Abstract
We newly developed a hybrid protein, tentatively named rMIKO-1, using gene technology. We herein investigated the effects of rMIKO-1 on activated macrophages and discussed its potential as a suppressor of experimental colitis. Fluorescent microscopy was used to observe the dynamic mobility of rMIKO-1 in macrophages. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis, Western blotting, fluorescent immunochemical staining, flow cytometry, enzyme-linked immunosorbent assays, a polymerase chain reaction/quantitative polymerase chain reaction, and hematoxylin and eosin staining were conducted to assess the potential activity of rMIKO-1. A large amount of bleeding was observed in rats treated with 5% dextran sulfate sodium (DSS) alone on day 8 after treatment initiation, but not in those treated with 5% DSS plus rMIKO-1. In the in vitro assay, rMIKO-1 rapidly bound to macrophages, immediately entered cells by an unknown mechanism, and then migrated inside the nucleus. This result suggests that rMIKO-1 plays important immunological roles in the nucleus. Despite the activation of macrophages by lipopolysaccharide, the mRNA expression of pro-inflammatory cytokines, such as tumor necrosis factor-α, interleukin-6, and interleukin-1β, was significantly suppressed in macrophages preliminarily treated with rMIKO-1 for 1 h. Complexes of rMIKO-1 with nuclear factor-kappa B (NF-κB)/p65 and β-actin formed in activated macrophages, which attenuated experimental colitis in rats. These results strongly suggest that rMIKO-1 negatively regulates excessively activated macrophages through the NF-κB/p65 signaling pathway. Therefore, rMIKO-1 is a novel suppressor of experimental colitis in rats through the negative regulation of activated macrophages.
KEY WORDS: macrophages, hybrid protein, NF-κB pathway, experimental colitis, COVID-19.
INTRODUCTION
S100A8 and S100A9 (S100 proteins) are calcium-binding proteins that belong to the S100 family, and mainly exist in the cytoplasm of immune cells of myeloid origin, such as macrophages and neutrophils [1–3]. S100 proteins are heavily involved in many inflammatory diseases, such as rheumatoid arthritis [1, 4], ulcerative colitis (UC) and Crohn’s disease (CD) [5–9], and acute hepatitis [10, 11]. In these diseases, S100 proteins may play important roles inside and/or outside of immune cells; however, their innate immunity has not yet been elucidated in detail. Macrophages are activated by many antigenic substances, but to some extent, similar to bacteria or their components in the large intestinal tract, via receptors, such as Toll-like receptor (TLR) 4 [6, 7, 12, 13] and the receptor for advanced glycation endproducts [14, 15]. S100 proteins participate in the regulation of abnormalities in macrophages to maintain homeostasis in the body by negatively regulating the excessive activation of cells induced by commensal bacteria, which contributes to the self-catalytic modulation of cells via known and/or undefined receptors [11]. A previous study reported that the activation of the signal transduction activator transcription 3 pathway in intestinal epithelial cells in the large colon by interleukin (IL)-6 resulted in the excessive secretion of S100A9 in the circulation, ultimately leading to the onset of inflammatory bowel diseases [9]. In contrast, the excessive secretion of S100A9 has been suggested to result in an accelerated inflammatory response in immune cells and/or epithelial cells of the large intestinal tract [16]. Despite extensive investigations in the past three decades, the mechanisms contributing to the potent immunity of S100A8 and S100A9 as immunological regulators remain unclear.
We recently detected an autocrine signal transduction pathway involving rat S100A8 (rS100A8) and rS100A9 in macrophages, and identified cluster of differentiation 68 (CD68) on the macrophages of rats as a receptor-like protein for rS100A8 and/or rS100A9 in the signal transduction pathway of these cells, with sugar chains on these molecules being closely involved in signal transduction [17–19]. The active center of proteins may be generally constructed on an amino acid (AA) sequence or its three-dimensional structure. We herein hypothesized that some immune functions of rS100A8 and rS100A9 depended on their AA sequences around the N- and C-terminal regions, respectively. To prove our hypothesis, we newly designed a hybrid protein (rat MIKO-1: rMIKO-1) and three derivatives (rMIKO-2 to rMIKO-4) by referring to the AA sequences of rS100A8 and rS100A9, and prepared them using gene technology.
In the present study, we investigated the immunological potency of rMIKO-1 to macrophages and discuss its potential as a therapeutic agent for experimental colitis in rats.
MATERIALS AND METHODS
Animals
All rats used in the present study (n = 60) were Wistar rats (male, 10 weeks old, 220–250 g) that were kept in captivity for approximately 1 week prior to the initiation of animal experiments and allowed free access to food and water.
Reagents
The pCold-1 vector and isopropyl β-d-1-thiogalactopyranoside (IPTG) were purchased from TAKARA-Bio Co., Ltd., Kyoto, Japan. Dextran sulfate sodium salt (DSS, MW: 36,000 ~ 50,000, MP Biochemicals) Colitis Grade was obtained from Wako Pure Chemical Industries, Ltd., Tokyo, Japan. Ni-agarose was obtained from Nacalai Tesque, Kyoto, Japan. The streptavidin (STA)-Cy5; anti-mouse IgG (horse)-horseradish peroxidase (HRP) or -fluorescein 5-isothiocyanate (FITC) conjugate and VECTASHIELD Mounting Medium were obtained from Vector Inc., Burlingame, CA, USA. Lipopolysaccharide (LPS) from Escherichia coli and an anti-β-actin monoclonal antibody were purchased from Sigma-Aldrich Co., LLC, Tokyo, Japan. N-Hydroxysuccinimidobiotin (EZ-Link™ NHS-Biotin) was from Thermo Fisher Scientific Co., Ltd., USA. The monoclonal antibody for nuclear factor kappa B (NF-κB) p65 (sc-8008) was purchased from Santa Cruz Biotechnology, Inc., Dallas, Texas 75,220, USA. The monoclonal antibody for Golgi bodies (GM130) was obtained from Medical & Biological Laboratories Co., Ltd., Tokyo, Japan. Monoclonal antibodies for rS100A8 (mAb2H6 and mAb8A6) and rS100A9 (mAb1D11 and mAb10D11) were provided by Yamasa Co., Ltd., Chiba, Japan. Clinical Thioglycollate Medium (E-MC17) was obtained from EIKEN Chemical Co., Ltd., Tokyo, Japan. Roswell Park Memorial Institute 1640 medium (RPMI-1640), diaminobenzidine tetrahydrochloride (DAB), and other reagents were obtained from Wakenyaku Co., Ltd., Kyoto, Japan.
Design of a Recombinant Protein (rMIKO-1) and Its Three Derivatives
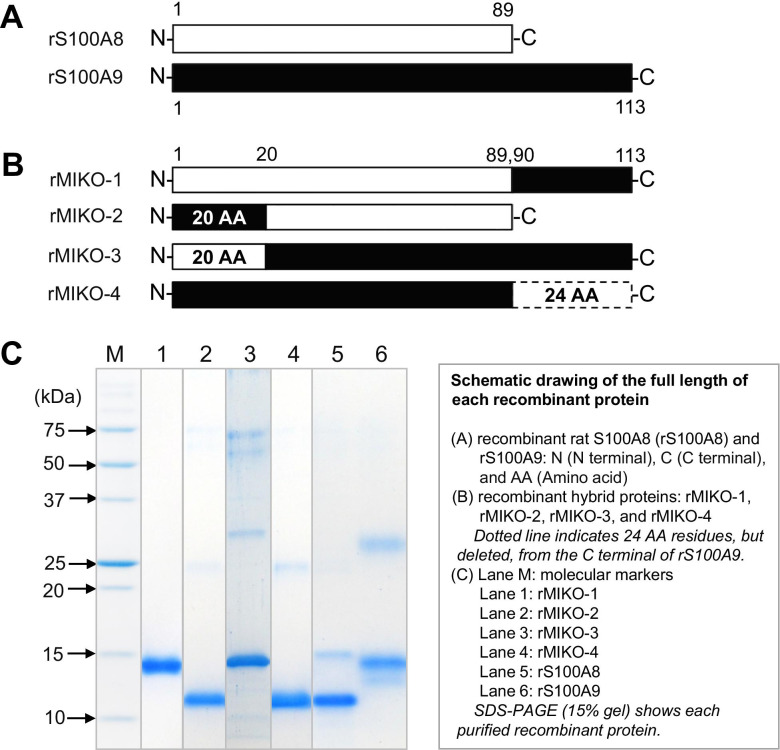
We newly designed a hybrid protein (rMIKO-1) and its three derivatives (rMIKO-2, -3, and -4) by referring to the AA sequences of rS100A8 and rS100A9, and artificially synthesized each cDNA for these proteins using gene technology (Fig. 1A and B) [1]. The whole frame of each rMIKO protein was schematically indicated in Fig. 1B. The frames of four rMIKO proteins were as follows: rMIKO-1, 24 AA residues from the C-terminal of rS100A9 were added to the C-terminal of rS100A8; rMIKO-2, 20 AA residues from the N-terminal of rS100A8 were exchanged with the N-terminal of rS100A9; rMIKO-3, 20 AA residues from the N-terminal of rS100A9 were exchanged with the N-terminal of rS100A8; rMIKO-4, 24 AA residues from the C-terminal of rS100A9 were deleted as indicated by the dotted line.
Fig. 1.
Design and development of a rMIKO-1 protein and its three derivatives. (A) The full-length amino acid (AA) sequences of rS100A8 and rS100A9 were schematically drawn by open and closed horizontal bars, respectively. (B) rMIKO-1: 24 AA residues from the C-terminal AA residue of rS100A9 were added to the C-terminal AA residue of rS100A8. rMIKO-2 and rMIKO-3: 20 AA residues from the N-terminal AA residue of rS100A8 and those of rS100A9 were replaced. rMIKO-4: 24 AA residues, as indicated by the dotted line, from the C-terminal AA residue of rS100A9 were deleted. (C) SDS-PAGE (15% gel) was performed using affinity-purified proteins in the presence of 2-mercaptoethanol. M: molecular weight markers. Lanes 1 to 6 indicate purified rMIKO-1, -2, -3, -4, rS100A8, and rS100A9, respectively.
Expression and Purification of rMIKO-1 and Its Three Derivatives
The expression of rMIKO-1 and its three derivatives in E. coli cells was performed as previously described [20]. Briefly, the cDNA of each MIKO protein was inserted into the pCold I vector (Takara Bio Co., Ltd., Kyoto, Japan) [21]. The four vectors of pCold I newly prepared were transformed in competent cells (E. coli cells, BL21). Recombinant cells were cultivated at 37 °C for 3 to 4 h. When the absorbance of the culture medium at 600 nm reached the range of 0.4 to 0.8, the temperature for cultivation was decreased to 15 °C and IPTG (1 mM as the final concentration) was added to a culture bottle followed by cultivation for 12 h at 15 °C while mixing. Recombinant cells were collected by centrifugation and washed once with 50 mM phosphate buffer/0.9% NaCl solution and then suspended in an adequate volume of binding buffer (pH8.0)/10 mM imidazole/0.3 M NaCl. After sonication, recombinant MIKO proteins were separately purified using a Ni-agarose column [17]. Each purified rMIKO protein was dialyzed against 50 mM phosphate buffer/0.9% NaCl by exchanging the buffer solution twice to mainly remove imidazole. Each recombinant rMIKO protein was then concentrated at an adequate concentration and frozen at − 80 °C until used. The concentration of endotoxin in rMIKO-1 samples was measured using a commercially provided kit (ToxinSensor™, Endotoxin Detection System, GenScript Inc., USA). The concentration of endotoxin in each sample was less than 0.6 EU/mg protein, indicating no influence on the activation of macrophages (data not shown).
Monoclonal Antibodies
Monoclonal antibodies for rMIKO-1 were prepared as previously described [22]. A part of each antibody was biotinylated using N-Hydroxysuccinimidobiotin according to the manufacturer’s instructions. The specificity and avidity of antibodies were confirmed by Western blotting, in which the anti-mouse IgG (horse)-HRP conjugate was used as a secondary antibody. These antibodies were frozen at − 80 °C until used. Monoclonal antibodies for rS100A8 (mAb2H6) and rS100A9 (mAb1D11) were used in the present study [17].
Animal Experiments
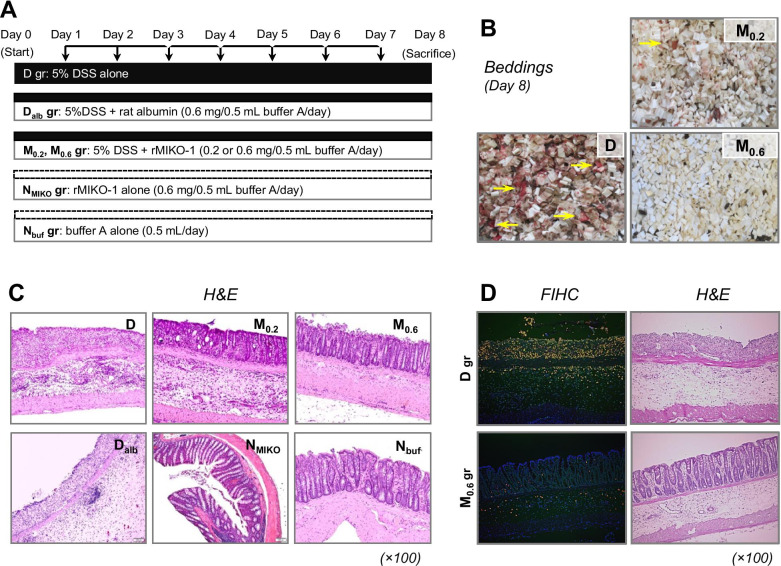
The protocol for animal experiments is shown in Fig. 2A. Intraperitoneal injections were performed under sterile condition as much as possible. The rats used were classified into the following groups: (1) D group: 5% DSS/distilled water (DW) alone was orally administered for 7 days to induce experimental colitis (n = 10); (2) M0.2 and M0.6 groups: rMIKO-1 (0.2 or 0.6 mg/50 mM phosphate buffer (pH7.4)/0.9% NaCl sterilized (0.5 ml)/rat) was injected into the abdominal cavity of each rat in the D group every morning (n = 10/group); (3) Dalb group: rat albumin (approx. 0.6 mg/50 mM phosphate buffer (pH7.4)/0.9% NaCl sterilized (0.5 ml)/rat) was injected into the abdominal cavity of each rat in the D group every morning during the experimental period (n = 5); (4) NMIKO group: rMIKO-1 alone (0.6 mg/50 mM phosphate buffer (pH7.4)/0.9% NaCl sterilized (0.5 ml)/rat) was injected into the abdominal cavity of each normal rat every morning during the experimental period (n = 5); (5) Nbuf group: 50 mM phosphate buffer (pH7.4)/0.9% NaCl sterilized (0.5 ml) alone was intraperitoneally injected into each normal rat (n = 5). Macrophages were obtained from the abdominal cavity of normal rats (n = 15). The large intestinal tract of each rat in all groups was then removed from the body and subjected to biochemical and histological examinations after its length had been measured.
Fig. 2.
Protocol of animal experiments and therapeutic effects of rMIKO-1 on experimental colitis in rats. (A) Animal experiments were performed using 5% DSS alone, 5% DSS plus rMIKO-1, or sterilized buffer A alone (50 mM phosphate buffer, pH7.4/0.9% NaCl). In the D group, the oral administration of 5% DSS alone to rats was conducted for eight days to induce experimental colitis. In the M group, rMIKO-1 (0.2 or 0.6 mg/0.5 ml buffer A/day/rat) was intraperitoneally administered, indicated by vertical arrows in the upper position, to the D group once a day every morning for seven days (n = 10 in each concentration). In the Dalb and M groups, rat albumin (0.6 mg/0.5 ml buffer A/day/rat) alone was intraperitoneally administered to the D group. In the NMIKO group, rMIKO-1 (0.6 mg/0.5 ml buffer A/day/rat) alone was intraperitoneally administered to each normal rat. In the Nbuf group, buffer A (0.5 ml/day/rat) alone was intraperitoneally administered to each normal rat. (B) The bleeding conditions of the D, M0.2, and M0.6 groups are shown, with heavy and slight bleeding being indicated by horizontal arrows (yellow). (C) H&E staining of the large colon (rectal tissue) was performed to evaluate tissue deterioration. (D) Fluorescent immunohistochemistry (FIHC), together with H&E staining, was performed using tissue sections of the large colon of rats in the D and M0.6 groups. rS100A8 and rS100A9 in these tissues were stained using mAb8H6-Cy5 and mAb1D11-FITC conjugates, respectively.
Extraction of Cytoplasmic and Nuclear Proteins from Macrophages
Rat macrophages were elicited by an intraperitoneal injection of 10 ml of sterilized 4% thioglycolate/DW [8]. After 3 days, 50 ml of 50 mM sterilized phosphate buffer solution (pH7.4)/0.9% NaCl (buffer A) was intraperitoneally injected into rats and macrophages were then carefully collected and suspended in buffer A. After centrifugation at 1500 rpm at 4 °C for 3 min, the cells collected were again suspended in RPMI-1640 medium supplemented with 10% fetal bovine serum (Biological Industries Ltd., Israel) (medium A). A total of 5×106 cells were plated in a Petri dish [100 mm (L) × 10 mm (D)] and incubated at 37 °C for 2 h in 5% CO2. After the incubation, non-adherent cells in each Petri dish were removed by washing the dish three times with buffer A. rMIKO-1 (approx. 50 μg/ml) was then added to the dish and further incubated at 37 °C for 2 h in 5% CO2. After the incubation, adherent cells were washed twice with buffer A and then collected in 1 ml of buffer A using a scraper. The cell suspension was slightly centrifuged at 1,500 rpm at 4 °C for 3 min and the supernatant was discarded. Cytoplasmic and nuclear proteins in macrophages from rats in the D and M groups were separately extracted using a Cytoplasmic & Nuclear Protein Extraction Kit (PO Box 1016, Mountain View, CA 94,042, USA) according to the manufacturer’s instructions. Extracted protein samples were frozen at − 80 °C until used.
Fluorescent Immunochemical Staining and Immunohistochemistry
Fluorescent immunochemical staining (FICS) was performed using the intraperitoneal macrophages of rats and the rMIKO-1-FITC conjugate, for which the rat albumin (50 μg/ml)-Cy5 conjugate was used as a negative control [23]. The dynamic mobility of rMIKO-1 in cells was observed every 10 min for 1 h under a fluorescent microscope (BIOREVO BZ-9000, KEYENCE Co., Ltd., Osaka). Immunohistochemistry was also performed as previously described, in which rS100A8 and rS100A9 were stained using mAb2H6 and mAb1D11, respectively, as the first antibody [17].
Enzyme-Linked Immunosorbent Assays
An enzyme-linked immunosorbent assay (ELISA) was performed to detect rMIKO-1-biotin and its complexes with NF-κB/p65, β-actin, and Golgi bodies in the cytoplasm and nucleus of macrophages using the STA-HRP conjugate as previously described. HRP activity was spectrophotometrically evaluated.
Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis and Western Blotting
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed as previously described [22, 24]. Western blotting was performed using the monoclonal antibodies of mAb2H6 and mAb1D11 for rS100A8 and rS100A9, respectively, as the primary antibody. Antibody-bound rS100A8 and rS100A9 were detected using the mAb8A6-HRP and mAb10D11-HRP conjugates, respectively, as the secondary antibody. Briefly, proteins were separated by SDS-PAGE in the presence of 2-mercaptoethanol and transferred to nitrocellulose. After the addition of Blocking One (Nacalai Co., Ltd., Kyoto), they were incubated at 4 °C for 1 h with the monoclonal antibody for β-actin (AC-74, Sigma-Aldrich Co., LLC) at a dilution of 1:1000. Membranes were then washed three times for 5 min with 10 mM Tris–HCl buffer (pH 7.4)/0.9% NaCl (buffer B), twice with buffer B/0.1% Tween 20, and once with buffer B, before being incubated with the anti-mouse IgG (horse)-HRP conjugate (the secondary antibody) diluted 500-fold at room temperature for 1 h. After washing the membranes, antibody-bound proteins were detected using DAB and hydrogen peroxide as the substrate. In ELISA, rMIKO-1-biotin, which was reacted with mAbC6 on the ELISA plate, was detected with STA-HRP. HRP activity was assessed using 1,2-phenylenediamine dihydrochloride (o–PD) and hydrogen peroxide as the substrates [22], with absorbance at 490 nm being tentatively measured using a microplate reader (Bio-Rad Co., Ltd., USA).
Immunoprecipitation
To detect possible complexes of rMIKO-1 with β-actin, immunoprecipitation was conducted using two specific monoclonal antibodies for rMIKO-1 (mAbC6 and mAbC5). Briefly, the two antibodies (5 μg/ml each as the final concentration) were added to each protein sample (0.5 ml), which was extracted from the cytoplasm and nucleus of macrophages treated with rMIKO-1 at 37˚C for 2 h in 5% CO2. The mixture was gently mixed at 4˚C overnight and then centrifuged at 3,500 rpm at 4˚C for 10 min. The resultant pellet was carefully washed twice with 50 mM phosphate buffer (pH7.4)/0.9% NaCl. After centrifugation as above, the precipitated proteins were subjected to SDS-PAGE and Western blotting using the specific monoclonal antibody for β-actin and the anti-mouse IgG (horse)-HRP conjugate as the first and second antibodies, respectively. Antibody-bound proteins were detected as previously described [22].
Possible Complexes of rMIKO-1 with Cytoplasmic or Nuclear Proteins in macrophages
The detection of possible complexes of rMIKO-1 with intracellular proteins in macrophages was attempted using an ELISA plate preliminarily coated with the monoclonal antibody for NF-κB (2 μg/ml), β-actin (2 μg/ml), or anti-Golgi bodies (2 μg/ml). Twenty-five microliters of the protein sample extracted from macrophages was added to each well of the plate preliminarily filled with 0.1 ml of the sample diluent (50 mM phosphate buffer (pH7.4)/0.9% NaCl) and then incubated at room temperature for 1 h after mixing. After washing three times with washing buffer, 0.1 ml of the STA-HRP conjugate (2 μg/ml) was added to each well of the plate followed by an incubation at room temperature for 30 min. After washing, HRP activity was assessed as described above.
Flow Cytometry
Flow cytometry was conducted as previously described [18]. Stained cells were detected and analyzed using Coulter Counter Navios (Beckman Coulter Co., Ltd., Tokyo, Japan) equipped with the AN50240 software.
Polymerase Chain Reaction and Quantitative Polymerase Chain Reaction
The semi-quantitative polymerase chain reaction (PCR) and quantitative PCR (qPCR) were conducted using a Thermal Cycler (C1000™, BIO-RAD) as previously described, in which all primers for PCR were the same as those previously described [17].
Histological Evaluation
Hematoxylin and eosin (H&E) staining was performed to evaluate the extent of tissue deterioration in the colons of rats in all groups. The severity of DSS-induced colitis in rats was also classified based on disease activity index scores [25].
Statistical Analysis
Pairwise comparisons with controls were performed using parametric tests. Significant differences between groups were identified using the Student’s t test (t test of the difference between the two mean values). Data are shown as mean±SD values. P values of < 0.05 were considered to be significant.
RESULTS
Expression and Purification of rMIKO Proteins
All rMIKO proteins (rMIKO-1 to rMIKO-4) were successfully expressed in E. coli cells. They were purified using a Ni-agarose column [26 mm (D) × 100 mm (L)] as described in the “Methods” section. In comparisons with the two protein bands of rS100A8 and rS100A9, the purity of each rMIKO protein was visually higher than 95% (Fig. 1C, lanes 1 to 4).
Preparation of Monoclonal Antibodies
Monoclonal antibodies for rMIKO-1 were prepared as previously described [22]. We obtained six clones (C5, C6, C7, C9, C10, and C20) of the antibody specific for rMIKO-1. The specificity of each antibody for rMIKO-1 was examined by Western blotting, and all antibodies cross-reacted with rS100A9, but not with rS100A8. These results suggest that these antibodies recognize specific epitopes for the AA sequence of rS100A9 or its three-dimensional structure. mAbC5 and mAbC6 reacted more strongly with rMIKO-1 than the other antibodies (data not shown).
Histological Evaluation of the Large Colon of Rats with Experimental Colitis
Bleeding from the large colon of rats in the D group began on day 5 after treatment initiation and increased during the last half of the experiment (Fig. 2B, panel D). Although slight bleeding was observed on day 6 in the M0.2 group (Fig. 2B, panel M0.2), bleeding was not detected on the same day in the M0.6 group (Fig. 2B, panel M0.6). In the Dalb group, experimental colitis in the large colon of rats in the D group was not suppressed by rat albumin (Fig. 2C, panel Dalb). This result suggests that rat albumin is not a negative regulator of experimental colitis. In the NMIKO group, upon the intraperitoneal administration of rMIKO-1 alone, no inflammatory changes were microscopically observed in large colon tissue (Fig. 2C, panel NMIKO). This result indicates that rMIKO-1 is not a toxic or antigenic molecule for rats. In the D group, the severe deterioration of the epithelial structure of the large colon (rectal tissue) was microscopically observed (Fig. 2C, panel D). In addition, numerous immune cells accumulated in the damaged tissue of the large colon of rats in the D group. In these cells, rS100A8 and rS100A9 microscopically appeared to be co-expressed; however, these cells almost disappeared in the colon tissue of rats treated with 5% DSS plus rMIKO-1 (Fig. 2D, FIHC). H&E staining showed the deterioration of large colon tissue in the D group, but not in the M0.6 group (Fig. 2D, H&E). The body weight of rats in the NMIKO group gradually increased after treatment initiation, whereas that of rats in the Dalb group markedly decreased after day 5 (data not shown). In addition, the length of the large colon of each rat in the D and Dalb groups appeared to be shorter than that prior to treatment initiation, whereas the length of the large colon in the other groups was similar to that in the Nbuf group (Table 1). These results suggest that rMIKO-1 protected against experimental colitis in the large colon of rats. A histological evaluation of the colon in each group was performed as previously described [25]. The results obtained are summarized in Table 1.
Table 1.
DAI score of DSS-induced colitis in rats treated with rMIKO-1, rat albumin, or PBS alone
| Bleeding and morphological changes (DAI score) | ||||||
|---|---|---|---|---|---|---|
| Groups (n) | Grading of colonic damage (0–3) | Infiltration of immune cells in the tissue (0–3) | Bleeding (0–3) | Depth of damage (0–3) | DAI score (total: 0–12) | Colon length (cm)* |
| Nbuf (n = 5) | ND | ND | ND | ND | ND | 17.4 |
| NMIKO (n = 5) | ND | ND | ND | ND | ND | 17.5 |
| Dalb (n = 5) | 3 | 3 | 3 | 3 | 12 | 12.3 |
| M0.2 (n = 10) | 0.5 | 0.5 | 0.2 | 0.2 | 0.4 | 17.0 |
| M0.6 (n = 10) | ND | ND | ND | ND | ND | 17.4 |
| D (n = 10) | 3 | 3 | 3 | 3 | 12 | 12.2 |
ND, not detected. The severity of DSS-induced colitis in rats was evaluated by depending to the scale reported by Konarska et al.
*Values are indicated by the average of the colon length of rats in each group
Immunological Potential of rMIKO-1 in Macrophages
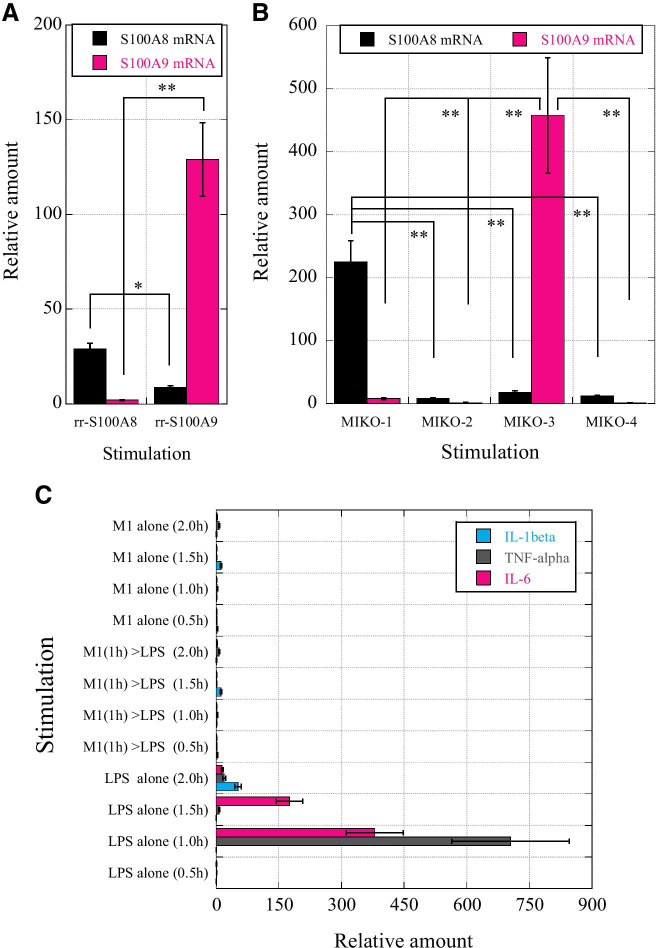
Recombinant rS100A8 (rrS100A8) and rrS100A9 strongly induced the mRNA expression of rS00A8 and rS100A9, respectively, in macrophages (Fig. 3A). The mRNA expression of rS100A8 and rS100A9 was induced in a limited manner by rMIKO-1 and rMIKO-3, but not by rMIKO-2 or rMIKO-4, respectively (Fig. 3B). These results strongly suggest that the short AA sequences around the N- and C-terminal regions of rMIKO-1 and rMIKO-3, respectively, are important for their effects on macrophages. Therefore, rMIKO-1 is a new functional molecule that combines two important AA regions of rS100A8 and rS100A9. Changes in tumor necrosis factor (TNF)-α, IL-1β, and IL-6 mRNA levels in macrophages treated with rMIKO-1 alone, LPS alone, and rMIKO-1 and LPS were examined after 0.5, 1.0, 1.5, and 2.0 h (Fig. 3C). After the stimulation with LPS (10 μg/ml) alone, TNF-α and IL-6 mRNA levels markedly increased after 1 h; however, rMIKO-1 did not significantly induce the mRNA expression of these inflammatory cytokines. These results indicate that rMIKO-1 is not an antigenic substance to macrophages. The mRNA expression of inflammatory cytokines was significantly downregulated in macrophages that were preliminarily treated with rMIKO-1 (10 μg/ml) for 1 h prior to the stimulation with LPS (10 μg/ml). These results suggest that rMIKO-1 is a novel functional molecule that plays an important role(s) in the nucleus of macrophages to negatively regulate the excessive activation of macrophages.
Fig. 3.
Molecular analysis of immune functions of rS100A8 and rS100A9 mRNAs and potential activity of rMIKO-1. rS100A8 and rS100A9 mRNA levels were measured using a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA 94,404, USA) to verify the immune responses of macrophages to rMIKO-1 and/or lipopolysaccharide (LPS) upon the stimulation of cells with rrS100A8 or rrS100A9. (A) rS100A8 and rS100A9 mRNA levels in macrophages were measured after the treatment with rrS100A8 (10 μg/ml) and rrS100A9 (10 μg/ml), respectively, at 37 C for 1 h in 5% CO2. (B) rS100A8 and rS100A9 mRNA levels in macrophages were measured after the treatment with rMIKO-1, -2, -3, and -4 (10 μg/ml each) at 37 °C for 1 h in 5% CO2. (C) Time-dependent changes in the mRNA levels of three inflammatory cytokines (IL-1β, TNF-α, and IL-6) in macrophages (0.5, 1.0, 1.5, and 2.0 h). The treatment groups were (1) rMIKO-1 (M1) alone (10 μg/ml), (2) LPS (10 μg/ml) alone after the treatment with rMIKO-1 (10 μg/ml), and (3) LPS alone (10 μg/ml). Error bars are shown as means ± SD values. *, P < 0.05; **, P < 0.01.
Flow Cytometry of rMIKO-1 and Its Dynamic Mobility in Macrophages
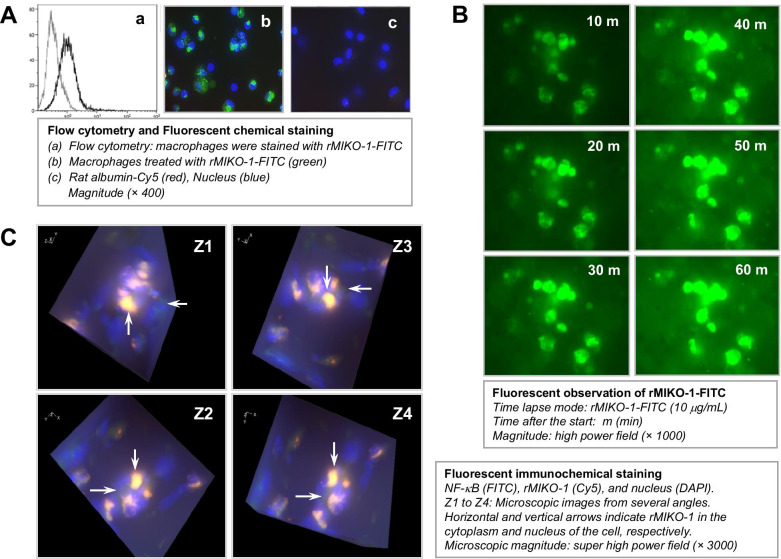
Flow cytometry and FICS showed that rMIKO-1 bound to macrophages in the in vitro study; however, rat albumin did not bind to these cells (Fig. 4A, panels a, b, and c). This result suggests the selective affinity of rMIKO-1 for macrophages. rMIKO-1 time-dependently bound to macrophages within 60 min, as shown by FICS, using fluorescent microscopy (BIOREVO BZ-9000, KEYENCE Co., Ltd., Osaka) (Fig. 4B). To observe the intracellular localization of rMIKO-1 in macrophages in more detail, microscopic images of rMIKO-1 and NF-κB p65 from several angles were observed using the Z-stack mode, which showed that rMIKO-1 appeared to localize in both the cytoplasm and nucleus of these cells (Fig. 4C, panels Z1 to Z4). These results indicate that rMIKO-1 plays an important, but still unknown role in the DNA chain in the nucleus of macrophages.
Fig. 4.
Microscopic observations of the dynamic mobility of rMIKO-1 in macrophages and its intracellular localization. Macrophages (approx. 2×106 cells) were used in the in vitro study. (A) Flow cytometry was performed using macrophages and the rMIKO-1-FITC conjugate. Cells were treated with the conjugate (10 μg/ml) on ice for 1 h, washed three times with 50 mM phosphate buffer (pH7.4)/0.9% NaCl, and re-suspended in an adequate volume of the same buffer. Stained cells were analyzed using Coulter Counter Navios (Beckman Coulter Co., Ltd., Tokyo, Japan) equipped with the AN50240 software. (A) Panel a indicates flow cytometry. Macrophages in panels b and c were treated with rMIKO-1-FITC and rat albumin-biotin, respectively, in which rat albumin-biotin was detected with the STA-Cy5 conjugate. (B) Macrophages (approx. 2×106 cells) were exposed to the rMIKO-1-FITC conjugate (10 μg/ml) in 5% CO2 at 37 C. After treatment initiation, the dynamic mobility of rMIKO-1-FITC in macrophages was immediately observed using a fluorescent microscopy (BIOREVO BZ-X700, KEYENCE Co., Ltd., Osaka, Japan) by the time-lapse mode at an interval of 10 min. Microscopic magnitude: super high power fields (×1,000). (C) Macrophages adhering to a thin cover glass were exposed to the rMIKO-1-FITC conjugate at 37 C for 1 h in 5% CO2. After fixation, NF-κB p65 was stained using the specific monoclonal antibody for NF-κB p65 (sc-8008) and the anti-mouse IgG (horse)-FITC conjugate, and rMIKO-1-biotin was detected by STA-Cy5. Microscopic images were taken from several angles to three-dimensionally observe the location of rMIKO-1 in the nuclei of cells, which were counterstained with DAPI. White horizontal arrows indicate NF-κB p65 in macrophages. White vertical arrows indicate the co-localization of rMIKO-1 and NF-κB p65 or their complexes in the nuclei of macrophages. Panels are shown by super high power fields (× 3,000). *, P<0.05; **, P<0.01.
Confirmation of rMIKO-1, rS100A8, and rS100A9 in the Cytoplasm and Nucleus of Macrophages by Western Blotting
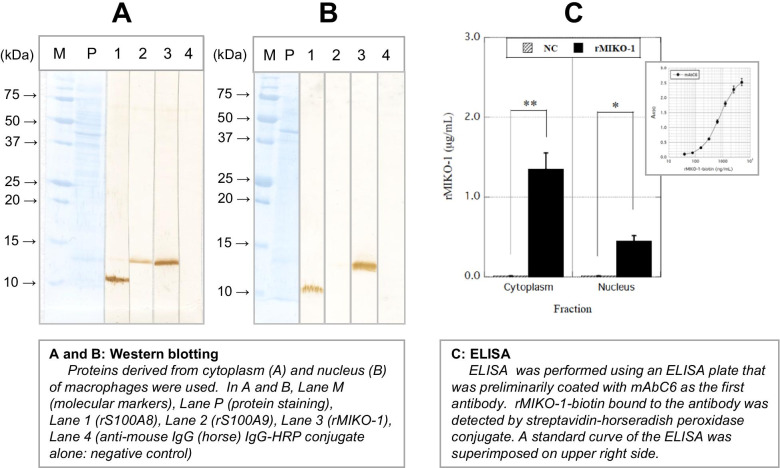
To detect rMIKO-1, rS100A8, and rS100A9 in macrophages, Western blotting was performed using the cytoplasmic and nuclear proteins of these cells. Western blotting revealed the presence of rMIKO-1, rS100A8, and rS100A9 in the cytoplasm of macrophages preliminarily treated with rMIKO-1 (Fig. 5A); however, rS100A9 was not significantly detected in the nucleus (Fig. 5B). These results strongly suggest that rMIKO-1 is capable of migrating inside the nucleus of macrophages. It was detected in the cytoplasm and nucleus of macrophages using preparative ELISA for rMIKO-1, in which standard curves for ELISA were superimposed (Fig. 5C). Therefore, the presence of rMIKO-1 in the cytoplasm and nucleus of macrophages was confirmed by both of these methods.
Fig. 5.
Detection of rMIKO-1 in the cytoplasm and nucleus of macrophages by Western blotting and ELISA. Western blotting was performed to detect rMIKO-1-biotin in the cytoplasm and nucleus of macrophages treated with rMIKO-1 at 37 C for 2 h in 5% CO2. (A) and (B) Lanes M, P, 1, 2, 3, and 4 indicate molecular markers, protein staining, rS100A8, rS100A9, rMIKO-1-biotin, and the negative control, respectively. rS100A8 and rS100A9 were stained using mAb8A6-HRP and mAb1D11-HRP conjugates, respectively. rMIKO-1-biotin was detected using the STA-HRP conjugate adequately diluted with a working solution of Blocking One (Nacalai Tesque, Kyoto, Japan), in which 3,3′-diaminobenzidine tetrahydrochloride n-hydrate and hydrogen peroxide were used as the substrates for color development. (C) Preparative ELISA for rMIKO-1 was performed to measure rMIKO-1-biotin using an ELISA plate that was coated with a monoclonal antibody of mAbC6 (5 μg/ml) as the first antibody. rMIKO-1-biotin bound to the first antibody was detected by the STA-HRP conjugate. HRP activity was spectrophotometrically assessed. A standard curve for rMIKO-1-biotin was superimposed on the upper right side. Error bars are shown as means±SD values. *, P<0.05; **, P<0.01.
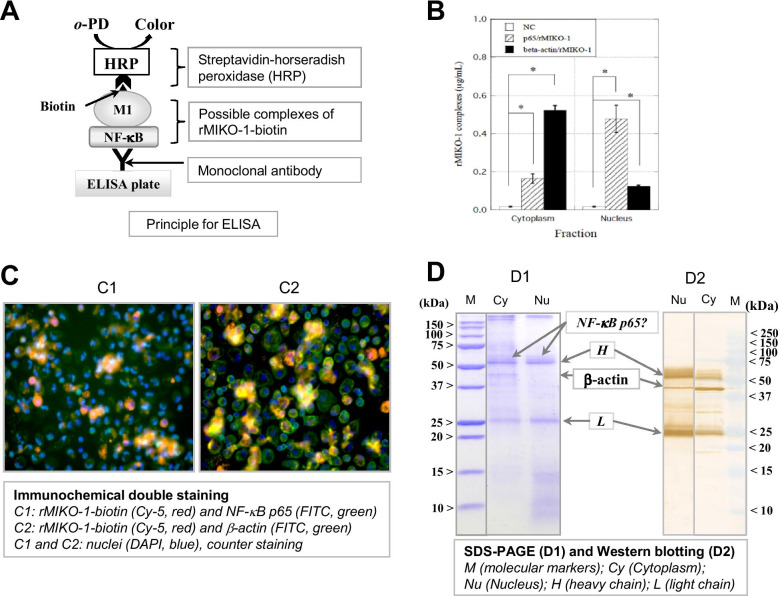
Possible Complexes of rMIKO-1 with NF-κB/p65 or β -actin in the Cytoplasm and Nucleus of Macrophages
ELISA for NF-κB p65 and β-actin was performed to detect complexes of rMIKO-1-biotin with NF-κB p65 or β-actin in the cytoplasm and nucleus of macrophages using a polycarbonate plate for ELISA, which was preliminarily coated with the monoclonal antibody for NF-κB p65 or β-actin (5 μg/ml) as the first antibody. rMIKO-1-biotin composed of complexes was detected by the STA-HRP conjugate (Fig. 6A). ELISA for NF-κB p65 and β-actin showed positive reactions (Fig. 6B). This result suggests that complexes of rMIKO-1 with at least NF-κB p65 or β-actin formed in macrophages treated with rMIKO-1. Fluorescent immunological double staining showed that rMIKO-1 microscopically shared its intracellular localization with NF-κB p65 or β-actin in these cells (Fig. 6C, panels C1 and C2). Collectively, these results suggest that rMIKO-1 forms a complex with NF-κB p65 or β-actin in the cytoplasm of macrophages. To detect complexes of rMIKO-1 with β-actin, immunoprecipitation was performed using two types of monoclonal antibodies for rMIKO-1 (mAbC6 and mAbC5). β-actin was detected in the main fraction of the cytoplasm and that of the nucleus, but only slightly in the fraction of macrophages (Fig. 6D, panel D2).
Fig. 6.
Possible complexes of rMIKO-1 with NF-κB p65 or β-actin in macrophages. (A) The principle of ELISA was schematically drawn in detail, in which M1, HRP, and o–PD indicate rMIKO-1, horseradish peroxidase, and o-phenylenediamine, respectively. (B) To detect the possible complex of rMIKO-1 with NF-κB p65 or β-actin, ELISA was performed using ELISA plates that were preliminarily coated with a specific antibody for NF-κB p65 or β-actin (5 μg/ml each) as the first antibody. rMIKO-1-biotin, a component of the complexes, was detected by the STA-HRP conjugate. HRP activity was assessed using o–PD and hydrogen peroxide as the substrates. (C) Fluorescent immunochemical double staining was performed using a specific monoclonal antibody for NF-κB p65 or β-actin (5 μg/ml) as the first antibody. Monoclonal antibodies and rMIKO-1-biotin were detected with anti-mouse IgG (horse)-FITC (secondary antibody, green) and STA-Cy5 (red) conjugates, respectively. C1: The colors of rMIKO-1, NF-κB p65, and the nucleus are red, green, and blue, respectively. C2: rMIKO-1 (red), β-actin (green), and the nucleus (blue). Microscopic images were shown using high power fields (×400). (D) SDS-PAGE and Western blotting were performed as previously described. Precipitated proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (D1). Proteins in the gel were transferred to a nitrocellulose membrane and Western blotting was performed using a specific monoclonal antibody for β-actin and the anti-mouse IgG (horse)-HRP conjugate as the first and second antibodies, respectively (D2), in which M indicates molecular markers. Cy and Nu indicate extracted protein fractions of the cytoplasm and nucleus, respectively, of macrophages. H and L indicate the heavy and light chains, respectively, of the first antibody. *, P<0.05.
DISCUSSION
In the present study, we described the potent ability of the new hybrid protein, rMIKO-1, and presented data to support our hypothesis that rMIKO-1 is a novel regulator of excessively activated macrophages. Our strategy is based on a difference in AA sequences between rS100A8 and rS100A9. We previously reported the potential activity of rS100A8 as a regulator of rS100A9 functions because of the promising roles of these two proteins in the immune system; however, limited information is currently available on their active centers. The biochemical and/or immunological activities of protein molecules are generally conserved in the AA sequences around the N- and/or C-terminal regions of each protein or in each three-dimensional structure.
Hybridization is a simple and effective strategy for developing novel therapeutic drugs, including peptides and proteins [26]. Based on our previous findings, we designed the frames of rMIKO-1 and its three derivatives to verify the active centers of rS100A8 and rS100A9, and successively produced them using gene technology (Fig. 1). The limited mRNA expression of rS100A8 or rS100A9 was induced by a stimulation with rMIKO-1 or rMIKO-3, respectively (Fig. 3A and B). Alternatively, rMIKO-2 and rMIKO-4 did not activate macrophages, as indicated by the lack of a significant increase in rS100A8 and rS100A9 mRNA levels, respectively (Fig. 3B). These results strongly suggest that at least two short AA regions close to the N- and C-terminals of rS100A8 and rS100A9, respectively, are specific AA regions for the expression of their immune functions in macrophages.
No bleeding and marked accumulation of many immune cells, such as macrophages and/or neutrophils, in the large colon of rats in the M0.6 group were observed at the terminal stage of the animal experiment (Fig. 2C, panel M0.6, Fig. 2D, panel FIHC). This observation strongly suggests that rMIKO-1 contributes to inhibiting the NF-κB signal transduction pathway in tissue macrophages in the large colon of rats in the M0.6 group. These results indirectly confirm our hypothesis that rMIKO-1 strongly suppresses the onset of experimental colitis by negatively regulating excessive activation of tissue macrophages in the large colon (Fig. 2B, panel M0.6). To prove our hypothesis, changes in the mRNA levels of inflammatory cytokines in macrophages are helpful for understanding the true function of rMIKO-1 in macrophages. The mRNA expression of inflammatory cytokines was significantly suppressed in macrophages preliminarily treated with rMIKO-1 for 1 h despite a subsequent stimulation with LPS (10 μg/mL) (Fig. 3C). In addition, the mRNA expression of inflammatory cytokines was not induced by rMIKO-1 alone, suggesting no antigenicity of rMIKO-1 to macrophages. These results support our hypothesis that rMIKO-1 functions as a negative regulator of the excessive activation of macrophages. In this context, rMIKO-1 may form a complex with NF-κB in the cytoplasm and/or nucleus of macrophages, or may form some components on the NF-κB motif in the nucleus to inhibit the mRNA transcription of inflammatory cytokines, as demonstrated by the presence of rMIKO-1 in the cytoplasm and nucleus of macrophages (Fig. 5A and B).
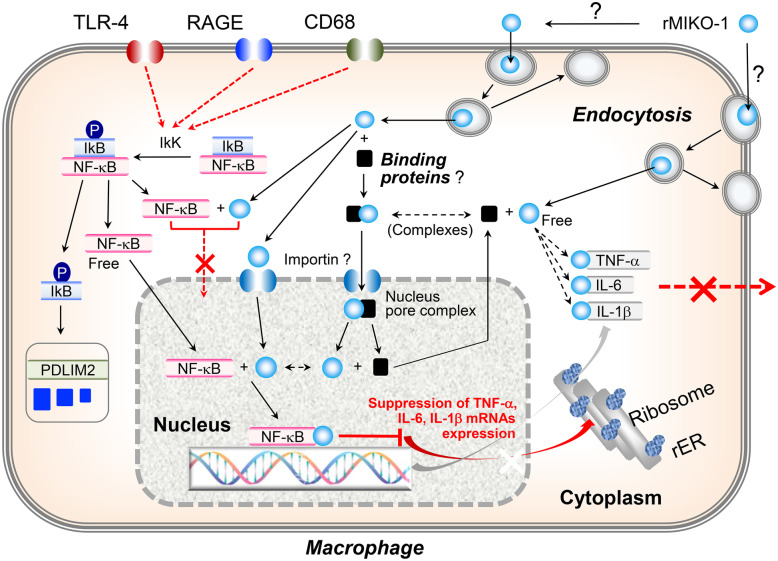
The dynamic mobility of rMIKO-1 in macrophages was important for understanding its potential activity in the immune system (Fig. 4). rMIKO-1 appeared to bind to activated macrophages in a specific manner and quickly migrated inside the nucleus (indicated by vertical arrows) through the cytoplasm (indicated by horizontal arrows); however, the mechanisms have not yet been elucidated in detail. Nevertheless, rMIKO-1 appears to play an important role(s) in the activated macrophages. To clarify the possibility, we performed Western blotting and found that rMIKO-1 was present in both the cytoplasm and nucleus of macrophages (Fig. 5), and that a possible complex of rMIKO-1 with NF-κB p65 or β-actin was detected in the cytoplasm and nucleus of macrophages using the preparative ELISA system (Fig. 6B). Furthermore, rMIKO-1 microscopically shared its intracellular localization in macrophages with NF-κB p65 or β-actin (Fig. 6C, panels C1 and C2). These results suggest that rMIKO-1 migrates on target DNA chains in the nucleus of macrophages to inhibit the mRNA transcription of inflammatory cytokines by concealing a specific region, e.g., the NF-κB motif. A schematic drawing of the intracellular mobility of rMIKO-1 in macrophages is tentatively shown in Fig. 7. Therefore, rMIKO-1 may inhibit intracellular signaling pathways, such as NF-κB, mitogen-activated protein kinase, and/or myeloid differentiation primary response 88. However, further investigations are warranted to elucidate the mechanisms underlying the dynamism of rMIKO-1 in more detail.
Fig. 7.
Schematic drawing of the proposed inhibitory mechanism suppressing the excessive activation of macrophages by rMIKO-1. NF-κB (nuclear factor-kappa B), TNF-α (tumor necrosis factor-α), IL-6 (interleukin-6), IL-1β (interleukin-1β), rER (rough endoplasmic reticulum), IκK (inhibitor kappa B kinase), IκB (inhibitor kappa B), p (phosphate), closed square (black, possible intracellular binding proteins), closed circle (blue, free rMIKO-1).
As suggested above, rMIKO-1 may contribute to the inhibition of some signal transduction pathways in order to induce inflammation in the rectal tissue of rats with DSS-induced experimental colitis. Among these pathways, the NF-κB p65 pathway via TLR4 may be a pivotal pathway, as shown in Fig. 7. We herein hypothesized that rMIKO-1 forms complexes with NF-κB p65 and/or β-actin in macrophages. We attempted to detect, albeit indirectly, these complexes using ELISA. A positive reaction was observed (Fig. 6B), which suggested the existence of a complex between rMIKO-1 and NF-κB p65 or β-actin. These results indicate that rMIKO-1 is capable of neutralizing the activity of NF-κB p65 in the cytoplasm and/or nucleus of macrophages leading to the downregulation of inflammatory cytokines’ mRNAs expression in cells. Although the mechanisms underlying the migration of these complexes inside the nucleus of macrophages remain unclear, β-actin in the cytoplasm may contribute to the transportation of rMIKO-1 inside the nucleus because β-actin was detected by immunoprecipitation (Fig. 6D, panel D2). Microscopic images showing that rMIKO-1 shared its intracellular localization with NF-κB p65 and β-actin in macrophages also appear to support our hypothesis (Fig. 6C, panels C1 and C2). Therefore, rMIKO-1 may inhibit a pivotal pathway, the NF-κB pathway, via TLR4 in cells.
The accumulation of many immune cells was not observed microscopically in the large colon of rats in the M0.6 group, which strongly suggests that rMIKO-1 prevented the migration of immune cells, such as macrophages, in the D group. Therefore, the limiting effects of rMIKO-1 on excessively activated macrophages may prevent further signal transduction to immune cells, such as neutrophils and lymphocytes, via other paracrine pathways. The effects of rMIKO-1 on immune cells with high division activity, such as reproductive and stem cells, have not yet been investigated. We speculate that rMIKO-1 ameliorates abnormalities in unusual macrophages in the large colon of rats with experimental colitis, but does not terminate their activity. The ability of macrophages to eliminate infectious agents from the body may be maintained, but not perfectly, in macrophages. rMIKO-1 may be digested by a complex mechanism after it has exerted its effects on macrophages. Although rMIKO-1 is not a mutagenic protein, its biochemical properties remain unclear. Therefore, further studies are warranted.
The number of individuals infected with coronavirus disease 2019 (COVID-19) continues to increase worldwide and is threatening humanity itself. This pandemic is contributing to the dissociation of medical systems in many countries. A large number of patients currently have severe pneumonia and/or respiratory failure and, despite treatment, may ultimately die. Although the cause of death by COVID-19 remains unclear, one reason may be due to the cytokine storm, which is derived from the excessive activation of immune cells, such as macrophages, lymphocytes, and neutrophils, in lung tissue. rMIKO-1 may effectively ameliorate the effects of the cytokine storm. Therefore, further studies on the effectiveness of rMIKO-1 to suppress the cytokine storm are underway. Collectively, the present results indicate that the specific properties of rMIKO-1 characterized by dynamic mobility in macrophages attenuate severe pneumonia associated with COVID-19.
CONCLUSIONS
We herein suggested that rMIKO-1 functions as a novel regulator of experimental colitis in rats through the negative regulation of the excessive activation of macrophages in the large colon. rMIKO-1 may be clinically beneficial for patients with inflammatory diseases, including inflammatory bowel diseases, such as UC and CD, and acute pneumonia, because its target cells are currently activated macrophages. The present results will contribute to the discovery of more effective medicines or therapeutic agents in the future.
Acknowledgements
Naoko Takeuchi and Akiko Satoh engaged in the study on S100 proteins in the first stage and technically supported us for approximately two years. We would like to thank Yamasa Shoyu Co., Ltd., Chiba, Japan, for providing monoclonal antibodies for rS100A8 (mAb2H6 and mAb8A6) and rS100A9 (mAb1D11 and mAb10D11) and their conjugates with HRP.
Author Contribution
MI and KO together conceived the study, participated in its design and coordination, and drafted the manuscript. MI and KO were mainly in charge of the protein assay and genetic analysis, respectively, in the present study. All authors have read and approved the final manuscript.
Funding
The present study was supported by a Grant-in-Aid for Scientific Research (C: 26460663) from the Ministry of Education, Science, Sports, and Culture of Japan to the corresponding author (Masaki Ikemoto, PhD). This was also supported by a Grant-in-Aid for Young Scientists (B: 21K15422) from the Ministry of Education, Science, Sports, and Culture of Japan to the first author (Kohki Okada, PhD).
Data Availability
We are ready to provide data and materials as required at any time.
DECLARATIONS
Ethics Approval and Consent to Participate
All experiments were performed in conformity with the WMA Declaration of Helsinki-Ethical Principles for Medical Research Involving Human Subjects (64th WMA General Assembly; Fortaleza, Brazil, October 2013). The corresponding author (Masaki Ikemoto, PhD) was approved by the Ethics Committee of Nagahama Institute of Bio-Science and Technology (permission number: 075). The first author (Kohki Okada, PhD) was also approved by the Ethics Committee of Medical Technology and Sciences, Faculty of Health Sciences, Kyoto Tachibana University (permission numbers: 20–03).
Consent for Publication
We agreed with the publication of this manuscript in Inflammation.
Competing Interests
The authors declare no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kohki Okada, Email: okada-ko@tachibana-u.ac.jp, Email: koukiokada11@yahoo.com.jp.
Masaki Ikemoto, Email: m_ikemoto@nagahama-i-bio.ac.jp, Email: i_masaki@gaia.eonet.ne.jp.
References
- 1.Odink K, Gerletti N, Bruggen J, Clerc RG, Tarcsay L, Zwadlo G, Gerhards G, Schiegel R, Sorg C. Two calcium-binding proteins in infiltrate macrophages of rheumatoid arthritis. Nature. 1987;330:80–82. doi: 10.1038/330080a0. [DOI] [PubMed] [Google Scholar]
- 2.Ryckman C, Vandal K, Rouleau P, Talbot M, Tessier PA. Proinflammatory activities of S100: Proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. The Journal of Immunology. 2003;170:3233–3242. doi: 10.4049/jimmunol.170.6.3233. [DOI] [PubMed] [Google Scholar]
- 3.Guignard F, Mauel J, Markert M. Identification and characterization of a novel human neutrophil protein related to the S100 family. Biochemical Journal. 1995;309:395–401. doi: 10.1042/bj3090395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imamichi T, Uchida I, Wahl SM, McCartney-Francis N. Expression and cloning of migration inhibitory factor-related protein (MRP)8 MRP14 in arthritis-susceptible rats. Biochemical and Biophysical Research Communications. 1993;194:819–825. doi: 10.1006/bbrc.1993.1895. [DOI] [PubMed] [Google Scholar]
- 5.Foell D, Wittkowski H, Ren Z, Turton J, Pang G, Daebritz J, Ehrchen J, Heidemann J, Borody T, Roth J, Clancy R. Phagocyte-specific S100 proteins are released from affected mucosa and promote immune responses during inflammatory bowel disease. The Journal of Pathology. 2008;216:183–192. doi: 10.1002/path.2394. [DOI] [PubMed] [Google Scholar]
- 6.Tibble JA, Bjarnason I. Non-invasive investigation of inflammatory bowel disease. World Journal of Gastroenterology. 2001;7:460–465. doi: 10.3748/wjg.v7.i4.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cario E, Podolsky DK. Differential alteration in intestinal epithelial cell expression of toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infection and Immunity. 2000;68:7010–7017. doi: 10.1128/IAI.68.12.7010-7017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshino T, Nakase H, Honzawa Y, Matsumura K, Yamamoto S, Takeda Y, Ueno S, Uza N, Masuda S, Inui K, Chiba T. Immunosuppressive effects of tacrolimus on macrophages ameliorate experimental colitis. Inflammatory Bowel Diseases. 2010;16:2022–2033. doi: 10.1002/ibd.21318. [DOI] [PubMed] [Google Scholar]
- 9.Lee, M.J., J.K. Lee, J.W. Choi, C.S. Lee, J.H. Sim, C.H. Cho, K.H. Lee, I.H. Cho, M.H. Chung, H.R. Kim, and S.K. Ye. 2012. Interleukin-6 induces S100A9 expression in colonic epithelial cells through STAT3 activation in experimental ulcerative colitis. PLos One 7: e38801. [DOI] [PMC free article] [PubMed]
- 10.Ikemoto M, Murayama H, Itoh H, Totani M, Fujita M. Intrinsic function of S100A8/A9 complex as an anti-inflammatory protein in liver injury induced by lipopolysaccharide in rats. Clinica Chimica Acta. 2007;376:197–204. doi: 10.1016/j.cca.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 11.Koike A, Arai S, Yamada A, Nagae A, Saita N, Itoh H, Uemoto S, Totani M, Ikemoto M. Dynamic mobility of immunological cells expressing S100A8 and S100A9 in vivo: A variety of functional roles of the two proteins as regulators in acute inflammatory reaction. Inflammation. 2012;35:409–419. doi: 10.1007/s10753-011-9330-8. [DOI] [PubMed] [Google Scholar]
- 12.Vogl T, Tenbrock K, Ludwig S, Leukert N, Ehrhardt C, van Zoelen MA, Nacken W, Foell D, van der Poll T, Sorg C, Roth J. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nature Medicine. 2007;13:1042–1049. doi: 10.1038/nm1638. [DOI] [PubMed] [Google Scholar]
- 13.Tsan MF, Gao B. Endogenous ligands of Toll-like receptors. Journal of Leukocyte Biology. 2004;76:514–519. doi: 10.1189/jlb.0304127. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh HL, Schafer BW, Sasaki N, Heizmann CW. Expression analysis of S100 proteins and RAGE in human tumors using tissue microarrays. Biochemical and Biophysical Research Communications. 2003;307:375–381. doi: 10.1016/S0006-291X(03)01190-2. [DOI] [PubMed] [Google Scholar]
- 15.Stern D, Yan SD, Yan SF, Schmidt AM. Receptor for advanced glycation endproducts: A multiligand receptor magnifying cell stress in diverse pathologic settings. Advanced Drug Delivery Reviews. 2002;54:1615–1625. doi: 10.1016/S0169-409X(02)00160-6. [DOI] [PubMed] [Google Scholar]
- 16.Foell D, Wittkowski H, Vogl T, Roth J. S100 proteins expressed in phagocytes: A novel group of damage-associated molecular pattern molecules. Journal of Leukocyte Biology. 2007;81:28–37. doi: 10.1189/jlb.0306170. [DOI] [PubMed] [Google Scholar]
- 17.Okada K, Arai S, Nakase H, Kohno H, Nakamura F, Takeda M, Toda Y, Itoh H, Adachi S, Ikemoto M. Autocrine pathways involving S100A8 and/or S100A9 that are postulated to regulate the immunological functions of macrophages in rats. Biochemical and Biophysical Research Communications. 2015;456:415–420. doi: 10.1016/j.bbrc.2014.11.099. [DOI] [PubMed] [Google Scholar]
- 18.Okada K, Arai S, Itoh H, Adachi S, Hayashida M, Nakase H, Ikemoto M. CD68 on rat macrophages binds tightly to S100A8 and S100A9 and helps to regulate the cells’ immune functions. Journal of Leukocyte Biology. 2016;100:1093–1104. doi: 10.1189/jlb.2A0415-170RRR. [DOI] [PubMed] [Google Scholar]
- 19.Okada K, Itoh H, Kamikubo Y, Adachi S, Ikemoto M. Establishment of S100A8 transgenic rats to understand innate property of S100A8 and its immunological role. Inflammation. 2018;41:59–72. doi: 10.1007/s10753-017-0664-8. [DOI] [PubMed] [Google Scholar]
- 20.Namura T, Arai S, Okawa K, Koike A, Yamada S, Saita N, Nagae A, Itoh H, Totani M, Uemoto S, Ikemoto M. Identification of serum proteins that bind with S100A8, S100A9 and S100A8/A9: Clinical significance of using proteins for monitoring the postoperative condition of liver recipients. Clinica Chimica Acta. 2010;411:1766–1773. doi: 10.1016/j.cca.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 21.Qing G, Ma LC, Khorchid A, Swapna GVT, Mal TK, Takayama MM, Xia B, Phadtare S, Ke H, Acton T, Montelione GT, Ikura M, Inouye M. Cold-shock induced high-yield protein production in Escherichia coli [Letter] Nature Biotechnology. 2004;22:877–882. doi: 10.1038/nbt984. [DOI] [PubMed] [Google Scholar]
- 22.Ikemoto M, Tanaka T, Takai Y, Murayama H, Tanaka K, Fujita M. New ELISA system for myeloid-related protein complex (MRP8/14) and its clinical significance as a sensitive marker for inflammatory responses associated with transplant rejection. Clinical Chemistry. 2003;49:594–600. doi: 10.1373/49.4.594. [DOI] [PubMed] [Google Scholar]
- 23.Kucharzik T, Maaser C, Lügering A, Kagnoff M, Mayer L, Targan S, Domschke W. Recent understanding of IBD pathogenesis: Implications for future therapies. Inflammatory Bowel Diseases. 2006;12:1068–1083. doi: 10.1097/01.mib.0000235827.21778.d5. [DOI] [PubMed] [Google Scholar]
- 24.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konarska K, Cieszkowski J, Warzecha Z, Ceranowicz P, Chmura A, Kusnierz-Cabala B, Galazka K, Kowalczyk P, Miskiewicz A, Konturek TJ, Pedziwiatr M, Dembinski A. Treatment with obestatin – a ghrelin gene-encoded peptide – reduces the severity of experimental colitis evoked by trinitrobenzene sulfonic acid. International Journal of Molecular Sciences. 2018;19:1643. doi: 10.3390/ijms19061643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Wei X, Zhang R, Petitte LN, Si D, Li Z, Cheng J, Du M. Design and development of a novel peptide for treating intestinal inflammation. Frontiers in Immunology. 2019;10:1841. doi: 10.3389/fimmu.2019.01841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We are ready to provide data and materials as required at any time.