ABSTRACT
During convergent differentiation, multiple developmental lineages produce a highly similar or identical cell type. However, few molecular players that drive convergent differentiation are known. Here, we show that the C. elegans Forkhead transcription factor UNC-130 is required in only one of three convergent lineages that produce the same glial cell type. UNC-130 acts transiently as a repressor in progenitors and newly-born terminal cells to allow the proper specification of cells related by lineage rather than by cell type or function. Specification defects correlate with UNC-130:DNA binding, and UNC-130 can be functionally replaced by its human homolog, the neural crest lineage determinant FoxD3. We propose that, in contrast to terminal selectors that activate cell type-specific transcriptional programs in terminally differentiating cells, UNC-130 acts early and specifically in one convergent lineage to produce a cell type that also arises from molecularly distinct progenitors in other lineages.
KEY WORDS: C. elegans, FoxD3, Cell lineage, Convergent differentiation, Glia, UNC-130
Summary: A transient transcriptional repressor is required in only one of three lineages that produce the same cell type.
INTRODUCTION
Development is the story of how a single cell divides to give rise to lineages that produce every cell type in the body. The standard framework for understanding this process is that cell lineages branch to produce increasingly divergent cell states, with each cell type produced exclusively by a single branch (Fig. 1A). An exception to this paradigm is convergent differentiation, a phenomenon in which multiple lineages produce identical or highly similar cell types (Fig. 1B). For example, mesoderm and neural crest lineages both produce the same type of heart cell (Dupin et al., 2018; Keyte and Hutson, 2012), and embryonic and extra-embryonic lineages both produce gut endoderm (Kwon et al., 2008). With the advent of single-cell RNA profiling coupled to lineage tracing, it is now appreciated that convergent differentiation is surprisingly prevalent in vertebrates (Chan et al., 2019; Liu et al., 2019; McKenna and Gagnon, 2019; Wagner et al., 2018). However, the molecular players that drive convergent differentiation remain unclear.
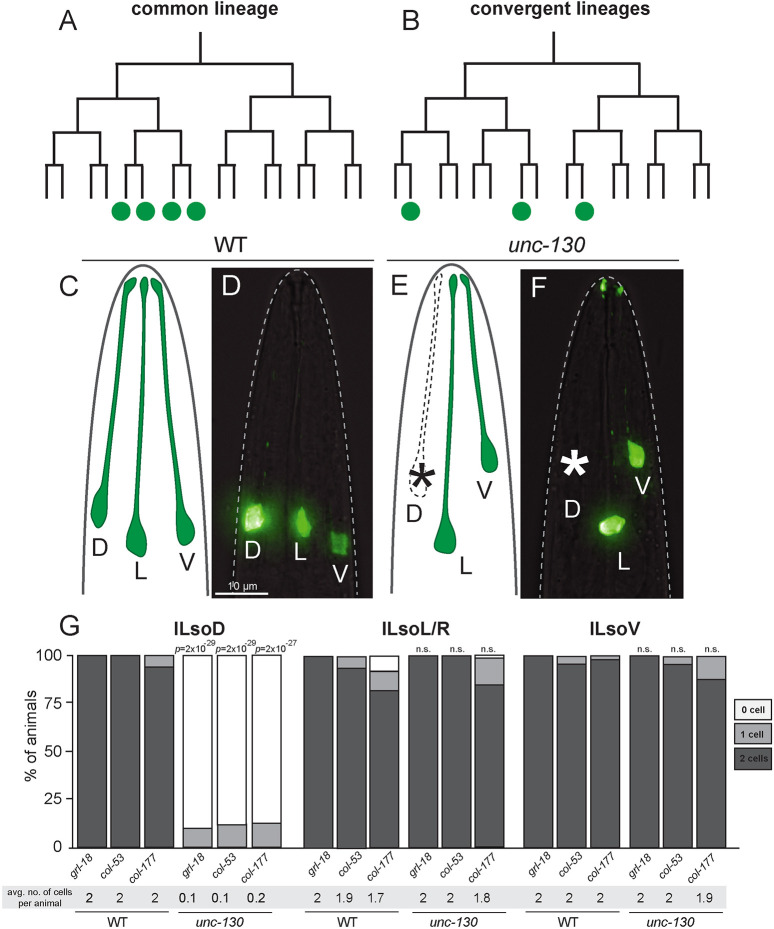
Fig. 1.
UNC-130 is required in one of three convergent lineages. (A,B) Schematic of a common lineage (A), in which shared progenitors give rise to a unique cell type, and a convergent lineage (B), in which distinct progenitors in divergent lineages produce a unique cell type. (C-F) Lateral views and schematics of wild-type (C,D) and unc-130 mutant (E,F) animals expressing an ILso marker, grl-18pro:YFP. Asterisks indicate missing cells. D, dorsal; L, lateral; V, ventral. (G) Percentage of wild-type and unc-130 mutant animals expressing ILso markers grl-18, col-53 and col-177 in zero, one or two dorsal (ILsoD), lateral (ILsoL/R) and ventral (ILsoV) glia. Average number of cells expressing each marker per animal is listed under each condition. n=50 animals per marker and genotype. P-values were calculated by Fisher's Exact test.
Importantly, in C. elegans, convergent differentiation has been appreciated for decades, ever since the complete developmental lineage was mapped (Sulston et al., 1983). A number of cell types that are present in symmetric anatomical regions – such as the dorsal and ventral equivalents of a given cell type – are produced through convergent differentiation by multipotent sublineages, which we will refer to as convergent lineages. A key issue for understanding convergent differentiation is to determine whether progenitors always follow the same path to produce a given cell type – that is, are shared or lineage-specific transcriptional trajectories employed across lineages that produce the same cell type? Intriguingly, several C. elegans mutants affect only a subset of convergently derived cells: for example, mls-2 mutations affect ventral, but not dorsal, CEP sheath glia; lin-32 mutations affect dorsal, but not ventral CEP neurons; and ceh-10 mutations affect dorsal, but not lateral or ventral RME neurons (Doitsidou et al., 2010; Forrester et al., 1998; Rojo Romanos et al., 2017; Yoshimura et al., 2008). These findings suggest that convergent lineages use distinct transcriptional trajectories to specify the same cell type. However, the mechanism by which transcription factors act across lineages to mediate convergent differentiation is not understood.
Most C. elegans glia are located in symmetrical groups of sense organs in the head, called the inner labial (IL), outer labial (OL), cephalic (CEP) and amphid (AM) sensilla. Each organ contains exactly two glial cell types – the sheath and socket. We focused on the development of the IL socket glia, which are sixfold radially symmetric, such that there is a dorsal, lateral and ventral pair of cells (ILsoD, ILsoL/R and ILsoV, respectively) (Mizeracka and Heiman, 2015). We recently identified specific markers for ILso glia and determined a role for these glia during dendrite extension of associated sensory neurons (Cebul et al., 2020). ILso glia develop via convergent differentiation – all three pairs of cells arise from distinct lineages that diverge at early stages of embryogenesis (Sulston et al., 1983). Importantly, all three pairs of ILso glia express the same reporter genes and appear as a uniform cluster in single-cell profiling experiments (Packer et al., 2019). In contrast, the three lineage-specific pairs of ILso parent cells cluster separately, suggesting that these progenitors are molecularly distinct. In single-cell RNA-sequencing studies, the ILso parent cells could not be identifiably linked to their terminal progeny, and thus were interpreted to develop through a ‘discontinuous’ transcriptional trajectory (Packer et al., 2019). This may be because the transcriptomes of ILso parent cells change quickly after their terminal divisions as they undergo convergent differentiation, thereby making it difficult to identify factors that are important for this process.
Here, we find a lineage-specific role for the conserved Forkhead transcription factor UNC-130 during the specification of a convergently derived glial cell type in C. elegans. We show that, during embryogenesis, UNC-130 is required for the specification of ILso cells that are derived from one convergent lineage, but is dispensable for the production of ILso cells derived from other lineages. Furthermore, consistent with previous work (Sarafi-Reinach and Sengupta, 2000), mutations in unc-130 perturb the specification of several cell types that are related by lineage, but not by function. We find that UNC-130 is a transiently expressed transcriptional repressor that acts at the time of birth of the ILsoD glia. The vertebrate homolog of UNC-130, FoxD3, also acts in a lineage-specific manner and is required for the specification of neural crest-derived cell types (Kos et al., 2001; Lister et al., 2006; Stewart et al., 2006). Intriguingly, we find that UNC-130 and FoxD3 share molecular features, including their preferred binding sites, and FoxD3 can functionally replace UNC-130. Lineage-specific regulatory factors such as UNC-130 and FoxD3 may represent an evolutionarily ancient mechanism that enables molecularly distinct progenitors in different lineages to produce the same cell type.
RESULTS
unc-130 mutants display defects in convergent differentiation
To identify genes controlling ILso glial fate, we performed a chemical mutagenesis screen and isolated a mutant strain in which one pair of ILso cells was missing marker expression (see Materials and Methods). Genetic mapping and sequencing revealed a causal mutation in the unc-130 gene, which encodes a conserved Forkhead transcription factor. We obtained the reference allele for this gene, the deletion strain ev505, and characterized sense organ perturbations in detail (Nash et al., 2000). In previous work, we identified the gene grl-18, which encodes a poorly characterized protein that contains a ground-like domain, as well as col-53 and col-177, both of which encode collagens, as highly specific markers for ILso glia (Cebul et al., 2020; Fung et al., 2020). Using these markers, we find that 100% of unc-130 mutants lost expression of all three known ILso markers (grl-18 pro, col-53 pro and col-177 pro) specifically in one or both dorsal ILso glia (ILsoD), but not in lateral (ILsoL/R) or ventral (ILsoV) glia (Fig. 1C-G, Table S1; see Materials and Methods). ILso glia are associated in sense organs with two neurons called IL1 and IL2, and the IL sheath glial cell (ILsh). We find that loss of marker expression was specific to ILsoD glia – unc-130 mutants occasionally have extra IL1 neurons and wild-type numbers of IL2 neurons (Table S2). Lack of specific reporters precluded examination of ILsh glia.
Loss of marker expression in ILsoD glia in unc-130 mutants could be due to defects in the cell division pattern of the lineage that normally produces these cells. For example, the progenitor cell could fail to divide, or the presumptive ILsoD cells could undergo cell death. To test these possibilities, we performed lineaging analysis on unc-130 mutants to track all cell divisions in this sublineage. In 8/8 unc-130 mutant embryos, we found that ILsoD progenitors divide to produce presumptive ILsoD glia and their sister cell, the skin cell hyp3 (Fig. S1). However, we found that the timing of this cell division was delayed compared with wild-type embryos. Cell cycle lengths of the other ILso progenitor cells were also slightly delayed in unc-130 mutants, although these cells were not as strongly affected as ILsoD progenitors (Fig. S1). These results provide evidence that the putative ILsoD cells are born at approximately the right time in unc-130 mutants, although the ILsoD progenitor may be abnormal, as reflected by its delayed cell division. We conclude that loss of marker expression in ILsoD glia in unc-130 mutants is likely due to a loss of identity, rather than to changes in the division patterns of the ILsoD lineage.
UNC-130 is required to produce cell types that are related by lineage, not by identity or function
We wanted to determine whether UNC-130 acts in ILsoD glia specifically or more broadly at the level of a sublineage. Previous work showed that UNC-130 is required for the specification of three sensory neurons (AWA, ASG and ASI) that are produced by an exclusively neuronal sublineage that is most closely related to the ILsoD sublineage (Fig. 2A) (Sarafi-Reinach and Sengupta, 2000). This left open the possibility that UNC-130 is specific to neuronal fates. In contrast, the ILsoD sublineage is multipotent, giving rise to two types of glia, a neuron and a skin cell (Fig. 2A) (Sulston et al., 1983).
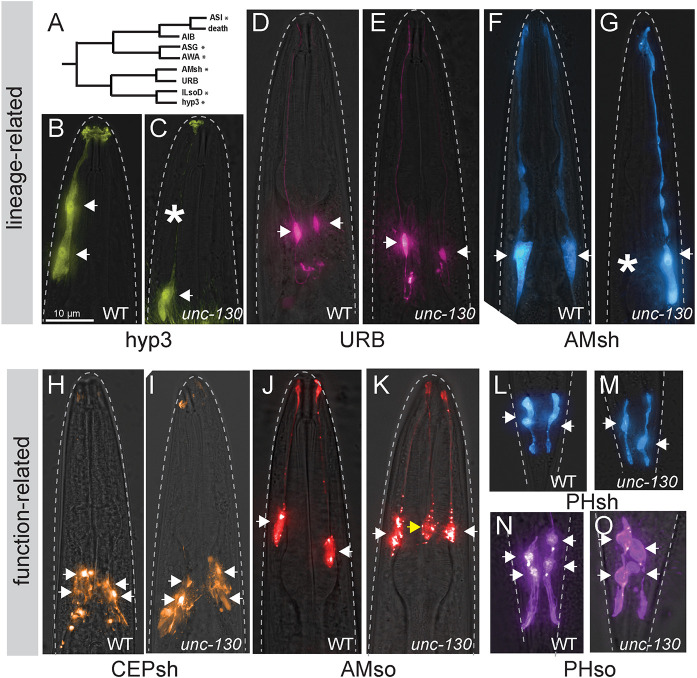
Fig. 2.
UNC-130 is required for the specification of lineage-related cell types. (A) Schematic of the ILsoD sublineage. Asterisks indicate cell types in which specification is affected in unc-130 mutants. (B-O) Wild-type (B,D,F,H,J,L,N) and unc-130 mutant (C,E,G,I,K,M,O) animals expressing a hyp3 marker (ceh-10pro:GFP; B,C), a URB marker (nlp-6pro:GFP; D,E), an AMsh marker (F16F9.3pro:mCherry; F,G), a CEPsh marker (hlh-17pro:GFP; H,I), an AMso marker (grl-2pro:mCherry; J,K), a PHsh marker (F16F9.3pro:mCherry; L,M) and a PHso marker (grl-2pro:YFP; N,O). Asterisks indicate missing cells. Arrows indicate cell bodies in all images. Yellow arrow indicates an extra AMso cell.
To systematically determine the requirement for UNC-130 in the ILsoD sublineage, we examined markers of each cell type: the hyp3 skin cell, the URB neuron and the amphid sheath (AMsh) glial cell (Fig. 2A). In wild-type animals, there are two hyp3 cells whose cell bodies are located dorsally and fuse to form a syncytium. By mining single-cell transcriptome and reporter expression data we identified ceh-10pro:GFP as a hyp3-specific marker (Packer et al., 2019; Wenick and Hobert, 2004; Fig. 2B). ceh-10 encodes a conserved ortholog of the transcription factor CHX10 that is also expressed in unrelated neurons (Altun-Gultekin et al., 2001; Forrester et al., 1998). We found that in 30% of unc-130 mutants, one or sometimes both hyp3 cells lacked marker expression (Fig. 2B,C, Table S3). Next, we used the same approach to identify a unique marker for the sensory neuron URB, the gene nlp-6, which encodes an uncharacterized neuropeptide (Packer et al., 2019, Fig. 2D). URB neurons form a bilateral pair on either side of the head (Fig. 2D). Specification of URB appeared unaffected in unc-130 mutant animals, as these cells exhibited unchanged expression of nlp-6pro:GFP (Fig. 2E, Table S3). In contrast, specification of AMsh glial cells, which are sister cells of URB and are found in the bilaterally symmetric amphid sense organ, was affected, with 38% of unc-130 mutants failing to express an AMsh-specific marker, F16F9.3, which encodes a secreted peptide, in one or occasionally both cells (Fig. 2F,G, Table S3). Loss of F16F9.3 expression was highly correlated with loss of two other AMsh glia-specific markers: T02B11.3pro:GFP and F53F4.13pro:GFP (Fig. S2A). Through transcriptional profiling experiments, these three markers have been determined to be highly specific for the AMsh, but their functions remain unknown (Bacaj et al., 2008; Fung et al., 2020; Wallace et al., 2016). Thus, we find that the specification of non-neuronal cell types that are derived from the ILsoD sublineage is affected in the absence of UNC-130.
Cell types that share an identity, e.g. neurons that express the same neurotransmitter, often express the same combination of transcription factors. To test whether UNC-130 affects cell types that are functionally related to ILso, we examined markers for other glial cell types. We found no defects in the specification of the four cephalic sheath glia (CEPsh); two phasmid sheath glia (PHsh), which are the functional equivalent of the AMsh glia in the tail and express many of the same markers; or four phasmid socket glia (PHso) (Fig. 2H,I,L-O, Table S3) (Fung et al., 2020; McMiller and Johnson, 2005; Yoshimura et al., 2008).
Surprisingly, we observed extra amphid socket (AMso) glia in unc-130 mutants, with 42% of mutant animals expressing the AMso-specific marker grl-2 in an extra cell (Fig. 2J,K, Table S3) (Hao et al., 2006). grl-2 encodes an uncharacterized protein that includes a ground-like domain (Hao et al., 2006). We found that expression of grl-2 in the extra cell was highly correlated with expression of two other AMso-specific markers: lin-48, which is the C. elegans ortholog of Ovo-like transcription factors in vertebrates; and itr-1, which encodes an inositol triphosphate receptor (Fig. S2C) (Fung et al., 2020; Heiman and Shaham, 2009; Johnson et al., 2001; Low et al., 2019). Because lin-48 encodes a highly conserved transcription factor that is expressed specifically in AMso, it is possible that it also plays a role in the specification of these cells. However, we found that mutations in lin-48 do not affect endogenous or ectopic AMso specification (Fig. S2D). We considered the possibility that the extra AMso glia could reflect duplication of the entire sublineage that normally gives rise to the AMso. Therefore, we examined specification of the CEM neuron, which is the sister cell of AMso glia in males, in wild-type and unc-130 animals. No extra CEM neurons were observed in mutant males, providing evidence that ectopic AMso do not arise from a sublineage duplication (Fig. S2B). Finally, we noted that, in contrast to endogenous AMso cells, which are positioned laterally, extra AMso cells were located dorsally, consistent with the position of the missing ILsoD and hyp3 cells (Fig. S2E). This raises the possibility that the extra AMso cells might arise by mis-specification of ILsoD or hyp3, or both, although, due to the timing of marker expression in the embryo, we were unable to test this directly.
Taken together, these observations suggest that, in the absence of UNC-130, several cell types – including neurons, glia, and skin – fail to be specified or possibly take on alternative fates. Importantly, defects in fate specification in unc-130 mutants are not solely related to the identity or function of a particular cell, but rather to cell types that share a lineage origin.
UNC-130 acts in lineage-specific progenitors and newly born precursor cells
Unlike terminal selectors, loss of UNC-130 affects several cell types, suggesting that it might act earlier in development. Indeed, previous work showed that UNC-130 is expressed in the immediate progenitors but not in the terminal cells of affected sensory neurons (Sarafi-Reinach and Sengupta, 2000). We examined a translational reporter strain in which UNC-130 was fused with GFP and acquired a time course of images during C. elegans embryogenesis (Sarov et al., 2006). Consistent with previous work and our lineaging experiments, UNC-130:GFP is brightly expressed at early stages of development (∼300 min to 500 min), and starts to wane as animals near hatching at 3-fold stages (550+ min) (Fig. 3A-G). In contrast, expression of the IL socket-specific marker grl-18 is not detectable at early stages, starts to be expressed in 2-fold embryos (500 min), and stays on into adulthood (Fig. 3H-N). These results provide evidence that unc-130 is expressed early in development, in progenitor cells and presumptive ILso cells after they are born, but its expression is downregulated as these cells differentiate.
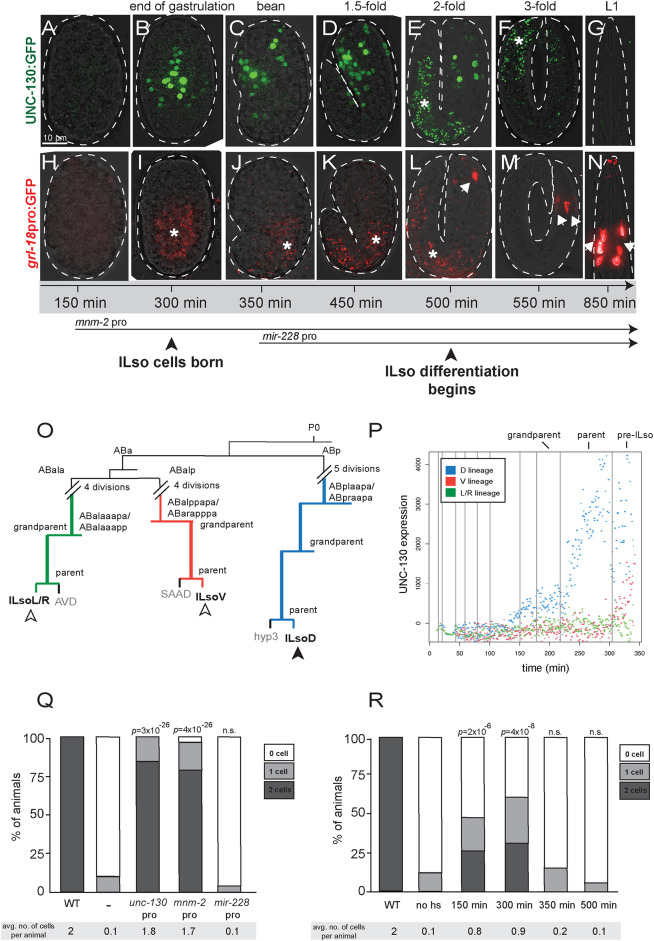
Fig. 3.
UNC-130 is expressed in a restricted lineage before the onset of differentiation. (A-G) Time course of embryos expressing UNC-130:GFP. (H-N) Time course of embryos expressing grl-18pro:GFP (pseudo-colored red). Asterisks indicate gut autofluorescence. Arrows indicate grl-18+ ILso cells. (O) Lineage diagram of a subset of embryonic cell divisions derived from the AB blastomere that gives rise to ILsoD (blue), ILsoV (red) and ILsoL/R (green) glia. (P) Levels of UNC-130:GFP signal in ILsoD (blue), ILsoV (red) and ILsoL/R (green) lineages over developmental time. Vertical lines indicate cell divisions. Subtraction of background fluorescence signal results in negative values for some cells. (Q) Heterologous promoters were used to drive expression of unc-130 in the unc-130 mutant strain, and the extent of rescue was assessed. Timing of mnm-2pro and mir-228pro expression is marked in A. Percentage of animals expressing grl-18pro:YFP in zero, one or two ILsoD glia in each condition. Average number of cells marked per animal is listed under each condition. n=50 animals per condition. P-values were calculated by Fisher's Exact test. (R) unc-130 mutant embryos expressing hsp:unc-130 were heat shocked at different time points and the extent of rescue was assessed. Percentage of animals expressing grl-18pro:YFP in zero, one or two ILsoD glia in each condition. Average number of cells marked per animal is listed under each condition. n=50 animals per condition. P-values were calculated by Fisher's Exact test.
We wanted to understand why mutation of unc-130 affects the specification of the ILsoD pair of glia, but not the equivalent lateral or ventral pairs. We noted that the lineages that give rise to the three pairs of ILso glia diverge at the 4- and 16-cell stages (Fig. 3O) (Sulston et al., 1983). Previous studies showed that unc-130 expression commences midway through embryogenesis in an ABp-derived lineage that gives rise to dorsal – but not lateral or ventral – ILso glia (Murray et al., 2012; Sarafi-Reinach and Sengupta, 2000). To better define this expression pattern, we performed UNC-130 lineaging experiments and extended the period of data acquisition to capture the birth of the presumptive ILso cells. We found that unc-130 is highly expressed in the ILsoD lineage and in the presumptive ILsoD cell after it is born, but not in the lineages that give rise to the ILsoL/R or ILsoV cells (Fig. 3P, Fig. S3). We did note low but increasing levels of expression in ILsoV glia after they are born. The significance of this is unclear, as ILsoV glia are specified normally in unc-130 mutants. Taken together, these findings show that UNC-130 is expressed at high levels in the progenitor that gives rise to the ILsoD glia around the time when the cells are born.
To functionally address when UNC-130 is required for fate specification, we used early and late promoters to drive unc-130 expression in unc-130 mutants and determined the extent of rescue of glial specification defects by scoring for the presence of grl-18+ cells in late larvae or young adults. For early rescue, we used a regulatory sequence we identified upstream of mnm-2, a gene that, similar to unc-130, is expressed early in the ILsoD sublineage but not in other lineages that normally express unc-130 (Fig. S4) (Murray et al., 2012). For late rescue, we used a sequence upstream of mir-228, which is expressed in all glial cells shortly after they are born (Fig. S4) (Fung et al., 2020; Pierce et al., 2008). We found that early unc-130 expression under the mnm-2 promoter in unc-130 mutants rescued specification of ILsoD glia almost as completely as expression driven by the unc-130 promoter (Fig. 3Q). In contrast, late expression of unc-130 driven by the mir-228 promoter did not rescue specification defects (Fig. 3Q). Intriguingly, expression of mir-228pro:unc-130 in unc-130 mutants resulted in a mild enhancement of specification defects such that a larger percentage of animals lacked grl-18 expression in ILsoD cells. This suggests that downregulation of unc-130 in newly born cells is important for fate specification.
In a complementary approach, we drove unc-130 expression using a heat-shock inducible promoter in unc-130 mutants to define a temporal window during which UNC-130 is required for fate specification. We found that heat-shock induction of UNC-130 at early stages of development resulted in a high lethality rate; therefore, we scored arrested embryos or larvae 12-18 h after heat shock. Heat shock-induced expression of unc-130 at ∼150 min (see Materials and Methods), before the birth of the ILsoD, resulted in moderate rescue, with 48% of animals expressing grl-18 in one or two ILsoD cells, compared with 12% of animals expressing grl-18 in one ILsoD cell in no heat-shock controls (Fig. 3R). Heat shock-induced expression of unc-130 at approximately the birth of ILsoD glia (300 min) resulted in the strongest rescue of fate specification defects, with 62% of animals expressing grl-18 in one or two ILsoD glia (Fig. 3R). Heat-shock induction after ∼350 min, which is after the ILsoD cells are born but before they start to differentiate, or any other later time points, did not result in significant rescue of specification defects when compared with no heat-shock controls (Fig. 3R).
Together, these results provide evidence that unc-130 acts transiently as ILsoD progenitors are dividing to produce ILsoD glia and shortly after these cells are born, but it cannot rescue specification defects after this temporal window has passed. This temporal requirement for UNC-130 before the onset of differentiation suggests that, rather than directly instructing cell fate, UNC-130 establishes competency for fate specification.
UNC-130 functions as a repressor
The UNC-130 homolog FoxD3 acts as a repressor in several developmental contexts, in some cases recruiting the Groucho repressor complex through a conserved engrailed homology (eh1) domain (Ono et al., 2014; Yaklichkin et al., 2007a). Paradoxically, FoxD3 also functions as a pioneer factor in embryonic stem cells and during early neural crest specification (Krishnakumar et al., 2016; Lukoseviciute et al., 2018). To determine whether UNC-130 acts as a repressor or an activator during convergent differentiation, we expressed synthetic constructs encoding the UNC-130 DNA-binding domain (DBD) fused to a canonical activator (VP64 – four copies of VP16 activator domain) or repressor (Drosophila Engrailed domain) under the unc-130 promoter in an unc-130 mutant strain and assessed rescue of glia specification defects by scoring for the presence of grl-18+ cells in late larvae or young adults (Fig. 4A,B). Expression of full-length UNC-130 fully rescued ILsoD glia defects (Fig. 4B). Expression of the UNC-130 DBD alone or a DBD-VP64 synthetic activator showed no rescue (Fig. 4B). In contrast, the synthetic repressor DBD-Engrailed substantially rescued ILsoD specification defects, suggesting that UNC-130 normally functions as a transcriptional repressor (Fig. 4B).
Fig. 4.
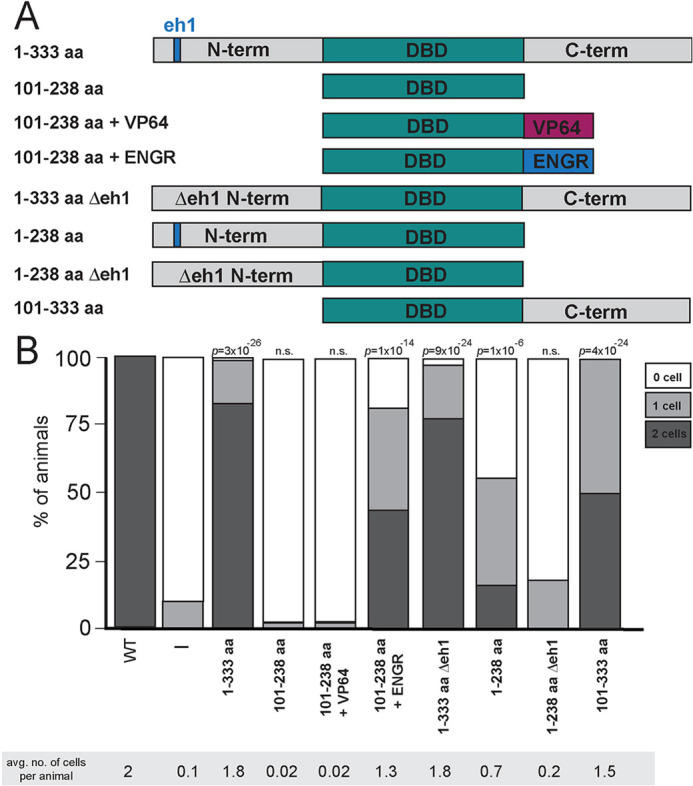
UNC-130 acts as a repressor to specify glial fates. (A) Schematic diagram of UNC-130 constructs used in rescue experiments to determine domain function. Full-length UNC-130 is 333 amino acids in length. (B) The unc-130 promoter was used to drive expression of individual constructs (schematized in A) in the unc-130 mutant strain and extent of rescue was assessed. Percentage of animals expressing grl-18pro:YFP in zero, one or two ILsoD glia in each condition. Average number of cells marked per animal is listed under each condition. n=50 animals per condition. P-values were calculated by Fisher's Exact test.
The vertebrate UNC-130 homolog FoxD3 contains an eh1 motif in its C-terminal domain that recruits the Groucho repressive complex. This region of UNC-130 is not conserved, but we identified a candidate eh1 sequence in the UNC-130 N-terminal domain and tested whether it might act similarly (Yaklichkin et al., 2007b) (Fig. 4A, Figs S5 and S6A). Surprisingly, deletion of the eh1 domain from full-length UNC-130 did not prevent rescue of specification defects, owing to a redundant function of the C terminus, as discussed below (Fig. 4, 1-333 aa Δeh1). We found that expression of a protein containing the UNC-130 N terminus and DBD partially rescued ILsoD defects, and the candidate eh1interaction motif was necessary for this rescuing function (Fig. 4, 1-238 aa and 1-238 aa Δeh1).
Interestingly, expression of the DBD with the C terminus also rescued all glial specification defects, which suggests it might also contain previously unidentified repressor motifs (Fig. 4, 101-333 aa). Using luciferase assays in cultured mammalian cells, we found that N-term:GAL4DBD:C-term and GAL4DBD:C-term have reduced reporter activity compared with GAL4 DBD alone (Fig. S5B,C), consistent with the C terminus harboring repressive activity.
We independently tested the role of the Groucho complex by examining mutants in unc-37, the sole C. elegans ortholog of Groucho. unc-37 mutants did not display any defects in ILso glial specification, which provides further evidence that UNC-130 harbors a redundant, Groucho-independent repressive motif (Fig. S5A). By contrast, we found that rescue with the N terminus alone shows a strong dependence on unc-37, providing evidence that the N-terminal domain acts through recruitment of the Groucho repressive complex akin to vertebrate Forkhead factors (Fig. S5A).
Thus, we find that UNC-130 promotes ILsoD fate by acting as a transient transcriptional repressor at the time when these glial cells are produced. One possibility is that UNC-130 directly represses genes that would activate alternative fates, allowing presumptive ILsoD glia to follow the correct transcriptional trajectory.
UNC-130 and the neural crest determinant FoxD3 share a conserved function
The UNC-130 DBD is highly conserved with its vertebrate homolog FoxD3 (Fig. 5A, Fig. S6A); thus, we wanted to determine whether they share DNA-binding specificities. Different Forkhead transcription factors can bind to a consensus primary motif RYAAAYA (FkhP), a related secondary motif AHAACA (FkhS) or an unrelated alternate motif GACGC (FHL) (Nakagawa et al., 2013). To determine its binding specificity, we incubated recombinant UNC-130-DBD protein with universal protein-binding microarrays (PBMs) containing all possible 10-mer double-stranded DNA sequences (Berger et al., 2006). We found that UNC-130-DBD preferentially binds [A/G][T/C]AAACA and AA[T/C]AACA sequences, variants of the primary and secondary Forkhead binding motifs, respectively, consistent with it behaving as a conserved Forkhead family member (Fig. 5B).
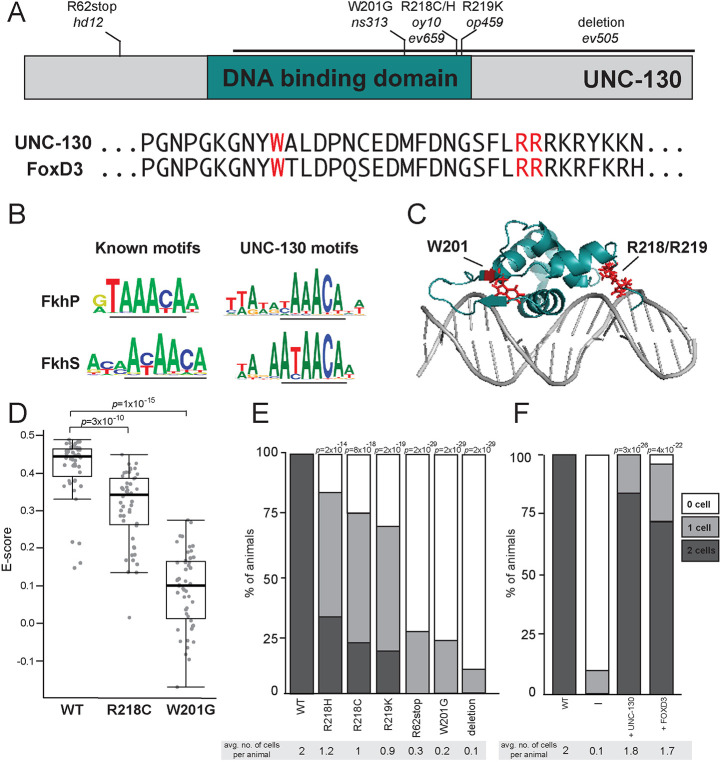
Fig. 5.
Severity of glial phenotypes correlate with UNC-130:DNA-binding defects. (A) Schematic diagram of UNC-130 protein with DNA-binding domain (turquoise) highlighted. Location of point mutations and a deletion are indicated. Alignment of UNC-130 and FoxD3 DBD with point mutations highlighted in red. (B) Logos of vertebrate primary (FkhP, top) and secondary (FkhS, bottom) Forkhead motifs and UNC-130 preferred DNA sequences that resemble primary (top) and secondary (bottom) motifs, as determined by this study. Core motifs are underlined. (C) Structure of FoxD3 DBD (turquoise) interacting with DNA (gray) (Protein Data Bank, https://www.rcsb.org; ID code 2HDC; Jin et al., 1999). W201, R218 and R219 residues are highlighted in red. (D) Scatter plot of E-scores for 8-mer DNA sequences matching [A/G][C/T]AAACA or AA[C/T]AACA from protein-binding microarray assays of wild-type, R218C and W201G mutant UNC-130 proteins. Black lines represent population median; top and bottom of boxes are 25th and 75th percentiles, respectively; and top and bottom of whiskers are either most extreme point or 1.5× the interquartile range. P-values were calculated by Mann–Whitney test. (E) Percentage of animals expressing grl-18pro:YFP in zero, one or two ILsoD glia in wild-type and unc-130 mutant strains. Average number of cells marked per animal is listed under each condition. n=50 animals per genotype. P-values were calculated by Fisher's Exact test. (F) The unc-130 promoter was used to drive expression of unc-130 and human FOXD3 separately in the unc-130 mutant strain and the extent of rescue was assessed. Percentage of animals expressing grl-18pro:YFP in zero, one or two ILsoD glia in each condition. Average number of cells marked per animal is listed under each condition. n=50 animals per condition. P-values were calculated by Fisher's Exact test.
From our mutant screen and previous studies, we assembled a collection of point mutations in the UNC-130 DBD: ns313 (W201); oy10 and ev659 (R218); and op459 (R219) (Nash et al., 2000; Sarafi-Reinach and Sengupta, 2000) (Fig. 5A, Fig. S6). Mapped onto the FoxD3 structure, W201 is located in a β-sheet that flanks the central helical bundle that inserts into the major groove of DNA, whereas the R218 and R219 residues are in a wing domain on the opposite side of the helix bundle (Fig. 5C) (Jin et al., 1999). To assess the effect of the W201G and R218C point mutations, we applied mutant UNC-130 recombinant proteins separately onto PBMs. We found that the R218C protein still preferentially binds DNA sequences recognized by wild-type UNC-130 but with lower affinity (Fig. 5D, Fig. S6), whereas the W201G mutant protein did not exhibit sequence-specific preferences for these or any other motifs, suggesting severely impaired DNA binding (Fig. 5D, Fig. S6). These binding defects strongly correlated with in vivo function. The W201G mutant exhibited defects in ILsoD specification that are nearly as strong as an early stop mutant (R62stop) or a deletion allele, such that >90% of ILsoD glia lose reporter expression (Fig. 5E). In contrast, in hypomorphic alleles, which include R218C, R218H and R219K, only 50% of ILsoD glia lose reporter expression (Fig. 5E). Thus, moderately impaired DNA binding appears to promote weak ILsoD glial defects, and severely impaired DNA binding results in strong ILsoD glial defects. We find that these highly conserved amino acid residues are crucial for UNC-130 function, and are likely to similarly disrupt FoxD3 function in vertebrates.
In vertebrates, FoxD3 is required for the specification of several cell types that arise from the neural crest, a multipotent lineage that participates in convergent differentiation (Dottori et al., 2001; Kos et al., 2001; Lister et al., 2006; Lukoseviciute et al., 2018; Sasai et al., 2001; Stewart et al., 2006; Teng et al., 2008). Although examination of UNC-130 and human FOXD3 protein sequences revealed little similarity outside of the highly conserved DBD and eh1 motif (Fig. 5A, Fig. S6), we found that expression of an unc-130 promoter::FOXD3 cDNA transgene almost completely rescues ILsoD glial specification defects in unc-130 mutants (Fig. 5F). This suggests that UNC-130 and FoxD3 functionality is conserved despite divergence at the primary sequence level.
In summary, although C. elegans lacks a neural crest, the similarities between FoxD3 and UNC-130 extend from their roles in lineage specification to their molecular mechanisms of action – including their preferred DNA-binding sites and their roles as transcriptional repressors, likely via an interaction with the Groucho repressive complex. More broadly, our results lead to the speculation that before the evolution of the neural crest, a FoxD3 precursor already existed that acted in a multipotent sublineage that exhibits convergent differentiation.
DISCUSSION
Unraveling the mechanisms that establish fate competence in progenitors and precursors is crucial to understanding how developmental lineage is coupled to cell fate – why do certain cell types arise only from particular lineages? The special case of convergent differentiation promises to offer important insights into this relationship.
An implication of our findings and previous studies is that different progenitors can take distinct transcriptional paths to produce the same cell type. We find that UNC-130 is only required in one of three convergent lineages to produce the ILso glial cell type. Although convergent differentiation was not their focus, previous studies provide additional examples. For example, loss of mls-2/Nkx affects mainly ventral, but not dorsal, CEPsh glia (Yoshimura et al., 2008). Similar to ILso glia, dorsal and ventral CEPsh glia are derived from lineages that diverge early in development. MLS-2 also appears to function at the lineage level, as other studies have determined that cell types related to CEPshV glia by lineage are mis-specified in mls-2 mutants (Abdus-Saboor et al., 2012; Kim et al., 2010). Other examples include the requirement for LIN-32 to specify dorsal but not ventral CEP neurons, and for CEH-10 to specify dorsal but not lateral or ventral RME neurons (Doitsidou et al., 2010; Forrester et al., 1998; Rojo Romanos et al., 2017). These observations provide tantalizing starting points for characterizing other factors that are involved in convergent differentiation.
Cell-type specification in terminally differentiating cells, which is mediated by master regulator transcription factors called terminal selectors, has been extensively studied in a number of cell types (Hobert and Kratsios, 2019). In contrast, factors that act transiently in progenitor cells to establish lineage-specific identity are less well understood. Some candidates for this type of factor include CEH-36 and UNC-30, which function redundantly to regulate progenitor cell cycle progression and cell position in a number of developmental lineages (Walton et al., 2015), and CND-1, which regulates mitotic progression and the establishment of neuronal fate in lineages that will give rise to neurons (Hallam et al., 2000). Another example is the lineage-restricted transcription factor TBX-37/38, which is expressed transiently early in embryogenesis to prime a locus that does not become active until several cell cycles later in the mature cell type (Charest et al., 2020; Cochella and Hobert, 2012). Similarly, in this study, we find that UNC-130, consistent with previous expression data from Sarafi-Reinach and Sengupta (2000), acts in lineage-related progenitor cells before these cells make terminal divisions and their progeny start to differentiate. Therefore, the function of these early acting factors is likely to establish competency for fate specification, a role that differs from that of terminal selectors.
How does UNC-130 establish fate competency in a specific lineage? UNC-130 is required broadly for the specification of several unrelated cell types produced by the ILsoD sublineage. It is possible that UNC-130 primes and represses cell type-specific loci in each progenitor cell based on cellular context and binding partners. Alternatively, it may not impart any specific cell type information, but rather functions to transition progenitor cells to a more restricted state through the repression of pluripotency genes. Alternatively, or in addition, it may block off paths to alternate fates by repressing target genes that need to remain inactive in a particular lineage.
The results presented in this study are highly relevant to understanding the relationship between lineage and cell fate in the vertebrate nervous system, an intricate structure comprising many diverse cell types arising from a myriad of lineages. Single-cell RNA profiling of the mammalian brain has shown that the glial classes of astrocytes and microglia, which had long been thought to consist of molecularly homogeneous cells, actually exhibit striking region-specific molecular heterogeneity (Hammond et al., 2019; John Lin et al., 2017; Marques et al., 2016; Masuda et al., 2019; Morel et al., 2017; Spitzer et al., 2019; Zeisel et al., 2015, 2018). In contrast, the molecular signatures of oligodendrocytes are highly similar (Marques et al., 2018; Zeisel et al., 2018). Because astrocytes and oligodendrocytes are thought to share a common progenitor, these observations suggest that regionally distinct progenitor cells may also undergo convergent differentiation in the mammalian nervous system. Elucidation of these pathways will require single-cell profiling methods to be combined with careful lineage tracing and functional perturbation of regulatory factors in vivo to ultimately achieve the level of resolution that is available in the far simpler nervous system of C. elegans.
MATERIALS AND METHODS
Strains
Strains were constructed in the N2 background and cultured under standard conditions (Brenner, 1974). Transgenic strains were generated with standard techniques (Mello and Fire, 1995) with injection of 100 ng/µl of DNA (5-50 µl per plasmid). Strains, plasmids and primers are listed in Tables S4-S7.
Isolation and mapping of unc-130 alleles
We isolated an allele of unc-130, ns313, from a genetic screen for sense organ abnormalities. Animals of genotype oyIs44 were mutagenized using 70 mM ethyl methanesulfonate (EMS, Sigma) at 20°C for 4 h. Nonclonal F2 progeny were examined on a fluorescence stereomicroscope and animals with sense organ defects were recovered. A mutant strain, ns313, exhibiting short amphid dendrites (14% penetrance) and amphid sheath migration defects (55% penetrance), was isolated.
With standard linkage mapping and SNP analysis (Wicks et al., 2001), ns313 was mapped to an interval between −6 cM and 15 cM on LG II. ns313 animals were crossed to the Hawaiian strain CB4856 and F2 progeny with the mutant phenotype were transferred to individual plates. All F3 recombinants were pooled and subjected to genomic DNA extraction and whole-genome sequencing for one-step mapping (Doitsidou et al., 2010). Analysis with CloudMap (Minevich et al., 2012) identified a linked region on LG II, including a point mutation in unc-130 [GAACTAT(T>G)GGGCGTGGA] (W201G).
Characterization of glial phenotypes
To score regional defects in IL sockets, we generated strains that co-expressed the ILso marker (hmnIs47 [grl-18pro:mApple]) with a marker for URX (ynIs48 [flp-8pro:GFP]), a dorsally located neuron whose dendrites fasciculate with the processes of the dorsal, but not lateral or ventral, ILso glia as a landmark. Glial specification defects were scored visually on either a Nikon SMZ1500 stereomicroscope with an HR Plan Apo 1.6× objective or a Deltavision Core imaging system (Applied Precision) with UApo/340 40×1.35NA objective (Olympus).
Fluorescence microscopy and image processing
Animals were mounted on 2% agarose pads in M9 buffer (Sulston et al., 1983) with 50-100 mM sodium azide depending on developmental stage, and imaged using a Deltavision Core imaging system (Applied Precision) with UApo/340 40×1.35NA, PlanApo 60×1.42NA and U-PlanApo 100×1.4NA objectives (Olympus) and CoolSnap HQ2 camera. Images were deconvolved using Softworx (Applied Precision) and maximum-brightness projections were obtained from contiguous optical sections using ImageJ.
Lineaging analysis
Cell lineage analysis was performed using the StarryNite/AceTree cell tracking system (Bao et al., 2006; Boyle et al., 2006; Santella et al., 2010). Embryos from strains RW11144 [ev505 ujIs113; wgIs476] and CHB3933 [ujIs113; wgIs746] were imaged on a Leica SP5 resonance-scanning confocal microscope with approximately 1.5 min time point spacing and 0.5 μm z-resolution as previously described (Richards et al., 2013). Computational cell tracking of histone-mCherry images was used to track cells across time and identify the time of all terminal divisions in the lineages leading to ILsoD [ABp(l/r)aapa], ILsoL/R (ABalaaap) and ILsoV (ABalppap and ABarappp).
Heat shock-induced expression
Strain CHB4158 (ev505; hmnEx2283 [hsp16-2pro:unc-130+hsp16-41pro:unc-130+grl-18pro:YFP+unc-122pro:RFP]) was grown at 20°C prior to heat-shock experiments. Age of embryos was determined by morphological state. Embryos at different developmental stages were sorted onto separate plates and heat-shocked at 34°C for 60 min to induce expression of UNC-130. After heat-shock, animals were grown at 20°C for 12-18 h, and embryos and larvae were scored for grl-18pro:YFP expression by fluorescence microscopy as described above.
PBM experiments and data analysis
PBM experiments were performed on universal ‘all-10-mer’ arrays in 8X60K format (Agilent, AMADID 030236) (Berger et al., 2006; Nakagawa et al., 2013). PBM experiments were performed at 500 nM protein concentration in the standard protein-binding reaction mixture, substituting buffer A for PBS [buffer A: 138 mM KGlu, 12 mM NaHCO3, 0.8 mM MgCl2 (pH 7.2)] in the standard PBM protocol (Berger and Bulyk, 2009). Protein binding was detected with an Alexa488-conjugated anti-GST antibody (Life Technologies A-11131), and arrays were scanned using a GenePix 4400A (Molecular Devices) microarray scanner. Binding was quantified using the Universal PBM Analysis Suite (Berger and Bulyk, 2009) to generate E-scores for each 8-mer. Motifs were derived using the Seed-and-Wobble algorithm (Berger et al., 2006; Berger and Bulyk, 2009). Two replicate experiments were performed, with replicate 1 having higher E-scores overall. Replicate 1 is shown in Fig. 5D; replicate 2 is shown in Fig. S6B.
Boxplots were generated in R, from the E-scores of the 8-mer sequences that match the UNC-130 FkhP motif [(AG)(CT)AAACA] or the FkhS motif [AA(CT)AACA]. Individual data points are displayed on the boxplots using the stripchart function in R. Significant differences in binding were evaluated using a one-sided Mann–Whitney test, with the Wilcox.test function in R (https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/wilcox.test).
Luciferase assays
HEK293T cells were cultured at 37°C and transfected with Fugene (Roche). 48 h post transfection, cells were collected in ice-cold 1×PBS and transferred into 96-well plates. Renilla and firefly luciferase activity were assayed according to manufacturer's instructions using the Dual-Glo assay (Promega), and bioluminescence was collected on a Molecular Devices Spectramax Paradigm plate reader. Firefly luciferase activity was normalized to renilla luciferase activity in each sample.
Supplementary Material
Acknowledgements
We thank Laura Moriarty for assistance with the genetic screen that isolated ns313; Oliver Hobert for whole genome sequencing; Piali Sengupta for unc-130 cDNA and the oy10 mutant strain; Joseph Culotti for unc-130 mutant strains; Raphael Bruckner for assistance with cell culture; Constance Cepko, Christopher Walsh, and the members of the Heiman laboratory for comments and advice on the manuscript; and WormBase. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Competing interests
M.L.B. is a co-inventor of patented PBM technology. All other authors declare no competing or financial interests.
Author contributions
Investigation: K.M., J.M.R., J.D.R.; Supervision: S.S., M.L.B., J.I.M., M.G.H.
Funding
This project was supported by a William Randolph Hearst fellowship from the Hearst Foundations to K.M.; by a Bioinformatics and Integrative Genomics training grant (T32HG002295) from the National Human Genome Research Institute and by a National Science Foundation Graduate Research fellowship to J.M.R.; by the National Institutes of Health/National Human Genome Research Institute (R01HG003985 and R01HG010501 to M.L.B.); by the National Institutes of Health/National Institute of Neurological Disorders and Stroke (R35NS105094 to S.S. and R01NS112343 to M.G.H.); by the National Institutes of Health/National Institute of General Medical Sciences (R35GM127093 to J.I.M.); and by a William F. Milton Award from Harvard University to M.G.H. Deposited in PMC for release after 12 months.
References
- Abdus-Saboor, I., Stone, C. E., Murray, J. I. and Sundaram, M. V. (2012). The Nkx5/HMX homeodomain protein MLS-2 is required for proper tube cell shape in the C. elegans excretory system. Dev. Biol. 366, 298-307. 10.1016/j.ydbio.2012.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun-Gultekin, Z., Andachi, Y., Tsalik, E. L., Pilgrim, D., Kohara, Y. and Hobert, O. (2001). A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development (Cambridge, England) 128, 1951-1969. 10.1242/dev.128.11.1951 [DOI] [PubMed] [Google Scholar]
- Bacaj, T., Tevlin, M., Lu, Y. and Shaham, S. (2008). Glia are essential for sensory organ function in C. elegans. Science 322, 744-747. 10.1126/science.1163074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, Z., Murray, J. I., Boyle, T., Ooi, S. L., Sandel, M. J. and Waterston, R. H. (2006). Automated cell lineage tracing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 103, 2707-2712. 10.1073/pnas.0511111103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, M. M. and Sternberg, P. W. (1999). A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature 401, 386-389. 10.1038/43913 [DOI] [PubMed] [Google Scholar]
- Berger, M. F. and Bulyk, M. L. (2009). Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat. Protoc. 4, 393-411. 10.1038/nprot.2008.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, M. F., Philippakis, A. A., Qureshi, A. M., He, F. S., Estep, P. W. and Bulyk, M. L. (2006). Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 24, 1429-1435. 10.1038/nbt1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle, T. J., Bao, Z., Murray, J. I., Araya, C. L. and Waterston, R. H. (2006). AceTree: a tool for visual analysis of Caenorhabditis elegans embryogenesis. BMC Bioinformatics 7, 275. 10.1186/1471-2105-7-275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. 10.1093/genetics/77.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebul, E. R., McLachlan, I. G. and Heiman, M. G. (2020). Dendrites with specialized glial attachments develop by retrograde extension using SAX-7 and GRDN-1. Development (Cambridge, England) 147, dev180448. 10.1242/dev.180448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, M. M., Smith, Z. D., Grosswendt, S., Kretzmer, H., Norman, T. M., Adamson, B., Jost, M., Quinn, J. J., Yang, D., Jones, M. G.et al. (2019). Molecular recording of mammalian embryogenesis. Nature 570, 77-82. 10.1038/s41586-019-1184-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest, J., Daniele, T., Wang, J., Bykov, A., Mandlbauer, A., Asparuhova, M., Röhsner, J., Gutiérrez-Pérez, P. and Cochella, L. (2020). Combinatorial Action of Temporally Segregated Transcription Factors. Dev. Cell 55, 483-499.e7. 10.1016/j.devcel.2020.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochella, L. and Hobert, O. (2012). Embryonic priming of a miRNA locus predetermines postmitotic neuronal left/right asymmetry in C. elegans. Cell 151, 1229-1242. 10.1016/j.cell.2012.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou, M., Poole, R. J., Sarin, S., Bigelow, H. and Hobert, O. (2010). C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS ONE 5, e15435. 10.1371/journal.pone.0015435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dottori, M., Gross, M. K., Labosky, P. and Goulding, M. (2001). The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development (Cambridge, England) 128, 4127-4138. 10.1242/dev.128.21.4127 [DOI] [PubMed] [Google Scholar]
- Dupin, E., Calloni, G. W., Coelho-Aguiar, J. M. and Le Douarin, N. M. (2018). The issue of the multipotency of the neural crest cells. Dev. Biol. 444 Suppl. 1, S47-S59. 10.1016/j.ydbio.2018.03.024 [DOI] [PubMed] [Google Scholar]
- Forrester, W. C., Perens, E., Zallen, J. A. and Garriga, G. (1998). Identification of Caenorhabditis elegans genes required for neuronal differentiation and migration. Genetics 148, 151-165. 10.1093/genetics/148.1.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung, W., Wexler, L. and Heiman, M. G. (2020). Cell-type-specific promoters for C. elegans glia. J. Neurogenet. 34, 335-346. 10.1080/01677063.2020.1781851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam, S., Singer, E., Waring, D. and Jin, Y. (2000). The C. elegans NeuroD homolog cnd-1 functions in multiple aspects of motor neuron fate specification. Development (Cambridge, England) 127, 4239-4252. 10.1242/dev.127.19.4239 [DOI] [PubMed] [Google Scholar]
- Hammond, T. R., Dufort, C., Dissing-Olesen, L., Giera, S., Young, A., Wysoker, A., Walker, A. J., Gergits, F., Segel, M., Nemesh, J.et al. (2019). Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253-271.e6. 10.1016/j.immuni.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, L., Johnsen, R., Lauter, G., Baillie, D. and Bürglin, T. R. (2006). Comprehensive analysis of gene expression patterns of hedgehog-related genes. BMC Genomics 7, 280. 10.1186/1471-2164-7-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman, M. G. and Shaham, S. (2009). DEX-1 and DYF-7 establish sensory dendrite length by anchoring dendritic tips during cell migration. Cell 137, 344-355. 10.1016/j.cell.2009.01.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert, O. and Kratsios, P. (2019). Neuronal identity control by terminal selectors in worms, flies, and chordates. Curr. Opin. Neurobiol. 56, 97-105. 10.1016/j.conb.2018.12.006 [DOI] [PubMed] [Google Scholar]
- Jin, C., Marsden, I., Chen, X. and Liao, X. (1999). Dynamic DNA contacts observed in the NMR structure of winged helix protein-DNA complex. J. Mol. Biol. 289, 683-690. 10.1006/jmbi.1999.2819 [DOI] [PubMed] [Google Scholar]
- John Lin, C.-C., Yu, K., Hatcher, A., Huang, T.-W., Lee, H. K., Carlson, J., Weston, M. C., Chen, F., Zhang, Y., Zhu, W.et al. (2017). Identification of diverse astrocyte populations and their malignant analogs. Nat. Neurosci. 20, 396-405. 10.1038/nn.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A. D., Fitzsimmons, D., Hagman, J. and Chamberlin, H. M. (2001). EGL-38 Pax regulates the ovo-related gene lin-48 during Caenorhabditis elegans organ development. Development (Cambridge, England) 128, 2857-2865. 10.1242/dev.128.15.2857 [DOI] [PubMed] [Google Scholar]
- Keyte, A. and Hutson, M. R. (2012). The neural crest in cardiac congenital anomalies. Differentiation 84, 25-40. 10.1016/j.diff.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K., Kim, R. and Sengupta, P. (2010). The HMX/NKX homeodomain protein MLS-2 specifies the identity of the AWC sensory neuron type via regulation of the ceh-36 Otx gene in C. elegans. Development (Cambridge, England) 137, 963-974. 10.1242/dev.044719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos, R., Reedy, M. V., Johnson, R. L. and Erickson, C. A. (2001). The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos. Development (Cambridge, England) 128, 1467-1479. 10.1242/dev.128.8.1467 [DOI] [PubMed] [Google Scholar]
- Krishnakumar, R., Chen, A. F., Pantovich, M. G., Danial, M., Parchem, R. J., Labosky, P. A. and Blelloch, R. (2016). FOXD3 regulates pluripotent stem cell potential by simultaneously initiating and repressing enhancer activity. Cell Stem Cell 18, 104-117. 10.1016/j.stem.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, G. S., Viotti, M. and Hadjantonakis, A.-K. (2008). The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev. Cell 15, 509-520. 10.1016/j.devcel.2008.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister, J. A., Cooper, C., Nguyen, K., Modrell, M., Grant, K. and Raible, D. W. (2006). Zebrafish Foxd3 is required for development of a subset of neural crest derivatives. Dev. Biol. 290, 92-104. 10.1016/j.ydbio.2005.11.014 [DOI] [PubMed] [Google Scholar]
- Liu, X., Chen, W., Li, W., Li, Y., Priest, J. R., Zhou, B., Wang, J. and Zhou, Z. (2019). Single-cell RNA-seq of the developing cardiac outflow tract reveals convergent development of the vascular smooth muscle cells. Cell Rep. 28, 1346-1361.e4. 10.1016/j.celrep.2019.06.092 [DOI] [PubMed] [Google Scholar]
- Low, I. I. C., Williams, C. R., Chong, M. K., McLachlan, I. G., Wierbowski, B. M., Kolotuev, I. and Heiman, M. G. (2019). Morphogenesis of neurons and glia within an epithelium. Development 146, dev171124. 10.1242/dev.171124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukoseviciute, M., Gavriouchkina, D., Williams, R. M., Hochgreb-Hagele, T., Senanayake, U., Chong-Morrison, V., Thongjuea, S., Repapi, E., Mead, A. and Sauka-Spengler, T. (2018). From pioneer to repressor: bimodal foxd3 activity dynamically remodels neural crest regulatory landscape in vivo. Dev. Cell 47, 608-628.e6. 10.1016/j.devcel.2018.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques, S., Zeisel, A., Codeluppi, S., van Bruggen, D., Mendanha Falcão, A., Xiao, L., Li, H., Häring, M., Hochgerner, H., Romanov, R. A.et al. (2016). Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science (New York, N.Y.) 352, 1326-1329. 10.1126/science.aaf6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques, S., van Bruggen, D., Vanichkina, D. P., Floriddia, E. M., Munguba, H., Väremo, L., Giacomello, S., Falcão, A. M., Meijer, M., Björklund, Å. K.et al. (2018). Transcriptional convergence of oligodendrocyte lineage progenitors during development. Dev. Cell 46, 504-517.e7. 10.1016/j.devcel.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda, T., Sankowski, R., Staszewski, O., Böttcher, C., Amann, L., Sagar, Scheiwe, C., Nessler, S., Kunz, P., van Loo, G.et al. (2019). Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388-392. 10.1038/s41586-019-0924-x [DOI] [PubMed] [Google Scholar]
- McKenna, A. and Gagnon, J. A. (2019). Recording development with single cell dynamic lineage tracing. Development 146, dev169730. 10.1242/dev.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMiller, T. L. and Johnson, C. M. (2005). Molecular characterization of HLH-17, a C. elegans bHLH protein required for normal larval development. Gene 356, 1-10. 10.1016/j.gene.2005.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello, C. and Fire, A. (1995). DNA transformation. Methods Cell Biol. 48, 451-482. 10.1016/S0091-679X(08)61399-0 [DOI] [PubMed] [Google Scholar]
- Minevich, G., Park, D. S., Blankenberg, D., Poole, R. J. and Hobert, O. (2012). CloudMap: A cloud-based pipeline for analysis of mutant genome sequences. Genetics 192, 1249-1269. 10.1534/genetics.112.144204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizeracka, K. and Heiman, M. G. (2015). The many glia of a tiny nematode: Studying glial diversity using Caenorhabditis elegans. Wiley Interdiscip. Rev. Dev. Biol. 4, 151-160. 10.1002/wdev.171 [DOI] [PubMed] [Google Scholar]
- Morel, L., Chiang, M. S. R., Higashimori, H., Shoneye, T., Iyer, L. K., Yelick, J., Tai, A. and Yang, Y. (2017). Molecular and functional properties of regional astrocytes in the adult brain. J. Neurosci. 37, 8706-8717. 10.1523/JNEUROSCI.3956-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, J. I., Boyle, T. J., Preston, E., Vafeados, D., Mericle, B., Weisdepp, P., Zhao, Z., Bao, Z., Boeck, M. and Waterston, R. H. (2012). Multidimensional regulation of gene expression in the C. elegans embryo. Genome Res. 22, 1282-1294. 10.1101/gr.131920.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, S., Gisselbrecht, S. S., Rogers, J. M., Hartl, D. L. and Bulyk, M. L. (2013). DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc. Natl. Acad. Sci. USA 110, 12349-12354. 10.1073/pnas.1310430110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash, B., Colavita, A., Zheng, H., Roy, P. J. and Culotti, J. G. (2000). The forkhead transcription factor UNC-130 is required for the graded spatial expression of the UNC-129 TGF-beta guidance factor in C. elegans. Genes Dev. 14, 2486-2500. 10.1101/gad.831500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, H., Kozmik, Z., Yu, J.-K. and Wada, H. (2014). A novel N-terminal motif is responsible for the evolution of neural crest-specific gene-regulatory activity in vertebrate FoxD3. Dev. Biol. 385, 396-404. 10.1016/j.ydbio.2013.11.010 [DOI] [PubMed] [Google Scholar]
- Packer, J. S., Zhu, Q., Huynh, C., Sivaramakrishnan, P., Preston, E., Dueck, H., Stefanik, D., Tan, K., Trapnell, C., Kim, J.et al. (2019). A lineage-resolved molecular atlas of C. elegans embryogenesis at single cell resolution. Science 365, eaax1971. 10.1126/science.aax1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce, M. L., Weston, M. D., Fritzsch, B., Gabel, H. W., Ruvkun, G. and Soukup, G. A. (2008). MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evol. Dev. 10, 106-113. 10.1111/j.1525-142X.2007.00217.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, J. L., Zacharias, A. L., Walton, T., Burdick, J. T. and Murray, J. I. (2013). A quantitative model of normal Caenorhabditis elegans embryogenesis and its disruption after stress. Dev. Biol. 374, 12-23. 10.1016/j.ydbio.2012.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo Romanos, T., Pladevall-Morera, D., Langebeck-Jensen, K., Hansen, S., Ng, L. and Pocock, R. (2017). LIN-32/atonal controls oxygen sensing neuron development in Caenorhabditis elegans. Sci. Rep. 7, 7294. 10.1038/s41598-017-07876-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santella, A., Du, Z., Nowotschin, S., Hadjantonakis, A.-K. and Bao, Z. (2010). A hybrid blob-slice model for accurate and efficient detection of fluorescence labeled nuclei in 3D. BMC Bioinformatics 11, 580. 10.1186/1471-2105-11-580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafi-Reinach, T. R. and Sengupta, P. (2000). The forkhead domain gene unc-130 generates chemosensory neuron diversity in C. elegans. Genes Dev. 14, 2472-2485. 10.1101/gad.832300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov, M., Schneider, S., Pozniakovski, A., Roguev, A., Ernst, S., Zhang, Y., Hyman, A. A. and Stewart, A. F. (2006). A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat. Methods 3, 839-844. 10.1038/nmeth933 [DOI] [PubMed] [Google Scholar]
- Sasai, N., Mizuseki, K. and Sasai, Y. (2001). Requirement of FoxD3-class signaling for neural crest determination in Xenopus. Development (Cambridge, England) 128, 2525-2536. 10.1242/dev.128.13.2525 [DOI] [PubMed] [Google Scholar]
- Spitzer, S. O., Sitnikov, S., Kamen, Y., Evans, K. A., Kronenberg-Versteeg, D., Dietmann, S., de Faria, O., Agathou, S. and Káradóttir, R. T. (2019). Oligodendrocyte progenitor cells become regionally diverse and heterogeneous with age. Neuron 101, 459-471.e5. 10.1016/j.neuron.2018.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, R. A., Arduini, B. L., Berghmans, S., George, R. E., Kanki, J. P., Henion, P. D. and Look, A. T. (2006). Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev. Biol. 292, 174-188. 10.1016/j.ydbio.2005.12.035 [DOI] [PubMed] [Google Scholar]
- Stout, R. F. and Parpura, V. (2011). Voltage-gated calcium channel types in cultured C. elegans CEPsh glial cells. Cell Calcium 50, 98-108. 10.1016/j.ceca.2011.05.016 [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., Schierenberg, E., White, J. G. and Thomson, J. N. (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64-119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Teng, L., Mundell, N. A., Frist, A. Y., Wang, Q. and Labosky, P. A. (2008). Requirement for Foxd3 in the maintenance of neural crest progenitors. Development (Cambridge, England) 135, 1615-1624. 10.1242/dev.012179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D. E., Weinreb, C., Collins, Z. M., Briggs, J. A., Megason, S. G. and Klein, A. M. (2018). Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science (New York, N.Y.) 360, 981-987. 10.1126/science.aar4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, S. W., Singhvi, A., Liang, Y., Lu, Y. and Shaham, S. (2016). PROS-1/prospero is a major regulator of the glia-specific secretome controlling sensory-neuron shape and function in C. elegans. Cell Rep. 15, 550-562. 10.1016/j.celrep.2016.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton, T., Preston, E., Nair, G., Zacharias, A. L., Raj, A. and Murray, J. I. (2015). The Bicoid class homeodomain factors ceh-36/OTX and unc-30/PITX cooperate in C. elegans embryonic progenitor cells to regulate robust development. PLoS Genet. 11, e1005003. 10.1371/journal.pgen.1005003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenick, A. S. and Hobert, O. (2004). Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev. Cell 6, 757-770. 10.1016/j.devcel.2004.05.004 [DOI] [PubMed] [Google Scholar]
- Wicks, S. R., Yeh, R. T., Gish, W. R., Waterston, R. H. and Plasterk, R. H. A. (2001). Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28, 160-164. 10.1038/88878 [DOI] [PubMed] [Google Scholar]
- Yaklichkin, S., Steiner, A. B., Lu, Q. and Kessler, D. S. (2007a). FoxD3 and Grg4 physically interact to repress transcription and induce mesoderm in Xenopus. J. Biol. Chem. 282, 2548-2557. 10.1074/jbc.M607412200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaklichkin, S., Vekker, A., Stayrook, S., Lewis, M. and Kessler, D. S. (2007b). Prevalence of the EH1 Groucho interaction motif in the metazoan Fox family of transcriptional regulators. BMC Genomics 8, 201. 10.1186/1471-2164-8-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, S., Murray, J. I., Lu, Y., Waterston, R. H. and Shaham, S. (2008). Mls-2 and vab-3 Control glia development, hlh-17/Olig expression and glia-dependent neurite extension in C. elegans. Development 135, 2263-2275. 10.1242/dev.019547 [DOI] [PubMed] [Google Scholar]
- Zeisel, A., Muñoz-Manchado, A. B., Codeluppi, S., Lönnerberg, P., La Manno, G., Juréus, A., Marques, S., Munguba, H., He, L., Betsholtz, C.et al. (2015). Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138-1142. 10.1126/science.aaa1934 [DOI] [PubMed] [Google Scholar]
- Zeisel, A., Hochgerner, H., Lönnerberg, P., Johnsson, A., Memic, F., van der Zwan, J., Häring, M., Braun, E., Borm, L. E., La Manno, G.et al. (2018). Molecular architecture of the mouse nervous system. Cell 174, 999-1014.e22. 10.1016/j.cell.2018.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.