Abstract
Background
Though massively parallel sequencing has been widely applied to noninvasive prenatal screen for common trisomy, the clinical use of massively parallel sequencing to noninvasive prenatal diagnose monogenic disorders is limited. This study was to develop a method for directly determining paternal haplotypes for noninvasive prenatal diagnosis of monogenic disorders without requiring proband’s samples.
Methods
The study recruited 40 families at high risk for autosomal recessive diseases. The targeted linked-read sequencing was performed on high molecular weight (HMW) DNA of parents using customized probes designed to capture targeted genes and single-nucleotide polymorphisms (SNPs) distributed within 1Mb flanking region of targeted genes. Plasma DNA from pregnant mothers also underwent targeted sequencing using the same probes to determine fetal haplotypes according to parental haplotypes. The results were further confirmed by invasive prenatal diagnosis.
Results
Seventy-eight parental haplotypes of targeted gene were successfully determined by targeted linked-read sequencing. The predicted fetal inheritance of variant was correctly deduced in 38 families in which the variants had been confirmed by invasive prenatal diagnosis. Two families were determined to be no-call.
Conclusions
Targeted linked-read sequencing method demonstrated to be an effective means to phase personal haplotype for noninvasive prenatal diagnosis of monogenic disorders.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-021-01091-x.
Keywords: Noninvasive prenatal diagnosis, Direct haplotype phasing, Targeted linked-read sequencing, Monogenic disease
Background
The discovery of cell-free fetal circulating DNA (cff-DNA) in maternal blood and the rapid advances of massively parallel sequencing (MPS) have provided an unprecedented opportunity to perform the prenatal genetic testing of common fetal aneuploidies and single-gene diseases. Though MPS has been widely applied to screen for fetal trisomy 21, 18 and 13 [1], the clinical use of MPS to diagnose monogenic disorders is limited [2]. Several studies have been conducted to develop noninvasive prenatal diagnosis (NIPD) for monogenic disease using various technologies such as real-time polymerase chain reaction (PCR), amplification at lower denaturation temperature-PCR, digital PCR, circulating single-molecule amplification and resequencing technology [3, 4] and MPS. These studies were confined to exclude paternally inherited [5] and detect de novo variants[6] based on variant-specific assays due to the strong interference of maternal background signal. The relative haplotype dosage approach has been demonstrated to detect parental inherited variants at the same time. Our group has employed a proband-based method for resolving parental haplotypes and successfully applied this method to NIPD of Duchenne muscular dystrophy (DMD) [7], congenital adrenal hyperplasia (CAH) [8], maple syrup urine disease (MSUD) [9], hyperphenylalaninemia [10] and spinal muscular atrophy (SMA) [11]. This phasing information makes it possible to measure the haplotype dosage imbalance in maternal plasma DNA. The advantage of relative haplotype dosage approach is that analysis is independent of variant types. While, the method needs proband’s samples to phase parental haplotypes, which hampers the application of NIPD to monogenic diseases in clinical practice. The haplotype phasing is a critical step for haplotype-based NIPD of monogenic disorders. Serval studies have reported specific haplotype building methods such as clone pool dilution sequencing [12], contiguity-preserving transposition sequencing [13], targeted locus amplification (TLA) [14], HaploSeq [15] and long fragment read (LFR) technology [16]. These approaches need complex experimental operations and are time consuming and associated with a low success rate. These limitations can be problematic for identifying single gene disorders. Population data-based personal haplotype phasing overcomes the above drawbacks. The population-based method is based on reference population with genotyping data of unrelated individuals and the accuracy of NIPD is only 80%, which is lower than the experimental methods [17]. In order to further improve the success rate and accuracy of haplotype phasing, microfluidics-based linked-read sequencing technology and TLA-based phasing were utilized to phase parental DNA directly [18, 19]. The former approach combined the whole-genome sequencing (WGS) and linked-read sequencing method and succeeded in predicting fetal inherited variants in 12 of 13 pregnancies. The informative sequencing depth (40x) of WGS and the expensive experimental reagents restricted its clinical practice for NIPD [18]. Targeted TLA-based phasing approach is also subject to the complex acquisition of TLA template and customized target kit for NIPD which is inconvenient. A customized probe which covers dozens of common single gene disorders in China is used for haplotype-based NIPD. Therefore, we speculated that the linked-read sequencing combined with targeted sequencing using the above probes would expand the list of single gene disorders and reduce the cost compared with the whole-genome sequencing.
In our previous study, we demonstrated direct haplotyping of NIPD based on linked‐read sequencing is accurate for the prediction of fetal pathogenic variants of DMD [20]. The objectives of this study are to investigate the feasibility and accuracy of targeted linked-read sequencing in six different types of autosomal recessive diseases. We analyzed 40 families at high risk for six kinds of autosomal recessive diseases and showed that direct haplotype phasing of parental high molecular weight (HMW) DNA is feasible using targeted linked-read sequencing of target genes. Targeted sequencing of maternal plasma DNA combined with the parental haplotype information were interpreted to determine the inherited variants in fetus. Our approach might be a cost-effective and applicable method for NIPD of autosomal recessive monogenic disorders in clinical settings.
Methods
Sample collection
We recruited 40 families at high risk for autosomal recessive diseases, including 13 methylmalonic acidemia (MMA) families, 12 β-thalassemia families, 8 phenylketonuria (PKU) families, 5 α-thalassemia families, 1 autosomal recessive polycystic kidney disease (ARPKD) family and 1 autosomal recessive deafness-1A (DFNB1A) family caused by pathogenic variants of GJB2 gene. The variants have been identified in all families (Table 1). All participants provided written informed consent to join in the study. The ethics committee of the participating hospitals and the Institutional Review Board of BGI approved the conduct of this study (BGI-IRB No 17080-T1).
Table 1.
Clinical information of the participating families
| Family | Disease | Gene | Genotypes of the Trios | GA | FF (%) | ||
|---|---|---|---|---|---|---|---|
| Mat | Pat | Fetus (Mat/Pat) | |||||
| F01 | β-thalassemia | HBB | c.316-197C>T /N | c.-78A>G/N | N/N | 12+4 | 9.3 |
| F02 | β-thalassemia | HBB | c.126_129delCTTT/N | c.126_129delCTTT/N | N/N | 20+5 | 15.9 |
| F03 | β-thalassemia | HBB | c.126_129delCTTT/N | c.-78A>G/N | c.126_129delCTTT/c.-78A>G | 12+3 | 15.4 |
| F04 | β-thalassemia | HBB | c.316-197C>T /N | c.126_129delCTTT/N | N/c.126_129delCTTT | 18 | 12.1 |
| F05 | β-thalassemia | HBB | c.126_129delCTTT/N | c.316-197C>T /N | c.126_129delCTTT/N | 13+6 | 20.6 |
| F06 | β-thalassemia | HBB | c.216_217insA/T/N | c.126_129delCTTT/N | c.216_217insA/T/ c.126_129delCTTT | 13+2 | 26.8 |
| F07 | β-thalassemia | HBB | c.79G>A/N | c.126_129delCTTT/N | N/c.126_129delCTTT | 11+3 | 12.3 |
| F08 | β-thalassemia | HBB | c.126_129delCTTT/N | c.316-197C>T/N | c.126_129delCTTT/N | 12+3 | 16.5 |
| F09 | β-thalassemia | HBB | c.52A>T/N | c.84_85insC/N | c.52A>T/N | 12+1 | 27.7 |
| F10 | β-thalassemia | HBB | c.126_129delCTTT/N | c.79G>A/N | c.126_129delCTTT/c.79G>A | 11+1 | 17.7 |
| F11 | β-thalassemia | HBB | c.126_129delCTTT/N | c.126_129delCTTT/N | c.126_129delCTTT/c.126_129delCTTT | 17 | 8.1 |
| F12 | β-thalassemia | HBB | c.126_129delCTTT/N | c.126_129delCTTT/N | N/c.126_129delCTTT | 17 | 9.7 |
| F13 | α-thalassemia | HBA | - -SEA/N | - -SEA/N | - -SEA/- -SEA | 13+3 | 15.7 |
| F14 | α-thalassemia | HBA | - -SEA/N | - -SEA/N | N/N | 11+6 | 13.7 |
| F15 | α-thalassemia | HBA | - -SEA/N | - -SEA/N | N/- -SEA | 12+4 | 17.5 |
| F16 | α-thalassemia | HBA | - -SEA/N | - -SEA/N | - -SEA/- -SEA | 11+3 | 23.5 |
| F17 | α-thalassemia | HBA | - -SEA/N | c.369C>G/N | - -SEA/c.369C>G | 18 | 6.7 |
| F18 | MMA | MMACHC | c.609G>A/N | c.609G>A/N | c.609G>A/N | 19 | 16.5 |
| F19 | MMA | MMACHC | c.656-658delAGA/N | c.609G>A/N | N/c.609G>A | 18 | 14.2 |
| F20 | MMA | MMACHC | c.609G>A/N | c.656-658delAGA/N | N/N | 16 | 12.8 |
| F21 | MMA | MMACHC | c.656-658delAGA/N | c.609G>A/N | N/N | 17 | 10.4 |
| F22 | MMA | MMACHC | c.80A>G/N | c.609G>A/N | c.80A>G/N | 17 | 10.2 |
| F23 | MMA | MMACHC | c.609G>A/N | c.441TG[2]/N | c.609G>A/c.441TG[2] | 17 | 10.1 |
| F24 | MMA | MMACHC | c.609G>A/N | c.609G>A/N | N/N | 18 | 17.8 |
| F25 | MMA | MMACHC | c.80A>G/N | c.609G>A/N | N/N | 17 | 13.7 |
| F26 | MMA | MMACHC | c.609G>A/N | c.658-660delAAG/N | c.609G>A/c.658-660delAAG | 17 | 9.8 |
| F27 | MMA | MMACHC | c.609G>A/N | c.445-446delTG/N | N/N | 17 | 10.4 |
| F28 | MMA | MMACHC | c.482G>A/N | c.445-446delTG/N | N/N | 17 | 8.2 |
| F29 | MMA | MMACHC | c.315C>G/N | c.609G>A/N | c.315C>G/N | 16 | 6.5 |
| F30 | MMA | MMACHC | c.609G>A/N | c.609G>A/N | N/N | 17+5 | 8.0 |
| F31 | PKU | PAH | c.1197A>T/N | c.764T>C/N | c.1197A>T/c.764T>C | 18 | 7.3 |
| F32 | PKU | PAH | c.992T>C/N | c.770G>T/N | N/c.770G>T | 17 | 7.5 |
| F33 | PKU | PAH | c.1045T>G/N | c.728G>A/N | N/N | 18 | 11.3 |
| F34 | PKU | PAH | c.728G>A/N | c.611A>G/N | N/N | 20 | 5.9 |
| F35 | PKU | PAH | c.977G>A/N | c.1238G>C/N | c.977G>A/N | 17 | 21.2 |
| F36 | PKU | PAH | c.473G>A/N | c.208_210delTCT | c.473G>A/c.208_210delTCT | 18 | 12.8 |
| F37 | PKU | PAH | c.1223G>A/N | c.727C>T/N | N/N | 12 | 8.5 |
| F38 | PKU | PAH | c.728G>A/N | c.721C>T/N | c.728G>A/c.721C>T | 12 | 7.2 |
| F39 | ARPKD | PKHD1 | c.11042T>G/N | c.5137G>T /N | N/c.5137G>T | 12+6 | 15.0 |
| F40 | DFNB1A | GJB2 | c.235delC/N | c.299-300delAT/N | c.235delC/N | 13+1 | 15.3 |
FF fetal fraction, GA gestational age, N Normal, PKU phenylketonuria, MMA methylmalonic academia, ARPKD autosomal recessive polycystic kidney disease, DFNB1A autosomal recessive deafness-1A
Target capture probe design
The targeted enrichment of DNA libraries was performed according to the custom-designed SeqCap EZ Choice Library (NimbleGen, Roche) protocol. The capture probes (NimbleGen, Roche) targeting the whole genes of HBB, HBA1, HBA2, and highly heterozygous SNPs within 1Mb flanking region of target genes were designed for NIPD of β-thalassemia and α-thalassemia. Another set of target capture probe was designed to cover the coding region and SNPs within 1Mb upstream and downstream regions of the interested genes, including MMACHC (MMA), PAH (PKU), PKHD1 (ARPKD) and GJB2 (DFNB1A).
Targeted linked-read sequencing
HMW genomic DNA (gDNA) was extracted from stored blood using the Mag Attract HMW Kit (Qiagen, Germany). The size of HMW gDNA should be more than 50kb according to the pulse electrophoresis results. Then gDNA was processed with Chromium™ Genome v2 libraries (10x Genomics, USA). Long gDNA strands were partitioned in barcoded gel beads through a microfluidic device. Barcoded oligonucleotides in a gel bead bind randomly onto the long molecules and generate short fragments with the same barcode. The chance that two molecules were covering the same genomic locus on each gel bead is low, and the short fragments with the same barcode were considered to come from the same long molecule. Libraries of the barcoded fragments were prepared and captured using the customized probe. The prepared DNA library was then sequenced using an Illumina HiSeq2500 sequencer with a paired-end format of 101 bp or 150 bp.
Variant calling and direct haplotype phasing
The barcoded libraries read were then processed with the Long Ranger pipeline (v.2.2.2) provided by 10x Genomics [21]. Reads associated with valid barcodes were aligned against the human genome 19 (Hg19) by using the Burrows-Wheeler Aligner (BWA) software [22]. Output files annotated with barcode and phasing information were generated and served as the reference haplotypes of the family for downstream analysis. The maternal plasma DNA sequencing reads were aligned against the reference hg19 using BWA. After duplicated reads were marked by the Picard Mark Duplicates tool, the GATK tools were applied to perform local realignment and base quality score recalibration [23].
The free Long Ranger (v.2.2.2) software was utilized to determine the parental haplotype in the interested region. Barcode information provides the clue to associate short reads to the original long input molecules. Variant-linked haplotype referred to those reads whose barcodes were consistent with the ones with variant alleles. In contrast, wild-linked haplotype denoted the reads carrying same barcode with the ones with wild-type alleles. The different haplotype blocks were linked with identified SNPs using the overlapping region. SNPs associated with the same haplotypes carrying the wild-type and variant alleles were used for the maternal plasma DNA analysis.
The estimation of fetal fraction and NIPD of monogenic disorders
The evaluation of fetal fraction could be conducted according to the procedure reported before [8]. The haplotype related to variant and wild alleles was constructed based on targeted linked-read sequencing. The informative SNPs that were heterozygous in the mother but homozygous in the father were analyzed for maternal inheritance. On the contrary, the paternal inheritance analysis followed the opposite strategy with maternal inheritance analysis. We used hidden Markov model (HMM) to predict the most likely inherited haplotype using our previously reported algorithm [24]. The probabilities that the fetus inherited the pathogenic and non-pathogenic alleles were evaluated using the number of reads in maternal plasma and then considered as the HMM emission probabilities. The genetic map from the National Center for Biotechnology Information provided the genetic position of the SNPs in centimorgan (cM) and recombination rates between SNPs, these probabilities were regarded as HMM transition probabilities. Lastly, the Viterbi algorithm was utilized to predict the inherited haplotype in the fetus.
Validation of NIPD
The samples obtained through invasive procedures including chorionic villus sampling (CVS) and amniocentesis were used for prenatal genetic diagnosis. After DNA extraction, Sanger sequencing, gap-PCR and reverse dot blot PCR for target variations were performed in a blind manner to further validate the accuracy of NIPD.
Results
Clinical information of the monogenic families
40 families at high risk for autosomal recessive diseases, including 13 MMA families, 12 β-thalassemia families, 8 PKU families, 5 α-thalassemia families, 1 ARPKD family and 1 DFNB1A family caused by pathogenic variants of GJB2 gene were recruited. The gestational age (GA) of 40 pregnant women varied from 11+1 to 20+5 weeks, with a median GA of 15.5 weeks. The clinical information, variant loci and variant status of the 40 families are presented in Table 1.
Targeted linked-reads sequencing
Targeted sequencing on the interested gene region was performed in plasma DNA samples from 40 pregnant women at different gestational weeks. The fetal fraction varied from 5.9 to 27.7%, with a mean fetal fraction of 13.2%, showing significant differences between individuals (Table 1). The targeted sequencing of gDNA samples showed the coverage was relatively consistent in the targeted genes, with a mean read depth of 402× (Additional file 1: Table S1). After data pre-processing and alignment, over 98% of the linked-reads were aligned to the hg19, an average of 50% of the bases were on-target (Additional file 1: Table S1). The summary statistics of alignment are presented in detail in Additional file 1: Table S1.
Direct haplotype phasing
The 10x genomics barcoding technology allowed us to obtain long-range information by linking the short sequencing reads produced. There were two haplotypes, the pathogenic haplotype (P) and normal haplotype (N). The former referred to the reads whose alleles or barcodes were in consistence with variant-supporting reads at heterozygous SNP positions. While the latter represented those reads whose alleles were opposite to the variant-supporting reads at heterozygous SNP positions. The two haplotypes of were directly determined by linking the haplotype blocks assembled by the barcoded reads for all parental gDNA. N50 phase-block length represents the contiguity achieved in the experimental haplotyping, the average length of N50 phase-block was 1Mb (range 413.04 kb~3.54 Mb). N50 phase block, phase block across the target region and longest phase block for the 40 families is depicted in Table 2 and Additional file 1: Table S1. The number of SNPs in the phase blocks used for phasing ranged from 3 to 2418 SNPs, with a mean of 1006 (Table 2). All variants carried by family members were initially detected by the targeted linked-read sequencing and verified to be concordant with those from the MPS data. The paternal haplotypes phasing of F27 and F36 failed, because the haplotype block cannot cover the pathogenic variants. Therefore, the NIPD analysis is not required for failed phasing individuals (pF27 and pF36).
Table 2.
Parental haplotypes phasing data
| Family | Sample | Gene | Phase block across target region | Phasing block size(kb) | No. of SNPs across the block |
|---|---|---|---|---|---|
| F01 | mat | HBB | chr11:4249489-6238960 | 1989.5 | 2367 |
| pat | HBB | chr11:4269280-5761797 | 1492.5 | 1469 | |
| F02 | mat | HBB | chr11:4366798-6246383 | 1879.6 | 1803 |
| pat | HBB | chr11:4366798-6237565 | 1870.8 | 1655 | |
| F03 | mat | HBB | chr11:4249238-5884595 | 1635.4 | 1716 |
| pat | HBB | chr11:4346064-6121271 | 1775.2 | 1972 | |
| F04 | mat | HBB | chr11:4587676-6243982 | 1656.3 | 2308 |
| pat | HBB | chr11:4905140-6216304 | 1311.2 | 1644 | |
| F05 | mat | HBB | chr11:5192535-5900085 | 707.6 | 955 |
| pat | HBB | chr11:4249095-5450493 | 1201.4 | 1359 | |
| F06 | mat | HBB | chr11:4852009-5555972 | 704.0 | 741 |
| pat | HBB | chr11:5196669-6082903 | 886.2 | 1541 | |
| F07 | mat | HBB | chr11:4697080-6239344 | 1542.3 | 1810 |
| pat | HBB | chr11:4306665-6246051 | 1939.4 | 2043 | |
| F08 | mat | HBB | chr11:4936613-6116142 | 1179.5 | 1544 |
| pat | HBB | chr11:4249126-5771915 | 1522.8 | 1369 | |
| F09 | mat | HBB | chr11:4436676-6239344 | 1802.7 | 1681 |
| pat | HBB | chr11:4249163-6090372 | 1841.2 | 2247 | |
| F10 | mat | HBB | chr11:4249271-6237565 | 1988.3 | 1666 |
| pat | HBB | chr11:4249031-6037803 | 1788.8 | 1733 | |
| F11 | mat | HBB | chr11:4345701-5647166 | 1301.5 | 1202 |
| pat | HBB | chr11:4389404-5719251 | 1329.8 | 1450 | |
| F12 | mat | HBB | chr11:4249095-6239344 | 1990.2 | 2301 |
| pat | HBB | chr11:4387760-6121428 | 1733.7 | 2418 | |
| F13 | mat | HBA | chr16:60185-679412 | 619.2 | 284 |
| pat | HBA | chr16:60185-1225628 | 1165.4 | 937 | |
| F14 | mat | HBA | chr16:186950-1216997 | 1030.0 | 606 |
| pat | HBA | chr16:132246-612607 | 480.4 | 251 | |
| F15 | mat | HBA | chr16:94080-1225184 | 1131.1 | 899 |
| pat | HBA | chr16:74039-1197612 | 1123.6 | 687 | |
| F16 | mat | HBA | chr16:79811-1223722 | 1143.9 | 883 |
| pat | HBA | chr16:60185-460830 | 400.6 | 339 | |
| F17 | mat | HBA | chr16:60185-1192620 | 1132.4 | 1045 |
| pat | HBA | chr16:60291-1225184 | 1164.9 | 1010 | |
| F18 | mat | MMACHC | chr1: 44966837-46952164 | 1985.3 | 1599 |
| pat | MMACHC | chr1: 44972309-46972958 | 2000.6 | 926 | |
| F19 | mat | MMACHC | chr1: 45513754-46973454 | 1459.7 | 440 |
| pat | MMACHC | chr1: 44979498-46975877 | 1996.3 | 831 | |
| F20 | mat | MMACHC | chr1: 45767431-46206444 | 439.0 | 119 |
| pat | MMACHC | chr1: 45386861-46503217 | 1116.3 | 247 | |
| F21 | mat | MMACHC | chr1: 45765523-46975294 | 1209.8 | 457 |
| pat | MMACHC | chr1:44967323-46975877 | 2008.6 | 812 | |
| F22 | mat | MMACHC | chr1:45762749-46722939 | 960.2 | 445 |
| pat | MMACHC | chr1:45701916-46097939 | 396.0 | 161 | |
| F23 | mat | MMACHC | chr1:45738336-46975450 | 1237.1 | 729 |
| pat | MMACHC | chr1:44967431-46975877 | 2008.4 | 1228 | |
| F24 | mat | MMACHC | chr1:45947353-46095125 | 147.8 | 27 |
| pat | MMACHC | chr1:45775550-46605728 | 830.2 | 609 | |
| F25 | mat | MMACHC | chr1:45765523-46053981 | 288.5 | 156 |
| pat | MMACHC | chr1:45765523-45982693 | 217.2 | 41 | |
| F26 | mat | MMACHC | chr1:45767431-46975877 | 1208.4 | 691 |
| pat | MMACHC | chr1:45762749-46975877 | 1213.1 | 684 | |
| F27 | mat | MMACHC | chr1:45683746-46645681 | 961.9 | 572 |
| pat | MMACHC | chr1:45962137-45974407 | 12.3 | 3 | |
| F28 | mat | MMACHC | chr1:44967323-45974520 | 1007.2 | 595 |
| pat | MMACHC | chr1:44967323-46691245 | 1723.9 | 1149 | |
| F29 | mat | MMACHC | chr1:45640368-46975877 | 1335.5 | 599 |
| pat | MMACHC | chr1:44973546-46975877 | 2002.3 | 1185 | |
| F30 | mat | MMACHC | chr1:44967825-46975877 | 2008.1 | 1082 |
| pat | MMACHC | chr1:45683419-46924563 | 1241.1 | 685 | |
| F31 | mat | PAH | chr12:103214192-104013534 | 799.3 | 301 |
| pat | PAH | chr12:102252463-104225303 | 1972.8 | 1299 | |
| F32 | mat | PAH | chr12:102241500-104309559 | 2068.1 | 1300 |
| pat | PAH | chr12:102240964-104261374 | 2020.4 | 1094 | |
| F33 | mat | PAH | chr12:102240964-103276441 | 1035.5 | 555 |
| pat | PAH | chr12:102241500-104173880 | 1932.4 | 1048 | |
| F34 | mat | PAH | chr12:102618568-104309712 | 1691.1 | 1095 |
| pat | PAH | chr12:102728895-104272113 | 1543.2 | 696 | |
| F35 | mat | PAH | chr12:102894838-103267467 | 372.6 | 136 |
| pat | PAH | chr12:103075411-104309383 | 1234.0 | 1069 | |
| F36 | mat | PAH | chr12:102248565-104275721 | 2027.2 | 1189 |
| pat | PAH | chr12:103105959-103274915 | 169.0 | 106 | |
| F37 | mat | PAH | chr12:102321986-103791220 | 1469.2 | 984 |
| pat | PAH | chr12:102710699-104300441 | 1589.7 | 1062 | |
| F38 | mat | PAH | chr12:102240964-103623855 | 1382.9 | 619 |
| pat | PAH | chr12:102240964-104304705 | 2063.7 | 1246 | |
| F39 | mat | PKHD1 | chr6:50968947-52950047 | 1981.1 | 1347 |
| pat | PKHD1 | chr6:50982112-52905592 | 1923.5 | 985 | |
| F40 | mat | GJB2 | chr13:20687773-20802900 | 115.1 | 93 |
| pat | GJB2 | chr13:20676993-21122165 | 445.2 | 279 |
Noninvasive prenatal diagnosis
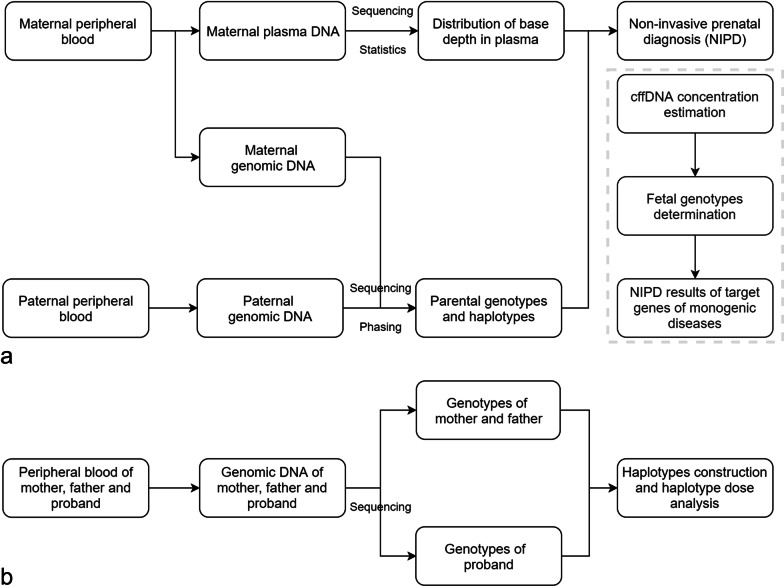
As shown in the NIPD flowchart (Figure 1), maternal and paternal haplotypes were first established using target-region sequencing data and the HMM and Viterbi algorithm was then applied to predict fetal haplotypes. Our goal was to precisely infer the fetal genotypes at pathogenic sites, not to correctly infer the haplotypes of all SNP markers flanking the target gene. Therefore, the specific rules [25] were set to determine the fetal genotype at the pathogenic site after obtaining the optimal path of the fetal haplotype block via the Viterbi algorithm. If the path contains only one halotype block (pathogenic or normal) and the block spans the target gene, the fetal genotype at the pathogenic site is the state of the haplotype block that spans the target gene. If the path contains two haplotype blocks (pathogenic and normal) and only one haplotype block spans the target gene, the fetal genotype at the pathogenic site is the state of the haplotype block that spans the target gene (for example, mF04 and mF06). If two haplotype block (pathogenic and normal) exists inside the target gene, the fetal genotype at the pathogenic site is determined as no-call (for example, mF36). A confidence score (CS) [25] was introduced into our algorithm to quantify the probability of obtaining the correct results for NIPD. The CS was calculated using the fetal fraction, sequencing depth of maternal plasma and number of parental informative SNPs as inputs for computational simulation. The detailed method can be referred to the published literature [25]. The condition that the CS was less than 0.99 was defined as no-call.
Fig. 1.
The flow charts of targeted linked-read sequencing and proband-based indirect phasing. a Parental genotypes and haplotype determination, prediction of fetal haplotype and noninvasive prenatal diagnosis of monogenic diseases using the targeted linked-read sequencing method. b Parental and proband's genotype and haplotype determination, prediction of fetal haplotype and noninvasive prenatal diagnosis of monogenic diseases using the proband-based indirect phasing method
The NIPD results exhibited that 38 fetuses had both alleles detected; of these 38 fetuses, 11 were affected, 15 were carriers and 12 were normal. (Table 3, Additional file 2: Figure S1, Additional file 3: Figure S2 and Additional file 4: Figure S3). For F27, only one normal haplotype inherited from mother can be inferred by NIPD. For F36, we cannot predict fetal haplotypes inherited from parents.
Table 3.
The NIPD results
| Family | Gene | No. of Maternal Informative SNPs | No. of Paternal Informative SNPs | CSmat (%) | CSpat (%) | NIPD (mat/pat) | Invasive prenatal diagnosis (mat/pat) |
|---|---|---|---|---|---|---|---|
| F01 | HBB | 1260 | 305 | 100 | 100 | N/N | N/N |
| F02 | HBB | 1073 | 607 | 100 | 100 | N/N | N/N |
| F03 | HBB | 521 | 566 | 100 | 100 | c.126_129delCTTT/c.-78A>G | c.126_129delCTTT/c.-78A>G |
| F04 | HBB | 394 | 317 | 100 | 100 | N/c.126_129delCTTT | N/c.126_129delCTTT |
| F05 | HBB | 255 | 555 | 100 | 100 | c.126_129delCTTT/N | c.126_129delCTTT/N |
| F06 | HBB | 268 | 453 | 100 | 100 | c.216_217insA/T/ c.126_129delCTTT | c.216_217insA/T/ c.126_129delCTTT |
| F07 | HBB | 697 | 695 | 100 | 100 | N/c.126_129delCTTT | N/c.126_129delCTTT |
| F08 | HBB | 636 | 442 | 100 | 100 | c.126_129delCTTT/N | c.126_129delCTTT/N |
| F09 | HBB | 669 | 553 | 100 | 100 | c.52A>T/N | c.52A>T/N |
| F10 | HBB | 908 | 594 | 100 | 100 | c.126_129delCTTT/c.79G>A | c.126_129delCTTT/c.79G>A |
| F11 | HBB | 603 | 380 | 100 | 100 | c.126_129delCTTT/c.126_129delCTTT | c.126_129delCTTT/c.126_129delCTTT |
| F12 | HBB | 1029 | 550 | 100 | 100 | N/c.126_129delCTTT | N/c.126_129delCTTT |
| F13 | HBA | 53 | 18 | 100 | 100 | - -SEA/- -SEA | - -SEA/- -SEA |
| F14 | HBA | 235 | 52 | 100 | 100 | N/N | N/N |
| F15 | HBA | 118 | 84 | 100 | 100 | N/- -SEA | N/- -SEA |
| F16 | HBA | 193 | 78 | 100 | 100 | - -SEA/- -SEA | - -SEA/- -SEA |
| F17 | HBA | 361 | 140 | 100 | 100 | - -SEA/c.369C>G | - -SEA/c.369C>G |
| F18 | MMACHC | 775 | 228 | 100 | 100 | c.609G>A/N | c.609G>A/N |
| F19 | MMACHC | 298 | 424 | 100 | 100 | N/c.609G>A | N/c.609G>A |
| F20 | MMACHC | 97 | 175 | 100 | 100 | N/N | N/N |
| F21 | MMACHC | 348 | 361 | 100 | 100 | N/N | N/N |
| F22 | MMACHC | 285 | 49 | 100 | 100 | c.80A>G/N | c.80A>G/N |
| F23 | MMACHC | 531 | 300 | 100 | 100 | c.609G>A/c.441TG[2] | c.609G>A/c.441TG[2] |
| F24 | MMACHC | 15 | 420 | 100 | 100 | N/N | N/N |
| F25 | MMACHC | 79 | 7 | 100 | 100 | N/N | N/N |
| F26 | MMACHC | 492 | 107 | 100 | 100 | c.609G>A/c.658-660delAAG | c.609G>A/c.658-660delAAG |
| F27 | MMACHC | 353 | NA | 100 | NA | N/NA | N/N |
| F28 | MMACHC | 474 | 457 | 100 | 100 | N/N | N/N |
| F29 | MMACHC | 319 | 469 | 100 | 100 | c.315C>G/N | c.315C>G/N |
| F30 | MMACHC | 776 | 42 | 100 | 100 | N/N | N/N |
| F31 | PAH | 69 | 321 | 100 | 100 | c.1197A>T/c.764T>C | c.1197A>T/c.764T>C |
| F32 | PAH | 362 | 185 | 100 | 100 | N/c.770G>T | N/c.770G>T |
| F33 | PAH | 161 | 147 | 100 | 100 | N/N | N/N |
| F34 | PAH | 262 | 95 | 100 | 100 | N/N | N/N |
| F35 | PAH | 13 | 174 | 100 | 100 | c.977G>A/N | c.977G>A/N |
| F36 | PAH | 188 | NA | 100 | NA | NC [*]/NA | c.473G>A/c.208_210delTCT |
| F37 | PAH | 261 | 164 | 100 | 100 | N/N | N/N |
| F38 | PAH | 406 | 561 | 100 | 100 | c.728G>A/c.721C>T | c.728G>A/c.721C>T |
| F39 | PKHD1 | 971 | 267 | 100 | 100 | N/c.5137G>T | N/c.5137G>T |
| F40 | GJB2 | 29 | 53 | 100 | 100 | c.235delC/N | c.235delC/N |
*NC no-call, NA not applicable, No. number, CSmat confidence score for fetal inheritance from maternal haplotype, CSpat confidence score for fetal inheritance from paternal haplotype
The fetal genotypes inferred by NIPD were compared with direct sequencing results of fetal gDNA extracted from CVS or amniotic fluid cells to further validate the accuracy of NIPD. The results of NIPD were in concordant with invasive diagnosis and the standard genotype of captured sequencing (Table 3).
Discussion
In our study, we applied the targeted linked-read sequencing method to resolve the parental haplotypes across a range of disease loci and successfully determined the fetal genotypes in 38 families, at risk for various single gene diseases. The previous method of NIPD needs the input of the genomics data of an affected family member and involves complex computational resources for indirectly phasing proband-based haplotype. As compared to the previous NIPD method, our targeted linked-read sequencing method may show certain advantages. Either genomics data from a proband or other family members may not be obligatory for deducing fetal variant status, or an additional capture probe. The new method may in particularly benefit the first pregnancy for those women carrying disease variants, due to lack of genomics information from other affected family members.
In recent years, several studies have utilized the direct haplotyping method to perform NIPD of single gene disorders [18, 23]. Hui et al conducted whole genome haplotyping method and resolved the parental haplotypes with the use of linked-read sequencing technology. They correctly deduced the fetal variant profiles in 12 out of 13 families at risk for a number of autosomal and X-linked diseases. However, the cost of whole genome haplotyping method is relatively high, which might limit its wide use in clinical settings. Vermeulen et al established the targeted locus amplification approach and phased heterozygous variants in selected genes, the method reduced the cost of whole genome haplotyping method and predicted fetal variant status with a high accuracy. Michael Parks utilized targeted capture enrichment of SNPs across a 6 Mb genomic window on chromosome 5 containing the SMN1 gene and successfully deduced fetal variants by relative haplotype dosage with 100% accuracy [11]. However, customizing the targeted region might be a complex task, due to population frequency difference of SNPs across different ethnicities [26]. Our method is advantageous to the above-mentioned 2 direct phasing methods with respect to the cost-effectiveness and recombination prediction. The current NIPD practically requires maternal, paternal DNA and proband’s DNA samples, therefore, the cost of the current proband-dependent method is approximately $830. The major advantage of our method is that it bypassed the availability of the proband’s DNA which considerably reduced the cost to $700. Moreover, multiplexing of a barcoded library further reduces the cost of linked-read sequencing. The turnaround time of linked-read sequencing is 3 weeks, that is more time-consuming than that of the proband-based method but is still affordable for noninvasive prenatal diagnosis. One potential application of our method is NIPD of cystic fibrosis variants which are more relevant to other ethnicity. As demonstrated in this study, the capture probes should cover the whole CF transmembrane regulator (CFTR) gene and highly heterozygous SNPs within 1Mb flanking region of CFTR. With reduced cost, the targeted linked-read sequencing method is capable of NIPD of a wide range of monogenic disorders independently of proband sample.
Despite the advantages as mentioned above, our method still has certain limitations. First, the average percentage of bases on target is approximately 50%, the low on-target rate is a potential limitation of this linked-read target sequencing and may increase the sequencing cost. However, as compared to two other studies, in which the authors reported mean on-target rates of 30.7% and 32% [7, 19], our linked-read target sequencing outperformed the previously published methods. Second, the design of target region and capture probe is critical to successfully conduct targeted linked-read sequencing. There is no existent recommended guideline on the design of capture probes. Additionally, it’s essential to evaluate recombination hot spots surrounding the target region and include the results in the recombination adjustment [27]. Given the clinical applicability of linked-read sequencing hasn’t fully characterized, more researches are required to validate the readiness and effectiveness of this technique in the future.
Conclusions
In summary, we have provided solid evidence that targeted linked-read sequencing method could be applied to the noninvasive assessment of a variety of fetal single gene diseases. The method is a cost-effective and could be widely adopted in clinical practice.
Supplementary Information
Additional file 1: Table S1. Summary statistics of alignment
Additional file 2: Figure S1. The NIPD results of α-thalassemia and β-thalassemia
Additional file 3: Figure S2. The NIPD results of MMA
Additional file 4: Figure S3. The NIPD results of PKU, ARPKD and DFNB1A
Acknowledgements
Not applicable
Abbreviations
- SNPs
Single-nucleotide polymorphisms
- cff-DNA
Cell-free fetal circulating DNA
- MPS
Massively parallel sequencing
- NIPD
Noninvasive prenatal diagnosis
- PCR
Polymerase chain reaction
- DMD
Duchenne muscular dystrophy
- CAH
Congenital adrenal hyperplasia
- MSUD
Maple syrup urine disease
- SMA
Spinal muscular atrophy
- TLA
Targeted locus amplification
- LFR
Long fragment read
- WGS
Whole-genome sequencing
- HMW
High molecular weight
- MMA
Methylmalonic acidemia
- PKU
Phenylketonuria
- ARPKD
Autosomal recessive polycystic kidney disease
- DFNB1A
Autosomal recessive deafness-1A
- gDNA
Genomic DNA
- BWA
Burrows–Wheeler Aligner
- HMM
Hidden Markov model
- cM
Centimorgan
- CVS
Chorionic villus sampling
- GA
Gestational age
- CS
Confidence score
Authors' contributions
JS and YY designed and directed the study. CC, LJ, FG, YZ, YW, HW, ZL performed the sequencing experiment and bioinformatic analysis. MC recruited single gene disorder families, conducted genetic counselling, validation of NIPD results. CC, JL and ZP participated in the writing and revision of the manuscript. All authors have read and approved the manuscript.
Funding
The work was supported by the National Natural Science Foundation of China (NSFC) (No. 81671470), the National Key Research and Development Program of China (2018YFC1004104), Major Technical Innovation Project of Hubei Province (No. 2017ACA097). The funding body played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets supporting the conclusions of this study are available from the corresponding author upon reasonable request. The raw datasets generated during the current study are not publicly available because it is possible that individual privacy could be compromised. The Human reference genome (Hg19) for alignment were obtained from UCSC (https://hgdownload.soe.ucsc.edu/goldenPath/hg19/bigZips/).
Declarations
Ethics approval and consent to participate
All the participants provided written informed consent. The study was approved by the Institutional Review Board of the Third Affiliated Hospital of Guangzhou Medical University and the Institutional Review Board of BGI (BGI-IRB No.17080-T1). This study was performed in accordance with the principles of the Helsinki Declaration.
Consent for publication
Not applicable.
Competing interests
ZP is on the editorial board for BMC Medical Genomics journal. No other potential conflict of interest relevant to this article was reported.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chao Chen, Min Chen, Yaping Zhu, Lu Jiang have contributed equally to this work.
Contributor Information
Yun Yang, Email: yangyun@genomics.cn.
Jun Sun, Email: sunjun@genomics.cn.
Reference
- 1.Chitty LS, Bianchi DW. Noninvasive prenatal testing: the paradigm is shifting rapidly. Prenat Diagn. 2013;33:511–3. doi: 10.1002/pd.4136. [DOI] [PubMed] [Google Scholar]
- 2.Jenkins LA, Deans ZC, Lewis C, Allen S. Delivering an accredited non-invasive prenatal diagnosis service for monogenic disorders and recommendations for best practice. Prenat Diagn. 2018;38:44–51. doi: 10.1002/pd.5197. [DOI] [PubMed] [Google Scholar]
- 3.Lench N, Barrett A, Fielding S, Mckay F, Hill M, Jenkins L, et al. The clinical implementation of non-invasive prenatal diagnosis for single-gene disorders: challenges and progress made. Prenat Diagn. 2013;33:555–62. doi: 10.1002/pd.4124. [DOI] [PubMed] [Google Scholar]
- 4.Drury S, Mason S, McKay F, Lo K, Boustred C, Jenkins L, et al. Implementing Non-Invasive Prenatal Diagnosis (NIPD) in a National Health Service Laboratory; From Dominant to Recessive Disorders BT - Circulating Nucleic Acids in Serum and Plasma – CNAPS IX. In: Gahan PB, Fleischhacker M, Schmidt B, editors. Cham: Springer; 2016. p. 71–5. [DOI] [PubMed]
- 5.Hill M, Twiss P, Verhoef TI, Drury S, Mckay F, Mason S, et al. Non-invasive prenatal diagnosis for cystic fibrosis: detection of paternal mutations, exploration of patient preferences and cost analysis. Prenat Diagn. 2015;35:950–8. doi: 10.1002/pd.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chitty LS, Mason S, Barrett AN, Mckay F, Lench N, Daley R, et al. Non-invasive prenatal diagnosis of achondroplasia and thanatophoric dysplasia: next-generation sequencing allows for a safer, more accurate, and comprehensive approach. Prenat Diagn. 2015;35:656–62. doi: 10.1002/pd.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoo SK, Lim BC, Byeun J, Hwang H, Kim KJ, Hwang YS, et al. Noninvasive prenatal diagnosis of Duchenne muscular dystrophy: comprehensive genetic diagnosis in carrier, proband, and fetus. Clin Chem. 2015;61:829–37. doi: 10.1373/clinchem.2014.236380. [DOI] [PubMed] [Google Scholar]
- 8.Ma D, Ge H, Li X, Jiang T, Chen F, Zhang Y, et al. Haplotype-based approach for noninvasive prenatal diagnosis of congenital adrenal hyperplasia by maternal plasma DNA sequencing. Gene. 2014;544:252–8. doi: 10.1016/j.gene.2014.04.055. [DOI] [PubMed] [Google Scholar]
- 9.You Y, Sun Y, Li X, Li Y, Wei X, Chen F, et al. Integration of targeted sequencing and NIPT into clinical practice in a Chinese family with maple syrup urine disease. Genet Med. 2014;16:594–600. doi: 10.1038/gim.2013.197. [DOI] [PubMed] [Google Scholar]
- 10.Ye J, Chen C, Yuan Y, Han L, Wang Y, Qiu W, et al. Haplotype-based noninvasive prenatal diagnosis of hyperphenylalaninemia through targeted sequencing of maternal plasma. Sci Rep. 2018;8:1–8. doi: 10.1038/s41598-017-18358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parks M, Court S, Bowns B, Cleary S, Clokie S, Hewitt J, et al. Non-invasive prenatal diagnosis of spinal muscular atrophy by relative haplotype dosage. Eur J Hum Genet. 2017;25:416–22. doi: 10.1038/ejhg.2016.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitzman JO, MacKenzie AP, Adey A, Hiatt JB, Patwardhan RP, Sudmant PH, et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat Biotechnol. 2011;29:59–64. doi: 10.1038/nbt.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amini S, Pushkarev D, Christiansen L, Kostem E, Royce T, Turk C, et al. Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nat Genet. 2014;46:1343–9. doi: 10.1038/ng.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Vree PJP, De Wit E, Yilmaz M, Van De Heijning M, Klous P, Verstegen MJAM, et al. Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat Biotechnol. 2014;32:1019–25. doi: 10.1038/nbt.2959. [DOI] [PubMed] [Google Scholar]
- 15.Selvaraj S, Dixon JR, Bansal V, Ren B. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nat Biotechnol. 2013;31:1111–8. doi: 10.1038/nbt.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters BA, Kermani BG, Sparks AB, Alferov O, Hong P, Alexeev A, et al. Accurate whole-genome sequencing and haplotyping from 10 to 20 human cells. Nature. 2012;487:190–5. doi: 10.1038/nature11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Browning SR, Browning BL. Haplotype phasing: existing methods and new developments. Nat Rev Genet. 2011;12:703–14. doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hui WWI, Jiang P, Tong YK, Lee WS, Cheng YKY, New MI, et al. Universal haplotype-based noninvasive prenatal testing for single gene diseases. Clin Chem. 2017;63:513–24. doi: 10.1373/clinchem.2016.268375. [DOI] [PubMed] [Google Scholar]
- 19.Jang SS, Lim BC, Yoo S-K, Shin J-Y, Kim K-J, Seo J-S, et al. Targeted linked-read sequencing for direct haplotype phasing of maternal DMD alleles: a practical and reliable method for noninvasive prenatal diagnosis. Sci Rep. 2018;8:8678. doi: 10.1038/s41598-018-26941-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen M, Chen C, Huang X, Sun J, Jiang L, Li Y, et al. Noninvasive prenatal diagnosis for Duchenne muscular dystrophy based on the direct haplotype phasing. Prenat Diagn. 2020;40:918–24. doi: 10.1002/pd.5641. [DOI] [PubMed] [Google Scholar]
- 21.Marks P, Garcia S, Barrio AM, Belhocine K, Bernate J, Bharadwaj R, et al. Resolving the full spectrum of human genome variation using linked-reads. Genome Res. 2019;29:635–45. doi: 10.1101/gr.234443.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Li X, Ge H-J, Xiao B, Zhang Y-Y, Ying X-M, et al. Haplotype-based approach for noninvasive prenatal tests of Duchenne muscular dystrophy using cell-free fetal DNA in maternal plasma. Genet Med. 2015;17:889–96. doi: 10.1038/gim.2014.207. [DOI] [PubMed] [Google Scholar]
- 25.Chen C, Li R, Sun J, Zhu Y, Jiang L, Li J, et al. Noninvasive prenatal testing of alpha-thalassemia and beta-thalassemia through population-based parental haplotyping. Genome Med. 2021;13(1):18. doi: 10.1186/s13073-021-00836-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vermeulen C, Geeven G, de Wit E, Verstegen MJAM, Jansen RPM, van Kranenburg M, et al. Sensitive monogenic noninvasive prenatal diagnosis by targeted haplotyping. Am J Hum Genet. 2017;101:326–39. doi: 10.1016/j.ajhg.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam KWG, Jiang P, Liao GJW, Chan KCA, Leung TY, Chiu RWK, et al. Noninvasive prenatal diagnosis of monogenic diseases by targeted massively parallel sequencing of maternal plasma: application to β-thalassemia. Clin Chem. 2012;58:1467–75. doi: 10.1373/clinchem.2012.189589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Summary statistics of alignment
Additional file 2: Figure S1. The NIPD results of α-thalassemia and β-thalassemia
Additional file 3: Figure S2. The NIPD results of MMA
Additional file 4: Figure S3. The NIPD results of PKU, ARPKD and DFNB1A
Data Availability Statement
The datasets supporting the conclusions of this study are available from the corresponding author upon reasonable request. The raw datasets generated during the current study are not publicly available because it is possible that individual privacy could be compromised. The Human reference genome (Hg19) for alignment were obtained from UCSC (https://hgdownload.soe.ucsc.edu/goldenPath/hg19/bigZips/).