Abstract
Posttranslational modification, in particular farnesylation, of Ras is crucial for activation of Saccharomyces cerevisiae adenylyl cyclase (CYR1). Based on the previous observation that association of CYR1 with cyclase-associated protein (CAP) is essential for its activation by posttranslationally modified Ras, we postulated that the associated CAP might contribute to the formation of a Ras-binding site of CYR1, which mediates CYR1 activation, other than the primary Ras-binding site, the leucine-rich repeat domain. Here, we observed a posttranslational modification-dependent association of Ras with a complex between CAP and CYR1 C-terminal region. When CAP mutants defective in Ras signaling but retaining the CYR1-binding activity were isolated by screening of a pool of randomly mutagenized CAP, CYR1 complexed with two of the obtained three mutants failed to be activated efficiently by modified Ras and exhibited a severely impaired ability to bind Ras, providing a genetic evidence for the importance of the physical association with Ras at the second Ras-binding site. On the other hand, CYR1, complexed with the other CAP mutant, failed to be activated by Ras but exhibited a greatly enhanced binding to Ras. Conversely, a Ras mutant E31K, which exhibits a greatly enhanced binding to the CYR1-CAP complex, failed to activate CYR1 efficiently. Thus, the strength of interaction at the second Ras-binding site appears to be a critical determinant of CYR1 regulation by Ras: too-weak and too-strong interactions are both detrimental to CYR1 activation. These results, taken together with those obtained with mammalian Raf, suggest the importance of the second Ras-binding site in effector regulation.
Ras proteins are small guanine nucleotide-binding proteins that cycle between the active GTP-bound and the inactive GDP-bound states. They are conserved from yeasts to mammals and play pivotal signaling roles in the regulation of cell growth and differentiation. Ras undergoes posttranslational lipid modifications, which are crucial for its membrane anchoring as well as for biological functions, at its C terminus (7, 8). In mammals, a serine/threonine kinase Raf-1 and its isoforms B-Raf and A-Raf are major effectors of Ras (9). In addition, recent searches have identified a number of mammalian Ras effectors and effector candidates, such as Ral guanine nucleotide dissociation stimulator (RalGDS), phosphoinositide 3-kinase, and protein kinase Cζ, all of which associate directly with the GTP-bound Ras (6, 26). However, the molecular mechanism of effector regulation by Ras is still unclear. It is established that the association with the posttranslationally modified Ras induces translocation of Raf-1 to the plasma membrane, where it is activated by membrane-bound factors (30, 44). The membrane recruitment role of Ras is also implicated in activation of RalGDS and phosphoinositide 3-kinase (33, 40). However, several recent studies have indicated that the mechanism of Raf activation by association with Ras seems to involve a number of complex processes in addition to the simple membrane recruitment (21, 36, 42, 46). In any of these cases, unavailability of in vitro systems, which could reconstitute the Ras-dependent effector regulation with the purified components only, hampered full elucidation of the Ras function other than the membrane recruitment.
On the other hand, in the budding yeast Saccharomyces cerevisiae, it has been established that adenylyl cyclase (CYR1) is a major downstream effector of RAS1 and RAS2, which are structural, functional, and biochemical homologues of mammalian Ras (3, 17). The Ras-CYR1 pathway has been implicated in transduction of a glucose-triggered signal to an intracellular environment where a protein phosphorylation cascade is initiated by cyclic AMP (cAMP). Since the Ras-dependent CYR1 activation could be reconstituted in vitro with the purified components only, the Ras-CYR1 system provided a unique opportunity to examine molecular mechanisms underlying Ras-dependent regulation of effector activities. CYR1 consists of 2,026-amino-acid residues that comprise at least four domains: the N-terminal, middle repetitive, catalytic, and C-terminal domains (23, 52). The middle repetitive domain is composed of a repetition of 23-amino-acid amphipathic leucine-rich motifs that have homology to the leucine-rich repeat (LRR) family of proteins (27). Genetic and biochemical studies demonstrated that the LRR domain contains a binding site for the GTP-bound Ras (35, 43, 49). Mammalian Ras can substitute for yeast RAS to activate CYR1 (4, 24). CYR1 forms a complex with the 60-kDa adenylyl cyclase-associated protein (CAP) (11, 12). CAP is a bifunctional protein: its N-terminal region binds to the C-terminal region of CYR1, and this association appears to be required for proper in vivo response to Ras, while its C-terminal region binds actin monomer and is somehow involved in the regulation of the actin cytoskeleton (13, 14, 16, 50). These two functions appear to be separable from each other (14). Although the mechanism of regulation of the Ras-CYR1 pathway by CAP was unknown, we have recently found a possible link between CAP and the CYR1 activation by the modified Ras.
By using an in vitro reconstituted system, we previously demonstrated that the posttranslational modification, in particular farnesylation, of Ras is required for efficient activation of CYR1 by Ras (28). However, we next observed that the posttranslational modification of Ras did not affect the association of Ras with the LRR domain of CYR1 and that the association with the CAP N-terminal region is essential for the efficient activation of CYR1 by the modified Ras (43). The effect of CAP was successfully reconstituted in vitro by the purified components only. These findings suggested that CAP might mediate the stimulatory effect of the modified Ras on CYR1 activation and reminded us of our past observation on mammalian Raf-1. We and others identified the cysteine-rich domain (CRD) of Raf-1 as another Ras-binding site than the primary binding site, the Ras-binding domain (5, 15, 21). The association of Ras with CRD is dependent on the posttranslational modification of Ras (21, 31) and is required for the efficient in vivo activation of Raf-1 by Ras (10, 21, 31, 38, 46). This led us to hypothesize that the associated CAP might constitute a Ras-binding site of CYR1, which mediates the CYR1 activation, other than the primary Ras-binding site LRR domain. Here we report that in fact CAP in complex with the CYR1 C-terminal region is capable of direct association with the posttranslationally modified Ras. By isolating mutants of CAP which are defective in association with Ras, we provide genetic evidence for the importance of this binding in the Ras-dependent CYR1 activation both in vivo and in vitro. Further, evidence is presented for the importance of the strength of this binding in the CYR1 activation.
MATERIALS AND METHODS
Strains and growth media.
The Saccharomyces cerevisiae strains used in this study are listed in Table 1. Replacement of the chromosomal CAP gene with its N-terminal deletion mutant CAPΔN-1 was carried out as described previously (43). The resulting yeast strain expressed only the C-terminal segment of CAP corresponding to residues 369 to 526 under control of the yeast alcohol dehydrogenase I (ADC1) promoter. Another N-terminal deletion allele, CAPΔN-2, was prepared similarly except that the HIS3 marker replaced the URA3 of CAPΔN-1. Yeast cells were grown in YPD (2% Bacto Peptone, 1% Bacto Yeast Extract, 2% glucose) or in yeast synthetic medium (0.67% yeast nitrogen base, 2% glucose) with appropriate auxotrophic supplements. Yeast cells bearing the cyr1-2 mutation were cultured at 30°C in the presence of 1 mM cAMP as described previously (34). Genetic manipulations of yeast cells were performed as described previously (41). Transformation into yeast cells was carried out with lithium acetate (22).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Extrachromosomal plasmid(s) |
|---|---|---|
| TK161-R2V (CAPΔN)a | MATa his3 leu2 trp1 ura3 ade8 ras2::LEU2 RAS2Val-19 cap::pCAPΔN-1 | |
| TK36-1a | MATα his3 leu2 trp1 ura3 ade8 cyr1-2 ras2::LEU2 | YEP24-ADC1-CYR1 |
| TS5 | MATα his3 leu2 trp1 ura3 ade8 cyr1-2 ras2::LEU2 | YEP24-ADC1-CYR1(606–1764) |
| FS1a | MATa his3 leu2 trp1 ura3 ade8 can1 cap::pCAPΔN-1 | |
| FS20 | MATa his3 leu2 trp1 ura3 ade8 can1 | pAD4-FLAG-CAP |
| FS56 | MATa his3 leu2 trp1 ura3 ade8 can1 cap::pCAPΔN-1 | pAD4-FLAG-CAP(L13P/E28V) |
| FS57 | MATa his3 leu2 trp1 ura3 ade8 can1 cap::pCAPΔN-1 | pAD4-FLAG-CAP(N12S/E28G) |
| FS55 | MATa his3 leu2 trp1 ura3 ade8 can1 cap::pCAPΔN-1 | pAD4-FLAG-CAP(R26G) |
| FS66 | MATa his3 leu2 trp1 ura3 ade8 can1 cap::pCAPΔN-1 | pAD4-FLAG-CAP, pAS2-1- GST-CYR1(1764–2026) |
| FS68 | MATa his3 leu2 trp1 ura3 ade8 can1 cap::pCAPΔN-1 | pAD4-FLAG-CAP(L13P/E28V), pAS2-1-GST-CYR1(1764–2026) |
| FS69 | MATa his3 leu2 trp1 ura3 ade8 can1 cap::pCAPΔN-1 | pAD4-FLAG-CAP(N12S/E28G), pAS2-1-GST-CYR1(1764–2026) |
| FS70 | MATa his3 leu2 trp1 ura3 ade8 can1 cap::pCAPΔN-1 | pAD4-FLAG-CAP(R26G), pAS2-1-GST-CYR1(1764–2026) |
| Y187–207 | MATα his3-200 leu2-3,112 trp1-901 ura3-52 ade2-101 canR gal4Δ met− gal80Δ ura3::GALUAS-GAL1tata-lacZ cap::pCAPΔN-2 | pGAD-CYR1(1898–2026) |
Random mutagenesis of CAP and construction of expression plasmids.
A DNA fragment corresponding to residues 1 to 77 of CAP, CAP(1–77), was subjected to an error-prone PCR (19) to introduce random mutations. A pool of the amplified DNA fragments were inserted into a yeast two-hybrid vector pGBT10 (2) for expression as fusions with GAL4 DNA-binding domain under control of the ADC1 promoter as described previously (37). pGAD-CYR1(1879–2026) was used for expression of residues 1879 to 2026 of CYR1, CYR1(1879–2026), as a GAL4 transactivation domain (GAD) fusion (37). A yeast expression vector pAD4-FLAG was constructed by insertion of an annealed pair of oligonucleotides, 5′-GATCATGGACTACAAGGACGACGATGACAG-3′ and 5′-GATCTCTTGTCATCGTCGTCCTTGTAGTCCAT-3′, encoding a FLAG epitope (DYKDDDDK), into a BglII cleavage site of pAD4 (12). The full-length wild-type CAP or its mutants were inserted into pAD4-FLAG for expression as FLAG epitope-fusions under control of the ADC1 promoter. An SphI fragment of pAD4-GST-CYR1(1764–2026) (50) bearing a glutathione S-transferase (GST) fusion CYR1(1764–2026) sandwiched between the ADC1 promoter and terminator was cloned into pAS2-1 (Clontech), which had been cleaved by SphI to remove the ADC1 promoter, the ADC1 terminator, and GAL4 GAD. pGEX-CYR1(1764–2026) was constructed by insertion of CYR1(1764–2026) into pGEX-2T (Amersham Pharmacia Biotech) and used for expression of GST-CYR1(1764–2026) in Escherichia coli. pAD4-GST-CYR1(606–1764) was used for expression of a GST fusion CYR1(606–1764), containing the whole LRR domain, in yeast cells (43).
Screening for CAP mutants which are functionally defective but retain the ability to interact with CYR1.
A yeast strain TK161-R2V(CAPΔN) bearing the RAS2Val-19 and the CAPΔN-1 genes was transformed with a pool of pGBT10-CAP(1–77) carrying mutations. The resulting Trp+ colonies were subjected to heat shock at 55°C for 5 min by a replica-plating method as described previously (47, 50). pGBT10-CAP(1–77) plasmids, which failed to restore the heat shock sensitivity, were recovered from the yeast clones which survived the heat shock treatment and again transformed into a yeast two-hybrid reporter strain Y187-207 carrying the chromosomal CAPΔN-2 gene and the episomal pGAD-CYR1(1898–2026). The resulting Leu+, Trp+, and His+ transformants were assayed for β-galactosidase activity by a filter assay as described previously (2). pGBT10-CAP(1–77) plasmids were isolated from the two-hybrid interaction-positive yeast clones and subjected to DNA sequence determination to reveal mutations in the CAP-coding sequence. Each of the identified mutations was transferred to the full-length CAP, which was inserted into pAD4-FLAG for expression as a FLAG fusion. CAP mutants defective in binding to the CYR1 C-terminal domain, CAP(L20R) and CAP(147–526), were described earlier (37).
Measurement of in vivo association between CAP and CYR1.
Yeast FS1 lacking the endogenous CAP was transformed with pAD5-FLAG-CAP carrying various mutations. The resulting transformants (Table 1) were grown to a density of 107 cells/ml in the yeast synthetic medium lacking leucine, harvested by centrifugation, and disrupted by shaking with glass beads in buffer C [50 mM 2-(N-morpholino)ethanesulfonic acid (pH 6.2), 0.1 mM MgCl2, 0.1 mM ethylenebis(oxyethylenenitrilo)tetraacetic acid, 2 mM dithiothreitol, 10% glycerol] as described previously (35, 50). The cytosol fraction was prepared by centrifugation at 100,000 × g for 60 min and subjected to immunoprecipitation with anti-FLAG antibody-conjugated resin M2 (Kodak). Subsequently FLAG-CAP proteins were eluted with TBS buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl) containing 100 μg of FLAG peptide per ml. Endogenous CYR1 copurified with FLAG-CAP was detected by Western immunoblotting with anti-CYR1CT antibody (37). Anti-FLAG antibody (Kodak) was used for detection of FLAG-CAP. Aliquots of these purified proteins were also used for adenylyl cyclase assays.
In vitro Ras binding assay.
YEP24-ADC1-GST-CYR1(1764–2026) was transformed into FS1 in combination with pAD5-FLAG-CAP wild type or its mutant. The resulting transformants (Table 1) were grown and lysed as described above, and the crude membrane fractions were prepared by centrifugation of the lysates at 27,000 × g for 80 min. GST-CYR1(1764–2026) was solubilized from the crude membrane fractions with buffer C containing 1% Lubrol PX, 0.5 M NaCl, and 1 mM phenylmethylsulfonyl fluoride and adsorbed onto glutathione-Sepharose resin (35). GST-CYR1(606–1764) was purified similarly from yeast TS5 harboring pAD4-GST-CYR1(606–1764). FLAG-CAP was purified from yeast FS20 by adsorption onto anti-FLAG antibody-conjugated resin. GST-CYR1(1764–2026) without any CAP bound was purified from Escherichia coli harboring pGEX-2T-CYR1(1764–2026) by adsorption onto glutathione-Sepharose. Aliquots (40 μl) of the resin carrying various proteins were incubated at 4°C for 2 h in a total volume of 100 μl of buffer A (20 mM Tris-HCl [pH 7.4], 40 mM NaCl, 5 mM MgCl2 1 mM EDTA, 1 mM dithiothreitol, 0.1% Lubrol PX) containing 150 nM Ha-Ras or its mutants in the posttranslationally modified or unmodified forms, which had been purified from Spodoptera frugiperda Sf9 cells infected with baculoviruses expressing the respective proteins (28, 39). The resin was washed, and the bound proteins were subsequently eluted with buffer A containing 20 mM glutathione. Ha-Ras in the eluate was detected by Western immunoblotting with anti-Ha-Ras monoclonal antibody F235 (Oncogene Science, Inc., New York, N.Y.).
Adenylyl cyclase assay.
Adenylyl cyclase activities of various CYR1 specimens were measured in the presence of 2.5 mM MgCl2 with the addition of various concentrations of purified Ha-Ras or its mutants, which had been loaded with 5′-O-(3-thiotriphosphate) (GTPγS) as described previously (43). For measurement of the Mn2+-dependent activity, 2.5 mM MnCl2 replaced MgCl2 and Ras.
Other methods.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western immunoblot analysis were performed as described earlier (29, 48). The enhanced chemiluminescence immunodetection system (Amersham Pharmacia Biotech) was used for signal development. GST-CYR1 and FLAG-CAP proteins were quantitated by densitometric estimation of the intensities of Coomassie brilliant blue-stained bands upon SDS-PAGE.
RESULTS
Posttranslational modification-dependent association of Ras with the CYR1-CAP complex.
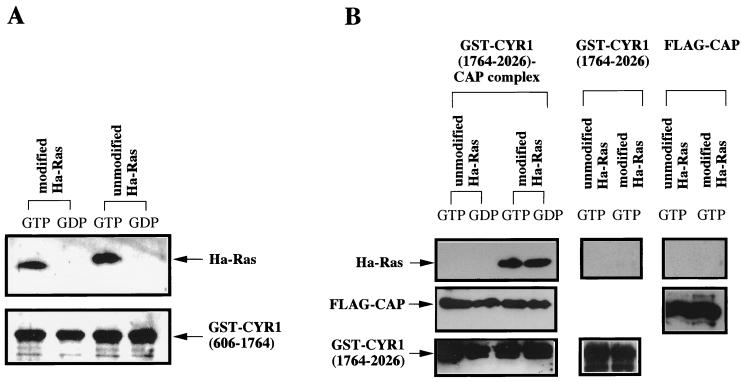
Based on the previous observation that association with CAP is essential for the efficient activation of CYR1 by the posttranslationally modified Ras (43), we postulated that the associated CAP might constitute a Ras-binding site of CYR1, which mediates the CYR1 activation, other than the primary Ras-binding site LRR domain. To prove this, we examined whether CAP alone or in complex with the CYR1 C-terminal region was capable of direct association with the modified Ras in vitro. GST-CYR1(1764–2026) complexed with FLAG epitope-fusion CAP was purified by adsorption onto glutathione-Sepharose resin from FS66 yeast cells (Table 1), expressing the two proteins, and examined for in vitro association with the posttranslationally modified and the unmodified forms of Ha-Ras, which had been loaded with GTPγS or guanosine 5′-O-(2-thiodiphosphate) (GDPβS), as described in Materials and Methods (Fig. 1B). The purified complex consisted only of GST-CYR1(1764–2026) and FLAG-CAP without any copurified protein, as examined by Coomassie brilliant blue staining (data not shown), as observed previously (43). GST-CYR1(606–1764), containing the whole LRR domain, was also examined for association with Ha-Ras (Fig. 1A). We observed a specific binding of the modified Ha-Ras to GST-CYR1(1764–2026) complexed with FLAG-CAP in a manner independent of the guanine nucleotide configuration of Ha-Ras. This finding was in striking contrast to the GTP-dependent association of GST-CYR1(606–1764) with Ha-Ras, which was unaffected by the posttranslational modification, as reported earlier (43). On the other hand, FLAG-CAP alone, purified from yeast FS20 by adsorption onto anti-FLAG antibody-conjugated affinity resin M2, as well as GST-CYR1(1764–2026) alone, purified from E. coli, did not exhibit any detectable association with the modified Ha-Ras when examined at similar amounts (Fig. 1B). These results suggested that the complex between CAP and the CYR1 C-terminal region might constitute a second Ras-binding site which mediates efficient activation of CYR1 by the modified form of Ras protein. However, it was impossible to obtain enough amounts of the CYR1-CAP complex to carry out more quantitative measurements for Ras association because CAP and GST-CYR1(1764–2026) coexpressed in E. coli were found to be unable to associate with each other for as-yet-unknown reasons. This led us to examine the significance of this association by means of the following genetic method.
FIG. 1.
Posttranslational modification of Ras is required for association with the CYR1-CAP complex but not for association with the LRR domain. (A) The modified and unmodified forms of Ha-Ras were loaded with GTPγS (GTP) or GDPβS (GDP) and incubated at 150 nM with approximately 0.1 μg of GST-CYR1(606–1764), which had been purified from yeast TS5 cells (Table 1) and immobilized on glutathione-Sepharose resin as described in Materials and Methods. The bound proteins were eluted with buffer A containing 20 mM glutathione, and GST-CYR1(606–1764) and Ha-Ras in the eluate were detected by Western immunoblotting with anti-GST polyclonal antibody (lower panel) and anti-Ha-Ras monoclonal antibody F235 (upper panel), respectively. (B) A complex of GST-CYR1(1764–2026) and FLAG-CAP, purified from yeast FS66 cells, and GST-CYR1(1764–2026), purified from E. coli, were immobilized on glutathione-Sepharose resin. FLAG-CAP, purified from yeast FS20 cells, was immobilized on anti-FLAG M2 resin. Aliquots (40 μl) of the resin carrying the various proteins were subjected to the in vitro binding assays with Ha-Ras as described in panel A. The top panels show the bound Ha-Ras. The middle panels show FLAG-CAP (approximately 0.1 μg) copurified with GST-CYR1(1764–2026), which was detected with anti-FLAG antibody (Kodak). The bottom panels show GST-CYR1(1764–2026) (approximately 0.4 μg). The experiments were repeated five times, yielding similar results.
Screening for CAP mutants which are defective in Ras signaling but retain the ability to associate with the CYR1 C-terminal region.
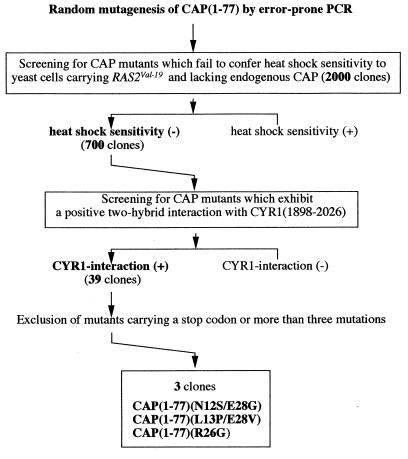
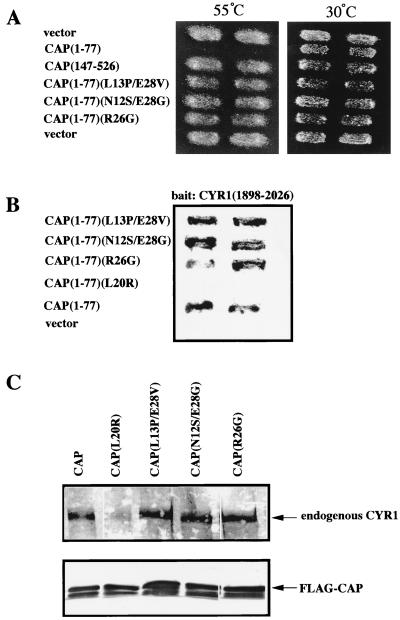
The N-terminal region of CAP is required not only for its function in Ras signaling (14) but also for the association with the CYR1 C-terminal region (50). The N-terminal function of CAP can be tested by its ability to confer heat shock sensitivity to yeast cells carrying the activated RAS2 gene, RAS2Val-19. We have recently shown that the N-terminal 36-residue region of CAP is sufficient for both of these functions (37). To establish a functional link between the observed Ras binding to the CYR1-CAP complex and the in vivo function of CAP, we carried out the following experiment. We reasoned that mutants of CAP defective in Ras binding might be obtained as those which lost the in vivo CAP function but retained the ability to associate with the CYR1 C-terminal region. To this end, we introduced random mutations into CAP(1–77) by an error-prone PCR (19). A pool of the mutated CAP(1–77) were cloned into pGBT vector, and the resulting 2,000 clones were examined for the activity to confer heat shock sensitivity to yeast TK161-R2V(CAPΔN) carrying the RAS2Val-19 gene and an N-terminal deletion of the chromosomal CAP gene as described in Materials and Methods (Fig. 2). Approximately 700 clones failed to confer heat shock sensitivity. pGBT-CAP(1–77) plasmids were isolated individually from the heat shock-resistant yeast clones and subsequently transformed into Y187-207 for examination of a two-hybrid interaction with CYR1(1898–2026) (Fig. 2). Finally, 39 mutants were found to retain the binding activity to CYR1. DNA sequence determination of these 39 mutants revealed that 9 carried a mutation involving Arg-26, 2 of which carried a single mutation of R26G, and that 5 carried a mutation involving Asn-12 and 7 carried a mutation involving Glu-28. After exclusion of the mutants carrying a stop codon or more than three mutated residues within residues 1 to 36, we finally obtained three kinds of CAP mutants: CAP(L13P/E28V), CAP(N12S/E28G), and CAP(R26G), in which combinations of the mutations L13P and E28V and of the mutations N12S and E28G appeared in more than one occasion. CAP(1–77) carrying these mutations failed to confer heat shock sensitivity to TK161-R2V(CAPΔN) cells (Fig. 3A) and exhibited a positive two-hybrid interaction with CYR1(1898–2026) (Fig. 3B). The association with CYR1 was further examined biochemically by using the full-length proteins. The L13P/E28V, N12S/E28G, and R26G mutations were transferred to the full-length CAP, which were expressed with a FLAG epitope-tag in yeast FS1 cells (Table 1) and examined for in vivo association with the endogenous CYR1 (Fig. 3C). The amounts of CYR1 copurified with FLAG-CAP carrying the mutations were almost equal to that copurified with FLAG-CAP wild type. On the other hand, FLAG-CAP carrying L20R mutation failed to bind CYR1 (Fig. 3B and C), as reported previously (37). In this experiment, the purified FLAG-CAP yielded two bands on the Western blot; the upper one corresponding to FLAG-CAP and the lower one presumably corresponding to immunoglobulin heavy chains eluted from the anti-FLAG resin.
FIG. 2.
Screening for CAP mutants defective in Ras signaling but retaining the ability to associate with CYR1.
FIG. 3.
In vivo function of CAP mutants obtained by the screening. (A) Heat shock sensitivity of yeast cells expressing the various CAP mutants. TK161-R2V(CAPΔN) yeast cells harboring pGBT10-CAP(1–77) carrying the indicated mutations were examined for heat shock sensitivity by a replica-plating method as described in Materials and Methods. Shown are photographs of the two replica plates, one subjected to a 55°C heat shock for 5 min (left panel) and the other without the heat shock treatment (right panel), after 2 days of growth at 30°C. (B) Yeast two-hybrid analysis for interaction of CAP(1–77) carrying the indicated mutations with CYR1(1898–2026) was performed as described in Materials and Methods. (C) FLAG-CAP carrying the indicated mutation was purified from yeast strains FS25, FS55, FS56, or FS57 (Table 1) as described in Materials and Methods. The endogenous CYR1, which was copurified with the FLAG-CAP mutants, was subjected to Western immunodetection. The upper panel shows the endogenous CYR1 detected with anti-CYR1CT antibody. The lower panel shows purified FLAG-CAP proteins (approximately 0.5 μg) detected with anti-FLAG antibody. All of the experiments were repeated three times and yielded similar results.
Characterization of the CAP mutants obtained by the screening.
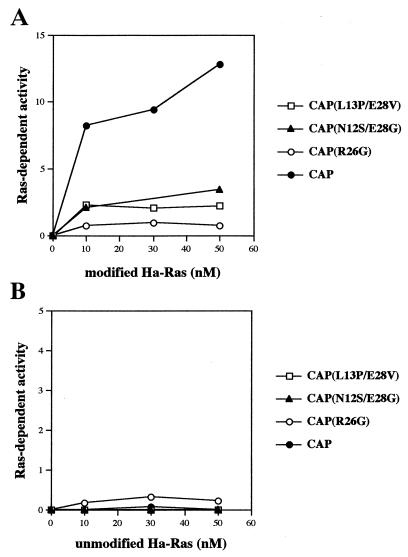
The three CAP mutants were examined for their activity to mediate the CYR1 activation by the modified Ras protein. The endogenous CYR1 copurified with FLAG-CAP carrying the L13P/E28V, N12S/E28G, or R26G mutation (Fig. 3C) was assayed for adenylyl cyclase activity in the presence of various concentrations of the GTPγS-bound form of the modified Ha-Ras (Fig. 4A) or of the unmodified Ha-Ras (Fig. 4B). One-tenth aliquots of the preparations shown in Fig. 3C, which contained roughly similar amounts of CYR1, were used for the assays. For more precise comparison of the activity levels among different CYR1 specimens, the Ras-dependent activity was presented as a ratio to the Mn2+-dependent activity of the same specimen. The Mn2+-dependent activity is unaffected by Ras and is proportional to the amount of the CYR1 protein. CYR1 complexed with FLAG-CAP wild type was activated by the modified Ha-Ras much more efficiently than by the unmodified form, and a 12-fold-higher activity was achieved by the modified Ha-Ras over the Mn2+-dependent activity. However, CYR1 complexed with any of the three CAP mutants exhibited a much-reduced activity dependent on the modified Ras (Fig. 4A). CYR1 complexed with CAP(R26G) was least active, and its modified-Ha-Ras-dependent activity managed to reach the level of the Mn2+-dependent activity (Fig. 4A). Thus, we successfully obtained CAP mutants which were defective in Ras signaling both in vivo and in vitro but retained the binding activity to CYR1.
FIG. 4.
In vitro activation of CYR1 complexed with the CAP mutants by the modified Ras. The endogenous CYR1 complexed with the FLAG-CAP mutants was purified as described in the legend to Fig. 3C and examined in vitro for stimulation of adenylyl cyclase activities by the GTPγS-bound forms of the modified (A) or the unmodified (B) Ha-Ras as described in Materials and Methods. The y axis shows the Ras-dependent adenylyl cyclase activity, which is presented as a ratio to the Mn2+-dependent activity of the same specimen. The Mn2+-dependent activities of the purified proteins ranged from 15 to 20 pmol of cAMP formed during 30 min of incubation at 30°C. Similar experiments performed on three occasions with different preparations of CYR1-CAP complex yielded equivalent results.
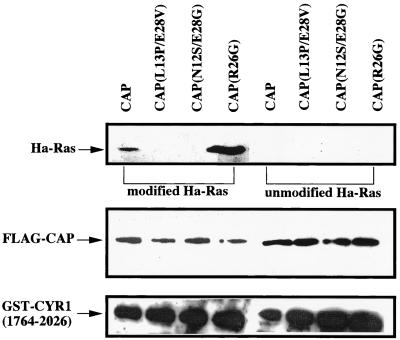
We next examined the effect of the CAP mutations on the activity to associate with the modified Ras. GST-CYR1(1764–2026) complexed with FLAG-CAP carrying the L13P/E28V, N12S/E28G, or R26G mutation was immobilized on glutathione-Sepharose resin and examined for in vitro association with the modified and the unmodified forms of Ha-Ras as described in Fig. 1 (Fig. 5). Both CAP(L13P/E28V) and CAP(N12S/E28G), complexed with CYR1(1764–2026), failed to exhibit any detectable binding to the modified Ha-Ras. Thus, two of the three CAP mutants, which were isolated by screening for those defective in Ras signaling but retained the CYR1-binding activity, turned out to exhibit a severely impaired ability to bind the modified Ras, providing genetic evidence for the importance of the physical association at the second Ras-binding site in Ras-dependent regulation of CYR1. On the other hand, CYR1 complexed with the other mutant CAP(R26G) exhibited a greatly enhanced ability to bind the modified Ras in sharp contrast to the other two CAP mutants (Fig. 5).
FIG. 5.
In vitro association of the CYR1(1764–2026)-mutant CAP complex with Ha-Ras. GST-CYR1(1764–2026) complexed with FLAG-CAP carrying the indicated mutations was purified from yeast strains FS66, FS68, FS69, and FS70 and examined for in vitro association with the modified and the unmodified forms of Ha-Ras as described in the legend to Fig. 1B. The top panel shows the bound Ha-Ras, and the middle panel shows FLAG-CAP (approximately 0.05 μg) copurified with GST-CYR1(1764–2026). The bottom panel shows GST-CYR1(1764–2026) (approximately 0.2 to 0.4 μg) eluted from the resin. The experiments were repeated three times, yielding equivalent results.
Too-strong interaction at the second Ras-binding site is detrimental to the CYR1 activation.
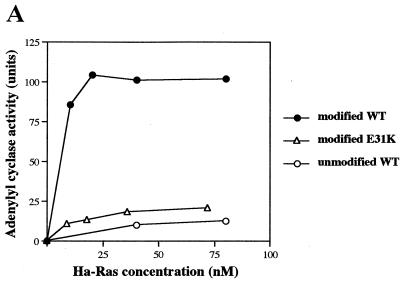
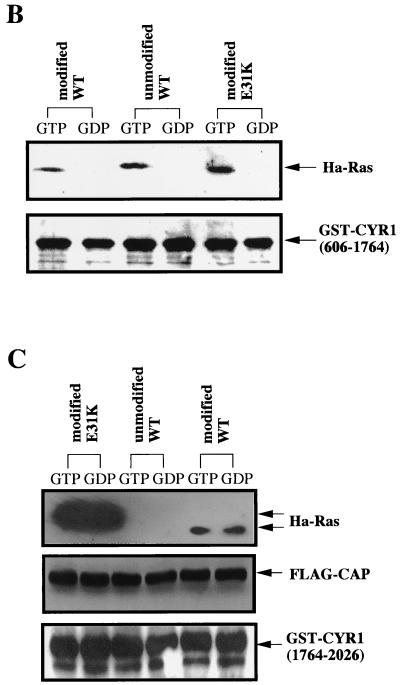
The observation that CYR1 complexed with CAP(R26G) was least responsive to Ras but exhibited an enhanced binding activity toward the modified Ras reminded us of our previous observation on the interaction of Raf-1 CRD with Rap1A (20). Rap1A, which lost the ability to activate Raf-1, possessed a greatly enhanced activity to associate with CRD compared to Ras. Ha-Ras protein carrying the Rap1A-type mutation E31K was also found to exhibit the same property (20). During examination of 50 Ha-Ras mutants for their abilities to activate CYR1, we had observed that Ha-Ras(E31K) was incapable of activating CYR1 efficiently (1). The level of CYR1 activity attained with the modified Ha-Ras(E31K) was so low as to be comparable to that with the unmodified wild-type Ha-Ras (Fig. 6A). However, Ha-Ras(E31K) possessed a binding activity comparable to that of the LRR domain of CYR1 with wild-type Ha-Ras (Fig. 6B). Thus, we next examined the effect of E31K mutation on the association with the CAP-CYR1(1764–2026) complex and found that Ha-Ras(E31K) exhibited a greatly enhanced binding activity compared to wild-type Ha-Ras (Fig. 6C). This result, taken together with that obtained with CAP(R26G), strongly suggests that too-strong interaction at the second Ras-binding site is detrimental to the CYR1 activation.
FIG. 6.
H-Ras(E31K) is incapable of activating CYR1 efficiently and exhibits a greatly enhanced binding to the CYR1-CAP complex. (A) The full-length CYR1 expressed in yeast TK36-1 cells (Table 1) was solubilized from the membrane fraction and was examined for activation by the modified and the unmodified forms of wild-type Ha-Ras (WT) and by the modified form of Ha-Ras(E31K) (E31K), all in the GTPγS-bound configurations, as described previously (43). One unit of activity is defined as 1 pmol of cAMP formed in 1 min of incubation with 1 mg of protein at 30°C. (B) The association of the indicated forms of Ha-Ras with GST-CYR1(606–1764) was examined in vitro as described in the legend to Fig. 1A. (C) The association of the indicated forms of Ha-Ras with GST-CYR1(1764–2026) complexed with FLAG-CAP were examined in vitro as described in the legend to Fig. 1B. Similar experiments performed on two occasions with different preparations of CYR1 and Ha-Ras yielded equivalent results.
DISCUSSION
The essential role of the posttranslational modification in the biological functions of Ras protein has been interpreted in terms of its membrane-anchoring function, which enables Ras to recruit the effector molecules to the plasma membrane. However, it was unclear whether the posttranslational modification has another function that is directly involved in the effector regulation. By using an in vitro reconstituted system, which enabled an analysis of such a Ras function separately from the membrane recruitment function, we were the first to show that the posttranslational modification, in particular farnesylation, of Ras is essential for efficient activation of CYR1 by Ras protein (28). Subsequently, the importance of the posttranslational modification, in particular farnesylation, of Ras was also shown for activation of B-Raf and Raf-1 by using crude in vitro reconstituted systems (39, 45, 51). Furthermore, discovery of the farnesylation-dependent association of Ras with Raf-1 CRD, which is crucial for the efficient Raf-1 activation by Ras, provided a clue to elucidating the molecular basis for the role of farnesylation on this process, although the precise role of this association in Raf-1 activation remains to be clarified (21, 31, 46).
Our previous observations (43) that association with the CAP N-terminal region was essential for the efficient activation of CYR1 by the posttranslationally modified Ras and that the posttranslational modification did not affect the ability of Ras to associate with the LRR domain of CYR1 led us to examine the possibility that CAP might constitute a second Ras-binding site of CYR1, mediating the CYR1 activation, which is analogous to Raf-1 CRD. In fact, here we have observed that CAP in complex with the CYR1 C-terminal region exhibits a direct association with Ras, which is dependent upon the posttranslational modification. By isolating mutants of CAP which are defective in association with the modified Ras, we have provided a genetic evidence for the importance of this binding in the cellular function of CAP, as tested by acquirement of heat shock sensitivity in the RAS2Val-19 background as well as in the in vitro activation of CYR1 by the modified Ras. Further, the isolation of a mutant, CAP(R26G), which is defective in mediating the CYR1 activation but exhibits a greatly enhanced binding at the second Ras-binding site, has hinted at the importance of the strength of this binding in the regulation of CYR1 activity. This is further supported by the observation that Ha-Ras protein carrying the Rap1A-type mutation E31K, which has lost the ability to activate CYR1 efficiently, possesses a greatly enhanced binding activity toward the CYR1-CAP complex. These observations suggest that too-weak and too-strong interactions at the second Ras-binding site are both detrimental to the CYR1 activation and are reminiscent of our observation on Raf that the strength of the interaction of Ras with the Raf CRD is a critical determinant of response of Raf to Ras: too-weak and too-strong interactions are both detrimental to the Raf activation (38). We presently do not have any mechanistic explanation for this phenomenon in molecular terms. The observation on Ha-Ras(E31K) is also similar to what we have observed for the same mutant upon examination of its abilities to activate Raf-1 and to bind to CRD, the second Ras-binding site of Raf-1 (20). Rap1A itself was known to possess the same properties as Ras(E31K) toward Raf-1, and we have shown that Rap1A also exerts the same activities toward CYR1 as did Ras(E31K) (data not shown). Thus, we have observed strikingly similar properties of the interaction with Ras in the two best-characterized effectors, Raf-1 and CYR1, which do not share any structural homology with each other. These findings lead us to propose that the presence of a second Ras-binding site, which mediates regulation of the effector activity by Ras, may represent a general property of the Ras effector proteins.
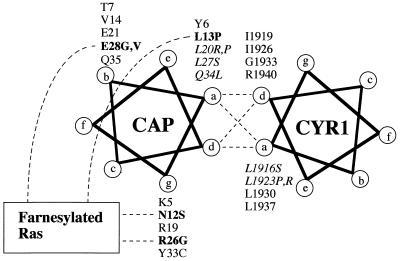
Isolation of the CAP mutants, which exhibit altered modes of association with Ras, has enabled us to propose a model for this interaction (Fig. 7). We showed previously that the N-terminal 36-residue region of CAP was sufficient for the association with the CYR1 C-terminal region, as well as for its function in the Ras-CYR1 pathway (37). Careful examination of the primary structures of the mutually interacting regions of CAP and CYR1 and the isolation of CAP mutants defective in the CYR1 binding indicated that CAP makes a coiled-coil interaction with CYR1 (37). Spinning-wheel representations of the mutually interacting regions indicated that residues at the a and d positions form a hydrophobic interface between the two α-helices (Fig. 7), of which Leu-20, Leu-27, and Val-30 of CAP, as well as Leu-1916 and Leu-1923 of CYR1, were identified as residues essential for the CYR1-CAP association (37). Of four residues which have been identified in the present study to be crucial for the association with the modified Ras, Asn-12 and Arg-26 occupy g positions and Glu-28 occupies a b position. These residues are located in a hydrophilic solvent-exposed surface of the coiled coils, which is commonly used as an interface for association with interacting proteins (32). On the other hand, Leu-13 is located at an a position. We have observed that mutation of Leu-13 has no effect on the association with CYR1 (data not shown). In addition, we have observed that the presence of both mutations is necessary for causing the functional defect of the CAP mutant carrying the double mutations N12S/E28G or L13P/E28V (data not shown). We speculate that Asn-12, Leu-13, Arg-26, and Glu-28 of CAP may contribute to formation of a binding site for the modified Ras as a complex with the CYR1 C-terminal region. It is presently unclear what structural characteristic of the modified Ras is recognized by the CYR1-CAP complex. It is likely that the C-terminal structure of Ras with the posttranslationally attached farnesyl group is a major determinant because the unmodified Ras has no binding activity. If this is the case, the absence of sequence conservation among the C-terminal regions of various Ras proteins limits the possible recognition site to the farnesylated and carboxymethylated Cys residue at the C terminus. This is supported by a recent nuclear magnetic resonance analysis of the tertiary structure of the guanine nucleotide dissociation inhibitor of Rho small GTPase, Rho-GDI, which revealed a hydrophobic isoprene binding pocket for interaction with the farnesylated Rho protein (18). Alternatively, the farnesylated C-terminal segment of Ras may fold back and come into close proximity to the other region of Ras, thereby altering its conformation to create a new epitope for the binding. This possibility also has a support from our present observation that the E31K mutation of Ha-Ras exerted an enhancing effect on the binding. Further studies are needed to elucidate the molecular mechanism by which the CYR1-CAP complex recognizes the modified Ras protein.
FIG. 7.
Model of interaction among CAP, CYR1, and Ras. Spinning-wheel representations of the mutually interacting segments of CAP and CYR1. The amino acids corresponding to the a, b, and g positions of CAP(1–36) and to the a and d positions of CYR1(1916–1940) are shown. The residues of CAP and CYR1 whose mutations abolished the CAP-CYR1 interaction are shown in italic type (data were taken from reference 37). The residues of CAP whose mutations resulted in gross alteration of the association with the modified Ha-Ras are shown in boldface type. The broken lines indicate possible interactions predicted from the mutational studies. See Discussion for a detailed explanation.
ACKNOWLEDGMENTS
We thank A. Seki and A. Kawabe for help in preparation of the manuscript.
This investigation was supported by grants-in-aid for scientific research on priority areas, for scientific research, and for JSPS fellows from the Ministry of Education, Science, Sports, and Culture of Japan and by grants from the Yamanouchi Foundation for Research on Metabolic Diseases and from Sankyo Foundation of Life Science. F. Shima is supported by a fellowship from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Akasaka K, Tamada M, Wang F, Kariya K, Shima F, Kikuchi A, Yamamoto M, Shirouzu M, Yokoyama S, Kataoka T. Differential structural requirements for interaction of Ras protein with its distinct downstream effectors. J Biol Chem. 1996;271:5353–5360. doi: 10.1074/jbc.271.10.5353. [DOI] [PubMed] [Google Scholar]
- 2.Bartel P L, Chien C-T, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley D A, editor. Cellular interactions in development: a practical approach. Oxford, United Kingdom: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- 3.Broach J R, Deschenes R J. The function of Ras in Saccharomyces cerevisiae. Adv Cancer Res. 1990;54:79–139. doi: 10.1016/s0065-230x(08)60809-x. [DOI] [PubMed] [Google Scholar]
- 4.Broek D, Samiy N, Fasano O, Fujiyama A, Tamanoi F, Northup J, Wigler M. Differential activation of yeast adenylate cyclase by wild-type and mutant RAS proteins. Cell. 1985;41:763–769. doi: 10.1016/s0092-8674(85)80057-x. [DOI] [PubMed] [Google Scholar]
- 5.Brtva T R, Drugan J K, Ghosh S, Terrell R S, Campbell-Burk S, Bell R M, Der C D. Two distinct Raf domains mediate interaction with Ras. J Biol Chem. 1995;270:9809–9812. doi: 10.1074/jbc.270.17.9809. [DOI] [PubMed] [Google Scholar]
- 6.Campbell S L, Khosravi-Far R, Rossman K L, Clark G J, Der C J. Increasing complexity of Ras signaling. Oncogene. 1998;17:1395–1413. doi: 10.1038/sj.onc.1202174. [DOI] [PubMed] [Google Scholar]
- 7.Casey P J. Lipid modification of G proteins. Curr Opin Cell Biol. 1994;6:219–225. doi: 10.1016/0955-0674(94)90139-2. [DOI] [PubMed] [Google Scholar]
- 8.Clark S. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu Rev Biochem. 1992;61:335–386. doi: 10.1146/annurev.bi.61.070192.002035. [DOI] [PubMed] [Google Scholar]
- 9.Daum G, Eisenmann-Tappe I, Fries H-W, Troppmair J, Rapp U R. The ins and outs of Raf kinases. Trends Biochem Sci. 1994;19:474–480. doi: 10.1016/0968-0004(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 10.Drugan J K, Khosravi-Far R, White M A, Der C J, Sung Y-J, Hwang Y-W, Campbell S L. Ras interaction with two distinct binding domains in Raf-1 may be required for Ras transformation. J Biol Chem. 1996;271:233–237. doi: 10.1074/jbc.271.1.233. [DOI] [PubMed] [Google Scholar]
- 11.Fedor-Chaiken M, Deschenes R J, Broach J R. SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell. 1990;61:329–340. doi: 10.1016/0092-8674(90)90813-t. [DOI] [PubMed] [Google Scholar]
- 12.Field J, Vojtek A, Ballester R, Bolger G, Colicelli J, Ferguson K, Gerst J, Kataoka T, Michaeli T, Powers S, Riggs M, Rodgers L, Wieland I, Wheland B, Wigler M. Cloning and characterization of CAP, the S. cerevisiae gene encoding the 70 kd adenylyl cyclase-associated protein. Cell. 1990;61:319–327. doi: 10.1016/0092-8674(90)90812-s. [DOI] [PubMed] [Google Scholar]
- 13.Freeman L N, Chen Z, Horenstein J, Weber A, Field J. An actin monomer binding activity localizes to the carboxyl-terminal half of the Saccharomyces cerevisiae cyclase-associated protein. J Biol Chem. 1995;270:5680–5685. doi: 10.1074/jbc.270.10.5680. [DOI] [PubMed] [Google Scholar]
- 14.Gerst J E, Ferguson K, Vojtek A, Wigler M, Field J. CAP is a bifunctional component of the Saccharomyces cerevisiae adenylyl cyclase complex. Mol Cell Biol. 1991;11:1248–1257. doi: 10.1128/mcb.11.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh S, Xie W Q, Quest A F G, Mabrouk G M, Strum J C, Bell R M. The cysteine-rich region of Raf-1 kinase contains zinc, translocates to liposomes, and is adjacent to a segment that binds GTP-Ras. J Biol Chem. 1994;269:10000–10007. [PubMed] [Google Scholar]
- 16.Gieselmann R, Mann K. ASP-56, a new actin sequestering protein from pig platelets with homology to CAP, an adenylate cyclase-associated protein from yeast. FEBS Lett. 1992;298:149–153. doi: 10.1016/0014-5793(92)80043-g. [DOI] [PubMed] [Google Scholar]
- 17.Gibbs J B, Marshall M. The ras oncogene—an important regulatory element in lower eukaryotic organisms. Microbiol Rev. 1989;53:171–185. doi: 10.1128/mr.53.2.171-185.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gosser Y Q, Nomanbhoy T K, Aghazadeh B, Manor D, Combs C, Cerione R A, Rosen M K. C-terminal binding domain of Rho GDP-dissociation inhibitor directs N-terminal inhibitory peptide to GTPases. Nature. 1997;387:814–819. doi: 10.1038/42961. [DOI] [PubMed] [Google Scholar]
- 19.Gram H, Marconi L-A, Barbas III C F, Collet T A, Lerner R A, Kang A S. In vitro selection and affinity maturation of antibodies from a native combinatorial immunoglobulin library. Proc Natl Acad Sci USA. 1992;89:3576–3580. doi: 10.1073/pnas.89.8.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu C-D, Kariya K, Kotani G, Shirouzu M, Yokoyama S, Kataoka T. Coassociation of Rap1A and Ha-Ras with Raf-1 N-terminal region interferes with Ras-dependent activation of Raf-1. J Biol Chem. 1997;272:11702–11705. doi: 10.1074/jbc.272.18.11702. [DOI] [PubMed] [Google Scholar]
- 21.Hu C-D, Kariya K, Tamada M, Akasaka K, Shirouzu M, Yokoyama S, Kataoka T. Cysteine-rich region of Raf-1 interacts with activator domain of post-translationally modified Ha-Ras. J Biol Chem. 1995;270:30274–30277. doi: 10.1074/jbc.270.51.30274. [DOI] [PubMed] [Google Scholar]
- 22.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kataoka T, Broek D, Wigler M. DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell. 1985;43:493–505. doi: 10.1016/0092-8674(85)90179-5. [DOI] [PubMed] [Google Scholar]
- 24.Kataoka T, Powers S, Cameron S, Fasano O, Goldfarb M, Broach J, Wigler M. Functional homology of mammalian and yeast RAS genes. Cell. 1985;40:19–26. doi: 10.1016/0092-8674(85)90304-6. [DOI] [PubMed] [Google Scholar]
- 25.Kataoka T, Powers S, McGill C, Fasano O, Strathern J, Broach J, Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984;37:437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- 26.Katz M E, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 27.Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 28.Kuroda Y, Suzuki N, Kataoka T. The effect of posttranslational modifications on the interaction of Ras2 with adenylyl cyclase. Science. 1993;259:683–686. doi: 10.1126/science.8430318. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:681–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Leevers S J, Paterson H F, Marshall C J. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature. 1994;369:411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- 31.Luo Z, Diaz B, Marshall M S, Avruch J. An intact Raf zinc finger is required for optimal binding to processed Ras and for Ras-dependent Raf activation in situ. Mol Cell Biol. 1997;17:46–53. doi: 10.1128/mcb.17.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 33.Matsubara K, Kishida S, Matsuura Y, Kitayama H, Noda M, Kikuchi A. Plasma membrane recruitment of RalGDS is critical for Ras-dependent Ral activation. Oncogene. 1999;18:1303–1312. doi: 10.1038/sj.onc.1202425. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto K, Uno I, Oshima Y, Ishikawa T. Isolation and characterization of yeast mutants deficient in adenylyl cyclase and cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1982;79:2355–2359. doi: 10.1073/pnas.79.7.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minato T, Wang J, Akasaka K, Okada T, Suzuki N, Kataoka T. Quantitative analysis of mutually competitive binding of human Raf-1 and yeast adenylyl cyclase to Ras proteins. J Biol Chem. 1994;269:20845–20851. [PubMed] [Google Scholar]
- 36.Mineo C, Anderson R G W, White M A. Physical association with Ras enhances activation of membrane-bound Raf (RafCAAX) J Biol Chem. 1997;272:10345–10348. doi: 10.1074/jbc.272.16.10345. [DOI] [PubMed] [Google Scholar]
- 37.Nishida Y, Shima F, Sen H, Tanaka Y, Yanagihara C, Yamawaki-Kataoka Y, Kariya K, Kataoka T. Coiled-coil interaction of N-terminal 36 residues of cyclase-associated protein with adenylyl cyclase is sufficient for its function in Saccharomyces cerevisiae Ras pathway. J Biol Chem. 1998;273:28019–28024. doi: 10.1074/jbc.273.43.28019. [DOI] [PubMed] [Google Scholar]
- 38.Okada T, Hu C-D, Jin T-G, Kariya K, Yamawaki-Kataoka Y, Kataoka T. The strength of interaction at the Raf cystein-rich domain is a critical determinant of response of Raf to Ras family small GTPases. Mol Cell Biol. 1999;19:6057–6064. doi: 10.1128/mcb.19.9.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okada T, Masuda T, Shinkai M, Kariya K, Kataoka T. Post-translational modification of H-Ras is required for activation of, but not for association with, B-Raf. J Biol Chem. 1996;271:4671–4678. doi: 10.1074/jbc.271.9.4671. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez-Viciana P, Warne P H, Vanhaesebroeck B, Waterfield M D, Downward J. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J. 1996;15:2442–2451. [PMC free article] [PubMed] [Google Scholar]
- 41.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 42.Roy S, Lane A, Yan J, McPherson R, Hancock J F. Activity of plasma membrane-recruited Raf-1 is regulated by Ras via the Raf zinc finger. J Biol Chem. 1997;272:20139–20145. doi: 10.1074/jbc.272.32.20139. [DOI] [PubMed] [Google Scholar]
- 43.Shima F, Yamawaki-Kataoka Y, Yanagihara C, Tamada M, Okada T, Kariya K, Kataoka T. Effect of association with adenylyl cyclase-associated protein on the interaction of yeast adenylyl cyclase with Ras protein. Mol Cell Biol. 1997;17:1057–1064. doi: 10.1128/mcb.17.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stokoe D, Macdonald S G, Cadwallander K, Symons M, Hancock J F. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 45.Stokoe D, McCormick F. Activation of c-Raf-1 by Ras and Src through different mechanisms: activation in vivo and in vitro. EMBO J. 1997;16:2384–2396. doi: 10.1093/emboj/16.9.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamada M, Hu C-D, Kariya K, Okada T, Kataoka T. Membrane recruitment of Raf-1 is not the only function of Ras in Raf-1 activation. Oncogene. 1997;15:2959–2964. doi: 10.1038/sj.onc.1201582. [DOI] [PubMed] [Google Scholar]
- 47.Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- 48.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 50.Wang J, Suzuki N, Nishida Y, Kataoka T. Analysis of the function of the 70-kilodalton cyclase-associated protein (CAP) by using mutants of adenylyl cyclase defective in CAP binding. Mol Cell Biol. 1993;13:4087–4097. doi: 10.1128/mcb.13.7.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamori B, Kuroda S, Shimizu K, Fukui K, Ohtsuka T, Takai Y. Purification of a Ras-dependent mitogen-activated protein kinase kinase kinase from bovine brain cytosol and its identification as a complex of B-Raf and 14-3-3 proteins. J Biol Chem. 1995;270:11723–11726. doi: 10.1074/jbc.270.20.11723. [DOI] [PubMed] [Google Scholar]
- 52.Yamawaki-Kataoka Y, Tamaoki T, Choe H-R, Tanaka H, Kataoka T. Adenylyl cyclases in yeast: a comparison of the gene from Schizosaccharomyces pombe and Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1989;86:5693–5697. doi: 10.1073/pnas.86.15.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]