Abstract
Posttranscriptional regulation of cancer gene expression programs plays a vital role in carcinogenesis; identifying the critical regulators of tumorigenesis and their molecular targets may provide novel strategies for cancer diagnosis and therapeutics. Highly conserved RNA-binding protein Pumilio-1 (PUM1) regulates mouse growth and cell proliferation, propelling us to examine its role in cancer. We found human PUM1 is highly expressed in a diverse group of cancer, including prostate cancer; enhanced PUM1 expression is also correlated with reduced survival among prostate cancer patients. Detailed expression analysis in twenty prostate cancer tissues showed enhanced expression of PUM1 at mRNA and protein levels. Knockdown of PUM1 reduced prostate cancer cell proliferation and colony formation, and subcutaneous injection of PUM1 knockdown cells led to reduced tumor size. Downregulation of PUM1 in prostate cancer cells consistently elevated cyclin-dependent kinase inhibitor 1B (CDKN1B) protein expression through increased translation but did not impact its mRNA level, while overexpression of PUM1 reduced CDKN1B protein level. Our finding established a critical role of PUM1 mediated translational control, particularly the PUM1-CDKN1B axis, in prostate cancer cell growth and tumorigenesis. We proposed that PUM1-CDKN1B regulatory axis may represent a novel mechanism for the loss of CDKN1B protein expression in diverse cancers and potential targets for therapeutics development.
Keywords: PUM1, prostate cancer, posttranscriptional regulation, CDKN1B
Introduction
Prostate cancer is the most common cancer and one of the leading causes of cancer death in men. Overexpression of RNA-binding proteins (RBPs) in prostate cancer (PCa) supports that posttranscriptional control plays a vital role in prostate carcinogenesis[1]. Only a few posttranscriptional regulators have been identified, and even fewer are mechanistically understood[2–3].
Human homologs of a significant proportion of mouse growth genes are also essential for tumorigenesis, either as oncogenes or tumor suppressors. Many of the growth regulators or tumorigenesis regulators are transcriptional regulators[4], the role of posttranscriptional regulation has begun to be appreciated[2]. Characterization of conserved RNA binding proteins in mammalian growth and tumorigenesis could help identify novel regulators and pathways important for cancer diagnosis and therapeutics development.
The PUF family (Pumilio and FBF), one of the highly conserved eukaryotic RBPs, functions by binding to Pumilio-binding elements (PBEs) on the 3′ UTR of their target mRNAs and play critical roles in cell fate decision and differentiation[2,5–7]. Recent genetic studies on members of the mammalian PUF family, Pum1 and Pum2 (the genes encoding RNA-binding protein Pumilio-1 [Pum1] and Pumilio-2 [Pum2]), uncovered the roles of PUM-mediated posttranscriptional regulation in growth, reproductive and neuronal development as well as diseases[8–16]. Pum mutant mice have reduced body weight and size, resulting from enhanced expression of cyclin-dependent kinase inhibitor 1B (CDKN1B) and reduced cell proliferation, raising the possibility that PUM-CDKN1B regulatory pathway may also be important in human growth and cell proliferation[15].
The CDKN1B (also known as p27Kip1, encoded by the gene CDKN1B) is a member of the Cip/Kip family of cyclin-dependent kinase (CDK) inhibitors that function to negatively control cell cycle progression by association with cyclin-dependent kinase 2 (CDK2) and cyclin E complexes to inhibit the transition from G1 to S phase[17]. Previous work has demonstrated that CDKN1B is a haploinsufficient tumor suppressor whose protein level has to be fine-tuned for its optimal function, and CDKN1B protein level may have prognostic and therapeutic implications[17–19]. While impaired synthesis, accelerated degradation, and mislocalization of CDKN1B have been studied[19], regulation of CDKN1B protein expression via translation in cancer has not been explored, and whether there exist key translational regulators remained unknown. We propose that Pum1 may exert control on human cell growth, in particular cancer cell growth, by repressing Cdkn1b expression[15].
Association of PUM with cancer cell growth has been suggested from studies on cancer cell lines[20–22], examination of PUM expression in human tumorigenesis and characterization of PUM mediated translation control directly in human cancer cells and the tumor is needed to reveal the extent and mechanism of PUM-mediated regulation during human tumorigenesis. Here, we report increased expression of PUM1 in various cancers from an extensive collection of human cancer datasets and twenty prostate tumor samples we collected from the clinic. We uncovered a role of PUM1-mediated translational repression of CDKN1B in prostate cancer cell proliferation and tumor formation. Our data underscore the importance of the PUM1-CDKN1B regulatory axis in human carcinogenesis and its potential for developing novel strategies in cancer diagnosis and therapeutics.
Material and methods
Patients and tissue samples
All human studies were approved by the Institutional Ethics Committee of Nanjing Medical University and performed after obtaining written informed consent. Twenty patients (mean age, 70.5 years; range, 61 to 76) underwent radical prostatectomy for prostate adenocarcinoma at the Second Affiliated Hospital of Nanjing Medical University. A summary of clinic and pathology parameters for the study cohort is included in Supplementary Table 1 (available online). For prostate cancer samples, the predominant tumor nodule and matched normal tissue were identified by histopathologists on fresh-frozen sections. Only foci with >80% purity of cancer cells were collected. Each prostate cancer and matched normal tissue were cut into three pieces. Among them, two parts were immediately frozen in liquid nitrogen and stored at –80 °C for later protein and RNA extraction, one piece of tissue was fixed in 10% neutral buffered formalin for 24 hours. The procedure for this research conforms to the provisions of the Declaration of Helsinki.
Immunohistochemistry
The histologic specimens were fixed in 10% formalin and routinely processed for paraffin embedding. Histologic sections, 5-μm thick, were stained with hematoxylin-eosin and reviewed by two pathologists to define the prostate cancer and matched normal tissues. Immunohistochemistry was performed on tumor paraffin sections after antigen retrieval using antibodies directed against PUM1 (Abcam, UK), according to the protocol described previously[15]. The specificity of PUM1 antibody has been confirmed using PUM1 immunostaining in testicular sections of wildtype and Pum1–/– mice[15].
Cell culture and transfection
The human prostate carcinoma cell lines (DU145, PC3, and LNCaP) were obtained from the American Type Culture Collection (Rockville, USA). Cells were cultured in DMEM supplemented with 10% fetal bovine serum. All cell lines were cultured in a humidified incubator at 37 °C with 5% carbon dioxide. The medium was replaced every 2 days. Cells at approximately 50% to 60% confluency were transfected for 24 hours with plasmids using Lipofectamine LTX Reagent (Invitrogen, USA).
Western blotting
Total protein was collected using the RIPA (Beyotime, China) and protease inhibitor cocktail (Roche, Switzerland). Standard Western blotting procedure[15] was followed with PVDF membrane (Bio-Rad, USA) used for protein transfer. Detection of HRP conjugated secondary antibody was performed with ECL (Vazyme, China). The antibodies used were as follows: rabbit anti-PUM1 (1:1000 dilution; Abcam), mouse anti-Actin (1:5000 dilution; Sigma, USA), rabbit anti-CDKN1B (1:1000 dilution; CST, USA), mouse anti-GAPDH (1:2000 dilution; Proteintech, USA), and mouse anti-α-Tubulin (1:5000 dilution; Santa Cruz, USA). The band intensity of specific proteins was quantified after normalization with that of α-Tubulin or GAPDH.
RNA extraction and real-time RT-PCR analysis
RNA was extracted by Trizol from tissues or cells stored at –80 °C and reverse transcribed for amplification of PUM1 cDNA. Real-time RT-PCR (qRT-PCR) analysis was performed using gene-specific sets of primers (Supplementary Table 2 , available online). The gene expression analysis was detected by ABI BioSystem StepOne plus. The gene expression level was quantified relative to the expression of ACTB, and the specificity of PCR products was confirmed by melting curve analysis. Each reaction filled up to an end volume of 20 μL containing two μL template cDNA, ten μL SYBR Premix Ex Taq buffer (TaKaRa, Japan), 0.4 μL ROX Reference Dye, eight pmol of each primer, and six μL ddH2O and was carried out in a standard 96-well plate. The cycling conditions consisted of an initial incubation at 95 °C for 3 minutes, followed by 45 cycles of 94 °C for 30 seconds, 60 °C for 30 seconds, and 72 °C for 30 seconds. A final incubation terminated the reaction at 95 °C for 15 seconds, 60 °C for 30 seconds, and 95 °C for 15 seconds. The expression level was calculated by the 2–ΔΔCt method to compare the relative expression.
RNA immunoprecipitation
A total of 2×107 prostate cancer cells (DU145 and PC3) were collected for each immunoprecipitation. The samples were lysed in polysome lysis buffer (0.5% NP40, 100 mmol/L KCl, 5 mmol/L MgCl2, 10 mmol/L HEPES, 1 mmol/L DTT, 100 units/mL RNaseOUT, and 1× protease inhibitor cocktail, pH 7.0) for 30 minutes and then centrifuged at 20 000 g for 20 minutes at 4 °C to remove the debris. The supernatant supplemented with RNase and protease inhibitor was immunoprecipitated with PUM1 antibody (Bethyl, USA) or goat IgG (Beyotime) for 6 hours at 4 °C and then incubated with Protein G Dynabeads (Invitrogen) overnight. Finally, the beads were divided into two portions. One portion was used to isolate protein for enrichment identification of PUM1, and the other portion was resuspended in 100 μL lysis buffer supplemented with RNase inhibitor. Then, 30 μg proteinase K was used to release the RNA at 55 °C for 30 minutes. The RNA was extracted using 1 mL TRIzol (Ambion, USA).
Luciferase reporter assay
CDKN1B 3′ UTR was subcloned into psiCHECK-2 vector (Promega, USA) using XhoI and PmeI restriction enzymes (New England Biolabs, USA). Wild-type PBE sequences 5′-TGTATATA-3′ was mutant to 5′-acaATATA-3′ as previous report[12]. Cells were co-transfected with pCMV6-hPUM1 and luciferase reporter plasmids, containing fragments or full-length 3′ UTRs. After 48 hours, cells were washed with PBS and lysed in Passive Lysis Buffer (Chroma-Glo Luciferase Assay System, Promega). Then, 20 μL of each lysate was analyzed using the Dual-Luciferase Reporter Assay System (Chroma-Glo Luciferase Assay System, Promega) in a 96 Microplate luminometer (BioTek, USA).
Sucrose gradient polysome fractionation
A total of 2×107 DU145 cells were collected, washed with PBS, and homogenized in 0.5 mL of MCB buffer. The lysate was centrifuged at 1300 g at 4 °C for 10 minutes. The supernatant was applied onto the top of a 15%–55% (W/W) linear sucrose gradient made by the Density Gradient Fractionation System (Teledyne ISCO Inc., USA). The gradient was centrifuged at 150 000 g for three hours (Beckman, USA). Fractions were collected and used for RNA extraction and analysis.
Colony formation
DU145 cells were generated by human PUM1 shRNA or scramble control lentiviral infection followed by puromycin selection. Cells were then trypsinized and counted by a hemocytometer. In total, 1×104 PUM1 knockdown or overexpressing DU-145 cells were seeded in complete growth media and allowed to grow for 14 to 21 days until visible colonies formed. Colonies were stained with 0.25% crystal violet in ddH2O, washed with PBS twice, and air-dried.
Xenograft model in vivo
Six-week-old male nude mice (BALB/c, Charles River Laboratories, USA) were used for the xenograft experiments. Cancer cells were trypsinized and harvested in PBS, then a total volume of 0.1 mL PBS was subcutaneously injected into the inguinal regions. DU-145 cells (5×105) transduced with sh-PUM1KD1 or Sh-Con were subcutaneously injected into the left and right inguinal regions of nude mice, respectively and the nude mice were monitored for 49 days. Tumor sizes were measured twice a week, starting at two weeks after cell injection using Vernier caliper. The animal study protocol was reviewed and approved by the institutional animal care and use committee of Nanjing Medical University.
Statistical analysis
All experiments were repeated at least three times. Statistical significance between two groups of data was evaluated by Student's t-test (two-tailed) comparison using GraphPad Prism 7 (GraphPad Software, USA).
Results
PUM1 expression was upregulated in various cancers, and overexpression of PUM1 was associated with poor prognosis in prostate cancer patients
To determine whether PUM1 has a role in tumorigenesis, we first examined PUM1 expression in The Cancer Genome Atlas (TCGA) database and found that PUM1 is widely expressed in diverse cancers, including prostate cancer (Fig. 1A). Given the reported correlation of loss of CDKN1B expression with prostate cancer survival[18], we hence investigated the role of PUM1 expression in prostate cancer. By focusing on 101 prostate samples from the TCGA database[23], we found that PUM1 mRNA was significantly higher in prostate carcinoma than that in benign tissues (P<0.05) (Fig. 1B). Compared with the normal prostate gland, a substantially higher level of PUM1 protein in prostate carcinoma was observed by immunohistochemical staining assays (Fig. 1C). Furthermore, the Kaplan-Meier curves indicated a significant correlation of PUM1 protein upregulation with poor patient survival (Fig. 1D). Hence, our analysis of TCGA cohorts suggested that PUM1 upregulation is associated with tumorigenicity, in particular, of prostate cancer, and poor outcomes in PCa patients.
Figure 1.
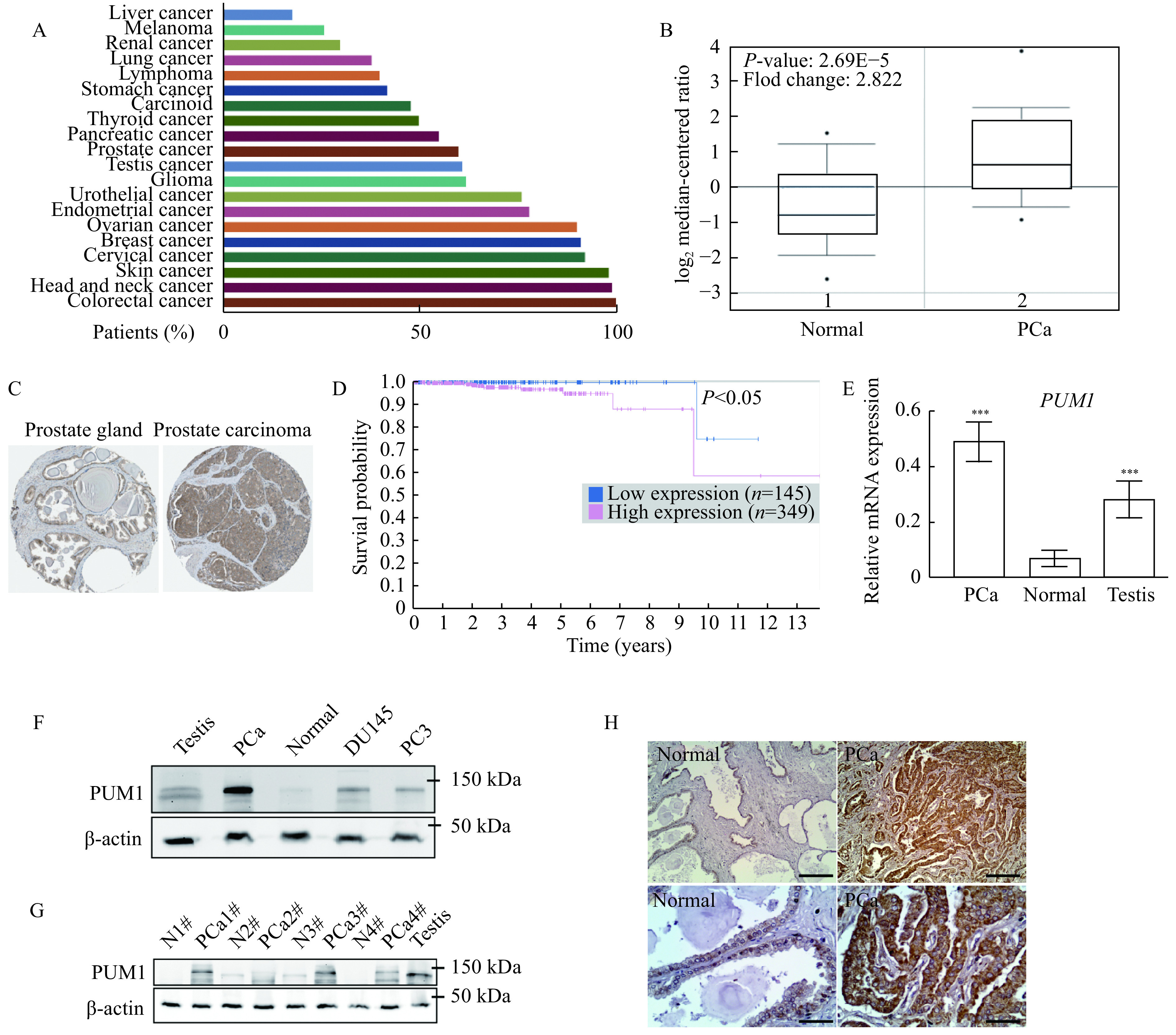
Overexpression of PUM1 was associated with poor prognosis in patients with PCa.
A: Graph showing the percentage of patients staining positive for PUM1 protein among TCGA cancer types using validated anti-PUM1 antibody from Human Protein Atlas. B: The box plot comparing PUM1 expression in the normal prostate gland (Normal) (n=29) and prostate carcinoma (PCa) (n=72) was derived from the Oncomine database (https://www.oncomine.org/). C: The expression of PUM1 in the normal prostate gland and prostate adenocarcinoma specimens. Representative images were taken from the Human Protein Atlas database. D: The Human Protein Atlas survival analysis for low and high expression levels of PUM1 on 494 prostate cancer patients with death outcome. E: The expression of PUM1 mRNA in prostate cancer tissue (PCa, n=5) and normal prostate tissues (Normal, n=5) was examined by qRT-PCR analysis. Human testis is a positive control since PUM1 is highly expressed in the testis. Data are presented as mean±SD. Comparisons between each two groups were analyzed using a two-tailed Student's t-test, ***P<0.001. F: PUM1 protein in prostate cancer cell lines (DU145 and PC3) and tumor tissue from one cancer patient was detected by Western blotting analysis. G: PUM1 protein level was detected in adjacent normal tissue (Normal,n=4) and prostate carcinoma tissue (PCa, n=4) of different cases. H: Immunohistochemistry of PUM1 protein in prostate carcinoma (PCa, n=10) and normal prostate gland (Normal, n=10). Scale bar: 200 μm (upper panel) and 50 μm (lower panel).
To further verify the correlation between PUM1 expression and PCa, we collected and analyzed PCa specimens from 20 prostate carcinoma patients in the clinic (Supplementary Table 2 , available online). qRT-PCR from total RNA obtained from PCa tissue and matched normal tissue confirmed the upregulation of PUM1 at the mRNA level in most of the patients examined (Fig. 1E). Western blotting analysis was performed on extracts obtained from the neoplastic tissue and the gland's contralateral part. We found that PUM1 protein was consistently upregulated in the neoplastic tissues, mostly basal cells (Fig. 1F and G). PUM1 protein was also detected in two types of prostate cancer cells (DU145 and PC3) (Fig. 1F). To further determine the localization of PUM1 protein in the prostate cancer tissue, we performed immunohistochemistry analysis on tissues of the same group of patients. PUM1 was expressed at low levels in the epithelial cells of matched normal prostate glands. By contrast, all epithelial cells of neoplastic glands were strongly positive for PUM1 (Fig. 1H), which confirmed that expression of PUM1 is indeed elevated in the neoplastic phenotype of prostate epithelial cells.
Downregulation of PUM1 in prostate cancer cells decreased proliferation and induced apoptosis
To understand the role of PUM1 in prostate carcinogenesis, we first interrogated the role of PUM1 in prostate cancer cell lines. We chose two commonly used prostate cancer cell lines representing different metastatic potentials of prostatic adenocarcinoma for our experiments, with PC3 from grade Ⅳ adenocarcinoma with high metastatic potential and DU145 from prostate carcinoma with moderated metastatic potential. Two PUM1 small hairpin knockdown shRNAs were designed against different parts of the PUM1 sequence and transfected into DU145 and PC3 cells. We found that PUM1 mRNAs were significantly reduced in DU145 and PC3 cells transduced with the PUM1 knockdown shRNA, confirming specific and efficient repression of PUM1 (Fig. 2A). Western blotting analysis confirmed the reduction of PUM1 protein in both DU145 and PC3 cells containing either knockdown construct (Fig. 2B). Consistent with the previous report in mouse cells[12], CCK8 assays indicated that downregulation of PUM1 caused a decrease in the proliferation rate of DU145 cells (Fig. 2C). Growth inhibition by PUM1 knockdown was also confirmed in PC3 cells (Fig. 2D). Annexin V/propidium iodide (PI) assay showed the population of apoptotic cells increased 1.41- and 1.35-fold in DU145, and 2.14- and 2.26-fold in PC3 when PUM1 was knocked down in these cells (Fig. 2E and F). Moreover, cleaved caspase-3 and cleaved caspase-9 were also increased in both cell lines upon PUM1 knockdown (Fig. 2G). The reduced cell growth was due to inhibition of G1/S transition: PUM1 knockdown significantly reduced DU145 cells in the S phase as assessed by incorporation of Edu (Fig. 2H) and DNA content (Fig. 2I). Taken together, our data demonstrated that an elevated PUM1 level is critical for the increased growth rate of prostate cancer cells.
Figure 2.
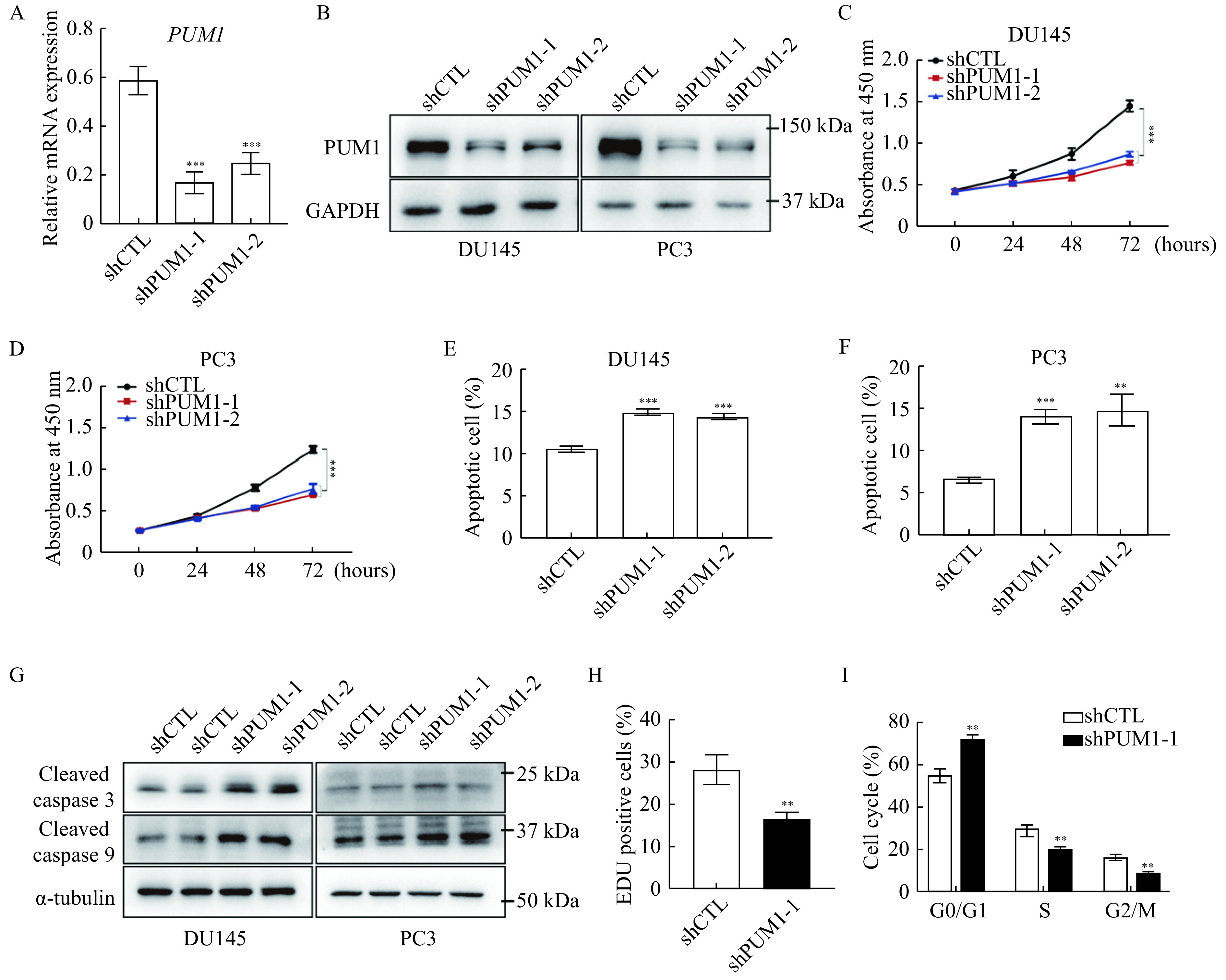
PUM1 knockdown reduced PCa cell proliferation and survival.
A: DU145 cells were transfected with PUM1 shRNAs (shPUM1-1 and shPUM1-2 targeting different PUM1 transcript) or non-targeting shRNA (shCTL). After 48 hours, the cells were harvested and PUM1 mRNA levels were examined by qRT-PCR analysis. ACTB was used as an internal control. Data obtained from three independent experiments are presented as mean±SD. P values were calculate by two-tailed Student's t-test analysis. ***P<0.001vs. shCTL group. B: Immunoblots showing the effect of shRNA-mediated PUM1 knockdown in DU145 and PC3 cells. C: CCK8 assay on DU145 cells transfected with the shCTL or PUM1 shRNAs as described in (A). Data are mean±SD of three independent experiments (unpaired t-test; ***P<0.001.) D: PC3 cells were transfected with CTL or PUM1 shRNAs, and the number of viable cells at the four time points was determined using CCK8 assay. Data are mean±SD of three independent experiments. Statistical analyses were performed by two-tailed Student'st-test. ***P<0.001. E and F: Apoptosis assay using flow cytometry after staining with annexin V-FITC/PI. Data are presented as mean±SD.**P<0.01,***P<0.001vs. shCTL group. G: Western blotting analysis of apoptosis-related proteins in PC3 and DU145 cells with PUM1 knockdown. H: EdU labeling of PUM1 knockdown DU145 cells revealed significantly reduced proliferation in comparison with control cells (two-tailed Student's t-test; **P<0.01vs. shCTL group). I: Cell cycle analysis of DU145 cells transfected with CTL or PUM1 shRNAs via flow cytometry. Data are presented as mean±SD (two-tailed Student's t-test; **P<0.01vs. shCTL group).
Overexpression of PUM1 promoted prostate cancer cell proliferation and colony formation
Since PUM1 level is positively correlated with prostate cancer cells' proliferation rate, we asked if overexpression of PUM1 could further promote the proliferation of prostate cancer cells. The expression levels of PUM1 in DU145 cells were manipulated by transfection of PUM1 knockdown construct and/or retroviral vector for PUM1 (Fig. 3A). PUM1 overexpression significantly increased the growth rate of DU145 (Fig. 3B). When PUM1 level was restored by retrovirus vector carrying PUM1 gene, the growth rate of PUM1-knockdown cells became comparable to that of control cells (shCTL + Empty retroviral vector), supporting PUM1 knockdown being responsible for reduced proliferation (Fig. 3B).
Figure 3.
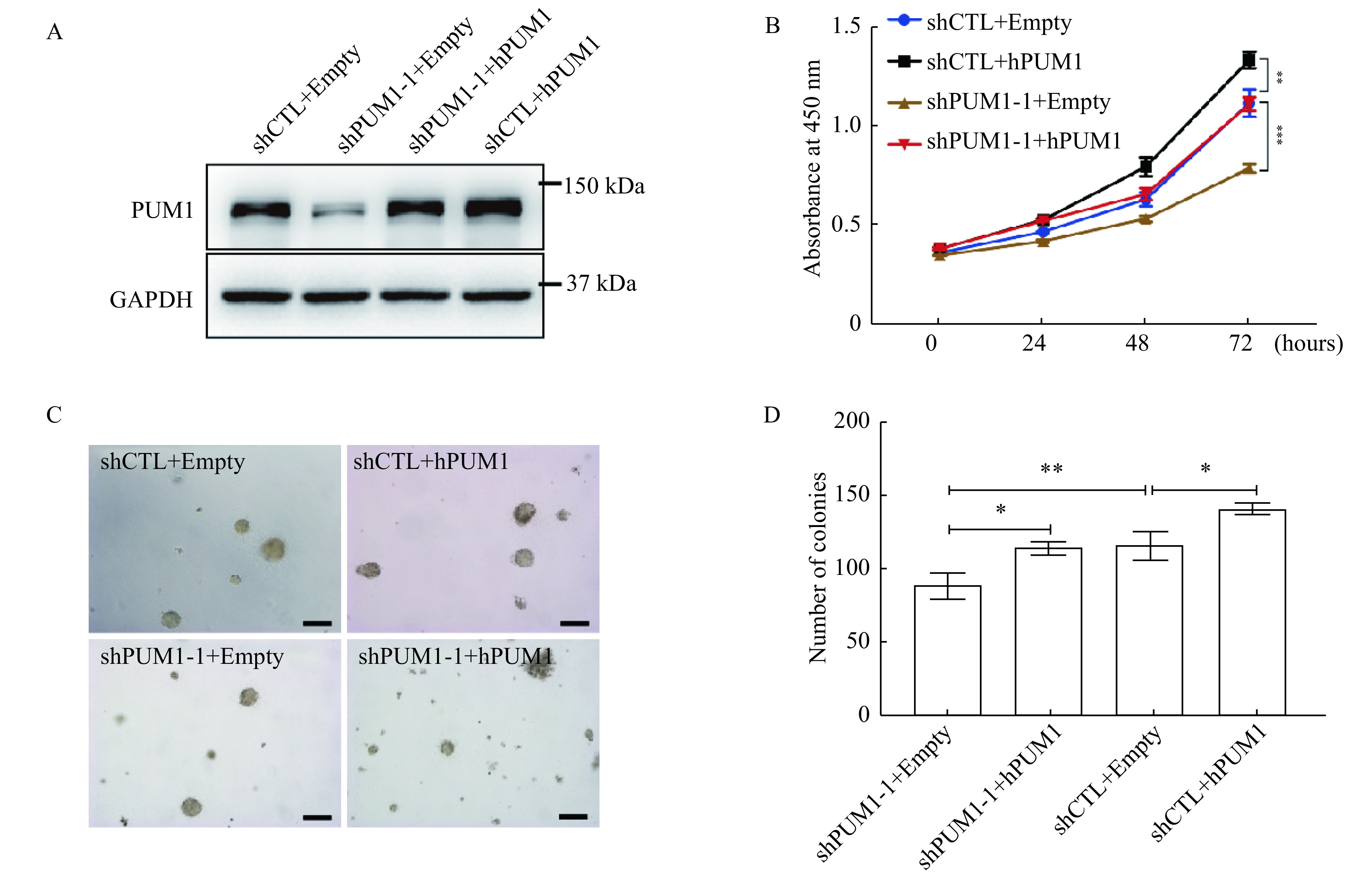
Overexpression of PUM1 promoted prostate cancer cell proliferation and colony formation.
A: Western blotting analysis of PUM1 protein expression from cells overexpressing PUM1 or PUM1 and shPUM1 at the same time. B: Cell proliferation assay on PUM1 overexpression and PUM1 knockdown. Data are presented as mean±SD (two-tailed Student's t-test; **P<0.01,***P<0.001). C: Colony assays were performed on the effect of overexpression ofPUM1 or knockdown of PUM1 on colony number and size. Scale bar: 100 μm. D: Colony number was counted for cells containing PUM1 overexpressing plasmids or knockdown plasmid, or both. Data are mean±SD of three independent experiments (Student's t-test; *P<0.05,**P<0.01).
We next determined the role of PUM1 in cell transformation by measuring the effect of PUM1 expression levels on the anchorage-independent growth of DU145 cells in soft agar assay. The total number of colonies was significantly reduced with PUM1-knockdown, while overexpression of PUM1 led to a substantially higher number of colonies (Fig. 3C and D). Overexpressing PUM1 in the cells with PUM1-knockdown rescued the ability of DU145 cells to form a comparable number of colonies, consistent with its rescue effect in cell proliferation. The positive correlation between PUM1 expression level and anchorage-independent growth further supports our hypothesis that PUM1 plays a critical role in prostate carcinogenesis.
Reduced PUM1 expression repressed prostate tumorigenesis in vivo
To test our hypothesis that PUM1 is essential for the growth of prostate cancer cells, we examined the effect of PUM1-knockdown on the tumorigenic capacity of DU145 cells in nude mice. DU145 cells transduced with PUM1 shRNA (sh-PUM1) or control shRNA (ShCTL) were injected into the left and right inguinal regions of the same nude mice (Fig. 4A and B). By both tumor weight and volumes, knockdown of PUM1 significantly reduced the tumor growth (Fig. 4C and D). These results confirmed the oncogenic activity of PUM1 in vivo and suggest that PUM1 may represent a potential target of treatment for prostate cancer.
Figure 4.
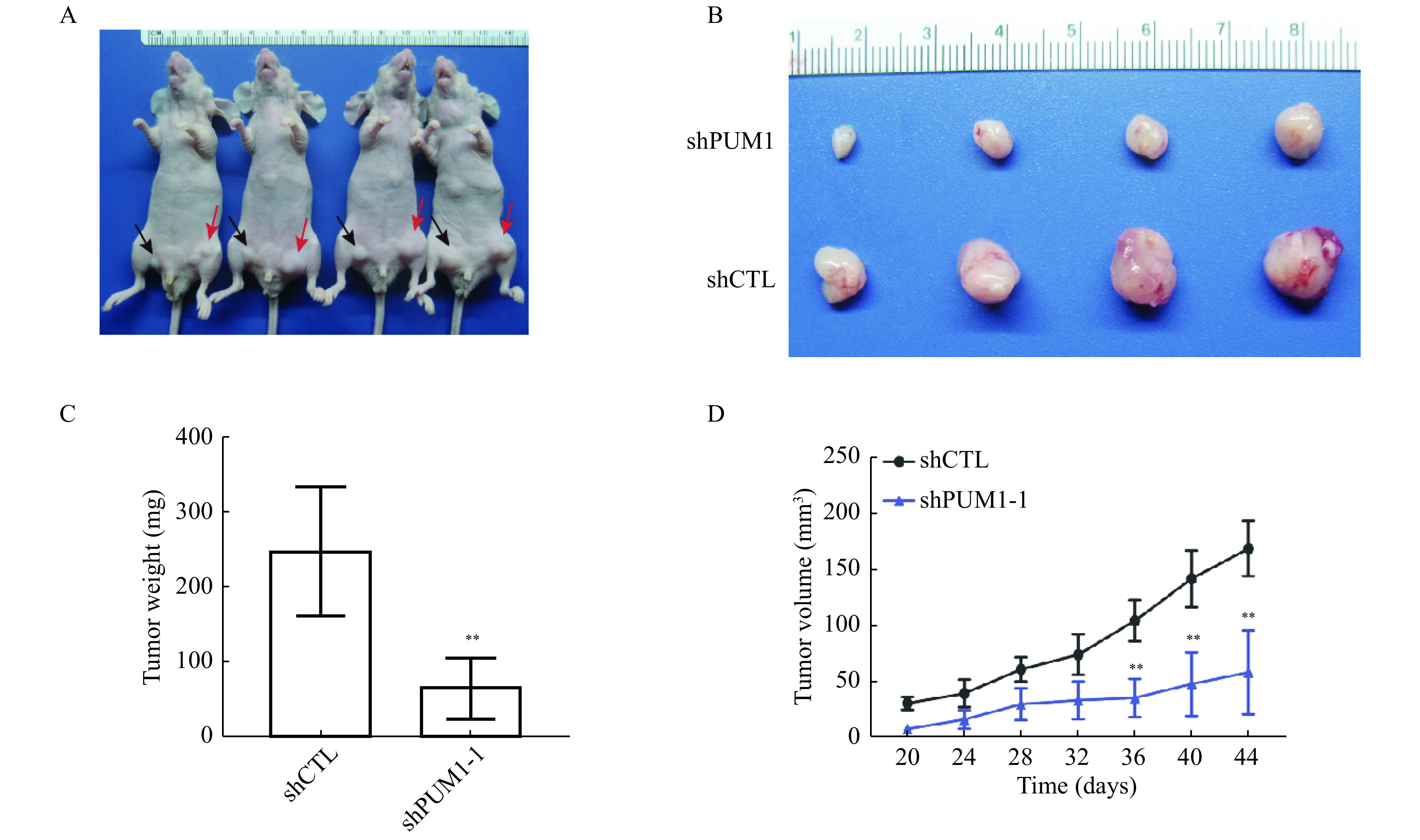
Knockdown PUM1 reduced tumorigenesis of DU145 cells in vivo.
A and B: Representative images of xenografted nude mice (A) and tumors formed from PUM1 knockdown cells and control cells (B). The black arrowhead indicates PUM1 knockdown cells, and the red arrow indicates control cells. C: Tumors from each group of mice were dissected and weighed for comparison of PUM1 knockdown and shCTL. Data are shown as mean±SD (n=4 mice in each group; unpaired t-test; **P<0.01vs. shCTL group). D: Tumor volumes from each group of mice were measured on the indicated days. Data are presented as mean±SD. Statistical analyses were performed using unpaired Student's t-test. **P<0.01vs. shCTL group.
PUM1 repressed the translation of tumor suppressor CDKN1B via binding to its PBE in prostate cancer cells
Genetic analyses established that Cdkn1b is a tumor suppressor in the prostate controlling prostatic epithelium growth[19]. According to TCGA data, CDKN1B expression was observed to be downregulated in prostate cancer compared to normal tissues (Fig. 5A). Base on previous reports, we wondered whether PUM1 is involved in tumorigenesis via regulating CDKN1B expression. We then evaluated the correlation between PUM1 and CDKN1B gene expression with the TCGA-based GEPIA tool, and the plots showed that PUM1 and CDKN1B expression is correlated in PCa with Pearson correlation coefficient being 0.63 (Fig. 5B). qRT-PCR showed CDKN1B mRNA was unaffected in both DU145 and PC3 cells with PUM1-knockdown (Fig. 5C).
Figure 5.
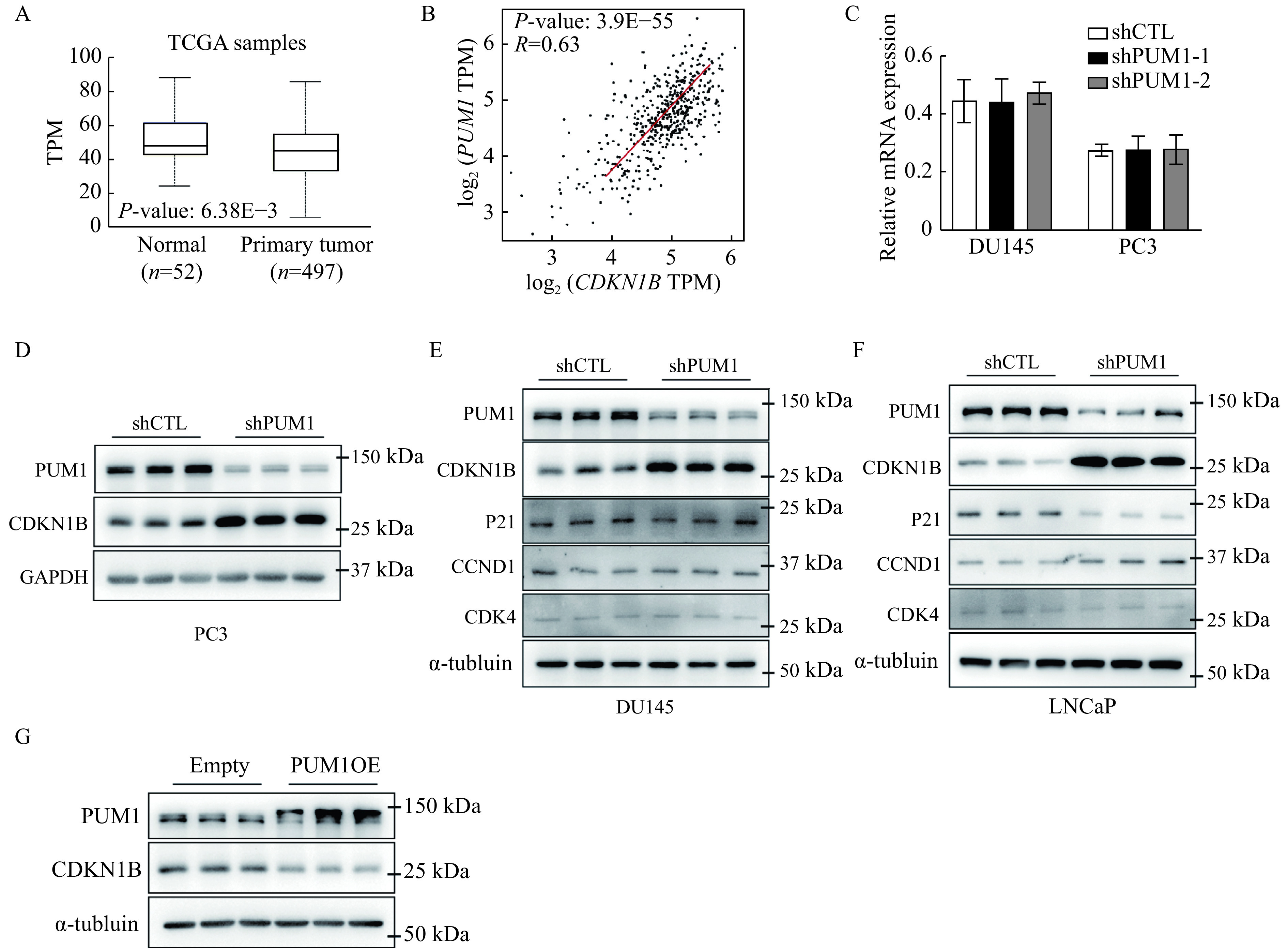
PUM1 repressed the translation of CDKN1B mRNA by binding to the PBE motif on its 3′ UTR.
A: Boxplots showed the expression of CDKN1B between normal prostate (n=52) and prostate carcinoma (n=497). Data were obtained from the GEPIA database (http://gepia.cancer-pku.cn/). B: The correlation analysis of PUM1 and CDKN1B in prostate cancer using the GEPIA tool. C: qRT-PCR analysis of CDKN1B mRNA levels in DU145 and PC3 cells with PUM1 knockdown. Data are expressed as mean±SD. Statistical analyses were performed by two-tailed Student's t-test for comparisons between each two groups. D–F: Cell cycle proteins expression levels in PC3, DU145, and LNCaP cells after 72 hours transfected with shPUM1-1 or shCTL were detected by Western blotting analysis. G: Western blotting analysis of CDKN1B protein level from DU145 cells transfected with human PUM1 (PUM1OE) or empty plasmids. TPM: transcripts per million.
To determine the molecular mechanism by which PUM1 contributed to cell proliferation and progression of prostate cancer, we analyzed the protein expression change of cell cycle regulators after inhibiting PUM1 expression. While other target cell cycle regulators exhibited variable changes or no changes in PUM1 knockdown cells, the protein expression of CDKN1B consistently increased upon knockdown of PUM1 in both PC3 and DU145 cell lines, supporting CDKN1B as a major cell cycle target of PUM1 protein (Fig. 5D and E).
To further determine if PUM1-mediated repression of CDKN1B is specific to androgen-insensitive DU145 and PC3 cell lines or general to all different types of prostate cancer cells, we chose LNCap cell line, which differs from DU145 and PC3 in that LNCap not only is from prostate carcinoma with low metastatic potential but also expresses androgen receptor and is androgen-sensitive. Knockdown of PUM1 led to a significant reduction of PUM1 expression. The examination of cell cycle regulators showed various degrees of protein expression changes, CDKN1B exhibited the highest increase in protein expression, supporting PUM1-mediated repression of CDKN1B may be a general mechanism among prostate cancer cells (Fig. 5F). CCND1 protein was slightly increased while p21 protein is slightly decreased (Fig. 5F ). These results suggested that the PUM1-CDKN1B axis may represent a major regulatory axis in both AR-positive and -negative PC cells.
Given that CDKN1B protein but not RNA increased in the PUM1 knockdown cells, we performed RNA immunoprecipitation and dual luciferase assay to test if PUM1 directly regulates the translation of CDKN1B via binding to the two PBEs of its 3′ UTR in prostate cancer cells. In the RNA immunoprecipitation experiment, we found that the pull-down fraction of PUM1 protein was significantly enriched with CDKN1B mRNAs, consistent with direct binding of PUM1 (Fig. 6A and B). Thus, CDKN1B mRNA is associated with PUM1 protein in DU145 and PC3 cells. Next, we constructed a luciferase reporter construct containing wildtype 3′ UTR or mutant 3′ UTR of CDKN1B mRNA with both PBE sites mutated (Fig. 6C). While PUM1 overexpression repressed expression of the reporter expression with wildtype CDKN1B 3′ UTR, mutations on PBEs negated the repressive effect of PUM1 on the reporter (Fig. 6C). These results supported that PBE sites on the 3′ UTR of CDKN1B are critical for PUM1 mediated translational repression in prostate cancer cells.
Figure 6.
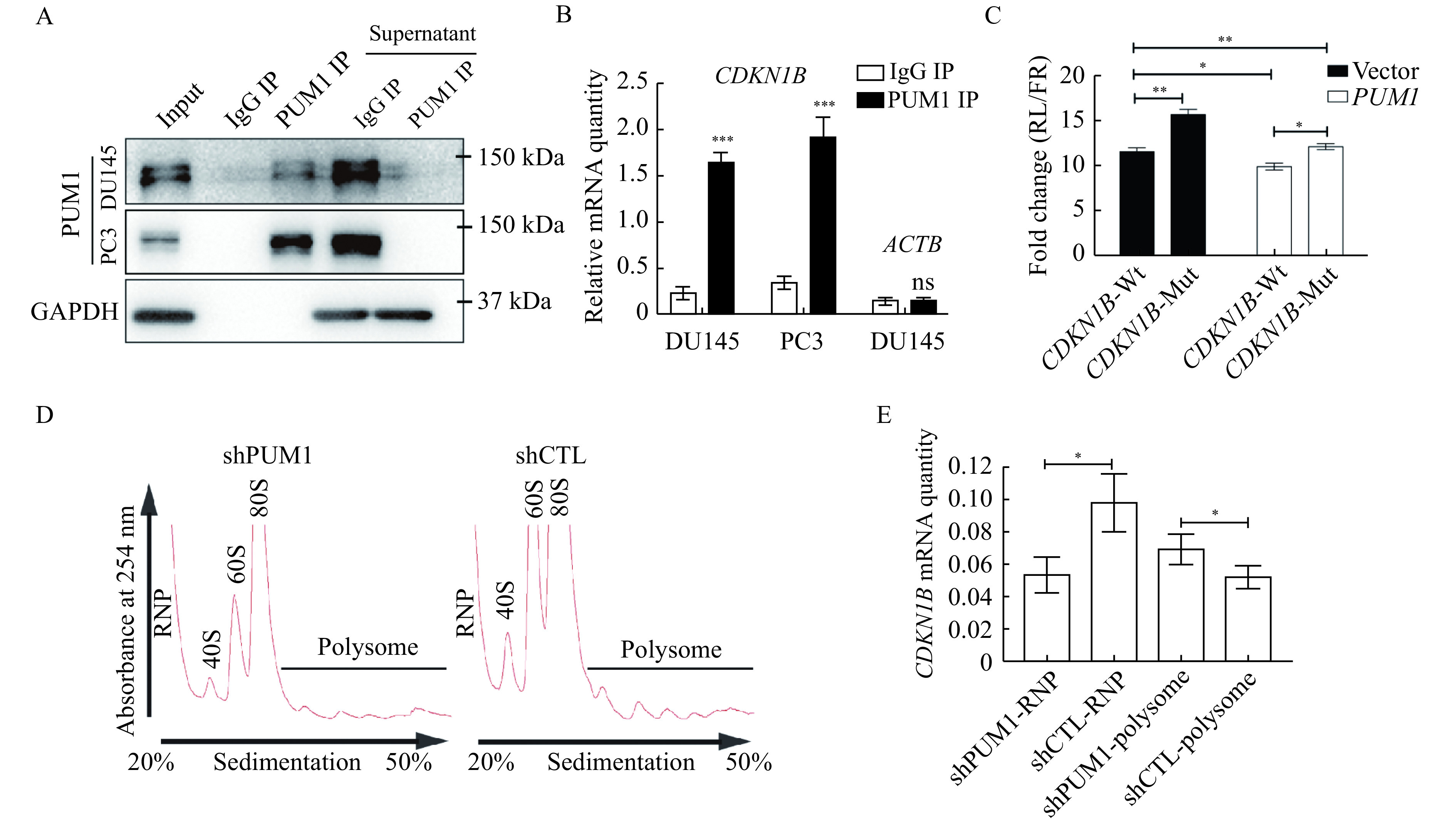
PUM1 repressed CDKN1B translation via binding to the 3′ UTR of CDKN1B mRNA.
A: Western blotting showing PUM1 IP efficiency in RNA immunoprecipitation from DU145 and PC3-cell lysates with anti-PUM1 antibody or IgG. B: RNA immunoprecipitation from DU145 and PC3 lysates with anti-PUM1 antibody or IgG. Enrichment of CDKN1B mRNAs were determined by qRT-PCR analysis. ACTB is a negative control for RIP. Data are shown as means±SD from three independent experiments (unpaired t-test; ***P<0.001, ns stands for not statistically significant). C: Diagram of wildtype and mutantCDKN1B 3′ UTR containing Pumilio-binding elements (PBEs) cloned in psiCHECK2 vector. Normalized luciferase activity expressed by the Luc-CDKN1B 3′ UTR constructs, co-transfected with the pCMV6-hPUM1 vs. pCMV6 (Vector) in 293T cells. Data are presented as mean±SD and compared using two-tailed unpaired t-tests (*P<0.05). D: Polysome profiles from polysome fractionation experiments of DU145 cells lysate ofPUM1 knockdown (shPUM1) and control (shCTL). E: qRT-PCR detected the distribution of CDKN1B mRNA in free RNP or polysome fractions. Data are presented as mean±SD. Comparisons between two groups were analyzed using a two-tailed Student's t-test. *P<0.05,**P<0.01,***P<0.001. RNP: ribonucleoprotein.
To test if PUM1 regulates the translation CDKN1B expression in prostate cancer cells, we performed a polysome fractionation experiment in DU145 cells transfected with or without a PUM1 knockdown construct. PUM1 knockdown significantly enriched CDKN1B mRNA in actively translating fraction, polysome fraction (Fig. 6D and E), indicating CDKN1B protein translation is promoted by PUM1 knockdown. Together, these results supported our hypothesis that increased PUM1 expression may contribute to the growth of prostate cancer cells via translational repression of cell cycle inhibitor—CDKN1B and PUM1-CDKN1B regulatory axis may be an important regulatory mechanism in cancer cell proliferation.
Discussion
In this study, we found PUM1 is frequently upregulated in human prostate carcinoma, and downregulation of PUM1 expression reduced PCa cell proliferation and survival. Our data from human cancer cells have unveiled a conserved translational regulation of cell cycle regulators by PUM1 in tumorigenesis and prostate cancer progression. Overexpression of PUM1 contributes to carcinogenesis by repressing the expression of negative cell cycle regulator CDKN1B, unveiling a novel mechanism for the loss of CDKN1B protein expression in tumorigenesis.
In cancer, the genetic control of the cell cycle is altered, resulting in unchecked growth and massive cell proliferation. CDKN1B is a negative regulator of the cell cycle and a tumor suppressor. CDKN1B is altered in 1.85% of all cancers with breast invasive ductal carcinoma, prostate adenocarcinoma, lung adenocarcinoma, colon adenocarcinoma, and testicular mixed germ cell tumor having the greatest prevalence of alterations[24]. Decrease, but not a complete loss of CDKN1B protein activity could stimulate tumorigenesis and is proposed to be an essential step in the development and maintenance of malignant prostatic epithelial cell phenotype[19,25]. Indeed down-regulation of CDKN1B is found in most human prostate cancer[25] and likely in other human tumors[26]. Posttranscriptional regulation of CDKN1B appears to be a primary mechanism for CDKN1B down-regulation during tumorigenesis[19,26–27]. Increased CDKN1B protein degradation via S-phase kinase-associated protein 2 or cyclin-dependent kinase subunit 1 mislocalization through posttranslational modification is shown to control CDKN1B protein abundance and its tumorigenic function[28]. While RNA binding protein HuD is implicated in reduced expression of CDKN1B in pancreatic cancer[29], but it is not known if translational regulation of CDKN1B abundance level is important for prostate cancer. Our finding that CDKN1B level is regulated at the translational level in prostate cancer cells by PUM1 protein revealed a novel mechanism regulating CDKN1B protein expression in prostate cancer, supporting the important roles of translational control in tumorigenesis.
Previously, it was reported the PUM1-mediated E2F transcription factor 3 post-transcription control may be necessary for the growth of cancer cell lines[21], and recently, PUM has been shown to be important for myeloid leukemia cell growth as well as hematopoietic stem cell growth[22]. Abnormal expression of PUM proteins is also shown to cause genomic instability[30–31]. However, it is unknown if PUM1 regulates tumorigenesis of solid tumors and to what extent PUM1-CDKN1B regulatory axis is important for cancer cell proliferation and tumor growth. Our findings support that PUM1 is a growth regulator in human prostate cancer and potentially a number of other cancers where PUM1 is highly expressed. We have identified CDKN1B as the main target of PUM1 in a range of prostate cancer cells with features of different aggressiveness and hormone dependency, including both androgen-insensitive and androgen-sensitive cells. Consistent with the down-regulation of CDKN1B in prostate cancer and its correlation with poor prognosis[32–33], our findings unveiled a novel regulatory mechanism of CDKN1B downregulation in cancer and a potential target for future therapeutics development.
Our previous study indicated PUM1 might regulate other cell cycle targets in mice[15–16]. PUM1 overexpression in cancer could elicit the growth-promoting effect via repression of targets other than CDKN1B. While other targets of PUM1 in prostate cancer are the subject of the future study, we have found that PUM1-CDKN1B repression appears to be robust and general among several cell cycle targets in different cancer cells we examined, and this axis may represent an important regulatory mechanism for cancer cell proliferation. Although the amplification of PUM1 and PUM2 locus is detected in a quarter of neuroendocrine prostate cancers[34], PUM alleles' gain is not associated with elevated mRNA expression. The mechanism underlying PUM1 overexpression in prostate cancers is also a subject of future study.
It has been reported that 3′ UTR of many key regulators of cancers tend to shorten or lost completely during the tumorigenesis[35], suggesting that dysregulation in posttranscriptional gene expression may be a common process that contributes to the pathogenesis of neoplasms. Our study of PUM1-mediated translational regulation of cell cycle regulators via binding to their 3′ UTR has demonstrated an example for such post-transcriptional regulation in cancer and argues for the importance of studying PUM1-CDKN1B regulatory axis in other cancers where the loss of CDKN1B protein expression is associated with tumorigenesis.
Our findings showed that PUM1 is a pro-proliferative factor in normal and neoplastic cells. It contributs to the emerging concept that post-transcription regulation represents a fundamental regulatory mechanism of cell growth. Further studies on the targets of PUM proteins and PUM protein expression regulation are required to fully elucidate their roles during tumorigenesis. The confirmation that PUM1 is essential for cell cycle progression and prostate tumorigenesis supports the potential role of PUM1 as a therapeutic target in PCa. Further examination of the PUM1-CDKN1B regulatory axis in diverse cancers could help understand how broadly such translational control contributes to tumorigenesis.
Acknowledgments
We would like to thank Dr. Yuan Ji at the University of Chicago for advice and discussion on the analysis of TCGA datasets and his lab for assistance. We appreciate comments from Drs. Jindan Yu and Takeshi Kurita as well as anonymous reviewers. This work was supported by the National Natural Science Foundation of China (Grant No. 31771652 and No. 81270737).
Footnotes
CLC number: R737.25, Ducument code: A
The authors reported no conflict of interests.
References
- 1.Wurth L, Gebauer F RNA–binding proteins, multifaceted translational regulators in cancer. Biochim Biophys Acta-Gene Regul Mech. 2015;1849(7):881–886. doi: 10.1016/j.bbagrm.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Abdelmohsen K, Gorospe M Posttranscriptional regulation of cancer traits by HuR. Wileys RNA. 2010;1(2):214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vellky JE, McSweeney ST, Ricke EA, et al RNA–binding protein DDX3 mediates posttranscriptional regulation of androgen receptor: a mechanism of castration resistance. Proc Natl Acad Sci U S A. 2020;117(45):28092–28101. doi: 10.1073/pnas.2008479117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradner JE, Hnisz D, Young RA Transcriptional addiction in cancer. Cell. 2017;168(4):629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstrohm AC, Hall TMT, McKenney KM Post–transcriptional regulatory functions of mammalian pumilio proteins. Trends Genet. 2018;34(12):972–990. doi: 10.1016/j.tig.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wickens M, Bernstein DS, Kimble J, et al A PUF family portrait: 3′ UTR regulation as a way of life. Trends Genet. 2002;18(3):150–157. doi: 10.1016/S0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 7.Zamore PD, Williamson JR, Lehmann R The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA–binding proteins. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1369583/ RNA. 1997;3(12):1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 8.Chen D, Zheng W, Lin AP, et al Pumilio 1 suppresses multiple activators of p53 to safeguard spermatogenesis. Curr Biol. 2012;22(5):420–425. doi: 10.1016/j.cub.2012.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gennarino VA, Singh RK, White JJ, et al Pumilio1 haploinsufficiency leads to SCA1–like neurodegeneration by increasing wild–type Ataxin1 levels . Cell. 2015;160(6):1087–1098. doi: 10.1016/j.cell.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mak W, Fang C, Holden T, et al An important role of pumilio 1 in regulating the development of the mammalian female germline. Biol Reprod. 2016;94(6):1–11. doi: 10.1095/biolreprod.115.137497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu EY, Chang R, Salmon NA, et al A gene trap mutation of a murine homolog of the Drosophila stem cell factor Pumilio results in smaller testes but does not affect litter size or fertility . Mol Reprod Dev. 2007;74(7):912–921. doi: 10.1002/mrd.20687. [DOI] [PubMed] [Google Scholar]
- 12.Zhang M, Chen D, Xia J, et al Post–transcriptional regulation of mouse neurogenesis by Pumilio proteins. Genes Dev. 2017;31(13):1354–1369. doi: 10.1101/gad.298752.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin K, Zhang S, Chen J, et al Generation and functional characterization of a conditional Pumilio2 null allele. J Biomed Res. 2017;32(6):434–441. doi: 10.7555/JBR.32.20170117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gennarino VA, Palmer EE, McDonell LM, et al A mild PUM1 mutation is associated with adult–onset ataxia, whereas haploinsufficiency causes developmental delay and seizures. Cell. 2018;172(5):924–936. doi: 10.1016/j.cell.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin K, Qiang W, Zhu M, et al Mammalian Pum1 and Pum2 control body size via translational regulation of the cell cycle inhibitor Cdkn1b . Cell Rep. 2019;26(9):2434–2450. doi: 10.1016/j.celrep.2019.01.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin K, Zhang S, Shi Q, et al Essential requirement of mammalian Pumilio family in embryonic development . Mol Biol Cell. 2018;29(24):2911–2968. doi: 10.1091/mbc.E18-10-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu IM, Hengst L, Slingerland JM The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008;8(4):253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 18.Kuczyk M, Machtens S, Hradil K, et al Predictive value of decreased p27Kip1 protein expression for the recurrence–free and long–term survival of prostate cancer patients . Br J Cancer. 1999;81(6):1052–1058. doi: 10.1038/sj.bjc.6690806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polyak K The p27Kip1 tumor suppressor gene: still a suspect or proven guilty? Cancer Cell. 2006;10(5):352–354. doi: 10.1016/j.ccr.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 20.Miles WO, Lembo A, Volorio A, et al Alternative polyadenylation in triple–negative breast tumors allows NRAS and c–JUN to bypass PUMILIO posttranscriptional regulation. Cancer Res. 2016;76(24):7231–7241. doi: 10.1158/0008-5472.CAN-16-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miles WO, Tschöp K, Herr A, et al Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 2012;26(4):356–368. doi: 10.1101/gad.182568.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naudin C, Hattabi A, Michelet F, et al PUMILIO/FOXP1 signaling drives expansion of hematopoietic stem/progenitor and leukemia cells. Blood. 2017;129(18):2493–2506. doi: 10.1182/blood-2016-10-747436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uhlen M, Zhang C, Lee S, et al A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352):eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 24.AACR Project GENIE Consortium AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017;7(8):818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Marzo AM, Meeker AK, Epstein JI, et al Prostate stem cell compartments: expression of the cell cycle inhibitor p27Kip1 in normal, hyperplastic, and neoplastic cells. Am J Pathol. 1998;153(3):911–919. doi: 10.1016/S0002-9440(10)65632-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koff A How to decrease p27Kip1 levels during tumor development . Cancer Cell. 2006;9(2):75–76. doi: 10.1016/j.ccr.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 27.Bencivenga D, Caldarelli I, Stampone E, et al p27Kip1 and human cancers: a reappraisal of a still enigmatic protein . Cancer Lett. 2017;403:354–365. doi: 10.1016/j.canlet.2017.06.031. [DOI] [PubMed] [Google Scholar]
- 28.Bochis OV, Irimie A, Pichler M, et al , Irimie, A., Pichler M, et al. (2015). The Role of Skp2 and its Substrate CDKN1B (p27) in Colorectal Cancer. J Gastrointestin Liver Dis. 2015;24(2):225–34. doi: 10.15403/jgld.2014.1121.242.skp2. [DOI] [PubMed] [Google Scholar]
- 29.Kim C, Jeong DE, Heo S, et al Reduced expression of the RNA–binding protein HuD in pancreatic neuroendocrine tumors correlates with low p27Kip1 levels and poor prognosis . J Pathol. 2018;246(2):231–243. doi: 10.1002/path.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee S, Kopp F, Chang TC, et al Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins . Cell. 2016;164(1-2):69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tichon A, Gil N, Lubelsky Y, et al A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat Commun. 2016;7(1):12209. doi: 10.1038/ncomms12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang BL, Zheng SL, Isaacs SD, et al A polymorphism in the CDKN1B gene is associated with increased risk of hereditary prostate cancer . Cancer Res. 2004;64(6):1997–1999. doi: 10.1158/0008-5472.CAN-03-2340. [DOI] [PubMed] [Google Scholar]
- 33.DeMarzo AM, Nelson WG, Isaacs WB, et al Pathological and molecular aspects of prostate cancer. Lancet. 2003;361(9361):955–964. doi: 10.1016/S0140-6736(03)12779-1. [DOI] [PubMed] [Google Scholar]
- 34.Beltran H, Prandi D, Mosquera JM, et al Divergent clonal evolution of castration–resistant neuroendocrine prostate cancer. Nat Med. 2016;22(3):298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayr C, Bartel DP Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138(4):673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]


