Abstract
Nucleic acid therapeutics are developing into precise medicines that can manipulate specific genes. However, the development of safe and effective delivery system for the target cells has remained a challenge. Lipid nanoparticles (LNPs) have provided a revolutionary delivery system that can ensure multiple clinical translation of RNA-based candidates. In 2018, Patisiran (Onpattro) was first approved as an LNP-based siRNA drug. In 2020, during the coronavirus disease 2019 (COVID-19) outbreak, LNPs have enabled the development of two SARS-CoV-2 mRNA vaccines, Tozinameran (Comirnaty or Pfizer-BioNTech COVID-19 vaccine) and Elasomeran (Spikevax or COVID-19 vaccine Moderna) for conditional approval. Here, we reviewed the state-of-the-art LNP technology employed in three approved drugs (one siRNA-based and two mRNA-based drugs) and discussed the differences in their mode of action, formulation design, and biodistribution.
Keywords: Lipid nanoparticles, Ionizable lipid, LNP, mRNA vaccine, COVID-19, Patisiran, Tozinameran, Elasomeran, COVID-19 vaccine moderna, SARS-CoV-2
1. Introduction
In the 21st century, nucleic acids are attracting attention as next generation modality beyond a small molecule and antibody. Since the approval of the world's first nucleic acid drug “fomivirsen” in 1998, a total of 16 nucleic acid medicines (9 antisense oligonucleotides (ASO), 4 short interfering RNAs (siRNA), 1 aptamer, and 2 messenger RNAs (mRNA)) have been approved in some areas of US, EU, and Japan, as of May 2021 [1]. More than 80% of these drugs (13 out of 16) have been approved since 2016. This rapid expansion of nucleic acid therapeutics has been triggered by scientific breakthrough in drug-delivery systems. Tremendous efforts toward the development of delivery carriers [[2], [3], [4], [5], [6], [7], [8], [9]] have identified lipid nanoparticles (LNPs) as a clinically validated platform technology that can deliver both long (i.e. mRNA [10,11]) and short nucleic acids (i.e. siRNA [12]) into target cells. Lipid nanoparticle technology has played a central role in the development of the world's first drugs associated with two completely independent phenomena, namely RNA interference by siRNA and vaccination by mRNA. In this review, we have focused on three approved drugs using lipid nanoparticle technology, namely Patisiran as a siRNA medicine, and Tozinameran and Elasomeranas mRNA vaccines. Their development history [13,14], recent clinical trials and perspectives [15], gene regulation [16], structure of ionizable lipid [17], and mRNA vaccine [[18], [19], [20]] have already been reported in previous studies. This review has partially referred to the information from an interview form archived in Pharmaceuticals and Medical Devices Agency (PMDA) [21,22,23].
Table 1 provides an overview of the three approved drugs. Patisiran (trade name: Onpattro, developed by Alnylam) was first approved in 2018 for the treatment of hereditary transthyretin (hATTR) amyloidosis. siRNA is designed to target a sequence within the untranslated region (3′UTR) of transthyretin (TTR) mRNA. LNP-formulated siRNA is administered intravenously to patients at a dose of 0.3 mg/kg every three weeks [24]. The risk of infusion reactions and adverse events often triggered by nanomedicine [25] are mitigated by two approaches, namely premedication and slow infusion [26]. Tozinameran (trade name: Comirnaty, developed by Pfizer/BioNTech) was granted Emergency Use Authorization (EUA) by the United States Food and Drug Administration (FDA) in December 2020 for the prevention of coronavirus disease 2019, COVID-19 [27]. mRNA is designed to encode the stabilized prefusion SARS-CoV-2 spike protein, a key molecule for eliciting neutralizing antibodies. LNP-formulated mRNA is administered intramuscularly to human at a dose of 30 μg in a series of two doses (0.3 mL each) three weeks apart. Elasomeran (trade name: Spikevax, developed by Moderna) was also granted EUA at approximately the same time in 2020 [28]. The two mRNA vaccines share multiple similarities, with slight differences in dosages and administration schedules; none required premedication. Differences in the storage conditions of all 3 drugs should, however, be noted. While the siRNA-LNP can be stored in a standard medical refrigerator (2 °C to 8 °C), the mRNA-LNP products need to be stored in a freezer (approximately −20 °C) or in ultra-cold storage (−80 °C to −60 °C).
Table 1.
Overview of the three approved drugs.
| Active ingredient | Patisiran | Tozinameran | Elasomeran | |
|---|---|---|---|---|
| Trade name | Onpattro | Comirnaty | Spikevax | |
| Company | Alnylam | Pfizer/BioNTech | Moderna | |
| Development code | ALN-TTR02 | BNT162b2 | mRNA-1273 | |
| First approval year | Aug-2018 | Dec-2020 (conditional) | Dec-2020 (conditional) | |
| Indication | hATTR amyloidosis | Prevention of COVID-19 | Prevention of COVID-19 | |
| Target protein | TTR | Spike of SARS-CoV-2a | Spike of SARS-CoV-2a | |
| LNP-formulated RNA | siRNA | mRNA | mRNA | |
| Administration route | Intravenous | Intramuscular | Intramuscular | |
| Administration schedule | every 3 weeks | 2 doses, 3 weeks apart | 2 doses, 4 weeks apart | |
| Dosage strength | 0.3 mg/kg siRNAb | 30 μg mRNA | 100 μg mRNA | |
| Dosage volume | Total 200 mLc | 0.3 mLd | 0.5 mL | |
| Administration time | over 70 min | immediately | immediately | |
| Premedication | required | Not required | Not required | |
| Storage condition | 2 °C to 8 °C (do not freeze) | −90 °C to −60 °C | −50 °C to −15 °C | |
The information is derived from the interview form archived in PMDA as of May 2021 [[21], [22], [23]]. hATTR, hereditary transthyretin; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID-19, coronavirus disease 2019; LNP, lipid nanoparticles.
Spike proteins are modified by 2 proline substitutions to produce prefusion-stabilized SARS-CoV-2 spike proteins, S–2P.
Body weight is less than 103 kg; 31.2 mg siRNA, when 104 kg or more.
Diluted by saline from 2 mg/mL siRNA in supplied vial.
Diluted by saline from 0.5 mg/mL mRNA in supplied vial to 0.1 mg/mL for injection.
2. Difference in mode of action
Since Tozinameran and Elasomeran have very similar modes of action, the 3 drugs have been classified into two categories in this section, namely siRNA-LNP and mRNA-LNP (Table 2 ).
Table 2.
Difference in mode of action.
| Active ingredient (Company) | Patisiran (Alnylam) | Tozinameran (Pfizer/BioNTech), Elasomeran (Moderna) |
|---|---|---|
| Drug substance | siRNA | mRNA |
| Strand | double strand | single strand |
| RNA length | 42 (21 + 21) | 4284 (Tozinameran)a |
| Base modification | 2ʹ-OMe-uridine, 2ʹ-OMe-cytosine | N1-methyl-pseudouridine |
| Purpose of base modification | suppress the innate immune response to mitigate off-target effect | suppress the innate immune response to increase protein production |
| Mode of action | stop production of target protein by degrading mRNA | initiate production of target protein by supplying mRNA, which triggers the adaptive immunity |
| Target protein | Transthyretin (TTR) | spike of SARS-CoV-2b |
| Endogenous machinery | RNA interference | translation |
| Location of machinery | cytoplasm | cytoplasm |
| Primary target cell of LNP | hepatocytes | antigen presenting cells (APCs) |
LNP, lipid nanoparticles; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Structure of Elasomeran is not described in detail.
Target spike proteins are modified by 2 proline substitutions to produce prefusion-stabilized SARS-CoV-2 spike protein.
2.1. Chemistry of siRNA and mRNA
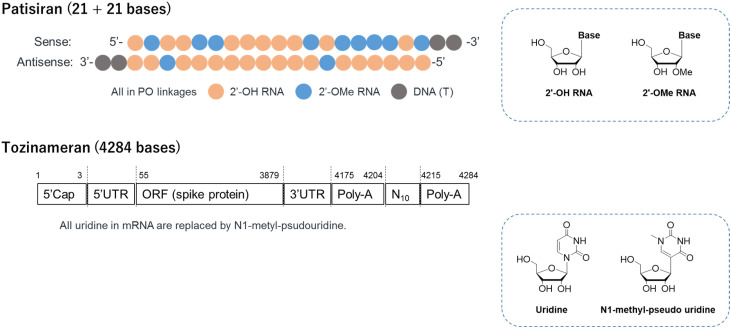
The siRNA in Patisiran is a short, double-stranded RNA with 21 bases in each strand while mRNA in the two vaccines is a long, single-stranded RNA with over 4000 bases (Fig. 1 ) [14]. Human body recognizes RNA as a toxic virus, not as medicines, via cellular sensors meant for biological defense against invading pathogens. Chemical modification can allow RNA to avoid this response triggered by pattern recognition receptors (PRRs), such as toll-like receptors (TLRs) and retinoic acid-inducible gene I receptors (RIG-I) [29,30]. In siRNA, uridine and cytosine are partially replaced by 2ʹ-O-methyl uridine and cytosine. In mRNA, uridine is fully replaced by N1-methyl pseudouridine [31] (Fig. 1). Since innate immune responses lead to downregulation of mRNA translation through phosphorylation of eIF2a by dsRNA-dependent protein kinase [32], N1-methyl pseudouridine modification enhances protein production, resulting in increased immunogenicity [31,33].
Fig. 1.
Structure of the drugs. For both mRNA designs, S–2P spike proteins were modified by 2 proline substitutions to produce prefusion-stabilized SARS-CoV-2 spike proteins. UTR, untranslated region; ORF, open reading frame.
In principle, siRNA can be chemically manufactured by phosphoramidite method using an automated solid-phase DNA/RNA synthesizer [34]. mRNA is enzymatically manufactured by in-vitro transcription (IVT) from a plasmid DNA template encoding mRNA sequences [35,36].
2.2. Mechanism of action of siRNA-LNP after administration
Intravenously administered LNP-formulated siRNA accumulates in the liver, the primary site of transthyretin (TTR) production. After the uptake of LNPs by hepatocytes in the liver, siRNA harnesses the endogenous RNA interference pathway, degrades TTR mRNA, and reduces the production of TTR protein.
2.3. Mechanism of action of mRNA-LNP after administration
The EMA assessment report has detailed the process associated with mRNA vaccine at the injection site as follows: “Intramuscular administration of LNP-formulated RNA vaccines results in transient local inflammation that drives recruitment of neutrophils and antigen presenting cells (APCs) to the site of delivery. Recruited APCs are capable of LNP uptake and protein expression (i.e. spike of SARS-CoV-2) by translation and can subsequently migrate to the local draining lymph nodes where T cell priming occurs [37].”
2.4. Delivery of LNP from extracellular to intracellular environment
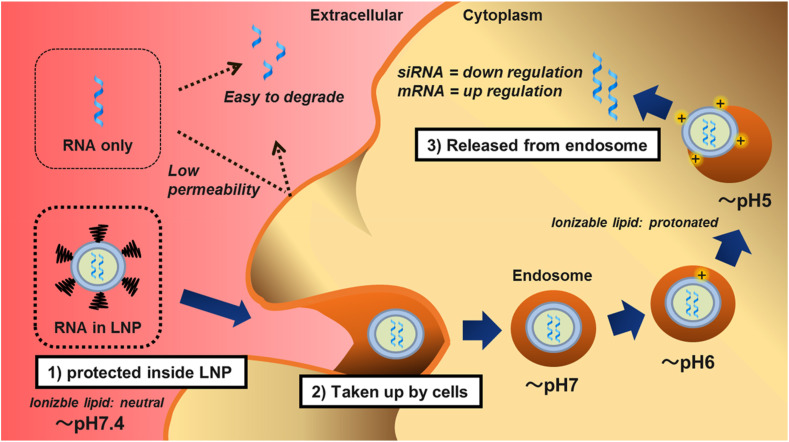
The biggest challenge for nucleic acid drugs is RNA delivery into the cytoplasm, where the endogenous machinery, for RNA interference and protein translation, is located. In principle, the LNP delivery mechanism is similar for siRNA [[38], [39], [40]] and mRNA [10,11,41]. The three main steps are as follows (Fig. 2 ). First, full encapsulation by LNP protects RNA from nuclease digestion. LNPs are neutral in physiological pH due to the ionizable lipid and polyethylene glycol (PEG)-lipid, thereby reducing non-specific interactions with serum proteins. Second, following dissociation of the PEG-lipid, cells take up LNPs via apolipoprotein E (ApoE)-dependent and/or ApoE-independent pathways. Finally, protonated LNPs, upon acidification in the endosome, induce hexagonal phase structures, disrupt the membranes, and release RNA molecules into the cytoplasm. The released RNA molecules lead to either down regulation (siRNA) or up regulation (mRNA) of target proteins. Details regarding the uptake of LNPs into hepatocytes have already been reported, revealing that LNPs are internalized via LDL receptors following the ApoE-dependent pathway [38]. Details regarding the uptake into antigen-presenting cells have not yet been clarified; however, efficient localization of nucleic acids in the cytoplasm of antigen presenting cells has been reported [42].
Fig. 2.
Delivery mechanism of lipid nanoparticles. First, full encapsulation by lipid nanoparticles (LNP) protects RNA from nuclease digestion. LNPs are neutral in physiological pH due to the ionizable lipid and PEG-lipid, thereby reducing non-specific interactions with serum proteins. Second, following dissociation of the PEG-lipid, cells take up LNPs via apolipoprotein E (ApoE)-dependent and/or ApoE-independent pathways. Finally, protonated LNPs, upon acidification in the endosome, induce hexagonal phase structures, disrupt the membranes, and release RNA molecules into the cytoplasm.
3. Difference in formulation
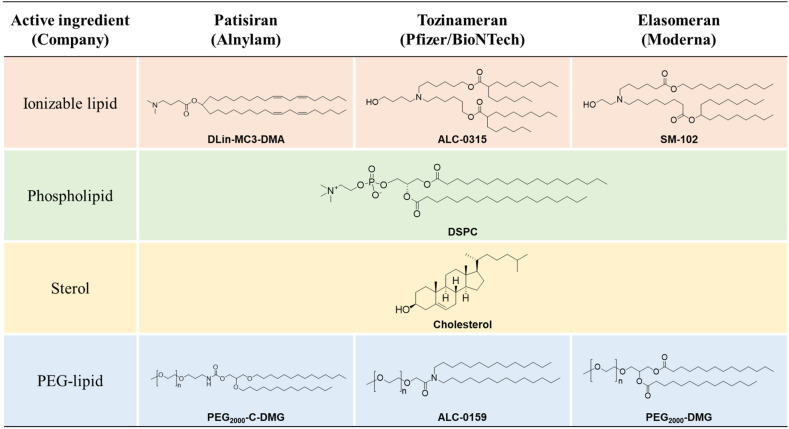
The three LNP-based drugs share multiple similarities in their formulation, and hence, behave similarly as nanoparticles in vivo. Importantly, all LNPs are composed of four types of lipids; ionizable lipid, phospholipid, cholesterol, and PEG-lipid (Fig. 3 ). All 3 ionizable lipids have tertiary amine group with pKa 6.0–6.7. These lipids switch its charge from neutral to cationic based on the neutral pH in the blood and the acidic pH in endosomes. The 3 PEG-lipids have dialkyl chains 14-carbon long, which are important for the rapid dissociation from the surface of LNPs once inside the body [43]. The biodegradable design of ALC-0315 [44] and SM-102 [11] is described later.
Fig. 3.
Chemical structure of lipids in lipid nanoparticles. ALC-0159 has PEG2000. All 3 ionizable lipids have tertiary amine groups, namely Dlin-MC3-DMA (MC3), pKa 6.44 [12] or pKa 6.35 [11]; ALC-0315, pKa 6.09 [44]; and SM-102, pKa 6.68 [11]. The related patents are as follows: Dlin-MC3-DMA, WO/2010/144740; ALC-0315, WO/2017/075531 (Lipid No. 3); and SM-102, WO/2017/049245 (Compound 25).
Lipid composition of the three drug products is summarized in Table 3 . All three LNPs were composed of four lipids in similar molar ratios. The major differences are in the storage condition of drug products. A recent review estimated that mRNAs, not LNPs, are the main factors for low storage stability of drug products, and thus mRNA vaccines need to be supplied at freezing (about −20 °C) or ultra-freezing (−80 to −60 °C) conditions to slow down mRNA hydrolysis [45]. Sucrose is used as a cryoprotectant to maintain the physical properties of LNPs during freeze-thaw cycles [46]. In contrast to the two mRNA vaccines, Patisiran needs to avoid freezing temperatures, probably because lipid nanoparticles dissolved in PBS, not in Sucrose, cannot tolerate freezing.
Table 3.
Composition of lipid nanoparticles.
| Active ingredient (Company) | Patisiran (Alnylam) | Tozinameran (Pfizer/BioNTech) | Elasomeran (Moderna) |
|---|---|---|---|
| Single dosage | 0.3 mg/kg siRNA | 0.030 mg mRNA | 0.10 mg mRNA |
| Unit | 5 mL | 0.45 mL | 0.5 mL |
| Drug substance | 10 mga | 0.225 mg | 0.10 mg |
| Ionizable lipid | 65.0 mg | 3.23 mg | 1.075 mg |
| Phospholipid | 16.5 mg | 0.7 mg | 0.275 mg |
| Cholesterol | 31.0 mg | 1.4 mg | 0.47 mg |
| PEG-lipid | 8.0 mg | 0.4 mg | 0.115 mg |
| Total lipid | 120.5 mg | 5.7 mg | 1.94 mg |
| Total lipid/RNA (wt/wt) | 12.1 | 25.5 | 19.4 |
| Drug concentration | 2.0 mg/mL | 0.5 mg/mL | 0.2 mg/mL |
| Sucrose in formulation | NA | 46 mg | 43.5 mg |
| Sucrose concentration | NA | 102 g/L | 87 g/L |
| pH | 6.4–7.5 | 6.9–7.9 | 7.0–8.0 |
| State of drug product | liquid | frozen suspension | frozen suspension |
| Shelf life of drug product (unopened vials)b | 27 months (2–8 °C, avoid freeze) | 6 months (−90 to −60 °C) | 6 months (−25 to −15 °C) |
The information is derived from the interview form archived in PMDA as of May 2021(21–23).
As Patisiran, without sodium salt.
Shelf life of each drug is updated by ongoing storage stability tests.
In principal, assembly of LNPs is achieved by rapid mixing of four lipids in ethanol with RNA molecules in an aqueous buffer (near pH 4) in a microfluidic or T-junction mixer [[47], [48], [49]].
4. Difference in biodistribution
The major difference in biodistribution of the three LNP-based drugs arises from the administration route (Table 4 ). Regarding Patisiran, a study on rats using 14C-labeled ionizable lipid (14C-MC3) had revealed that with a single intravenous dose of siRNA-LNP, approximately 90% of the administered radioactivity was detected in the liver 4 h after administration [21]. Regarding Tozinameran, a study on rats using 3H-labeled cholesteryl hexadecyl ether (3H-CHE) had revealed that with a single intramuscular dose of mRNA-LNP (luciferase mRNA as surrogate), radioactivity concentration was the highest at the injection site between 15 min and 48 h (i.e. the entire measurement period) after administration [22]. Besides that in the administration site, the total radioactivity recovery rate for the dose was highest in the liver (up to 18%) 8–48 h after administration [22]. Subcutaneously administered liposomes (approximately <0.1 μm) did not enter the blood capillary owing to their size; they remained at the injection site and were drained through lymphatic capillaries to nearby lymph nodes [50]. The innate biodistribution of nanoparticles is favorable for mRNA vaccines, since 1) adaptive immune response occurs at lymph nodes, and 2) unwanted systemic exposure is reduced. A detailed study had been conducted with intramuscularly administered mRNA-LNPs in non-human primates [51].
Table 4.
Topics related to the pharmacokinetics of lipid nanoparticles.
| Active ingredient (Company) |
Patisiran (Alnylam) |
Tozinameran (Pfizer/BioNTech) |
|---|---|---|
| Administration route | intravenous | intramuscular |
| Primary biodistribution | liver | injection site |
| Biodegradable property in ionizable lipid | no | yes |
| Anti-drug antibody (ADA) assessments | anti-PEG antibody | not reported |
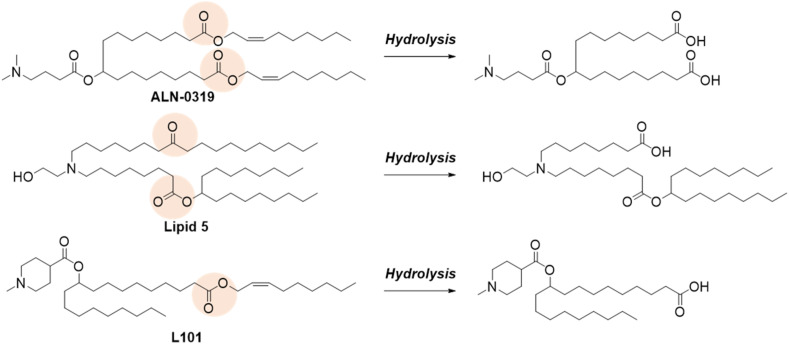
Ideal drug carriers are rapidly eliminated from the body once their purpose is served. Various LNPs incorporated biodegradable designs in an ionizable lipid as a means to facilitate their elimination (Fig. 4 ) [10,52,53]. In contrast to MC3, ALC-0315 and SM-102 have ester linkages in the lipid tail. Intramuscular injection of mRNA-LNPs composed of SM-102 or MC3 resulted in faster clearance of SM-102, along with improved tolerability, over MC3 at the injection site [11]. The two ester linkages in ALC-0315 were also hydrolyzed in vivo [22].
Fig. 4.
Biodegradable design in ionizable lipid. Pink circle in each chemical structure shows the hydrolysis site. Hydrolysis of ester linkage facilitates rapid elimination and improved tolerability. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Biologics, modified with PEG, produce anti-drug antibodies (ADA), more specifically anti-PEG antibodies, in vivo [54]. siRNA-LNP [55] and mRNA-LNP [56] have been shown to generate anti-PEG antibodies in mice, leading to accelerated blood clearance (ABC) upon repeated administration. In the phase 3 APOLLO trial, ADAs to Patisiran were evaluated by measuring antibodies specific to PEG2000-C-DMG that is exposed on the surface of LNPs [57]. The overall incidence of treatment-emergent ADAs was 3.4% (5 of 145 patients) in the Patisiran group and 1.3% (1 of 77 patients) in the placebo group. Treatment-emergent ADAs to PEG2000-C-DMG occurred at a low frequency, were transient and appeared to have no effect on PK, PD, safety or efficacy [57]. To the best of our knowledge, no study has been reported on anti-PEG antibody yet, which causes accelerated blood clearance due to intramuscularly administered LNPs.
In relation to anti-PEG antibody, a severe allergic reaction, named anaphylaxis, has been reported to occur (though rarely; a couple of cases per million doses) due to the administration of two mRNA vaccines [58]. The trigger of anaphylaxis is considered to be due to anti-PEG IgE against PEG-lipid [59,60].
5. Conclusion
This review focused on the differences in LNP technology across three approved drugs. With the initial success of siRNA therapeutics [14], the technology platform has become well-established and its scope has expanded from siRNA to mRNA [15]. In early 2020, two pioneering candidates challenged to fight against COVID-19 outbreak, namely mRNA-1273 by Moderna [61] and BNT162 by Pfizer/BioNTech [62]. Following their examples, multiple mRNA vaccines leveraging LNP technology are currently in clinical trials, including CVnCoV by CureVac/BAYER [63,64], ARCT-021 by Arcturus Therapeutics [65], ARCoV by Walvax Biotechnology/Suzhou Abogen Biosciences [66], LNP-nCoVsaRNA by Imperial College London [67], and DS-5670 by Daiichi Sankyo/The University of Tokyo [68]. Recent clinical study of CVnCoV using mRNA with unmodified nucleotides may reveal the importance of mRNA chemistry [64]. A detailed understanding of these LNPs and RNA chemistry would expand the platform technology and open up pathways to next-generation therapeutics.
Author contributions
Y.S. wrote manuscript. H.I. reviewed manuscript.
Declaration of competing interest
The authors declare no conflicts of interest associated with this manuscript.
References
- 1.Takakusa H., Iwazaki N., Nishikawa M., Yoshida T., Obika S., Inoue T. Drug metabolism & pharmacokinetics of oligonucleotide therapeutics : profiles and evaluation approaches. Pharmaceutical and Medical Device Regulatory Science. 2021;52(2):76–84. [Google Scholar]
- 2.Love K.T., Mahon K.P., Levins C.G., Whitehead K.A., Querbes W., Dorkin J.R., et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci USA. 2010;107(5):1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato Y., Hatakeyama H., Sakurai Y., Hyodo M., Akita H., Harashima H. A pH-sensitive cationic lipid facilitates the delivery of liposomal siRNA and gene silencing activity in vitro and in vivo. J Contr Release. 2012;163(3):267–276. doi: 10.1016/j.jconrel.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Dong Y., Love K.T., Dorkin J.R., Sirirungruang S., Zhang Y., Chen D., et al. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc Natl Acad Sci USA. 2014;111(11):3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akita H., Noguchi Y., Hatakeyama H., Sato Y., Tange K., Nakai Y., et al. Molecular tuning of a vitamin E-scaffold pH-sensitive and reductive cleavable lipid-like material for accelerated in vivo hepatic siRNA delivery. ACS Biomater Sci Eng. 2015;1(9):834–844. doi: 10.1021/acsbiomaterials.5b00203. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki Y., Hyodo K., Tanaka Y., Ishihara H. siRNA-lipid nanoparticles with long-term storage stability facilitate potent gene-silencing in vivo. J Contr Release. 2015;220:44–50. doi: 10.1016/j.jconrel.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 7.Hajj K.A., Ball R.L., Deluty S.B., Singh S.R., Strelkova D., Knapp C.M., et al. Branched-tail lipid nanoparticles potently deliver mRNA in vivo due to enhanced ionization at endosomal pH. Small. 2019;15(6) doi: 10.1002/smll.201805097. [DOI] [PubMed] [Google Scholar]
- 8.Kuboyama T., Yagi K., Naoi T., Era T., Yagi N., Nakasato Y., et al. Simplifying the chemical structure of cationic lipids for siRNA-lipid nanoparticles. ACS Med Chem Lett. 2019;10(5):749–753. doi: 10.1021/acsmedchemlett.8b00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajappan K., Tanis S.P., Mukthavaram R., Roberts S., Nguyen M., Tachikawa K., et al. Property-driven design and development of lipids for efficient delivery of siRNA. J Med Chem. 2020;63(21):12992–13012. doi: 10.1021/acs.jmedchem.0c01407. [DOI] [PubMed] [Google Scholar]
- 10.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol Ther. 2018;26(6):1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol Ther Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing InVivo. Angew Chem Int Ed. 2012;51(34):8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yonezawa S., Koide H., Asai T. Recent advances in siRNA delivery mediated by lipid-based nanoparticles. Adv Drug Deliv Rev. 2020;154–155:64–78. doi: 10.1016/j.addr.2020.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akinc A., Maier M.A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat Nanotechnol. 2019;14(12):1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 15.Samaridou E., Heyes J., Lutwyche P. Lipid nanoparticles for nucleic acid delivery: current perspectives. Adv Drug Deliv Rev. 2020;154–155:37–63. doi: 10.1016/j.addr.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Witzigmann D., Kulkarni J.A., Leung J., Chen S., Cullis P.R., van der Meel R. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv Drug Deliv Rev. 2020;159:344–363. doi: 10.1016/j.addr.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel P., Ibrahim N.M., Cheng K. The importance of apparent pKa in the development of nanoparticles encapsulating siRNA and mRNA. Trends Pharmacol Sci. 2021;42(6):448–460. doi: 10.1016/j.tips.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson N.A.C., Kester K.E., Casimiro D., Gurunathan S., DeRosa F. The promise of mRNA vaccines: a biotech and industrial perspective. NPJ vaccine. 2020;5(1):11. doi: 10.1038/s41541-020-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buschmann M.D., Carrasco M.J., Alishetty S., Paige M., Alameh M.G., Weissman D. Nanomaterial delivery systems for mRNA vaccines. Vaccines. 2021;9(1) doi: 10.3390/vaccines9010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onpattro Interview form (ver.4), archived in PMDA. 2021. https://www.info.pmda.go.jp/go/interview/2/112773_1290400A1024_2_4F.pdf. Feb-2021
- 22.Comirnaty Interview form (ver.1), archived in PMDA. 2021. https://www.info.pmda.go.jp/go/interview/1/672212_631341DA1025_1_1F.pdf. Apr-2021
- 23.COVID-19 Vaccine Moderna, interview form (ver.1), archived in PMDA. 2021. https://www.info.pmda.go.jp/go/interview/1/400256_631341EA1020_1_001_1F.pdf. May-2021
- 24.Adams D., Gonzalez-Duarte A., O'Riordan W.D., Yang C.C., Ueda M., Kristen A.V., et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med. 2018;379(1):11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 25.Szebeni J., Simberg D., González-Fernández Á., Barenholz Y., Dobrovolskaia M.A. Roadmap and strategy for overcoming infusion reactions to nanomedicines. Nat Nanotechnol. 2018;13(12):1100–1108. doi: 10.1038/s41565-018-0273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams D., Suhr O.B., Dyck P.J., Litchy W.J., Leahy R.G., Chen J., et al. Trial design and rationale for APOLLO, a Phase 3, placebo-controlled study of patisiran in patients with hereditary ATTR amyloidosis with polyneuropathy. BMC Neurol. 2017;17(1):181. doi: 10.1186/s12883-017-0948-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Judge A.D., Bola G., Lee A.C., MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13(3):494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23(2):165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 31.Svitkin Y.V., Cheng Y.M., Chakraborty T., Presnyak V., John M., Sonenberg N. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 2017;45(10):6023–6036. doi: 10.1093/nar/gkx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.García M.A., Meurs E.F., Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89(6–7):799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16(11):1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caruthers M.H. The chemical synthesis of DNA/RNA: our gift to science. J Biol Chem. 2013;288(2):1420–1427. doi: 10.1074/jbc.X112.442855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlake T., Thess A., Fotin-Mleczek M., Kallen K.J. Developing mRNA-vaccine technologies. RNA Biol. 2012;9(11):1319–1330. doi: 10.4161/rna.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson J., Sorensen E.W., Mintri S., Rabideau A.E., Zheng W., Besin G., et al. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci Adv. 2020;6(26) doi: 10.1126/sciadv.aaz6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.EMA 2020. https://www.ema.europa.eu/en/documents/assessment-report/comirnaty-epar-public-assessment-report_en.pdf
- 38.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther. 2010;18(7):1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki Y., Ishihara H. Structure, activity and uptake mechanism of siRNA-lipid nanoparticles with an asymmetric ionizable lipid. Int J Pharm. 2016;510(1):350–358. doi: 10.1016/j.ijpharm.2016.06.124. [DOI] [PubMed] [Google Scholar]
- 40.Wittrup A., Ai A., Liu X., Hamar P., Trifonova R., Charisse K., et al. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat Biotechnol. 2015;33(8):870–876. doi: 10.1038/nbt.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maugeri M., Nawaz M., Papadimitriou A., Angerfors A., Camponeschi A., Na M., et al. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat Commun. 2019;10(1):4333. doi: 10.1038/s41467-019-12275-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basha G., Novobrantseva T.I., Rosin N., Tam Y.Y.C., Hafez I.M., Wong M.K., et al. Influence of cationic lipid composition on gene silencing properties of lipid nanoparticle formulations of siRNA in antigen-presenting cells. Mol Ther. 2011;19(12):2186–2200. doi: 10.1038/mt.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mui B.L., Tam Y.K., Jayaraman M., Ansell S.M., Du X., Tam Y.Y., et al. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol Ther Nucleic Acids. 2013;2(12):e139. doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novel lipids and lipid nanoparticle formulations for delivery of nucleic acids. WO2017075531A1; 2017. [Google Scholar]
- 45.Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G., Jiskoot W., et al. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int J Pharm. 2021;601:120586. doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ball R.L., Bajaj P., Whitehead K.A. Achieving long-term stability of lipid nanoparticles: examining the effect of pH, temperature, and lyophilization. Int J Nanomed. 2017;12:305–315. doi: 10.2147/IJN.S123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gindy M.E., Leone A.M., Cunningham J.J. Challenges in the pharmaceutical development of lipid-based short interfering ribonucleic acid therapeutics. Expet Opin Drug Deliv. 2012;9(2):171–182. doi: 10.1517/17425247.2012.642363. [DOI] [PubMed] [Google Scholar]
- 48.Bailey-Hytholt C.M., Ghosh P., Dugas J., Zarraga I.E., Bandekar A. Formulating and characterizing lipid nanoparticles for gene delivery using a microfluidic mixing platform. JoVE : JoVE. 2021;168 doi: 10.3791/62226. [DOI] [PubMed] [Google Scholar]
- 49.Terada T., Kulkarni J.A., Huynh A., Chen S., van der Meel R., Tam Y.Y.C., et al. Characterization of lipid nanoparticles containing ionizable cationic lipids using design-of-experiments approach. Langmuir : ACS J Surfaces Colloids. 2021;37(3):1120–1128. doi: 10.1021/acs.langmuir.0c03039. [DOI] [PubMed] [Google Scholar]
- 50.Oussoren C., Storm G. Liposomes to target the lymphatics by subcutaneous administration. Adv Drug Deliv Rev. 2001;50(1–2):143–156. doi: 10.1016/s0169-409x(01)00154-5. [DOI] [PubMed] [Google Scholar]
- 51.Liang F., Lindgren G., Lin A., Thompson E.A., Ols S., Röhss J., et al. Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in rhesus macaques. Mol Ther. 2017;25(12):2635–2647. doi: 10.1016/j.ymthe.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol Ther. 2013;21(8):1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki Y., Hyodo K., Suzuki T., Tanaka Y., Kikuchi H., Ishihara H. Biodegradable lipid nanoparticles induce a prolonged RNA interference-mediated protein knockdown and show rapid hepatic clearance in mice and nonhuman primates. Int J Pharm. 2017;519(1):34–43. doi: 10.1016/j.ijpharm.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 54.Kozma G.T., Shimizu T., Ishida T., Szebeni J. Anti-PEG antibodies: properties, formation, testing and role in adverse immune reactions to PEGylated nano-biopharmaceuticals. Adv Drug Deliv Rev. 2020;154–155:163–175. doi: 10.1016/j.addr.2020.07.024. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki T., Suzuki Y., Hihara T., Kubara K., Kondo K., Hyodo K., et al. PEG shedding-rate-dependent blood clearance of PEGylated lipid nanoparticles in mice: faster PEG shedding attenuates anti-PEG IgM production. Int J Pharm. 2020;588:119792. doi: 10.1016/j.ijpharm.2020.119792. [DOI] [PubMed] [Google Scholar]
- 56.Besin G., Milton J., Sabnis S., Howell R., Mihai C., Burke K., et al. Accelerated blood clearance of lipid nanoparticles entails a biphasic humoral response of B-1 followed by B-2 lymphocytes to distinct antigenic moieties. Immuno Horizon. 2019;3(7):282–293. doi: 10.4049/immunohorizons.1900029. [DOI] [PubMed] [Google Scholar]
- 57.Zhang X., Goel V., Attarwala H., Sweetser M.T., Clausen V.A., Robbie G.J. Patisiran pharmacokinetics, pharmacodynamics, and exposure-response analyses in the phase 3 APOLLO trial in patients with hereditary transthyretin-mediated (hATTR) amyloidosis. J Clin Pharmacol. 2020;60(1):37–49. doi: 10.1002/jcph.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimabukuro T.T., Cole M., Su J.R. Reports of anaphylaxis After receipt of mRNA COVID-19 vaccines in the US-december 14, 2020-january 18, 2021. Jama. 2021;325(11):1101–1102. doi: 10.1001/jama.2021.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Banerji A., Wickner P.G., Saff R., Stone C.A., Jr., Robinson L.B., Long A.A., et al. mRNA vaccines to prevent COVID-19 disease and reported allergic reactions: current evidence and suggested approach. J allergy Clin Immunol In Pract. 2021;9(4):1423–1437. doi: 10.1016/j.jaip.2020.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Z.H., Stone C.A., Jr., Jakubovic B., Phillips E.J., Sussman G., Park J., et al. Anti-PEG IgE in anaphylaxis associated with polyethylene glycol. J allergy Clin Immunol In Pract. 2021;9(4):1731–1733. doi: 10.1016/j.jaip.2020.11.011. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laczkó D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., et al. A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity. 2020;53(4):724–732. doi: 10.1016/j.immuni.2020.07.019. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rauch S., Roth N., Schwendt K., Fotin-Mleczek M., Mueller S.O., Petsch B. mRNA-based SARS-CoV-2 vaccine candidate CVnCoV induces high levels of virus-neutralising antibodies and mediates protection in rodents. NPJ vaccine. 2021;6(1):57. doi: 10.1038/s41541-021-00311-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dolgin E. CureVac COVID vaccine let-down spotlights mRNA design challenges. Nature. 2021;594(7864):483. doi: 10.1038/d41586-021-01661-0. [DOI] [PubMed] [Google Scholar]
- 65.de Alwis R., Gan E.S., Chen S., Leong Y.S., Tan H.C., Zhang S.L., et al. A single dose of self-transcribing and replicating RNA-based SARS-CoV-2 vaccine produces protective adaptive immunity in mice. Mol Ther. 2021;29(6):1970–1983. doi: 10.1016/j.ymthe.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang N.N., Li X.F., Deng Y.Q., Zhao H., Huang Y.J., Yang G., et al. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182(5):1271–1283. doi: 10.1016/j.cell.2020.07.024. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., et al. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat Commun. 2020;11(1):3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kobiyama K., Imai M., Jounai N., Nakayama M., Hioki K., Iwatsuki-Horimoto K., et al. Optimization of an LNP-mRNA vaccine candidate targeting SARS-CoV-2 receptor-binding domain. bioRxiv. 2021:2021. 03.04.433852. [Google Scholar]