Abstract
The EH domain proteins Pan1p and End3p of budding yeast have been known to form a complex in vivo and play important roles in organization of the actin cytoskeleton and endocytosis. In this report, we describe new findings concerning the function of the Pan1p-End3p complex. First, we found that the Pan1p-End3p complex associates with Sla1p, another protein known to be required for the assembly of cortical actin structures. Sla1p interacts with the first long repeat region of Pan1p and the N-terminal EH domain of End3p, thus leaving the Pan1p-End3p interaction, which requires the second long repeat of Pan1p and the C-terminal repeat region of End3p, undisturbed. Second, Pan1p, End3p, and Sla1p are also required for normal cell wall morphogenesis. Each of the Pan1-4, sla1Δ, and end3Δ mutants displays the abnormal cell wall morphology previously reported for the act1-1 mutant. These cell wall defects are also exhibited by wild-type cells overproducing the C-terminal region of Sla1p that is responsible for interactions with Pan1p and End3p. These results indicate that the functions of Pan1p, End3p, and Sla1p in cell wall morphogenesis may depend on the formation of a heterotrimeric complex. Interestingly, the cell wall abnormalities exhibited by these cells are independent of the actin cytoskeleton organization on the cell cortex, as they manifest despite the presence of apparently normal cortical actin cytoskeleton. Examination of several act1 mutants also supports this conclusion. These observations suggest that the Pan1p-End3p-Sla1p complex is required not only for normal actin cytoskeleton organization but also for normal cell wall morphogenesis in yeast.
The actin cytoskeleton participates in a wide range of processes in eukaryotic cells. In the yeast Saccharomyces cerevisiae, phenotypic analysis of cells carrying mutations in the gene encoding actin, ACT1, has implicated the actin cytoskeleton in polarized cell growth, cell wall synthesis, endocytosis, and a variety of other processes (for a recent review, see reference 6). The yeast actin cytoskeleton consists of two types of structure visible by fluorescence light microscopy: the cortical actin patches and the cytoplasmic actin cables. Both the actin patches and actin cables undergo extensive reorganization throughout the cell cycle (1, 19). In unbudded G1 cells that grow isotropically, the cortical actin patches are distributed randomly over the entire cell surface. During the budding period, the actin patches congregate first at the nascent bud site and later inside the bud with the actin cables aligned toward them. The pattern of actin patch distribution during the cell cycle correlates explicitly with that of localized cell surface growth (1, 17). This has led to the widely held view that the actin cytoskeleton may specify the site of cell surface expansion, possibly by directing secretory vesicles to the cell surface for new material deposition. In support of this hypothesis, some mutations in ACT1 and other genes that result in an abnormal distribution of the cortical actin patches also lead to delocalized cell surface growth and aberrant cell wall morphologies (23, 26).
Endocytosis, a process of vesicle trafficking from the cell surface, has also been suggested to be actin cytoskeleton dependent. The same allele of ACT1 (act1-1) that confers aberrant cell wall morphologies also confers defects in membrane receptor endocytosis (18). The null mutant of SAC6, which is deficient in the yeast homologue of the vertebrate actin filament-bundling protein fimbrin (18), is also defective in endocytosis. The connection between the actin cytoskeleton and endocytosis is further strengthened by the isolation and characterization of many mutants that exhibit defects simultaneously in both functions (5, 24, 32, 41, 43). Despite the wealth of information gathered so far concerning the involvement of the actin cytoskeleton in endocytosis and cell wall deposition, the fundamental mechanisms underlying the function of the actin cytoskeleton in these two processes are still not understood.
The characteristic cell wall abnormalities exhibited by the act1-1 mutant include unusually thick cell walls that appear to consist of multiple layers, with each layer of the thickness of a normal cell wall (23). In addition, the multilayered cell wall is confined to the mother cell of budded cells only, as the bud always exhibits wild-type wall morphology. It is not clear how actin cytoskeleton dysfunction can lead to such cell wall abnormalities, if it is indeed the causal factor. One speculation is that the actin cytoskeleton may play a role in cell wall deposition through its role in endocytosis. It is conceivable, for example, that cell surface proteins, such as cell wall-synthesizing enzymes, have to be internalized via endocytosis after their tasks are accomplished. Defects in endocytosis, as observed in act1, sla2, and other actin-defective mutants, may thus result in continuous deposition of cell wall materials, thereby giving rise to the multilayered cell walls (23). This hypothesis, however, is not supported by the phenotypes of some mutants (such as sac6) that display strong defects in endocytosis but only minor defects in cell wall morphology (18, 23).
The Pan1p-End3p complex has been found to play essential roles in both the organization of the actin cytoskeleton and endocytosis (5, 40–43). Immunofluorescent staining reveals that Pan1p colocalizes with cortical actin patches (40, 41). Mutations in PAN1 result in defects in the organization of actin cytoskeleton and in endocytosis (40, 41, 43). Structurally, Pan1p contains two repeats of the EH domain, a ca. 70-amino-acid motif present in a family of proteins including the mammalian epidermal growth factor receptor tyrosine kinase substrate Eps15 (45). End3p, which associates with Pan1p and also contains an EH domain, is known to be required for both endocytosis and actin cytoskeleton organization (5, 41).
In addition to the two EH domains, Pan1p contains a motif named the Sla1 homology domain (40) because of its sequence similarity with Sla1p, a protein involved in assembly of the cortical actin cytoskeleton (15). Sla1p was originally identified as a protein required for viability of the abp1 null mutant (15). It contains three SH3 domains at the N terminus and a repeated motif in the C-terminal region with a core sequence of TGGAMMP. The Sla1 homology domain of Pan1p shares this TGGAMMP repeat (15, 40). Recently, it has been demonstrated that a region containing the third SH3 domain of Sla1p is important for the protein's function in maintaining normal actin cytoskeleton organization, while the C-terminal repeats of Sla1p are required for the rescue of ABP1 dependency (2). Like Pan1p, Sla1p has been reported to associate with the cortical actin patches (2, 3, 11). The notion that Pan1p and Sla1p may be involved in a common function arises from the observation that the two mutations (pan1-4 and sla1Δ) are synthetically lethal at a temperature permissive to both single mutations (40).
In this study, the relationship between Sla1p and the Pan1p-End3p complex is explored in greater detail. Our results indicate that the three proteins are able to interact with each other and form a heterotrimeric complex. The region of Sla1p involved in the interaction with Pan1p and End3p has been mapped to the C-terminal repeats. The findings that the pan1-4, end3Δ, and sla1Δ mutants, as well as wild-type cells overproducing the Sla1p repeats, all exhibit severe cell wall defects suggest that the Pan1p-End3p-Sla1p complex is required not only for normal actin cytoskeleton organization but also for normal cell wall morphogenesis.
MATERIALS AND METHODS
Strains, media, and general methods.
The yeast strains used in this study are listed in Table 1. Yeast cells were propagated in rich medium (YPD) or synthetic complete medium (SC) or SC lacking the appropriate amino acids for plasmid maintenance (34). For testing cell wall defects in the mutants, calcofluor white M2R (fluorescent brightener 28; Sigma) was added to YPD at a final concentration of 1 mg/ml. In experiments requiring the expression of genes under the GAL1 promoter, galactose instead of dextrose was used as the carbon source. Genetic and recombinant DNA manipulations were done according to standard methods (34, 37).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| SFY526 | MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112 canr gal4-542 gal80-538 URA3::GAL1-lacZ | Clontech Laboratories, Inc. |
| CRY1 | MATa can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-52 | 41 |
| YHT99 | MATa can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-52 pan1-4 | 41 |
| YHT167 | MATa can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-52 end3Δ::LEU2 | 41 |
| YMC437 | MATa can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-52 sla1Δ::HIS3 | This study |
| YMC438 | MATa can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-52 pan1-4 sla1Δ::HIS3 pRS316-PAN1 | This study |
| YMC439 | MATa/α can1-100/+ ade2-1/+ his3-11,15/+ leu2-3,112/+ trp1-1/+ ura3-52/+ end3Δ::LEU2/+ sla1Δ::HIS3/+ | This study |
| YMC440 | MATa can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-52 sla1Δ::HIS3 pRS316-HA-PAN1 | This study |
| YMC441 | MATa can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-52 sla1Δ::HIS3 end3Δ::LEU2 pRS316-HA-PAN1 | This study |
| YMC442 | MATa can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-52 sla1Δ::HIS3 pan1Δ::HIS3 pRS314-PAN1-HA pRS315-Myc-SLA1 | This study |
| YMC443 | MATa can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-52 pan1Δ::HIS3 pRS314-PAN1-HA | This study |
| YMC444 | MATa can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-52 sla1Δ::HIS3 pRS315-Myc-SLA1 | This study |
| DDY335 | MATa his3Δ200 leu2-3,112 ura3-52 tub2-201 act1-3(act1-1) | 14 |
| DDY338 | MATa his3Δ200 leu2-3,112 ura3-52 can1-1 tub2-201 cry1 act1-101::HIS3 | 44 |
| DDY342 | MATα his3Δ200 leu2-3,112 ura3-52 can1-1 tub2-201 act1-113::HIS3 | 44 |
| DDY346 | MATa his3Δ200 leu2-3,112 ura3-52 ade2-101(am) tub2-201 cry1 act1-119::HIS3 | 44 |
| DDY349 | MATα his3Δ200 leu2-3,112 ura3-52 tub2-201 act1-124::HIS3 | 44 |
Plasmid and strain constructions.
The plasmids used in this study are described in Table 2. The pRS series of shuttle vectors was used throughout this study (8, 39). The 4.3-kb XhoI/SacII fragment containing the SLA1 gene was obtained by PCR using a primer 407 bp upstream of the start codon and another 198 bp downstream of the stop codon of SLA1. The fragment, when cloned into pRS314, complemented the sla1Δ strain and the pan1-4 sla1Δ synthetic lethality.
TABLE 2.
Plasmid constructs used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pRS314 | CEN6 TRP1 | 39 |
| pRS316 | CEN6 URA3 | 39 |
| pRS314-PAN1 | Pan1p; BamHI/EcoRI fragment in pRS314 | 41 |
| pRS316-PAN1 | Pan1p; BamHI/EcoRI fragment in pRS316 | 41 |
| pRS316-HA-PAN1 | HA-Pan1p; HA-tagged PAN1 in pRS316 | 41 |
| PRS314-PAN1-HA | PCR was used to generate an AscI site immediately before the stop codon of Pan1p in pRS314-PAN1, and a cassette containing three copies of the HA epitope together with the ADH1 terminator sequences was ligated to the AscI site | |
| pGAL-HA-LR1 | HA-Pan1p(1–385) containing the first long repeat; generated by PCR and cloned in frame with the HA epitope under GAL1 promoter control in pRS316 | |
| pGAL-HA-LR2 | HA-Pan1p(386–846) containing the second long repeat; generated by PCR and cloned in frame with the HA epitope under GAL1 promoter control in pRS316 | |
| pRS424 | 2 μm TRP1 | 8 |
| pRS425 | 2 μm LEU2 | 8 |
| pRS424-END3 | End3p; XbaI/ClaI fragment in pRS424 | 41 |
| pRS425-END3 | End3p; XbaI/ClaI fragment in pRS425 | 41 |
| pGAL-END3 | END3 was generated by PCR and placed under GAL1 promoter control in pRS314 | |
| pRS314-SLA1 | Sla1p; 4.3-kb XhoI/SacII fragment in pRS314 | |
| pRS316-SLA1 | Sla1p; 4.3-kb XhoI/SacII fragment in pRS316 | |
| pRS424-SLA1 | Sla1p; 4.3-kb XhoI/SacII fragment in pRS424 | |
| pRS424-SH3 | Sla1p(1–443) containing only the three SH3 domains. Sequence downstream of BglII in pRS424-SLA1 was removed (BglII/EcoRV) and replaced with the ADH1 terminator of pGBT9 (BamHI/EcoRV). | |
| pRS424-SLA1ΔSR | Sla1p(1–854) without the C-terminal Sla1p repeats. Sequence downstream of BamHI in pRS424-SLA1 was removed (BamHI/EcoRV) and replaced with the ADH1 terminator of pGBT9 (BamHI/EcoRV) | |
| pRS424-SR | Sla1p(856–1244) containing the C-terminal Sla1p repeats; generated by PCR and placed under SLA1 promoter control in pRS424 | |
| pGAL-SH3 | Sla1p(1–440) construct containing the three SH3 domains; generated by PCR and placed under GAL1 promoter control in pRS316 | |
| pGAL-SR | Sla1p(856–1244) construct containing the Sla1p repeats; generated by PCR and placed under GAL1 promoter control in pRS316 | |
| pGST-SH3 | GST-Sla1p SH3 domains (2–440); generated by PCR and cloned into pGEX-4T-1 | |
| pGST-SR | GST-Sla1p repeats (856–1244); generated by PCR and cloned into pGEX-4T-1 | |
| pRS315-Myc-SLA1 | Myc-Sla1p. SLA1 open reading frame was generated by PCR and cloned in frame after three copies of the Myc epitope under SLA1 promoter control in pRS315 | |
| pGBT9 | Gal4(1–147) DNA-binding domain, TRP1 | 4 |
| pGAD424 | Gal4(768–881) activation domain, LEU2 | 4 |
| pPAN1-LR1 | Pan1p(98–385) construct containing the first long repeat in pGBT9; generated by PCR | |
| pPAN1-LR2 | Pan1p(384–846) construct containing the second long repeat in pGAD424; identical to pPAN1.2 | 41 |
| pPAN1-CT | Pan1p(902–1480) construct containing the C-terminal proline-rich region in pGBT9; 2.3-kb NcoI/SalI PAN1 fragment was cloned into SmaI/SalI sites of pGBT9 | |
| pEND3 | End3p(1–349) in pGBT9; generated by PCR | |
| pEND3-EH | End3p(1–114) construct containing the N-terminal EH domain in pGBT9; generated by PCR | |
| pEND3-ER | End3p(254–349) construct containing the C-terminal repeats in pGBT9; identical to pEND3.4 | 41 |
| pSLA1ΔSR | Sla1p(1–854) construct lacking the C-terminal Sla1p repeats in pGAD424. A BamHI site was first generated in front of the ATG codon in pRS424-SLA1ΔSR. SLA1, without the C-terminal repeats, was then removed by BamHI digestion and moved to pGAD424 | |
| pSLA1-SR | Sla1p(856–1244) construct containing only the C-terminal Sla1p repeats in pGAD424; generated by PCR | |
| pSLA1-SRΔNPF | Sla1p(856–1238) construct containing the C-terminal Sla1p repeats without the NPF motif in pGAD424; generated by PCR |
Gene disruptions for PAN1 (pan1-Δ::HIS3), SLA1 (sla1Δ::HIS3), and END3 (end3Δ::LEU2) have been described previously (40, 41). YMC439 was generated by crossing YHT151 (41) with YMC437.
Coimmunoprecipitation and glutathione S-transferase (GST) fusion protein binding experiments.
Yeast extracts were prepared as described previously (41). For immunoprecipitation of Pan1 tagged with hemagglutinin (HA), approximately 1 mg of cell lysates was incubated with rabbit polyclonal anti-HA antibody Y-11 (Santa Cruz Biotechnology, Inc.) for 1 h at 4°C and then with protein A-Sepharose beads (Pharmacia) for another hour. The beads were then washed five times with lysis buffer (1% Triton X-100, 100 mM NaCl, 0.5% sodium deoxycholate, 50 mM Tris-HCl [pH 7.2], 1 mM phenylmethylsulfonyl fluoride, protease inhibitors), and the immunocomplexes were released by boiling in loading buffer for 5 min. The precipitated proteins were separated on sodium dodecyl sulfate (SDS)–8% polyacrylamide gels to detect Pan1-HA or Myc-Sla1p and on 10% gels to detect End3p. The gels were electroblotted onto Hybond-C Extra nitrocellulose membranes (Amersham) and probed with mouse monoclonal anti-HA (12CA5; Boehringer Mannheim), rabbit polyclonal anti-Myc (A-14; Santa Cruz Biotechnology), and rabbit anti-End3 (5) antibodies to detect Pan1-HAp, Myc-Sla1p, and End3p, respectively. Immunoprecipitation of Myc-Sla1p was performed as described above, using anti-Myc antibody A-14.
To make GST fusion proteins, the coding regions of Sla1p SH3 domains (amino acid residues 2 to 440) and the Sla1p C-terminal repeats (amino acid residues 856 to 1244) were obtained by PCR and fused in frame to bacterial GST expression vector pGEX-4T-1 (Pharmacia). Plasmids pGST-SH3 and pGST-SR were transformed into Escherichia coli BL21, and the transformants were grown in 200 ml of Luria-Bertani medium containing 100 μg of ampicillin (Sigma) per ml to an optical density at 600 nm of 0.5. The expression of GST fusion proteins was induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside (Life Technologies, Inc.) at 37°C for 4 h. Cells were collected by centrifugation, resuspended in cold phosphate-buffered saline (PBS), and sonicated on ice to lyse the cells. Lysates were centrifuged at high speed for 10 min, and the supernatants were incubated with 200 μl (bed volume) of glutathione-Sepharose 4B beads (Pharmacia) for 1 h at 4°C. The beads containing the GST fusion proteins were washed five times with PBS and finally resuspended in an equal volume of PBS. For precipitation, approximately 1 mg of yeast extracts was incubated with 25 μl of GST fusion protein-bound beads for 2 h at 4°C and then washed five times with lysis buffer. The bound proteins were released by boiling in 30 μl of loading buffer for 5 min, and 10 μl was loaded per lane on SDS-polyacrylamide gels. The gels were then transferred to nitrocellulose membranes and probed with either anti-HA antibody 12CA5 or anti-End3p antibody.
Yeast two-hybrid assays.
The two-hybrid assays were performed as described previously (41). DNA fragments of PAN1, SLA1, and END3 were fused to the Gal4 activation domain of pGAD424 or the DNA binding domain of pGBT9 as indicated in Table 2. Plasmids were cotransformed into S. cerevisiae SFY526, and β-galactosidase activities were measured as instructed by the manufacturer (Clontech).
Electron microscopy analysis.
Yeast cells were grown in liquid culture to early log phase in the conditions described and prepared for electron microscopy by a modification of the method of Numata et al. (27). Twenty-five milliliters of cell suspension was prefixed with 1.5 ml of 50% glutaraldehyde solution for 2 h at 24°C. After being washed three times with distilled water, cells were fixed in 2% fresh potassium permanganate for 2 h at 24°C. After several washes with water, cells were dehydrated in a graded series of ethanol and then embedded in low-viscosity Spurr resin (Sigma). Ultrathin sections were stained with uranyl acetate and lead citrate and examined in a JEOL 1200EX electron microscope.
Actin staining and lucifer yellow uptake.
The actin cytoskeleton was visualized as described previously (40). Cells were grown in liquid medium in the conditions described above and fixed with formaldehyde at a final concentration of 3.7% for 10 min. Cells were collected by centrifugation, resuspended in PBS containing 3.7% formaldehyde, and incubated for another hour. Cells were then washed three times in PBS and stained with rhodamine-conjugated phalloidin (Molecular Probes Inc.) for 2 h at 24°C. Cells were subsequently washed five times with PBS and resuspended in 90% glycerol containing p-phenylenediamine. Cells were observed and photographed with a Zeiss Axioplan microscope.
For lucifer yellow assay, cells were grown at 24°C in liquid media containing 2% galactose until early log phase (optical density at 260 nm of ∼0.3). Lucifer yellow was added to a final concentration of 5 mg/ml, and cells were incubated for 2.5 h at 24°C. Cells were then washed five times in PBS containing 10 mM sodium azide and 50 mM sodium fluoride and observed under the microscope.
RESULTS
Genetic interactions of PAN1, END3, and SLA1.
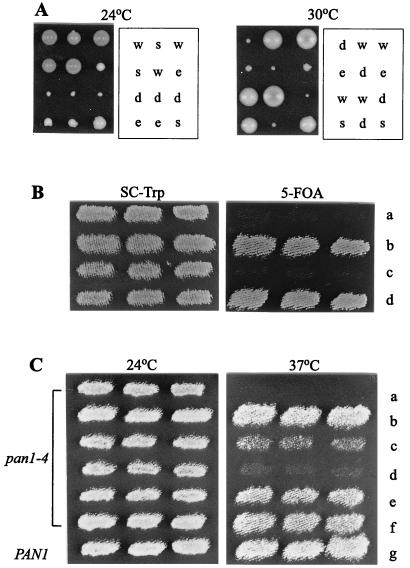
We noted previously that pan1-4 in combination with either the end3 or sla1 null mutation conferred synthetic lethality (40, 41). Further genetic analysis showed that unlike the pan1-4 sla1Δ and pan1-4 end3Δ double mutants, both of which were dead at all temperatures tested, the sla1Δ end3Δ double mutant was viable, albeit very weakly, at both 24 and 30°C (Fig. 1A). This result indicates that SLA1 and END3 genes are not essential for cell viability as long as the wild-type PAN1 gene is present.
FIG. 1.
Genetic interactions between PAN1, END3, and SLA1. (A) The end3Δ sla1Δ heterozygous diploid, YMC439, was sporulated, dissected, and grown for 5 days at 24°C and 30°C. Letters to the right of each panel indicate the identity of each colony determined from the segregation of the disruption markers: w, wild type; e, end3Δ; s, sla1Δ; d, end3Δ sla1Δ. (B) Suppression of the pan1-4 sla1Δ synthetic lethality. YMC438 (pan1-4 sla1Δ, pRS316-PAN1) cells transformed with pRS314 (a), pRS314-SLA1 (b), pRS424-END3 (c), and pRS314-PAN1 (d), were patched onto an SC-Trp plate and replica plated onto a 5-fluoro-orotic acid plate. All plates were incubated at 24°C. (C) Effects of SLA1 overexpression in the pan1-4 mutant. YHT99 (pan1-4) cells doubly transformed with pRS425 plus pRS424-SLA1 (a), pRS425-END3 plus pRS424 (b), pRS425-END3 plus pRS424-SLA1 (c), pRS425-END3 plus pRS424-SR (d), pRS425-END3 plus pRS424-SH3 (e), and pRS425-END3 plus pRS424-SLA1ΔSR (f) were patched onto an SC-Leu,-Trp plate at 24°C and replica plated at 37°C. As a control, the wild-type CRY1 was also transformed with pRS425 plus pRS424-SR (g) and analyzed in parallel.
As reported previously, overexpression of END3 was sufficient to restore growth of the pan1-4 mutant at 37°C (41), suggesting that it rescued those essential functions of Pan1p that are defective in the mutant. As an initial step to test whether Sla1p plays a role in the function of the Pan1p-End3p complex, we examined whether the suppression of pan1-4 by END3 overexpression requires the cooperation of Sla1p. As shown in Fig. 1B, overexpression of END3 was unable to support the growth of the pan1-4 sla1Δ double mutant at 24°C, a temperature permissive to growth of the sla1Δ single mutant. The suppression of pan1-4 by multicopy END3, therefore, takes place only in the presence of the wild-type SLA1 gene. A possible explanation for this result is that Sla1p may be required for the full function of the Pan1p-End3p complex. Alternatively, Sla1p may act in a parallel pathway and share some functions with Pan1p and End3p.
To further investigate Sla1p in relation to the function of Pan1p, we examined the effect of SLA1 overexpression in the pan1-4 mutant. Interestingly, overexpression of SLA1 not only was unable to suppress the temperature sensitivity of pan1-4 but also antagonized the suppression of pan1-4 by END3, as the pan1-4 cells containing both genes in multiple copies did not grow well at 37°C (Fig. 1C). We wished to find out which region in SLA1 is responsible for this effect. The conspicuous structural features of Sla1p are the three SH3 domains at the N terminus and the TGGAMMP repeats (Sla1p repeats) at the C terminus (Fig. 2D) (15). The SH3 region of SLA1 (pRS424-SH3) had no negative effects on the suppression of pan1-4 by END3, nor did the SLA1 construct with the Sla1p repeat region deleted (pRS424-SLA1ΔSR). However, when a construct containing only the C-terminal Sla1p repeats (pRS424-SR) was present, multicopy END3 could no longer support the growth of the pan1-4 mutant at 37°C (Fig. 1C). The antagonistic effect of Sla1p or Sla1p repeats on the suppression of pan1-4 by END3 is not due to some general toxicity of their overexpression, since neither pRS424-SLA1 nor pRS424-SR resulted in lethality to wild-type cells (Fig. 1C and data not shown). Rather, this effect may come about as a result of the specific interactions of these three proteins (see below).
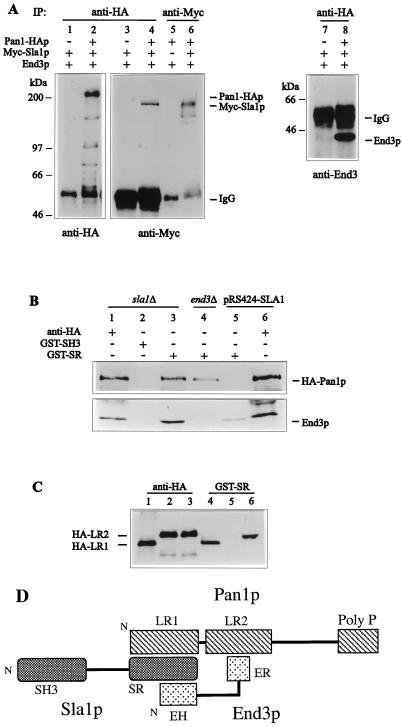
FIG. 2.
Physical interactions between Pan1p, End3p, and Sla1p. (A) In vivo coimmunoprecipitation of Pan1p-HA, Myc-Sla1p, and End3p. Equal amount of yeast extracts prepared from YMC442 (lanes 2, 4, 6, and 8), YMC443 (lane 5), and YMC444 (lanes 1, 3, and 7) were subjected to anti-HA or anti-Myc immunoprecipitations (IP) as indicated and analyzed, after electrophoresis, by immunoblotting using anti-HA (left), anti-Myc (middle), and anti-End3p (right) antibodies. To detect the directly immunoprecipitated proteins (lanes 1, 2, 5, and 6) 4 μl of each sample was loaded. All other lanes were loaded with 25 μl. (B) Binding of GST-SR fusion protein to HA-Pan1p and End3p. Equal amounts of yeast extracts derived from YMC440 (sla1Δ, pRS316-HA-PAN1; lanes 1 to 3), YMC441 (sla1Δ end3Δ, pRS316-HA-PAN1; lane 4), and YMC440 transformed with pRS424-SLA1 (lanes 5 and 6) were incubated with anti-HA, GST-SH3, or GST-SR as indicated. The precipitates were separated by SDS-PAGE and immunoblotted with anti-HA (top) and anti-End3p (bottom) antibodies to visualize HA-Pan1p and End3p, respectively. The lesser amount of HA-Pan1p observed in lane 4 is due to the slower growth of the YMC441 cells, as less HA-Pan1p was also detected in the cell extract with anti-HA antibody (data not shown). (C) Interaction between Pan1p long repeats and Sla1p C-terminal repeats. CRY1 cells containing pGAL-HA-LR1 (lanes 1 and 4), pGAL-HA-LR2 (lanes 2 and 5), and pGAL-HA-LR2 together with pGAL-END3 (lanes 3 and 6) were grown in galactose-containing liquid medium to early log phase, and extracts were prepared. Equal amounts of the extracts were incubated with anti-HA (lanes 1 to 3) or GST-SR (lanes 4 to 6) as indicated. The precipitates were subjected to SDS-PAGE and immunoblotted with anti-HA to detected the HA-tagged constructs. (D) Formation of the Pan1p-End3p-Sla1p complex. The diagram shows the structural domains of Pan1p, Sla1p, and End3p and the interactions among them. Pan1p LR2 is involved in the interaction with the End3p C-terminal repeats (ER), whereas LR1 of Pan1p and the N-terminal EH domain of End3p bind to the Sla1p C-terminal repeats (SR). Based on this interaction pattern, Pan1p, End3p, and Sla1p are able to exist as a heterotrimeric complex. N, N terminus.
Physical interactions of Sla1p, Pan1p, and End3p.
Physical interactions of Pan1p, Sla1p, and End3p were first examined by the coimmunoprecipitation experiments. The detection of Sla1p was achieved by tagging the protein with three copies of the c-Myc epitope at its N terminus, whereas Pan1p was identified by having three copies of the HA epitope at its C terminus. The resultant constructs, pRS315-Myc-SLA1 and pRS314-PAN1-HA, were able to complement the sla1Δ and pan1Δ mutants, respectively (data not shown). To detect the proteins, cell extracts prepared from YMC442 were incubated with an anti-HA (Y-11) or anti-Myc (A-14) polyclonal antibody, and the immunocomplexes were collected by using protein A-Sepharose beads. Upon analysis by Western blotting, Pan1-HA and Myc-Sla1p were detected as bands migrating at about 200 and 150 kDa, respectively (Fig. 2A, lanes 2 and 6). The bands were not observed in the extracts from cells with untagged Pan1p and Sla1p (Fig. 2A, lanes 1 and 5).
To determine whether Pan1p and Sla1p associate with each other in vivo, the anti-HA immunoprecipitates from YMC442 extracts were loaded on SDS-polyacrylamide gels and blotted onto membranes. As we reported previously (41), End3p was readily detected in the Pan1-HAp immunocomplex with the anti-End3 antibody (Fig. 2A, lane 8). When the anti-HA immunoprecipitates were probed with anti-Myc antibody A-14, Myc-Sla1p was also found to be present in the Pan1-HA immunocomplex (Fig. 2A, lane 4). This result demonstrates that Pan1p and Sla1p associate with each other in vivo. This conclusion was further confirmed by a reciprocal immunoprecipitation experiment, where Pan1-HA was detected in the anti-Myc immunoprecipitates (data not shown). Similar results were also obtained with His-tagged Sla1p used in place of Myc-Sla1p (data not shown).
Following the demonstration of the in vivo association of Sla1p with the Pan1p-End3p complex, we next sought to determine the region of Sla1p involved in the interaction. GST fusion proteins containing different regions of Sla1p were generated for this purpose. GST fusion proteins produced in E. coli were absorbed on glutathione-Sepharose 4B beads, and their levels of production were similar as assessed by SDS-polyacrylamide gel electrophoresis (PAGE) (data not shown). Equal amounts of the beads containing the immobilized GST fusion proteins were incubated with equal amounts of cell extract derived from YMC440, the sla1Δ mutant harboring plasmid pRS316-HA-PAN1. After binding and washing, the bound proteins were separated by SDS-PAGE and probed with either anti-HA antibody 12CA5 or anti-End3p antibody. As shown in Fig. 2B, both HA-Pan1p and End3p could be precipitated by the GST fusion protein containing only the C-terminal Sla1p repeats (GST-SR), and neither of them interacted with the GST fusion protein containing only the SH3 domains of Sla1p (GST-SH3) (Fig. 2B, lanes 2 and 3). These results show that Sla1p is able to interact with the Pan1p-End3p complex through its C-terminal repeats.
Sla1p and End3p can each interact with Pan1p independently of the other.
To determine whether Sla1p is required for formation of the Pan1p-End3p complex, the immunoprecipitation experiment was performed with extracts made from YMC440 (sla1Δ) cells. The Pan1p-End3p complex was still present in the sla1Δ mutant, indicating that the Pan1p-End3p interaction can occur in the absence of Sla1p (Fig. 2B, lane 1). Similarly, End3p was found to be not essential for Pan1p-Sla1p interaction, as HA-Pan1p could still be precipitated by GST-SR from extracts derived from the YMC441 (sla1Δ end3Δ) cells (Fig. 2B, lane 4). We conclude, therefore, that Sla1p and End3p can each interact with Pan1p independently of the other. Whether the Sla1p-End3p complex exists in the absence of Pan1p could not be tested, as the null mutation of PAN1 is lethal.
When the binding assay was performed with YMC440 cells that had been transformed with pRS424-SLA1, GST-SR could no longer precipitate HA-Pan1p from the extracts, and the amount of End3p precipitated was also drastically reduced (Fig. 2B, lane 5). This implies that overproduction of Sla1p saturated the Sla1p binding sites on HA-Pan1p and End3p in vivo and thereby blocked binding of the exogenous GST-SR to these two proteins. As described earlier, overexpression of SLA1 and the Sla1p repeats abrogated the suppression of pan1-4 by multicopy END3. To determine whether this was a result of interference in the Pan1p-End3p interaction, the same cell extract of YMC440 containing pRS424-SLA1 was incubated with anti-HA antibody to precipitate HA-Pan1p. When probed with anti-End3p antibody, End3p was still coprecipitated in the presence of pRS424-SLA1 (Fig. 2B, lane 6). The same results were obtained when pRS424-SR was used instead of pRS424-SLA1 (data not shown). These data show that overproduction of Sla1p (and the Sla1p repeats) does not disrupt the Pan1p-End3p interaction. It is likely, therefore, that different regions in Pan1p are involved in interactions with Sla1p and End3p.
Analysis of the interacting domains in Sla1p, Pan1p, and End3p.
Using the two-hybrid system, we investigated the regions in Sla1p and Pan1p that are involved in the interaction. Pan1p contains two long repeats at the N terminus, each of which embodies an EH domain (Fig. 2D) (40). The second long repeat (LR2) is essential for viability, whereas the first one (LR1) is dispensable for cell growth (35). LR2 is also the main target that End3p interacts with (41). As shown in Table 3, interactions were detected only between constructs containing LR1 of Pan1p (pPAN1-LR1) and the C-terminal repeats of Sla1p (pSLA1-SR) (Table 3). No interactions were observed with the construct lacking the Sla1p repeats (pSLA1ΔSR). Both LR2 of Pan1p and the C-terminal repeats of Sla1p induced β-galactosidase activity strongly by themselves when fused to the Gal4 DNA binding domain, and therefore we could not test their interaction by this assay (data not shown). To determine whether these two regions interact, we resorted to the binding assay using GST fusion proteins as described above. Two HA-tagged Pan1p constructs, containing either LR1 (pGAL-HA-LR1) or LR2 (pGAL-HA-LR2), were generated and placed under GAL1 promoter control. Extracts from wild-type cells expressing these constructs were incubated with the GST-SR beads, and the bound proteins were separated by SDS-PAGE and probed with anti-HA antibody 12CA5. We found that GST-SR could efficiently precipitate HA-LR1 but not HA-LR2 (Fig. 2C, lanes 4 and 5). When the HA-LR1-containing extract was incubated with GST-SH3, the HA-tagged protein was not precipitated (data not shown). These results show that Sla1p interacts with Pan1p through binding of its C-terminal repeats to LR1 of Pan1p.
TABLE 3.
Two-Hybrid interactions between Pan1p, End3p, and Sla1pa
| Plasmid | β-Galactosidase activity (U)
|
||||
|---|---|---|---|---|---|
| pGAD424 | pSLA1ΔSR | pSLA1-SR | pSLA1-SRΔNPF | pPAN1-LR2 | |
| pGBT9 | <1 | <1 | <1 | <1 | <1 |
| pPAN1-LR1 | <1 | <1 | 17 ± 8 | 14 ± 6 | ND |
| pPAN1-CT | <1 | <1 | <1 | ND | ND |
| pEND3 | <1 | <1 | 58 ± 12 | 42 ± 9 | 216 ± 23 |
| pEND3-EH | <1 | <1 | 93 ± 15 | 79 ± 19 | <1 |
| pEND3-ER | <1 | <1 | <1 | ND | 136 ± 18 |
The indicated pairs of plasmids were introduced into SFY526 cells, and β-galactosidase activity (average ± standard deviations for at least three transformants) was determined as described in Materials and Methods. ND, not determined.
The region of End3p capable of interacting with Sla1p was also examined in the two-hybrid assay. When fused to the Gal4 DNA binding domain, full-length End3p could interact strongly with pSLA1-SR but not with pSLA1ΔSR (Table 3), confirming the result obtained with GST fusion proteins. End3p contains two important domains, an EH domain at the N terminus and a repeated region bearing limited homology to α-actinin at the C terminus (Fig. 2D) (5). As shown in Table 3, it was the EH domain (pEND3-EH), not the C-terminal repeats (pEND3-ER), of End3p that displayed a strong interaction with the Sla1 repeats. On the other hand, it was pEND3-ER, not pEND3-EH, that interacted strongly with LR2 of Pan1p (Table 3), reconfirming our previous results (41).
EH domains from a number of proteins have been demonstrated to interact with the Asn-Pro-Phe (NPF) motif (7, 29, 36, 42, 46). Since Sla1p contains a single NPF motif at the C-terminal end of the protein (residues 1240 to 1242), we tested whether this motif is involved in the interaction with Pan1p and End3p. A Sla1p construct containing the C-terminal repeats but without the NPF motif (pSLA1-SRΔNPF) was generated and tested for interaction with pPAN1-LR1 and pEND3-EH in the yeast two-hybrid assay. As shown in Table 3, removal of the NPF motif from the Sla1p repeats did not abolish the interaction with LR1 of Pan1p or with the EH domain of End3p.
As the GST-SR fusion could precipitate HA-LR1 but not HA-LR2 of Pan1p, we realized that this interaction assay could be used to test whether Pan1p, End3p, and Sla1p can from a ternary complex. If these proteins could form a heterotrimeric complex, one would expect GST-SR to precipitate HA-LR2 after End3p, which interacts with LR2 through its C-terminal region and with the Sla1p repeats through its EH domain, was produced to a level similar to that of LR2. To test this idea, a plasmid containing END3 under the control of the GAL1 promoter (pGAL-END3) was introduced into cells harboring pGAL-HA-LR2. When protein extract prepared from these cells was incubated with GST-SR, HA-LR2 could be precipitated (Fig. 2C, lane 6). This experiment demonstrates that Pan1p, End3p, and Sla1p can indeed form a heterotrimeric complex.
To summarize, we find that Pan1p interacts with Sla1p through LR1 and with End3p through LR2, that End3p interacts with Sla1p through the N-terminal EH domain and with Pan1p through the C-terminal repeats, and that Sla1p interacts with both Pan1p and End3p through the Sla1p repeats. This pattern of interactions allows the three proteins to form a stable heterotrimeric complex, as illustrated in Fig. 2D.
Overexpression of the Sla1p repeats.
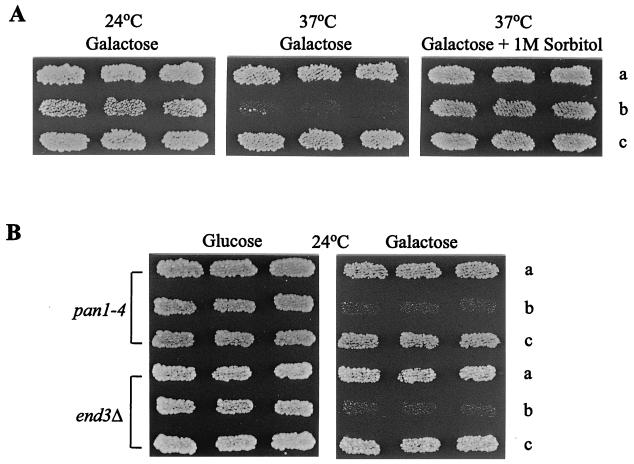
The fact that the C-terminal repeats of Sla1p are essential for interaction with both Pan1p and End3p prompted us to further study the cellular function of the Sla1p repeats by examining the effect of Sla1p repeat overexpression. Since placing the Sla1p repeats under the SLA1 promoter on a multicopy plasmid did not result in any noticeable phenotype in wild-type cells, as mentioned above, we chose to use the GAL1 promoter for overexpression. Wild-type cells carrying the Sla1p repeats on a centromere plasmid under GAL1 promoter control (pGAL-SR) grew normally at either 24 or 37°C in medium containing glucose or raffinose as the sole carbon source (data not shown). Following induction by galactose, however, the growth rate of the cells at 24°C was reduced. Galactose induction also resulted in heavy flocculation at 24°C (data not shown), indicating that these cells had some defects in the cell wall. At 37°C, cells containing pGAL-SR were inviable in galactose-containing medium (Fig. 3A). Viability could be restored to these cells by addition of 1 M sorbitol to the medium (Fig. 3A).
FIG. 3.
Effects of Sla1p C-terminal repeat overexpression. (A) Wild-type (CRY1) cells transformed with pRS316 (a), pGAL-SR (b), and pGAL-SH3 (c) were patched onto a galactose-containing SC-Ura plate at 24°C and then replica plated to fresh plates, with or without 1 M sorbitol, at 37°C. (B) YHT99 (pan1-4) and YHT167 (end3Δ) cells transformed with pRS316 (a), pGAL-SR (b), and pGAL-SH3 (c) were patched onto a glucose-containing SC-Ura plate and replica plated to a galactose-containing SC-Ura plate. Both plates were incubated at 24°C.
The deleterious effects of Sla1p repeat overexpression became more evident in the pan1-4 and end3Δ mutants, as pGAL-SR induced lethality in both mutants at 24°C (Fig. 3B). The galactose-induced cell lethality is a specific effect of the Sla1p repeats, since the SH3 domains of Sla1p overexpressed in the same way (pGAL-SH3) resulted in no detectable phenotypes in any of these cells (Fig. 3). In addition, introduction of pGAL-SH3 into cells harboring pGAL-SR had no effect on the flocculation and temperature-sensitive phenotypes (data not shown), suggesting that the SH3 domains of Sla1p could not interfere with the activity of the Sla1p repeats.
Cell wall morphology of the cells overexpressing the Sla1p repeats.
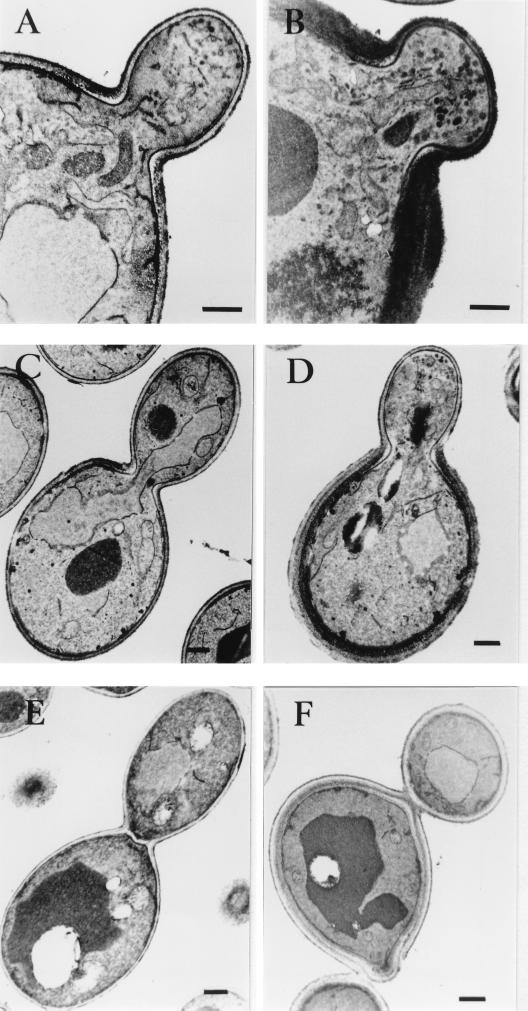
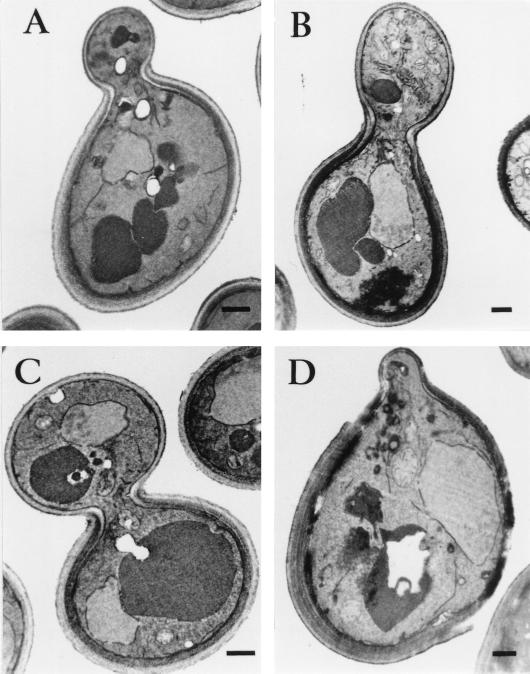
As the Sla1p repeat-induced lethality can be rescued by an adjustment in osmotic pressure of the medium, it is possible that Sla1p repeat overexpression affects cell wall integrity. Electron microscopy was used to investigate this possibility. At 24°C, all of the wild-type cells displayed a single layer of cell wall throughout the cell cycle, with no apparent difference between the mother and the bud (Fig. 4A, C, and E). Cells overexpressing the Sla1p repeats, on the other hand, exhibited strikingly aberrant cell wall morphologies. As shown in Fig. 4, the cell wall of the mother was multilayered and appeared much thicker than that of the bud (Fig. 4B, D, and F). Quantitative analysis indicated that 48% of the budded cells exhibited the abnormal cell wall morphology. These cell wall abnormalities are in fact similar, if not identical, to those observed in act1-1 and sla2 mutants (23). Analysis of cells representative of all cell cycle stages revealed that the abnormal cell wall morphology was always restricted to the mother, with the bud remaining single layered from early stages of bud formation to septation (Fig. 4B, D, and F).
FIG. 4.
Cell wall morphology of cells overexpressing the Sla1p C-terminal repeats. Wild-type (CRY1) cells, with or without plasmid pGAL-SR, were grown in galactose-containing liquid medium at 24°C and processed for electron microscopy analysis as described in Materials and methods. In cells without pGAL-SR, both the mother and bud, from small budded (A) to large budded (C) and until septation had occurred (E), had a single thin cell wall layer. A slight thickening of the cell wall was observed only at the mother-bud junction. In cells overexpressing the Sla1p C-terminal repeats (B, D, and F), the mother cell displayed an aberrant cell wall that was thicker and consisted of more than one layer. The bud, however, remained as a monolayer through out the cell cycle. Bars, 500 nm.
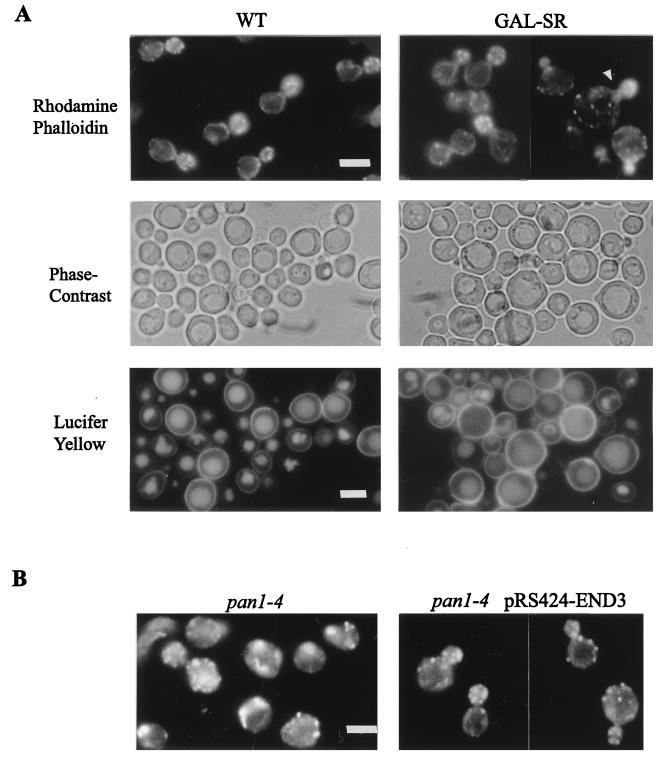
SLA1 is known to be important for the assembly of cortical actin cytoskeleton, and the sla1 null mutant exhibits gross disorganization of the cortical actin cytoskeleton at 37°C (15). Based on this knowledge, the abnormal cell wall morphology of the cells overexpressing the Sla1p repeats may just be another example of cell wall defects caused by the actin cytoskeleton disorganization. This turned out not to be the case. At 24°C, the actin cytoskeleton organization in most of the cells overexpressing the Sla1p repeats was like that in wild-type cells. Cortical actin patches were concentrated in the bud, and by adjusting the focus, we could observe actin cables aligning toward the bud as in wild-type cells (Fig. 5A). Only a minor population of the cells (less than 30%) appeared to have a slightly abnormal morphology, being larger and more round than wild-type cells. The cortical actin patches were still polarized to the bud in these cells, although the actin cables were not apparent (Fig. 5A). These phenotypes, however, could not be the cause of the multilayer cell wall morphology, as the cell wall abnormalities were more prevalent and were observed in many cells that were similar in size and shape to wild-type cells (Fig. 4). These results show that cell wall abnormalities in cells overexpressing the Sla1p repeats are not associated with disorganization of the cortical actin cytoskeleton.
FIG. 5.
(A) Fluorescence micrographs of wild-type CRY1 cells containing pRS316 (WT) or pGAL-SR (GAL-SR) grown in galactose-containing liquid medium at 24°C and stained with rhodamine-phalloidin (top). The arrowhead indicates an example of the enlarged cells. The cells were also stained with lucifer yellow and visualized by phase contrast (middle) to highlight the vacuole and with fluorescein isothiocyanate fluorescence optics (bottom) to visualize lucifer yellow. (B) Rhodamine-phalloidin staining of YHT99 (pan1-4) cells and YHT99 cells containing pRS424-END3 incubated for 3 h at 37°C. Bars, 5 μm.
Since the Sla1p repeats were involved in interactions with Pan1p and End3p, both of which are required for endocytosis, the effects of Sla1p repeat overexpression on endocytosis were also examined. After staining with the endocytic marker lucifer yellow, some of the enlarged cells overexpressing the Sla1p repeats were found to be defective in lucifer yellow uptake (Fig. 5A). Nevertheless, the majority of the cells were able to accumulate the dye in the vacuole at 24°C, indicating that they were largely competent in fluid-phase endocytosis (Fig. 5A).
Cell wall morphology in the sla1Δ, pan1-4, and end3Δ mutants.
The cell wall abnormalities induced by Sla1p repeat overexpression are dominant over the wild-type SLA1 gene. As the repeats are the binding sites for both Pan1p and End3p, overexpression of the repeats will likely prevent these two proteins from interacting with Sla1p. It follows then that the cell wall abnormalities induced by Sla1p repeat overexpression may be a common phenotype of the sla1Δ, pan1-4, and end3Δ mutants. Indeed, even at the permissive temperature of 24°C, all three mutants exhibited prominent cell wall abnormalities similar to that in the Sla1p repeat-overexpressing cells (Fig. 6). Quantitative analysis of the budded cells indicated that 77, 63, and 73% of the sla1Δ, end3Δ, and pan1-4 cells, respectively, displayed the abnormal cell wall phenotype at 24°C. After a temperature shift to 37°C for 3 h, the cell wall defects in these mutants were exacerbated, as the outer layers of the multilayered cell wall of the mother were often seen to be torn apart (Fig. 6D). Again, the aberrant cell wall morphology was not likely a direct result of the cortical actin cytoskeleton disorganization, as both pan1-4 and sla1Δ mutants maintained apparently normal cortical actin cytoskeleton distribution at 24°C (15, 40).
FIG. 6.
Cell wall morphology of pan1-4, end3Δ, and sla1Δ mutants. Electron microscopy analysis of the cell wall ultrastructure in YMC437 (sla1Δ; A), YHT167 (end3Δ; B), and YHT99 (pan1-4; C) cells grown in YPD liquid medium at 24°C. (D) Electron micrograph showing the damaged cell wall of strain YHT99 after 3 h of incubation at 37°C. Such a phenotype was also observed in sla1Δ and end3Δ mutants at 37°C (data not shown). Bars, 500 nm.
Overexpression of END3 could not correct the cell wall defect in the pan1-4 mutant.
Overexpression of END3, as reported previously, suppressed the lethality of the pan1-4 mutant at 37°C (41). Whether END3 overexpression simultaneously suppressed other defects in the mutant has not been thoroughly investigated. We analyzed the pan1-4 mutant carrying multicopy END3 in more detail. Examination of cell wall morphology indicated that multicopy END3 made no significant difference to the number of budded cells displaying cell wall abnormalities in the pan1-4 mutant at either 24 or 37°C (Table 4). Therefore, the cell wall defect in the mutant was not affected by multicopy END3. On the other hand, overexpression of END3 greatly improved the actin cytoskeleton organization in the mutant. Although delocalization of the cortical actin patches was still observed occasionally, the majority of the pan1-4 cells containing pRS424-END3 displayed a polarized distribution of the cortical actin patches. Furthermore, the abnormal actin structures such as large or curved actin aggregates in the pan1-4 mutant were no longer detectable in the presence of multicopy END3 (Fig. 5B) (40, 47). The actin cables, however, remained difficult to visualize in these cells. The apparent absence of actin cables could not be the cause of the abnormal cell wall morphology, as the act1-101 mutant, whose actin cables were similarly undetectable, did not exhibit any abnormal cell wall phenotype (see below). These results indicate that END3 is more efficient in suppressing the defect of the actin cytoskeleton than those of cell wall in the pan1-4 mutant and that the deficiencies of cell wall morphology may not be a consequence of the cortical actin cytoskeleton disorganization.
TABLE 4.
Overexpression of END3 could not rescue the cell wall defect of pan1-4 cellsa
| Strains (temp [°C]) | % of budded cells with abnormal cell wall |
|---|---|
| CRY1 (24) | <1 |
| YHT99 (24) | 73 |
| YHT99 (37) | 95 |
| YHT99 with pRS424-END3 (24) | 75 |
| YHT99 with pRS424-END3 (37) | 90 |
Cells were grown in SC or SC-Trp liquid medium at the temperature indicated and processed for electron microscopy as described in Materials and Methods. At least 100 cells were examined in each case.
The cell wall abnormalities are an allele-specific phenotype of act1 mutants.
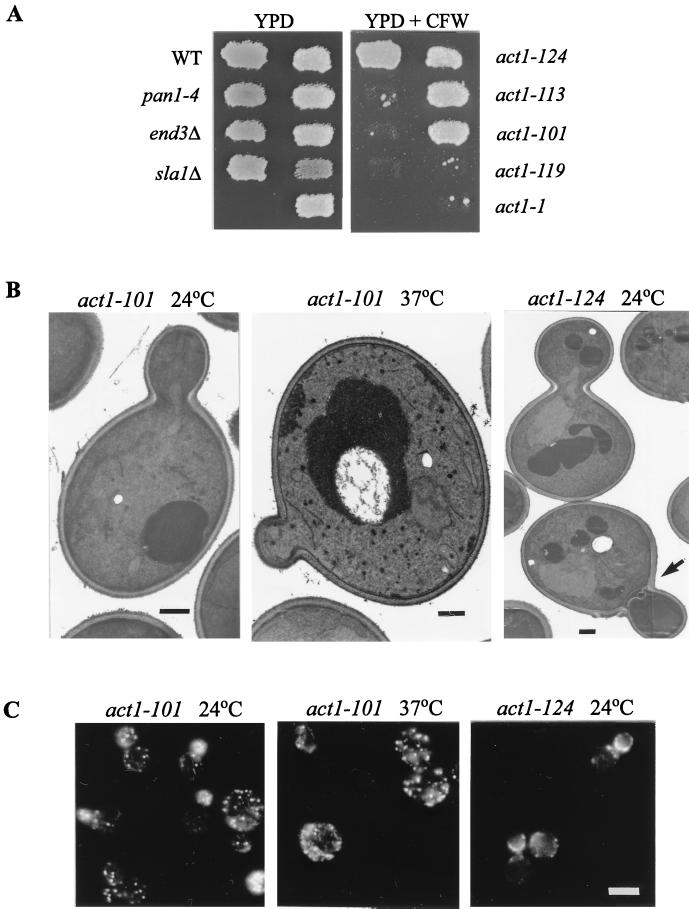
To further demonstrate that the cell wall and actin cytoskeleton defects are separable, we examined various act1 mutants for their cell wall morphology. A convenient method for detecting cell wall defects is to test the sensitivity to calcofluor white, a fluorescent dye that binds to the chitin component and interferes with the organization of the cell wall (13, 21, 31). Mutants with a defective cell wall have been shown to be inviable at concentrations of the dye that do not affect wild-type cells (21, 25, 31). In agreement with their defects in cell wall synthesis as described above, the pan1-4, sla1Δ, and end3Δ mutants all failed to grow in the presence of calcofluor white at 24°C (Fig. 7A). The act1-1 mutant, which has been shown to bear similarly defective cell walls (23), was also sensitive to calcofluor white (Fig. 7A), as was the act1-119 mutant. On the other hand, the act1-101, act1-113, and act1-124 mutants were not sensitive to calcofluor white (Fig. 7A). The act1-124 mutant, upon further replica plating to fresh calcofluor white-containing plates, eventually became partially sensitive (data not shown).
FIG. 7.
Cell wall defects in various act1 mutants. (A) Calcofluor white sensitivity. Strains CRY1 (wild type [WT]), YHT99 (pan1-4), YHT167 (end3Δ), YMC437 (sla1Δ), DDY335 (act1-1), DDY338 (act1-101), DDY342 (act1-113), DDY346 (act1-119), and DDY349 (act1-124) were patched onto a YPD plate as indicated and then replica plated onto a YPD plate supplemented with calcofluor white (CFW); 1 mg/ml). Both plates were incubated at 24°C. (B) Electron micrographs of act1-101 (left) and act1-124 (right) mutants grown in YPD liquid medium at 24°C. The middle panel shows the act1-101 mutant after 2 h of incubation at 37°C. The arrow in the right-hand panel indicates a cell with defective septum. Bars, 500 nm. (C) Fluorescence micrographs of act1-101 (left) and act1-124 (right) mutants grown in YPD liquid medium at 24°C and stained with rhodamine-phalloidin. The middle panel shows the actin distribution of the act1-101 mutant after 2 h of incubation at 37°C. Bar, 5 μm.
The fact that all five alleles of the act1 mutation caused a temperature-sensitive phenotype (44) but differed in sensitivity to calcofluor white shows that the cell wall deficiencies are not an unavoidable consequence of the actin cytoskeleton defects. This observation was further confirmed by electron microscopy examination of the cell wall morphologies of the act1-101 and act1-124 mutants. Consistent with its insensitivity to calcofluor white, the act1-101 mutant exhibited a single-layered cell wall similar to that of wild-type cells at 24°C (Fig. 7B), even though a significant portion (about 25%) of these cells displayed a delocalized distribution of cortical actin patches (Fig. 7C). In addition, the majority of the cell population did not show visible actin cables (Fig. 7C). Upon incubation at 37°C for 2 h, the cortical actin patches in the act1-101 cells became randomly distributed while their cell wall morphology remained normal (single layered) (Fig. 7B and C). The actin cytoskeleton organization in the act1-124 mutant was similar to that of the act1-101 mutant at 24°C, except that about 20% of the cells contained buds that had cortical actin patches polarized towards the mother-bud junction (Fig. 7C), but contained no DNA (data not shown). This phenotype is indicative of premature cytokinesis. In agreement with this finding, electron microscopy analysis indicated that the act1-124 mutant displayed a defect in septation but no defect in the mother cell wall morphology (Fig. 7B). These results demonstrate that the cell wall abnormalities are an allele-specific phenotype of the actin mutants, and they support the notion that cell wall deposition may not depend on the actin cytoskeleton per se but more likely depends on some specific cell surface proteins that interact with the actin cytoskeleton.
DISCUSSION
The actin cytoskeleton is one of the most dynamic and complex systems in eukaryotic cells, and its proper function depends on the participation of numerous actin-associated or actin-regulatory factors. These factors are likely to be required for performing specific tasks in actin-related processes such as polarized growth, endocytosis, and cytokinesis. Studies of these factors, therefore, hold the key to understanding this system. It has been previously reported that a complex containing EH domain proteins Pan1p and End3p plays a dual role in organization of the actin cytoskeleton and endocytosis (5, 40, 41, 43). Now we have found that this complex contains an additional factor, Sla1p, previously known to be required for cortical actin cytoskeleton assembly. Our results suggest that Pan1p, End3p, and Sla1p are components of a complex whose functions are required not only for normal actin cytoskeleton organization but also for normal cell wall morphogenesis.
Sla1p interacts with the Pan1p-End3p complex.
The interaction between Sla1p and the Pan1p-End3p complex is supported by genetic as well as biochemical evidence. It is first inferred from the synthetic lethality of the pan1-4 and sla1Δ mutations. This synthetic lethality is likely not a nonspecific effect of combining two temperature-sensitive mutations, because both single mutants have good viability at 24°C. In addition, overexpression of END3, which suppresses the temperature sensitivity of pan1-4 at 37°C, could not support growth of the pan1-4 sla1Δ cells at 24°C. Therefore, this synthetic lethality must result from some overlapping defects jointly conferred by pan1-4 and sla1Δ mutations.
Interactions at the protein level are confirmed by a variety of methods including coimmunoprecipitation, binding of GST fusion proteins, and yeast two-hybrid assays. Both End3p and Sla1p can be detected in Pan1-HA immunoprecipitates, although it appears that the amount of Myc-Sla1p coimmunoprecipitated with Pan1-HAp was less than the amount of End3p detected in the Pan1-HA immunocomplex. This may suggest that not all of the Pan1p-End3p complex contained Sla1p. The implications of this finding on the regulation of the function of the Pan1p-End3p-Sla1p complex are not clear at present. Using the two-hybrid assay and the GST fusion protein binding assay, we found that each of the three proteins could interact with the others directly and that the interactions between Pan1p and either Sla1p or End3p could take place in the absence of End3p or Sla1p, respectively. Whether the Sla1p-End3p interaction can occur in the absence of Pan1p in vivo remains to be determined. Such an association may not be critical because loss of both proteins does not lead to cell lethality (at 30°C and below) in the presence of wild-type Pan1p. The interacting regions of each protein have also been elucidated. Sla1p was found to bind to the LR1 domain of Pan1p and the N-terminal region of End3p, thus leaving the Pan1p-End3p interaction, which requires mainly the LR2 region of Pan1p and the C-terminal region of End3p (41), undisturbed. Although Sla1p uses the same region, the C-terminal repeats, for its interactions with both Pan1p and End3p, the 390-amino-acid region comprising 16 repeated elements must be large enough to accommodate simultaneous binding of multiple factors. It is interesting that the Sla1p repeats and the two long repeats of Pan1p contain multiple sequence elements recently identified as the phosphorylation sites for Prk1p, a serine/threonine kinase involved in regulation of the actin cytoskeleton organization in yeast (47). This raises the possibility that the formation or activity of the Pan1p-Sla1p-End3p complex may be modulated by Prk1p phosphorylation.
The C-terminal region of Sla1p contains an NPF motif located outside the Sla1p repeat region. Although a number of proteins containing this motif have been shown to interact with EH domains from different sources, deletion of the NPF motif from Sla1p had no effect on the interaction with Pan1p-End3p assayed by the two-hybrid system. In a recent attempt to identify ligands of mammalian and yeast EH domains by screening a nonapeptide display library, the End3p EH domain was found to bind peptides containing the HT/SF or SWG motif (29). No peptides containing the NPF motif were selected with the End3p EH domain as a probe. In addition, the first EH domain of Pan1p did not select any peptide whereas the second EH domain could bind to peptides containing the NPF motif (29). These data support our observation that the NPF motif of Sla1p is not involved in interactions with either the End3p EH domain or LR1 of Pan1p. It is noteworthy that the Sla1p repeats contain no HT/SF or SWG motifs, suggesting that other motifs may be involved in the interactions with End3p and Pan1p.
The direct evidence to support the formation of a ternary complex by Pan1p, End3p, and Sla1p comes from the finding that LR2 of Pan1p can be precipitated by GST-fused Sla1p repeats only when End3p is similarly expressed as LR2. This is because End3p, which associates with LR2 through its C-terminal region and with the Sla1p repeats through its EH domain, provides a linkage between LR2 and the Sla1p repeats. Although the three proteins can exist as a trimeric complex, other forms of interaction cannot be ruled out. Moreover, additional factors may be involved in the formation of these complexes in vivo. Examples of such factors are Bee1p, a protein shown to interact with Sla1p (20), and yAP180, a protein reported to bind Pan1p (42). Whether all of these proteins are present in the same complex for the same function or in different complexes for different functions remains to be determined.
Recently, a novel group of EH domain proteins which contain multiple SH3 domains was isolated (28, 33, 38, 46). This group of proteins, known as intersectins, has been found in mammals, Xenopus, and Drosophila (28, 33, 38, 46). The yeast S. cerevisiae does not have intersectin-like proteins (29, 43). Intersectins contain two EH domains at the N termini, a central coiled-coil domain, and four or five SH3 domains at the C termini (28, 33, 38, 46). Interestingly, the mammalian homologue of intersectin, Ese1, is constitutively associated with the EH domain protein Eps15 through its central coiled-coil domain (38). Therefore, the Ese1/Eps15 complex may be viewed as the counterpart of the Pan1p-End3p-Sla1p complex. Both complexes are capable of bringing EH- and SH3-binding proteins into a macromolecular complex.
Role of the Pan1p-End3p-Sla1p complex in cell wall morphology.
Cell wall defects were first observed in cells overexpressing the C-terminal repeats of Sla1p, which induced heavy flocculation and thermosensitivity correctable by addition of the osmotic stabilizer sorbitol to the media. Examination by electron microscopy confirmed the abnormal wall morphology of these cells.
While searching for additional factors interacting with Sla1p, we found that the sla1Δ mutation was synthetically lethal with either the hoc1 or fks1 mutation (data not shown). This lends further support to the role of the Pan1p-End3p-Sla1p complex in cell wall morphogenesis. HOC1 is a suppressor of the cell lysis phenotype of a pkc1 mutant (25) and has therefore been implicated in cell wall synthesis. FKS1 encodes an integral membrane protein, a subunit of β-1,3-glucan synthase, involved in synthesis of the cell wall component β-1,3-glucan (9). Interestingly, Fks1p activity is regulated by the GTP-binding protein Rho1p, which is also known to play a role in actin cytoskeleton organization (10, 16, 22, 30).
During the preparation of this report, Sla1p was reported to be required for the proper localization of Rho1p and Sla2p, two proteins involved in cell wall morphogenesis and the actin cytoskeleton (2). These authors also found that the sla1 mutant displays cell wall abnormalities similar to that reported previously for act1 and sla2 mutants (2). Our present finding are in agreement with the function of Sla1p in normal cell wall morphogenesis described by Ayscough et al. (2).
Uncoupling cell wall defects from the actin cytoskeleton organization.
The cell wall abnormalities exhibited by sla1Δ, pan1-4, and end3Δ cells as well as by cells overexpressing the Sla1p repeats resemble the abnormal cell wall morphology observed in the act1-1 and sla2 mutants (2, 23). Since both act1-1 and sla2 mutants have constitutive and temperature-independent delocalization of cortical actin patches, the cell wall defects observed in these mutants are thought to be a consequence of the depolarized cortical actin patches (23). This would be consistent with the knowledge that the Pan1p, Sla1p, and End3p proteins are all required for normal actin cytoskeleton organization. However, evidence provided in this report supports the conclusion that deficiencies in cell wall morphology do not necessarily come from actin cytoskeleton delocalization.
Uncoupling of cell wall abnormalities from delocalization of actin patches was evident in a number of cases. First, the majority of cells overexpressing the Sla1p repeats had a wild-type pattern of the actin cytoskeleton distribution while displaying prominent cell wall abnormalities. In addition, the pan1-4 and sla1Δ mutants both had defective cell wall morphology at 24°C, a temperature at which they exhibited normal cortical actin cytoskeleton distribution (15, 40). More interestingly, overexpression of END3 in the pan1-4 mutant could rescue the defect of the actin organization of the mutant but not that of the cell wall. These results indicate that the cell wall defects described above are not simply a consequence of the delocalization of actin cytoskeleton.
This conclusion is further supported by analysis of different alleles of the act1 mutants. As measured by sensitivity to calcofluor white and in some cases confirmed by electron microscopy, different alleles of act1 conferred different patterns of cell wall deficiency: act1-1 and act1-119 mutants were sensitive to calcofluor white and hence cell wall defective, while act1-101 and act1-113 mutants were not and the act1-124 mutant was intermediate in cell wall defectiveness. Both act1-101 and act1-124 cells could maintain normal cell wall morphology even though delocalization of the cortical actin cytoskeleton was observed. These results indicate, again, that defects in the actin cytoskeleton do not inevitably cause cell wall abnormalities.
Instead, it is more reasonable to suggest that the actin cytoskeleton and cell surface activities, including cell wall morphogenesis and endocytosis, are connected through functions of some cell surface-membrane protein complexes. These complexes achieve various activities through a combination of concerted functions of the complex as a whole and the specialized functions of some of its components. The Pan1p-End3p-Sla1p complex is an ideal candidate for such functions. Each of the Pan1, End3, and Sla1 proteins has now been found to be required for normal actin cytoskeleton organization (5, 15, 40), endocytosis (5, 41, 43; the role of Sla1p in endocytosis is cited in reference 5), and cell wall morphogenesis (this report and reference 2). Further studies of these proteins will provide valuable insights into regulation of the actin cytoskeleton and related processes.
ACKNOWLEDGMENTS
We are grateful to David Drubin for various act1 mutants. Alan Munn and Catherine Pallen are thanked for critical reading of the manuscript. We also thank Jun Wang for general technical assistance and other members of Cai laboratory for helpful discussions.
This work was supported by the Singapore National Science and Technology Board.
REFERENCES
- 1.Adams A E M, Pringle J R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayscough K R, Eby J J, Lila T, Dewar H, Kozminsk K G, Drubin D G. Sla1p is a functionally modular component of the yeast cortical actin cytoskeleton required for correct localization of both Rho1p-GTPase and Sla2p, a protein with Talin homology. Mol Biol Cell. 1999;10:1061–1075. doi: 10.1091/mbc.10.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayscough K R, Stryker J, Pokala N, Sanders M, Crews P, Drubin D G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel P L, Chien C T, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley D A, editor. Cellular interactions in development: a practical approach. Oxford, England: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- 5.Benedetti H, Raths S, Crausaz F, Riezman H. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994;5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botstein D, Amberg D, Mulholland J, Huffaker T, Adams A, Drubin D, Stearns T. The yeast cytoskeleton. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: cell cycle and cell biology. Vol. 3. New York, N.Y: Cold Spring Harbor Press; 1997. pp. 1–90. [Google Scholar]
- 7.Chen H, Fre S, Slepnev V I, Capua M R, Takei K, Butler M H, Di Fiore P P, De Camilli P. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature. 1998;394:793–797. doi: 10.1038/29555. [DOI] [PubMed] [Google Scholar]
- 8.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 9.Douglas C M, Foor F, Marrinan J A, Morin N, Nielsen J B, Dahl A M, Mazur P, Baginsky W, Li W, El-Sherbeini M, Clemas J A, Mandala S M, Frommer B R, Kurtz M B. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-β-d-glucan synthase. Proc Natl Acad Sci USA. 1994;91:12907–12911. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drgonova J, Drgon T, Tanaka K, Kollar R, Chen G C, Ford R A, Chan C S M, Takai Y, Cabib E. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science. 1996;272:277–279. doi: 10.1126/science.272.5259.277. [DOI] [PubMed] [Google Scholar]
- 11.Drubin D G, Nelson W J. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- 12.Dunn B, Wobbe C R. Preparation of protein extracts from yeast. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons; 1993. pp. 13.13.1–13.13.9. [DOI] [PubMed] [Google Scholar]
- 13.Elorza M V, Rico H, Sentandreu R. Calcofluor White alters the assembly of chitin fibrils in Saccharomyces cerevisiae and Candida albicans cells. J Gen Microbiol. 1983;129:1577–1582. doi: 10.1099/00221287-129-5-1577. [DOI] [PubMed] [Google Scholar]
- 14.Holtzman D A, Wertman K F, Drubin D G. Mapping actin surfaces required for functional interactions in vivo. J Cell Biol. 1994;126:423–432. doi: 10.1083/jcb.126.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holtzman D A, Yang S, Drubin D G. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J Cell Biol. 1993;122:635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilmartin J V, Adams A E M. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubler E, Riezman H. Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 1993;12:2855–2862. doi: 10.1002/j.1460-2075.1993.tb05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lew D J, Reed S I. Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 20.Li R. Bee1, a yeast protein with homology to Wiscott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J Cell Biol. 1997;136:649–658. doi: 10.1083/jcb.136.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lussier M, White A M, Sheraton J, di Paolo T, Treadwell J, Southard S B, Horenstein C I, Chen-Weiner J, Ram A F J, Kapteyn J C, Roemer T W, Vo D H, Bondoc D C, Hall J, Zhong W W, Sdicu A M, Davies J, Klis F M, Robbins P W, Bussey H. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazur P, Baginsky W. In vitro activity of 1,3-β-d-glucan synthase requires the GTP-binding protein Rho1. J Biol Chem. 1996;271:14604–14609. doi: 10.1074/jbc.271.24.14604. [DOI] [PubMed] [Google Scholar]
- 23.Mulholland J, Wesp A, Riezman H, Botstein D. Yeast actin cytoskeleton mutants accumulate a new class of golgi-derived secretory vesicle. Mol Biol Cell. 1997;8:1481–1499. doi: 10.1091/mbc.8.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munn A L, Stevenson B J, Geli M I, Riezman H. end5, end6, and end7: Mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1721–1742. doi: 10.1091/mbc.6.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neiman A M, Mhaiskar V, Manus V, Galibert F, Dean N. Saccharomyces cerevisiae HOC1, a suppressor of pkc1, encodes a putative glycosyltransferase. Genetics. 1997;145:637–645. doi: 10.1093/genetics/145.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novick P, Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- 27.Numata K, Ueki T, Naito N, Yamada N, Kamasawa N, Oki T, Osumi M. Morphological changes of Candida albicans induced by BMY-28864, a highly water soluble Pramidicin derivative. J Electron Microsc. 1993;42:147–155. [PubMed] [Google Scholar]
- 28.Okamoto M, Schoch S, Sudhof T C. EHSH1/Intersectin, a protein that contains EH and SH3 domains and binds to dynamin and SNAP-25. J Biol Chem. 1999;274:18446–18454. doi: 10.1074/jbc.274.26.18446. [DOI] [PubMed] [Google Scholar]
- 29.Paoluzi S, Castagnoli L, Lauro I, Salcini A E, Coda L, Fre S, Confalonieri S, Pelicci P G, Di Fiore P P, Cesareni G. Recognition specificity of individual EH domains of mammals and yeast. EMBO J. 1998;17:6541–6550. doi: 10.1093/emboj/17.22.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qadota H, Python C P, Inoue S B, Arisawa M, Anraku Y, Zheng Y, Watanabe T, Levin D E, Ohya Y. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science. 1996;272:279–281. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- 31.Ram A F J, Wolters A, Ten Hoopen R, Klis F M. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to Calcofluor White. Yeast. 1994;10:1019–1030. doi: 10.1002/yea.320100804. [DOI] [PubMed] [Google Scholar]
- 32.Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roos J, Kelly R B. Dap160, a neural-specific Eps15 homology and multiple SH3 domain-containing protein that interacts with Drosophila dynamin. J Biol Chem. 1998;273:19108–19119. doi: 10.1074/jbc.273.30.19108. [DOI] [PubMed] [Google Scholar]
- 34.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring HarborNew York, N.Y.: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 35.Sachs A B, Deardorff J A. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- 36.Salcini A E, Confalonieri S, Doria M, Santolini E, Tassi E, Minenkova O, Cesarini G, Pelicci P G, Di Fiore P P. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 1997;11:2239–2249. doi: 10.1101/gad.11.17.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring HarborNew York, N.Y.: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Sengar A S, Wang W, Bishay J, Cohen S, Egan S E. The EH and SH3 domain Ese proteins regulate endocytosis by linking to dynamin and Eps15. EMBO J. 1999;18:1159–1171. doi: 10.1093/emboj/18.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang H-Y, Cai M. The EH domain-containing protein Pan1 is required for normal organization of the actin cytoskeleton in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4897–4914. doi: 10.1128/mcb.16.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang H-Y, Munn A, Cai M. EH domain proteins Pan1p and End3p are components of a complex that plays a dual role in organization of the cortical actin cytoskeleton and endocytosis in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4294–4304. doi: 10.1128/mcb.17.8.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wendland B, Emr S D. Pan1p, yeast esp15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J Cell Biol. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wendland B, McCaffery J M, Xiao Q, Emr S D. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian esp15. J Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wertman K F, Drubin D G, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong W T, Schumacher C, Salcini A E, Romano A, Castagnino P, Pelicci P G, Di Fiore P P. A protein-binding domain, EH, identified in the receptor tyrosine kinase substrate Eps15 and conserved in evolution. Proc Natl Acad Sci USA. 1995;92:9530–9534. doi: 10.1073/pnas.92.21.9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamabhai M, Hoffman N G, Hardison N L, McPherson P S, Castagnoli L, Cesarini G, Kay B K. Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J Biol Chem. 1998;273:31401–31407. doi: 10.1074/jbc.273.47.31401. [DOI] [PubMed] [Google Scholar]
- 47.Zeng G, Cai M. Regulation of the actin cytoskeleton organization in yeast by a novel serine/threonine kinase Prk1p. J Cell Biol. 1999;144:71–82. doi: 10.1083/jcb.144.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]