Abstract
We report a case of Leptotrichia species bacteremia in a patient undergoing treatment for acute myelogenous leukemia. Like previously reported Leptotrichia species, this is a gram-variable, pleomorphic rod that is catalase negative and utilizes glucose and sucrose. However, it is more fastidious than previously reported isolates of Leptotrichia and may represent a novel species.
A 50-year-old white male with acute myelogenous leukemia received chemotherapy with idarubicin and cytarabine. He developed neutropenic fever without an obvious source and was treated with ofloxacin, gentamicin, and metronidazole. The following week, he developed transient candidemia secondary to an infected Hickman catheter, was treated with amphotericin B, and became afebrile. After the catheter was removed and a new central line was inserted, the patient received reinduction chemotherapy and the metronidazole therapy was discontinued. Six days later, the second catheter was also removed and a peripherally inserted central catheter (PICC) was placed in the patient’s right upper arm. On the same day, the patient developed a new fever, despite treatment with ofloxacin, gentamicin, and amphotericin B. The leukocyte count was 200 cells per μl and the absolute neutrophil count (ANC) was zero. He had been neutropenic (ANC < 500 cells/μl) for approximately 3 weeks. A physical exam revealed that the patient was chronically ill, with a temperature of 101.2°F and stable vital signs. There were no skin or oral lesions, a cardiac exam showed no murmur, and the PICC insertion site had no erythema or tenderness.
On days 7 and 9 after chemotherapy reinduction, two sets of blood samples were drawn from the PICC and cultured. Both sets of cultures grew a coagulase-negative Staphylococcus sp., and vancomycin was added to the antibiotic regimen. However, three sets of follow-up blood samples drawn from the PICC on days 10 and 11 after reinduction and cultured grew an apparent gram-positive rod from the anaerobic bottle only. Vancomycin therapy was discontinued on day 13 but then restarted on day 16, when cultures of new blood samples, drawn from the PICC on day 15, grew a coagulase-negative Staphylococcus sp. On day 14, several small ulcers were noted on the patient’s lips. These ulcers were attributed to herpes simplex virus, and acyclovir therapy was initiated. High-grade fevers up to 103.1°F persisted, and cultures of two blood sample sets, drawn from the peripheral vein on days 18 and 19, again grew an apparent gram-positive rod in the anaerobic bottles only. The PICC was removed on day 19, and a culture of the catheter tip was negative. Two subsequent blood cultures, from samples collected by peripheral venipuncture on days 21 and 22, were positive for the gram-positive rod. Ofloxacin, gentamicin, and vancomycin therapy was discontinued on day 23, and imipenem therapy was initiated. On the following day, the fever subsided, and 3 days later, the patient’s leukocyte count increased to 1,700 cells per μl, with an ANC of 1,003 cells per μl. The patient was discharged on day 28 after reinduction. Imipenem, amphotericin B, and acyclovir therapy was discontinued, and the patient left the hospital on amoxicillin-clavulanic acid therapy. Cultures of blood collected 7 days after the patient was discharged were negative. Subsequent identification of the blood culture isolate showed that the organism was most closely related to Leptotrichia buccalis.
Blood cultures were performed with the BACTEC 9240 system with PLUS AEROBIC/F and PLUS ANAEROBIC/F bottles (Becton Dickinson Microbiology Systems, Sparks, Md.). All biochemical tests were performed by RapID NH and RapID ANA (Innovative Diagnostic Systems, Norcross, Ga.) commercial identification kits according to the manufacturer’s directions, except that the organism was grown on blood agar in an 8 to 10% CO2 atmosphere for 72 h. β-Lactamase production was detected with Cefinase disks (BBL Microbiology Systems, Cockeysville, Md.).
Whole-cell fatty acids were extracted and analyzed as previously reported (14), except that the organisms were grown on blood agar (BBL; Becton Dickinson) for 4 days at 35°C in 8 to 10% CO2. Analysis was performed with an automated Hewlett-Packard HP 5890 II microbial identification system (MIDI, Inc., Newark, Del.). Fatty acid profiles were compared with those from a library of cellular fatty acid profiles of clinically relevant bacteria, generating a similarity index (8).
The MicroSeq 16S rRNA gene kit (PE Applied Biosystems, Foster City, Calif.) was used to amplify and sequence the entire 16S rRNA gene. Amplification and sequencing were performed according to the manufacturer’s instructions. DNA sequencing utilized dRhodamine dye-labeled terminators and provided on average three to four overlapping strands of data when 12 sequencing primers were used. The sequence data was analyzed and assembled with Auto Assembler software (PE Applied Biosystems). Bacterial identifications based on 16S rRNA gene sequence data, generation of a dendrogram, and calculation of the percent difference between sequences were performed with the MicroSeq microbial identification and analysis software (PE Applied Biosystems).
For examination by electron microscopy, 5-day-old colonies orbiting around a Staphylococcus aureus streak were fixed in phosphate-buffered 2% glutaraldehyde, postfixed in 1% phosphate-buffered osmium tetroxide, and embedded in Epon 812 resin. The colonies were sectioned at 30 μm, stained with uranyl acetate and lead citrate, and viewed with a JEOL 1200 transmission electron microscope.
Seven of the 10 cultures of blood obtained from the patient over a 12-day period grew the gram-positive rod, and the rod was detected in all by the BACTEC 9240 system after approximately 48 h of incubation at 35°C. Only the anaerobic bottle supported growth of the bacterium. Gram stains from positive bottles initially revealed gram-positive rods, but subcultures from the bottle to a variety of conventional laboratory media failed to yield growth in either aerobic or anaerobic environments. Specifically, this bacterium would not grow on Trypticase soy agar with 5% sheep blood, chocolate agar, brucella agar supplemented with hemin and vitamin K, buffered charcoal-yeast extract agar, or thioglycolate broth (Table 1). In addition, blood agar with a variety of nutritional supplements, including X and V factor strips, pyridoxal hydrochloride (20-g disk), and paper disks saturated with olive oil or 10% Tween 80, would not support growth. Likewise, the bacterium would not grow in thioglycolate broth supplemented with 5% human serum or 5% laked horse blood. However, the bacterium grew when subcultured to a BACTEC PLUS ANAEROBIC/F bottle or to the anaerobic and aerobic Bac-T-alert FAN bottles and routine anaerobic and aerobic Bac-T-alert bottles (Organon Teknika, Durham, N.C.). In addition, the bacterium exhibited orbiting around an S. aureus (ATCC 25923) streak on brain heart infusion media with blood, Schaedler’s sheep blood, and Trypticase soy blood agar with 5% sheep blood, all incubated at 35 to 37°C. After the second subculture, an inoculum from the zone around the S. aureus streak grew on chocolate agar and on the above-listed blood-containing media without the S. aureus streak.
TABLE 1.
Summary of phenotypic properties for a novel Leptotrichia species
| Property or characteristic | Comments |
|---|---|
| Fastidious growth | No growth on Trypticase soy agar with 5% sheep blood, chocolate agar, or brucella agar with hemin and vitamin K upon primary subculture. Orbits S. aureus on Trypticase soy agar with 5% sheep blood. |
| Facultative anaerobe | Grows equally well in anaerobic and CO2-supplemented atmospheres. |
| Gram variable | Stains gram positive in young cultures. Cell wall resembles that of gram-negative organisms by electron microscopy. Cellular fatty acid profile is consistent with that of gram-negative organisms. |
| Biochemically active | Catalase negative. Hydrolyzes substitution-containing pyrrolidonyl, phenylalanine, α-d-glucoside, phosphate, glucose, and sucrose. |
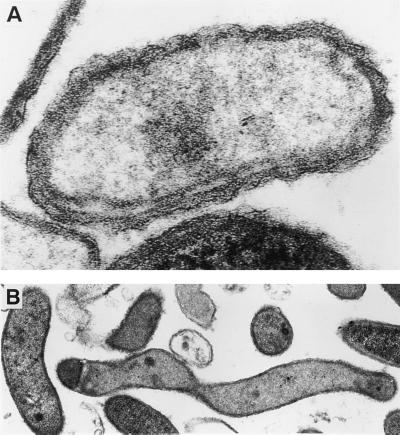
Biochemical testing indicated that the organism was catalase negative and hydrolyzed substitution-containing pyrrolidonyl, phenylalanine, α-d-glucoside, and phosphate (RapID ANA code no. 020122), as well as glucose and sucrose (RapID NH code no. 0300). The cellular fatty acid profile of this isolate was not significantly similar (i.e., the similarity indices were >0.3) to that of any species in the MIDI database, but data analysis indicated that the isolate was most closely related to L. buccalis, with a similarity index of 0.2 (8). The major peaks were 16:0 (30.3%), 14:0 (24.8%), 18:1c11/t9/t6 (23.4%), and 14:0-3-OH (8.0%). The high level of the 14:0 fatty acid is typical of a gram-negative organism. The 16S rRNA gene sequence did not match any sequence in the MicroSeq database; however, using the GenBank library, we found that there was a 95% (1,204/1,265) sequence identity with L. buccalis. Gram stains on positive blood cultures showed gram-positive rods, but gram stains of subcultured bacteria were difficult to interpret. Both gram-positive and gram-negative rods, 0.3 to 0.5 by 3.0 to 15 μm, were seen. Electron microscopy showed that the organisms were wavy, with a cell wall resembling that of a gram-negative bacterium (Fig. 1).
FIG. 1.
Electron micrograph of the novel isolate. (A) Magnification of a single cell demonstrating the cell wall structure; (B) lower magnification of several pleomorphic cells.
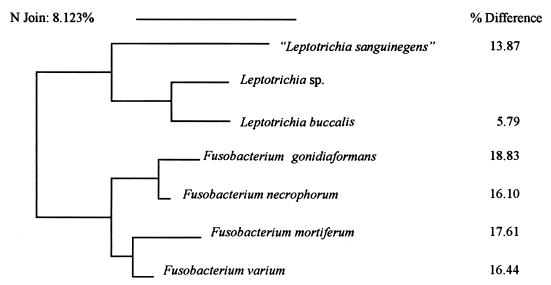
This bacterium may represent a previously undescribed member of the genus Leptotrichia, in which there is now only one species, L. buccalis. Like the organism described here, L. buccalis has a gram-negative cell wall structure (but it can stain gram positive when young), is catalase negative, and utilizes glucose and sucrose (1). However, L. buccalis is much less fastidious than the bacterium described here. Initially, cultures of L. buccalis grow only under anaerobic conditions on a variety of routine laboratory media, including brucella agar and Trypticase soy agar with 5% sheep blood. After several passages, many isolates of L. buccalis can adapt to growth under aerobic conditions with 5 to 10% CO2. In contrast, upon initial subculture, the organism in this study failed to grow in any media other than blood culture broth and blood-containing solid media with an S. aureus streak. It grew equally well in anaerobic and CO2-supplemented atmospheres. The strongest evidence that this bacterium is a unique species is the difference between the 16S rRNA gene sequences of L. buccalis and this isolate (Fig. 2). In general, a >1% difference between sequences is sufficient to characterize a bacterium as a unique species, and the difference between the sequence of this bacterium and that of L. buccalis is 5% (11). The fastidious nature of the isolate described here resembles that of an isolate described by Hanff et al. (5) with the proposed name Leptotrichia sanguinegens. However, both phenotypic evidence and molecular evidence indicate that these bacteria represent distinct species; the addition of serum facilitates growth of “L. sanguinegens” but not of the isolate described here, and the sequences of the 16S rRNA genes from these isolates are significantly different (Fig. 2).
FIG. 2.
Dendrogram depicting the degree of difference between 16S rRNA gene sequences. The dendrogram was generated by the neighbor-joining pair group method (12). The sequences of L. buccalis and “L. sanguinegens” are from GenBank (accession no. L37788 and L37789, respectively) (5). The sequences of the Fusobacterium spp. are part of the MicroSeq database. These are genomic sequences of the American Type Culture Collection type strain for each species (ATCC 25563, ATCC 25286, ATCC 2557, and ATCC 8501).
Antimicrobial susceptibility testing could not be performed on this isolate because of its poor growth characteristics, but we predict that its profile may be similar to that of L. buccalis. Typically, L. buccalis is susceptible to β-lactam drugs, imipenem, clindamycin, metronidazole, rifampin, tetracycline, and chloramphenicol. It is resistant to erythromycin, vancomycin, aminoglycosides, and fluoroquinolones. The patient’s bacteremia persisted for 12 days, despite previous and continuing therapy with ofloxacin, gentamicin, and vancomycin, but it resolved upon imipenem therapy and subsequent amoxicillin-clavulanic acid therapy. This clinical response is consistent with the L. buccalis antimicrobial susceptibility profile.
The isolation of a Leptotrichia sp. from seven blood samples during a 12-day period, four of which were obtained through peripheral venous access, supports the role of this organism as an opportunistic pathogen in this patient. No other source for the persistent fevers was found. Although a coagulase-negative Staphylococcus sp. was isolated on multiple occasions (five sets over a 10-day period), these isolates were likely to represent contaminants (or line-related colonization), since all of the samples culture positive for this organism were obtained through venous catheters. Most cases of Leptotrichia sp. bacteremia reported in the literature are associated with mucositis, suggesting an oral portal of entry (2, 6, 9, 10, 13). Although this patient did not suffer from mucositis, herpes simplex virus oral lesions were noted 3 days after the first positive blood samples were drawn. These lesions may have been the portal of entry.
Reports on 15 cases of serious infections with a Leptotrichia sp. were found in a search of English language literature. Like the case reported here, the majority of these infections were primary bacteremias in neutropenic patients (2, 7, 10, 13). In all but three of these patients, hematologic malignancy was the underlying disease. The three remaining patients had ovarian carcinoma, osteogenic sarcoma, or idiopathic medullary aplasia (9, 13). Also worth noting, oral lesions or gingivitis was found in nine of the patients shortly before or during the bacteremia.
Neutropenia does not appear to be a necessary risk factor for bacteremia with a Leptotrichia sp. (3, 4, 6). L. buccalis bacteremia associated with a hepatic abscess was reported for an apparently immunocompetent patient with severe parodontitis. In addition, three reported cases of endocarditis with a Leptotrichia sp. al. occurred in immunocompetent patients with either a congenital heart defect or a prosthetic valve (3, 4). However, in the case reported here, the patient’s fever subsided when his neutropenia resolved. This defervescence also coincided with the switch to what is probably more appropriate antibiotic therapy. Therefore, it is not clear whether this infection resolved as a result of host factors, antibiotic therapy, or both.
To our knowledge, this is the first reported case of a bacteremia with an isolate that resembles a Leptotrichia sp. but failed to grow on conventional laboratory media. However, this case is similar to previous cases in that the infection occurred in a patient suffering from neutropenic fever which was treated with non-β-lactam antibiotics.
Acknowledgments
Jean Baldus Patel is the William Pepper/Becton Dickinson Fellow in Clinical Microbiology.
REFERENCES
- 1.Anonymous. Gram-negative, anaerobic, straight, curved, and helical bacteria. In: Holt J G, Krieg N R, Sneath P H A, Staley J T, Williams S T, editors. Bergey’s manual of determinative bacteriology. Baltimore, Md: Williams & Wilkins Co.; 1994. pp. 291–334. [Google Scholar]
- 2.Baquero F, Fernandez J, Dronda F, Erice A, Perez de Oteiza J, Reguera J A, Reig M. Capnophilic and anaerobic bacteremia in neutropenic patients: an oral source. Rev Infect Dis. 1990;12(Suppl. 2):S157–S160. doi: 10.1093/clinids/12.supplement_2.s157. [DOI] [PubMed] [Google Scholar]
- 3.Duperval R, Beland S, Marcoux J A. Infective endocarditis due to Leptotrichia buccalis: a case report. Can Med Assoc J. 1984;130:422–424. [PMC free article] [PubMed] [Google Scholar]
- 4.Hammann R, Iwand A, Brachmann J, Keller K, Werner A. Endocarditis caused by a Leptotrichia buccalis-like bacterium in a patient with a prosthetic aortic valve. Eur J Clin Microbiol Infect Dis. 1993;12:280–282. doi: 10.1007/BF01967258. [DOI] [PubMed] [Google Scholar]
- 5.Hanff P A, Rosol-Donoghue J, Spiegel C A, Wilson K H, Moore L H. Leptotrichia sanguinegens sp. nov., a new agent of postpartum and neonatal bacteremia. Clin Infect Dis. 1995;20(Suppl. 2):S237–S239. doi: 10.1093/clinids/20.supplement_2.s237. [DOI] [PubMed] [Google Scholar]
- 6.Messiaen T, Lefebvre C, Geubel A. Hepatic abscess likely related to Leptotrichia buccalis in an immunocompetent patient. Liver. 1996;16:342–343. doi: 10.1111/j.1600-0676.1996.tb00758.x. [DOI] [PubMed] [Google Scholar]
- 7.Morgenstein A A, Citron D M, Orisek B, Finegold S M. Serious infection with Leptotrichia buccalis. Report of a case and review of the literature. Am J Med. 1980;69:782–785. doi: 10.1016/0002-9343(80)90452-0. [DOI] [PubMed] [Google Scholar]
- 8.Paisley R. Training manual: MIS whole cell fatty acid analysis by gas chromatography. Newark, Del: MIDI, Inc.; 1998. [Google Scholar]
- 9.Reig M, Baquero F, García-Campello M, Loza E. Leptotrichia buccalis bacteremia in neutropenic children. J Clin Microbiol. 1985;22:320–321. doi: 10.1128/jcm.22.2.320-321.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz D N, Schable B, Tenover F C, Miller R A. Leptotrichia buccalis bacteremia in patients treated in a single bone marrow transplant unit. Clin Infect Dis. 1995;20:762–767. doi: 10.1093/clinids/20.4.762. [DOI] [PubMed] [Google Scholar]
- 11.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 12.Studier J A, Keppler K J. A note on the neighbor-joining algorithm of Saitou and Nei. Mol Biol Evol. 1988;5:729–731. doi: 10.1093/oxfordjournals.molbev.a040527. [DOI] [PubMed] [Google Scholar]
- 13.Weinberger M, Wu T, Rubin M, Gill V J, Pizzo P A. Leptotrichia buccalis bacteremia in patients with cancer: report of four cases and review. Rev Infect Dis. 1991;13:201–206. doi: 10.1093/clinids/13.2.201. [DOI] [PubMed] [Google Scholar]
- 14.Welch D F. Applications of cellular fatty acid analysis. Clin Microbiol Rev. 1991;4:422–438. doi: 10.1128/cmr.4.4.422. [DOI] [PMC free article] [PubMed] [Google Scholar]