Abstract
The extracellular matrix (ECM) is a three-dimensional network of macromolecules that provides a microenvironment capable of supporting and regulating cell functions. However, only a few research organisms are available for the systematic dissection of the composition and functions of the ECM, particularly during regeneration. We utilized the free-living flatworm Schmidtea mediterranea to develop an integrative approach consisting of decellularization, proteomics, and RNAi to characterize and investigate ECM functions during tissue homeostasis and regeneration. ECM-enriched samples were isolated from planarians, and their proteomes were characterized by LC–MS/MS. The functions of identified ECM components were interrogated using RNA interference. Using this approach, we found that heparan sulfate proteoglycan is essential for tissue regeneration. Our strategy provides an experimental approach for identifying both known and novel ECM components involved in regeneration.
Keywords: extracellular matrix, planarians, regeneration, proteomics, stem cells
Abbreviations: dNSAF, distributed normalized spectral abundance factor; dpa, day postamputation; ECM, extracellular matrix; ER, endoplasmic reticulum; FA-ECM, formaldehyde ECM; FDR, false discovery rate; GO, Gene Ontology; hspg, heparan sulfate proteoglycan; MMP-3, matrix metalloproteinase-3; MW, molecular weight; NAC-ECM, N-acetyl cysteine pretreatment ECM; NP-ECM, no pretreatment ECM; PVDF, polyvinylidene difluoride; RT, room temperature; tll, tolloid-like protein 1; WA, nondecellularized whole animal
Graphical Abstract

Highlights
-
•
Decellularization method developed to isolate ECM from complete animals.
-
•
Optimization of decellularization method for protein MS compatibility.
-
•
Identification of known and novel ECM components for their function in tissue regeneration.
In Brief
The ECM is a three-dimensional network of macromolecules that supports and regulates cell functions. Yet, only a few research organisms are available for the systematic dissection of the composition and functions of the ECM, particularly during regeneration. In this study, we report an integrative approach involving whole animal decellularization, protein mass spectrometry (LC–MS/MS), and RNA interference-based loss-of-function assays to identify both known and novel ECM components involved in regeneration.
Tissue regeneration is an essential process for many organisms and can be activated during embryogenesis, throughout life during the constant physiological renewal of tissues, and in the dramatic restoration of missing body parts following injury or amputation (1). The complexity of tissue regeneration is reflected by the numerous and overlapping molecular and cellular activities underpinning the restoration and integration of missing body parts (2). In order for the organism to maintain and repair physiological form and function during and after tissue regeneration, cells need to communicate flawlessly with their microenvironment (3, 4, 5).
The extracellular matrix (ECM), which is a collection of dynamically secreted and modified macromolecules occupying intercellular space, plays a central role in effecting cell–environmental communication (6, 7). Bidirectional crosstalk between cells and the ECM via secretion and selective degradation creates microenvironmental conditions capable of modulating cell proliferation, migration, differentiation, and ultimately, the homeostatic maintenance of tissues throughout the lifetime of an organism (8, 9, 10, 11). Of particular interest are the microenvironmental conditions governing stem cell biology (12, 13, 14).
Several animal models have been used for investigating how the ECM is involved in tissue regeneration processes, such as in Hydra vulgaris (15), axolotls (16), and zebrafish (17). Although progress has been made in these organisms implicating the ECM in regeneration, the inherent biology of these animals makes it challenging to systematically dissect ECM composition and to functionally study their possible roles in regeneration. For example, although Hydra is able to regenerate a whole animal from a clump of dissociated cells (18), whereas axolotls (16) and zebrafish (17) can regenerate missing body parts, it is still difficult to carry out large-scale loss-of-function screens in these adult organisms. Therefore, in an effort to systematically interrogate how ECM may contribute to whole-body and/or tissue regeneration, we chose to study the free-living freshwater planarian flatworm Schmidtea mediterranea (19), which has extraordinary regenerative capacities and has been shown to be amenable to large-scale genetic interrogation (20).
Because our current knowledge of ECM biology in planarians is limited (21, 22, 23), it is first necessary to develop a comprehensive and optimized workflow to characterize and study the planarian ECM. A recent study has characterized the transcriptional landscape of ECM components in planarians by constructing an in silico dataset (24). However, this has not revealed the actual protein composition and distribution of these molecules. Similarly, Sonpho et al. (25) successfully developed a simple technique for characterizing the morphology of isolated ECM by whole organism decellularization of a different planarian species. However, a complete systematic workflow for studying ECM biology in planarians has not yet been established.
Here, we propose an integrative workflow to systematically characterize the S. mediterranea ECM. This workflow consists of three core components: decellularization, proteomics, and RNAi of identified ECM components. First, decellularization of S. mediterranea was optimized for the isolation of planarian ECM. Second, we subjected the decellularized fraction to biochemical characterization by LC coupled to MS (LC–MS/MS), which allowed us to determine the protein components from ECM-enriched samples derived from planarians. Third, RNAi screening was performed by using a set of candidate proteins from our ECM MS to confirm whether our workflow was practical for discovering novel genes relevant for tissue regeneration. Finally, we identified one ECM protein that plays an important role in tissue regeneration. In summary, by combining decellularization, proteomics, and RNAi screening, we provide proof-of-concept experimental evidence illustrating the potential of this workflow to discover and study ECM composition, function, and dynamics in an adult regeneration-competent organism. Our approach also lays the foundation for a systematic and functional dissection of the role that the ECM may play in regulating stem cell behavior and function during both animal homeostasis and regeneration in planarians.
Experimental Procedures
Animal Husbandry
S. mediterranea asexual clonal line CIW4 was maintained in 1× Montjuic salt solution, as described previously for static culture (26). S. mediterranea were fed with beef liver once a week. The animals were starved for at least 1 week before experiments.
Animal Decellularization
Whole body planarian decellularization has been optimized based on a previous publication (25). We optimized three protocols for decellularization, which we refer to as no pretreatment ECM (NP-ECM), formaldehyde ECM (FA-ECM), and N-acetyl cysteine (NAC) pretreatment (NAC-ECM). All worms were collected from static culture. All three protocols were designed for 20 planarians.
For NP-ECM, planarians were incubated in 40 ml of 0.08% SDS decellularization solution for 18 h at 4 °C. ECM was harvested carefully using plastic transfer pipettes and collected into a microtube. Decellularization solution was removed and replaced with 1 ml of wash solution for 30 min, three times, on ice. The ECM pellet was centrifuged at 14,000g for 10 min.
For FA-ECM, planarians were fixed and stabilized by incubating in 0.8% FA solution at 4 °C for 1 h without shaking. After stabilization, the solution was replaced with 40 ml of 0.7% SDS decellularization solution for 18 h at 4 °C. To ensure complete decellularization, the solution was replaced with 40 ml 0.08% SDS decellularization solution for 2 h at 4 °C.
For NAC-ECM, planarians were first incubated with 5% NAC (Sigma–Aldrich) solution in 1× PBS for 10 min at room temperature (RT) on a seesaw rocker. Afterward, planarians were transferred into 1× PBS for a brief wash. Then, planarians were transferred into 40 ml of 0.08% SDS decellularization solution for 18 h at 4 °C. Subsequently, the solution was replaced with 40 ml of 0.04% SDS decellularization solution for 2 h at 4 °C. All the solution components for S. mediterranea decellularization are reported in supplemental Table S1A.
Quantification of Nucleotide Contamination
Nucleic acid material remaining in ECM preparations was isolated using a DNeasy Blood & Tissue Kit (Qiagen). The amount of DNA was quantified using Qubit fluorometric quantification.
Protein Quantitation and Western Blot Analysis
Amounts of proteins obtained from all three ECM methods were quantified by Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) and reported in supplemental Table S1B. All samples were processed for separation on SDS-PAGE (supplemental Fig. S1) and analyzed by Western blot. Briefly, ECM was harvested and solubilized by adding radioimmunoprecipitation buffer (Abcam) and Laemmli sample buffer. Equal amounts of proteins were prepared and loaded per lane. Polyvinylidene difluoride (PVDF) membrane was activated by incubating in 100% methanol for 5 min before washing in the transfer buffer. Proteins were transferred from the gel to the PVDF membrane with 100 V for 1 h, and then the PVDF membrane was washed with TBS–Tween 0.05% and blocked with 5% milk in TBS–Tween 0.05%, for 1 h at RT. A dilution of primary antibody was prepared in a fresh blocking solution as follows: rabbit antihistone H3 antibody (Abcam; 1791) diluted at 1:4000 and rabbit anti–matrix metalloproteinase-3 (MMP-3) antibody (Signaling Technology; D7F5B) diluted at 1:1000. The blocking solution was replaced with the primary antibody solution overnight at RT. Membrane was then washed with TBS-Tween 0.05%, four times for 15 min each. The swine anti-rabbit-horseradish peroxidase secondary antibody (DAKO; P0399) was diluted at 1:5000 in 1% milk. The membrane was incubated with the secondary antibody for 1 h at RT. The membrane was washed with TBS–Tween 0.05%, four times for 15 min each. The signal of horseradish peroxidase was visualized via SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific). The membrane was developed until the signal appeared. Exposures for antihistone H3 and anti-MMP-3 antibodies were 1 min and 2 h, respectively.
Histological Staining
Worms were processed using a Delta Pathos hybrid tissue processor (Milestone Medical). Briefly, after fixing planarians in 1% nitric acid, 50 μM MgSO4, and 0.8% FA overnight, tissues were rinsed and dehydrated in 70% ethanol. The following steps were automatically performed by the Delta Pathos processor: 4 min rinse with 70% ethanol; 100% ethanol for 10 min at 65 °C; and 100% isopropanol for 30 min at 68 °C. For paraffin infiltration, the first bath of paraffin was set at 70 °C for 8 min, and then, fresh paraffin was replaced for an additional 6 min, followed by a final step of fresh paraffin incubation for another 20 min at 65 °C (Tissue Infiltration Medium by Surgipath available through Leica). The paraffin-embedded tissue was sectioned in Richard–Allan type 9 paraffin (Thermo Fisher Scientific). Tissue was sectioned at 5 μm thickness, dried, heated, and then stained with Masson's trichrome stain. This procedure is derived from Histotechnology, a Self-Instructional Text (27).
Scanning Electron Microscopy
To prepare samples for observation under a scanning electron microscope, collected planarian ECM was incubated in fixative solution composed of 2.5% glutaraldehyde, 2% para-FA, 1 mM calcium chloride, and 1% sucrose in 50 mM sodium cacodylate buffer. Afterward, the sample was stained with tannic acid, osmium, and thiocarbohydrazide, as previously described (28). At the end of dehydration, the samples were critical point dried in a Tousimis Samdri 795 critical point dryer and then mounted on stubs and coated with 5 nm of gold palladium in a Leica ACE600 sputter coater. The samples were imaged using a Zeiss Merlin scanning electron microscope at 20 kV and 400 pA.
Proteomics Experimental Design and Statistical Rationale
Three biological replicate preparations of FA-ECM and NAC-ECM and two biological replicates of NP-ECM samples were processed in parallel and analyzed independently. Three biological replicate protein preparations from nondecellularized whole animal (WA) were also prepared for comparison by freezing them in liquid nitrogen. All decellularized samples were centrifuged at 16,900g at 4 °C to recover the ECM pellets. ECM and WA proteins were resuspended in 120 μl of 100 mM Tris–HCl, pH 8.5, and 8 M urea. The samples were vortexed vigorously to dissolve pellets. To reduce disulfide bonds, Tris(2-carboxyethyl)phosphine hydrochloride (Pierce) was added to 5 mM (0.6 μl of 1 M of Tris(2-carboxyethyl)phosphine hydrochloride) and incubated at RT for 30 min. Reduced cysteines were alkylated by adding 2.4 μl of a freshly made 0.5 M chloroacetamide stock solution (Sigma) and incubated at RT in the dark for 30 min. Samples were deglycosylated with PNGase F by adding 5 μl of a solution consisting of 18 μl of glycobuffer 2 (10×) and 27 μl of PNGaseF (NEB; P0708S). To ensure complete digestion of proteins, a two-step digestion was performed. First, endoproteinase Lys-C (Promega), 5 μl at 1 μg/μl, was added, and samples were incubated at 37 °C overnight while shaking (29). Next, solutions were diluted to 2 M urea by adding 360 μl of 100 mM Tris–HCl and 2 μl of CaCl2, followed by 10 μl of trypsin (Promega) at 0.1 μg/μl. Samples were incubated at 37 °C overnight while shaking. Reactions were quenched by adding 90% formic acid to a final concentration of 5%. After digestion, peptide concentrations were quantitated by the Pierce Quantitative Colorimetric Peptide Assay (Thermo Fisher Scientific).
Peptides were diluted with buffer A (5% acetonitrile and 0.1% formic acid). Then, samples were injected on an in-house–packed 50 μm inner diameter microcapillary column packed with 10 cm of 1.9 μm Aqua C18 resin (Dr Maisch GmbH). The samples were injected in 100% buffer A and subsequently eluted using an Ultimate 3000 UPLC (Dionex) at a flow rate of 0.120 μl/min for 240 min from 10 to 40% buffer B (80% acetonitrile and 0.1% formic acid) before ramping to 95% B in 25 min. The high organic concentration was maintained for 15 min before re-equilibration and the reinjection with the next sample. Peptides were ionized by the application of a 2.5 kV positive voltage applied distally to the peptides eluting into a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific). Full MS spectra were recorded on the eluting peptides over a 350 to 1700 m/z range at 70,000 resolving power. Automated gain control target was 1 × 106 with a maximum injection time set to 50 ms. The top 15 peptides with charges 2 to 5 were fragmented by high-energy collision dissociation at 27% normalized collision energy, and the MS/MS fragmentation was collected at 17,500 resolving power, with an automated gain control target of 1 × 105 and a maximum injection time set to 150 ms. Dynamic exclusion was enabled for 30 s (30). Mass spectrometer scan functions and HPLC solvent gradients were controlled by the XCalibur data system (Thermo Fisher Scientific).
Database Search Parameters and Acceptance Criteria for Identifications
RAW files were extracted to peak lists in.ms2 file format (31) using an in-house software, RawDistiller, version 1.0 (Washburn Lab/Proteomics at the Stowers Institute; https://github.com/tzw-wen/kite/tree/master/windowsapp/RawDistiller) (32). RawDistiller settings were used to abstract MS1 scan profiles by Gaussian fitting and to implement dynamic offline lock mass using six background polydimethylcyclosiloxane ions as internal calibrants (32). MS/MS spectra were first searched using ProLuCID, version 1.3.3 (Yates lab at the Scripps Research Institute) (33), with a peptide mass tolerance of 10 and 25 ppm for peptide and fragment ions, respectively. Trypsin specificity was imposed on both ends of candidate peptides with up to three missed cleavages. The S. mediterranea protein sequences were downloaded from the PlanMine database (https://planmine.mpibpc.mpg.de/planmine/aspect.do?name=Gene%20Predictions) on September 1, 2019 and contained 30,863 predicted protein sequences, as well as 483 usual contaminants, such as human keratins, immunoglobulins, and proteolytic enzymes. To estimate false discovery rates (FDRs), each protein sequence was randomized (keeping the same amino acid composition and length), and the resulting “shuffled” sequences were added to the database, for a total search space of 62,564 amino acid sequences. A mass shift of 57.0215 Da was statically added to cysteine residues to account for alkylation by chloroacetamide, mass shifts of +15.9949 was differentially added to methionine, lysine, and proline residues to account for oxidation or hydroxylation, and +0.98401 Da was differentially added to asparagine residues to account for deglycosylation by PNGaseF resulting in asparagine deamidation.
DTASelect, version 1.9 (Yates lab at the Scripps Research Institute) (34), was used in combination with an in-house software, swallow, version 1.0 (https://github.com/tzw-wen/kite), to select and sort peptide–spectrum matches to FDRs of less than 1% at the peptide and protein levels. Results from the worm without decellularization (three replicates of WA), FA-treated (three replicates of FA-ECM), NAC-treated (three replicates of NAC-ECM), and not pretreated (two replicates of NP-ECM) ECMs were merged and compared using CONTRAST, version 1.9 (Yates lab at the Scripps Research Institute) (34) in combination with sandmartin, version 1.0, an in-house software (Washburn Lab/Proteomics at the Stowers Institute; https://github.com/tzw-wen/kite/tree/master/kitelinux), to select proteins at FDRs less than 5% when combining all runs. Proteins that were subsets of others were removed using the parsimony option in DTASelect on the proteins detected after merging all runs. Proteins that were identified by the same set of peptides were grouped together, and one accession number was arbitrarily considered as representative of each protein group.
NSAF7, version 0.0.1 (35) (Washburn Lab/Proteomics at the Stowers Institute; https://github.com/tzw-wen/kite/tree/master/windowsapp/NSAF7x64), was used to create the final reports on all detected peptides and nonredundant proteins identified across the different runs and calculate label-free distributed normalized spectral abundance factor (dNSAF) quantitative values for all detected protein/protein groups. Spectral and protein level FDRs were, on average, 0.29 ± 0.12% and 1.3 ± 0.7%, respectively. The full list of proteins identified in each sample type is provided in supplemental Table S2A, along with their peptide counts (Pep), spectral counts (SpC|total, unique, shared, and distributed), % sequence coverage, and label-free quantitation (dNSAF). No proteins identified by a single peptide–spectrum match are reported here. Modified peptide spectrum matches that mapped to proteins detected from WA and ECM preparations are provided in supplemental Table S2B, along with the identification of the matches with the highest cross-correlation scores; parameters used to assess the quality of the matches (peptide's charge state, cross-correlation score, normalized difference in Xcorrs, observed precursor ion mass, calculated precursor ion mass, calculated mass error in part per million, rank based on Sp score, SEQUEST preliminary score, and percentage of peptide's fragment ions matched to experimental tandem mass spectrum); the number of different spectra matched to each peptide/charge state combination; the sequence of each peptide mapped to protein/protein group, position of peptide's first residue within the protein, peptide's tryptic status, whether the peptide is unique to this protein/protein group or can be mapped to multiple proteins whose accession numbers are provided; the number of post-translational modifications in peptide sequence, and the computationally assigned position(s) of the modified amino acid residue(s) within the protein sequence followed by post-translational modification symbol(s) (@ for hydroxylation/oxidation and ∼ for deglycosylation). No ambiguous assignments are reported, and fully annotated spectra of all modified peptides are provided as supplemental File S1.
Statistical Analysis of Differential Protein Expression
QPROT (Nesvizhskii Lab at the University of Michigan) (36) was used to calculate log2 fold changes and FDRs to identify proteins enriched in replicate analyses of each of the three ECM preparations, compared with the abundance of proteins as extracted from WAs (supplemental Table S2C). Volcano significance plots were generated in R software by plotting the log2 fold change (X-axis) and the −log10(FDR) from QPROT (supplemental Fig. S6A).
Gene Ontology Enrichment
S. mediterranea gene models were assigned Gene Ontology (GO) (37) terms (supplemental Table S2, D–G) by combining the GO annotations from PlanMine (38, 39), BLAST2GO (version 5.2) (40), AHRD (version 3.3.3), Uniprot/SWISS-PROT best BLAST hits (param: −evalue = 0.001; db date: 20180501) (41), InterProScan (version 5.32-71.0) (42), and PANTHER (43). GO enrichment was performed using TopGO (version 2.34.0) (44).
RNAi Food Preparation and Feeding
The RNAi food was prepared as previously described (45). The primers used for cloning each gene are shown in supplemental Table S3. The colored liver puree was added to the bacteria pellet and fed to planarians cultured in an automatic water exchanger system (46). About ten feeding cycles were implemented in both primary screening and secondary screening prior to use in following experiments. The essential transcription factor nkx-2.2 was used as a positive control (47), whereas the Caenorhabditis elegans gene unc-22 was used as a negative control for RNAi treatment (20).
Homeostasis and Regeneration Assays
Phenotypes of the targeted ECM components in tissue homeostasis were evaluated by observing unamputated planarians at 2 days after completion of the RNAi feeding. Amputations were performed by arranging planarians on a wet filter paper placed on a cold aluminum plate buried in ice and cutting with a surgical blade. The surgical blade was cleaned with 70% ethanol between amputations. Any signs of abnormal body plan or lysis were considered phenotypes. Worms were evaluated 10 days after sagittal amputation for morphological regeneration phenotypes. Any signs of incomplete or lack of newly regenerated tissue (blastema) were considered a defective phenotype. Worms that showed no touch response were considered dead.
Whole-Mount in Situ Hybridization and Animal Radiation
A GammaCell 40 Exactor irradiator was used to expose planarians to 6000 rad for lethal irradiations. For the whole-mount in situ hybridization experiment, the protocol was as previously described (48).
Determination of Cell Type–Specific Expression of Genes of Interest
To access the published single-cell RNA-Seq database, we used the planaria single cell sequencing online tool Digiworm (Reddien Lab at the Whitehead Institute for Biomedical Research) (49). The partial or complete contig of gene IDs was entered into the search box. For matching genes to the planarian transcriptome, the transcriptome assembly was mapped to the dd_v4 transcriptome as previously indicated (50). The transcriptome assembly ID (dd_v4) was entered into the search box to generate Seurat maps.
Statistical Analysis of Data
Statistical analysis of DNA contents and survival rates after amputation were carried out by using the R software, version 3.14. A two-way ANOVA was performed for WA and ECMs in all experiments. The protected least significant difference post hoc test (a = 0.05) was used for multiple comparison tests between WA and ECMs.
Results
Decellularized Structure Can Be Successfully Isolated From S. mediterranea
In order to establish a systematic workflow for studying ECM in S. mediterranea, we devised a combination of whole animal decellularization, proteomics, and RNAi screening (Fig. 1A). Although a whole-body planarian decellularization procedure for Dugesia japonica has recently been described, this protocol had not been tested for the isolation of ECM from S. mediterranea (25). Therefore, we explored detergent-based tissue decellularization at low temperature for isolating the ECM in S. mediterranea by incubating planarians with a low concentration of SDS at 4 °C (Fig. 1B). Compared with our WA (Fig. 1C), the results were similar to those from D. japonica (25) in that the decellularization without any pretreatment (NP-ECM) could maintain the animal's overall morphology, yet, the external structure of the decellularized animals was tremendously compromised (Fig. 1D). Moreover, internal structures of decellularized animals were not well defined. As in D. japonica, pretreating the planarians with FA prior to decellularization did preserve the external and internal integrity of isolated decellularized structure in S. mediterranea (Fig. 1E) (25). The resultant decellularized animals (FA-ECM) had a translucent appearance with intact gross external structures and maintained the organization of internal organs such as the digestive tract branches, eye spots, and pharynx (Fig. 1E). These results suggest that the decellularization protocol for D. japonica has the potential to be applied to S. mediterranea since the resultant decellularized structures were similar.
Fig. 1.
Isolation of Schmidtea mediterranea ECM by tissue decellularization.A, an overview of our pipeline for characterizing planarian ECM to study regeneration, development, and homeostasis. B, the detailed protocol for ECM isolation. Overall morphology of (C) nondecellularized whole animal (WA) and ECMs isolated from different pretreatment procedures, including (D) no pretreatment (NP-ECM), (E) formaldehyde-treated ECM (FA-ECM), and (F) N-acetyl cysteine–treated ECM (NAC-ECM). C–F, the scale bars represent 1 mm. G, amount of total DNA removed by the decellularization procedure. The error bars represent standard deviation. The data were calculated from three independent experiments, ANOVA (∗p < 0.05). H, Western blot against MMP-3 and histone H3. Equal amount of total protein was loaded into each lane. I, gross morphology and histological sections of decellularized planarians stained with Masson's trichrome. Decellularizing FA-ECMs were collected at 18 h. Black arrows indicate the layer of the collagen sheet. Insets demonstrate the magnification of black square area. Red asterisk indicates the location of the pharynx. The scale bars represent 500 μm. J, the external surface topology of planarians and FA-ECM were visualized by scanning electron microscopy. The scale bars represent 10 μm. ECM, extracellular matrix; MMP-3, matrix metalloproteinase-3.
Since FA crosslinking could decrease protein solubility in downstream applications, we next sought a different method to preserve the intact structures of the decellularized animals. We selected NAC to pretreat animals prior to decellularization because this reagent is commonly used in S. mediterranea to remove mucus and euthanize the animal before fixation. Interestingly, our results indicate that NAC pretreatment prior to decellularization (NAC-ECM) could also preserve decellularized structures but to a lesser extent than FA-ECM (Fig. 1F). Therefore, we concluded that pretreatment of animals either by FA or NAC prior to whole-body decellularization is imperative for successfully isolating intact decellularized structures from whole planarians.
In addition, pretreatment of animals via FA fixation yields an external structure that is better preserved than either NAC-ECM or NP-ECM. Furthermore, the decellularization process was also captured by time-lapse movies for providing the real-time dynamics of whole animal decellularization (supplemental Video S1). The footage displayed the reduction of animal size during tissue decellularization in all three decellularization protocols. There was a small change in size from FA-ECM, whereas there was a dramatic change in NAC-ECM. Meanwhile, NP-ECM displayed a thoroughly compromised external structure. In summary, we successfully implemented three approaches for decellularizing whole S. mediterranea planarians.
Cellular Components Are Depleted in Decellularized Samples
We tested depletion of cellular components by measuring the amount of DNA present in the decellularized samples. The results indicated that the amount of DNA per worm in the three decellularization preparations was at least two orders of magnitude lower when compared with WA (Fig. 1G). The absence of cells was further validated by Western blot analysis. The estimated amount of proteins from three decellularization techniques is reported in supplemental Table S1B. Differential SDS-PAGE banding patterns between WA and ECM samples indicated that proteomic profiles between WA and ECM could be different (supplemental Fig. S1), where certain proteins might be enriched by decellularization, whereas others might be depleted. An antibody against histone H3 was used as the marker for intracellular proteins, and MMP-3 was used as the marker of ECM. The results indicated that histone H3 was detected in the WA, whereas FA-ECM showed a very faint band of histone H3 (Fig. 1H). NAC-ECM and NP-ECM showed no visible band of histone H3. In addition, the antibody against MMP-3 revealed the presence of MMP-3 in both WA and NP-ECM. The intensity of the MMP-3 band in the WA was much fainter than the analogous band in NP-ECM, suggesting an enrichment of this marker in the ECM fractions by decellularization. Taken together, the absence of detectable DNA and histone H3 suggests that the three decellularization protocols are sufficient to reduce intracellular components.
Collagen Layer is Maintained in Decellularized Planarians
As an additional means of testing for the presence of expected decellularized animal structures, we performed two morphological characterizations on the most structurally preserved decellularized animal, FA-ECM. First, the preservation of the collagen layer postdecellularization was assessed by Masson's trichrome staining. The result demonstrated that decellularization was able to infiltrate epithelial cells and remove mesenchymal cells based on the depletion of pink and purple staining in decellularized animal as compared with the WA whilst maintaining the integrity of the collagen layer, indicated in blue at the outermost region of ECM labeled with black arrows (Fig. 1I). The results also confirmed that internal structures were preserved during decellularization, as evidenced by the intact pharynx. Second, the external surfaces of decellularized animals were examined by scanning electron microscopy. The dorsal and ventral sides of WA were covered with a mucus layer and a ciliated surface, respectively (Fig. 1J). However, for FA-ECM, a fibrous surface was observed on the dorsal side, whereas a nonciliated surface was observed ventrally. This suggests that the decellularization process removes the mucus layer and epithelial cells as well as the ventral ciliated surface. On the whole, the two independent morphological characterizations we performed confirm the cell-free environment of our FA-ECM as well as the preservation of decellularized planarians integrity with an intact collagen layer.
Decellularization Methods Are Compatible With Protein MS
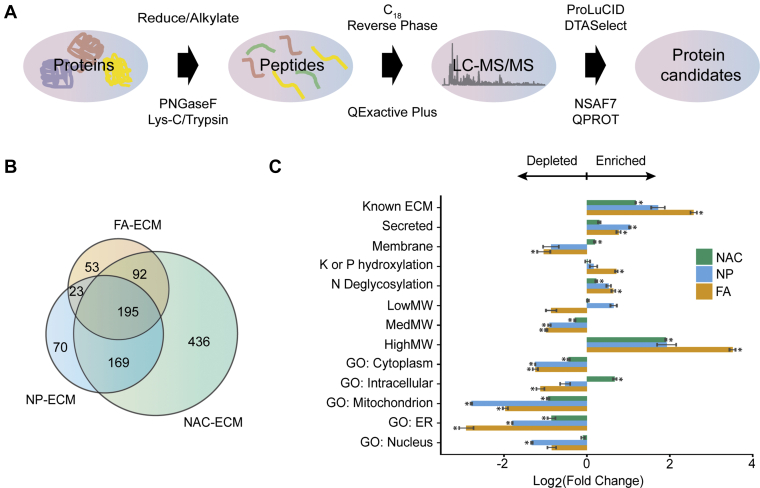
Decellularizing the planarian provided us with a sample that is enriched for extracellular components. We utilized this opportunity to identify the protein composition of decellularized animals via MS approaches as illustrated in Figure 2A. We first needed to determine whether our decellularized animal samples were compatible with protein MS. MS/MS spectra were detected from all ECM preparation methods and WA (supplemental Fig. S2 and supplemental Table S2A). As expected for samples from which most intracellular components were removed, the numbers of matched MS/MS spectra, sequenced peptides, and proteins detected from the three types of decellularized samples were lower than the numbers observed from WA analyzed with the same LC gradient and MS setting (supplemental Fig. S2 and supplemental Table S2A). Moreover, the similarity and differences of protein identifications among three decellularization protocols were analyzed by Venn diagram (Fig. 2B), in which 195 proteins were shared between all three ECM isolation methods, whereas 53, 436, and 70 proteins were exclusively detected in FA-ECM, NAC-ECM, and NP-ECM, respectively. Therefore, we demonstrated that proteins were able to be identified from our three decellularization procedures.
Fig. 2.
Proteomics analysis reveals a complex and enriched ECM profile from decellularized planarians.A, workflow for ECM bottom–up proteomics analysis. B, Venn diagram representing the overlapping proteins identified from three different ECM isolation procedures. C, the fold change of mean sum of dNSAF protein abundance measured in each decellularization technique and WA. The “known ECM” category represents genes annotated with an ECM GO term, genes reported by Cote et al. (24), and/or genes identified as ECM in MatrixDB. “Secreted” represents genes with predicted extracellular export signal peptide from SignalP and genes identified as secretory proteins in MatrixDB. “Membrane” represents genes predicted with membrane protein topology by TMHMM and genes identified as transmembrane proteins in MatrixDB. “K or P hydroxylation” and “N deglycosylation” represent genes with protein modifications as reported in supplemental Table S2B. “HighMW,” “MedMW,” and “LowMW” represent proteins with molecular weights ≤20 kDa, between 20 kDa and 100 kDa, and >100 kDa, respectively. Mean sum of dNSAF values was calculated from three (FA-ECM, NAC-ECM, and WA) and two independent experiments (NP-ECM), respectively. Statistically supported differences between mean sum of dNSAF values measured in ECM samples compared with WA were assessed using Student's t test (∗p < 0.01). Detailed p values calculated for each pair-wise comparison are reported in supplemental Table S2H. dNSAF, distributed normalized spectral abundance factor; ECM, extracellular matrix; GO, Gene Ontology; WA, nondecellularized whole animal.
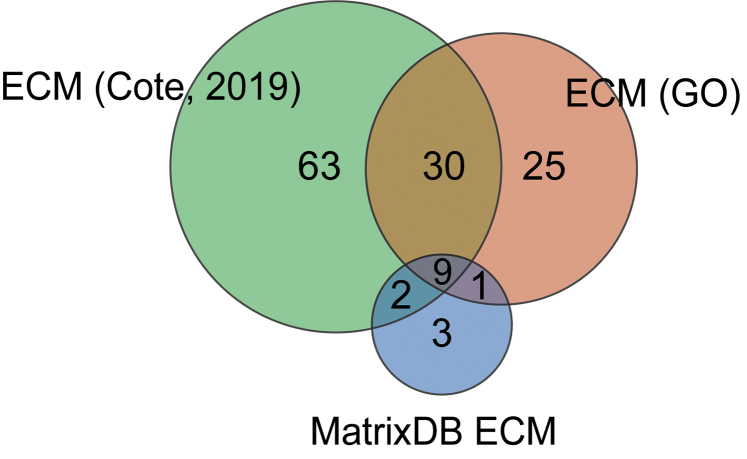
Next, we assessed whether the tissue decellularization processes we performed resulted in the expected increase in abundance of known ECM proteins over WA. The mean of the sum of dNSAF values for proteins falling into different categories of functional annotations or attributes, such as known ECM proteins, were calculated in all four sample types. Then, those values were used to calculate a fold change of enrichment or depletion between the three different decellularization procedures and WA as a baseline (Fig. 2C). Because of the difficulty of inferring ECM protein by homology in an organism evolutionarily distant from any organism with a well-characterized ECM (39) and the lack of a comprehensive annotation of planarian ECM proteins, we created a list of “known ECM” proteins. To create this list, we combined the datasets from Cote et al. (24) and MatrixDB (51) with our own list called “ECM (GO),” which consists of genes annotated with the GO term “Extracellular Matrix” or any of its child terms. Using a combination of three functional annotations instead of one should maximize our ability to identify ECM proteins in our analysis. It should be noted that each of these annotation methods could differently label the proteins found in our samples (supplemental Fig. S3). A clear enrichment of “Known ECM” was observed in all three decellularization samples (Fig. 2C), suggesting that planarian decellularization increases the likelihood of identifying ECM proteins compared with WA.
To confirm this finding, we analyzed additional hallmarks of ECM proteins, such as signal peptides since ECM proteins are secreted into the extracellular milieu (52). We therefore utilized MatrixDB and SignalP (53) to determine if proteins found in decellularized samples were known or predicted to be secreted or have signal peptides for secretion. We found that all the three decellularized samples are also enriched for secreted proteins or proteins with extracellular signaling peptide (Fig. 2C). Furthermore, there are reports that many ECM proteins are heavily glycosylated and some ECM proteins, such as collagen, are hydroxylated at prolyl and lysyl residues (54). Hence, we calculated the fold change of proteins for which we observed N-deglycosylation and K/P-hydroxylation (supplemental Table S2B) in the proteomics datasets of the samples from three decellularization procedures (Fig. 2C). The results showed that a significantly higher fraction of proteins have been modified with hydroxylation and deglycosylation in three decellularization samples compared with proteins extracted from the WA. Finally, it has been reported that most ECM proteins have high molecular weight (MW) (55). We classified planarian proteins into three bins based on the distributions of MWs in the entire S. mediterranea protein database: low MW (≤20 kDa), medium MW, and high MW (>100 kDa) (supplemental Fig. S4). We found that high MW proteins were recovered at higher abundance in all three decellularization procedures compared with WA. Examples of well-known ECM proteins that were empirically identified in our decellularized samples are nidogen 2 (SMESG000016992.1) (56), mucin-2 like (SMESG000041532.1) (57), and collagen alpha (IV) chain (SMESG000066546.1) (58). The detail of the enrichment analysis of proteins detected from WA and ECM preparations is provided in supplemental Table S2H.
Our findings indicate that our protocols are compatible with MS applications. Interestingly, the results suggested that our decellularization procedures enrich for ECM since they increase the abundance of ECM identifiers in the sample relative to the undecellularized sample. More importantly, the presence of well-reported ECM proteins confirms the success of our decellularization procedure to isolate genuine ECM material from planarians and, thus, we now call the product from decellularized planarians an ECM-enriched sample.
Different Decellularization Protocols Enrich Different Types of ECM Proteins
Previously, we showed that decellularization methods could be used for ECM identification in planarians (supplemental Fig. S2). Moreover, proteomic profiles among samples (FA-ECM, NAC-ECM, NP-ECM, and WA) could be different (Fig. 2B and supplemental Fig. S5). We tested next whether the three decellularization procedures had differences and caveats by analyzing the enrichment of proteins in each condition.
We found that some ECM components or unannotated proteins with signal peptides were specifically enriched by our decellularization procedures compared with WA extraction, as they fell on the right side of volcano plot plotting the fold change over WA baseline (supplemental Fig. S6A), such as collagen alpha (IV) chain (SMESG000066546.1) (supplemental Table S2A). This confirms that our ECM isolation procedures were able to enrich ECM. Moreover, it is worth noting that although several ECM proteins were identified in ECM-enriched samples, they were not significantly enriched compared with protein extraction from WA, such as MMP-1 (SMESG000040780.1) and MMP-2 (SMESG000014326.1) (supplemental Table S2A). Furthermore, some ECM proteins were significantly decreased in ECM-enriched samples including lectin 2c (SMESG000056364.1) and mannose-binding protein C (SMESG000046618.1) (supplemental Table S2A). Moreover, GO enrichment was analyzed to confirm the enrichment of ECM in all ECM-enriched samples. We found that all our three decellularization procedures significantly enriched proteins with ECM GO term (GO: 0031012), but WA does not (supplemental Table S2, D–G). These results indicate that although our ECM isolation procedures were able to enrich certain types of ECM proteins, our dataset might have limitations in that not every ECM protein will be enriched or some ECM proteins are lost during intensive tissue decellularization processes.
As we previously found that there are similarities and differences of ECM profiles between the three decellularization procedures, we then asked whether or not different decellularization procedures could result in enriching specific types of ECM proteins. We analyzed the distribution of MW of peptides found in each decellularization procedure (Fig. 2C). The results demonstrated that FA-ECM significantly enriched high MW proteins, whereas NP-ECM significantly enriched low MW proteins (Fig. 2C). To illustrate this point, hemicentin 1 (SMESG000021248.1) was only detected in FA-ECM and NAC-ECM, whereas the short protein cystatin-B-like (SMESG000034246.1) was only detected and significantly enriched in NP-ECM samples (supplemental Table S2C). This finding suggested that different decellularization procedures might be complementary and enrich specific types of ECM proteins.
Having demonstrated that our decellularization protocols could enrich ECM proteins over WA, we subsequently evaluated the presence of intracellular proteins in ECM-enriched samples and WA. Although we experimentally verified the depletion of one marker for intracellular proteins by Western blot (Fig. 1H), we decided to perform bioinformatics verification by comparing protein abundance from all conditions that have GO terms as cytoplasm, intracellular, mitochondrion, endoplasmic reticulum (ER), nucleus, and proteins with predicted transmembrane structure by TMHMM (DTU Health Tech) (59) (Fig. 2C). We found that the abundances of proteins labeled with GO terms as “cytoplasm,” “mitochondrion,” and “ER” were significantly decreased in all ECM-enriched samples over WA. These results demonstrated that our ECM-enriched samples are depleted for cytosolic, ER, and mitochondrial proteins. Moreover, the result indicated that GO: Nucleus is decreased in all ECM-enriched samples with statistical difference in only NP-ECM. However, when we analyzed abundances of proteins with predicted transmembrane domains or with “intracellular” GO annotations, we found that they were significantly increased in NAC-ECM. Overall, while, as a group, intracellular components are mostly depleted from the FA-ECM and NP-ECM fractions, some individual proteins are still present at significant levels, in particular in the NAC-ECM sample, which appears to be the least different from the WA samples as shown by the lowest measured fold changes compared with WA in all protein categories.
To confirm our analysis in Figure 2C and supplemental Fig. S6A, we calculated the ratio of different types of proteins (known ECM, unannotated proteins without/with signal peptide, and intracellular proteins) in the population of proteins that have been either significantly enriched or depleted in all three ECM preparation procedures compared with WA (supplemental Fig. S6B). These pie charts demonstrated that 44%, 64%, and 58% of proteins that had been enriched in NAC-ECM, NP-ECM, and FA-ECM, respectively, were “known ECM” or unannotated proteins with predicted signal peptides combined. More importantly, it showed that greater than 60% of proteins that had been depleted in all three ECM preparations were intracellular proteins. This analysis then confirms our conclusion from previous analysis in Figure 2C and supplemental Fig. S6A in that the majority of enriched proteins in NP-ECM and FA-ECM are potential ECM components, whereas the majority of depleted proteins in all three ECM preparations are intracellular proteins. In conclusion, we were able to identify bona fide ECM proteins in our enriched samples. However, it is important to recognize that our ECM proteomic inventory might not be complete because of the unavoidable loss of certain proteins during sample preparation, insufficiently deep sampling, and unavoidable intracellular contamination. Nevertheless, we demonstrated that our MS data uncovered several reported ECM proteins as well as unknown proteins of high MW and predicted to be secreted, glycosylated, and/or hydroxylated, that might be unreported ECM proteins.
Functional Analysis of Identified Proteins via RNAi
We completed our workflow by testing whether the list of proteins identified from the decellularized planarian could be used in RNAi experiments for discovering the role of ECM in both tissue homeostasis and regeneration (Fig. 3A). The criteria for selecting gene candidates for RNAi were that they should be presented in either FA-ECM, NAC-ECM, or NP-ECM, and their gene identity was not necessarily defined as ECM gene by only Cote et al. or our ECM (GO). These criteria allowed for the possibility to uncover unannotated ECM genes that function in tissue regeneration. A summary of the data for each targeted gene and their ECM status is elaborated in supplemental Table S3.
Fig. 3.
RNAi screening of empirical ECM dataset discovers two genes of interest involved in tissue homeostasis and/or regeneration.A, workflow for dsRNA feeding for RNAi screening. B, dsRNA feeding schedule. C, cartoon illustrating the time line for homeostasis and regeneration assays. D, characteristic phenotypes of planarians in homeostasis assay. The scale bars represent 500 μm. Phenotype of planarians with RNAi against unc-22 (RNAi) as negative control. Phenotype of planarians with RNAi against tolloid-like protein 1 (tll). E, characteristic phenotype of planarians with RNAi against unc-22 (RNAi) as negative control. Phenotype of planarians with RNAi against basement membrane heparan sulfate proteoglycan (hspg). The red arrow indicates the hump-like structure. Phenotype of planarians with RNAi against tll. The yellow arrow indicates a rough dorsal surface. F, survival rate of RNAi-fed planarians 1, 3, 5, 7, 9, and 10 days after sagittal amputation. The error bars represent standard deviation. Stars indicate statistically significant differences, as calculated by ANOVA (∗p < 0.05), compared with unc-22 as the negative control. Biological replicates of RNAi-treated worms of unc-22, hspg, and tll worms were 3, 6, and 5, respectively. ECM, extracellular matrix.
Thirty nine proteins were selected for analysis via RNAi as well as one previously published ECM protein (tolloid-like protein 1 [tll]) as a positive control for known ECM proteins that were not present in our inventory (60). The worms were fed with bacteria expressing dsRNA of the target gene every 3 days for a total of ten feedings (Fig. 3B). Upon completion of the RNAi feeding schedule, planarians were subjected to two assays to examine the role of the genes of interest during homeostasis and regeneration (Fig. 3C). In the regeneration assay, worms were amputated sagittally to avoid any biases because of the unknown distribution of ECM proteins along anterior–posterior axis.
After 1 month of RNAi, the animals were indistinguishable from control RNAi animals except for the positive control tll RNAi (Fig. 3D). However, after animals were amputated sagittally to score for regeneration, we identified roles for one gene encoding the basement membrane heparan sulfate proteoglycan (hspg) (Fig. 3E). Negative controls showed normally regenerated tissue, as indicated by the presence of eye spots on the new tissue with an unpigmented blastema. In contrast, the hspg-knockdown worms showed a bulging structure at the pharyngeal position, indicated with the red arrow, whereas the regenerated blastema and eye spots were normal. As expected, tll, which previously showed a phenotype in the homeostasis assay (Fig. 3D), was also found to have a phenotype in the regeneration assay (Fig. 3E). The worms failed to produce a blastema and eye spots, indicated by a lack of unpigmented tissue at the wounded edge; the dorsal surface of the planarians also appeared to be rough, indicated with the yellow arrow. Moreover, the tll(RNAi) animals were ventrally curled, particularly at the tail region, in a manner that was similar to the analogous worms at homeostasis. In total, one ECM gene was positively identified from regeneration assays, and the reduction of gene expression in all RNAi conditions was confirmed by quantitative polymerase chain reaction (supplemental Fig. S7).
Because some animals died during the regeneration assay, we then conducted a survival assay to quantify the lethality of these two RNAi conditions. hspg(RNAi) animals showed reduced survival during regeneration, whereas 100% of the negative control animals fully regenerated and survived (Fig. 3F). Only 15% of tll-knockdown worms survived at 1 day postamputation (dpa), whereas the percentage of survival for hspg-knockdown worms was 68% at the same time point. At the endpoint, the survival of RNAi-treated worms at 10 dpa was 67% and 10% for hspg and tll, respectively. These results indicated that the knockdown worms of hspg and tll genes were sensitive to injury than negative control animals since their survival rates dropped sharply on the first day after amputation. This suggests that these genes could be required for the survival of planarians at the early stage of tissue injury.
To summarize, we conclude that hspg is important for tissue regeneration. Because we were able to replicate the previously reported phenotype for the predicted ECM gene tll, we conclude that our RNAi assays are reliable and could provide insights on contribution of a particular ECM gene in homeostasis and regeneration. We therefore anticipated that the list generated from decellularized planarian MS could be used for RNAi screening.
Preliminary Characterization of hspg Identified From Our Proposed Workflow
In order to further investigate the putative function of hspg, the protein domain composition of hspg was bioinformatically predicted and compared with its human homolog (supplemental Fig. S8). The domain analysis showed that the HSPG protein contained several domains such as immunoglobulin, low-density lipoprotein receptor domain class A, laminin B domain, and epidermal growth factor–like domain. For the ECM positive control gene, the TLL protein possesses a zinc-dependent metalloprotease domain with several repeats of CUB domains and calcium-binding epidermal growth factor–like domain. These results suggest the potential function of each protein behind our discovered phenotypes in knockdown animals: hspg likely encodes a large core matrix structural protein.
In addition, because we discovered roles for hspg in regeneration, we therefore sought to determine whether these genes were expressed in stem cells. We used γ-irradiation to eliminate neoblasts, followed by whole-mount in situ hybridization to visualize the gene expression patterns (Fig. 4A). Prior to irradiation, hspg is expressed all over the body of planarians, especially at the pharynx. The expression of tll was also found to be well distributed in every part of the worm body. Interestingly, while irradiation eliminated the probe signal for our neoblast marker, piwi-1, it did not eradicate the probe signals for hspg and tll, suggesting that they are enriched in differentiated cells. In order to pinpoint the cell type responsible for expressing hspg, we consulted the planarian single cell RNA-Seq database Digiworm (49). The results showed that hspg and tll were highly expressed in muscle cells (Fig. 4B). Both whole-mount in situ hybridization and single-cell RNA-Seq suggest that hspg and tll are mainly expressed in differentiated cells, such as gut, muscle, or epidermis.
Fig. 4.
Spatiotemporal characterization of genes of interest during tissue homeostasis and regeneration.A, in situ hybridization of genes of interest on unamputated planarians prior to, and 2 days after, irradiation. The scale bars represent 500 μm. B, the reference atlas of RNA single-cell sequencing published by Fincher et al. (49) and the relative expression of hspg and tll in each cell type in the RNA single-cell sequencing. Different dot colors indicate differences in level of gene expression. C and D, in situ hybridization of (C) hspg and (D) tll on sagittally amputated planarians regenerated for 1, 3, 7, and 13 days. All scale bars represent 500 μm. hspg, heparan sulfate proteoglycan; tll, tolloid-like protein 1.
As hspg affects tissue regeneration in amputated planarians, we used in situ hybridization to determine if the expression patterns of hspg genes change during regeneration. While the original pattern of hspg remained, after 1 dpa, there was an increase in expression at the location of the original pharynx (Fig. 4C). At 3 dpa, hspg global expression was reduced, whereas prominent expression of hspg remained in the newly formed pharynx area. The signal of hspg expression in the pharynx was persistent throughout the regeneration process. Finally, the expression of tll decreased globally at 1 dpa, and only a slight signal of tll expression was observed at the edge of the wound (Fig. 4D). However, the signal reemerged in the newly regenerated blastema at 3 dpa, and its expression was persistent particularly at the newly regenerated blastema from 7 dpa onward. Our results suggest that hspgs are expressed mostly in differentiated cells such as muscle or epithelial cells. Moreover, hspg responds to tissue amputation as shown by dynamic changes in their gene expression patterns following wounding. Taken together, these results indicate the role of hspg in the physiological response of planarian regeneration.
Ultimately, we have demonstrated the potential of our newly described decellularization procedures for enriching ECM from planarians for use in many downstream applications. Here, we showed that the isolated ECM can be used for a protein MS pipeline, from which the first experimentally based ECM proteomic database has been established. This work empowers systematic study of planarian ECM to characterize its function in tissue homeostasis and regeneration.
Discussion
The study of ECM in highly regenerative systems is currently limited, preventing us from understanding how the ECM may or may not contribute to tissue regeneration (21, 22, 23). To overcome this limitation, we established an integrative workflow consisting of tissue decellularization, proteomics, and RNA-mediated genetic interference (RNAi) for the purification and characterization of planarian ECM.
The quality of isolated ECM is a significant parameter for its downstream characterization. Our decellularization procedure resulted in the production of a full-body ECM scaffold from S. mediterranea possessing both little residual genomic DNA and a well-defined collagen sheet in the case of FA-ECM (Fig. 1). Both these parameters are generally used to gauge the quality of purified ECM after intensive decellularization (61, 62, 63, 64). We were able to optimize the published method for D. japonica for use with S. mediterranea by increasing the concentration of SDS used in the decellularization solution (25). Not only does our procedure produce an enriched-ECM sample but also it suggests that a single protocol with a few customizations may ultimately be applicable to a broad diversity of planarians. We have developed three separate ECM sample preparation methods that can be used for various downstream applications. Samples from all three sample methods were subjected to protein MS. This allowed us to characterize the protein composition of each sample preparation method as well as identify bona fide planarian ECM proteins.
We found that all our three decellularization techniques could be used for protein MS (supplemental Fig. S2). The proteins observed among the different methods employed for decellularization are in good agreement with previously published analyses (65, 66). Moreover, our collective results suggest that each ECM preparation has their own advantages and disadvantages. Considering the completeness of tissue structure after tissue decellularization, we anticipated that FA-ECM could be the best option since it captures nearly complete anatomical structure of planarians and may contain nearly complete ECM proteins as well (Fig. 1E). In fact, enrichment analysis revealed that FA-ECM enriched all queried properties for ECM (known ECM, secreted, hydroxylation, and deglycosylation) as well as high MW proteins (Fig. 2C). While it has been reported that FA crosslinks are labile and cleavable during the MS fragmentation process (67), the number of protein counts for FA-ECM was the lowest among the three ECM preparations (supplemental Fig. S2). Nevertheless, the intact structure of FA-ECM makes it to be the best procedure for preserving ECM in their natural state. The drawbacks of the FA-ECM decellularization procedure could be further addressed by optimizing sample preparation procedure prior to MS. This will make FA-ECM decellularization procedure a promising tool to interrogate ECM in planarians. For the NAC-ECM and NP-ECM preparations, we found that they provided a high number of protein counts (supplemental Fig. S2). We hypothesized that since they do not have FA crosslinking involved, protein digestion and solubilization treatments are more complete. Although these seem to be suitable methods for proteomic analysis, we anticipated that since these decellularized animals have less-intact structure compared with FA-ECM (Fig. 1, D–F), they might not be the best option to capture the whole ECM landscape in planarians. Protein enrichment analysis showed NAC-ECM and NP-ECM might not be suitable for the enrichment of high MW proteins (Fig. 2C). Nevertheless, it is worth pointing out that our decellularization did not contain many protease inhibitors. It might be possible that a large MW ECM could be partially cleaved during ECM extraction. Overall, although each of our three planarian decellularization methods for proteomic analysis have their own advantages and disadvantages, our data show that they are all able to enrich ECM proteins (Fig. 2C and supplemental Fig. S6). The choice of ECM decellularization procedure therefore depends on the purpose of the experiment and downstream applications.
It is important to note that the list of proteins we identified from ECM-enriched samples (supplemental Table S2A) might not fully capture the complete matrisome because of several caveats. First, there are unintended losses of ECM proteins during the sample preparation process, in particular during the extensive decellularization with high concentration of SDS and washing steps (68). Some well-defined ECM proteins in the computationally derived ECM catalog (24), such as lysyl oxidase 3, netrin 5, and fibrillin 2, are missing from our dataset. Moreover, although the laminin subunit β (SMESG000062589.1) was detected in our ECM-enriched samples, the other laminin subunits were not detected. Nevertheless, we found a strong degree of overlap between our dataset and the computationally derived ECM catalog, including proteins such as MMP (SMESG000049445.1), neurogenic locus notch-1; Tenascin (SMESG000041513.1), and SPARC-like protein (SMESG000024280.1) (24). Second, there might be some residual intracellular protein contaminants in our ECM preperations that might interfere with our analysis. Some examples of known intracellular proteins found in our dataset are nuclear lamin (SMESG000009886.1), dynein light chain (SMESG000007775.1), and tubulin alpha chain (SMESG000009951.1). Third, the incomplete annotation of the planarian genome and proteome led to the identification of several unannotated proteins in our dataset. Interestingly, one unknown protein (SMESG000081459.1) was only abundant in all three ECM preparation samples and not in the WA. This suggests that our ECM dataset potentially reveals novel and uncharacterized components of the planarian ECM. Our research group is looking forward to characterizing those unknown proteins in a future work. Finally, the high abundance of major ECM proteins such as collagen might interfere with the detection of a minor population of ECM proteins by MS (69). We discovered different types of collagen protein such as collagen alpha-1 (II) chain (SMESG000026651.1) and collagen alpha-2 (I) chain (SMESG000024244.1). It would be possible to address this in a future experiment via collagenase treatment after decellularization or prior to proteomic analysis.
The proteomics of decellularized planarians offered the possibility of initiating an RNAi screen to discover novel ECM genes responsible for tissue homeostasis and regeneration in planarians (Fig. 3A). Our workflow identified new roles in planarian tissue regeneration for a homolog of a known mammalian ECM gene, hspg (70, 71) (Fig. 3E). The expression of hspg during both homeostasis and regeneration is mostly localized to muscle, particularly in the pharynx that is mainly composed of muscle cells (72). Based on the RNAi phenotype (Fig. 3E), we hypothesized that hspg likely plays a role in pharynx regeneration. hspg is a long carbohydrate chain with repeating units of disaccharide and acts as a component of the basement membrane (73). We hypothesized that hspg might also be a component of the planarian basement membrane as well. Interestingly, planarian hspg has a similar mRNA expression pattern to megf6 and hemicentin in that the gene expression is mainly located at the pharynx of the animal and low levels across the rest of the body (74). Moreover, they demonstrated that megf6 and hemicentin are important for maintaining basement membrane integrity. The disruption of membrane causes hyperproliferation, which results in ectopic lesions. Our hspg(RNAi) knockdown planarians also showed ectopic tissue growth as well. Therefore, we hypothesize that hspg might also function with megf6 and hemicentin in maintaining the basement membrane and preventing ectopic tissue growth outside the basement membrane. Further investigation is needed to verify this hypothesis.
tll has been selected as a positive control gene for RNAi screening because it has been previously predicted as an ECM protein (24), and it has a reproducible phenotype (60). tll is essential for dorsoventral polarity regulation throughout the animal kingdom (75). In ECM biology, tll might serve as a secreted ECM-modification protein in addition to participating in the bone morphogenetic protein pathway. We selected tll as a positive control to confirm whether our RNAi screening scheme is able to identify phenotypes from dsRNA feeding of ECM genes (Fig. 3, B and C). Expectedly, knockdown of tll resulted in obvious and strong phenotypes (Fig. 3D). Therefore, our RNAi screening and phenotyping method is reproducible and should be able to screen ECM genes essential for tissue homeostasis and regeneration in planarians.
Because our target gene list was generated directly from decellularized planarian MS, we believe that any gene candidates successfully identified from our workflow deserve to be characterized in detail. Therefore, it might be possible to discover novel ECM genes that have been bioinformatically missed since they do not share a well-annotated ECM domain by computational prediction. We anticipated that the unannotated proteins with potential signal peptides are a good source of potentially novel and planaria-specific ECM proteins, especially if those proteins also contain N-glycosylation and K or P hydroxylation sites such as unknown protein (SMESG000076927.1) enriched in NAC-ECM (supplemental Table S2C). As a whole, the discovery of hspg, which was identified and enriched in FA-ECM and NP-ECM (supplemental Table S3), with promising functions and obvious knockdown phenotypes supports the validity of our decellularization-based ECM MS approach to target gene candidates for RNAi screening in planarians.
Apart from the proof-of-concept workflow proposed in this article, our three optimized ECM decellularization strategies also have the potential to empower new directions in planarian research. Feasible applications for each type of ECM are predicted as follows. FA-ECM can be used for imaging purposes such as histological section staining (Fig. 1I) and electron microscopy (Fig. 1J) because of its well-preserved structure. Moreover, FA-ECM offers a promising avenue to investigate spatial organization of the planarian matrisome through immunological assays as well, since it has been reported that ECM is not evenly distributed throughout the tissue and organism (76). In particular, this would provide the opportunity to investigate the distribution of planarian ECM across body axes to study its role in controlling polarity in an animal. NAC-ECM also yields a decent structure without toxic FA pretreatment. This structure may be of use in developing, much needed, techniques to culture planarian neoblasts in vitro (77). NAC-ECM could be utilized as a biological scaffold for neoblast seeding sites as illustrated by neoblast injection into FA-ECM isolated from D. japonica (25). Because of the fact that NAC-ECM does not have FA-pretreatment steps, this would likely improve the survival rate of injected neoblasts. In the case of NP-ECM, it could be applied in neoblast cell culture as well by adding NP-ECM in the culture media as a supplementary ingredient.
In conclusion, our proposed workflow is practical for discovering ECM protein function in planarian tissue regeneration and homeostasis. The protein components of decellularized planarians include putative ECM targets that can be used for testing gene function by in vivo RNAi screening. Our work thus sets the stage for investigating the involvement of different ECM components in complex biological processes, offering a unique approach for examining in detail the effects the cellular microenvironment may play in stem cell fate regulation and determination during tissue homeostasis and whole-body regeneration. More importantly, our work highlights the potential of planarians as an emerging model system for the study of ECM biology and their remodeling in the process of tissue regeneration.
Data Availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium with identifier PXD024753 and may be accessed through the MassIVE partner repository via FTP with username MSV000087054. Original data underlying this article may also be accessed after publication from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1407.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
We are grateful for the support received from members of the research core facilities at the Stowers Institute for Medical Research including all planaria core facility personnel; Yan Hao, proteomics center; Nancy Thomas, histology facility; Dan Bradford and Michael Peterson, molecular biology facility; Melainia McClain, electron microscopy center; and Cindy Maddera, microscopy center. We also appreciate constructive comments from Lauren Cote following our preprint publication. We appreciate the valuable input from every ASA laboratory member. Works in Ounjai laboratory are supported by the Center of Excellence on Environmental Health and Toxicology, Faculty of Science, Mahidol University, and the Thailand Science Research and Innovation. Moreover, we also thank Ekarat Sonpho for editing the video material, Jirawat Salungyu for assistance in statistical analysis, Nattarat Punyasu for her idea in the graphical abstract, and Sirapob Phootsri for the beautiful artworks.
Funding and additional information
We acknowledge the financial support to E. S. from Junior Research Scholarship Program, Fulbright Program during his stay at the Stowers Institute for Medical Research, USA. A. S. A., E. J. R., and F. G. M., Jr, are supported by Howard Hughes Medical Institute.
Author contributions
E. S. and A. S. A. conceptualization; E. J. R. software; M. L., E. J. R., L. F., and A. S. formal analyses; F. G. M., Jr, M. L., C. G.-H., L. F., A. S., and V. D. investigation; M. L., E. J. R., L. F., and A. S. data curation; E. S., L. F., V. D., and P. O. writing–original draft; F. G. M., Jr, L. F., V. D., and P. O. writing–review and editing; C. G.-H. visualization; P. O. and A. S. A. supervision; A. S. A. project administration; A. S. A. funding acquisition.
Footnotes
Present address for Michaella Levy: KCAS, Shawnee, Kansas 66126, USA.
Present address for Anita Saraf: University of Kansas, Lawrence, Kansas 66047, USA.
Supplemental Data
Supplemental Figure S1.
Supplemental Figure S2.
Supplemental Figure S3.
Supplemental Figure S4.
Supplemental Figure S5.
Supplemental Figure S6.
Supplemental Figure S7.

Supplemental Figure S8.
References
- 1.Galliot B., Crescenzi M., Jacinto A., Tajbakhsh S. Trends in tissue repair and regeneration. Development. 2017;144:357–364. doi: 10.1242/dev.144279. [DOI] [PubMed] [Google Scholar]
- 2.Poss K.D. Getting to the heart of regeneration in zebrafish. Semin. Cell Dev. Biol. 2007;18:36–45. doi: 10.1016/j.semcdb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Wang J., Karra R., Dickson A.L., Poss K.D. Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev. Biol. 2013;382:427–435. doi: 10.1016/j.ydbio.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lukjanenko L., Jung M.J., Hegde N., Perruisseau-Carrier C., Migliavacca E., Rozo M., Karaz S., Jacot G., Schmidt M., Li L., Metairon S., Raymond F., Lee U., Sizzano F., Wilson D.H. Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nat. Med. 2016;22:897–905. doi: 10.1038/nm.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godwin J.W., Debuque R., Salimova E., Rosenthal N.A. Heart regeneration in the salamander relies on macrophage-mediated control of fibroblast activation and the extracellular landscape. NPJ Regen. Med. 2017;2:22. doi: 10.1038/s41536-017-0027-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daley W.P., Peters S.B., Larsen M. Extracellular matrix dynamics in development and regenerative medicine. J. Cell Sci. 2008;121:255–264. doi: 10.1242/jcs.006064. [DOI] [PubMed] [Google Scholar]
- 7.Schultz G.S., Wysocki A. Interactions between extracellular matrix and growth factors in wound healing. Wound Repair Regen. 2009;17:153–162. doi: 10.1111/j.1524-475X.2009.00466.x. [DOI] [PubMed] [Google Scholar]
- 8.Koochekpour S., Merzak A., Pilkington G.J. Extracellular matrix proteins inhibit proliferation, upregulate migration and induce morphological changes in human glioma cell lines. Eur. J. Cancer. 1995;31A:375–380. doi: 10.1016/0959-8049(94)00476-l. [DOI] [PubMed] [Google Scholar]
- 9.Bosnakovski D., Mizuno M., Kim G., Takagi S., Okumura M., Fujinaga T. Chondrogenic differentiation of bovine bone marrow mesenchymal stem cells (MSCs) in different hydrogels: Influence of collagen type II extracellular matrix on MSC chondrogenesis. Biotechnol. Bioeng. 2006;93:1152–1163. doi: 10.1002/bit.20828. [DOI] [PubMed] [Google Scholar]
- 10.Zhen G., Cao X. Targeting TGFβ signaling in subchondral bone and articular cartilage homeostasis. Trends Pharmacol. Sci. 2014;35:227–236. doi: 10.1016/j.tips.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonnans C., Chou J., Werb Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.You J., Zhang Y., Li Z., Lou Z., Jin L., Lin X. Drosophila perlecan regulates intestinal stem cell activity via cell-matrix attachment. Stem Cell Reports. 2014;2:761–769. doi: 10.1016/j.stemcr.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgner J., Ghatak S., Jakobi T., Dieterich C., Aumailley M., Wickström S.A. Integrin-linked kinase regulates the niche of quiescent epidermal stem cells. Nat. Commun. 2015;6:8198. doi: 10.1038/ncomms9198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seyedhassantehrani N., Otsuka T., Singh S., Gardiner D.M. The axolotl limb regeneration model as a discovery tool for engineering the stem cell niche. Curr. Stem Cell Rep. 2017;3:156–163. doi: 10.1007/s40778-017-0085-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarras M.P. Components, structure, biogenesis and function of the hydra extracellular matrix in regeneration, pattern formation and cell differentiation. Int. J. Dev. Biol. 2012;56:567–576. doi: 10.1387/ijdb.113445ms. [DOI] [PubMed] [Google Scholar]
- 16.Phan A.Q., Lee J., Oei M., Flath C., Hwe C., Mariano R., Vu T., Shu C., Dinh A., Simkin J., Muneoka K., Bryant S.V., Gardiner D.M. Positional information in axolotl and mouse limb extracellular matrix is mediated via heparan sulfate and fibroblast growth factor during limb regeneration in the axolotl (Ambystoma mexicanum) Regeneration (Oxf.) 2015;2:182–201. doi: 10.1002/reg2.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sánchez-Iranzo H., Galardi-Castilla M., Sanz-Morejón A., González-Rosa J.M., Costa R., Ernst A., Sainz de Aja J., Langa X., Mercader N. Transient fibrosis resolves via fibroblast inactivation in the regenerating zebrafish heart. Proc. Natl. Acad. Sci. U. S. A. 2018;115:4188–4193. doi: 10.1073/pnas.1716713115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogg M.C., Galliot B., Tsiairis C.D. Model systems for regeneration: Hydra. Development. 2019;146 doi: 10.1242/dev.177212. [DOI] [PubMed] [Google Scholar]
- 19.Reddien P.W., Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 20.Reddien P.W., Bermange A.L., Murfitt K.J., Jennings J.R., Sánchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell. 2005;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isolani M.E., Abril J.F., Saló E., Deri P., Bianucci A.M., Batistoni R. Planarians as a model to assess in vivo the role of matrix metalloproteinase genes during homeostasis and regeneration. PLoS One. 2013;8 doi: 10.1371/journal.pone.0055649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seebeck F., März M., Meyer A.W., Reuter H., Vogg M.C., Stehling M., Mildner K., Zeuschner D., Rabert F., Bartscherer K. Integrins are required for tissue organization and restriction of neurogenesis in regenerating planarians. Development. 2017;144:795–807. doi: 10.1242/dev.139774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan A., Ma S., Pearson B.J., Chan D. Collagen IV differentially regulates planarian stem cell potency and lineage progression. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2021251118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cote L.E., Simental E., Reddien P.W. Muscle functions as a connective tissue and source of extracellular matrix in planarians. Nat. Commun. 2019;10:1592. doi: 10.1038/s41467-019-09539-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonpho E., Wootthichairangsan C., Ishida M., Inoue T., Agata K., Maleehuan A., Charngkaew K., Chomanee N., Moonsom S., Wongtrakoongate P., Chairoungdua A., Ounjai P. ECM-body: A cell-free 3D biomimetic scaffold derived from intact planarian body. Zoolog. Sci. 2020;37:307–313. doi: 10.2108/zs190135. [DOI] [PubMed] [Google Scholar]
- 26.Newmark P.A., Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev. Biol. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- 27.Carson F.L., Hladik C. 3rd Ed. ASCP Press; Chicago, IL: 2009. Histotechnology: A Self-Instructional Text; pp. 162–165. [Google Scholar]
- 28.Jongebloed W.L., Stokroos I., Van Der Want J.J., Kalicharan D. Non-coating fixation techniques or redundancy of conductive coating, low kV FE-SEM operation and combined SEM/TEM of biological tissues. J. Microsc. 1999;193:158–170. doi: 10.1046/j.1365-2818.1999.00430.x. [DOI] [PubMed] [Google Scholar]
- 29.Washburn M.P., Wolters D., Yates J.R. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Wen Z., Washburn M.P., Florens L. Effect of dynamic exclusion duration on spectral count based quantitative proteomics. Anal. Chem. 2009;81:6317–6326. doi: 10.1021/ac9004887. [DOI] [PubMed] [Google Scholar]
- 31.McDonald W.H., Tabb D.L., Sadygov R.G., MacCoss M.J., Venable J., Graumann J., Johnson J.R., Cociorva D., Yates J.R. MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 2004;18:2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y., Wen Z., Washburn M.P., Florens L. Improving proteomics mass accuracy by dynamic offline lock mass. Anal. Chem. 2011;83:9344–9351. doi: 10.1021/ac201867h. [DOI] [PubMed] [Google Scholar]
- 33.Xu T., Park S.K., Venable J.D., Wohlschlegel J.A., Diedrich J.K., Cociorva D., Lu B., Liao L., Hewel J., Han X., Wong C.C.L., Fonslow B., Delahunty C., Gao Y., Shah H. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteomics. 2015;129:16–24. doi: 10.1016/j.jprot.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabb D.L., McDonald W.H., Yates J.R. DTASelect and contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Wen Z., Washburn M.P., Florens L. Refinements to label free proteome quantitation: How to deal with peptides shared by multiple proteins. Anal. Chem. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 36.Choi H., Kim S., Fermin D., Tsou C.C., Nesvizhskii A.I. QPROT: Statistical method for testing differential expression using protein-level intensity data in label-free quantitative proteomics. J. Proteomics. 2015;129:121–126. doi: 10.1016/j.jprot.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rozanski A., Moon H., Brandl H., Martín-Durán J.M., Grohme M.A., Hüttner K., Bartscherer K., Henry I., Rink J.C. PlanMine 3.0-improvements to a mineable resource of flatworm biology and biodiversity. Nucleic Acids Res. 2019;47:D812–D820. doi: 10.1093/nar/gky1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grohme M.A., Schloissnig S., Rozanski A., Pippel M., Young G.R., Winkler S., Brandl H., Henry I., Dahl A., Powell S., Hiller M., Myers E., Rink J.C. The genome of Schmidtea mediterranea and the evolution of core cellular mechanisms. Nature. 2018;554:56–61. doi: 10.1038/nature25473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Götz S., García-Gómez J.M., Terol J., Williams T.D., Nagaraj S.H., Nueda M.J., Robles M., Talón M., Dopazo J., Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bateman A. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., Pesseat S., Quinn A.F., Sangrador-Vegas A., Scheremetjew M., Yong S.Y. InterProScan 5: Genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas P.D., Kejariwal A., Campbell M.J., Mi H., Diemer K., Guo N., Ladunga I., Ulitsky-Lazareva B., Muruganujan A., Rabkin S., Vandergriff J.A., Doremieux O. PANTHER: A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alexa A., Rahnenfuhrer J. R package version 2.44.0; 2018. topGO: Enrichment Analysis for Gene Ontology. [Google Scholar]
- 45.Rouhana L., Weiss J.A., Forsthoefel D.J., Lee H., King R.S., Inoue T., Shibata N., Agata K., Newmark P.A. RNA interference by feeding in vitro-synthesized double-stranded RNA to planarians: Methodology and dynamics. Dev. Dyn. 2013;242:718–730. doi: 10.1002/dvdy.23950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arnold C.P., Merryman M.S., Harris-Arnold A., McKinney S.A., Seidel C.W., Loethen S., Proctor K.N., Guo L., Sánchez Alvarado A. Pathogenic shifts in endogenous microbiota impede tissue regeneration via distinct activation of TAK1/MKK/p38. Elife. 2016;5 doi: 10.7554/eLife.16793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forsthoefel D.J., James N.P., Escobar D.J., Stary J.M., Vieira A.P., Waters F.A., Newmark P.A. An RNAi screen reveals intestinal regulators of branching morphogenesis, differentiation, and stem cell proliferation in planarians. Dev. Cell. 2012;23:691–704. doi: 10.1016/j.devcel.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearson B.J., Eisenhoffer G.T., Gurley K.A., Rink J.C., Miller D.E., Sánchez Alvarado A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev. Dyn. 2009;238:443–450. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fincher C.T., Wurtzel O., de Hoog T., Kravarik K.M., Reddien P.W. Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science. 2018;360 doi: 10.1126/science.aaq1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brandl H., Moon H., Vila-Farré M., Liu S.Y., Henry I., Rink J.C. PlanMine--a mineable resource of planarian biology and biodiversity. Nucleic Acids Res. 2016;44:D764–D773. doi: 10.1093/nar/gkv1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clerc O., Deniaud M., Vallet S.D., Naba A., Rivet A., Perez S., Thierry-Mieg N., Ricard-Blum S. MatrixDB: Integration of new data with a focus on glycosaminoglycan interactions. Nucleic Acids Res. 2019;47:D376–D381. doi: 10.1093/nar/gky1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Unlu G., Levic D.S., Melville D.B., Knapik E.W. Trafficking mechanisms of extracellular matrix macromolecules: Insights from vertebrate development and human diseases. Int. J. Biochem. Cell Biol. 2014;47:57–67. doi: 10.1016/j.biocel.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Almagro Armenteros J.J., Tsirigos K.D., Sønderby C.K., Petersen T.N., Winther O., Brunak S., von Heijne G., Nielsen H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 54.Rodriguez-Pascual F., Slatter D.A. Collagen cross-linking: Insights on the evolution of metazoan extracellular matrix. Sci. Rep. 2016;6:1–7. doi: 10.1038/srep37374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goddard E.T., Hill R.C., Barrett A., Betts C., Guo Q., Maller O., Borges V.F., Hansen K.C., Schedin P. Quantitative extracellular matrix proteomics to study mammary and liver tissue microenvironments. Int. J. Biochem. Cell Biol. 2016;81:223–232. doi: 10.1016/j.biocel.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kohfeldt E., Sasaki T., Göhring W., Timpl R. Nidogen-2: A new basement membrane protein with diverse binding properties. J. Mol. Biol. 1998;282:99–109. doi: 10.1006/jmbi.1998.2004. [DOI] [PubMed] [Google Scholar]
- 57.Ciborowski P., Finn O.J. Non-glycosylated tandem repeats of MUC1 facilitate attachment of breast tumor cells to normal human lung tissue and immobilized extracellular matrix proteins (ECM) in vitro: Potential role in metastasis. Clin. Exp. Metastasis. 2002;19:339–345. doi: 10.1023/a:1015590515957. [DOI] [PubMed] [Google Scholar]
- 58.Ricard-Blum S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krogh A., Larsson B., Von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 60.Reddien P.W., Bermange A.L., Kicza A.M., Sánchez Alvarado A. BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development. 2007;134:4043–4051. doi: 10.1242/dev.007138. [DOI] [PubMed] [Google Scholar]
- 61.Gilbert T.W., Freund J.M., Badylak S.F. Quantification of DNA in biologic scaffold materials. J. Surg. Res. 2009;152:135–139. doi: 10.1016/j.jss.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hynes R.O. The extracellular matrix: Not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ross E.A., Williams M.J., Hamazaki T., Terada N., Clapp W.L., Adin C., Ellison G.W., Jorgensen M., Batich C.D. Embryonic stem cells proliferate and differentiate when seeded into kidney scaffolds. J. Am. Soc. Nephrol. 2009;20:2338–2347. doi: 10.1681/ASN.2008111196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakayama K.H., Batchelder C.A., Lee C.I., Tarantal A.F. Decellularized rhesus monkey kidney as a three-dimensional scaffold for renal tissue engineering. Tissue Eng. Part A. 2010;16:2207–2216. doi: 10.1089/ten.tea.2009.0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Didangelos A., Yin X., Mandal K., Baumert M., Jahangiri M., Mayr M. Proteomics characterization of extracellular space components in the human aorta. Mol. Cell. Proteomics. 2010;9:2048–2062. doi: 10.1074/mcp.M110.001693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coronado R.E., Somaraki-Cormier M., Natesan S., Christy R.J., Ong J.L., Halff G.A. Decellularization and solubilization of porcine liver for use as a substrate for porcine hepatocyte culture: Method optimization and comparison. Cell Transplant. 2017;26:1840–1854. doi: 10.1177/0963689717742157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tayri-Wilk T., Slavin M., Zamel J., Blass A., Cohen S., Motzik A., Sun X., Shalev D.E., Ram O., Kalisman N. Mass spectrometry reveals the chemistry of formaldehyde cross-linking in structured proteins. Nat. Commun. 2020;11:3128. doi: 10.1038/s41467-020-16935-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Q., Uygun B.E., Geerts S., Ozer S., Scalf M., Gilpin S.E., Ott H.C., Yarmush M.L., Smith L.M., Welham N.V., Frey B.L. Proteomic analysis of naturally-sourced biological scaffolds. Biomaterials. 2016;75:37–46. doi: 10.1016/j.biomaterials.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Canty E.G., Kadler K.E. Procollagen trafficking, processing and fibrillogenesis. J. Cell Sci. 2005;118:1341–1353. doi: 10.1242/jcs.01731. [DOI] [PubMed] [Google Scholar]
- 70.Christianson H.C., Belting M. Heparan sulfate proteoglycan as a cell-surface endocytosis receptor. Matrix Biol. 2014;35:51–55. doi: 10.1016/j.matbio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 71.Park H., Kim M., Kim H.J., Lee Y., Seo Y., Pham C.D., Lee J., Byun S.J., Kwon M.H. Heparan sulfate proteoglycans (HSPGs) and chondroitin sulfate proteoglycans (CSPGs) function as endocytic receptors for an internalizing anti-nucleic acid antibody. Sci. Rep. 2017;7:14373. doi: 10.1038/s41598-017-14793-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kobayashi C., Kobayashi S., Orii H., Watanabe K., Agata K. Identification of two distinct muscles in the planarian Dugesia japonica by their expression of myosin heavy chain genes. Zoolog. Sci. 1998;15:861–869. [Google Scholar]
- 73.Halfter W., Dong S., Schurer B., Cole G.J. Collagen XVIII is a basement membrane heparan sulfate proteoglycan. J. Biol. Chem. 1998;273:25404–25412. doi: 10.1074/jbc.273.39.25404. [DOI] [PubMed] [Google Scholar]
- 74.Lindsay-Mosher N., Chan A., Pearson B.J. Planarian EGF repeat-containing genes megf6 and hemicentin are required to restrict the stem cell compartment. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tucker J.A., Mintzer K.A., Mullins M.C. The BMP signaling gradient patterns dorsoventral tissues in a temporally progressive manner along the anteroposterior axis. Dev. Cell. 2008;14:108–119. doi: 10.1016/j.devcel.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu P., Takai K., Weaver V.M., Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lei K., McKinney S.A., Ross E.J., Lee H.C., Sánchez Alvarado A. Cultured pluripotent planarian stem cells retain potency and express proteins from exogenously introduced mRNAs. bioRxiv. 2019 doi: 10.1101/573725. [preprint] [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium with identifier PXD024753 and may be accessed through the MassIVE partner repository via FTP with username MSV000087054. Original data underlying this article may also be accessed after publication from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1407.