Abstract
Spx is a global transcriptional regulator in Gram-positive bacteria and has been inferred to efficiently activate transcription upon oxidative stress by engaging RNA polymerase (RNAP) and promoter DNA. However, the precise mechanism by which it interacts with RNAP and promoter DNA to initiate transcription remains obscure. Here, we report the cryo-EM structure of an intact Spx-dependent transcription activation complex (Spx–TAC) from Bacillus subtilis at 4.2 Å resolution. The structure traps Spx in an active conformation and defines key interactions accounting for Spx-dependent transcription activation. Strikingly, an oxidized Spx monomer engages RNAP by simultaneously interacting with the C-terminal domain of RNAP alpha subunit (αCTD) and σA. The interface between Spx and αCTD is distinct from those previously reported activators, indicating αCTD as a multiple target for the interaction between RNAP and various transcription activators. Notably, Spx specifically wraps the conserved –44 element of promoter DNA, thereby stabilizing Spx–TAC. Besides, Spx interacts extensively with σA through three different interfaces and promotes Spx-dependent transcription activation. Together, our structural and biochemical results provide a novel mechanistic framework for the regulation of bacterial transcription activation and shed new light on the physiological roles of the global Spx-family transcription factors.
INTRODUCTION
Most bacteria rely heavily on transcription activation for precise response to changing environmental cues. Such regulation occurs mostly at the transcription initiation step catalyzed by RNA polymerase and plays critical regulatory roles in gene expression (1–3). For canonical promoters that carry the conserved –35 and –10 elements, transcription initiation can be orchestrated solely by bacterial RNAP holoenzyme, which comprises a promoter-specific factor σ and core bacterial RNA polymerase (α2ββ'ω). Bacterial RNAP holoenzyme binds the promoter DNA duplex to form the RNAP-promoter closed complex (RPc), isomerizes to RNAP-promoter open complex (RPo), and finally initiates RNA synthesis (4–10). For non-canonical promoters, transcription initiation is stimulated by specific activators that typically bind to distinctive promoters and/or interact with bacterial RNAP holoenzyme, forming an initiation-competent transcription activation complex (TAC) (11–15). Besides the universally conserved five subunits (α2ββ′ω), Bacillus subtilis RNAP consists of another two small auxiliary subunits, δ subunit encoded by rpoE and ϵ subunit encoded by rpoY (16,17). δ subunit has been reported to play multiple regulation roles in transcription initiation, elongation and termination, and is required for maintaining cellular fitness and certain virulence gene expression (18–20). While, ϵ subunit has been indicated to be involved in RNAP stability and protection from phage infection (21,22).
Spx, initially named as suppressor of clpP and clpX, is highly conserved among low GC-content Gram-positive bacteria and is widely distributed in the phylum Firmicutes, including some disseminated human pathogens (23,24). Genetic and biochemical studies have shown that B. subtilis Spx is a pleiotropic transcriptional regulator that responds to various stresses (such as oxidative stress, thermo damage, and cell wall-active antibiotics) and serves as a prototype for a large group of stress-response regulators (25–29). Although it is known to inhibit transcription by acting as an ‘anti-α factor’, numerous biochemical and structural analyses have painted a gradually clear outline of Spx as a global transcription activator by forming Spx-dependent transcription activation complex (Spx–TAC):
Spx-dependent transcription activation is typically induced by thiol-specific oxidative stress, and the conserved N-terminal cysteine-x-x-cysteine (CXXC) motif of Spx functions as a reversible redox-sensing switch in response to cellular redox stress (24–26). Previous genomic and transcriptomic analyses showed that expression of the 275 probable disulfide stress-response genes, including those encoding thioredoxin (trxA) and thioredoxin reductase (trxB), relies mostly on oxidized Spx (30,31).
Spx-dependent transcription activation requires the direct interaction between Spx and the C-terminal domain of the alpha subunit of RNAP (αCTD), which typically interacts with the specific UP-elements (AT-rich DNA regions locate upstream of the promoter, approximate from –40 to –60) of the promoter (1,32). Both in vivo and in vitro experiments have revealed that a Spx monomer engages RNAP, and disruption of key interactions between Spx and αCTD causes severe defects in Spx-dependent transcription activation. Residue G52 of Spx and the ‘‘261’’ determinant of αCTD are particularly important (33–37). Though analysis of the Spx-αCTD structure can readily account for the repression activity of Spx, it is difficult to explain how such interaction can also enhance transcription initiation. The conformational transition between the reduced and oxidized Spx-αCTD structures may provide a feasible explanation.
Unlike canonical DNA-binding transcriptional activators, Spx shows no intrinsic DNA-binding activity on its own. However, in combination with αCTD, Spx can recognize the conserved –44 element (A/TGCA/T motif; numbered relative to the transcription start site) (31,36–38). Protein-DNA cross-linking results showed that interactions between RNAP-bound Spx and the –44 element causes RNAP σA to reposition from an upstream –35-like element to the downstream –36 position of trxB promoter (39), indicative of conformational changes of Spx–TAC upon oxidative stress and Spx binding. Interactions between Spx and σA may also occur during this process to enhance transcription activation by Spx.
Thus, Spx represents a unique family of bacterial transcription activators. While the canonical bacterial transcription activators have been extensively characterized (11,13,15,40), the distinct molecular mechanism and structural basis of Spx-dependent global transcription activation are still obscure, partially because of a lack of precise structural information for B. subtilis RNAP in complex with Spx. The specific interactions among Spx, RNAP and the promoter DNA remain to be elucidated.
In the present study, we determined the cryo-EM structure of an intact Spx-dependent transcription activation complex (Spx–TAC) at 4.2 Å resolution. The structure traps an active conformation of Spx in complex with B. subtilis RNAP holoenzyme and the promoter DNA. Strikingly, a single oxidized Spx monomer engages B. subtilis RNAP by simultaneously interacting with RNAP αCTD and σA. The interface between Spx and αCTD is distinct from those of the previously reported transcription activators. Notably, oxidized Spx changes conformation and specifically binds different faces of the conserved –44 element of promoter DNA, facilitating remodeling of RNAP and stabilization of Spx–TAC. In addition, the three interfaces observed between Spx and σA may play different roles in promoting Spx-dependent transcription activation. Our results unveil the functional contacts that mediate Spx-dependent transcription activation, provide a new mechanistic framework for the regulation of bacterial transcription activation, and support further exploration on the physiological roles of Spx-family transcription factors.
MATERIALS AND METHODS
Plasmid construction
Plasmid pET28a-TEV-Spx encoding N-terminal His6-TEV tagged B. subtilis Spx under the control of T7 promoter was synthesized by Sangon Biotech, Inc. B. subtilis σA and α subunits were amplified from B. subtilis genomic DNA and cloned into pET-28a using homologous recombination (ToloBio, Inc.), respectively. Linear trxA_mango DNA fragment corresponding to –75 to + 78 of B. subtilis trxA promoter followed by Mango III coding sequence (41–45) was generated by de novo PCR using primers of trxA-mango F1, F2, R1 and R2 (Supplementary Table S1), purified using the QIAquick PCR Purification Kit (Qiagen, Inc.), and stored at –80°C. Similarly, linear rrnJ P1_mango DNA fragment corresponding to –94 to + 66 of B. subtilis rrnJ P1 promoter followed by Mango III coding sequence was prepared by using primers of rrnJ P1-mango F1, F2, R1 and R2 (Supplementary Table S1) and purified. The sequences of trxA-mango or rrnJ P1-mango promoter DNA are provided in Supplementary Figure S1. Plasmids carrying Spx amino acid substitutions were constructed using site-directed mutagenesis (QuikChange Site-Directed Mutagenesis Kit, Agilent, Inc.). Primers of Spx mutants and others used in this study are shown in Supplementary Table S1.
Spx protein purification
Escherichia coli strain BL21(DE3) (Invitrogen, Inc.) was transformed with plasmid pET28a-TEV-Spx or pET28a-TEV-Spx derivatives. Single colonies of the resulting transformants were used to inoculate 1 L LB broth containing 50 μg/ml kanamycin, and cultures were incubated at 37°C with shaking until OD600 reached 0.6. Protein expression was induced by addition of IPTG to 0.5 mM, and cultures were incubated 14 h at 20°C. The cells were harvested by centrifugation (5000 rpm; 10 min at 4°C), resuspended in 20 ml buffer A (20 mM Tris–HCl, pH 8.0, 0.2 M NaCl, 5% glycerol), and lysed using an ATS AH-10013 cell disrupter (ATS, Inc.). The lysate was centrifuged (12 000 rpm; 40 min at 4°C), and the supernatant was loaded onto a 3 ml Ni-NTA agarose column (Qiagen, Inc.) equilibrated with buffer A. The column was subsequently washed with 15 ml buffer A containing 0.02 M, 0.04 M imidazole and eluted with 15 ml buffer A containing 0.2 M imidazole. The eluate was loaded onto a HiLoad 16/600 Superdex 200 column (GE Healthcare, Inc.) equilibrated in buffer B (20 mM Tris–HCl, pH 8.0, 75 mM NaCl, 5 mM MgCl2), and the column was eluted with 120 ml of the same buffer. Fractions containing B. subtilis Spx were pooled and stored at –80°C. B. subtilis Spx derivatives were expressed and purified using the same procedure as the wild type protein. The yields were ∼3 mg/l, and the purities were >95%. To test that each B. subtilis Spx derivative remains folded properly and as stable as the wild-type Spx, the thermal stability assays were performed for wild-type B. subtilis Spx and its derivatives according to the user manual of Tycho NT.6 system (NanoTemper Technologies GmbH, Inc.).
B. subtilis σA purification
B. subtilis σA was prepared as described (21) with some modifications. Briefly, E. coli strain BL21(DE3) (Invitrogen, Inc.) was transformed with plasmid pET-28a-σA. Single colonies of the resulting transformants were used to inoculate 1 l LB broth containing 50 μg/ml kanamycin, and cultures were incubated at 37°C with shaking until OD600 reached 0.8. Protein expression was induced by addition of IPTG to 0.5 mM, and cultures were incubated 14 h at 18°C. Then cells were harvested, resuspended in buffer C (50 mM Tris–HCl, pH 8.0, 0.3 M NaCl, 5% v/v glycerol), and lysed using an ATS AH-10013 cell disrupter (ATS, Inc.). The lysate was centrifuged (12 000 rpm; 40 min at 4°C), and the supernatant was loaded onto a 2 ml Ni-NTA column (Smart Lifesciences Inc., China), which was washed with buffer C containing 20 mM imidazole and eluted with buffer C containing 300 mM imidazole. The eluted fractions were diluted with TGED buffer (10 mM Tris–HCl, pH 8.0, 5% glycerol, 1 mM DTT and 1 mM EDTA). The sample was loaded onto a 5 ml HiTrap Heparin HP column (GE Healthcare Life Sciences) equilibrated by buffer D (50 mM Tris–HCl, pH 8.0, 50 mM NaCl, 5% v/v glycerol, 1 mM DTT), and eluted with a 100 ml linear gradient of 0.05–1 M NaCl in buffer D. Fractions containing B. subtilis σA were concentrated using an Amicon Ultra-15 centrifugal filter (10 kDa MWCO; Merck Millipore, Inc.). The purity of the eluted fractions containing B. subtilis σA was >95%, and the yield was ∼5 mg/l.
B. subtilis α purification
Briefly, E. coli strain Rosetta 2 (Invitrogen, Inc.) was transformed with plasmid pET-28a-α. Single colonies of the resulting transformants were used to inoculate 1 l LB broth containing 50 μg/ml kanamycin, and cultures were incubated at 37°C with shaking until OD600 reached 0.8. Protein expression was induced by addition of IPTG to 0.5 mM, and cultures were incubated 14 h at 18°C. Then cells were harvested, resuspended in resuspended in 20 ml buffer A (20 mM Tris–HCl, pH 8.0, 0.2 M NaCl, 5% glycerol), and lysed using an ATS AH-10013 cell disrupter (ATS, Inc.). The lysate was centrifuged (12 000 rpm; 40 min at 4°C), and the supernatant was loaded onto a 2 ml Ni-NTA agarose column (Qiagen, Inc.) equilibrated with buffer A. The column was subsequently washed with 10 ml buffer A containing 0.02 M, 0.04 M imidazole and eluted with 10 ml buffer A containing 0.2 M imidazole. The eluate was loaded onto a HiLoad 16/600 Superdex 200 column (GE Healthcare, Inc.) equilibrated in buffer B (20 mM Tris–HCl, pH 8.0, 75 mM NaCl, 5 mM MgCl2), and the column was eluted with 120 ml of the same buffer. Fractions containing B. subtilis α were pooled and concentrated to ∼12 mg/l, and the purities were >95%.
B. subtilis RNAP holoenzyme purification
Endogenous B. subtilis RNAP holoenzyme that contains σA was prepared from B. subtilis (strain 168) as described (21) with some modifications. After incubation in LB broth at 37°C with shaking for 22 h, cells were harvested, resuspended in buffer E (40 mM Tris–HCl, pH, 8.0, 200 mM NaCl, 5% glycerol, 2 mM EDTA, 2 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and lysed using an ATS AH-10013 cell disrupter (ATS, Inc.). The lysate was centrifuged (12 000 rpm; 40 min at 4°C), and the supernatant was precipitated with polyethylenimine (0.7% final) at 4°C for 15 min. After centrifugation, the pellet was washed three times and then resuspended with TGED buffer with an additional 1 M NaCl. The supernatant was then precipitated by ammonium sulfate (0.3 g/ml final), and the resulting pellet was collected and dissolved with TGED buffer. The supernatant was loaded onto a 5 ml HiTrap Heparin HP column (GE Healthcare Life Sciences) equilibrated in buffer E (40 mM Tris–HCl, pH 8.0, 0.1 M NaCl, 5% v/v glycerol, 1 mM DTT, and 0.1 mM EDTA), and eluted with a 100 ml linear gradient of 0.1–1 M NaCl in buffer E. Fractions containing B. subtilis RNAP holoenzyme were further loaded onto a Mono Q 10/100 GL column (GE Healthcare Life Sciences) and eluted with a 120 ml linear gradient of 0.1–0.6 M NaCl in buffer E. The fractions containing pure B. subtilis RNAP holoenzyme were collected, concentrated, and stored at –80°C. The yield of RNAP holoenzyme was 0.4 mg/l, and the purity was > 95%.
Assembly of B. subtilis Spx–TAC
DNA oligonucleotides trxA scaffold (sequences are shown in Figure 1A) were synthesized (Sangon Biotech, Inc.) and dissolved in nuclease-free water to 1 mM. Template strand DNA and nontemplate strand DNA were annealed at 1:1 ratio in 10 mM Tris–HCl, pH 7.9, 0.2 M NaCl. Then, B. subtilis Spx–TAC was assembled by incubating B. subtilis RNAP holoenzyme, B. subtilis σA, trxA scaffold and B. subtilis Spx in a molar ratio of 1:0.5: 1.1:10 at 4°C overnight. Subsequently, the mixture was applied to a HiLoad 16/600 Superdex 200 pg column (GE Healthcare Life Sciences) equilibrated in buffer B. After identification by SDS-PAGE and electrophoretic mobility shift assay (EMSA), the fractions containing B. subtilis Spx–TAC were concentrated to 13 mg/ml using Amicon Ultra centrifugal filters (10 kDa MWCO, Merck Millipore, Inc.).
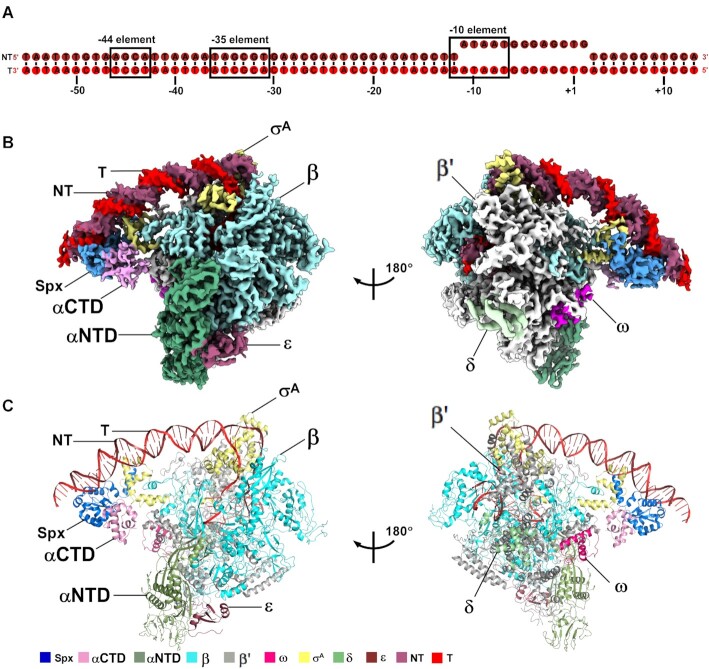
Figure 1.
The overall structure of B. subtilis Spx–TAC. (A) DNA scaffold used in structure determination of B. subtilis Spx–TAC. (B, C) two views of the cryo-EM density map (B) and structure model (C) of B. subtilis Spx–TAC. The EM density maps and cartoon representations of Spx–TAC are colored as indicated in the color key. NT, non-template-strand promoter DNA; T, template-strand promoter DNA.
Cryo-EM grid preparation
Immediately before freezing, 4 mM CHAPSO was added to the freshly purified B. subtilis Spx–TAC. C-flat grids (CF-1.2/1.3-4C; Protochips, Inc.) were glow-discharged for 60 s at 15 mA prior to the application of 3 μl of B. subtilis Spx–TAC complex, then plunge-frozen in liquid ethane using a Vitrobot (FEI, Inc.) with 95% chamber humidity at 10°C.
Cryo-EM data acquisition and processing
The grids with B. subtilis Spx–TAC were imaged using a 300 kV Titan Krios (FEI, Inc.) equipped with a K2 Summit direct electron detector (Gatan, Inc.). Images were recorded with Serial EM (46) in counting mode with a physical pixel size of 1.307 Å and a defocus range of 1.5–2.5 μm. Data were collected with a dose of 10 e/pixel/s. Images were recorded with a 10 s exposure and 0.25 s subframes to give a total dose of 59 e/Å2. Subframes were aligned and summed using MotionCor2 (47). The contrast transfer function was estimated for each summed image using CTFFIND4 (48). From the summed images, ∼10 000 particles were manually picked and subjected to 2D classification in RELION (49). 2D averages of the best classes were used as templates for auto-picking in RELION. Auto-picked particles were manually inspected, then subjected to 2D classification in RELION. Poorly populated classes were removed, resulting in a dataset of 308 440 particles. These particles were 3D-classified in RELION using a map of B. subtilis BmrR-TAC (EMD-30390, with BmrR omitted) (21) low-pass filtered to 40 Å resolution as a reference. 3D classification resulted in four classes. Particles in Classes 2 and 4 were 3D auto-refined, and the best-resolved class containing 153 870 particles was post-processed in RELION. The Gold-standard Fourier-shell-correlation analysis indicated a mean map resolution of 4.2 Å (Supplementary Figure S3-S4B, E).
Cryo-EM model building and refinement
The model of B. subtilis RNAP holoenzyme and the downstream DNA from the structure of B. subtilis BmrR-TAC (PDB ID:7CKQ) (21), B. subtilis RNAP-HelD (PDB ID:6ZCA) (19) and the crystal structure of Spx in complex with B. subtilis RNAP αCTD (PDB ID:1Z3E) (34) were manually fitted into the cryo-EM density map in Coot (50), followed by adjustment of main- and side-chain conformations in Coot and real-space refinement using Phenix. The models of upstream promoter DNA and Spx helix α7 were built manually in Coot. The coordinates were real-space refined with secondary structure restraints in Phenix.
In vitro transcription assay
In vitro Mango-based transcription assays were carried out by incubating B. subtilis RNAP holoenzyme, B. subtilis σA, trxA-mango scaffold or rrnJ P1-mango scaffold, and B. subtilis Spx or its derivatives. Transcription assay was performed in a 96-well microplate format. Reaction mixtures (40 μl) contained: 0 or 3 μM Spx or Spx derivatives or reduced Spx, 0.1 μM B. subtilis RNAP holoenzyme, 0.1 μM B. subtilis σA, 50 nM trxA-mango scaffold or rrnJ P1-mango scaffold, 1 μM TO1-Biotin, 0.1 mM NTP mix (ATP, UTP, GTP and CTP), 40 mM Tris–HCl, pH 8.0, 50 mM NaCl, 10 mM MgCl2. First, B. subtilis RNAP holoenzyme, B. subtilis σA, and trxA-mango scaffold were incubated for 10 min at 37°C, then B. subtilis Spx or mutant derivatives was added into the mixture and incubated for 30 min at 37°C. Following incubation of NTP mix and TO1-biotin for another 10 min at 37°C, fluorescence emission intensities were measured using a multimode plate reader (EnVision, PerkinElmer Inc.; excitation wavelength = 510 nm; emission wavelength = 535 nm). Relative transcription activities of Spx derivatives were calculated using:
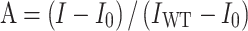 |
(1) |
where IWT and I are the fluorescence intensities in the presence of Spx and Spx derivatives; I0 is the fluorescence intensity in the absence of Spx.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays (EMSA) of B. subtilis Spx were performed in reaction mixtures (20 μl) containing: 10 μM Spx (with or without DTT) or Spx derivatives, 10 μM B. subtilis RNAP α subunit, 0.1 μM trxA-mango scaffold or rrnJ P1-mango scaffold in EMSA buffer (20 mM Tris–HCl, pH 8.0, 50 mM NaCl, 5 mM MgCl2, 5% glycerol). Reduced Spx was treated with 5mM DTT for 60 min at 25°C. Spx or Spx derivatives was incubated with B. subtilis RNAP α subunit for 10 min at 25°C and then incubated with DNA scaffold for 15 min at 25°C. The reaction mixtures were applied to 5% polyacrylamide slab gels (29:1 acrylamide/bisacrylamide), electrophoresed in 90 mM Tris-borate, pH 8.0, and 0.2 mM EDTA, and stained with 4S Red Plus Nucleic Acid Stain (Sangon Biotech, Inc.) according to the procedure of the manufacturer.
To further clarify whether Spx derivatives affect the formation of Spx–TAC or not, we also carried out electrophoretic mobility shift assays in reaction mixtures (20 μl) containing: 3 μM Spx or Spx derivatives, 0.1 μM B. subtilis RNAP, 0.012 μM trxA-mango promoter DNA in EMSA buffer B (40 mM Tris–HCl, pH 8.0, 100 mM NaCl, 10 mM MgCl2, 5% glycerol). B. subtilis RNAP was firstly incubated with trxA-mango promoter DNA for 10 min at 37°C, then incubated with Spx or Spx derivatives for 20 min at 37°C. Reaction mixtures were supplemented with 0.02 mg/ml heparin and incubated 2 min at 22°C. The reaction mixtures were finally applied to 5% polyacrylamide slab gels (29:1 acrylamide/bisacrylamide), electrophoresed in 90 mM Tris-borate, pH 8.0, and 0.2 mM EDTA, and stained with 4S Red Plus Nucleic Acid Stain (Sangon Biotech, Inc.) according to the procedure of the manufacturer.
Data analysis
Data for in vitro transcription assays are means of three technical replicates. Error bars represent mean ± SEM of n = 3 experiments.
RESULTS
Overall structure of Spx–TAC
To obtain a structure of Spx–TAC, we used a nucleic-acid scaffold corresponding to positions –55 to + 13 of a thioredoxin gene trxA promoter (positions numbered relative to the transcription start site; Figure 1A). The scaffold contains a presumed Spx binding site (–44 element centered around position –44), a –35 element, and a 13-bp non-complementary transcription bubble (–11 to + 2) with a consensus –10 element, which was designed to stabilize the conformation of RNAP and downstream DNA (Figure 1A). In vitro Mango-based transcription assay with the purified endogenous B. subtilis RNAP holoenzyme and trxA-mango promoter showed that the purified recombinant B. subtilis Spx significantly activated transcription of trxA, indicating that the purified proteins had expected biological functions (Supplementary Figure S2A and B). Thus, we assembled Spx–TAC using B. subtilis RNAP holoenzyme, trxA promoter DNA scaffold, and B. subtilis Spx. Gel-filtration chromatography and SDS-PAGE confirmed that all expected protein components are present in the complex with stoichiometric levels, and native gel analysis of the purified complex verified the DNA scaffold is included, too (Supplementary Figure S2A, C and D).
Subsequently, we froze the complex and collected data on a Titan Krios cryo-EM. 328,348 particles picked from 1816 movies were sequentially processed and classified into 4 classes after 3D classification. Classes 2 and 4 showed extra density of Spx and may represent Spx–TAC, and were subjected to CTF refinement, Bayesian polishing, 3D auto-refinement and post-processing, and finally the structure was determined at a nominal resolution of 4.22 Å (Figure 1B and C; Supplementary Figures S3, S4 and S5; Table 1). Local resolution calculation exhibits a central core at ∼3.5–4.5 Å and a peripheral Spx at ∼5.5–7 Å (Supplementary Figure S4C). Intriguingly, the cryo-EM density map of Spx–TAC shows a clear signal for Spx monomer, 68 bp promoter DNA (–55 to + 13), and B. subtilis RNAP holoenzyme carrying σA, δ and ϵ subunits (Figure 1B and C). It is worth noting that the Spx monomer functionally engages RNAP, σA, RNAP αCTD and promoter DNA in a manner distinct from other reported transcription activators (11,13–15,40).
Table 1.
Single particle cryo-EM data collection, processing, and model building for Bacillus subtilis Spx–TAC
| Data collection and processing | |
| Microscope | Titan Krios |
| Voltage (kV) | 300 |
| Detector | K2 summit |
| Electron exposure (e/Å2) | 59 |
| Defocus range (μm) | 1.5–2.5 |
| Data collection mode | counting |
| Physical pixel size (Å/pixel) | 1.307 |
| Symmetry imposed | C1 |
| Initial particle images | 328 348 |
| Final particle images | 153 870 |
| Map resolution (Å)a | 4.22 |
| Refinement | |
| Map sharpening B-factor (Å) | –104 |
| Root-mean-square deviation | |
| Bond length (Å) | 0.007 |
| Bond angle (º) | 1.013 |
| MolProbity statistics | 2.06 |
| Clashscore | 5.96 |
| Rotamer outliers (%) | 1.83 |
| Cβ outliers (%) | 0.0 |
| Ramachandran plot | |
| Favored (%) | 90.57 |
| Outliers (%) | 0.09 |
aGold-standard FSC 0.143 cutoff criteria.
Structural analysis shows that the conformation of B. subtilis RNAP and σA in Spx–TAC is similar to the recently reported structures of B. subtilis RNAP-BmrR-TAC (21) and E. coli RNAP-promoter open complex (RPo) (PDB ID: 6CA0) (9) with root mean square deviation (RMSD) of 2.263 Å (2987 Cαs aligned) and 1.680 Å (2527 Cαs aligned), respectively. ϵ subunit presents a similar conformation with that of RNAP-BmrR-TAC, which locates in a cavity created by RNAP-α, -β, and -β′ subunits at the base of RNAP core enzyme. While δ subunit locates far away from DNA and make extensive interactions with β’ subunit. This indicates that the two subunits may play regulation roles in assembly or function of RNAP core enzyme as previously reported (16,21–22). Furthermore, region 2 (σAR2) and region 4 (σAR4) of σA interact with the consensus –10 element, β’ coiled-coil, and –35 element in the same way as those in RPo (Supplementary Figure S6A and B). Moreover, the crystal structure of B. subtilis Spx in complex with αCTD could be well fitted into the map beneath the upstream DNA. Because the last 13 residues (119–131) of Spx were not resolved in the Spx-αCTD binary complex structure, we fitted and built them into the unambiguous density map near σAR4 as a long helix by rigid body refinement in Phenix program, revealing a novel interface between Spx and σAR4 (Figure 1B and C; Supplementary Figure S6C and D). In addition, another visible electron density which connects with residue E1153 of the C terminal portion of RNAP β subunit, twists, and finally touches the electron densities from σAR4.1 (σA H293-V306, corresponding to σ70 S530-V549) and from a helix (G1105-A1112) of β subunit, could be ascribed to the C-terminal domain of RNAP β subunit (βCTD), which may be involved in transcription regulation by stabilizing σAR4.1 (51,52). (Supplementary Figure S7).
Overall, in Spx–TAC, Spx not only interacts with αCTD but also displays substantial contacts with the promoter DNA and σAR4 (Figure 1B and C; Figure 2A). This unique interaction pattern renders the upstream promoter DNA in a unique orientation toward the β’ subunit side of RNAP, which is different from those of the other transcription activation complexes (Supplementary Figure S8) (13,15,53).
Figure 2.
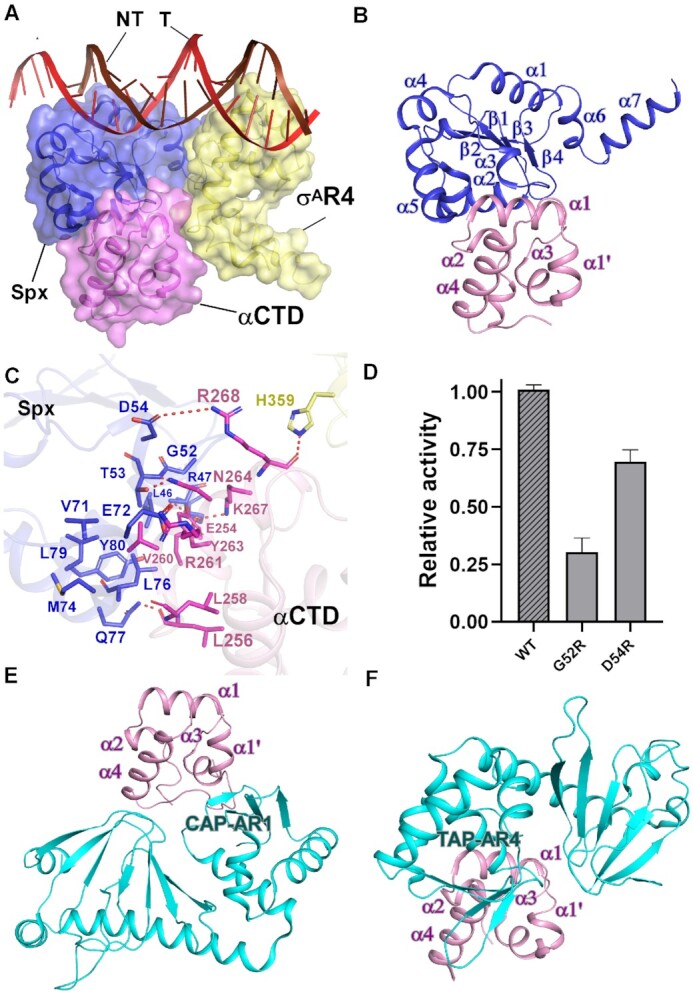
The interactions between Spx and RNAP αCTD. (A) Relative locations of Spx, σAR4, B. subtilis RNAP αCTD, and the upstream double-stranded DNA. (B) Spx interacts with B. subtilis RNAP αCTD. (C) Detailed interactions between B. subtilis RNAP αCTD and Spx. Salt-bridges are shown as red dashed lines. (D) Substitutions of Spx residues involved in interactions with RNAP αCTD decreased in vitro transcription activity. (E) Interface between E. coli CAP (catabolite activator protein) and E. coli RNAP αCTD. (F) Interface between T. thermophilus TAP (transcription activator protein TTHB099) and T. thermophilus RNAP αCTD.
Spx interacts with RNAP αCTD with a novel interface
Previous studies have predicted that B. subtilis RNAP αCTD may interact with Spx and the promoter DNA to facilitate Spx-dependent transcription activation (36,39). However, in Spx–TAC, RNAP αCTD shows no interaction with the upstream promoter DNA, and the main body of Spx (α1–α6, β1–β4), except the newly resolved helix α7, packs against α1 and the preceding loop of αCTD using the α2 helix, α5 helix, and a loop region between α2 and α3 helices, burying a large surface area of 1,311 Å2 (Figure 2A and B). Two types of interactions are mainly involved in the interface between αCTD and Spx (Figure 2C and Supplementary Figure S6C): (1) residues R47, T53, and D54 located on the N terminus of α2 and α3 from Spx form hydrogen bonds with residues K267, N264 and R268 of α1 from αCTD, respectively. In addition, E72 and L76 of Spx form hydrogen bonds with the assumed UP-element binding residues R261 and L258 of αCTD, preventing αCTD from contacting promoter DNA. Q77 and Y80 from the N terminus of α5 of Spx interact with residues L256 and E254 of αCTD by hydrogen bonds, respectively. (2) L46, G52, V71, M74, L76 and L79 of Spx cluster to form a hydrophobic center and make van der Waals contacts with V260 and the highly conserved Y263 residue present in all low GC-content Gram-positive bacteria. This is supported by our Mango-based transcription assay with Spx mutants (Figure 2D) and a series of genetic and mutational studies performed both in vivo and in vitro, reflecting the importance of such interactions for Spx-dependent transcription activation, especially for G52 (34–37). Furthermore, the hydrogen bond between αCTD residue N264 and σA residue H359 also contributes to retain αCTD at a position unable to contact promoter DNA.
Besides the above observations, comparative analysis of Spx and the prototypical class I and class II activators (11,13,15) reveals remarkable differences in the interfaces between αCTD and transcription activators (Figure 2E and F; Supplementary Figure S8). In class I CAP-TAC (catabolite activator protein-transcription activation complex, PDB ID: 6B6H), the activating region 1(AR1) of one CAP monomer makes contact with the 287 determinant of α3 and the C-terminal loop of αCTD, burying a small interfacial area of 76.6 Å2 (Figure 2E; Supplementary Figure S8B). In class II TAP-TAC (transcription activator protein TTHB099-transcription activation complex, PDB ID: 5I2D), the activating region 4 (AR4) of one TAP monomer interacts with 9 αCTD residues located at the N-terminus of α4 helix, the loop connecting α3 and α4, and R264 from α1, burying a surface area of ∼300 Å2 (Figure 2F, Supplementary Figure S8C). The above distinct interaction patterns demonstrate that RNAP αCTD serves as a central target for various activators to engage RNAP and regulate transcription. Since it is widely conserved among all multi-subunit RNA polymerases (1,54,55), RNAP αCTD of other organisms may interact with different regulators to remodel RNAP, as well.
Oxidized Spx monomer specifically interacts with the conserved upstream promoter element
As expected, residues C10 and C13 of the Spx redox-sensing center form a disulfide bond in Spx–TAC, exhibiting an oxidized Spx with an intact α4 helix, consistent with the oxidized Spx structures in Spx-αCTD binary complexes (34–37). Strikingly, structural analysis shows that oxidized Spx contacts different faces of the upstream promoter DNA at the extended –44 element (positions –46 to –37) (Figure 3A and B, Supplementary Figure S6D). In contrast, the prototypical Class I and Class II activators bind DNA from only one direction (11,13,15). In one direction, the conserved positively charged residue R60 from the loop connecting helices α3 and α4 stretches to interact with DNA backbone phosphate of the non-template strand; S58, K62, and Q65 from α4 helix interact with DNA backbone phosphate of the template strand with K62 protruding into the DNA major groove. Notably, beneath the extended –44 element, R91 from the conserved linker region of Spx forms hydrogen bond with DNA backbone phosphate, and R92 makes hydrogen bonds with –45C (N4) of the template DNA to allow sequence readout. In another direction, S12 forms a hydrogen bond with –43T of the template DNA; T11 specifically interact with –45G of the non-template DNA. Meanwhile, R14 from helix α1 and K15, Y105, N106, E107 and D108 from the loop connecting β4 and α6 helix contact the DNA backbone phosphates to restrain the DNA conformation. Accordingly, mutations of the key residues confer severe defects in Spx–TAC formation and transcription activation activity (Supplementary Figure S9, Figure 3C). This is in agreement with previous mutational studies (36,39). Thermal stability assay results also showed that the thermostability curve of Spx mutants is similar to that of wild-type Spx, revealing that these mutants share similar stabilities to that of the wild-type Spx and are folded properly except N106A which may confer defect in the overall conformation of Spx (Supplementary Figure S10). Thus, the data presented here and reported previously evidently reveal the physiological importance of these interactions in stabilizing the DNA conformation, and further activating Spx-dependent transcription.
Figure 3.
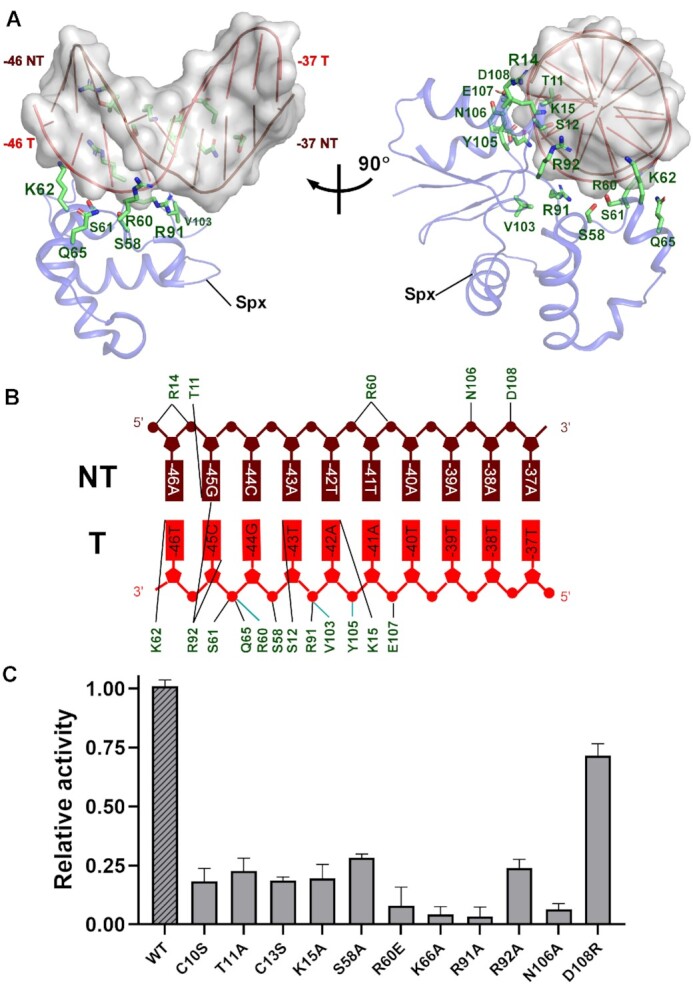
The interactions between Spx and the upstream extended -44 element of promoter DNA. (A) Structure of Spx in complex with the upstream consensus double-stranded DNA. Spx and DNA are represented as cartoons; Spx residues that contact DNA are shown as sticks. (B) Summary of the interactions between consensus extended –44 element and Spx. Cyan lines indicate residues that contact DNA with backbone atoms; Black lines indicate residues that contact DNA with side-chain atoms. (C) Substitutions of residues involved in Spx-DNA interactions suppressed in vitro transcription activity. Data for in vitro transcription assays are means of three technical replicates. Error bars represent mean ± SEM of n = 3 experiments.
Oxidative stress and promoter –44 element are required for Spx-dependent transcription activation
To further clarify the potential mechanism underlying DNA binding upon oxidative stress, we superimposed the reduced Spx from the Spx-αCTD binary complex onto the oxidized Spx in our Spx–TAC (36). Comparative structural analysis of the reduced Spx reported and the oxidized one showed that the most striking differences between them are the conformations of helix α4 and its adjacent loop (Figure 4A). Helix α4 retains an intact helix conformation in the structure of oxidized Spx instead of being disordered in the reduced form, which is in good accordance with the previous structural and biochemical analysis (36). The conserved residues (R60, K62, R92) from disordered helix α4 and its adjacent loop of reduced Spx may lose the stabilizing interactions with the –44 element, in addition, side chains of residues S9, R60 and Y105 may also cause steric clashes with the promoter DNA (Figure 4A). These data reveal that oxidative stress is required to trigger positive conformation changes in these DNA binding residues, enhance DNA binding, and further promote Spx-dependent transcription activation. When being reduced, Spx–αCTD binary complex will show no obvious DNA binding activity and this confers defects in transcription activity (36).
Figure 4.
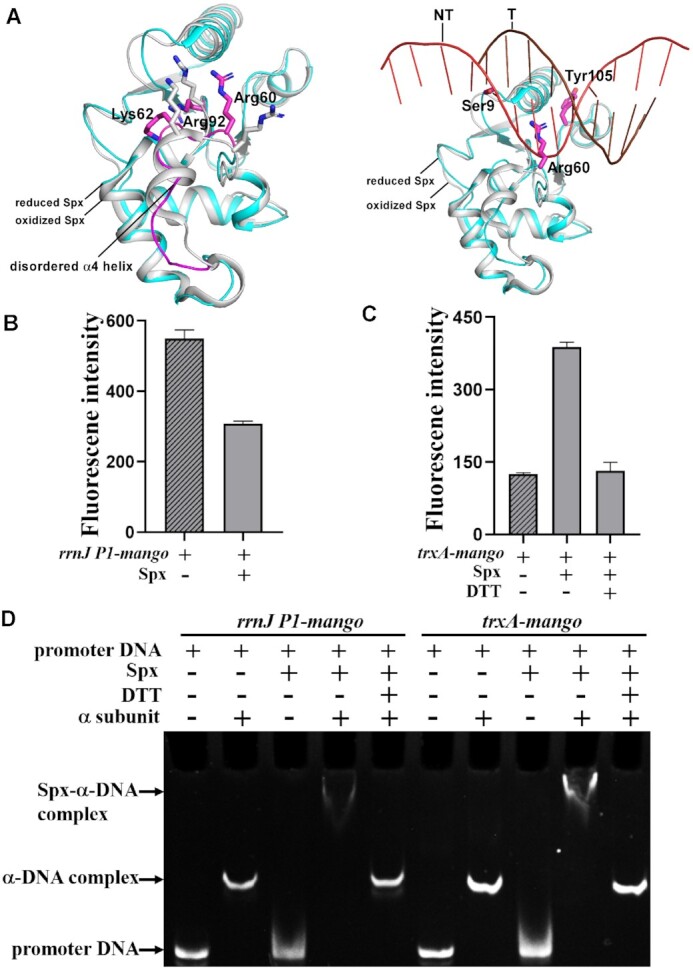
The –44 element of promoter DNA and oxidative stress are required for Spx-dependent transcription activation. (A) Structural superimposition of reduced Spx (PDB ID: 3IHQ) onto the oxidized Spx from Spx–TAC. Reduced Spx is shown as cyan cartoon and oxidized Spx as gray cartoon. Disordered helix α4 from reduced Spx is highlighted in magenta cartoon. Residues that have conformation differences between reduced or oxidized Spx (left panel) are shown in magenta or gray sticks, respectively. Residues from reduced Spx that clash with DNA (right panel) are colored in magenta. (B–D) Mango-based transcription assay and electrophoretic mobility shift assay demonstrate that both –44 element of promoter DNA and oxidative stress are required for Spx-dependent transcription activation activity. Promoter rrnJ P1-mango does not contain –44 element, while promoter trxA-mango consists of –44 element. Reaction conditions are described in detail in Materials and Methods. Data for in vitro transcription assays are means of three technical replicates. Error bars represent mean ± SEM of n = 3 experiments.
Previous transcriptomic and chromatin immunoprecipitation analyses showed that the conserved –44 element of Spx regulons is correlated with Spx-dependent transcription activation activity (25,31). To determine whether the interactions between Spx and DNA are sequence-specific, we performed in vitro Mango-based transcription assays in the presence or absence of the –44 element. As expected, our results showed that Spx displayed no transcription activation activity for rrnJ P1-mango promoter with an UP-element instead of a –44 element (Figure 4B). Spx could efficiently activate transcription of the –44 element-containing trxA-mango promoter, but it almost lost its entire activation activity when being reduced by DTT (Figure 4C). In accordance, further EMSA showed that oxidized Spx (without DTT) alone had no DNA binding activity, while in the presence of B. subtilis RNAP α subunit, Spx formed more stable Spx-α–DNA complexes with trxA-mango promoter than with rrnJ P1-mango promoter (Figure 4D). Once Spx was reduced, the band of Spx-α-DNA complexes disappeared, indicating Spx may be in free state. These results demonstrate that both oxidative stress and the –44 element of promoter DNA are required for Spx-dependent transcription activation, and Spx inhibits transcription of UP-element containing promoters possibly by influencing the productive interactions of αCTD with the UP elements as previously suggested (27,31) and form fewer transcription initiation competent complex than Spx–TAC.
Spx interacts extensively with σA
Previous pull-down assay and site-directed mutational studies implied that Spx may interact with σA to facilitate Spx-dependent transcription activation, and the 13 C-terminal residues are required for RNAP engagement (33,39). Our cryo-EM structure of Spx–TAC is consistent with these predictions. In Spx–TAC, Spx interacts extensively with σA and buries a large surface area of 288.6 Å2 (Figure 5A–C, Supplementary Figure S7D). σA R4 inserts into the major groove of the –35 promoter element in a manner similar to that in RPo (56). Helix α7 dissociates from the main body of Spx, rotates and protrudes onto the hydrophobic surface of σA R4, creating van der Waals interactions between the conserved residue L129 and σAR4 residue V338 (Figure 5B and C). R119 of Spx α7 contacts E315 of σAR4 by a salt bridge; Q122 and L123 of Spx α7 form hydrogen bonds with R313 and E316 of σAR4, respectively. Two other interfaces are involved in mediating the Spx-σAR4 interaction (Figure 5C): first, in the αCTD-Spx-σA interface, E50 and D51 located on the loop connecting helices α2 and α3 of Spx interact with H359 of σA via hydrogen bond; Second, in the Spx α6-σA R4 interface, a patch of conserved positively charged residues in Spx α6 and the nearby loops (K99, R100, R111, R112, P115, R116 and R119) cluster to interact with the negatively charged surface of σA (T309, D308, T311 and D312) via a hydrophilic network of hydrogen bonds and salt bridges. Accordingly, the thermal stability assay results showed that the thermostability curves of Spx mutants are similar to that of wild-type Spx, revealing that these mutants share similar stabilities to that of the wild-type Spx and are folded properly (Supplementary Figure S10), mutations of these conserved residues in our EMSA and Mango-based transcription assay affect Spx–TAC formation and suppressed transcription activation (Figure 5D), indicating the functional importance of these interactions in stabilizing σAR4 for promoting Spx-dependent transcription activation, especially interactions involved in Spx α6–σA R4 interface.
Figure 5.
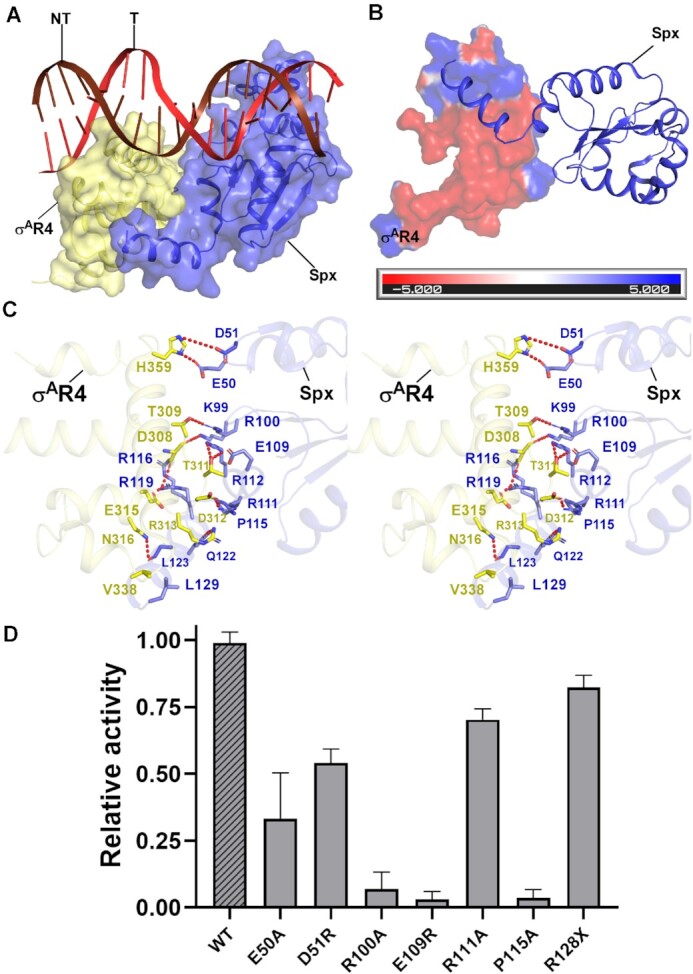
The interactions between Spx and σAR4. (A) Relative locations of Spx, σAR4, and upstream double-stranded DNA in B. subtilis Spx–TAC. (B) Spx interacts with the negatively charged surface of σA. The electrostatic potential surface of σAR4 was generated using PyMOL program. Spx is represented as blue cartoon. (C) Detailed interactions between σAR4 and Spx (stereo view). Hydrogen bonds are shown as red dashed lines. (D) Substitutions of the residues involved in Spx-σA interactions reduced in vitro transcription activity. Data for in vitro transcription assays are means of three technical replicates. Error bars represent mean ± SEM of n = 3 experiments.
DISCUSSION
Spx has been established as a global transcription activator for stress-response genes, such as those alleviating oxidative stresses. However, a detailed structural model that accounts for the molecular mechanism of Spx-dependent transcription activation remained elusive. Our Spx–TAC structure and biochemical experiments offer four new pieces of information: (1) In Spx–TAC, a single monomer of oxidized Spx simultaneously engages RNAP holoenzyme and the promoter DNA. The interaction mode is supported by previous pull-down assays (33) and is different from the reported transcription activators, which function in dimers (11,13,15). (2) Spx interacts with RNAP αCTD with a new interface, revealing αCTD as a central RNAP target for multiple transcription activators. (3) One oxidized Spx monomer specifically binds different faces of the conserved –44 promoter element, undergoes conformation changes and promotes the formation of Spx–TAC. (4) Spx interacts extensively with σA via three interfaces. A collection of evidence reveals that the last 28 residues of Spx are targets for recognition by the adaptor protein YjbH and proteolysis by ClpXP, which play an essential role in post-translational control of cytoplasmic concentration of Spx (24,57,58). However, in Spx–TAC, these residues form two helices and are mostly involved in extensive interactions with σA, thereby potentially protecting Spx from ClpXP proteolysis and preserving its physiological functions. Besides, similar to the acidic regions of CAP in CAP–TAC, the other two interfaces may also play important roles in isomerization of RPc and stabilization of Spx–TAC (11,13,15).
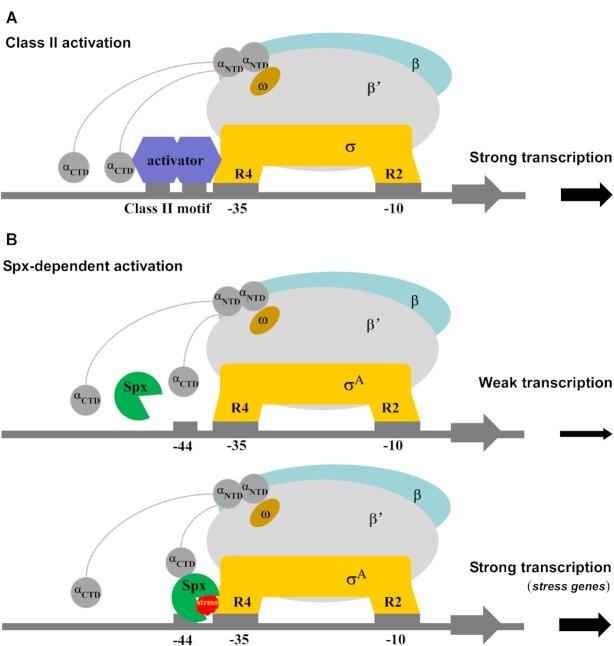
Canonical bacterial transcription activators are generally categorized into two classes based on whether the DNA binding site is located upstream to or overlapping the –35 element (59,60). Considering the overlapping promoter binding sites of Spx and σA, Spx has been proposed to be similar to class II transcription activators (Figure 6A). Based on our results and previous reports, we propose a plausible model for Spx-dependent transcription regulation (Figure 6B): the reduced form of Spx cannot bind RNAP α subunit or promoter DNA with –44 element and cannot activate transcription efficiently (Figure 4, Figure 6B, top panel). Once being oxidized, Spx binds close to the UP-element binding surface of αCTD via a large interface and converts itself into a primary DNA binding determinant. If the promoter DNA does not contain a conserved –44 element (regardless of the presence of an UP-element), in most cases, Spx-αCTD does not bind the promoter DNA with high affinity and fails in initiating transcription. This is confirmed by our Mango-based transcription assay with the rrnJ P1-mango promoter that contains an UP-element but not the conserved –44 element (Figure 4). This is also consistent with transcription inhibition assays of Spx with ResD or ComA (61). When the conserved –44 element and oxidative stress are both present, the redox-sensing switch of Spx triggers conformational changes in Spx to form substantial interactions with DNA and σA, leading σA to reposition to the –35 element and stabilizing this activation-competent conformation as suggested (31,38). Such remodeling of RNAP further promotes the isomerization of Spx-RPc to an initiation-competent Spx–TAC and efficiently initiates transcription of the downstream stress-response genes (Figure 6B, bottom panel). From this perspective, Spx functions as a versatile transcription activator possibly through two types of activation mechanisms. On one hand, in contrast to other classic transcriptional activators that recruit RNAP to promoter DNA, Spx only acquires DNA binding activity after binding to RNAP, it likely firstly activates transcription by a ‘pre-recruitment’ mechanism (11,62). On the other hand, when bound to RNAP and –44 element, Spx can redirect σA from the upstream promoter DNA to the downstream –35 element and –10 element, and further enhance interactions between RNAP and DNA as the previous protein-DNA crosslinking studies showed (38,39), indicating that Spx may subsequently activate transcription by promoting DNA opening of RNAP to form transcription-competent Spx–TAC.
Figure 6.
Proposed models for class-II-activator-dependent and Spx-dependent transcription activation. (A) Proposed working model for class-II-activator-dependent transcription activation. (B) Proposed working model for Spx-dependent transcription activation. Spx shows weak transcription activation activity in the absence of Spx-RNAP interactions, and activates the transcription of most genes with the promoter containing consensus –44 element under oxidative stress.
Since the discovery of B. subtilis Spx, a number of Spx homologs emerged and constitute a new family of transcription factors. Sequence alignment shows that most of these Spx-family factors exhibit the conserved CXXC motif, G52, and RPI motif (Supplementary Figure S11). Consistent with previous reports, our data confirm that the CXXC motif acts as an oxidative stress-sensing switch and G52 plays an essential role in stabilizing Spx-αCTD interactions. The conserved RPI motif in the Spx family has multiple roles: R92 interacts with the template DNA to stabilize Spx–TAC, P93 is probably involved in modulating the reactivity of C10 within the redox-sensing switch, and I94 may stabilize the unique structure of Spx through van der Waals contacts with the proximal hydrophobic residues. These evolutionarily conserved motifs likely play similar regulation roles in the physiological activities of Spx-family factors.
Moreover, the above transcription regulation mechanism of Spx may provide a new paradigm for the Spx-family global transcription factors and other stress-response transcription factors. This is reminiscent of the most recently reported iron-sulfur cluster-containing transcription factor WhiB7 (63). Similar to Spx, WhiB7 plays regulatory roles in redox homeostasis regulation in Mycobacterium tuberculosis (M. tuberculosis), and shows DNA binding activity. However, there exists significant differences between them. In comparison with Spx, it is considered that αCTD is not indispensable for the physiological function of WhiB7, and WhiB7 activates transcription mainly through simultaneously interacting with σA-holoenzyme and an AT-rich DNA element by its AT-hook, which is rare in bacteria.
DATA AVAILABILITY
Accession number for cryo-EM density map: EMD-31485 (Electron Microscopy Data Bank). Accession number for atomic coordinates: 7F75 (Protein Data Bank).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Yu Zhang (Chinese Academy of Sciences, Shanghai Institute of Plant Physiology and Ecology) for providing the Bacillus subtilis strain and Shenghai Chang at the Center of Cryo-Electron Microscopy in Zhejiang University School of Medicine and National Center for Protein Science, Shanghai for help with cryo-EM sample preparation and data collection. We thank the Core Facilities, Zhejiang University School of Medicine for technical support. We thank the Experiment Center for Science and Technology, Nanjing University of Chinese Medicine for experimental assistance.
Author contributions: J.S., F.F.L., A.J.W., L.W., F.L.W., Y.L.J. performed the experiments. A.J.W., S.J. performed cryo-EM sample preparations and data collections. F.Y., W.L. performed cryo-EM structure determination. J.S., F.F.L. performed biochemical experiments. J.S., W.L. designed the study, J.S., W.L., L.B.Y. and Y.F. analyzed data, and wrote the paper.
Contributor Information
Jing Shi, Department of Pathogen Biology, School of Medicine & Holistic Integrative Medicine, Nanjing University of Chinese Medicine, Nanjing, China.
Fangfang Li, Department of Pathogen Biology, School of Medicine & Holistic Integrative Medicine, Nanjing University of Chinese Medicine, Nanjing, China.
Aijia Wen, Department of Biophysics, Zhejiang University School of Medicine, Hangzhou, China; Department of Pathology of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China.
Libing Yu, Institute of Materials, China Academy of Engineering Physics, Mianyang, China.
Lu Wang, Department of Pathogen Biology, School of Medicine & Holistic Integrative Medicine, Nanjing University of Chinese Medicine, Nanjing, China.
Fulin Wang, Department of Pathogen Biology, School of Medicine & Holistic Integrative Medicine, Nanjing University of Chinese Medicine, Nanjing, China.
Yuanling Jin, Department of Pathogen Biology, School of Medicine & Holistic Integrative Medicine, Nanjing University of Chinese Medicine, Nanjing, China.
Sha Jin, Department of Biophysics, Zhejiang University School of Medicine, Hangzhou, China; Department of Pathology of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China.
Yu Feng, Department of Biophysics, Zhejiang University School of Medicine, Hangzhou, China; Department of Pathology of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China.
Wei Lin, Department of Pathogen Biology, School of Medicine & Holistic Integrative Medicine, Nanjing University of Chinese Medicine, Nanjing, China; State Key Laboratory of Natural Medicines, China Pharmaceutical University, Nanjing, China; Jiangsu Collaborative Innovation Center of Chinese Medicinal Resources Industrialization, Nanjing 210023, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [82072240, 81903526, 81991523 to W.L., 32000025 to J.S.]; Jiangsu Province of China [BK20190798 to W.L., BK2021020389 to J.S.]; Open Project of State Key Laboratory of Natural Medicines [SKLNMKF202004 to W.L.]; Open Project of Chinese Materia Medica First-Class Discipline of Nanjing University of Chinese Medicine [2020YLXK008 to W.L., 2020YLXK016 to J.S.]; Fok Ying Tung Education Foundation; Jiangsu Specially-Appointed Professor Talent Program (to W.L.). Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Decker K.B., Hinton D.M.. Transcription regulation at the core: similarities among bacterial, archaeal, and eukaryotic RNA polymerases. Annu. Rev. Microbiol. 2013; 67:113–139. [DOI] [PubMed] [Google Scholar]
- 2. Browning D.F., Busby S.J.. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016; 14:638–650. [DOI] [PubMed] [Google Scholar]
- 3. Chen J., Boyaci H., Campbell E.A.. Diverse and unified mechanisms of transcription initiation in bacteria. Nat. Rev. Microbiol. 2021; 19:95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feklistov A., Darst S.A.. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell. 2011; 147:1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Feklistov A., Sharon B.D., Darst S.A., Gross C.A.. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 2014; 68:357–376. [DOI] [PubMed] [Google Scholar]
- 6. Saecker R.M., Record M.T., Dehaseth P.L.. Mechanism of bacterial transcription initiation: RNA polymerase-promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J Mol Bio. 2011; 412:754–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Y., Feng Y., Chatterjee S., Tuske S., Ho M.X., Arnold E., Ebright R.H.. Structural basis of transcription initiation. Science. 2012; 338:1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bae B., Feklistov A., Lass-Napiorkowska A., Landick R., Darst S.A.. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. Elife. 2015; 4:e08504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Narayanan A., Vago F.S., Li K., Qayyum M.Z., Yernool D., Jiang W., Murakami K.S.. Cryo-EM structure of Escherichia coli sigma(70) RNA polymerase and promoter DNA complex revealed a role of sigma non-conserved region during the open complex formation. J. Biol. Chem. 2018; 293:7367–7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen J., Chiu C., Gopalkrishnan S., Chen A.Y., Olinares P.D.B., Saecker R.M., Winkelman J.T., Maloney M.F., Chait B.T., Ross W.et al.. Stepwise promoter melting by bacterial RNA polymerase. Mol. Cell. 2020; 78:275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feng Y., Zhang Y., Ebright R.H.. Structural basis of transcription activation. Science. 2016; 352:1330–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi J., Wen A., Zhao M., You L., Zhang Y., Feng Y.. Structural basis of sigma appropriation. Nucleic Acids Res. 2019; 47:9423–9432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi W., Jiang Y.N., Deng Y.B., Dong Z.G., Liu B.. Visualization of two architectures in class-II CAP-dependent transcription activation. PLoS Biol. 2020; 18:e3000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi J., Wen A.J., Jin S., Gao B., Huang Y., Feng Y.. Transcription activation by a sliding clamp. Nat. Commun. 2021; 12:1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu B., Hong C., Huang R.K., Yu Z., Steitz T.A.. Structural basis of bacterial transcription activation. Science. 2017; 358:947–951. [DOI] [PubMed] [Google Scholar]
- 16. Weiss A., Shaw L.N.. Small things considered: the small accessory subunits of RNA polymerase in Gram-positive bacteria. FEMS Microbiol. Rev. 2015; 39:541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spiegelman G.B., Hiatt W.R., Whiteley H.R.. Role of the 21,000 molecular weight polypeptide of Bacillus subtilis RNA polymerase in RNA synthesis. J. Biol. Chem. 1978; 253:1756–1765. [PubMed] [Google Scholar]
- 18. Rabatinova A., Sanderova H., Jirat Matejckova J., Korelusova J., Sojka L., Barvik I., Papouskova V., Sklenar V., Zidek L., Krasny L.. The delta subunit of RNA polymerase is required for rapid changes in gene expression and competitive fitness of the cell. J. Bacteriol. 2013; 195:2603–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pei H.H., Hilal T., Chen Z.A., Huang Y.H., Gao Y., Said N., Loll B., Rappsilber J., Belogurov G.A., Artsimovitch I.et al.. The delta subunit and NTPase HelD institute a two-pronged mechanism for RNA polymerase recycling. Nat. Commun. 2020; 11:6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xue X., Tomasch J., Sztajer H., Wagner-Dobler I.. The delta subunit of RNA polymerase, RpoE, is a global modulator of Streptococcus mutans environmental adaptation. J. Bacteriol. 2010; 192:5081–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fang C., Li L., Zhao Y., Wu X., Philips S.J., You L., Zhong M., Shi X., O’Halloran T.V., Li Q.et al.. The bacterial multidrug resistance regulator BmrR distorts promoter DNA to activate transcription. Nat. Commun. 2020; 11:6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keller A.N., Yang X., Wiedermannova J., Delumeau O., Krasny L., Lewis P.J.. epsilon, a new subunit of RNA polymerase found in gram-positive bacteria. J. Bacteriol. 2014; 196:3622–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Turlan C., Prudhomme M., Fichant G., Martin B., Gutierrez C.. SpxA1, a novel transcriptional regulator involved in X-state (competence) development in Streptococcus pneumoniae. Mol. Microbiol. 2009; 73:492–506. [DOI] [PubMed] [Google Scholar]
- 24. Rojas-Tapias D.F., Helmann J.D.. Roles and regulation of Spx family transcription factors in Bacillus subtilis and related species. Adv. Microb. Physiol. 2019; 75:279–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakano S., Kuster-Schock E., Grossman A.D., Zuber P.. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:13603–13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakano S., Erwin K.N., Ralle M., Zuber P.. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol. Microbiol. 2005; 55:498–510. [DOI] [PubMed] [Google Scholar]
- 27. Schafer H., Heinz A., Sudzinova P., Voss M., Hantke I., Krasny L., Turgay K.. Spx, the central regulator of the heat and oxidative stress response in B. subtilis, can repress transcription of translation-related genes. Mol. Microbiol. 2019; 111:514–533. [DOI] [PubMed] [Google Scholar]
- 28. Rojas-Tapias D.F., Helmann J.D.. Induction of the Spx regulon by cell wall stress reveals novel regulatory mechanisms in Bacillus subtilis. Mol. Microbiol. 2018; 107:659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rojas-Tapias D.F., Helmann J.D.. Stabilization of Bacillus subtilis Spx under cell wall stress requires the anti-adaptor protein YirB. PLoS Genet. 2018; 14:e1007531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leichert L.I.O., Scharf C., Hecker M.. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 2003; 185:1967–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rochat T., Nicolas P., Delumeau O., Rabatinova A., Korelusova J., Leduc A., Bessieres P., Dervyn E., Krasny L., Noirot P.. Genome-wide identification of genes directly regulated by the pleiotropic transcription factor Spx in Bacillus subtilis. Nucleic Acids Res. 2012; 40:9571–9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ross W., Gosink K.K., Salomon J., Igarashi K., Zou C., Ishihama A., Severinov K., Gourse R.L.. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993; 262:1407–1413. [DOI] [PubMed] [Google Scholar]
- 33. Lin A.A., Zuber P.. Evidence that a single monomer of Spx can productively interact with RNA polymerase in Bacillus subtilis. J. Bacteriol. 2012; 194:1697–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Newberry K.J., Nakano S., Zuber P., Brennan R.G.. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15839–15844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lamour V., Westblade L.F., Campbell E.A., Darst S.A.. Crystal structure of the in vivo-assembled Bacillus subtilis Spx/RNA polymerase alpha subunit C-terminal domain complex. J. Struct. Biol. 2009; 168:352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakano M.M., Lin A., Zuber C.S., Newberry K.J., Brennan R.G., Zuber P.. Promoter recognition by a complex of Spx and the C-terminal domain of the RNA polymerase alpha subunit. PLoS One. 2010; 5:e8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Birch C.A., Davis M.J., Mbengi L., Zuber P.. Exploring the amino acid residue requirements of the RNA polymerase (RNAP) alpha subunit C-terminal domain for productive interaction between Spx and RNAP of Bacillus subtilis. J. Bacteriol. 2017; 199:e00124-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reyes D.Y., Zuber P.. Activation of transcription initiation by Spx: formation of transcription complex and identification of a Cis-acting element required for transcriptional activation. Mol. Microbiol. 2008; 69:765–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lin A.A., Walthers D., Zuber P.. Residue substitutions near the redox center of Bacillus subtilis Spx affect RNA polymerase interaction, redox control, and Spx-DNA contact at a conserved cis-acting element. J. Bacteriol. 2013; 195:3967–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shi J., Wen A.J., Zhao M.X., You L.L., Zhang Y., Feng Y.. Structural basis of sigma appropriation. Nucleic. Acids. Res. 2019; 47:9423–9432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Autour A., S C.Y.J., A D.C., Abdolahzadeh A., Galli A., Panchapakesan S.S.S., Rueda D., Ryckelynck M., Unrau P.J.. Fluorogenic RNA mango aptamers for imaging small non-coding RNAs in mammalian cells. Nat. Commun. 2018; 9:656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jeng S.C., Chan H.H., Booy E.P., McKenna S.A., Unrau P.J.. Fluorophore ligand binding and complex stabilization of the RNA mango and RNA spinach aptamers. RNA. 2016; 22:1884–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Trachman R.J., Demeshkina N.A., Lau M.W.L., Panchapakesan S.S.S., Jeng S.C.Y., Unrau P.J., Ferre-D’Amare A.R.. Structural basis for high-affinity fluorophore binding and activation by RNA Mango. Nat. Chem. Biol. 2017; 13:807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shi J., Gao X., Tian T., Yu Z., Gao B., Wen A., You L., Chang S., Zhang X., Zhang Y.et al.. Structural basis of Q-dependent transcription antitermination. Nat. Commun. 2019; 10:2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang F., Shi J., He D., Tong B., Zhang C., Wen A., Zhang Y., Feng Y., Lin W.. Structural basis for transcription inhibition by E. coli SspA. Nucleic Acids Res. 2020; 48:9931–9942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005; 152:36–51. [DOI] [PubMed] [Google Scholar]
- 47. Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y.F., Agard D.A.. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017; 14:331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rohou A., Grigorieff N.. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015; 192:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scheres S.H.W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 51. Borukhov S., Severinov K.. Role of the RNA polymerase sigma subunit in transcription initiation. Res. Microbiol. 2002; 153:557–562. [DOI] [PubMed] [Google Scholar]
- 52. Urbauer J.L., Adelman K., Urbauer R.J., Simeonov M.F., Gilmore J.M., Zolkiewski M., Brody E.N.. Conserved regions 4.1 and 4.2 of sigma(70) constitute the recognition sites for the anti-sigma factor AsiA, and AsiA is a dimer free in solution. J. Biol. Chem. 2001; 276:41128–41132. [DOI] [PubMed] [Google Scholar]
- 53. Feng Y., Zhang Y., Ebright R.H.. Structural basis of transcription activation. Science. 2016; 352:1330–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lloyd G.S., Niu W., Tebbutt J., Ebright R.H., Busby S.J.. Requirement for two copies of RNA polymerase alpha subunit C-terminal domain for synergistic transcription activation at complex bacterial promoters. Genes Dev. 2002; 16:2557–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lara-Gonzalez S., Birktoft J.J., Lawson C.L.. Structure of the Escherichia coli RNA polymerase alpha subunit C-terminal domain. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:806–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Narayanan A., Vago F.S., Li K., Qayyum M.Z., Yernool D., Jiang W., Murakami K.S.. Cryo-EM structure of Escherichia coli sigma(70) RNA polymerase and promoter DNA complex revealed a role of sigma non-conserved region during the open complex formation. J. Biol. Chem. 2018; 293:7367–7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chan C.M., Hahn E., Zuber P.. Adaptor bypass mutations of Bacillus subtilis spx suggest a mechanism for YjbH-enhanced proteolysis of the regulator Spx by ClpXP. Mol. Microbiol. 2014; 93:426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Awad W., Al-Eryani Y., Ekstrom S., Logan D.T., Wachenfeldt C.. Structural basis for YjbH adaptor-mediated recognition of transcription factor Spx. Structure. 2019; 27:923–936. [DOI] [PubMed] [Google Scholar]
- 59. Lee D.J., Minchin S.D., Busby S.J.W.. Activating transcription in bacteria. Annu. Rev. Microbiol. 2012; 66:125–152. [DOI] [PubMed] [Google Scholar]
- 60. Busby S.J.W. Transcription activation in bacteria: ancient and modern. Microbiology (Reading). 2019; 165:386–395. [DOI] [PubMed] [Google Scholar]
- 61. Nakano S., Nakano M.M., Zhang Y., Leelakriangsak M., Zuber P.. A regulatory protein that interferes with activator-stimulated transcription in bacteria. P Natl Acad Sci USA. 2003; 100:4233–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Griffith K.L., Shah I.M., Myers T.E., O’Neill M.C., Wolf R.E. Jr. Evidence for ‘pre-recruitment’ as a new mechanism of transcription activation in Escherichia coli: the large excess of SoxS binding sites per cell relative to the number of SoxS molecules per cell. Biochem. Biophys. Res. Commun. 2002; 291:979–986. [DOI] [PubMed] [Google Scholar]
- 63. Wan T., Horova M., Beltran D.G., Li S., Wong H.X., Zhang L.M.. Structural insights into the functional divergence of WhiB-like proteins in Mycobacterium tuberculosis. Mol. Cell. 2021; 81:2887–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession number for cryo-EM density map: EMD-31485 (Electron Microscopy Data Bank). Accession number for atomic coordinates: 7F75 (Protein Data Bank).