Abstract
MCM1 is an essential gene in the yeast Saccharomyces cerevisiae and is a member of the MADS-box family of transcriptional regulatory factors. To understand the nature of the protein-DNA interactions of this class of proteins, we have made a series of alanine substitutions in the DNA-binding domain of Mcm1 and examined the effects of these mutations in vivo and in vitro. Our results indicate which residues of Mcm1 are important for viability, transcriptional activation, and DNA binding and bending. Substitution of residues in Mcm1 which are highly conserved among the MADS-box proteins are lethal to the cell and abolish DNA binding in vitro. These positions have almost identical interactions with DNA in both the serum response factor-DNA and α2-Mcm1-DNA crystal structures, suggesting that these residues make up a conserved core of protein-DNA interactions responsible for docking MADS-box proteins to DNA. Substitution of residues which are not as well conserved among members of the MADS-box family play important roles in contributing to the specificity of DNA binding. These results suggest a general model of how MADS-box proteins recognize and bind DNA. We also provide evidence that the N-terminal extension of Mcm1 may have considerable conformational freedom, possibly to allow binding to different DNA sites. Finally, we have identified two mutants at positions which are critical for Mcm1-mediated DNA bending that have a slow-growth phenotype. This finding is consistent with our earlier results, indicating that DNA bending may have a role in Mcm1 function in the cell.
The yeast Mcm1 protein is a member of the MADS-box family of transcriptional regulatory factors. This growing class of proteins is named after Mcm1, Arg80, Agamous, Deficiens, and serum response factor (SRF), which were among the first proteins isolated containing this DNA-binding and dimerization motif (5, 16, 18, 23, 32). This class of proteins has at least 60 members and can be found in organisms which range from the single-cell yeast Saccharomyces cerevisiae (MCM1, ARG80, and RLM1) to Xenopus (SRF), Drosophila (DRSF and MEF2), plants (Ag, Def, and Glo) and humans (SRF) (2, 5, 13, 15, 16, 18, 20, 23, 28, 32). Although the DNA-binding domains are highly conserved among this family of proteins, their function varies greatly, ranging from regulatory factors that control basic cellular processes to those required for the development and determination of cell type (2, 11, 27, 28). Understanding how this class of proteins functions is therefore important for understanding the mechanism of regulation of many cellular and developmental processes.
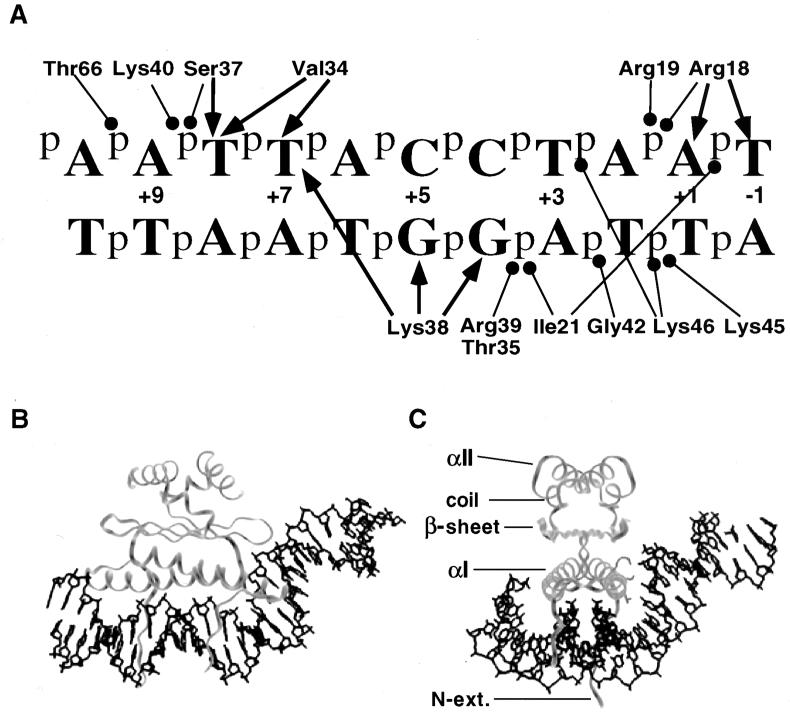
The cocrystal of SRF protein in complex with DNA revealed the structure of the MADS domain for the first time (19). More recently, the structure of another member of the MADS-box family, the yeast Mcm1 protein, indicated that the overall fold of the protein and many of the DNA contacts are remarkably conserved between the yeast and human proteins (24). These structures show the MADS-box domain forms a protein dimer composed of three layers (Fig. 1). The first layer consists of antiparallel coiled-coil alpha-helices, one from each monomer, positioned above the minor groove at the center of the recognition site. Residues in these helices make numerous phosphate and base-specific contacts with the major groove. The middle layer consists of a hydrophobic four-stranded antiparallel β-sheet, formed from two β-strands of each monomer and is aligned roughly parallel to the coiled-coil alpha-helices. The β-loop linking the strands of one monomer is involved in a phosphate contact at a position outside of the conserved CArG-box [CC(A/T)6GG] binding site. The top layer of the protein consists of an extended coil region, followed by an alpha-helix of three turns. This second alpha-helix of each monomer packs against the corresponding face of the other monomer, adding to the dimerization interface. In addition to the three layers of the protein, there is also an N-terminal extension from the first alpha-helix of each monomer that makes several base-specific and phosphate backbone contacts with positions at the center of the recognition site. Interestingly, the position of the extension is very different in the two proteins. In the SRF structure this region follows a path through the minor groove in the center of the site. In the Mcm1 structure, this region crosses over the minor groove and makes base-specific contacts in the major groove in the center of the site.
FIG. 1.
Mcm1 contacts to a consensus binding site DNA. (A) The contacts shown are predicted from the observed contacts in the α2-Mcm1-DNA ternary complex and are to the consensus symmetric Mcm1 binding site used in the studies described here. One-half of the site is shown with the axis of symmetry between +1 and −1 bp. The contacts shown are for one Mcm1 monomer, except for Lys46, which is from the adjacent monomer in the complex. The arrows indicate base-specific contacts; the circles indicate phosphate or sugar backbone contacts. (B and C) Models of the Mcm1 dimer binding to its site derived from the coordinates of the crystal structure of the Mcm1 ternary complex (24). Two different views of the complex are shown, along with the positions of the different layers of the protein. Side chains of residues required for DNA bending, K40, S37, and V34 (top to bottom), are shown in the right monomer in panel C.
Biochemical and structural studies have shown that many MADS-box proteins produce a large bend in the DNA upon binding to their recognition sites (1, 8, 19–22, 24, 30). Circular permutation assays suggest that SRF produces a significant bend in the DNA when binding to its site. These results agree with the SRF crystal structure, in which the DNA is bent on either side of the CArG recognition site towards the protein to produce an overall bend angle of 72° (19). In the Mcm1 structure, the DNA is also bent around the protein, but at the edge of the site there is a second DNA bend away from the protein in a plane perpendicular to the first bend (24). This is unlike the DNA in the SRF structure, which lies predominantly within one plane. Mutations at positions outside of the conserved CArG site affect the degree of bending by Mcm1 (1). These base-pair substitutions also have a large effect on transcriptional activation, suggesting there may be a possible link between the DNA-bending activity of Mcm1 and transcriptional regulation by this MADS-box protein.
To obtain a deeper understanding of the protein-DNA interactions and the requirements for DNA-bending by the Mcm1 protein, we have constructed a series of alanine substitutions in the DNA-binding domain and assayed the effects of these mutations both in vivo and in vitro. Our results identify residues which are critical for supporting viability in the cell and measure the contributions of each side chain toward DNA binding and transcriptional activation. The results presented here, together with the SRF-DNA and α2-Mcm1-DNA crystal structures, help define a conserved set of protein-DNA interactions critical for DNA binding by MADS-box proteins. Our results also identify the residues required for sequence-specific DNA bending by Mcm1. We show that there does not appear to be a strict correlation between simple DNA bending and transcriptional activation by Mcm1 on its own. However, mutants with altered DNA bending do have a slow growth phenotype, indicating that it may have an important role in Mcm1 function in the cell.
MATERIALS AND METHODS
Plasmids.
Individual alanine substitutions at each position in the DNA-binding region of Mcm1 were constructed by cloning double-stranded oligonucleotides containing the desired codon changes into pJM231, a derivative of the pRS413 (CEN HIS3) vector which contains an engineered MCM1 gene expressed from its native promoter. Unique restriction sites, KpnI (bp 24), SacI (bp 51), MluI (bp 91), SpeI (bp 100), MfeI (bp 140), and StuI (bp 170), were engineered into the region coding for the DNA-binding domain of Mcm1 by introducing silent mutations at the indicated nucleotide positions by using recursive PCR as described previously (14). The sequences of the inserts and the adjacent regions were determined to verify the presence of the specific base-pair substitutions and to ensure that other mutations were not introduced during the cloning process. The engineered MCM1 construct complements an mcm1 null mutant for viability and activates transcription at the same level as the unmodified gene, indicating the codon changes did not significantly alter the expression or activity of the protein.
Strains and in vivo assays.
The ability of the MCM1 mutants to maintain cell viability and activate transcription was measured in strain YY2052 [MATa P(PAL)-lacZ::FUS1 leu2-3,112 trp1-1 ura3 his3-11,15 ade2-1 can1-100 mcm1::LEU2/pSL1574-CEN/ARS MCM1 URA3] which was provided by G. Sprague (4). Cells were transformed with the mutant derivatives of pJM231 (CEN HIS3 MCM1) and colonies that have lost the wild-type copy of MCM1 on the plasmid pSL1574 were selected by plating on synthetic medium containing 5-fluoroorotic acid (5-FOA) in the absence of histidine and leucine. Mcm1-dependent transcriptional activation was measured in strain YY2052 by assaying for lacZ expression from the integrated CYC1-lacZ reporter promoter with a P(PAL) Mcm1 binding site by liquid β-galactosidase assays (4). To assay the effects of mutations in the Mcm1 protein in combination with mutant binding sites, mutant derivatives of pJM231 were transformed into strain JM02 (MATa leu2-3,112 trp1-1 ura3 his3-11,15 ade2-1 can1-100 mcm1::LEU2/pSL1574-CEN/ARS MCM1 URA3), and the loss of the plasmid pSL1574, containing the wild-type gene, was selected on medium containing 5-FOA. These strains were then transformed with derivatives of pTBA43, a URA3, 2μm, CYC1-lacZ transcription reporter vector in which the CYC1 upstream activation sequence UAS elements have been removed and replaced with a consensus Mcm1 binding site containing symmetric base-pair substitutions (1). The activation results shown in the tables are based on the averages of three independent transformants for each mutant and are presented as the percent activity of the wild-type protein (see Table 1) or units of β-galactosidase activity (see Table 2 and Fig. 6).
TABLE 1.
Activity of the Mcm1 alanine substitutions
| Mutanta | Growthb | % Activationc | Relative decrease in DNA- binding affinityd (fold) |
|---|---|---|---|
| Wild type | + | 100 | 1 |
| Q14A | + | 104 | 1.5 |
| Q15A | + | 103 | 1.4 |
| K16A | + | 80 | 2.5 |
| E17A | + | 107 | 1.5 |
| R18A | + | 100 | 1.6 |
| R19A | + | 21 | 4 |
| K20A | + | 80 | 3 |
| I21A | + | 53 | 4.5 |
| E22A | + | 87 | 1.4 |
| I23A | + | 34 | 4.3 |
| K24A | + | 110 | 0.8 |
| F25A | − | NDB | |
| I26A | − | NDB | |
| E27A | + | 98 | 2 |
| N28A | + | 87 | 20 |
| K29A | + | 100 | 0.8 |
| T30A | + | 100 | |
| R31A | + | 70 | |
| R32A | − | 110 | |
| H33A | + | 120 | 0.25 |
| V34A | S | 9 | 2.7 |
| T35A | + | 82 | 3 |
| F36A | − | NDB | |
| S37A | S | 66 | 2.1 |
| K38A | − | NDB | |
| R39A | − | NDB | |
| K40A | − | 125 | |
| H41A | + | 100 | 0.12 |
| G42A | − | NDB | |
| I43A | − | 250 | |
| M44A | − | >1,000 | |
| K45A | − | >1,000 | |
| K46A | − | NDB | |
| A47 | + | 100 | 1 |
| F48A | + | 34 | 0.8 |
| E49A | S | 24 | 48 |
| S51A | + | 99 | 1.0 |
| V52A | + | 91 | 1.2 |
| L53A | − | NDB | |
| T54A | − | NDB | |
| T66A | + | 77 | 3.1 |
DNA contacts are in boldface (based on the α2/Mcm1/DNA crystal structure).
+, Normal growth after swapping out the wild-type copy of MCM1; −, no growth after 5 days; S, slow-growth phenotype.
The level of activation is based upon lacZ expression in comparison to the wild type.
The relative decrease in DNA-binding affinity in comparison to the wild-type protein. NDB, no detectable binding was observed in the EMSA.
TABLE 2.
Activation of the N-terminal extension mutants with wild-type and mutant DNA sitesa
| Site | β-Gal activity (U)
|
|||
|---|---|---|---|---|
| WT | E17A | R18A | R19A | |
| None | 0.4 | 0.3 | 0.4 | 0.4 |
| WT | 5.5 | 4.6 | 4.9 | 0.3 |
| A1C | 1.4 | 0.9 | 0.9 | 0.9 |
Values shown are the units of β-galactosidase activity (β-Gal) with standard deviations of <10%. WT, wild type.
FIG. 6.
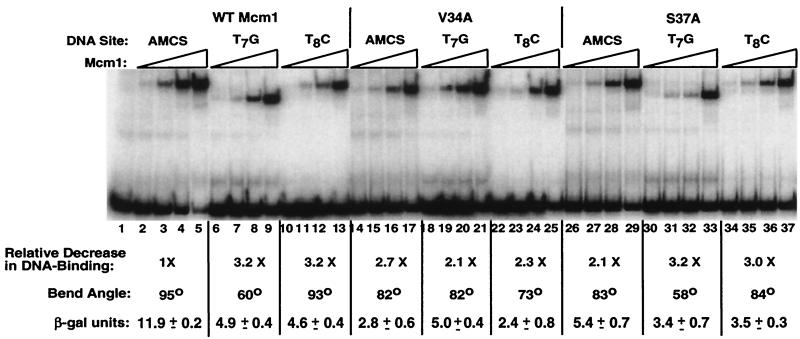
DNA-binding affinity, DNA bending, and transcriptional activation of wild-type, V34A, and S37A proteins to wild-type and mutant binding sites. DNA binding to a symmetric consensus site by wild-type Mcm1 (lanes 2 to 13), V34A (lanes 14 to 25), and S37A (lanes 26 to 37) to the wild-type site (lanes 2 to 5, 14 to 17, and 26 to 29), the T7G site (lanes 6 to 9, 18 to 21, and 30 to 33), and T8C site (lanes 10 to 13, 22 to 25, and 34 to 37). The DNA-binding affinity of each protein for each site was calculated from the binding curves and shown as the fold decrease in affinity relative to the wild-type protein. The apparent bend angle of each of the proteins for the different sites are derived from the data shown in Fig. 4 and 5. The levels of transcriptional activation in vivo by the proteins for each of the sites driving the expression of lacZ and measured as described in Materials and Methods are shown. Values shown are the units of β-galactosidase activity and are the average of three independent isolates.
Protein purification.
Mutant Mcm11–96 proteins used in the in vitro DNA-binding assays were purified from Escherichia coli BL21 cells which were transformed with pHA227, a protein expression vector in which the sequences coding for Mcm1 residues 1 to 96 were fused in frame to the gene coding for the maltose-binding protein. The mutant fusion proteins were purified from 100-ml cultures as described earlier (1). The proteins isolated by this procedure were >95% homogenous and consisted of two nonnative N-terminal amino acids, Gly-Ser, followed by the native Mcm1 residues 1 to 96. The protein concentration of each mutant was determined by Bradford assays (Bio-Rad), normalized, and verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels stained with Coomassie blue.
EMSAs.
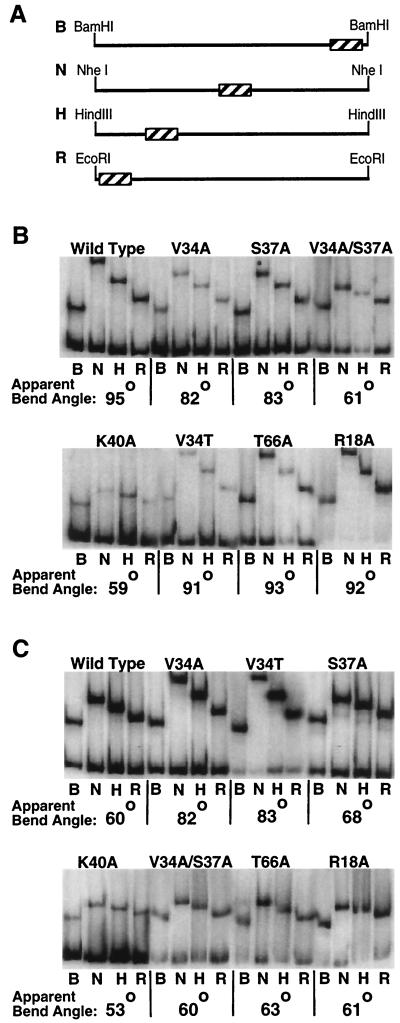
The DNA fragments utilized in the electrophoretic mobility shift assays (EMSAs) (see Fig. 2 and Fig. 6) were synthesized through PCR amplification of wild-type and mutant α2/Mcm1 operator sites from transcription reporter plasmids. As described previously, oligonucleotides W340 (5′-CACGCCTGGCGGATCTGC-3′), which was [γ-32P]ATP end labeled and purified on a Sephadex G-25 spin column, and W341 (5′-GCCCACGCTAGGCAATC-3′) were annealed in the CYC1 promoter region on either side of the Mcm1 binding site and were used to amplify a 120-bp fragment containing the Mcm1 recognition site (1, 33). The PCR-generated fragments were then purified through native PAGE. The circular permutation assays (see Fig. 4 and Fig. 5) were performed with BamHI-, NheI-, HindIII-, or EcoRI-generated end-labeled fragments from derivatives of pGD579, which contains a tandem repeat of a 375-bp fragment separated by a 24-bp polylinker containing wild-type or mutant Mcm1 binding sites (1).
FIG. 2.
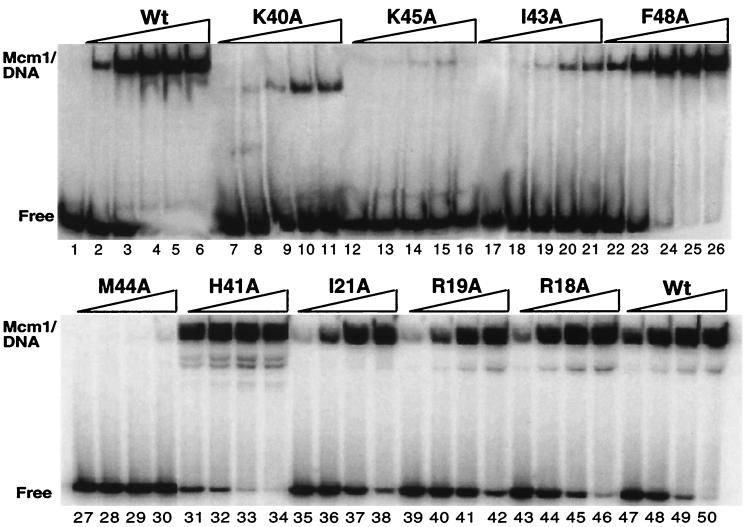
EMSA analysis of Mcm1 substitutions. DNA binding to a symmetric consensus site by wild-type Mcm1 (lanes 2 to 6 and 47 to 50), K40A (lanes 7 to 11), K45A (lanes 12 to 16), I43A (lanes 17 to 21), F48A (lanes 22 to 26), M44A (lanes 27 to 30), H41A (lanes 31 to 34), I21A (lanes 35 to 38), R19A (lanes 39 to 42), and R18A (lanes 43 to 46). All proteins span residues 1 to 98 of Mcm1, which contains the entire MADS-box domain. Fivefold titrations of Mcm1 were performed, increasing from a concentration of 2.2 × 10−10 M (lanes 2, 7, 12, 17, and 22) or 2.0 × 10−9 (lanes 27, 31, 35, 39, 43, and 47). The relative binding affinity calculated for each mutant in comparison to wild-type is shown in Table 1.
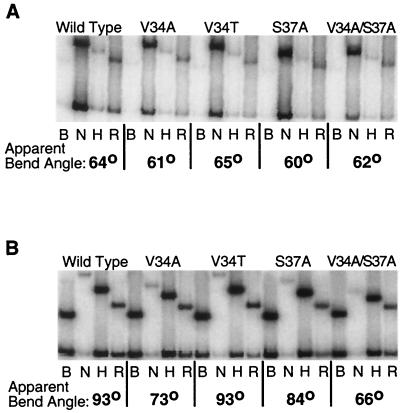
FIG. 4.
Substitutions in Mcm1 that reduce DNA bending to the wild-type and T7G sites. EMSAs were performed by utilizing the position-permuted fragments shown in panel A containing the wild-type (B) or T7G (C) binding sites with wild-type, V34A, S37A, V34A-S37A, K40A, V34T, T66A, and R18A purified proteins as indicated. Lanes labeled B, N, H, and R denote labeled fragments that were generated with BamHI, NheI, HindIII, and EcoRI enzymes, respectively, as shown in panel A. The bend angle for each protein-DNA complex is shown below and was calculated based on the Thompson and Landy relationship (26).
FIG. 5.
The effects of Mcm1 mutants on DNA bending of T8G and T8C mutant sites. EMSAs were performed by utilizing position-permuted fragments containing the T8G (A) and T8C (B) binding sites with wild-type, V34A, V34T, S37A, and V34A-S37A purified proteins as described in Fig. 4.
The purified mutant Mcm11–96 proteins were incubated for 1 h at room temperature with the labeled DNA fragment in 20 mM Tris (pH 7.6), 200 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA, 0.1% NP-40, 10% glycerol, and 10 μg of herring sperm DNA per ml. The circular permutation EMSA was performed with a 6% polyacrylamide gel in 1× Tris-borate-EDTA (TBE) at 200 V, while all other EMSAs were performed with 6% polyacrylamide gel in 0.5× TBE. EMSA gels were dried and visualized by using a Molecular Dynamics 425E PhosphorImager. The relative binding affinity of the Mcm1 mutants was calculated by linear regression and compared against the wild-type protein.
Cross-linking assays.
Oligonucleotides used in the cross-linking experiments were synthesized by using the photoreactive reagent 5-bromo-2′-deoxyuridine-β-cyanoethylphosphoramidite (Glen Research, Inc.) in place of thymine phosphoramadite at position +1 in the top DNA strand of the Mcm1 symmetric consensus site (5′-GCCCTACCTAABrUTAGGTACCCG-3′). The complementary bottom strand (5′-CGGGTACCTAATTAGGTAGGGC-3′), which did not contain 5-bromouracil (BrU), annealed to the BrU containing oligonucleotides to produce a double-stranded recognition site. Cross-linking experiments were performed with purified proteins as described earlier (1).
RESULTS
Conserved residues in Mcm1 are essential for viability.
To determine which residues in Mcm1 are important for its activity, each residue in the DNA-binding region of the MADS-box domain was individually substituted with alanine. MCM1 is an essential gene in yeast, and therefore the alanine mutants were first assayed for their ability to support growth of the cell (18). In order to accomplish this, a plasmid shuffle technique was used to introduce the mcm1 mutants into yeast and to remove the wild-type copy of the gene. The yeast strain YY2052 contains a chromosomal mcm1::LEU2 null mutation and is maintained by a plasmid containing wild-type MCM1 (4). This plasmid also contains URA3, and thus cells that have lost this plasmid can be selected by plating on media containing the drug 5-FOA. Yeast cells were transformed with derivatives of plasmid pJM231 containing the mcm1 alanine substitution mutants and tested for the ability to support growth after plating on 5-FOA (3). Transformants that fail to grow after the wild-type MCM1 has been lost indicates that the alanine substitution has a significant effect on Mcm1 activity and that the mutant is unable to complement the mcm1 null mutation.
The results of the alanine scanning mutagenesis indicate that 14 of the 40 side chains in the DNA-binding region of the Mcm1 MADS-box domain are essential for the ability to support growth of the cell (Table 1). Twelve of the lethal mutations are in the alpha-helix which forms a coiled-coil interface between the monomers and contacts the DNA. Many of these residues contact the DNA in both the SRF and Mcm1 crystal structures and are highly conserved among MADS-box proteins (19, 24) (Fig. 1). Alanine substitutions of residues K38, R39, K40, G42, K45, and K46, which make direct contacts with DNA in the Mcm1 crystal structure, do not support cell viability. Substitutions at other conserved positions in the alpha-helices that are not in contact with the DNA, such as R32, F36, I43, M44, L53, and T54, also fail to complement the mcm1 null mutant and are lethal to the cell. These residues are positioned along the dimer interface, and one possible explanation for their inability to support growth in the cell is that these side chains may be critical for protein stability and proper folding. Only two mutations in the N-terminal extension, F25A and I26A, failed to complement the null mutant. Both of these positions are located at the beginning of the N-terminal extension, where it folds back onto the coiled-coil alpha-helices and forms part of the dimer interface. These substitutions may also affect protein stability. Three mutations, V34A, S37A, and E49A, are viable but have a slow-growth phenotype, suggesting that they only weakly complement the mcm1 null mutant.
Effects of alanine substitutions on Mcm1-mediated transcriptional activation.
Although many of the mutants fail to complement the mcm1 null mutation and are therefore lethal to the cell, a large number of the alanine substitutions are viable. To determine whether these substitutions affect Mcm1 activity, the viable mutants were assayed for their ability to activate transcription in vivo. This assay used an integrated lacZ reporter gene under the control of a P(PAL) Mcm1 binding site (Table 1) (4, 10). Many of the viable mutants have wild-type or near-wild-type levels of transcriptional activation of the reporter promoter, indicating that they do not dramatically alter the structure or function of the protein. Most of these are substitutions of residues on the surface of the protein that do not contact the DNA. As expected, substitutions of residues which do not contact the DNA, such as R19, I21, S37, and T66, have reduced levels of activation. Interestingly, the slow-growth mutant V34A, which also contacts DNA, has the lowest level of activation of any viable mutant. Alanine substitutions of residues I23, F48, and E49 also show reduced levels of activation. These residues are near the dimer interface, and it is possible that alanine substitutions at these positions alter the stability of the protein, causing the decrease in transcriptional activation. Although most of the results correlate with the predictions based on the Mcm1 crystal structure, some of our results were unexpected (24). For example, residue R18 makes several base-specific and phosphate backbone contacts with the base pairs at the center of the site and therefore might be expected to have an important role in the function of the protein. However, the alanine substitution at this position is viable and activates transcription at wild-type levels, indicating that the R18 side chain is not critical for Mcm1 activity at this site.
Effects of the alanine substitutions on DNA-binding affinity in vitro.
A significant number of mutants with alanine substitutions at residues involved in protein-DNA contacts fail to complement the mcm1 null mutant for cell viability. To determine if these substitutions affect the DNA-binding activity of the protein, the mutants were cloned into a bacterial expression vector and purified, and their DNA-binding affinity was assayed by EMSAs. In addition, all of the viable mutants were expressed and purified in this manner to determine if the reductions in Mcm1-mediated activation in vivo are a function of decreased DNA-binding affinity. An example of the assays is shown in Fig. 2, and the relative binding affinity calculated for each mutant is listed in Table 1.
In general, the results of the DNA-binding affinity analysis are in good agreement with the in vivo viability and activation data and correlate with predictions based on the Mcm1 crystal structure (24). A number of the alanine substitutions that do not support viability, including F25A, I26A, R32A, F36A, I43A, M44A, L53A, and T54A, are at residues which form the hydrophobic core of the protein or the dimer interface. During the purification of these mutants from bacteria, many of the proteins were present at lower levels in the lysates, exhibited decreased affinity for the heparin column, and were more susceptible to degradation. This finding is consistent with a model that these substitutions affect folding of the protein, therefore making them less stable and more susceptible to proteolysis. All of these mutants show a >100-fold decrease in DNA-binding affinity. These results suggest that the loss of viability with these mutants is a function of their inability to produce proteins that are competent for DNA binding.
The remaining nonviable substitutions, K38A, R39A, K40A, G42A, K45A, and K46A, are on the binding face of the coiled-coil alpha-helices, and all of these residues contact DNA in the crystal structure (24). Many of these mutants show a >1,000-fold decrease in DNA-binding affinity in vitro, indicating that these residues have an essential role in DNA binding by Mcm1. Our results and the fact that these residues are highly conserved among all MADS-box proteins underscores their importance for DNA binding and supports a model with a conserved mechanism of DNA recognition by this family of proteins.
Many of the viable mutants also show decreases in DNA-binding affinity, but in general they have less of an effect on the DNA-binding affinity than the nonviable mutants. Alanine substitutions of other residues which contact the DNA in the crystal structure, such as R18, R19, I21, T35, S37, and T66, show modest decreases in DNA-binding affinity in vitro, which correlate well with the observed decreases in activation by these mutants in vivo. These residues are somewhat less conserved among the MADS-box proteins than K38, R39, G42, K45, and K46, and although not essential for viability these residues clearly contribute to the DNA-binding affinity and transcriptional activation. Additionally, some of these residues may have a role in determining the specificity of DNA binding.
Two of the mutants with a slow-growth phenotype, V34A and S37A, produce protein-DNA complexes with higher mobility than the wild-type protein (see Fig. 6). These mutations also cause significant decreases in transcriptional activation, with V34A resulting in the lowest β-galactosidase activity of all the viable mutants. In contrast, other mutations, such as N28A and I21A, have greater decreases in binding affinity than V34A but do not have similar decreases in the levels of transcriptional activation. These results suggest that V34A and S37A may affect other aspects of Mcm1 function.
Loss of base-specific contacts by mutations in the N-terminal extension.
One of the most striking differences between the SRF and the Mcm1 crystal structures is the location of the N-terminal extension of each protein. In the Mcm1 structure, the R18 side chain is making a set of contacts in the major groove, while in the SRF structure the analogous residue is making a series of contacts in the minor groove of the DNA (19, 24). This difference was predicted in the analysis of the SRF structure and is supported by differences in protein-DNA cross-linking of SRF and Mcm1 to base pairs at the center of the recognition site (1, 19). Additionally, mutational analysis of the DNA site shows that substitutions of the central base pairs produce a large decrease in Mcm1-mediated activation, indicating that there are sequence-specific contacts at these positions (1). Residue R18 makes an extensive set of base-specific and phosphate backbone contacts to the center of the site in the crystal structure (24). However, an alanine substitution at position R18 does not significantly alter transcriptional activation in vivo or DNA-binding affinity in vitro. This result was unexpected, since other positions which are involved in base-specific contacts have significantly larger effects on viability and activation. In comparison, a substitution at the adjacent residue, R19, has a significantly larger effect on transcriptional activation and DNA-binding affinity.
To further investigate the contributions of these residues in the N-terminal extension on specificity and activation, we performed loss-of-contact experiments (6) in which the E17A, R18A, and R19A mutations were assayed for their ability to activate transcription of a reporter promoter containing an A1C base-pair substitution in the Mcm1 site. In comparison to the wild-type site, the A1C substitution causes a fourfold reduction in the level of transcriptional activation by the wild-type Mcm1 protein (1). If the R18A mutant shows similar transcriptional activation for both the wild-type and the mutant sites, then it would indicate that the alanine substitution has removed the base-specific contacts to that position in the DNA, since the protein is unable to discriminate between the wild-type and mutant sites. However, if the mutant protein shows a further decrease in activity to the mutant site in comparison to the wild-type site, then it would suggest that the base-specific contacts at these positions are mediated by other residues. The R18A mutation shows a decrease in activation with A1C binding site mutation in comparison to the wild-type site (Table 2). This reduction in activation by the R18A mutant from the reporter with the A1C mutation is similar to the reduction by the wild-type protein, suggesting that other residues are contacting these base pairs. The E17A mutant also displays similar activity. On the other hand, the R19A mutation does not have a reduction in activation with A1C compared with the wild-type binding site. In fact the activation of this mutant with the A1C binding site is slightly increased, suggesting there may be steric interference between this Arg side chain and the mutant DNA site. These results raise the possibility that R19 may contact the central base pairs of the consensus symmetric binding site used in our experiments.
Amino acid residues of Mcm1 required for BrU-mediated cross-linking to the center of the site.
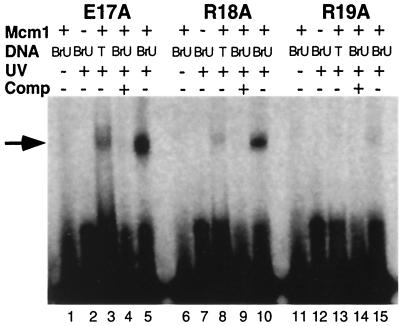
BrU is a photoactivatable, isosteric analog of thymine. Irradiation of BrU with UV light leads to a reactive species which may form a covalent bond with other groups that are in close proximity (17). We have previously used this property of BrU to show that the Mcm1 protein contacts the thymine methyl groups in the major groove of a base pair at a position in the center of the DNA recognition site (1). In the Mcm1 crystal structure the NH1 group of R18 is in close proximity with the C-5 methyl group of T−1 and is the likely target for cross-linking (24). To test which amino acid from the N-terminal extension is involved in a base-specific contact with the thymine at the center of the recognition site, BrU cross-linking assays were performed with the Mcm1 alanine substitution mutants. The E17A and R18A proteins form a cross-link with the DNA (Fig. 3). This result suggests these side chains are not involved in a direct contact with the center bases of the site we have used in our experiments.
FIG. 3.
Formation of cross-linked complexes with Mcm1 mutants. Urea-denaturing PAGE of the BrU-mediated photo-cross-linking reactions of the E17A, R18A, and R19A mutants with labeled site containing a BrU substitution for the thymine at base −1. Lanes 1, 6, and 11 are control reactions omitting UV radiation. Lanes 2, 7, and 12 are control reactions omitting the Mcm1 protein. Lanes 3, 8, and 13 are control reactions with labeled Mcm1 binding site without BrU (denoted by T in the figure). Lanes 4, 9, and 14 are control reactions in which a 10-fold excess of unlabeled competitor was added. Lanes 5, 10, and 15 are the cross-linking reactions. The arrow indicates the position of the cross-linked Mcm1-DNA products.
The R19A mutant, however, did not form a significant level of BrU-mediated cross-linked band, suggesting that this residue is required for interaction with the thymine methyl group at position −1. The small amount of cross-linking at this position may, in part, be the result of the background level of Mcm1 photo-cross-linking observed even in the absence of BrU-substituted DNA (Fig. 3, lanes 3, 8, and 13). It is also possible that the R19A substitution alters the structure of the N-terminal extension, thereby not allowing another residue to interact with the BrU functional group. However, we have previously shown that an Mcm1 fragment missing the first 17 residues of the protein, Mcm118–96, forms a cross-linked product with this DNA position (1). This result indicates that first 17 residues of the N-terminal extension are not involved in the cross-link. The cross-linking results and the loss-of-contact data are in agreement and suggest that R19 may be involved in the major groove contact with the thymine methyl group at position +1 or −1 in the site we have used in our experiments.
Mcm1 mutants with altered bending activity.
Base-pair substitutions at positions +7 or −7 in the Mcm1 DNA-binding site cause a large decrease in DNA bending without a corresponding effect on DNA-binding affinity (1). Interestingly, mutations at this position also result in a significant decrease in transcriptional activation. In the Mcm1 crystal structure, residues V34 and K38 make base-specific contacts to this position in the DNA site (24). Along with residue S37, V34 also makes base-specific contacts at position 8 of the site. To investigate if mutations at these residues affect the DNA-bending activity of Mcm1, circular permutation EMSAs were performed with these mutants and the apparent bend angles were calculated (Fig. 4). The V34A and S37A mutants show significant decreases in DNA bending compared to the wild-type protein. These results support predictions based on the crystal structure and show that these residues are important for producing the Mcm1-dependent DNA bend. Interestingly, the decrease in bending by each of the mutants is about one-half the decrease in bending resulting from the T7G mutant in the DNA (1). One possibility for the intermediate effect of the protein mutants is that each side chain is contributing to the bending. To test this model, the V34A-S37A double mutant was constructed and assayed for viability and bending. Unlike the single mutants, the V34A-S37A double mutant is unable to complement the mcm1 null mutant. The V34A-S37A mutant has an apparent bend angle of 61°, more than double the observed decrease from each of the single mutants, and it is comparable to the bending by the wild-type protein to the T7G site. This result suggests that the large drop in bending observed for the T7G base-pair substitution is similar to the loss of contacts by residues V34 and S37.
In addition to the base-specific contacts by V34 and S37, residues K40, T66, and to a lesser degree K29 and H33, are close to this region of the DNA and possibly contribute to DNA bending (24, 29). The K40A mutation was not able to support viability in the cell, and the DNA-binding affinity of this mutant is decreased over 125-fold compared to the wild-type protein. The circular permutation analysis shows that DNA bending is decreased by 36° (Fig. 4B). This decrease is equivalent to the decrease in bending observed with the V34A-S37A double mutant and indicates that this residue has a critical role in DNA bending. In contrast, we found that alanine substitutions of K29, H33, and T66 had little or no effect on DNA binding, bending, or transcriptional activation (Table 1, Fig. 4B; DNA bending data are not shown for K29A and H33A). These results show that these residues do not make a large contribution to DNA binding or bending by the protein.
To determine if other residues in the DNA-binding region of Mcm1 are required for DNA bending, we assayed each alanine substitution in an EMSA with the NheI fragment used in the circular permutation experiments. This fragment shows the largest differences in shift mobility with mutations that affect DNA bending and serves as a sensitive diagnostic assay for identifying mutants with altered bending. With the exception of the mutants described above, none of the other mutant proteins that bind DNA appear to alter DNA bending (data not shown). It is possible other side chains, such as K38, may also contribute to DNA bending. However, since they failed to bind DNA under our assay conditions, we were unable to measure their effect on bending.
Sequence specificity of DNA bending by Mcm1.
In the crystal structure, residues V34A, S37A, and K40A make base-specific and phosphate backbone contacts to positions 7 and 8 of the DNA site, and we have shown that alanine substitutions of these residues cause decreases in the degree of DNA bending by the protein (24). To examine the specificity required for bending, we tested the ability of the Mcm1 mutants to bend DNA with mutant binding sites. If the V34A and S37A mutants show the same degree of bending to a mutant site as to the wild-type site, then it would suggest that these residues are contacting that particular base pair and that no additional base-specific contacts to that position are lost. However, if there is a further reduction in bending by a mutant protein to a mutant site, then it would indicate that other base-specific contacts are being made to that base pair. The V34A substitution has the same apparent degree of DNA bending with the T7G and wild-type sites, providing evidence that the decrease in bending by the mutant site is caused by a loss of contact with residue V34 (Fig. 4C). This result suggests that the K38 contact to the thymine at this base pair is not critical for bending. On the other hand, the T7G mutation causes a significant decrease in DNA bending with the wild type (35°) and R18A (29°), S37A (25°), and T66A (32°) mutants (Fig. 4C). This result indicates that these residues are not in contact with this base pair. As expected, the V34A-S37A double mutant does not show a further decrease in bending to the T7G mutation.
The SRF protein binds to the Mcm1 site with moderate affinity and produces a significant bend in the DNA. However, unlike Mcm1, DNA bending by SRF is not affected by the T7G base-pair substitution (1). This finding suggests that even though most residues in contact with the DNA are identical between Mcm1 and SRF, the mechanism of bending is significantly different. One difference between the two proteins is that SRF contains a threonine at the position that is analogous to V34 of Mcm1, and this side chain makes contacts to position 8 of the DNA but not to position 7 (19). To test if the differences in the specificity of bending between the proteins is due to the presence of a threonine at position 34, we constructed the V34T substitution in Mcm1 and assayed its effects on transcription in vivo and DNA binding and bending in vitro. The V34T mutant complements the mcm1 null mutant and has near-wild-type levels of transcriptional activation from the P(PAL) site in vivo, showing that threonine can functionally substitute for valine at this position in Mcm1 (data not shown). The V34T mutant also binds with almost wild-type affinity and produces a bend of the wild-type site that is similar to the wild-type protein (Fig. 4B). However, unlike SRF, the V34T mutation exhibits a decrease in apparent bend angle with the T7G substitution (Fig. 4C). These results indicate that the difference in the specificity of bending between the two proteins is not solely due to the different amino acids at position 34 of the MADS-box domain and that the mechanism of bending by this mutant is different from either wild-type Mcm1 or SRF. Furthermore, the results suggest that the mechanisms used by the proteins to bend DNA differs more significantly than would be expected for two proteins with near identity in this region of the protein. Interestingly, the magnitude of bending by V34T to the T7G site is similar to V34A binding this mutant DNA site. Neither of these mutants show as severe a decrease in bending as the wild-type protein to this site. The T7G substitution may sterically interfere with the wild-type valine side chain at residue 34, and substitution of this residue to alanine may relieve this interference, permitting increased bending in comparison to the wild-type protein. It is unlikely that the V34T substitution relieves the steric interference, and therefore this residue may make an alternative set of contacts to partially restore bending.
Residues V34 and S37 contact the C-5 methyl group of the thymine at position 8 in the DNA recognition site, and these interactions may also contribute to Mcm1-dependent DNA bending (24). A T8G substitution in the Mcm1 binding site results in a complete loss of transcriptional activation in vivo (1). This base-pair substitution causes a large decrease in DNA bending by wild-type and mutant proteins, indicating that there is sequence specificity at this position for DNA bending (Fig. 5A). In contrast to the T8G substitution, a T8C mutation in the Mcm1 binding site only causes a modest reduction in the level of activation by the wild-type protein (1). Interestingly, even though the thymine contacted by the V34 and S37 residues has been removed in the T8C substitution, this site shows normal levels of DNA bending by the wild-type protein (Fig. 5B). This result suggests that alternate contacts are made by the protein to the mutant site which restore bending. These alternate contacts are still dependent on the V34 and S37 side chains because alanine substitutions of these residues cause a decrease in DNA bending of the T8C site (Fig. 5B).
In vivo and in vitro DNA sequence specificity of the altered bending mutants.
The V34A mutant has decreased levels of DNA bending and transcriptional activation, suggesting that DNA bending may be important for the transcriptional activation by this protein. To further examine the possible role and sequence specificity of DNA bending, the wild type and V34A and S37A mutants were assayed in combination with the T7G and T8C DNA sites for transcriptional activation in vivo and DNA binding in vitro (Fig. 6). The wild-type protein shows similar decreases in transcriptional activation and DNA-binding affinity with both the T7G and T8C DNA sites. However, the T7G mutant causes a large decrease in DNA bending, while the T8C mutant does not. These results are not consistent with a model directly linking the degree of DNA bending with activation.
The S37A protein has decreased levels of DNA bending and transcriptional activation with all three binding sites. However, decreases in activation appear to correspond with changes in the binding affinity by this protein. Interestingly, we find the binding affinity of the wild-type and S37A proteins for the mutant DNA sites are identical, but the activation levels of the mutant are reduced compared to the wild-type protein. The V34A protein, much like the S37A protein, has decreased bending and activation with each of the sites, and the levels of activation roughly correspond to changes in binding affinity. Although the DNA-binding affinity of V34A is greater than the wild-type protein for the mutant DNA sites, the levels of activation by V34A through these sites is equal or less than for the wild-type protein. Taken together, these results indicate that DNA-binding affinity is an important component in transcriptional activation by Mcm1 and that there is not a simple correlation between the absolute degree of DNA bending and activation. However, the fact that, in comparison to the wild-type protein, V34A and S37A have decreased activation with the mutant sites while binding these sites as well as or better than the wild-type protein suggests that mutations at these residues affect a characteristic of Mcm1 that is important for transcriptional activation that does not strictly correlate with DNA-binding affinity.
DISCUSSION
The determination of the crystal structures of SRF and Mcm1 bound to DNA identified residues of MADS-box proteins that make base-specific and phosphate backbone contacts with the DNA (19, 24). We have performed alanine scanning mutagenesis of the DNA-binding region of Mcm1 to examine the relative importance of each of these contacts to transcriptional activation by Mcm1 in vivo and DNA binding in vitro. Alanine substitution of 14 of the 40 residues that were mutated results in a loss of cell viability, indicating that these side chains are crucial for the proper function of Mcm1 in the cell. As expected, a number of these side chains, residues F25, I26, R32, F36, I43, and M44, are buried in the hydrophobic core of the protein or are at the dimer interface. Although these residues are not directly involved in protein-DNA contacts in the crystal structure, they do not have detectable levels of DNA binding in vitro, which suggests that they may be important for stability or protein folding.
Many of the lethal mutations are at positions which are highly conserved among all MADS-box proteins and contact the DNA in both the SRF and Mcm1 crystal structures (19, 24). These residues, K38, R39, G42, K45, and K46, are unable to complement the mcm1 null mutant and do not have detectable DNA-binding activity in vitro. There are other residues that are highly conserved among the MADS-box proteins that make similar contacts to the DNA in the Mcm1 and SRF crystal structures. However, the results from the mutagenesis of these residues suggest that these side chains do not make critical contributions to the function of the protein. For example, residue T35 of Mcm1 is at a position that is conserved in nearly all MADS-box proteins, and this residue makes identical contacts to the phosphate backbone at G4 of the DNA in both the Mcm1 and SRF crystal structures (19, 24). However, an alanine substitution of this residue in Mcm1 complements the mcm1 null mutant and has near-wild-type levels of transcriptional activation; also, the DNA-binding affinity of the mutant is only threefold less than that of the wild-type protein. Likewise, residue T66 is also highly conserved among MADS-box proteins, but the alanine substitution of this residue has little or no effect on viability, activation, or DNA-binding affinity. These results suggest that although these contacts are highly conserved their relative importance is not as great as the other contacts common among the MADS-box proteins.
The results of the alanine scanning mutagenesis of Mcm1 and the structural determination of SRF and Mcm1 indicates which protein-DNA interactions have essential roles in recognition and binding by a MADS-box protein. The conserved base-specific contacts made by K38, along with the phosphate backbone contacts made by K39, G42, K45, and K46, may provide the proper general framework for DNA binding by MADS-box proteins. The conserved mechanism of DNA binding among MADS-box proteins is very similar to what has been observed with homeodomain proteins (7). Among homeodomain proteins, the majority of the contacts with the DNA are by a highly conserved alpha-helix that lies in the major groove. The N51 residue, along with several Lys and Arg residues in this helix, are conserved among virtually all homeodomain proteins, and these side chains make a set of base-specific and phosphate backbone contacts that are nearly identical in every homeodomain-DNA structure that has been determined. Differences in sequence-specific binding among the homeodomains appear to be a result of base-specific contacts by nonconserved residues in the helix and by contacts from an N-terminal arm with the minor groove of the DNA. Based upon the similarity of the contacts in the SRF and Mcm1 crystal structures, along with the high degree of conservation of residues that we have shown are essential for DNA binding, it appears that MADS-box proteins also use a conserved mechanism to bind the DNA. Residue K38 in Mcm1, which is absolutely conserved among all MADS-box proteins, may function in a manner similar to N51 in homeodomain proteins by making conserved base-specific contacts to the DNA. Other side chains which are also highly conserved among the MADS-box family of proteins, such as residues R39, G42, K45, and K46, make similar contacts to the DNA phosphate backbone in the SRF and Mcm1 crystal structures and are essential for DNA binding. These residues may have a similar role in docking MADS-box proteins to DNA as do the conserved lysines in the recognition helix of homeodomain proteins, which appear to position and anchor the helix for base-specific recognition.
Although MADS-box proteins may contain a conserved mechanism to bind DNA, site selection and mutagenesis experiments have shown that there are significant differences in sequence specificity (9, 31). The differences in specificity between the different MADS-box proteins appear to be largely due to different contacts by the N-terminal extension with the center of the recognition site and residues of the coiled-coil alpha-helices which interact with base pairs outside the conserved CArG site. For example, the position and DNA contacts by the N-terminal extension in the center of the site is very different in the SRF and Mcm1 crystal structures and contribute to differences in specificity (19, 24). We have also shown that side chains at residues 34 and 37 of Mcm1 contribute to the sequence specificity at positions outside of the conserved CArG site. In addition, although many of the residues contacting the DNA outside of the CArG box are only involved in contacts with the phosphate backbone, these contacts may contribute to the sequence specificity of binding. For example, many of the phosphate backbone contacts made by Mcm1 would only occur if the DNA is bent, and our results have shown that the ability to bend is sequence specific. Differences in sequence specificity may be responsible for the broad range of DNA bending observed among the various MADS-box proteins (29).
The effects of many of the mutations we have made correlate with the Mcm1 crystal structure (24). However, our results on the effects of mutations in the N-terminal extension were unexpected. In the Mcm1 crystal structure residue R18 is making a base-specific contact with the center of the site. Our in vivo transcription data and in vitro cross-linking results suggest that R18 has a limited role in contacting this base pair. In contrast, our data show that R19 is required for, and may be making, the contact in the major groove at the center of the site. One possible explanation for these results is that the structural determination of the Mcm1 MADS-box was solved in complex with the α2 protein. The DNA contacts of the N-terminal extensions and other parts of the proteins may be altered by the presence of α2. Indeed, in the α2-Mcm1-DNA structure, the N-terminal extension from the trans Mcm1 monomer (interacting with an α2 monomer from another DNA strand) makes minor groove contacts as a consequence of the interactions of the trans α2 monomer (24). Another possible explanation for this result is that the site we have used in our experiments varies at several bases in the center of the site from the one used in the crystal structure. It is possible that the N-terminal extension adopts different conformations to make optimal contacts at the different sites. The fact that the position and contacts of N-terminal extensions of SRF and Mcm1 differ greatly and that the position of the extensions in the two Mcm1 monomers in the crystal structure are also significantly different suggests that this region may have considerable conformational freedom. The conformational freedom of the extension may be important for allowing MADS-box proteins to recognize different sites alone or in complex with other cofactors. Interestingly, amino acid side chains in the first 17 residues of Mcm1 are phosphorylated under specific conditions in vivo, and it was hypothesized that these different isoforms of the protein may recognize different target sites (12).
Alanine scanning mutagenesis identified three mutants that have altered DNA-bending activity: V34A, S37A, and K40A. These mutants were then used to study the mechanism of the sequence-specific DNA bending. Our results examining the DNA bending of the V34A mutant in combination with the T7G site suggests that V34 contacts position 7 and that this interaction contributes to Mcm1-dependent DNA bending. The DNA-bending activity of the S37A mutant is not further decreased by the T8C mutation, a result consistent with the model that the interaction between S37 and T8 contributes to the DNA-bending activity of Mcm1. These results correlate perfectly with the crystal structure of Mcm1 and reveal the relative importance of each of these interactions in producing Mcm1-dependent DNA bending. Interestingly, the bending associated with the K40A mutant does not appear to have sequence specificity, since bending of the wild-type and mutant sites are identical. This finding is supported by the Mcm1 crystal structure, which shows this side chain is involved in a phosphate contact at position T8. It is possible that V34 and S37 dictate the specificity of bending and that K40 helps provide the necessary change in free energy to stabilize the DNA in this altered conformation. Without the contributions of all three side chains wild-type levels of DNA bending cannot occur. Predictions based on the Mcm1 crystal structure and work with other MADS-box proteins suggest that K29 and T66 may also contribute to DNA bending (24, 29). However, our results indicate that these residues contribute very little to Mcm1 function in vivo or Mcm1-dependent DNA bending in vitro.
Our results show that for the wild-type protein and the majority of the mutants, there is a good correlation between the levels of transcriptional activation and DNA-binding affinity. However, the observation that V34A and S37A, mutants which alter DNA bending with little or no effect on DNA-binding affinity, have slow-growth phenotypes suggests that DNA bending by Mcm1 may be important for an essential function of this protein. The fact that other mutations in Mcm1 with similar or greater decreases in DNA-binding affinity do not result in slow growth or have similar reductions in transcriptional activation supports this idea. However, our results show that there is not a simple correlation between the apparent degree of Mcm1-mediated DNA bending and transcriptional activation. The ternary complex crystal structure shows that Mcm1 produces two different bends in the DNA: one around the protein and one which alters the DNA path away from the protein (24). It is possible that one of these specific bends has a role in transcriptional activation. The calculations of the DNA bend angle from the circular permutation assay are based on the assumption that the protein produces a single bend in one plane. Therefore, this assay, although sensitive to relative conformational changes in the DNA, is not able to distinguish between contributions of each type of bend towards the calculated angle. The V34A and S37A mutants clearly alter the conformation of the Mcm1-DNA complex, but with this assay we cannot determine whether they affect the bend around the protein, away from the protein, or a combination of both of these. However, these mutants clearly show that DNA-binding affinity alone is not the sole determinant of Mcm1-mediated transcriptional activation.
An alternative explanation for the effects of the V34A and S37A mutants is that the DNA contacts, and therefore the DNA bend, may affect the position and conformation of these side chains. It has been shown that Mcm1 alters its conformation when binding DNA in complex with the α1 protein (25). Mutations at residues H33 and H41 have little or no effect on DNA-binding affinity in complex with α1 and yet result in decreased levels of α1-Mcm1-dependent transcriptional activation (4). These observations have led to the suggestion that this region of the protein may have a role in the α1-induced conformational change of Mcm1 (4). It is possible that the V34 and S37 contacts with the DNA provide a similar type of change in the structure of the protein and that this altered conformation is important for transcriptional activation by the protein.
It is important to keep in mind that we address here the activation and DNA-binding activity of Mcm1 alone. In the cell, Mcm1 interacts with many different proteins to regulate diverse pathways. Although our results suggest that bending of the DNA is not an overriding factor in transcriptional activation by Mcm1 on its own, DNA bending may play a more important role when it is in complex with other factors to regulate different pathways. As has been observed for SRF, bending by Mcm1 may be needed for the recruitment of these other protein factors to the promoter regions of its target genes (21). Future studies utilizing these mutants may help provide a deeper understanding of the importance of DNA bending in transcriptional activation or to other functions of Mcm1 in the cell.
ACKNOWLEDGMENTS
We thank George Sprague for providing strains and plasmids and Tim Richmond and Song Tan for providing us with the coordinates of the α2-Mcm1-DNA crystal structure and for many helpful discussions.
A.M.S. is a recipient of a Howard Hughes Medical Institute Undergraduate Summer Research Fellowship. This research was supported by a grant from the National Institutes of Health (GM49265) to A.K.V.
REFERENCES
- 1.Acton T B, Zhong H, Vershon A K. DNA-binding specificity of Mcm1: operator mutations that alter DNA bending and transcriptional activities by a MADS box protein. Mol Cell Biol. 1997;17:1881–1889. doi: 10.1128/mcb.17.4.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Affolter M, Montagne J, Walldorf U, Groppe J, Kolter U, LaRosa M, Gehring W J. The Drosophila SRF homolog is expressed in a subset of tracheal cells and maps within a genomic region required for tracheal development. Development. 1994;120:743–753. doi: 10.1242/dev.120.4.743. [DOI] [PubMed] [Google Scholar]
- 3.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 4.Bruhn L, Sprague G F J. MCM1 point mutants deficient in expression of α-specific genes: residues important for interaction with α1. Mol Cell Biol. 1994;14:2534–2544. doi: 10.1128/mcb.14.4.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubois E, Messenguy F. Integration of the multiple controls regulating the expression of the arginase gene CAR1 of Saccharomyces cerevisiae in response to different nitrogen signals: role of Gln3p, ArgRp-Mcm1p, and Ume6p. Mol Gen Genet. 1997;253:568–580. doi: 10.1007/s004380050359. [DOI] [PubMed] [Google Scholar]
- 6.Ebright R H. Identification of amino acid-base pair contacts by genetic methods. Methods Enzymol. 1991;208:620–640. doi: 10.1016/0076-6879(91)08032-d. [DOI] [PubMed] [Google Scholar]
- 7.Gehring W J, Qian Y Q, Billeter M, Furukubo-Tokunaga K, Schier A F, Otting G, Wuthrich K. Homeodomain-DNA recognition. Cell. 1994;78:211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 8.Gustafson T A, Taylor A, Kedes L. DNA bending is induced by a transcription factor that interacts with the human c-fos and alpha-actin promoters. Proc Natl Acad Sci USA. 1989;86:2162–2166. doi: 10.1073/pnas.86.7.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H, Mizukami Y, Hu Y, Ma H. Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene Agamous. Nucleic Acids Res. 1993;21:1089–1095. doi: 10.1093/nar/21.20.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarvis E E, Clark K L, Sprague G F. The yeast transcription activator PRTF, a homolog of the mammalian serum response factor, is encoded by the MCM1 gene. Genes Dev. 1989;3:936–945. doi: 10.1101/gad.3.7.936. [DOI] [PubMed] [Google Scholar]
- 11.Keleher C A, Goutte C, Johnson A D. The yeast cell-type-specific repressor α2 acts cooperatively with a non-cell-type-specific protein. Cell. 1988;53:927–936. doi: 10.1016/s0092-8674(88)90449-7. [DOI] [PubMed] [Google Scholar]
- 12.Kuo M H, Nadeau E T, Grayhack E J. Multiple phosphorylated forms of the Saccharomyces cerevisiae Mcm1 protein include an isoform induced in response to high salt concentrations. Mol Cell Biol. 1997;17:819–832. doi: 10.1128/mcb.17.2.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lilly B, Galewsky S, Firulli A B, Schulz R A, Olson E N. D-MEF2: a MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc Natl Acad Sci USA. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mead J, Zhong H, Acton T B, Vershon A K. The yeast α2 and Mcm1 proteins interact through a region similar to a motif found in homeodomain proteins of higher eukaryotes. Mol Cell Biol. 1996;16:2135–2143. doi: 10.1128/mcb.16.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohun T J, Chambers A E, Towers N, Taylor M V. Expression of genes encoding the transcription factor SRF during early development of Xenopus laevis: identification of a CArG box-binding activity as SRF. EMBO J. 1991;10:933–940. doi: 10.1002/j.1460-2075.1991.tb08027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55:989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- 17.Ogata R T, Gilbert W. Contacts between the lac repressor and thymines in the lac operator. Proc Natl Acad Sci USA. 1977;74:4973–4976. doi: 10.1073/pnas.74.11.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Passmore S, Maine G T, Elble R, Christ C, Tye B-K. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MATα cells. J Mol Biol. 1988;204:593–606. doi: 10.1016/0022-2836(88)90358-0. [DOI] [PubMed] [Google Scholar]
- 19.Pellegrini L, Tan S, Richmond T J. Structure of serum response factor core bound to DNA. Nature. 1995;376:490–498. doi: 10.1038/376490a0. [DOI] [PubMed] [Google Scholar]
- 20.Reichmann J L, Wang M, Meyerowitz E M. DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMAOUS. Nucleic Acids Res. 1996;24:3134–3141. doi: 10.1093/nar/24.16.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharrocks A D, Shore P. DNA bending in the ternary nucleoprotein complex at the c-fos promoter. Nucleic Acids Res. 1995;23:2442–2449. doi: 10.1093/nar/23.13.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith D L, Desai A B, Johnson A D. DNA bending by the a1 and α2 homeodomain proteins from yeast. Nucleic Acids Res. 1995;23:1239–1243. doi: 10.1093/nar/23.7.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sommer H, Beltran J-P, Huijser P, Pape H, Lonig W-E, Saedler H, Schwarz-Sommer Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J. 1990;9:605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan S, Richmond T J. Crystal structure of the yeast MATalpha2/MCM1/DNA ternary complex. Nature. 1998;391:660–666. doi: 10.1038/35563. [DOI] [PubMed] [Google Scholar]
- 25.Tan S, Richmond T J. DNA binding-induced conformational change of the yeast transcriptional activator PRTF. Cell. 1990;62:367–377. doi: 10.1016/0092-8674(90)90373-m. [DOI] [PubMed] [Google Scholar]
- 26.Thompson J F, Landy A. Empirical estimation of protein-induced DNA-bending angles: applications to site-specific recombination complexes. Nucleic Acids Res. 1988;16:9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46:567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- 28.Trobner W, Ramirez L, Motte P, Hue H, Huijser P, Lonnig W-E, Saedler H, Sommer H, Schwarz-Sommer Z. GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogensis. EMBO J. 1992;11:4693–4704. doi: 10.1002/j.1460-2075.1992.tb05574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West A G, Sharrocks A D. MADS-box transcription factors adopt alternative mechanisms for bending DNA. J Mol Biol. 1999;286:1311–23. doi: 10.1006/jmbi.1999.2576. [DOI] [PubMed] [Google Scholar]
- 30.West A G, Shore P, Sharrocks A D. DNA binding by MADS-box transcription factors: a molecular mechanism for differential DNA bending. Mol Cell Biol. 1997;17:2876–2887. doi: 10.1128/mcb.17.5.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wynne J, Treisman R. SRF and MCM1 have related but distinct DNA binding specificities. Nucleic Acids Res. 1992;20:3297–3303. doi: 10.1093/nar/20.13.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yanofsky M F, Ma H, Bowman J L, Drews G N, Feldmann K A, Meyerowitz E M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcriptional factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- 33.Zhong H, Vershon A K. The yeast homeodomain protein Matα2 shows extended DNA-binding specificity in complex with Mcm1. J Biol Chem. 1997;272:8402–8409. doi: 10.1074/jbc.272.13.8402. [DOI] [PubMed] [Google Scholar]