Abstract
The initial step of simian virus 40 (SV40) DNA replication is the binding of the large tumor antigen (T-Ag) to the SV40 core origin. In the presence of Mg2+ and ATP, T-Ag forms a double-hexamer complex covering the complete core origin. By using electron microscopy and negative staining, we visualized for the first time T-Ag double hexamers bound to the SV40 origin. Image processing of side views of these nucleoprotein complexes revealed bilobed particles 24 nm long and 8 to 12 nm wide, which indicates that the two T-Ag hexamers are oriented head to head. Taking into account all of the biochemical data known on the T-Ag–DNA interactions at the replication origin, we present a model in which the DNA passes through the inner channel of both hexamers. In addition, we describe a previously undetected structural domain of the T-Ag hexamer and thereby amend the previously published dimensions of the T-Ag hexamer. This domain we have determined to be the DNA-binding domain of T-Ag.
The protein-DNA interactions that take place at origins of DNA replication (ori) in eukaryotes are poorly characterized. A useful model for studying these interactions is the binding of the simian virus 40 (SV40)-encoded large tumor antigen (T-Ag) to the SV40 origin (ori). During the initial steps of replication, T-Ag binds to specific sequences within the SV40 ori. The core of this DNA region (SV40 ori core) has a length of 64 bp and consists of three domains (8): (i) a central 27-bp region, site II, with a perfect palindrome that includes four GAGGC pentanucleotides, which are the specific binding sites for T-Ag; (ii) an AT-rich domain upstream of site II; and (iii) an imperfect inverted repeat, the early palindrome, downstream of site II (12).
During binding to the core region, T-Ag multimerizes into a bilobed structure that has been described as a double hexamer (5, 25, 34). The assembly of the T-Ag double hexamer requires ATP (2) and binds to a head-to-head-oriented pair of the four pentanucleotides of site II at the core origin of replication (16). The protein thus assembles around the DNA (7), and the dodecamer formed has been shown by DNase I digestion to protect a 74-bp DNA fragment that spans the entire SV40 ori core (2). Subsequently, the DNA is unwound bidirectionally by the helicase activity of T-Ag hexamers migrating in the 3′-to-5′ direction along the DNA leading strand; the reaction is driven by ATP hydrolysis (31). Each oligomer can be visualized by electron microscopy at the forks of unwound double strand (11). In addition, SV40 replication requires multiple interactions among T-Ag hexamers, the eukaryotic single-stranded binding protein RP-A, and polymerase α-primase (reviewed in reference 3).
Formation of the T-Ag dodecamers at the SV40 ori core depends on ATP binding but not on ATP hydrolysis (6). In solutions of purified T-Ag, ATP alone (or ADP or nonhydrolyzable ATP analogues) suffices to trigger T-Ag oligomerization into hexamers. The size and general shape of both the protein itself and the nucleoprotein complexes of the two types of structures formed, double and single hexamers, have been studied by using various techniques, including scanning transmission microscopy (25), transmission electron microscopy (29, 34), and atomic force microscopy (26). In the presence of nucleotides, but in the absence of DNA, T-Ag builds up a hexameric propeller-shaped particle with a maximum diameter of 12 nm with an open longitudinal channel that runs through the entire particle (29). The reconstructed volume of this particle shows a clear vorticity that could provide the basis for the known polarity in DNA unwinding.
Additional insights into the T-Ag structure came from the nuclear magnetic resonance solution structure of T-Ag-OBD131–260 (21), a T-Ag derivative containing amino acids 131 to 260 of the protein, the domain responsible for the specific binding to the SV40 ori region (1). When the DNA encoding this domain is cloned and expressed independently, the T-Ag derivative synthesized preserves its specific DNA binding activity. One pair of GAGGC pentanucleotides arranged in a head-to-head orientation and separated by approximately one turn of the DNA double helix is required for binding (15). In addition, this domain has been proposed to mediate the interactions between hexamers within the double hexamer (33). Nevertheless, there was no information prior to this work on where this domain is located within the quaternary structure of the protein or the hexamer.
The structural characterization of the proteins involved is one of several key factors in understanding eukaryotic replication. Previous work has already established the existence of T-Ag double hexamers at the SV40 ori and their role in the first steps of replication, but the approaches used did not allow the study of crucial aspects, such as the orientation and alignment of T-Ag hexamers within the nucleoprotein complexes (25, 26). In this work, we used electron microscopy of negatively stained specimens, together with two-dimensional digital image processing and analysis of single particles, to visualize unambiguously T-Ag double hexamers assembled at the SV40 origin of replication. Further characterization of these nucleoprotein complexes resulted in a model for T-Ag–ori interaction that effortlessly accommodates all the known biochemical data available. Furthermore, we detected and characterized a structural domain within T-Ag hexamers, one not resolved in our previous study, that we have proved to be the DNA-binding domain. Additionally, the C terminus has also been localized in the structure.
MATERIALS AND METHODS
DNA fragment purification and labelling.
HindIII-NcoI double digestion of plasmid pOR1 (10) resulted in a DNA fragment of about 80 bp containing only the SV40 T-Ag binding site II. Fragments were separated by agarose gel electrophoresis, eluted, and purified. To obtain the DNA fragment labelled with 32P, plasmid pOR1 was used as template in PCR reactions with Pfu DNA polymerase (Stratagene), [α-32P]dATP, and the oligonucleotides 5′-GGTACCGACTGATTAAAAAAAATA and 5′-TTCGAAAGAGTGATGAAGACC as primers. The manufacturer's protocol was followed. Labelled DNA was separated on agarose gels and electroeluted.
T-Ag–DNA complex purification.
Immunopurified T-Ag (30), 60 μg in 200 μl of a buffer containing 20 mM Tris-HCl (pH 7.8), 50 mM NaCl, 5 mM KCl, and 2 mM Mg2Cl, was incubated with 2 μg of the 80-bp DNA fragment containing the SV40 ori core and 20 ng of the 32P-labeled DNA fragment. After 15 min at 37°C, ADP was added to a final concentration of 2 mM; the reaction was allowed to proceed for 1 h at the same temperature. The final reaction mixture was directly loaded onto a Superose 6 HRa gel filtration column (Pharmacia, Stockholm, Sweden) equilibrated with the same buffer as before but supplemented with 2 mM ADP and was run in a high-pressure liquid chromatography system.
Monoclonal antibody decoration of T-Ag–DNA complexes.
Labelling with either monoclonal antibody Pab220 (13, 14) or Pab101 (27) was performed directly on the T-Ag–DNA reaction mixtures, prepared as described above, by the addition of the corresponding antibody solutions at a molar ratio of 1:2/5 (IgG/T-Ag hexamer) and further incubation for 2 h at 37°C. Monoclonal antibody Pab220 was a kind gift of E. Fanning.
Electron microscopy and image processing.
The material that eluted in the different chromatographic fractions was directly adsorbed onto carbon-collodion-coated copper grids, which were previously glow discharged. The samples were negatively stained with 2% uranyl acetate and visualised in a JEOL 1200 EX II electron microscope. The micrographs were taken at a ×60,000 magnification under a low electron dosage and were digitized in an EIKONIX IEEE 488 camera at 3.8 Å/pixel. Alternatively, samples from the monoclonal antibody decoration reaction mixtures were prepared as before for electron microscopy visualization, and individual images of complexes were collected by using a slow scan charge-coupled device with a low electron dosage. The images were processed by using the XMIPP program package (23) and aligned by cross-correlation and pattern-free alignment methods (24, 28). Heterogeneities were analyzed by a self-organizing map algorithm (17, 22). For each final average image, the resolution was estimated by the spectral signal-to-noise ratio method (32), with the threshold set at a value of 4.
RESULTS
Analysis of T-Ag–DNA complexes.
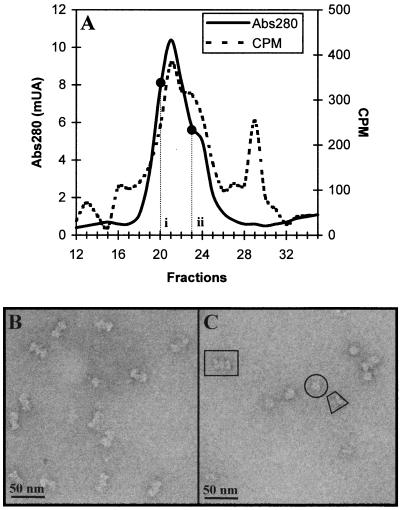
Immunopurified T-Ag (30) was incubated for 1 h at 37°C in the presence of 2 mM ADP with an 80-bp DNA fragment containing the SV40 core origin of replication (see Materials and Methods). ADP was present in the reaction mixture instead of ATP because the latter triggers the helicase activity of the T-Ag hexamers, resulting in the dissembling of the dodecameric complexes because of the bidirectional migration of the hexamers along their corresponding DNA strand. Therefore, in the presence of ATP far fewer double-hexamer complexes are observed (results not shown). Nucleoprotein complexes were separated from free DNA and free protein by Superose-6 gel filtration (Fig. 1A). T-Ag–DNA complexes were found in fractions 19 to 25. These fractions contained at least two different types of nucleoprotein complexes, one centered at fraction 21 (position i in Fig. 1A; the expected position for T-Ag dodecamers) and a minor population that formed a shoulder associated with the main peak (position ii in Fig. 1A). Protein-free DNA eluted in fractions 28 to 31.
FIG. 1.
Analysis of T-Ag–DNA complexes. (A) Gel filtration chromatography on a Superose 6 HRa column of the reaction mixture containing T-Ag, SV40 ori DNA, and ADP. The absorbance at 280 nm (solid line) and the radioactivity due to 32P-labeled DNA (dashed line) were monitored for each fraction. (B) Electron micrograph of a sample prepared from fraction 20 (position i in panel A). A homogeneous population of side-view projections of the T-Ag double-hexamer complexes can be seen. (C) Electron micrograph of a sample prepared from fraction 23 (position ii in panel A). Three types of projections can be seen: front-on (example in a circle) and side (example in a trapezoid) views of single T-Ag hexamers and side views of the double T-Ag hexamer (example in a rectangle). Samples in panels B and C were negatively stained with 2% uranyl acetate.
Aliquots from fractions 20 and 23 were negatively stained and visualized by electron microscopy (Fig. 1B and 1C, respectively). The sample in Fig. 1B displays a homogeneous population of elongated bilobed particles. Particles from fraction 23, however, are clearly heterogeneous, and they differ both in shape and size (Fig. 1C). Three different types were identified in this latter sample: large bilobed particles, which were very similar to those of fraction 20 (a representative particle is encased in a rectangle in Fig. 1C); globule-shaped particles (a representative particle is circled); and nonglobular and asymmetric particles (an example is enclosed in a trapezoid).
The chromatographic elution profile and the electron micrographs of the selected fractions suggest that the reaction mixture contains a mixture of nucleoprotein complexes made up of double or single T-Ag hexamers bound to the SV40 origin of replication, together with some DNA-free T-Ag hexamers. We extracted sets of single particles from electron micrographs and then analyzed the structures of each set by digital image processing.
Image processing of T-Ag double hexamers bound to the SV40 core origin.
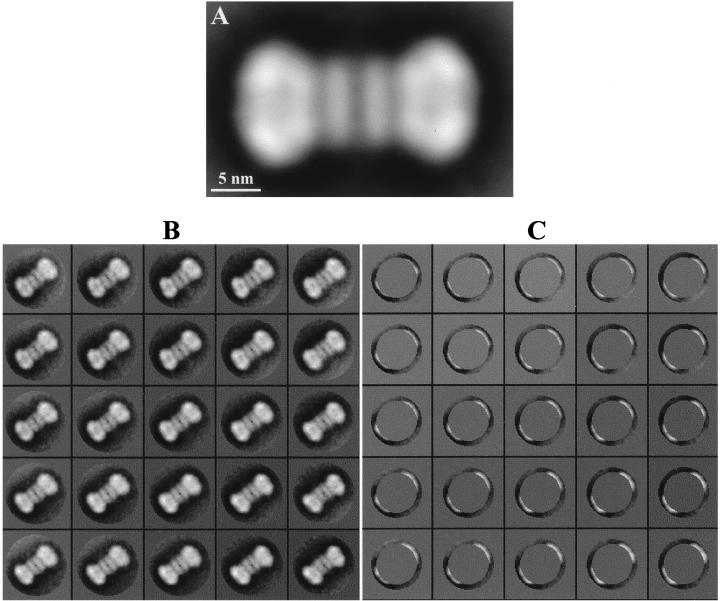
Particles from the main peak (position i in Fig. 1A), which comprise just one type of particle, large and bilobed, were analyzed first. A total of 1,768 single, nonoverlapping particles were selected from homogeneously stained regions and were subjected to image processing by using the XMIPP program package (23). The average image is shown in Fig. 2A. The structure is elongated, with a length of 23 to 24 nm and a width that varied along the length of the particle from about 8 to 12 nm. Interestingly, this structure exhibits a twofold symmetry along a perpendicular axis midpoint to the main longitudinal axis. Thus, it seems that the large structure is made up of two identical smaller structures placed in a head-to-head orientation. These smaller structures (11 to 12 nm in length) each have two distinct regions: a wide region at the distal end of the particle (12 nm wide and 8 to 9 nm long) and a narrow region located at the center of the larger structure (8 to 9 nm wide and approximately 3 nm long). These wide and narrow regions within each smaller structure seem to be structurally independent from each other, as indicated by the noticeable decrease in the intensity of the area located between these two regions, which actually looks like a gap in the projection image. Taking into account the available biochemical data (3, 7, 25), along with the results of the image processing, we conclude that these larger structures are side-view projection images of double hexamers of T-Ag formed at the SV40 replication origin, arising from two single T-Ag hexamers positioned at the replication core in a head-to-head orientation.
FIG. 2.
Digital image processing of the T-Ag double hexamer at the SV40 origin of replication. (A) Refined average image from a total of 1,010 particles. The image was filtered to the calculated resolution of 2.8 nm. (B and C) Analysis of heterogeneities in the T-Ag double-hexamer–DNA complexes. A self-organizing map of code vectors is shown. (B) The whole double-hexamer image was taken into account. (C) An external crown of the images containing only the distal domain of each of the hexamers in the double-hexamer complex was analyzed.
To characterize the T-Ag double hexamers bound to SV40 DNA in more detail, we searched for heterogeneities within this set of particles, such as in the length of the nucleoprotein complexes and their straightness, by using a neuronal-network-based self-organizing map (SOM) algorithm (22). SOM is a powerful classification tool that requires no a priori knowledge of the initial population. SOM maps the input data (images in this case) into a two-dimensional array of nodes or code vectors. The code vectors at each node of the network represent the main trend of variability, and associated with them is a cluster of similar images. We analyzed images of T-Ag double hexamers by SOM by using two different types of input: one in which all the pixels of the whole double-hexamer images were considered and another in which only the pixels within a crown restricted to contain the distal regions of the double hexamer were analyzed. The corresponding, independent, 5×5 code-vector maps are depicted in Fig. 2B and C. When the complete particle was considered (Fig. 2B), the main variability was detected along the longitudinal axis. Thus, while some of the double-hexamer complexes were straight as a rod, there were some subgroups that exhibited various degrees of axial curvature. This feature is particularly obvious in the subsets shown in the lower-right and upper-left corners of the maps in Fig. 2B. Some of these kink double hexamers showed a deviation of just one hexamer with respect to the midpoint of the longitudinal axis, while in others the kink in the complex was more pronounced because the two hexamers were actually deviating. We also detected variability in the length of the double hexamers, although this variability was less pronounced in general since it was detected only when the outer crowns of the particles were analyzed, as illustrated in Fig. 2C. The amount of distal regions extracted with a centered fixed mask increased slightly from the top-left code vectors to the bottom-right code vectors in Fig. 2C, whereas each of the two hexamers contributed equally to the extracted crown (judged by the crescents being identical in each of the code vectors). At our working resolution of 2.8 nm, no other structural differences were detected, either between double hexamers or between hexamers within dodecamers.
Visualization of single T-Ag hexamers bound to SV40 DNA.
In addition to T-Ag double hexamers, two different types of particles (globule- and nonglobule-shaped) were detected in fraction 23 (position ii in Fig. 1A). By comparison with the results obtained by our groups previously (29), we consider the globule-shaped particles to be front-view projections of single T-Ag hexamers. The nonglobular asymmetrical particles are thought to be side-view projections of single T-Ag hexamers because of their similarity to the smaller structures that make up the side views of the T-Ag double hexamer discussed in the previous section.
Analysis of single T-Ag hexamer front views.
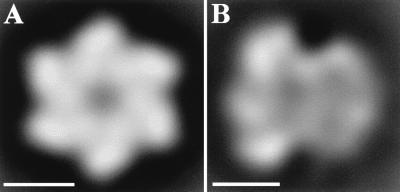
A total of 1,060 front-view particles were extracted from electron micrographs of negatively stained material (a representative micrograph is shown in Fig. 1C). The initial set of image particles was processed as described in Materials and Methods, and the resulting average image is presented in Fig. 3A. This image is similar to that obtained previously by our groups (29). The particle contains six density maxima arranged around a central region where the staining agent penetrates. The outer diameter of this particle is approximately 12 nm, and the diameter of the inner channel is approximately 2 nm. This particle exhibits the distinctive structural vorticity first described by San Martín et al. (29).
FIG. 3.
Digital image processing of the T-Ag single hexamers of fraction 23. (A) Sixfold-symmetrized average image of the entire population of front views of the T-Ag hexamers, showing sixfold symmetry. (B) Average image of the side views of T-Ag hexamers. Images in panels A and B were filtered to their calculated resolution of 2.5 nm. The bars represent 5 nm.
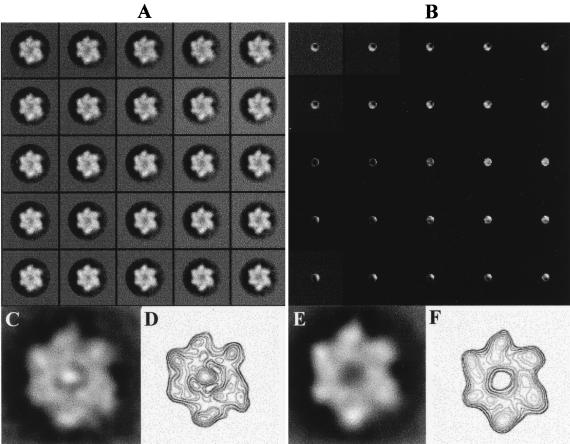
The structural heterogeneities of these particles were studied by subjecting the whole particle to SOM classification. The output map of code vectors (Fig. 4A) showed that only the subset of particles placed at the bottom-right quadrant of the map (approximately 250 of 1,060 particles) could be described as being homogeneously well stained and matching very closely the features described previously for the T-Ag hexamer by our groups (29). The remaining particles, which were poorly stained and therefore faulty, were either elliptical or had half of the ring ill defined.
FIG. 4.
Analysis of heterogeneities in the T-Ag single hexamers. (A and B) Self-organizing map of code vectors. (A) The whole particle image was used. (B) A central crown with a radius of five pixels was analyzed in order to focus on the central region of the particle. An average image of a subset of particles that were well stained at the center (C) and its corresponding contour map (D) are also shown. (E) Average image of those particles that were classified as not being stained at the center. (F) Contour level map corresponding to panel E. Both subsets were reprocessed independently.
Since double-stranded DNA was present in the sample, we also searched for heterogeneities in the central, stain-penetrating channel, which could be the entrance for the nucleic acid. A central crown with a radius of 5 pixels extracted from the images was analyzed with the SOM algorithm. From the output map (Fig. 4B), it is clear that a great heterogeneity exists in this central region. In a subset of the particles, the staining agent is very noticeably excluded from the central region of the particle. This set of particles was reprocessed independently, and the average refined image is presented in Fig. 4C, where an area of high density can be observed at the center of the particle. This feature is highlighted in the corresponding contour map shown in Fig. 4D; a very prominent density maximum is found at the center of the particle, surrounded by six other maxima of variable densities attributable to each of the six subunits of the T-Ag hexamer. The remaining particles of Fig. 4B showed some degree of staining around the central region, but the size of the stained spot and its position were both variable. A subset of particles having a sizeable amount of staining in the center was reprocessed independently. The averaged image is shown in Fig. 4E, and its corresponding contour map is shown in Fig. 4F. In both presentations it is clear that the center of the particle is void of material.
We also processed images taken from a reaction mixture lacking the DNA probe. We found the entire population of T-Ag single hexamers head-on views to be completely homogeneous (results not shown), with the central channel always perfectly well stained, in full agreement with our previous work on T-Ag (28), where occlusion of the central channel was never observed.
Analysis of single T-Ag hexamer side views.
Side-view particles (878 total) were extracted and image processed as described above. The averaged refined image obtained for these particles shows a nonglobular asymmetric structure that is 11 nm long and possesses wider and narrower domains 12 and 9 nm wide, respectively. A gap-like region of very low density (Fig. 3B) separates these domains. The resulting image is quite similar in the general construction and dimensions to each of the two halves of the T-Ag double hexamer (Fig. 2A), thus supporting the interpretation that these nonglobular views are side views of a T-Ag hexamer. It should be noted, however, that these side views of the T-Ag hexamer are more poorly defined than each of the two halves of the double hexamer, and a possible interpretation will be presented in the Discussion.
Monoclonal antibody decoration of T-Ag–DNA complexes.
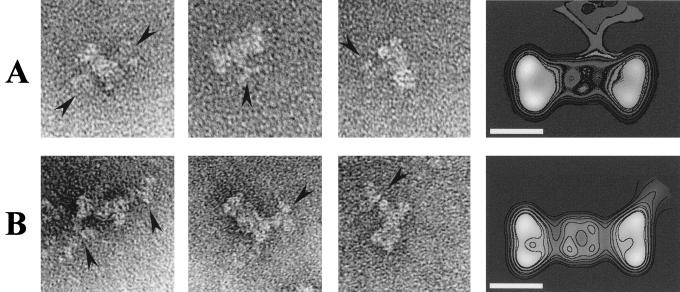
We have used monoclonal antibodies Pab220 (13, 14), whose epitope mapped within the T-Ag DNA binding domain (residues 130 to 246 in the T-Ag sequence), and Pab101 (27), directed against the last eight amino acids at the T-Ag C terminus (residues 701 to 708), to localize these two regions within the double-hexamer projection image. Figure 5A shows a short gallery of individual images, as well as the average image (from a total of 87 particles), obtained for the immunocomplex formed by Pab220. The corresponding contour level map has been superposed for clarity. As can be seen in the figure, Pab220 binds to the narrow region located at the center of the T-Ag double hexamer and somehow slightly distorts the general outlook of the latter. Similarly, Fig. 5B presents the results obtained with Pab101. In this case the average image comes from a total of 204 individual images, and Pab101 binds to the wide region at the distal end of the double hexamer with no apparent effect on the general morphology of the nucleoprotein complex.
FIG. 5.
Localization of the DNA-binding domain and the C terminus of T-Ag. (A) Immunolabelling with Pab220 (13, 14), a monoclonal antibody that recognizes T-Ag DNA-binding domain. (B) Immunolabelling with Pab101 (27), a monoclonal antibody against the eight C-terminal amino acids of the T-Ag sequence. The first image in each gallery shows a T-Ag double hexamer with two immunoglobulin G antibodies bound at opposite sides of the nucleoprotein complex. The arrowheads point to the antibodies. The average images of the immunocomplex were filtered to their calculated resolutions of 4.8 nm (A) and 3.6 nm (B). The bar represents 10 nm.
DISCUSSION
T-Ag is the only viral protein required for SV40 DNA replication and plays a key role in both the recognition and the unwinding of the cognate DNA (12). In the presence of the SV40 replication origin (SV40 ori core) and nucleotides, T-Ag assembles into a double-hexamer complex and bidirectionally unwinds the DNA in an ATP-dependent process (7).
The interactions between T-Ag dodecamers and SV40 ori have been subjects of intense research (7, 16, 25, 34). Previous studies to characterize the double-hexamer structure at the replication origin used approaches of a rather limited resolution and therefore failed to provide detailed features of the nucleoprotein macromolecular organization (25, 26, 34).
We applied transmission electron microscopy coupled with advanced image-processing methods to the study of nucleoprotein complexes consisting of T-Ag double hexamers bound to SV40 ori core in an effort to increase the structural accuracy of the studies on the T-Ag–DNA complexes.
Analysis of the views of the dodecameric nucleoprotein complexes.
Complexes of double hexamers of T-Ag bound to DNA were always visualized as side-view projection images. In the average image of these complexes, which shows an apparent mirror plane at its center, the mass at each side of this plane corresponds to a side view of one of the hexamers, and the two hexamers are arranged head-to-head around this plane. This interpretation is in accordance with the facts that SV40 ori DNA is unwound bidirectionally (19) and that the structure of each of the hexamers presents a marked polarity (29).
Some variability among the views of these complexes in the length along their major axis, as well as in the degree of straightness relative to the apparent mirror plane, was detected. This reflects flexibility in the nucleocomplexes: a double-stranded DNA fragment holding together in close vicinity two independent hexamers is not likely to form a completely rigid complex. Slight variations in the distance separating the two hexamers in the complex, affecting the total length of the double hexamer, and a smooth kink around the middle of the DNA fragment would not be surprising. No further structural variability was detected among these views.
Analysis of the views of single-hexamer complexes.
Single hexamers were visualized as both side- and head-on views. The amount of the coeluting DNA-associated radioactivity was clearly in excess of that trapped in the residual double hexamers, which only exist as nucleoprotein complexes, present in this fraction. At least a fraction of the T-Ag single hexamers had to be bound to the 80-bp DNA probe.
The average image of the side views of the single hexamers is very similar, in dimensions and general shape, to that of each of the two masses at each side of the apparent mirror plane in the views of the double hexamers. This similarity strongly reinforces our interpretation of the views of the double hexamers presented above. This is the first time that such types of views have been reported for the T-Ag hexamers. We think that because of the interaction with the DNA, the single hexamer tends to lie along its main longitudinal axis. The side views of single hexamers were more heterogeneous than the side views of the double hexamers, and the structures were therefore more poorly defined. This could be due to the lack of a unique metastable position of the particle on the carbon film and the subsequent variable rocking.
Our current front-view particles were clearly more heterogeneous than those reported previously in our study of single T-Ag hexamers prepared in the absence of DNA (29). Only a small subset of our side-view particles were homogeneously stained and round; the remainder were poorly stained or of elliptical shape due to rocking of the hexamer on the grid. If these single hexamers are interacting with the DNA, then the DNA could affect how these particles lay flat on the carbon support of the grid. Greater heterogeneity was found when we restricted our search to the central region of the particles. In some, the staining agent penetrated through the central region to the same extent as reported previously (29), while in others the stain was nearly completely excluded from this central region and an area of density at the center of the particle could be seen. This would be compatible with the DNA passing through the inner channel of the T-Ag hexamer, giving rise to a projection image in which the central region is occupied by stain-excluding material. However, we are studying negatively stained particles at a relatively low resolution, and we cannot unambiguously assert that the DNA is actually threading through the channel of the single T-Ag hexamer. Nevertheless, this remains the most likely possibility.
The height of the T-Ag hexamers measured along the major axis on the side views was previously greatly underestimated. Our current estimation of the height of the T-Ag hexamer is approximately 12 nm, which strongly differs from the height of 3.8 nm (29) shown in our previous three-dimensional reconstruction by negative stain of single hexamers, in which we explicitly reported that only 54% of the molecular mass was accounted for in the reconstructed volume. The flattening of the specimen in these preparations, which was probably caused by the negative stain and the dehydration, is at its most devastating when the T-Ag hexamers lie on their base on the support film of the grid. In addition, it is generally accepted that above a certain height limit, negative staining rarely succeeds in contrasting the whole structure and, as a consequence, portions of the macromolecule may not be visualized. Therefore, we conclude that a structural domain of the T-Ag hexamer was not reconstructed in our previous work (29). This newly described domain is located at the far end of the wide base of the propeller-shaped particle (29); in the double-hexamer side view, this domain corresponds to the vertical densities at the center of the complex—the regions of the two hexamers that are closest to each other.
Localization of T-Ag DNA-binding and C-terminal domains.
The use of a monoclonal antibody raised against the DNA-binding domain, Pab220 (13, 14), has allowed us to identify this newly described structural domain as the T-Ag DNA-binding domain. Most of the immunocomplexes observed corresponded to single-labelled specimens, but we did see a few double-labelled complexes in which two Pab220 molecules bound to the double hexamer at opposite sides (binding of two antibodies on the same side would be sterically greatly disfavored or even impossible). Binding of Pab220 to its epitope on the double-hexamer T-Ag complexes induces an outward displacement of the wider domain of the T-Ag hexamer, as if the antibody was wedging itself between the narrow and wide regions. This observation may indicate that the recognized region, the DNA-binding domain, benefits from a certain detachment from the rest of the macromolecule. Additionally, we have found that the last eight amino acids of the T-Ag polypeptide sequence are placed at the very edge of the wide base of the propeller-shaped T-Ag hexameric particle. These two amino acids sequences are, therefore, spatially quite apart within the quaternary structure of the T-Ag multimer.
A model of the dodecameric T-Ag complex with the SV40 ori.
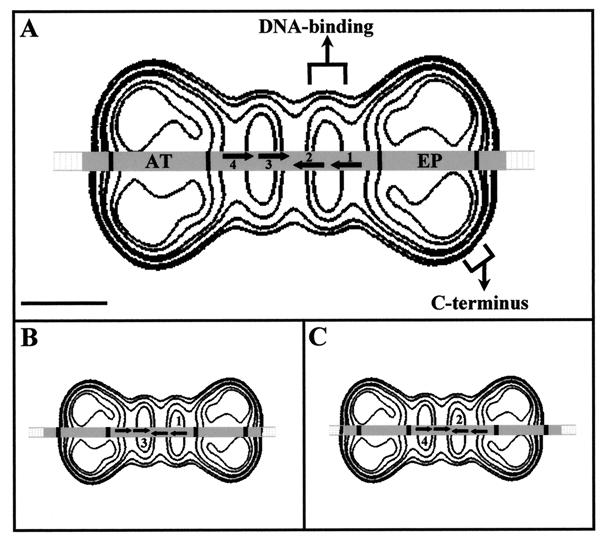
The calculated length of the 74-bp SV40 DNA fragment protected against DNase I digestion (2) matches very well with that of the double T-Ag hexamer (24 nm). In Fig. 6A, we show a contour level map of the T-Ag double hexamer in which the 80-bp DNA fragment present in the reaction mixture is drawn passing through the longitudinal channels of both T-Ag hexamers. The relative dimensions of the protein and the DNA are scaled. Nearly the entire 74-bp DNA (in gray) is covered by the double hexamer. All the data obtained here support a model in which the oligomerization of T-Ag around the DNA results in a double-hexamer structure in which the DNA traverses simultaneously through the inner channel of the two hexamers. This model does not require any other type of protein-DNA interactions to account for all the biochemical data known regarding the T-Ag–ori interaction.
FIG. 6.
Model of the interaction of the T-Ag double hexamer with the SV40 ori DNA. (A) The 80-bp DNA fragment used in the reaction mixture is shown superimposed on the contour map of the T-Ag double hexamer; the DNA passes through the inner channel of each hexamer. The DNA corresponding to the 74 bp previously shown to be protected against DNase I digestion is shaded gray. The four GAGGC pentanucleotides necessary for the dodecamer formation are numbered 1 to 4, and arrows indicate their positions and orientations. Flanking this perfect palindrome is the AT-rich region (AT) and the early palindrome (EP). The length of the protected DNA matches that of the T-Ag double hexamer (24 nm). Each of the pentanucleotides in any of the active pairs could interact with the proximal (narrower) domain of the corresponding T-Ag hexamer. The bar represents 5 nm. (B and C) Models illustrating the interactions based on the pairs 1 and 3 (B) or 2 and 4 (C). In both cases, the position of the pentanucleotides perfectly matches the density maxima of the proximal domains, where the DNA-binding domains map. All of the representations are scaled.
The formation of the double hexamer is preferentially supported by the interaction with the pentanucleotide pair 1 and 3 and pair 2 and 4 (depicted in Fig. 6B and C) and, to a minor extent, with the pair 1 and 4 (16). Each hexamer binds to one-half of the perfect palindrome at the center of the SV40 core ori (25). This indicates that the same region of each of the hexamers in the double hexamers must specifically recognize the DNA. In our scaled model in Fig. 6A, the region of T-Ag hexamers that would be close enough to interact with the pentanucleotides corresponds to the smaller, narrower domain, where the DNA-binding domain was mapped by antibody labelling. When the DNA is moved slightly to the left (Fig. 6B) or to the right (Fig. 6C) with respect to the contour map of the double hexamer, each of the vertical elliptical density maxima representing the DNA-binding domain in each T-Ag hexamer fall fully in register to define perfectly the interaction based on the pentanucleotide pair 1/3 (Fig. 6B) or the pair 2 and 4 (Fig. 6C). In our model, the regions of the hexamers that are close to each other are these smaller structural domains where the DNA-binding domains map. This is in perfect agreement with the recent finding that the hexamer-hexamer interaction within the double hexamer is mediated through the DNA-binding domain, which is also needed for double-hexamer assembly and SV40 ori DNA unwinding (33). The DNA-binding domain is known to be a functionally independent region that when isolated exhibits an activity similar to that of the intact T-Ag hexamer (15). We propose that this functional independence correlates with a structural independence of the domain.
ACKNOWLEDGMENTS
This work was partially supported by grants CAM 07B/0027/1997 from Comunidad de Madrid and BIO98-0761 from Comisión Interministerial de Ciencia y Tecnología to J.M.C. M.V. is recipient of a Postdoctoral Fellowship from Comunidad de Madrid. L.E.D. is supported by a contract from the Ministerio de Educación y Cultura.
We are very grateful to E. Fanning for antibody Pab220 and to O. Llorca for his expert advice and careful reading of the manuscript. The help of Y. Robledo and M. Bárcena is also appreciated. Karen A. Brune is acknowledged for editing the manuscript.
REFERENCES
- 1.Arthur A K, Höss A, Fanning E. Expression of simian virus 40 T antigen in Escherichia coli: localization of T-antigen origin DNA-binding domain to within 129 amino acids. J Virol. 1998;62:2204–2208. doi: 10.1128/jvi.62.6.1999-2006.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borowiec J A, Hurwitz J. ATP stimulates the binding of simian virus 40 (SV40) large tumour antigen to the SV40 origin of replication. Proc Natl Acad Sci USA. 1998;85:64–68. doi: 10.1073/pnas.85.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borowiec J A, Dean F B, Bullock P A, Hurwitz J. Binding and unwinding. How T antigen engages the SV40 origin of DNA replication. Cell. 1990;60:181–184. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- 4.Braithwaite A, Stürzbeches H, Addison C, Palmer C, Rudge K, Jenkins J R. Mouse p53 inhibits SV40 origin-dependent DNA replication. Nature. 1987;329:458–460. doi: 10.1038/329458a0. [DOI] [PubMed] [Google Scholar]
- 5.Dean F B, Dodson M, Echols H, Hurwitz J. ATP-dependent formation of a specialized nucleoprotein structure by simian virus 40 (SV40) large tumor antigen at the replication origin. Proc Natl Acad Sci USA. 1987;84:8981–8985. doi: 10.1073/pnas.84.24.8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean F B, Hurwitz J. Simian virus 40 large T antigen untwist DNA at the origin of DNA replication. J Biol Chem. 1991;266:5062–5071. [PubMed] [Google Scholar]
- 7.Dean F B, Borowiec J A, Eki T, Hurwitz J. The simian virus T antigen double hexamer assembles around the DNA at replication origin. J Biol Chem. 1992;267:14129–14137. [PubMed] [Google Scholar]
- 8.Deb S, De Lucía A L, Banr C-P, Koff A, Tegtmeyer P. Domain structure of the simian virus 40 core origin of replication. Mol Cell Biol. 1986;6:1663–1670. doi: 10.1128/mcb.6.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Caprio J A, Ludlow J W, Figge J, Shew J Y, Huang C M, Lee W H, Marsillo E, Pancha E, Livingston D M. SV40 large tumour antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 10.De Lucia A L, Deb S, Partin K, Tegtmeyer P. Functional interactions of the simian virus 40 core origin of replication with flanking regulatory sequences. J Virol. 1986;57:138–144. doi: 10.1128/jvi.57.1.138-144.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dodson M, Dean F B, Bullock P, Echols H, Hurwitz J. Unwinding of duplex DNA from the SV40 origin of replication by T antigen. Science. 1987;238:964–967. doi: 10.1126/science.2823389. [DOI] [PubMed] [Google Scholar]
- 12.Fanning E, Knippers R. Structure and function of simian virus 40 large tumour antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 13.Gurney E G, Harrison R O, Fenno J. Monoclonal antibodies against simian virus 40 T antigens: evidence for distinct subclasses of large T antigens and for similarities among nonviral T antigens. J Virol. 1980;34:752–763. doi: 10.1128/jvi.34.3.752-763.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gurney E G, Tamowsky D, Deppert W. Antigenic binding sites of monoclonal antibodies specific for simian virus large T antigen. J Virol. 1986;57:1168–1172. doi: 10.1128/jvi.57.3.1168-1172.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joo W S, Luo X, Dennis D, Kim H Y, Rainey G J, Jones C, Sreekumar K R, Bullock P A. Purification of the simian virus 40 (SV40) T antigen DNA-binding domain and characterization of its interactions with the SV40 origin. J Virol. 1997;71:3972–3985. doi: 10.1128/jvi.71.5.3972-3985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joo W S, Kim H Y, Purviance J D, Sreekumar K R, Bullock P A. Assembly of T-antigen double hexamers on the simian virus 40 core origin requires only a subset of the available binding sites. Mol Cell Biol. 1998;18:2677–2687. doi: 10.1128/mcb.18.5.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohonen T. The self-organizing map. Proc IEEE. 1990;78:1464–1480. [Google Scholar]
- 18.Lewine A J. Viruses. New York, N.Y: Scientific American Library; 1992. The DNA tumour viruses; pp. 87–111. [Google Scholar]
- 19.Li J J, Kelly T J. Simian virus 40 DNA replication in vitro: specificity of initiation and evidence for bidirectional replication. Mol Cell Biol. 1985;11:2108–2115. doi: 10.1128/mcb.5.6.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J J, Peden K W C, Dixon R A F, Kelly T. Functional organization of the simian virus 40 origin of DNA replication. Mol Cell Biol. 1986;6:1117–1128. doi: 10.1128/mcb.6.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo X, Sanford D G, Bullock P A, Bachovchin W W. Solution structure of the origin DNA-binding domain of SV40 T-antigen. Nat Struct Biol. 1996;3:1034–1039. doi: 10.1038/nsb1296-1034. [DOI] [PubMed] [Google Scholar]
- 22.Marabini R, Carazo J M. Pattern recognition and classification of images of biological macromolecules using artificial neural networks. Biophys J. 1994;66:1804–1814. doi: 10.1016/S0006-3495(94)80974-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marabini R, Masegosa I M, San Martín M C, Marco S, Fernández J J, de la Fraga L G, Vaquerizo C, Carazo J M. XMIPP: an image processing package for electron microscopy. J Struct Biol. 1996;116:237–240. doi: 10.1006/jsbi.1996.0036. [DOI] [PubMed] [Google Scholar]
- 24.Marco S, Chagoyen M, de la Fraga L G, Carazo J M, Carrascosa J L. A variant to the random approximation of the reference-free alignment algorithm. Ultramicroscopy. 1996;66:5–10. [Google Scholar]
- 25.Mastrangelo I A, Hough P V C, Wall J S, Dodson M, Dean F B, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 26.Mastrangelo I A, Bezanilla M, Hansma P K, Hough P V C, Hansma H G. Structures of large T antigen at the origin of SV40 DNA replication by atomic force microscopy. Biophys J. 1994;66:293–298. doi: 10.1016/s0006-3495(94)80800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mole S E, Gannon J V, Ford M J, Lane D P. Structure and function of large T antigen. Philos Trans R Soc Lond Ser B. 1987;317:455–469. doi: 10.1098/rstb.1987.0072. [DOI] [PubMed] [Google Scholar]
- 28.Penzeck P, Radermacher M, Frank J. Three-dimensional reconstructions of single particle embedded in ice. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- 29.San Martín M C, Gruss C, Carazo J M. Six molecules of SV40 large T antigen assemble in a propeller-shaped particle around a channel. J Mol Biol. 1997;268:15–20. doi: 10.1006/jmbi.1997.0952. [DOI] [PubMed] [Google Scholar]
- 30.Simanis V, Lane D P. An immunoaffinity purification procedure for SV40 large tumour antigen. Virology. 1985;144:88–100. doi: 10.1016/0042-6822(85)90308-3. [DOI] [PubMed] [Google Scholar]
- 31.Tsurimoto T, Melendy T, Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990;346:534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- 32.Unser M, Trus B, Steven A C. A new resolution criterion based on spectral signal-to-noise ratios. Ultramicroscopy. 1987;23:39–52. doi: 10.1016/0304-3991(87)90225-7. [DOI] [PubMed] [Google Scholar]
- 33.Weisshart K, Taneja P, Jenne A, Herbig U, Simmons D T, Fanning E. Two regions of simian virus 40 T antigen determine cooperativity of double-hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J Virol. 1999;73:2201–2211. doi: 10.1128/jvi.73.3.2201-2211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wessel R, Schweizer J, Stahl H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]