Abstract
Pulsed-field gel electrophoresis (PFGE) of 61 Salmonella enterica serotype Paratyphi C isolates from six countries gave five distinct clusters. Twenty-four isolates from five countries were susceptible to 10 antimicrobials tested and gave similar restriction endonuclease digest patterns of the 38-MDa plasmid. In contrast, plasmid and PFGE profiles of 37 multidrug-resistant isolates from Zaire were different from those from other countries.
Although bacteriophage typing, antibiotic susceptibility, and plasmid profiles have previously been found to be useful in epidemiological studies of outbreaks of salmonella infections, these properties fail to reflect the evolutionary relationships of bacterial strains (9). In the present study, antibiotic susceptibility testing, plasmid profiling, and restriction endonuclease fragment patterns (REFPs) of plasmids were used to investigate Paratyphi C isolates obtained from different parts of the world.
Isolates were initially characterized by biochemical tests on triple sugar iron, motility indole ornithine, urea, oxidase, salicin, inositol, dulcitol, and mucate and serotyped using salmonella-agglutinating antisera (Murex Diagnostics, Dartford, United Kingdom). Paratyphi C isolates were tested for susceptibility to antimicrobials by a controlled disk diffusion technique on diagnostic sensitivity test agar plates containing 5% lysed horse blood. Escherichia coli ATCC 25922 was used as the susceptible control. The antibiotic discs (Oxoid Ltd., Basingstoke, United Kingdom) used were ampicillin (10 μg), tetracycline (30 μg), trimethoprim (5 μg), sulfamethoxazole (100 μg), chloramphenicol (30 μg), streptomycin (10 μg), gentamicin (10 μg), co-amoxiclav (20 μg of amoxicillin; 10 μg of clavulanic acid), ciprofloxacin (3 μg), and nalidixic acid (10 μg). MICs were determined by using doubling dilutions of antimicrobials (Adatabs; Mast Laboratories, Liverpool, United Kingdom) in nutrient agar according to the National Committee for Clinical Laboratory Standards guidelines (8). Chromosomal DNA was prepared in agarose plugs as described previously by Thong et al. (11). The plugs were digested with 25 U of XbaI or 20 U of SpeI. Pulsed-field gel electrophoresis (PFGE) of agarose plug inserts was then performed on a CHEF-DR II system (Bio-Rad Laboratories) for 20 h at 6 V/cm, with a pulse time of 1 to 40 s at 14°C. A lambda DNA digest consisting of a ladder (ca. 22 fragments) of increasing size from 48.5 kbp to approximately 1,000 kbp was included as a DNA size standard. The REFPs were interpreted according to guidelines described by Tenover et al. (10). Plasmid DNA was isolated as described previously (4). Conjugation experiments were performed for drug-resistant isolates only, with E. coli K-12 as a recipient as described previously (5). Purified plasmid DNA from transconjugants and from sensitive strains was subjected to restriction endonuclease digestion using PstI, HindIII, and EcoRI (Gibco-BRL, Paisley, United Kingdom).
A total of 61 Paratyphi C isolates were studied. Thirty-seven isolates from Zaire (1992 and 1996) were obtained from children with clinical cases of bacteremia from two regions of the country. Isolates from 1916 to 1949 were obtained from various other countries: two isolates came from Iraq, first obtained in 1918 and 1941; two isolates came from East Africa, isolated in 1939 and 1949; and one isolate came from Greece, isolated in 1916 (3). Fifteen isolates from India were taken from patients during outbreaks of enteric fever which occurred within one year (1983). Four other isolates from 1960 to 1976 from India were from patients with sporadic cases of enteric fever. All Paratyphi C isolates from Iraq, India, and East Africa were found to be fully susceptible to all the antimicrobials tested. For these isolates the highest MIC at which 90% of isolates were inhibited (MIC90) was 8 μg/ml (for streptomycin) and the lowest MIC90 was <0.5 μg/ml (for tetracycline). In contrast, all isolates from Zaire were multiply resistant to a combination of the commonly available antimicrobials, including tetracycline, ampicillin, sulfamethoxazole, trimethoprim, streptomycin, and chloramphenicol. Only two isolates from Zaire were resistant to two antimicrobials, while the rest were resistant to five or more antimicrobials. The MICs of ampicillin for all the isolates from Zaire were particularly high (MIC50 and MIC90 > 16 μg/ml). However, the MICs of chloramphenicol for isolates from 1996 (MIC50 and MIC90 > 32 μg/ml) were significantly higher than the MICs for isolates from 1992 (MIC50 = 4 and MIC90 = 32 μg/ml) (P <0.001). Furthermore, 12% of isolates from 1996 were resistant to co-amoxiclav (MIC90 = 16 μg/ml), whereas all isolates from 1992 were fully susceptible. Gentamicin, ciprofloxacin, and nalidixic acid were the only antimicrobials to which each of the isolates from Zaire was susceptible.
Eight distinct PFGE clusters were obtained from 57 out of the 61 Paratyphi C isolates used in this study. Chromosomal DNA from four isolates from India failed to give discrete bands when either XbaI or SpeI was used. REFPs for isolates from Zaire from 1992 revealed they were closely related; they fell into a single cluster (cluster 1) (Fig. 1), while those from 1996 showed greater diversity, as they fell into five different clusters. Isolates from India from the 1983 epidemic fell into a single cluster (cluster 2), but they were clearly distinct from isolates from the earlier outbreaks from 1960 to 1976. The PFGE clusters of other Paratyphi C isolates are shown in Table 1. Of 37 isolates from Zaire, 7 had 60- and 10-MDa plasmids; 21 isolates had 60-MDa plasmids. Plasmids could not be obtained from two isolates from India, while one isolate had plasmids of 4 and 2 MDa. From conjugation tests, both the 60- and 45-MDa plasmids from three isolates from Zaire transferred multidrug resistance to E. coli K-12. The 45-MDa plasmid was also self-transferable in all other multidrug-resistant isolates from Zaire. The most common resistance phenotypes transferred to E. coli K-12 were ampicillin, tetracycline, and chloramphenicol resistance phenotypes. No attempt was made to transfer the 38-MDa plasmid from antibiotic-susceptible isolates. PstI digestion of the 38- to 60-MDa plasmids produced a total of sixteen to eight fragments, ranging in size from 800 bp to 4.2 kb. HindIII and EcoRI digests of the 38-MDa plasmid produced identical patterns, of five and six bands, respectively. The REFPs for plasmids from the Paratyphi C isolates are shown in Table 2.
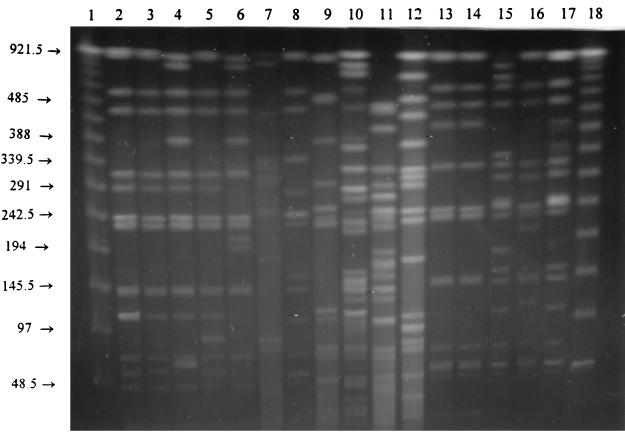
FIG. 1.
XbaI REFPs of representative Paratyphi C isolates from various countries. Lanes 1 and 18, 48.5-kb-ladder molecular size standard (sizes [in kilobases] are given to the left of the gel); lane 2, E48; lane 3, E49; lane 4, E51; lane 5, E52; lane 6, E53 in PFGE cluster 1; lane 7, E2 from Zaire in PFGE cluster 2; lane 8, E19 from Zaire in PFGE cluster 3; lane 9, E54 from Zaire in PFGE cluster 4; lane 10, E41 from Zaire in PFGE cluster 5; lane 11, E56 from Zaire in PFGE cluster 6; lane 12, E414 from India in PFGE cluster 7; lane 13, E1160 from India in PFGE cluster 3; lane 14, E1060 India in from PFGE cluster 3; lane 15, E701 from Iraq in PFGE cluster 8; lane 16, E699 from East Africa in PFGE cluster 3; lane 17, E700 from Greece in PFGE cluster 8. Strain types in lanes 2 to 6 were laboratory references from Zaire.
TABLE 1.
PFGE clusters of 57 Paratyphi C isolates
| PFGE cluster | No. of isolates | Source | Yr(s) of isolation |
|---|---|---|---|
| 1 | 8 | Zaire | 1992 |
| 2 | 8 | Zaire | 1996 |
| 3 | 7 | Zaire | 1996 |
| 2 | East Africa | 1939, 1949 | |
| 12 | India | 1983 | |
| 4 | 4 | Zaire | 1996 |
| 5 | 6 | Zaire | 1996 |
| 6 | 4 | Zaire | 1996 |
| 7 | 3 | India | 1976 |
| 8 | 1 | Greece | 1916 |
| 2 | Iraq | 1918, 1941 |
TABLE 2.
REFPs of plasmids from Paratyphi C isolates from various countries
| Plasmid size (MDa) | No. of isolates | Plasmid source | No. of fragments obtained by using:
|
||
|---|---|---|---|---|---|
| PstI | HindIII | EcoRI | |||
| 60 | 28 | Zaire | 16–18 | 8 | 7 |
| 45 | 37 | Zaire | 16–17 | 6–8 | 8 |
| 38 | 15 | India | 17 | 5 | 6 |
| 38 | 1 | India | 17 | 5 | 7 |
| 38 | 2 | Iraq | 17 | 5 | 7 |
| 38 | 2 | East Africa | 17 | 5 | 7 |
This study observed a high prevalence of multidrug resistance among recent Paratyphi C isolates from Zaire for nearly all of the commonly available antimicrobials, including cotrimoxazole, ampicillin, streptomycin, chloramphenicol, and tetracycline. Paratyphi C isolates from 1996 from Zaire were resistant to five or all six of these antimicrobials and were generally more resistant than the isolates from 1992. This may be the result of the increased antibiotic pressure over the course of time. As has been previously shown by other workers (1, 7), multidrug resistance to commonly available antimicrobials seriously compromises the ability of health workers to treat and effectively control disease outbreaks in developing countries. In Zaire, for instance, it was necessary to use ciprofloxacin in order to treat cases of multidrug-resistant non-typhi salmonellae (2), while in neighboring Rwanda expanded-spectrum generation cephalosporins were used (6). These antimicrobials, though effective against multidrug-resistant Paratyphi C isolates, would normally be too expensive for or unavailable to the general public.
By PFGE, Paratyphi C strains responsible for cases of bacteremia in 1992 in children from Zaire were found to be closely related, as they formed one cluster. However, the PFGE digest patterns of isolates from 1996 from Zaire were diverse and significantly different from isolates from 1992. This may indicate a significant mutational change in the strains responsible for the outbreak in 1996 from Zaire but which left the 45-MDa R plasmids conserved within the strains. Furthermore, the plasmid content and REFPs of plasmids were also diverse, revealing that the isolates were unlikely to be related. Of the 19 Paratyphi C isolates from India, 12 formed a single cluster. In addition, these Paratyphi C isolates were of the same serotype and had a common 38-MDa plasmid, which yielded similar PstI, HindIII, and EcoRI REFPs. These isolates are likely to be subtypes of the same strain that caused the outbreak of enteric fever in 1983 in India but were unlikely to be related to the strains responsible for the outbreak in 1976. Recently, Selander et al. (9) observed that the majority of Paratyphi C from various parts of the world analyzed by multilocus enzyme electrophoresis formed a total of nine electrophoretic types, representing seven clones and subclones of strains that bore significant genetic relatedness. This is in general agreement with the findings of the present study; Paratyphi C isolates fell into eight different PFGE clusters.
In conclusion, the present study has demonstrated that outbreaks of Paratyphi C from 1983 from India were likely to be from a similar strain unrelated to the 1976 outbreak and to the outbreaks from either Iraq or East Africa, despite possessing similar REFPs of plasmids and antimicrobial susceptibility patterns. This study has also revealed that the Paratyphi C isolates from Zaire were highly diverse, multidrug resistant, and unrelated to those from the Indian subcontinent; recent isolates from Zaire from 1996 were generally more resistant than earlier isolates from 1992.
Acknowledgments
S. K. thanks the Director of the Kenya Medical Research Institute for permission to publish.
This study was supported by a Research Development Award of the Wellcome Trust.
REFERENCES
- 1.Cohen M L. Epidemiology of drug resistance: implications for a post-antimicrobial era. Science. 1992;234:964–969. doi: 10.1126/science.257.5073.1050. [DOI] [PubMed] [Google Scholar]
- 2.Green S D R, Ilunga F M, Numbi A, Cheesbrough J S, Tillotson G S. An open study of ciprofloxacin for the treatment of proven or suspected extra intestinal salmonellosis in African children: a preliminary report. Adv Antimicrob Antineoplastic Chemother. 1992;11:181–187. [Google Scholar]
- 3.Hirschfeld L. A new germ of paratyphoid. Lancet. 1919;i:296. [Google Scholar]
- 4.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kariuki S, Gilks C, Corkill J, Kimari J, Benea A, Waiyaki P, Hart C A. Multi-drug resistant non-typhi salmonellae in Kenya. J Antimicrob Chemother. 1996;33:425–434. doi: 10.1093/jac/38.3.425. [DOI] [PubMed] [Google Scholar]
- 6.Lepage P, Bogaerts J, Nsengumuremyi F, Hitimana D G, Van Goethem C, Vandepitte J, Butzler J P. Severe multiresistant Salmonella typhimurium systemic infections in central Africa—clinical features and treatment in a paediatric department. J Antimicrob Chemother. 1984;14:153–159. doi: 10.1093/jac/14.suppl_b.153. [DOI] [PubMed] [Google Scholar]
- 7.Murray B E. Problems and dilemmas of antimicrobial resistance. Pharmacology. 1992;12:86–93. [PubMed] [Google Scholar]
- 8.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 9.Selander R K, Beltran P, Smith N H, Helmuth R, Rubin F A, Kopecko D J, Ferris K, Tall B D, Cravioto A, Musser J M. Evolutionary genetic relationships of clones of Salmonella serovars that cause human typhoid and other enteric fevers. Infect Immun. 1990;58:2262–2275. doi: 10.1128/iai.58.7.2262-2275.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thong K L, Ngeow Y F, Altwegg P N, Pang T. Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J Clin Microbiol. 1995;33:1070–1074. doi: 10.1128/jcm.33.5.1070-1074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]