Abstract
We analysed immunological response during vaccination by using quantitative anti-spike IgG antibodies (qAbs) and Interferon-gamma (IFNγ) production by SARS-CoV-2-specific CD4+ and CD8+ T cells (QuantiFERON® assay). Blood samples were collected at four time points: a day before the reception of first (T0) and second (T1) BNT162b2 doses, 14 (T2) and 28 days (T3) after second dose. Fifty individuals were included: 34 previously infected by SARS-CoV-2 (CoV2+) and 16 that were not (CoV2-). Among CoV2+, we only observed significant differences after the first dose in both qAbs and IFNγ+ T cells. CoV2- showed differences after each dose, and the response was lower than CoV2+. Older people presented a higher response in CoV2+, while in CoV2, young people responded best. Our results suggest that the second BNT162b2 vaccine dose is not a priority in people with previous COVID-19. QuantiFERON® is a good option to monitor T-cell immunity to SARS-CoV-2.
Keywords: mRNA vaccine, COVID-19, Immune response, SARS-CoV-2, IFNγ release assay
1. Introduction
The primary goal of SARS-CoV-2 vaccines is to stop transmission and prevent infection and disease, mainly focusing on those who are at higher risk of severe complications, as is the case for older people. Rapid vaccination implementation is now the world´s health care priority. The first vaccines that have been approved—BNT162b2 (Pfizer-BioNTech) and mRNA-1273 (Moderna)—rely on a new vaccine approach, the introduction of lipid nanoparticles containing messenger RNA (mRNA) molecules encoding for the spike (S) protein of the virus, which is the main target of neutralizing antibodies (NABS) against SARS-CoV-2 (Krammer, 2020). Studies in non-human primates have shown that infection with wild-type viruses prevents subsequent reinfection, thus indicating the existence of protective immunity, which has been associated with the production of NABS (Chandrashekar et al., 2020; Deng et al., 2020). Prevention of reinfection has also been observed in human epidemiological studies (Addetia et al., 2020), indicating that sufficient NABS may be a clue for protection against COVID-19. The first vaccine trials have demonstrated good immunogenicity and similar levels of neutralizing and receptor-binding domain (RBD) antibodies in vaccinated compared to infected individuals (Krammer, 2020; Walsh et al., 2020).
Some pre-clinical studies in non-human primates have shown that vaccines also elicit a T cell response, which may help protect against challenge with the virus (Chandrashekar et al., 2020; Corbett et al., 2020). Furthermore, the first trials in humans with mRNA vaccines have concluded that they are able to induce a Th1-polarized cell response after a 2-dose schedule (Sahin et al., 2020) rather than a Th2 response, which has been related to complications after SARS and MERS vaccinations in animal models (Graepel et al., 2020; Graham et al., 2013). What is clear is that a study of the combination of the humoral and cellular response is needed to increase knowledge of the efficacy of the vaccines—including, most importantly, whether they can protect from infection and severe cases of COVID-19, and if so what levels of surrogate markers may indicate protection. It has also been demonstrated that a robust CD4+ and CD8+ T cell activation is reached after natural SARS-CoV-2 infection (Grifoni et al., 2020; Sekine et al., 2020), even in the absence of circulating antibodies (Gallais et al., 2021; Sekine et al., 2020). Moreover, an adaptive immune response can be detected long after a natural infection (Sherina et al., 2021; Wang et al., 2021). But, limited research has been conducted regarding the immune response, especially the T cell response, to vaccination after a previous natural infection (Krammer, 2020), and this situation is common, given that a large number of people have already been exposed to the virus. And, if a single dose is able to boost protection sufficiently, perhaps the vaccination schedule could be modified, as has been suggested (Ewer et al., 2020; Krammer et al., 2021).
Different assays have been used to assess T cell response to various pathogens, including viruses, by using different platforms and technologies, such as intracellular cytokine staining followed by flow cytometry, peptide-major histocompatibility complex multimer technologies, or Interferon-gamma (IFNγ) production detected either by ELISpot or ELISA (Tischer et al., 2014). The QuantiFERON® SARS-CoV-2 RUO (Qiagen) consists of the detection of IFNγ production by either CD4+ or CD4+ and CD8+ T cells by an ELISA assay, benefiting from the previous experience with the tuberculosis assay (Zhou et al., 2020), highly expanded and standardized.
Our goal is to provide data so that public health policies can implement the best vaccination strategies to optimize available vaccines and achieve high vaccination coverage quickly. Therefore, we have investigated immunological responses after the first and second BNT162b2 doses by using quantitative anti-spike IgG antibodies and IFNγ production by CD4+ and CD8+ T cells with the new commercial IFNγ release assay (IGRA), QuantiFERON® SARS-CoV-2 RUO (Qiagen), as markers of humoral and cellular immunity, respectively.
2. Materials and methods
2.1. Study population
We included fifty individuals from a nursing home: 27 residents, all over 60 years old, and 23 socio-sanitary staff, all under 60 years old, scheduled for BNT162b2 vaccination in January 2021. Previous COVID-19 infection in March–April 2020 was confirmed in 34 people (CoV2+), 31 of them by RNA RT-PCR and three by seroconversion. All of the cases were classified as asymptomatic or mild, with only two of them requiring hospital admission. Serum and whole blood samples of participants were drawn at four time points: the day before the administration of the first (T0) and second (T1) BNT162b2 doses; then, 14 (T2) and 28 days (T3) after the second dose. Clinical data were not recorded as this was not the objective of the study. All the procedures of the study were carried out in accordance with the ethical standards described in the Helsinki Declaration. Confirmed consent was obtained from all participants. The study was approved by our ethics committee (140/2020).
2.2. Quantitative IgG antibodies detection
SARS-CoV-2 specific antibodies were detected with the quantitative IgG (S) enzyme-linked immunosorbent assay (ELISA) (Euroimmun, Lübeck, Germany), followed by additional 1:10 and 1:100 sample dilutions, as recommended by the manufacturer. A result >10 RU/ml is considered positive.
2.3. Commercial IFNγ release assay
Cellular immune response was assessed with the QuantiFERON® SARS-CoV-2 RUO (Qiagen) commercial assay. This test is based on in vitro CD4+ and CD8+ lymphocytes stimulation in heparinized whole blood with a combination of specific SARS-CoV-2 antigens covering the S protein, followed by measurement in plasma of IFNγ production by ELISA (QuantiFERON® ELISA). Briefly, the assay has two sets with two different tubes each, which are designed to draw 1 ml of blood per tube for stimulation. The QuantiFERON SARS-CoV-2 Starter Set Blood Collection Tubes (BCT) consist of two tubes, named Ag1 and Ag2, which detect IFNγ production by CD4+ (IFNγ+CD4+) T cells and CD4+ and CD8+ (IFNγ+CD4+&CD8+) T cells together, respectively. The Ag1 tube contains CD4+ epitopes derived from the S1 subunit (Receptor Binging Domain, RBD), and the Ag2 tube contains CD4+ and CD8+ epitopes from the S1 and S2 subunits. The Control Tube Set contains negative (Nil) and positive (Mitogen) control tubes. All tubes were incubated at 37 ºC for 16 to 24 hours, then centrifuged for 15 minutes at 2500 g to harvest the plasma. Plasma samples can be stored for up to 28 days at 2 to 8 ºC, so that they can be processed together in batches for the detection of IFNγ by ELISA, thus allowing an easy implementation of the assay in the laboratory workflow. Then, the IFNγ ELISA was performed in a Dynex DS2® automated ELISA system. Final IFNγ values (IU/ml) for CD4+, CD4+&CD8+, and mitogen were obtained by subtracting the Nil value from the raw data. As cutoff has yet to be determined by the manufacturer, in an attempt to establish a cutoff value for the present study we added three standard deviation to the mean IFNγ value obtained from 25 unvaccinated and COVID-19 negative healthy donors (HD).
2.4. Statistical analysis
Statistical analysis was performed with SPSS Statistics (version 26.0) and Statgraphics Centurion XVII. Comparison between groups was performed with the chi-square or Fisher's exact tests for categorical variables, and the Wilcoxon signed-rank and Mann-Whitney U tests for dependent and independent continuous variables, respectively. Figures were created with GraphPad Prism (version 9.02). A P-value <0.05 was considered statistically significant.
3. Results
A total of 50 participants were included. Their median age was 67 years (range, 19–97; 54% >60 years); 34 (68%) with (CoV2+) and 16 (32%) without (CoV2-) previous documented COVID-19. In the CoV2+ group, 94% were female compared to 75% in the CoV2- (P = 0.074). Differences in median age between the two cohorts were statistically significant (P = 0.01): 77 (range, 19–97) vs 43 (range, 26–91) years in CoV2+ and CoV2-, respectively.
From the 25 HD samples, the calculated cutoff values for IFNγ+CD4+ and IFNγ+CD4+&CD8+ T cells were of approximately 0.05 (median: 0.00, mean: 0.0036, SD: 0.0132, range: 0.00–0.06) and 0.20 (mean: 0.0304, SD: 0.0554, range: 0.00–0.25) IU/ml, respectively. In order to include all the obtained values from the cohort, and have 100% specificity, we finally established more restrictive cutoffs: 0.10 and 0.25 IU/ml for CD4+ and CD4+&CD8+ T cells, respectively. The results obtained from the 50 participants in the study are shown in Table 1 .
Table 1.
SARS-CoV-2 specific IFNγ production by CD4+ and CD4+&CD8+ T cells measured by the QuantiFERON® assay, according to the proposed cutoffa at the different time points of the study (T0-T3), in previously COVID-19-infected (CoV2+) vs uninfected (CoV2-) individuals.
| Time point | T0 | T1 | T2 | T3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IFNγ+ T cells (IU/ml) | CD4 (IU/ml)(%) |
CD4&CD8 (IU/ml) (%) |
Totalb (IU/ml) (%) |
CD4 (IU/ml)(%) |
CD4&CD8 (IU/ml) (%) |
Totalb (IU/ml) (%) |
CD4 (IU/ml) (%) |
CD4&CD8 (IU/ml) (%) |
Totalb (IU/ml) (%) |
CD4 (IU/ml)(%) |
CD4&CD8 (IU/ml) (%) |
Totalb (IU/ml) (%) |
|
| CoV2- | <60 yrs n = 10 |
0 (0%) | 0 (0%) | 0 (0%) | 9 (90%) | 9 (90%) | 9 (90%) | 10 (100%) | 10 (100%) | 10 (100%) | 9 (90%) | 9 (90%) | 10 (100%) |
| >60 yrs n = 6 |
0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 2 (33%) | 2 (33%) | 3 (50%) | 2 (33%) | 3 (50%) | 3 (50%) | |
| CoV2+ | <60 yrs n = 13 |
2 (15%) | 2 (15%) | 3 (23%) | 13 (100%) | 11 (85%) | 13 (100%) | 12 (92%) | 9 (69%) | 12 (92%) | 11 (85%) | 11 (85%) | 12 (92%) |
| >60 yrs n = 21 |
14 (67%) | 13 (62%) | 15 (71%) | 20 (95%) | 20 (95%) | 20 (95%) | 19 (90%) | 18 (86%) | 19 (90%) | 20 (95%) | 20 (95%) | 21 (100%) | |
| Overall n = 50 |
16 (32%) | 15 (30%) | 18 (36%) | 42 (84%) | 40 (80%) | 42 (84%) | 43 (86%) | 39 (78%) | 44 (88%) | 38 (76%) | 43 (86%) | 46 (92%) | |
The proposed cut-off values are 0.10 and 0.25 IU/ml IFNγ+CD4+ and IFNγ+CD4+&CD8+ T cells, respectively.
A total positive result was considered when at least one of the IFNγ+CD4+ or IFNγ+CD4+&CD8+ T cells was above the cut-off.
At T0, 17/34 (50%) seropositive individuals were detected in the CoV2+ group, while 18/34 (53%) had at least one detectable QuantiFERON parameter of specific T cell response. Among the seropositive participants, 4 out of 17 did not reach positive levels in the QuantiFERON test; thus, 5 people had a negative anti-spike IgG result that had a detectable specific T cell response. The IFNγ-producing T cells were statistically higher among >60 years. There was no humoral nor T-cell response in the CoV2- group.
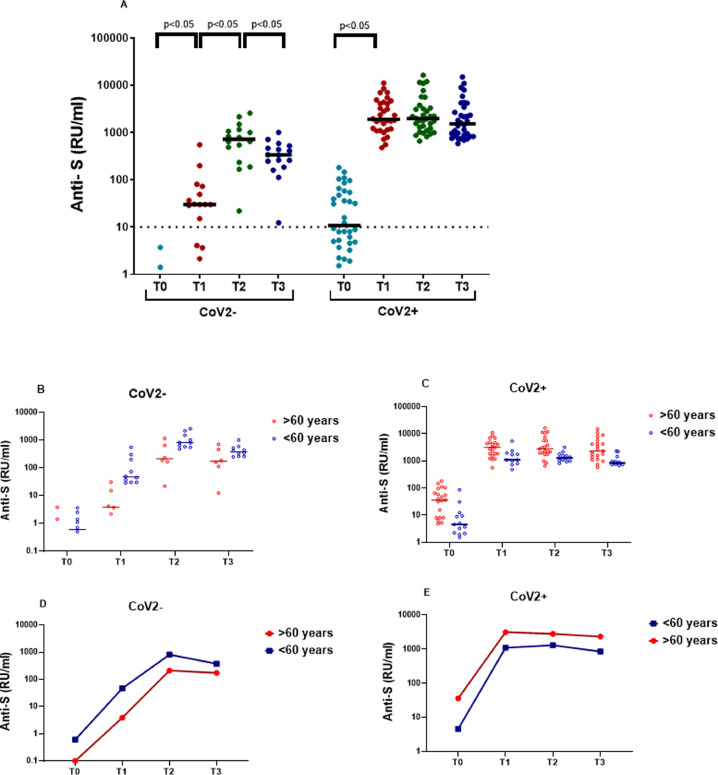
Anti-spike IgG quantification after vaccination in CoV2- individuals showed statistically significant increases after each dose (Fig. 1 A, Table 2 ). Eleven participants presented positivity at T1, but 4 (25%) did not reach levels above the assay cutoff. After the second dose (T2 and T3) all the CoV2- participants were seropositive, although one of them close to the cutoff. Regarding the cellular response, results for the naïve individuals showed a statistically significant increase after the first (T1) and the second doses (T2) in both T-cell subsets, and a month after the vaccination schedule ended, a tendency to decrease (P > 0.05) was seen, also in both T-cell subsets (Fig. 2 A-B). In 3 individuals, all aged >60 years, IFNγ was not detected in any of the lymphocyte populations studied (Table 1), coinciding in T3 with the lowest antibodies values. Humoral (Table 2) and cellular responses (Table 3 ) in the CoV2- study were weaker in participants over 60 years compared to those younger, significantly so in most cases.
Fig. 1.
Quantitative anti-spike IgG at the different time points in the cohort study (n = 50). (A) Comparison between individuals in the CoV2- and CoV2+ groups. (B-E) Antibody kinetics between less than and more than 60 years old individuals in the 2 previous groups, CoV2- and CoV2+, respectively. The horizontal black solid lines indicate median anti-spike IgGs in each cohort. Statistically significant (P < 0.05) differences between paired results are added in the figure.
Table 2.
Median anti-spike IgG antibodies results in the different groups of the study at the four time points.
| Median anti-spike IgG (RU/ml)(IQR) |
|||||
|---|---|---|---|---|---|
| T0 | T1 | T2 | T3 | ||
| CoV2- n = 16 |
Total | 0 (0–1) |
30 (12–55) |
645 (424–1039) |
340 (253–494) |
| <60 yrs n = 6 |
0.6 (0–1.4) |
47 (31–168) |
813 (596–1375) |
374 (279–501) |
|
| >60 yrs n = 10 |
0 (0–1) |
3.8 (2–12.3) |
211 (172–540) |
185 (160–465) |
|
| P-valuea | 0.337 | 0.005 | 0.017 | 0.195 | |
| CoV2+ n = 34 |
Total | 11 (5–53) |
1902 (1182–4163) |
1977 (1546–3281) |
1526 (856–3260) |
| <60 yrs n = 13 |
5 (2–9.3) |
1100 (1022–1857) |
1300 (1016–1558) |
848 (743–958) |
|
| >60 yrs n = 21 |
36 (8–58) |
2802 (1762–4055) |
2340 (1988–3308) |
2159 (1335–3454) |
|
| P-valuea | 0.003 | 0.016 | 0.005 | 0.001 | |
The P-values were obtained by the U Mann-Whitney test between <60 yrs and >60 yrs groups at each time point.
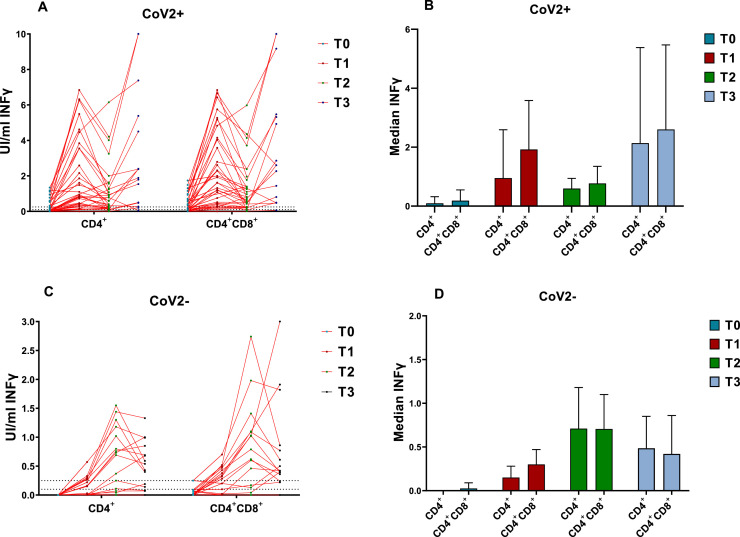
Fig. 2.
IFNγ production by CD4+ and CD4+&CD8+ T lymphocytes after stimulation with a combination of specific SARS-CoV-2 antigens (QuantiFERON) at different time points (T0-T3) for previously COVID-19 infected (CoV2+, n = 34) vs not infected (CoV2-, n = 16) participants. The Wilcoxon test was used to compare the results in CoV2- and CoV-2+ among the 4 time points. In CoV2-, all total values showed significant differences (P < 0.05) except between T2 and T3 for IFNγ+CD4+ (P = 0.107) and IFNγ+CD4+&CD8+ (P = 0.149) T cells. In CoV2+, all total values were significantly different except between T1 and T3 for IFNγ+CD4+ (P = 0.945) and IFNγ+CD4+&CD8+ (P = 0.761). IFNγ = Interferon-gamma.
Table 3.
Median IFNγ production by CD4+ (A) and CD4+&CD8+ (B) T lymphocytes by the QuantiFERON assay at the different time points of the study (T0-T3), in <60 and >60 yrs old in previously infected (CoV2+) vs uninfected (CoV2-) individuals.
| Time point | T0 |
T1 |
T2 |
T3 |
|||||
|---|---|---|---|---|---|---|---|---|---|
| IFNγ+ T cells (IU/ml) | CD4(IQR) | CD4&CD8(IQR) | CD4(IQR) | CD4&CD8(IQR) | CD4(IQR) | CD4&CD8(IQR) | CD4(IQR) | CD4&CD8(IQR) | |
| CoV2- n = 16 | Total | 0.00 (0.00–0.00) | 0.02 (0.00–0.07) | 0.15 (0.00–0.28) | 0.3 (0.04–0.46) | 0.71 (0.08–1.27) | 0.7 (0.14–1.33) | 0.48 (0.09–0.81) | 0.42 (0.27–0.84) |
| <60 yrs n = 6 |
0.00 (0.00–0.00) | 0.01 (0.00–0.05) | 0.24 (0.15–0.31) | 0.4 (0.31–0.50) | 1.1 (0.72–1.47) | 1.06 (0.75–2.09) | 0.68 (0.51–0.99) | 0.69 (0.41–1.84) | |
| >60 yrs n = 10 |
0.00 (0.00–0.00) | 0.06 (0.00–0.13) | 0 (0.00–0.015) | 0.01 (0.00–0.12) | 0.05 (0.00–0.14) | 0.1 (0.00–0.49) | 0.07 (0.00–0.15) | 0.29 (0.00–0.40) | |
| P-valueª | 1.000 | 0.212 | 0.004 | 0.001 | 0.001 | 0.003 | 0.001 | 0.008 | |
| CoV2+ n = 34 | Total | 0.09 (0.03–0.56) | 0.18 (0.05–0.66) | 0.95a (0.42–3.55) | 1.92 (0.52–4.15) | 0.59 (0.20–1.25) | 0.77 (0.26–1.49) | 0.79a (0.27–2.38) | 1.34 (0.45–3.38) |
| <60 yrs n = 13 |
0.03 (0.02–0.09) | 0.06 (0.03–0.17) | 0.45 (0.29–0.86) | 0.49 (0.28–1.29) | 0.22 (0.13–0.37) | 0.41 (0.21–0.73) | 0.29 (0.15–0.85) | 0.49 (0.32–1.25) | |
| >60 yrs n = 21 |
0.32 (0.05–0.85) | 0.41 (0.16–1.06) | 1.61 (0.78–4.46) | 2.61 (1.32–5.18) | 0.89 (0.40–1.30) | 1.25 (0.46–2.38) | 1.54 (0.63–4.46) | 1.98 (0.94–5.27) | |
| P-valueª | 0.005 | 0.004 | 0.017 | 0.003 | 0.008 | 0.018 | 0.009 | 0.012 | |
P-values were obtained by the U Mann-Whitney test between <60 yrs and >60 yrs groups at each time point.
In the previously-naturally-infected group (CoV2+), a significant increase was observed only after the first dose, in both anti-S IgGs and IFNγ-producing CD4+ and CD4+&CD8+ T-cells; and all median values in the different immune subsets were higher than in the CoV2- group. After the second dose, there was a slight increase (P < 0.05) in antibody levels (Fig. 1C), and, unlike the COV2- group, those over 60 years reached higher IgGs values at all-time points compared to those under 60 years (Fig. 1D). The T response after the first dose (T1) was much greater than in the CoV2- group, and only in one individual was IFNγ not detected in the QuantiFERON assay. Two weeks after the second dose (T2), a significant decrease of IFNγ production by T cells was observed, followed by a subsequent increase in T3 to levels comparable to those obtained in T1 (Fig. 2C-D). All the CoV2+ participants showed QuantiFERON positive results at some time point, but at the end of the study, one individual <60 years was above the cutoff. When the cellular response was compared between CoV2+ age groups, all the values were significantly higher in the >60 year group (Table 3).
4. Discussion
In the present study, we have observed two substantially different kinetics in the adaptive immune responses (humoral and cellular) to a 2-dose schedule mRNA vaccine among persons with and without previous COVID-19.
All the individuals who had a proved COVID-19 infection approximately 10 months before experienced, after the first vaccine dose, a rapid and elevated increase in the production of anti-spike IgG levels, as observed in other recent studies (Krammer et al., 2021; Prendecki et al., 2021), requiring a 1:100 additional dilution of samples. This was accompanied by a potent activation of virus-specific CD4+ and CD8+ T effector memory cells, seen through the increase of IFNγ cytokine production, and that was previously detected in other studies of cellular immune response follow-up after COVID-19 infection (Dan et al., 2021; Rodda et al., 2021).
In fact, what we have observed is a booster effect following the first vaccine dose, as the individuals were exposed to the same antigen twice (Lu, 2009). Another study has shown a similar response after one dose of the same vaccine (Prendecki et al., 2021). The second vaccine dose does not improved either humoral or cellular immune responses, as no significant increases in anti-spike IgG levels or specific IFNγ-producing T cells was detected, as stated by others (Krammer et al., 2021). Additionally, we observed a significant decrease in IFNγ production by T cells early (2 weeks) after the second vaccine dose, which may be related to a downregulation of the response, potentially related to regulatory T cells (Treg). Antiviral vaccination has been shown to stimulate Treg, which control exacerbated responses and prevent inflammatory damage, but that can limit the efficacy of the vaccines (De Wolf et al., 2017; Lin et al., 2018). Improvements are being made with vaccine adjuvants in order to inhibit Treg, thus enhancing T cell response (Bayry, 2014; De Wolf et al., 2017). In any event, 1 month after the second dose IFNγ production by T cells increased over this previous time point (14 days after second dose), to levels similar to those achieved after the first dose, meaning that the downregulation could be transient, as suggested by others (De Wolf et al., 2017). More investigation of that point is needed.
Surprisingly, when we analysed results by age groups, we found that older people (>60 years) had more robust immune responses, displaying more than twice the serum antibody titres and IFNγ levels of CoV2+ people aged <60 years. This difference may be explained by a more exacerbated natural primary response in older people, as has been previously reported, that, in addition to implying more severity in this age-group (Etard et al., 2020; Martínez-Serrano et al., 2020; Verity et al., 2020), also generates a greater T-cell memory production, allowing a more intense booster effect, even though the members of our studied cohort did not had severe COVID-19.
In contrast to participants with previous COVID-19, individuals with no previous natural infection showed a slower and lower increase in both the humoral and cellular response markers. After the first dose, specific antibody levels and IFNγ production by T cells were strikingly poorer, remarkably so in older people, making the second dose essential. This may be explained by immunological senescence (Lang and Aspinall, 2012) that makes vaccination less effective in the elderly, as happens with other active immunisation processes. Even if the response may seem weak in the CoV2- group, more studies are assured in order to examine the time evolution of this immune subsets, considering also other markers that have not been studied here. In a recent study, an mRNA vaccine elicited a Th1-biased response, being the production of TNFα and IL-2 higher than IFNγ, which may in part explain why our results are not very high (Jackson et al., 2020). Also, it is important to note that the local immune response may differ from the peripheral blood compartment.
Here, we demonstrate that a commercial IGRA, the QuantiFERON SARS-CoV-2 assay, is a valuable tool to analyse T cell response in large numbers of samples, being easy to implement in a routine laboratory workflow. To our knowledge, no other studies have been published using this specific assay. Similar approximation work has preceded ours (Aiello et al., 2021; Murugesan et al., 2020; Petrone et al., 2021). We have proposed a cutoff for a specific T-cell response to SARS-CoV-2 of at least one result above 0.10 and 0.25 IU/ml for CD4+ and CD4+&CD8+ T cells, respectively. In a previous work, Murugesan et al. (Murugesan et al., 2020) developed an in-house IGRA assay for detection of SARS-CoV-2, and median IFNγ results in healthy donors was 0.01 (IQR 0–0.01) IU/ml, comparable to ours (0.00 IU/ml). They found that stimulation with the entire SARS-CoV-2 proteome showed higher results in COVID-19 convalescent people, which is not our case (the vaccination target is the S protein). Another study found a better IFNγ response after stimulation with a spike mega-pool compared to a remainder-antigens mega-pool (Petrone et al., 2021), probably because the latter did not include the S-region. Even so, for purposes other than vaccine response, it is important to note that T cell response to SARS-CoV-2 has been proven to bind a number of epitopes in the non-S region (Grifoni et al., 2020).
In our study, 5 out of 17 participants who had been previously infected with SARS-CoV-2 (29%) had a detectable T cell response by QuantiFERON with negative anti-spike results, demonstrating that the assay may be also useful in detecting seronegative individuals with previous infection, as suggested by others (Gallais et al., 2021; Sekine et al., 2020). This aspect is of relevance because it can help to better estimate the exposed / immune population, and to evaluate the efficacy of the vaccines, taking into account the potential waning of antibody responses over time, and the fact that specific CD4+ and CD8+ T cells appear to confer broad and long-lasting protection to severe reinfection (Sekine et al., 2020). A SARS-CoV-2 IGRA test has been shown to anticipate the antibody response in some COVID-19 acute cases (Aiello et al., 2021), thus it may also be of interest in diagnosis.
It is also notable that, like us, some published data did not detect a non-specific response by using an IGRA test (Murugesan et al., 2020; Petrone et al., 2021), contrary to what others have found with flow cytometry assays (Sekine et al., 2020), which may be related to the nature of the test. In fact, we detected some “residual response” in HD, which was higher for the IFNγ+CD4+&CD8+ T cell subset than for the IFNγ+CD4+ T cell one, which may also been explained by the different epitopes used to stimulate each subset, being RBD and S1/S2, respectively. It has been published by others (Braun et al., 2020), that the CD4+ T cell reactivity to SARS-CoV-2 in HD was mainly due to the C-terminal S epitopes -corresponding to the S2 subunit-, which have a higher homology to the S proteins of other common human coronaviruses; in comparison to N-terminal epitopes -where the RBD is situated-. In order to detect specific T-cell response to SARS-CoV-2 vaccine, we raised the cutoff values of the tests above the highest value obtained from the uninfected donors.
Another study detected IFNγ production by ELISpot (Sahin et al., 2020), showing that a 2-dose BNT162b2 schedule successfully primed a strong CD4+ T cell response (better than that of CD8+, in accordance with Murugesan et al. (Murugesan et al., 2020)), which was higher for a two dose than a single schedule. According to our data, the CoV2- group benefits equally from the second dose. To sum up, even though more investigation is needed, the QuantiFERON assay could help in the future as a surrogate marker of protective immunity to SARS-CoV-2 or severe COVID-19.
Our study is robust because we were able to assess samples from all participants at all interval times, permitting an optimal follow-up of immune response until 1 month after the second dose of the mRNA vaccine. However, there are limitations in the present work: the total number of participants in each group is low, a female bias exists in both CoV2-/Cov2+ groups, and there are statistically differences in the age between both groups (due to the characteristics of the studied cohort), thus limiting the generalisation of our results. Also, the studied immune subsets, the stimulating epitopes and the detected cytokine (IFNγ) may underrepresent the global response.
5. Conclusions
Our work supports the strategy that a second vaccine dose is not a priority in people with previous COVID-19. This approach would make it possible to optimise the available doses of vaccines without increasing the risk of infection, allowing a faster expansion of vaccination coverage, which is now imperative. For this strategy to work, however, it is necessary to know with certainty who has previously had COVID-19. Thus, we recommend a pre-vaccination epidemiological survey and/or serological investigation in doubtful cases. Alternatively, SARS-CoV-2 serology and T-cell response could be scheduled after the first dose, to demonstrate the described booster effect and assist in deciding if a second dose is advisable. The implementation of more automated assays, such as ELISA and QuantiFERON, may help in generalising the proposed scheme.
Author contribution statement
Nuria Tormo and David Navalpotro: Conceptualization, Methodology, Data curation, Software, Validation, Formal analysis, Writing-original draft preparation. María Martínez: Conceptualization, Methodology, Data curation, Validation, Writing – Reviewing and Editing. Marta Moreno: Data Curation, Investigation, Visualization. Fernando Grosson: Conceptualization, Project Administration. Irene Tur: Investigation, Resources, Data Curation. Maria Remedios Guna: Methodology, Data Curation, Validation, Writing – Reviewing and Editing. Pepa Soriano and Ana Tornero: Project Administration, Data Curation, Resources. Concepción Gimeno: Conceptualization, Supervision, Writing – Reviewing and Editing.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
Acknowledgments
We thank to the staff of the Microbiology Department which have participated in the processing of samples, especially Silvia Navarro and Leticia Bueno.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- Addetia A, Crawford KHD, Dingens A, Zhu H, Roychoudhury P, Huang ML, et al. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with a high attack rate. J Clin Microbiol. 2020;58(11) doi: 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello A, Fard SN, Petruccioli E, Petrone L, Vanini V, Farroni C, et al. Spike is the most recognized antigen in the whole-blood platform in both acute and convalescent COVID-19 patients. Int J Infect Dis. 2021 doi: 10.1016/j.ijid.2021.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayry J. Regulatory T cells as adjuvant target for enhancing the viral disease vaccine efficacy. VirusDis. 2014;25(1):18–25. doi: 10.1007/s13337-013-0187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020:1–8. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Chandrashekar A, Liu J, Martinot AJ, McMahan K, Mercado NB, Peter L, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science (80-.) 2020;369(6505):812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N Engl J Med. 2020 doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J, Mateus J, Kato Y, Hastie K, Dawen Yu E, Faliti C, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science (80-.) 2021 doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wolf ACMT, Van Aalst S, Ludwig IS, Bodinham CL, Lewis DJ, Van Der Zee R, et al. Regulatory T cell frequencies and phenotypes following anti-viral vaccination. PLoS One. 2017;12(6) doi: 10.1371/journal.pone.0179942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Bao L, Liu J, Xiao C, Liu J, Xue J, et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science (80-.) 2020;369(6505):818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etard J-F, Vanhems P, Atlani-Duault L, Ecochard R. Potential lethal outbreak of coronavirus disease (COVID-19) among the elderly in retirement homes and long-term facilities, France, March 2020. Eurosurveillance. 2020;25(15) doi: 10.2807/1560-7917.ES.2020.25.15.2000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewer KJ, Barrett JR, Belij-Rammerstorfer S, Sharpe H, Makinson R, Morter R, et al., “T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial,” Nat Med 2020, doi: 10.1038/s41591-020-01194-5. [DOI] [PubMed]
- Krammer F, Srivastava K, Alshammary H, Amoaka, Awawda MH, Beach KF, et al., “Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine,” 2021. doi: 10.1056/NEJMc2101667. [DOI] [PMC free article] [PubMed]
- Gallais F, Velay A, Nazon C, Wendling MJ, Partisani M, Sibilia J, et al. Intrafamilial exposure to SARS-CoV-2 associated with cellular immune response without Seroconversion, France. Emerg Infect Dis. 2021;27(1):113–121. doi: 10.3201/EID2701.203611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graepel KW, Kochhar S, Clayton EW, Edwards KE. Balancing expediency and scientific rigor in severe acute respiratory syndrome coronavirus 2 vaccine development. J Infect Dis. 2020;222(2):180–182. doi: 10.1093/infdis/jiaa234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RL, Donaldson EF, and Baric RS, “A decade after SARS: strategies for controlling emerging coronaviruses,” 2013, doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed]
- Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher C, et al. Targets of T cell responses to SARS-CoV-2 Coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, et al. An mRNA vaccine against SARS-CoV-2 — Preliminary Report. N Engl J Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. | Nat |. 2020;586:516. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Lang PO, Aspinall R. Immunosenescence and herd immunity: With an ever-increasing aging population do we need to rethink vaccine schedules? Expert Rev Vaccines. 2012;11(2):167–176. doi: 10.1586/erv.11.187. [DOI] [PubMed] [Google Scholar]
- Lin P-H, Wong WI, Wang YL, Hsieh MP, Lu CW, Liang CY, et al. Vaccine-induced antigen-specific regulatory T cells attenuate the antiviral immunity against acute influenza virus infection. Mucosal Immunol. 2018;11:1239–1253. doi: 10.1038/s41385-018-0004-9. [DOI] [PubMed] [Google Scholar]
- Lu S. Heterologous prime–boost vaccination. Curr Opin Immunol. 2009;21(3):346–351. doi: 10.1016/j.coi.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugesan K, Jagannathan P, Pham TD, Pandey S, Bonilla HF, Jacobson K, et al. Interferon-gamma release assay for accurate detection of SARS-CoV-2 T cell response. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrone L, Petruccioli E, Vanini V, Cuzzi G, Najafi Fard S, Alonzi T, et al. A whole blood test to measure SARS-CoV-2-specific response in COVID-19 patients. Clin Microbiol Infect. 2021;27(2):286.e7–286.e13. doi: 10.1016/j.cmi.2020.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendecki M, Clarke C, Brown J, Cox A, Gleeson S, Guckian M, et al. Effect of previous SARS-CoV-2 infection on humoral and T-cell responses to single-dose BNT162b2 vaccine. The Lancet. 2021;397(10280):1178–1181. Elsevier B.V. doi: 10.1016/S0140-6736(21)00502-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda LB, Netland J, Shehata L, Pruner KB, Morawski PA, Thouvenel CD, et al. Functional SARS-CoV-2-Specific immune memory persists after mild COVID-19. Cell. 2021;184(1):169–183.e17. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M, et al. COVID-19 vaccine BNT162b1 elicits human antibody and T H 1 T cell responses. | Nat |. 2020;586:594. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Sekine T, Perez-Potti A, Rivera-Ballesteros O, Strålin K, Gorin JB, Olsson A, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1):158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Serrano M, Navalpotro D, Tormo N, Olmos R, Moreno-Córdoba M, Ocete MD, et al. Comparison of commercial lateral flow immunoassays and ELISA for SARS-CoV-2 antibody detection. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherina N, Piralla A, Du L, Wan H, Kumagai-Braesch M, Andréll J, et al. Persistence of SARS-CoV-2 specific B- and T-cell responses in convalescent COVID-19 patients 6-8 months after the infection. Med. 2021;2(3):281–295.e4. doi: 10.1016/j.medj.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer S, Dieks D, Sukdolak C, Bunse C, Figueiredo C, Immenschuh S, et al. Evaluation of suitable target antigens and immunoassays for high-accuracy immune monitoring of cytomegalovirus and Epstein-Barr virus-specific T cells as targets of interest in immunotherapeutic approaches. J Immunol Methods. 2014;408:101–113. doi: 10.1016/j.jim.2014.05.011. [DOI] [PubMed] [Google Scholar]
- Verity R, Okell LC, Dorigatti I, Winskill P, Whittaker C, Imai N, et al., “Articles estimates of the severity of coronavirus disease 2019: a model-based analysis,” 2020. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed]
- Walsh EE, et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li J, Li H, Lei P, Shen G, Yang C. Persistence of SARS-CoV-2-specific antibodies in COVID-19 patients. Int Immunopharmacol. 2021;90 doi: 10.1016/j.intimp.2020.107271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Luo Q, Luo S, Teng Z, Ji Z, Yang J, et al. Interferon-γ release assays or tuberculin skin test for detection and management of latent tuberculosis infection: a systematic review and meta-analysis. Lancet Infect Dis. 2020;20(12):1457–1469. doi: 10.1016/S1473-3099(20)30276-0. [DOI] [PubMed] [Google Scholar]