screening assay to assess ligand/DNA junction interactions in vitro. Nucleic Acids Res. 2018; 46:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Duskova K., Lamarche J., Amor S., Caron C., Queyriaux N., Gaschard M., Penouilh M.-J., de Robillard G., Delmas D., Devillers C.H.et al.. Identification of three-way DNA junction ligands through screening of chemical libraries and validation by complementary in vitro assays. J. Med. Chem. 2019; 62:4456–4466. [DOI] [PubMed] [Google Scholar]
- 61. Duskova K., Lejault P., Benchimol É., Guillot R., Britton S., Granzhan A., Monchaud D.. DNA junction ligands trigger DNA damage and are synthetic lethal with DNA repair inhibitors in cancer cells. J. Am. Chem. Soc. 2020; 142:424–435. [DOI] [PubMed] [Google Scholar]
- 62. Ambrus A., Chen D., Dai J.X., Jones R.A., Yang D.Z.. Solution structure of the biologically relevant g-quadruplex element in the human c-MYC promoter. implications for g-quadruplex stabilization. Biochem. 2005; 44:2048–2058. [DOI] [PubMed] [Google Scholar]
- 63. Chung W.J., Heddi B., Hamon F., Teulade-Fichou M.P., Phan A.T.. Solution structure of a G-quadruplex bound to the bisquinolinium compound Phen-DC3. Angew. Chem. 2014; 126:1017–1020. [DOI] [PubMed] [Google Scholar]
- 64. Thiviyanathan V., Luxon B.A., Leontis N.B., Illangasekare N., Donne D.G., Gorenstein D.G.. Hybrid-hybrid matrix structural refinement of a DNA three-way junction from 3D NOESY-NOESY. J. Biomol. NMR. 1999; 14:209–221. [DOI] [PubMed] [Google Scholar]
- 65. Amrane S., De Cian A., Rosu F., Kaiser M., De Pauw E., Teulade-Fichou M.-P., Mergny J.-L.. Identification of trinucleotide repeat ligands with a FRET melting assay. ChemBioChem. 2008; 9:1229–1234. [DOI] [PubMed] [Google Scholar]
- 66. Novotna J., Laguerre A., Granzhan A., Pirrotta M., Teulade-Fichou M.-P., Monchaud D.. Cationic azacryptands as selective three-way DNA junction binding agents. Org. Biomol. Chem. 2015; 13:215–222. [DOI] [PubMed] [Google Scholar]
- 67. Granzhan A., Monchaud D., Saettel N., Guédin A., Mergny J.-L., Teulade-Fichou M.-P.. ‘One ring to bind them all’—Part II: identification of promising G-quadruplex ligands by screening of cyclophane-type macrocycles. J. Nucleic Acids. 2010; 2010:460561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Malina J., Hannon M.J., Brabec V.. Recognition of DNA three-way junctions by metallosupramolecular cylinders: gel electrophoresis studies. Chem. Eur. J. 2007; 13:3871–3877. [DOI] [PubMed] [Google Scholar]
- 69. Chou T.-C., Talalay P.. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme. Regul. 1984; 22:27–55. [DOI] [PubMed] [Google Scholar]
- 70. Chou T.-C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010; 70:440–446. [DOI] [PubMed] [Google Scholar]
- 71. De Rache A., Mergny J.-L.. Assessment of selectivity of G-quadruplex ligands via an optimised FRET melting assay. Biochimie. 2015; 115:194–202. [DOI] [PubMed] [Google Scholar]
- 72. Haudecoeur R., Stefan L., Monchaud D.. Multitasking water-soluble synthetic G-quartets: from preferential RNA-quadruplex interaction to biocatalytic activity. Chem. Eur. J. 2013; 19:12739–12747. [DOI] [PubMed] [Google Scholar]
- 73. Ragazzon P.A., Garbett N.C., Chaires J.B.. Competition dialysis: a method for the study of structural selective nucleic acid binding. Methods. 2007; 42:173–182. [DOI] [PubMed] [Google Scholar]
- 74. Lin C., Dickerhoff J., Yang D.. NMR studies of G-quadruplex structures and G-quadruplex-interactive compounds. G-Quadruplex Nucleic Acids. 2019; Springer; 157–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Haider S.M., Neidle S., Parkinson G.N.. A structural analysis of G-quadruplex/ligand interactions. Biochimie. 2011; 93:1239–1251. [DOI] [PubMed] [Google Scholar]
- 76. Ropp P.J., Spiegel J.O., Walker J.L., Green H., Morales G.A., Milliken K.A., Ringe J.J., Durrant J.D.. Gypsum-DL: an open-source program for preparing small-molecule libraries for structure-based virtual screening. J. Cheminformatics. 2019; 11:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J.. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009; 30:2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hess B., Kutzner C., van der Spoel D., Lindahl E.. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008; 4:435–447. [DOI] [PubMed] [Google Scholar]
- 79. Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M.R., Smith J.C., Kasson P.M., van der Spoel D.et al.. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013; 29:845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Páll S., Abraham M.J., Kutzner C., Hess B., Lindahl E.. Tackling Exascale Software Challenges in Molecular Dynamics Simulations with GROMACS. 2014; Springer; 3–27.International Conference on Exascale Applications and Software. [Google Scholar]
- 81. Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E.. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015; 1-2:19–25. [Google Scholar]
- 82. Jourdan M., Garcia J., Lhomme J., Teulade-Fichou M.-P., Vigneron J.-P., Lehn J.-M.. Threading bis-intercalation of a macrocyclic bisacridine at Abasic sites in DNA: nuclear magnetic resonance and molecular modeling study. Biochem. 1999; 38:14205–14213. [DOI] [PubMed] [Google Scholar]
- 83. Cudic P., Vigneron J.-P., Lehn J.-M., Cesario M., Prangé T.. Molecular recognition of azobenzene dicarboxylates by acridine-based receptor molecules, crystal structure of the supramolecular inclusion complex of trans-3,3′-azobenzene dicarboxylate with a cyclo-bis-intercaland receptor. Eur. J. Org. Chem. 1999; 1999:2479–2484. [Google Scholar]
- 84. Jourdan M., Granzhan A., Guillot R., Dumy P., Teulade-Fichou M.-P.. Double threading through DNA: NMR structural study of a bis-naphthalene macrocycle bound to a thymine–thymine mismatch. Nucleic. Acids. Res. 2012; 40:5115–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Amendola V., Bergamaschi G., Miljkovic A.. Azacryptands as molecular cages for anions and metal ions. Supramol. Chem. 2018; 30:236–242. [Google Scholar]
- 86. La Cognata S., Miljkovic A., Mobili R., Bergamaschi G., Amendola V.. Organic cages as building blocks for mechanically interlocked molecules: towards molecular machines. ChemPlusChem. 2020; 85:1145–1155. [DOI] [PubMed] [Google Scholar]
- 87. Alibrandi G., Amendola V., Bergamaschi G., Fabbrizzi L., Licchelli M.. Bistren cryptands and cryptates: versatile receptors for anion inclusion and recognition in water. Org. Biomol. Chem. 2015; 13:3510–3524. [DOI] [PubMed] [Google Scholar]
- 88. Soule H., Vazquez J., Long A., Albert S., Brennan M.. A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl. Cancer Inst. 1973; 51:1409–1416. [DOI] [PubMed] [Google Scholar]
- 89. Levenson A.S., Jordan V.C.. MCF-7: the first hormone-responsive breast cancer cell line. Cancer Res. 1997; 57:3071–3078. [PubMed] [Google Scholar]
- 90. Cailleau R., Young R., Olive M., Reeves W. Jr. Breast tumor cell lines from pleural effusions. J. Natl. Cancer Inst. 1974; 53:661–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Skehan P., Storeng R., Scudiero D., Monks A., McMahon J., Vistica D., Warren J.T., Bokesch H., Kenney S., Boyd M.R.. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990; 82:1107–1112. [DOI] [PubMed] [Google Scholar]
- 92. Vichai V., Kirtikara K.. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006; 1:1112. [DOI] [PubMed] [Google Scholar]
- 93. Bonner W.M., Redon C.E., Dickey J.S., Nakamura A.J., Sedelnikova O.A., Solier S., Pommier Y.. γH2AX and cancer. Nat. Rev. Cancer. 2008; 8:957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Burma S., Chen B.P., Murphy M., Kurimasa A., Chen D.J.. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001; 276:42462–42467. [DOI] [PubMed] [Google Scholar]
- 95. Paull K., Shoemaker R., Hodes L., Monks A., Scudiero D., Rubinstein L., Plowman J., Boyd M.. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J. Natl. Cancer Inst. 1989; 81:1088–1092. [DOI] [PubMed] [Google Scholar]
- 96. Shoemaker R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006; 6:813–823. [DOI] [PubMed] [Google Scholar]
- 97. Hampel S.M., Pepe A., Greulich-Bode K.M., Malhotra S.V., Reszka A.P., Veith S., Boukamp P., Neidle S.. Mechanism of the antiproliferative activity of some naphthalene diimide G-quadruplex ligands. Mol. Pharmacol. 2013; 83:470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Saleh M.M., Laughton C.A., Bradshaw T.D., Moody C.J.. Development of a series of bis-triazoles as G-quadruplex ligands. RSC Adv. 2017; 7:47297–47308. [Google Scholar]
- 99. Larsen A.K., Grondard L., Couprie J., Desoize B., Comoe L., Jardillier J.-C., Riou J.-F.. The antileukemic alkaloid fagaronine is an inhibitor of DNA topoisomerases I and II. Biochem. Pharmacol. 1993; 46:1403–1412. [DOI] [PubMed] [Google Scholar]
- 100. Olivieri M., Cho T., Álvarez-Quilón A., Li K., Schellenberg M.J., Zimmermann M., Hustedt N., Rossi S.E., Adam S., Melo H.et al.. A genetic map of the response to DNA damage in human cells. Cell. 2020; 182:481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bruno P.M., Lu M., Dennis K.A., Inam H., Moore C.J., Sheehe J., Elledge S.J., Hemann M.T., Pritchard J.R.. The primary mechanism of cytotoxicity of the chemotherapeutic agent CX-5461 is topoisomerase II poisoning. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:4053–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bossaert M., Pipier A., Riou J.F., Noirot C., Nguyễn L.-T., Serre R.-F., Bouchez O., Defrancq E., Calsou P., Britton S.et al.. Transcription-associated topoisomerase activities control DNA-breaks production by G-quadruplex ligands. eLife. 2021; 10:e65184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kawatani M., Takayama H., Muroi M., Kimura S., Maekawa T., Osada H.. Identification of a small-molecule inhibitor of DNA topoisomerase II by proteomic profiling. Chem. Biol. 2011; 18:743–751. [DOI] [PubMed] [Google Scholar]
- 104. Jensen P.B., Sehested M.. DNA topoisomerase II rescue by catalytic inhibitors: a new strategy to improve the antitumor selectivity of etoposide. Biochem. Pharmacol. 1997; 54:755–759. [DOI] [PubMed] [Google Scholar]
- 105. Treangen T.J., Salzberg S.L.. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 2012; 13:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Cañeque T., Müller S., Rodriguez R.. Visualizing biologically active small molecules in cells using click chemistry. Nat. Rev. Chem. 2018; 2:202–215. [Google Scholar]
- 107. Biffi G., Tannahill D., McCafferty J., Balasubramanian S.. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013; 5:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Britton S., Coates J., Jackson S.P.. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell Biol. 2013; 202:579–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Koningsbruggen S.V., Gierliński M., Schofield P., Martin D., Barton G.J., Ariyurek Y., Dunnen J.T., Lamond A.I.. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol. Biol. Cell. 2010; 21:3735–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Németh A., Conesa A., Santoyo-Lopez J., Medina I., Montaner D., Péterfia B., Solovei I., Cremer T., Dopazo J., Längst G.. Initial genomics of the human nucleolus. PLoS Genet. 2010; 6:e1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Bizhanova A., Kaufman P.D.. Close to the edge: heterochromatin at the nucleolar and nuclear peripheries. Biochim. Biophys. Acta (BBA) - Gene Regul. Mech. 2021; 1864:194666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Caburet S., Conti C., Schurra C., Lebofsky R., Edelstein S.J., Bensimon A.. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 2005; 15:1079–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang J.C. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002; 3:430–440. [DOI] [PubMed] [Google Scholar]
- 114. Wang G., Vasquez K.M.. Non-B DNA structure-induced genetic instability. Mutat. Res. 2006; 598:103–119. [DOI] [PubMed] [Google Scholar]
- 115. Vasquez K.M., Wang G.. The yin and yang of repair mechanisms in DNA structure-induced genetic instability. Mut. Res.-Fundam. Mol. Mech. Mutagen. 2013; 743:118–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Froelich-Ammon S., Gale K.C., Osheroff N.. Site-specific cleavage of a DNA hairpin by topoisomerase II. DNA secondary structure as a determinant of enzyme recognition/cleavage. J. Biol. Chem. 1994; 269:7719–7725. [PubMed] [Google Scholar]
- 117. Jonstrup A.T., Thomsen T., Wang Y., Knudsen B.R., Koch J., Andersen A.H.. Hairpin structures formed by alpha satellite DNA of human centromeres are cleaved by human topoisomerase IIα. Nucleic Acids Res. 2008; 36:6165–6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Atkin N.D., Raimer H.M., Wang Y.-H.. Broken by the cut: a journey into the role of topoisomerase II in DNA fragility. Genes. 2019; 10:791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Szlachta K., Manukyan A., Raimer H.M., Singh S., Salamon A., Guo W., Lobachev K.S., Wang Y.-H.. Topoisomerase II contributes to DNA secondary structure-mediated double-stranded breaks. Nucleic Acids Res. 2020; 48:6654–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Nitiss J.L. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer. 2009; 9:338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Delgado J.L., Hsieh C.-M., Chan N.-L., Hiasa H.. Topoisomerases as anticancer targets. Biochem. J. 2018; 475:373–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Pilati P., Nitti D., Mocellin S.. Cancer resistance to type II topoisomerase inhibitors. Curr. Med. Chem. 2012; 19:3900–3906. [DOI] [PubMed] [Google Scholar]
- 123. Peltonen K., Colis L., Liu H., Trivedi R., Moubarek M.S., Moore H.M., Bai B., Rudek M.A., Bieberich C.J., Laiho M.. A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell. 2014; 25:77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Chodosh L.A., Fire A., Samuels M., Sharp P.A.. 5,6-Dichloro-1-β-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J. Biol. Chem. 1989; 264:2250–2257. [PubMed] [Google Scholar]
- 125. Spadari S., Sala F., Pedrali-Noy G.. Aphidicolin: a specific inhibitor of nuclear DNA replication in eukaryotes. Trends Biochem. Sci. 1982; 7:29–32. [Google Scholar]
- 126. Leonard C.J., Berns K.I.. Cohn W.E., Moldave K.. Adeno-associated virus type 2: a latent life cycle. Progress in Nucleic Acid Research and Molecular Biology. 1994; 48:Academic Press; 29–52. [DOI] [PubMed] [Google Scholar]
- 127. Berns K.I. The unusual properties of the AAV inverted terminal repeat. Hum. Gene Ther. 2020; 31:518–523. [DOI] [PubMed] [Google Scholar]
- 128. Shaheen M., Allen C., Nickoloff J.A., Hromas R.. Synthetic lethality: exploiting the addiction of cancer to DNA repair. Blood. 2011; 117:6074–6082. [DOI] [PubMed] [Google Scholar]
- 129. Leahy J.J., Golding B.T., Griffin R.J., Hardcastle I.R., Richardson C., Rigoreau L., Smith G.C.. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg. Med. Chem. Lett. 2004; 14:6083–6087. [DOI] [PubMed] [Google Scholar]
- 130. Hickson I., Zhao Y., Richardson C.J., Green S.J., Martin N.M., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C.. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004; 64:9152–9159. [DOI] [PubMed] [Google Scholar]
- 131. Huang F., Motlekar N.A., Burgwin C.M., Napper A.D., Diamond S.L., Mazin A.V.. Identification of specific inhibitors of human RAD51 recombinase using high-throughput screening. ACS Chem. Biol. 2011; 6:628–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Tang J., Wennerberg K., Aittokallio T.. What is synergy? The Saariselkä agreement revisited. Front. Pharmacol. 2015; 6:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Rodriguez R., Mueller S., Yeoman J.A., Trentesaux C., Riou J.-F., Balasubramanian S.. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 2008; 130:15758–15758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. De Cian A., DeLemos E., Mergny J.-L., Teulade-Fichou M.-P., Monchaud D.. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007; 129:1856–1857. [DOI] [PubMed] [Google Scholar]
- 135. De Cian A., Cristofari G., Reichenbach P., De Lemos E., Monchaud D., Teulade-Fichou M.-P., Shin-Ya K., Lacroix L., Lingner J., Mergny J.-L.. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:17347–17352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Pennarun G., Granotier C., Gauthier L.R., Gomez D., Boussin F.D.. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene. 2005; 24:2917–2928. [DOI] [PubMed] [Google Scholar]
- 137. Granotier C., Pennarun G., Riou L., Hoffschir F., Gauthier L.R., De Cian A., Gomez D., Mandine E., Riou J.F., Mergny J.L.et al.. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res. 2005; 33:4182–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.