Abstract
Colistin-resistant multidrug-resistant (MDR), extensively drug-resistant (XDR), and pan-drug-resistant (PDR) bacteria are highly lethal and many researchers have tried hard to combat these microorganisms around the world. Infections caused by these bacteria are resistant to the last resort of antibiotic therapy and have posed a major challenge in clinical and public health. Since the production of new antibiotics is very expensive and also very slow compared to the increasing rate of antibiotic resistance, researchers are suggesting the use of natural substances with high antibacterial potential. Bacteriophages are one of the most effective therapeutic measures that are known to exist for use for incurable and highly resistant infections. Phages are highly taken into consideration due to the lack of side effects, potential spread to various body organs, distinct modes of action from antibiotics, and proliferation at the site of infection. Although the effects of phages on MDR and XDR bacteria have been demonstrated in various studies, only a few have investigated the effect of phage therapy on colistin-resistant isolates. Therefore, in this review, we discuss the problems caused by colistin-resistant MDR and XDR bacteria in the clinics, explain the different mechanisms associated with colistin resistance, introduce bacteriophage therapy as a powerful remedy, and finally present new studies that have used bacteriophages against colistin-resistant isolates.
Introduction
In the fight between microorganisms and antibiotics, microorganisms have won; thus, challenging doctors and researchers in treating antibiotic-resistant microorganisms. Various studies have reported that antibiotic-resistant bacteria use various mechanisms to resist different antibiotics, even those considered as last-line antibiotics [1–4]. The emergence of multidrug-resistant (MDR) bacteria, followed by extensively drug resistant (XDR) bacteria, has posed a threat to the lives of patients with infectious diseases. Over the past few years, reports of pan-drug-resistant bacteria (PDR), which are resistant to all classes of antibiotics, have brought up extensive concerns [5]. About 25,000 patients in Europe die each year from infectious diseases caused by MDR bacteria, and more than 50% of cases die due to infections caused by XDR bacteria. Antibiotic-resistant infectious diseases impose a heavy annual cost of about 20 billion US dollars on the worldwide [5]. Today, the world is facing a growing and challenging threat with the emergence of bacteria which are almost resistant to the remaining effective antibiotics [6, 7]. Unfortunately, due to the limited activity of pharmaceutical industries in producing new antibiotics, physicians have been urged to use older highly toxic antibiotics such as colistin. For decades, colistin has been used in clinics, mainly against infections caused by MDR and XDR Gram-negative pathogens [8, 9]. Resistance to antibiotics have gradually developed in bacteria following their overuse and in the last decade, these microorganisms have even become resistant to last-line antibiotics such as carbapenem and colistin [10–12]. Resistance to colistin as a last-line antibiotic has been observed in Gram-negative bacteria, and in addition to chromosomal mutations, various other mechanisms have been shown to be involved in colistin resistance that can be transferred from one bacterium to another by horizontal transmission [13]. Therefore, the main problem is that final options for treating resistant pathogens, especially Gram-negative bacteria that cause nosocomial and community-acquired infections, are running out. Hence, it is predicted that antibiotic resistance could become an uncontrollable global catastrophe and the use of alternatives with significant antimicrobial activity, especially specific bacteriophages, has become a priority for researchers [5, 14]. The use of phages is so important that the US Food and Drug Administration (FDA) has approved bacteriophage cocktails as food additives to prevent foodborne bacterial contamination [15]. Phages have been reported to be used for prophylaxis and treatment of antibiotic-resistant bacteria, and a wide range of lytic phages have been tested for their therapeutic potentials against bacterial infections in animals and humans [7]. The results have been promising, raising the attention of researchers to this therapeutic method for eradicating MDR, XDR and PDR Gram-negative bacteria [16]. This review focuses on increased rate of MDR and XDR colistin-resistant Gram-negative bacteria as well as the subsequent challenges associated with these microorganisms in the clinic. Then we review the mechanisms that confer colistin resistance and discuss phage therapy as a promising option to eradicate MDR and XDR colistin-resistant Gram-negative bacteria.
MDR and XDR Challenge
It has been almost a decade since the World Health Organization (WHO) declared "combat drug resistance: no action today, no cure tomorrow". Over time, the development of antibiotic resistance in bacteria has become inevitable. Infections caused by MDR and XDR Gram-negative bacteria contribute to the exacerbation of the infection due to resistance to last-line antibiotics has been reported [12, 17]. MDR isolates are bacteria that are resistant to at least one antibiotic within three or more classes of antibiotics. XDR isolates are bacteria that are resistant to all but one or two classes of antibiotics [18]. The important issue about infections caused by MDR and XDR bacteria is that in addition to employing known mechanisms of antibiotic resistance, repeated or long-term administration of antibiotics could enhance the development of resistance to new compounds [19]. In addition, many of the different mechanisms of antibiotic resistance in these MDR and XDR Gram-negative bacteria are due to the presence of resistance genes that are located on the mobile genetic elements and can be transmitted to other bacteria, leading to the spread of resistance [12]. According to various studies, a high mortality rate is observed due to infections by MDR and XDR isolates. Different mechanisms of antibiotic therapy failure include the external barriers (preventing the entry of drugs into bacteria), genetic transmission of resistance (through plasmids, integrons, transposons, and other mobile genetic elements), natural mutations in antibiotic targets, enzyme-dependent drug alterations, and efflux pumps [5]. According to recent reports, antibiotic-resistant bacterial infections in hospitals account for approximately 400,000 deaths annually [20]. Mortality rate varies depending on the bacterial spp., type of infection and the geographical region. The mortality rate of MDR Pseudomonas aeruginosa isolates, as one of the main causes of nosocomial infections is estimated to be 18–61% [21, 22]. On the other hand, the mortality rate of infections caused by MDR and XDR Acinetobacter baumannii isolates is 21.2% due to bacteremia, 5% in different wards of the hospital (general ward), and 54% in intensive care units (ICU) [23, 24]. Therefore, MDR and XDR bacteria are important factors in increasing mortality and morbidity of patients as well as the high cost of medical care. Antimicrobial drug resistance is reported to add 30–100 billion US dollars to health care costs per year [25]. Moreover, infections caused by MDR and XDR bacteria pose unavoidable problems such as the delayed treatment process, prolonged hospitalization time, and the need to use more toxic antibiotics [12]. Therefore, there is a clear and vital need to develop a new approach for treating infections caused by MDR and XDR pathogens. Although the use of a combination of several antibiotics and the production of new antibiotics have become a priority in combating infections caused by these bacteria, there have not been much success [5]. Therefore, researchers have conducted serious research on non-antibiotic drugs for the treatment of MDR and XDR bacterial infections, and the use of specific phages has been introduced as a main alternative to antibiotics.
Colistin-Resistant MDR and XDR Bacteria
Colistin was discovered about 80 years ago (1940s), however, its use was discontinued by doctors for decades due to having many side effects and the discovery of other less toxic antibiotics. In recent years, the emergence of MDR and XDR bacteria as well as the lack of efficient antibiotics have led to the reuse of colistin against infections [12]. There are currently two commercially available types of colistin, including colistimethate sodium and colistin sulfate [26]. One monotherapy option against MDR and XDR Gram-negative bacteria isolated from nosocomial infections is colistin. Although colistin has still a good effect on antibiotic-resistant isolates in many places, it has many side effects [27]. After re-use of colistin in the clinic, the main noticed side effect was nephrotoxicity. Studies have confirmed that nephrotoxicity varies from 6 to 58% after intravenous colistin administration. In patients with normal renal function, nephrotoxicity was observed in 10% of cases, but in abnormal kidneys, nephrotoxicity was observed in 27–58% of subjects [28, 29]. Widespread use of colistin in livestock around the world has gradually led to the emergence of colistin-resistant bacteria in livestock, which can be transmitted from animals to humans through food. Increased resistance to this antibiotic has led to a ban on its use in livestock, especially in developed countries, to prevent further development of resistance to colistin [12]. Accordingly, severe side effects of colistin and emergence of colistin-resistant MDR and XDR Gram-negative bacteria have made their employment even more challenging as resistance to last-line antibiotics poses a major challenge for physicians. There have been various reports of colistin-resistant MDR and XDR isolates in some parts of the world, but there no exact pattern of prevalence and rate of resistance to this antibiotic has been demonstrated in all countries. Today, colistin resistance in K. pneumoniae isolates is increasing, and mortality rate due to severe infections caused by these isolates range from 25 to 71% [30]. Colistin-resistant MDR, XDR, and pandrug-resistant (PDR] Acinetobacter spp. have also been associated with high mortality rate and nephrotoxicity [27]. The emergence of a highly resistant clone of K. pneumoniae ST14 with high colistin resistance rate of 37.1% has been reported, posing a serious concern [31]. The emergence of the mcr-3 gene (a plasmid-mediated colistin resistance gene) was detected in both mcr-1 and mcr-2 negative Escherichia coli isolates. This plasmid-borne mcr-3 was transferred to an E. coli recipient by conjugation. This resistance plasmid was also identified in K. pneumoniae isolate from Thailand, E. coli isolate from Malaysia, and Salmonella enterica serovar Typhimurium isolate from the USA. According to the ubiquitousness of Aeromonas in the environment and the transmission of mcr-3 between Aeromonas and Enterobacteriaceae isolates, the broad dissemination of mcr-3 might be underestimated. As colistin has been widely used in veterinary medicine and is increasingly prescribed for humans, monitoring plasmid-borne colistin resistance in colistin-resistant Gram-negative bacteria is imperative for prevention and control of the spread of mcr genes [32]. Colistin resistance has been reported in 21.3% of MDR and XDR P. aeruginosa isolates. Among colistin-resistant isolates, a protein profile belonging to OprH as an efflux pump has been reported which differs from the LPS profile [33]. In various studies, mcr genes (mcr-1 to mcr-10), which are involved in colistin resistance in MDR and XDR isolates, have been reported in different bacteria [34–36]. In another study, a novel mcr gene, mcr-10, was identified in a clinical Enterobacter roggenkampii isolate. When plasmid-borne mcr-10 was cloned into a colistin-susceptible E. roggenkampii strain, MIC of colistin showed fourfold increase (from 1 to 4 mg/L) [37]. Colistin resistance in 646 V. parahaemolyticus isolates was screened in China, among which 25 (2.5%) showed colistin resistance. The mcr-1 gene was found in one colistin-resistant isolate. Class A β-lactamase gene blaCARB-17 and the plasmid-mediated quinolone resistance (PMQR) gene qnrVC5 were accompanied by mcr-1 gene in a colistin-resistant V. parahaemolyticus isolate [38]. One study identified an MDR Stenotrophomonas spp. with high levels of resistance to colistin and meropenem, which was of great concern as no resistance plasmid was found after genome sequencing; however, mcr-5.3, mcr-8.2 and four β-lactamase genes were widely observed. In addition, 12 genes associated with seven types of efflux pumps have been identified which are thought to play a major role in the acquisition and transmission of colistin resistance [39]. Two colistin-resistant and nine heteroresistant P. aeruginosa isolates were identified, and among these, the two colistin-resistant isolates showed mutations in PmrB. Also, heteroresistant P. aeruginosa showed alterations in the PmrAB regulatory system. The conversion of heteroresistance to resistance must be noted in future clinical surveillance [40]. Two colistin-resistant E. coli sequence type (ST) 131 isolates were obtained from peritoneal fluid and abscess of the surgical wound. Genome sequencing revealed the presence of the plasmid-mediated AmpC β-lactamase gene blaCMY-2 and the mcr-1 gene [41]. Importantly, colistin resistance in MDR and XDR isolates can be associated with the presence of predominant resistance genes, such as carbapenemases, beta-lactamases, and metallobetalactamases. The co-transfer of multiple important antimicrobial resistance genes pose a certain challenge for healthcare settings as resistance to colistin and other antibiotics could enhance the menace of infections resistant to all classes of antibiotics [38, 39, 41, 42]. Above studies show that resistance to colistin has been observed in a wide range of Gram-negative bacteria, especially MDR and XDR isolates, and transmission of resistance through transferable mobile genes can increase the resistance rate in Enterobacteriaceae and other Gram-negative bacteria. Therefore, it is very important to come up with new solutions to deal with these troublesome resistant bacteria.
The Mechanism of Colistin Resistance
Authoritative literature has introduced five mechanisms of action for colistin. The direct antibacterial activity of colistin is such that the cationic diaminobutyric acid residues of colistin bind to anionic phosphate of lipid A moiety of LPS in the outer membrane via electrostatic bonds, thereby affecting the outer and inner membranes of bacteria and leading to cell lysis. Colistin also has anti-endotoxin activity, such that it binds to LPS molecules and inhibits the activity of lipid A endotoxin [43, 44]. This leads to the release of tumor necrosis factor-alpha (TNF-α) and interleukin-8 (IL-8), thereby inhibiting shock induction. Vesicle-vesicle contact pathway is another mechanism by which colistin acts on Gram-negative bacteria. Colistin binds to anionic phospholipid vesicles after transiting to the outer membrane, causing the fusion of the inner leaflet of the outer membrane with the outer leaflet of the cytoplasmic membrane, subsequently leading to the loss of phospholipids and bacteria death. In the mechanism of the hydroxyl radical death pathway, colistin leads to the release of reactive oxygen species. This pathway is known as the Fenton reaction, which damages DNA, lipids and proteins, eventually leading to bacterial death. Inhibition of respiratory enzymes by colistin is another mechanism of action in which colistin exerts antibacterial activity by inhibiting vital respiratory enzymes [44]. Unfortunately, MDR, XDR, and PDR isolates of Gram-negative bacteria such as P. aeruginosa, E. coli, A. baumannii, K. pneumoniae, and other Enterobacteriaceae are globally found to harbor multiple resistance mechanisms. Also, Morganella morganii, Providencia spp., Serratia marcescens, Proteus spp., Vibrio cholera, Brucella, Edwardsiella spp., Legionella, Chromobacterium, Neisseria spp., anaerobic Gram-negative cocci, some Aeromonas spp., Burkholderia cepacia, Campylobacter, eukaryotic microbes, and mammalian cells possess intrinsic colistin resistance [44–46]. Mechanisms of colistin resistance may be due to chromosomal mutations and transmissible plasmid genes called mcr (mcr-1 to mcr-10) which are able to spread through horizontal transfer between bacteria [26]. At the present time, PCR-based techniques are the most extensively adopted to identified mcr genes, however, owing to the presence of various genotypes of mcr genes and their capability to undergo horizontal gene transfer; PCR techniques are limited in clinical practice, containing the basic quarantine stations and the primary-care hospitals [47]. The mcr genes in Gram-negative bacteria have been identified on various plasmids, including IncY, IncF, IncI2, IncP, IncHI2, ColE10-like ones, and IncX4 [48]. The plasmid-borne mcr gene has an interesting mechanism. The product of this gene transfers phosphoethanolamine residues to the main target of colistin which is the lipid moiety of LPS. This leads to alterations in the LPS of Gram-negative bacteria, thereby reducing the bacterial affinity to react with colistin and thus the effectiveness of the drug. It has been reported that the mcr gene can also alter the susceptibility of Gram-negative bacteria to antimicrobial peptides and other available antibiotics [49, 50]. In addition, high expression of the mcr gene contribute to changes in fitness, growth rate and structural integrity of the outer membrane, which are attributed to the integration of mcr into the bacterial membrane and changes in lipid A through its enzymatic activity [50]. In addition to the plasmid gene, another mechanism of resistance to colistin is chromosomal mutations that occur in lipid A synthesis genes including lpxO2, lpxD, lpxC, and lpxA, causing incomplete LPS production. It has also been shown that in case the ISAba11 sequence is inserted into the LPS synthesis genes (lpxC and lpxA), the bacterium loses the ability of LPS production and therefore, a high level of resistance occurs [51, 52]. In case Gram-negative bacteria are deficient in LPS, less negative charges are present on bacterial surface, leading to the reduced affinity to colistin [53]. Positive charges are suggested as major factors for lipid A changes which are induced by the addition of galactosamine (naxD), phosphoethanolamine (pEtN, mediated by chromosomally encoded eptA or plasmid-borne mcr), and 4-amino-4-deoxy-L-arabinose (L-Ara4N, mediated by arnBCADTEF-ugd operon) to lipid A, which decrease the ability of colistin to bind and disrupt the outer membrane [12]. Two-component systems (PhoPQ and PmrAB) are known to be encoded by chromosomes and are responsible for the intrinsic resistance to colistin [54, 55]. The two-component PmrAB system consists of several constituents, including histidine kinase and a response regulator that responds to environmental stimuli. This system enables Gram-negative bacteria to sense and respond appropriately to a variety of environmental conditions, including different pH levels as well as the presence of Mg2+, Fe3+, and Al3+ ions. The two-component PmrAB system can confer colistin resistance in Gram-negative bacteria by affecting the expression of genes involved in alterations of lipid A [56–59]. Point mutations in the pmrA and pmrB genes have been shown to increase the expression level of pmrAB, which subsequently leads to altering the bacterial outer membrane, followed by decreased membrane permeability and resistance to colistin [58, 60]. The two-component PhoPQ system is stimulated by environmental factors, such as Mg2 +, Ca2 +, and cationic antimicrobial peptides (polymyxin, indolicidin, and LL-37), and plays a major role in the virulence and alteration of LPS, as well as increased colistin resistance [12]. One of the most common mechanisms of colistin resistance is mutation of the mgrB gene, which regulates negative feedback of the PhoPQ two-component system, activates PhoPQ, and directly increases the expression of the arnBCADTEF operon [61]. One of the causes of missense or nonsense mutations in mgrB gene is insertion sequence (IS) truncation, among which IS102, IS5 family, IS3-like, IS5-like, and ISKpn14 were related with mgrB mutations in clinical samples [12]. Capsule formation is another mechanism of colistin resistance as polymyxin reacts with the capsule polysaccharides by anionic interactions, leading to polymyxin attenuation [62]. Also, regulators of capsule production, such as conjugative pilus expression (Cpx), and regulator of capsule synthesis (Rcs), are capable of activating efflux pumps that confer resistance to colistin (KpnEF for Cpx and PhoPQ for Rcs regulon) [54, 62]. Colistin resistance by efflux pumps belonging to the RND (resistance-nodulation-cell division) family has been reported. These efflux pumps are composed of membrane fusion protein (encoded by adeA gene), which acquires substrate and transports it from cytoplasm or within phospholipid bilayer to the extracellular medium (encoded by adeB), and an outer membrane protein channel (encoded by adeC) regulated by the adeR gene [63]. Other efflux pumps that have been implicated in colistin resistance include MexXY–OprM, CarO, sapABCDF, acrAB–tolC, kpnEF, and emrAB; however, the mechanisms of colistin resistance by these efflux pumps are not yet clear [61, 64, 65]. Colistin resistance could be obtained via colistin-heteroresistant bacteria. Colistin heteroresistance is an intermediate condition which exhibits a phenotype characterized by the presence of resistant subpopulations among a sensitive population. Colistin heteroresistance phenotypes may account for the unexplained treatment failures and are frequently detected among MDR A. baumannii, P. aeruginosa, and K. pneumoniae isolates. There are different mechanisms attributed to colistin heteroresistance which include mutations in lipid A biosynthesis genes, soxRS-regulated overexpression of the acrAB-tolC efflux pump, putrescine/YceI communication, activation of two-component regulatory systems PmrAB, PhoPQ, ParRS, CprRS, and ColRS, as well as biofilm formation [40, 44]. The lptD is a vital locus for insertion of the newly produced LPS into the outer membrane, and removal of this locus leads to the complete loss of LPS and increased polymyxin resistance [66]. Deletion mutation in the locus responsible for biotin synthesis has been implicated in polymyxin resistance as biotin is a main co-factor of lipid metabolism. Therefore, higher biotin levels responsible for the increased production of lipid A can increase colistin susceptibility [67, 68]. Another factor contributing to colistin resistance is the overexpression of the outer membrane protein OprH, which binds to negatively charged phosphate groups, ultimately inhibiting the binding of polymyxin. Noteworthy, downregulation of OprD (a porin) is also attributable to polymyxin resistance in P. aeruginosa [44]. Intriguingly, a single mutation in vacJ contributes to the emergence of colistin-resistant phenotype [69]. The sodB and sodC genes also contribute to colistin resistance, most probably via the detoxification of reactive oxygen species [70]. Acylation of lipid A portion, which contributes to colistin resistance, could be regulated by lpxM. Inactivation of lpxM inhibits L-Ara4N modifications and leads to the decreased polymyxin resistance [71]. Bmul_2133 and Bmul_2134 were found to be putative hopanoid biosynthesis genes in Burkholderia multivorans which are essential for the stabilization of outer membrane permeability. Therefore these two genes are responsible for polymyxin resistance via a mechanism which is independent of LPS-binding activity [44]. DedA family protein (a membrane transporter) in Burkholderia thailandensis is required for colistin resistance. DedA leads to alterations in lipopolysaccharide (LPS) lipid A and the subsequent colistin resistance [72]. For a better comprehension, a summary of the mechanisms of colistin resistance in Gram-negative bacteria is shown in Table 1.
Table 1.
A summary of the different mechanisms that Gram-negative bacteria use for colistin resistance
| Mode of action of colistin | Mechanism of colistin resistance | Function |
|---|---|---|
|
Direct antibacterial activity [43, 44] Anti-endotoxin activity [43, 44] Vesicle-vesicle contact pathway [44] Hydroxyl radical death pathway [44] Inhibition of respiratory enzymes [44] |
The mcr plasmid | Leads to changes in the LPS of Gram-negative bacteria that reduce the affinity to react with colistin and thus leading to the effectiveness of the drug [13, 37, 73] |
| Mutations that occur in lipid A synthesis genes | Cause incomplete LPS production and induce less negative charges on the surface [51, 52] | |
| Insertion of ISAba11 into the LPS synthesis genes | Induces the loss of LPS production and high level of resistance [51, 52] | |
| Positive charges | Majorly alter lipid A, leading to the decreased ability of colistin binding and prevention of the disruption of outer membrane[12] | |
| Two-component systems (PhoPQ and PmrAB] |
PmrAB leads to colistin resistance by affecting the expression of genes involved in lipid A alterations [56–59] Point mutations in the pmrA and pmrB genes decrease the membrane permeability and resistance to colistin [58, 60] PhoPQ plays a major role in the virulence and alteration of LPS, and its mutations increase colistin resistance [12] |
|
| Mutation in the mgrB gene | Directly increases the expression of the arnBCADTEF operon and leads to lipid A changes [61] | |
| Capsule formation | Leads to polymyxin attenuation [62] | |
| Efflux pumps | Transport colistin from cytoplasm or within the phospholipid bilayer to the extracellular medium [63] | |
| Miscellaneous chromosomally encoded colistin resistance genes | These include the lptD locus, biotin synthesis locus, OprH protein, OprD porin, vacJ locus, sodB and sodC genes, lpxM gene, Bmul_2133 and Bmul_2134, as well as DedA family protein [44, 67–70] | |
| Colistin heteroresistance phenotype | Mutations in lipid A biosynthesis genes, soxRS-regulated overexpression of the acrAB-tolC efflux pump, putrescine/YceI communication, activation of PmrAB and PhoPQ two-component regulatory systems, ParRS, CprRS, and ColRS two-component regulatory systems, and biofilm formation [40, 44] |
Bacteriophage
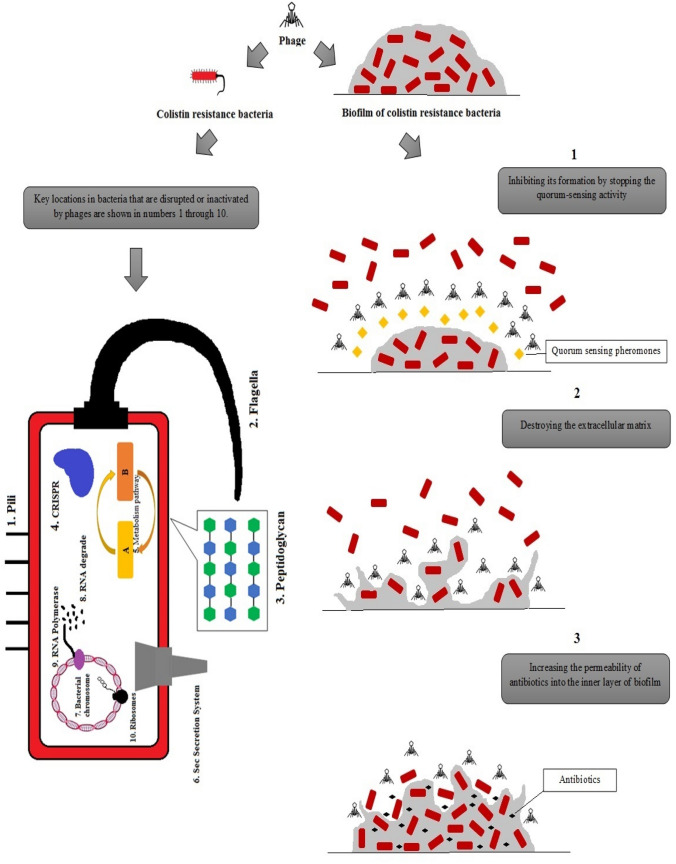
Bacteriophages or phages are a group of viruses that act specifically on bacteria and can be used as a therapeutic agent to treat bacterial infections. The basis of phage treatment in infections is the binding of specific phage to the bacterium, which results in rapid lysis of the bacterial cell [5]. In 1896, the British researcher Hankin discovered a biological source in the Indian River Ganges and Jumna that changed the culture of the bacterium that causes cholera. This discovery was possibly the first report of bacteriophage activity [5]. Further reports described bacteriophage as an "anti-Shiga microbe" as it was found in the feces of patients with shigellosis. The curiosity of other researchers led them to believe that the bacterial virus was responsible for the destruction of bacteria [5]. Nevertheless, the use of phages as antimicrobial agents to fight bacterial infections was first introduced almost 90 years ago [74]. It was also suggested that environmental phage counts which have been common heavy greater than 106/ml effecting a single host bacterial strain to arrive the rates of bacterial loss [75]. In the last few decades, the production of new antibiotics has no longer been cost-effective as resistance to them has occurred after production. In recent years, with the advent of MDR, XDR, and PDR bacteria as well as the low production of new antibiotics, the reuse of bacteriophages has been noticed [16, 76]. Unique properties of lytic phages in general include absence of inherent toxicity, lack of cross-resistance with antibiotic classes, high selectivity, bactericidal activity, and the ability of proliferation in the presence of pathogenic resistant bacteria [5]. As phages are highly specific to bacteria, unlike broad-spectrum antibiotics, they do not kill the commensal microbiota, which are vital for patients with malnutrition and immunodeficiency. Phages can be stored in a dry powder formulation without the need for a cold chain [77]. Since phages are widely found in a variety of environments, such as sewage effluent, soil, water, hospital effluent, fecal materials, as well as the gastrointestinal tract of humans and animals, they can be rapidly isolated. Consequently, isolation of phages is more cost-effective than the production of antibiotics because phage found in a variety of environments even in special unusual environments such as hot springs [78]. In addition, one of the other factors that put phages under focus is that they can be administered through respiratory, parenteral, gastrointestinal, and topical routes [77, 79]. The absence of side effects, decreased inflammatory responses, and potentially distribution of phages all over the body are the most promising aspects of phage therapy [5]. Different mechanisms of action of phages on both Gram-negative and Gram-positive antibiotic resistant bacteria include the inhibition of peptidoglycan synthesis, engaging the host cell secretion machinery, interference with cellular motility, metabolism, transcription and translation, DNA silencing, RNA degradation, and clustered regularly interspaced short palindromic repeats (CRISPR)-mediated immunity [80, 81]. Phages can eradicate biofilms resistant to antibiotic and environmental stresses by destroying the extracellular matrix, preventing the quorum-sensing mechanism, and increasing the permeation of antibiotics into the inner layers of biofilm structures (Fig. 1) [82]. Phages can also be used as a cocktail, with the benefit of having greater effects on target bacteria and reducing the formation of phage-resistant bacteria. This is because different types of phages can target same species and strains of bacteria [83]. In addition, phages have the ability to destroy biofilms (accumulation of bacterial cells that attach to a surface or to each other and are placed in a self-generated matrix with a high resistance to antibiotics). All of these unique properties of phages mentioned in this section have urged researchers and physicians to consider phage therapy as a potential effective treatment approach. Also, by using the potential of bacteriophages, researchers have achieved significant success in food safety, veterinary medicine, agriculture, industry, and clinical diagnosis (detection and typing of bacteria in human infections) [5]. To date, the use of phages in in vitro, animals, and even humans with various infections such as antibiotic-resistant chronic rhinosinusitis, necrotizing pancreatitis, and septicemia have been used with promising results. The use of phages against human antibiotic-resistant infections in the clinic proves that phages are suitable weapons against resistant bacteria and can be considered as a promising solution for the treatment of infections caused by bacteria resistant to last-resort antibiotics [5].
Fig. 1.
Shows the effect of phages on antibiotic-resistant gram-negative bacteria in planktonic and biofilm modes of growth. The effect of phage on antibiotic-resistant biofilms is through three mechanisms including 1) destroying the extracellular matrix, 2) preventing the quorum-sensing mechanism, and 3) increasing the permeation of antibiotics into the inner layers of biofilm structures as illustrated in the right. The release of the planktonic form of antibiotic-resistant bacteria leads to bacterial death by phages through the disruption of 1) pili, 2) flagella, 3) peptidoglycan, 4) CRISPR, 5) metabolic pathway, 6) sec secretion system, 7) bacterial chromosome, 8) RNA degradation, 9) RNA polymerase, 10) ribosomes of bacteria, as marked with numbers on the left side of the figure
Bacteriophages Against Colistin Resistance
Colistin has been mentioned as the latest treatment option for bacteria with high levels of antibiotic resistance, as sometimes is the only effective antibiotic. The global development of colistin resistance in Gram-negative bacteria is alarmingly increasing. Colistin-resistant bacteria are usually resistant to other antibiotics, and they therefore pose a challenge in clinical treatment, public health, and medical interventions [84, 85]. The use of various strategies by researchers to overcome these challenging bacteria has been considered, the most important of which are combination antibiotic therapy, antimicrobial peptides, monoclonal antibodies, nanoparticles, natural compounds, herbal extracts, and phages [12]. In the meantime, of all these treatment options, phage therapy has been used in the clinic, especially in cases infected with MDR, XDR, and PDR superbugs. Accordingly, researchers have identified bacteriophages as a viable alternative to antibiotics in the treatment of infections with superbugs [5]. In a study by Ebrahimi, IsfAB78 lytic phage was examined against colistin-resistant MDR A. baumannii clinical isolates. Their results showed that the IsfAB78 phage was able to significantly lyse MDR A. baumannii cultures after 40 min and reduced the number of these resistant bacteria in biofilm structures for about 19–87% [86]. In another study, Hao et al. induced colistin resistance in carbapenem-resistant K. pneumoniae (colistin-resistant CRKP) isolates by horizontally transferring mcr-1 gene and inactivating the mgrB chromosomal gene. They investigated the effect of lytic phage ɸNJS1 on these isolates. The results showed that the reduction of negative charges on bacterial surface due to colistin resistance led to the increased ɸNJS1 phage adhesion and subsequent infection. Colistin-resistant bacteria were also shown to be more sensitive to the ɸNJS1 phage compared to the wild-type strains when grown in biofilms or moth larvae and during mammalian colon colonization [87]. Shokri et al. aimed at detecting specific phages for the selected MDR, XDR, and PDR clinical isolates of P. aeruginosa from waste waters and hospital sewages. They found that phage cocktails (Psu1, Psu2, and Psu3) had antibacterial activity against all MDR, XDR and PDR isolates of colistin-resistant P. aeruginosa and completely destroyed the bacterial cells [88]. In another study, Abdelkader et al. used the PMK34 bacteriophage (which encoded an endolysin with potent muralytic activity and was isolated from raw sewage water) against colistin-resistant A. baumannii. Two PDR colistin-resistant A. baumannii strains exhibited a similar susceptibility to PMK34 phage. Their results indicated that the combination of LysMK34 and colistin decreased the MIC of colistin up to 32-fold, and that colistin-resistant strains were resensitized in Mueller–Hinton broth. Therefore, LysMK34 can be used to maintain the applicability of colistin as a last-resort antibiotic [89]. Manohar et al. investigated the therapeutic features and efficacy of specific phages (from sewage water samples) against E. coli, K. pneumoniae, and Enterobacter species. The three bacteriophages included myPSH2311 (infecting E. coli), myPSH1235 (infecting K. pneumoniae), and myPSH1140 (infecting four different Enterobacter spp.). Interestingly, phage cocktail decreased the bacterial load from 106 to 103 CFU/mL within 2 h. All the three characterized phages were detected to have a broad host range activity and the phage cocktail was effective against meropenem- and colistin- (two last-resort antibiotics) resistant bacteria [90]. Some studies have reported the excellent effectiveness of the combination of phage and phage-derived protein with colistin against Gram-negative bacteria with high levels of antibiotic resistance [91, 92]. Blasco et al. investigated the efficacy of ElyA1 and ElyA2 endolysins (from Ab1051Φ and Ab1052Φ phages, respectively) alone and combination with colistin against clinical isolates of MDR Gram-negative bacteria. According to their results, ElyA1 showed antibacterial activity against 25 MDR P. aeruginosa, 25 MDR A. baumannii, and 17 carbapenemase-producing K. pneumoniae isolates (three colistin-resistant strains: A. baumannii SOF004b, P. aeruginosa AUS034 and K. pneumoniae KP2). No antibacterial activity was found in ElyA2. The combination of colistin and ElyA1 decreased the MIC of colistin in all isolates (thus reducing the associated toxicity), except in K. pneumoniae [93]. Bernasconi et al. investigated the antibacterial activity of three phages (PYO, INTESTI, and Septaphage) against MDR E. coli and Proteus spp. isolates from humans, food, and animals. Only four isolates were colistin-resistant, one of which harbored the mcr-1 plasmid gene. Although Septaphage had no antibacterial activity, 5 of 8 carbapenemase producers and 3 of 4 colistin-resistant isolates were susceptible to PYO and INTESTI, respectively [94]. In another study, Aslam et al. used bacteriophage therapy in two and one lung transplant recipients with life‐threatening MDR infections caused by P. aeruginosa and Burkholderia dolosa, respectively. Two ventilator‐dependent lung transplant recipients with large airway complications and refractory MDR P. aeruginosa pneumonia (intermediate colistin resistance) received phage therapy. Both cases responded clinically and were discharged from the hospital without the need for ventilator support. Although the third patient showed recurrent B. dolosa (susceptible to colistin) infection following transplant, the improved consolidative opacities and ventilator weaning were demonstrated via phage therapy. No phage therapy‐related side effects were detected in the studied patients. Phage therapy was well related to clinical recovery of lung transplant recipients with MDR Gram-negative bacterial infections that were not responsive to antibiotics [95]. In 2020, researcher screened two phages including vB_AbaM_ISTD and vB_AbaM_NOVI (isolated from wastewaters) for their potential in eradicating 103 clinical A. baumannii isolates (three isolates were colistin-resistant). Both phages had fast adsorption rates, high depolymerizing activity, proper growth rates, broad host range, and antibacterial effectiveness against planktonic and biofilm-associated bacteria. Among carbapenem-resistant A. baumannii isolates, two colistin-resistant isolates were also sensitive to both NOVI and ISTD phages [96]. Schirmeier et al. measured the inhibitory and bactericidal effects of Artilysin Art-175 (endolysin encoded by bacteriophage) against colistin-resistant mcr-1-positive E. coli isolates. They observed that Art-175 had a high antimicrobial activity against all mcr-1 colistin-resistant E. coli isolates. Overall, the number of mcr-1-positive colistin-resistant bacteria reduced. Also, they demonstrated no cross-resistance between colistin and Art-175 [97]. In another study, Art-175 was used to treat MDR A. baumannii isolates. According to the results, Art-175 had high bactericidal properties against all isolates, even those resistant to colistin [98]. Table 2 shows a summary of the above studies confirming the promising effects of phages on colistin-resistant bacteria. Although studies on the miraculous effect of phages and phage-derived protein against colistin-resistant Gram-negative isolates are limited, the presented studies in this review could pave the way for attracting more attention on discovering different phages to counteract colistin-resistant bacteria. Due to the loss of colistin efficiency on these superbugs, doctors have limited treatment options; therefore, phages as new treatment options could be a new hope in the fight against these infections in the future. Not only colistin-resistant isolates are usually resistant to all antibiotics, the ability of biofilm formation in these isolates can exacerbate the challenges associated with their treatment. However, these studies show that phages can show a great potential in destroying biofilms.
Table 2.
A summary of the therapeutic properties of bacteriophage against infections caused by colistin-resistant Gram-negative bacteria
| Author name | Year | Phage | Organism | Result | Ref. |
|---|---|---|---|---|---|
| Ebrahimi et al | 2020 | IsfAB78 | Colistin-resistant MDR A. baumannii | IsfAB78 phage was able to significantly lyse culture and biofilm of MDR A. baumannii | [86] |
| Hao et al | 2019 | ɸNJS1 | Colistin-resistant CRKP | Colistin-resistant CRKP was also shown to be more susceptible to ɸNJS1 phage compared to wild-type bacteria when grown in biofilms or moth larvae and during mammalian colon colonization | [87] |
| Shokri et al | 2017 | Psu1, Psu2, and Psu3 | MDR, XDR and PDR isolates of colistin-resistant P. aeruginosa | Phage cocktails showed antibacterial activity and completely destroyed the bacterial cells | [88] |
| Abdelkader et al | 2020 | PMK34 | PDR colistin-resistant A. baumannii | PDR colistin-resistant A. baumannii strains were susceptible to the PMK34 phage | [89] |
| Manohar et al | 2019 | myPSH2311, myPSH1235, and myPSH1140 | Meropenem and colistin-resistant E. coli, K. pneumoniae, and Enterobacter spp. | There was a reduction in bacterial load and efficacy was observed against all bacteria | [90] |
| Blasco et al | 2019 | ElyA1 and ElyA2 endolysins [from phage Ab1051Φ and Ab1052Φ] | Colistin-resistant MDR P. aeruginosa, MDR A. baumannii, and carbapenemase-producing K. pneumoniae isolates | ElyA1 illustrated antibacterial activity and combination of colistin and ElyA1 decreased the MIC of colistin in all isolates | [93] |
| Bernasconi et al | 2017 | PYO, INTESTI, and Septaphage | MDR carbapenemase-producers, colistin-resistant E. coli and Proteus spp. isolates | Although Septaphage had no antibacterial activity, carbapenemase producers and colistin-resistant isolates were susceptible to PYO and INTESTI | [94] |
| Aslam et al | 2019 | AB‐PA01, AB‐PA01 m1, Navy phage cocktail1, and Navy phage cocktail2 for patient one, AB‐PA01 for patient two, and single lytic phage for patient three | Colistin-resistant MDR P. aeruginosa and B. dolosa | Phage therapy was well related to clinical recovery in lung transplant recipients with MDR Gram-negative bacterial infections | [95] |
| Vukotic et al | 2020 | vB_AbaM_ISTD and vB_AbaM_NOVI | Colistin-resistant A. baumannii | Two colistin-resistant isolates were also sensitive to both NOVI and ISTD phages | [96] |
| Schirmeier et al | 2018 | Artilysin Art-175 | Colistin-resistant mcr-1-positive E. coli isolates | Art-175 showed a high antimicrobial activity against all mcr-1 colistin-resistant E. coli isolates and the number of bacteria was reduced | [97] |
| Defraine et al | 2016 | Artilysin Art-175 | MDR colistin susceptible and resistant A. baumannii | Art-175 had high bactericidal properties against all isolates, even colistin-resistant isolates | [98] |
Conclusion
Colistin resistance is a critical issue to deal with nowadays. Researchers have introduced novel alternatives to colistin with better efficiency and stability. Bacteriophages, either alone or in a cocktail, have lysing properties against various MDR, XDR, and PDR colistin-resistant Gram-negative bacteria and reduce the bacterial load in both planktonic and biofilm modes of growth. In addition, phage-derived enzymatic proteins have antibacterial activity against Gram-negative bacteria that are resistant to last-line antibiotics. Therefore, phages, with different mechanisms from antibiotics, can be considered as good alternatives to colistin and can even be used along with colistin to fight superbugs in the future due to having synergistic effects. This review represents an important step in the introduction of novel phages that may be considered in the successful treatment of diseases caused by colistin-resistant pathogens. Introducing phages as new agents for last-line antibiotic resistant pathogens requires a strong collaboration among scientists around the world and not only this but additionally the implementation of the required infrastructures.
Acknowledgements
None.
Authors' contributions
Majid Taati Moghadam developed the idea, wrote, and drafted the paper. Mahsa Shahbandeh and Samane Mohebi provided the figures and tables. Sajad Babakhani, Maryam Mirshekar, and others developed the article.
Funding
No financial support was provided relevant to this article.
Data availability
Not applicable.
Code Availability
Not applicable.
Declarations
Conflict of interest
The authors declare no conflicts of interest relevant to this article.
Ethical approval
Not required.
Informed consent
All authors have read and agreed to the published version of the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Moghadam M, Shariati A, Mirkalantari S, Karmostaji A. The complex genetic region conferring transferable antibiotic resistance in multidrug-resistant and extremely drug-resistant Klebsiella pneumoniae clinical isolates. New Microbes New Infections. 2020;36:100693. doi: 10.1016/j.nmni.2020.100693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiaei S, Moradi M, Nave HH, Hashemizadeh Z, Taati-Moghadam M, Kalantar-Neyestanaki D. Emergence of co-existence of bla NDM with rmtC and qnrB genes in clinical carbapenem-resistant Klebsiella pneumoniae isolates in burning center from southeast of Iran. Folia Microbiol. 2019;64(1):55–62. doi: 10.1007/s12223-018-0630-3. [DOI] [PubMed] [Google Scholar]
- 3.Shariati A, Dadashi M, Moghadam MT, van Belkum A, Yaslianifard S, Darban-Sarokhalil D. Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Sci Rep. 2020;10(1):1–16. doi: 10.1038/s41598-020-69058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hadizadeh M, Norouzi A, Taghadosi R, Mohebi S, Mohammadi M, Hasanzade A, et al. Prevalence of qnr, intI, and intII genes in extendedspectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from clinical samples in Iran. Trop J Pharm Res. 2017;16(1):141–147. doi: 10.4314/tjpr.v16i1.18. [DOI] [Google Scholar]
- 5.Moghadam MT, Amirmozafari N, Shariati A, Hallajzadeh M, Mirkalantari S, Khoshbayan A, et al. How phages overcome the challenges of drug resistant bacteria in clinical infections. Infection and Drug Resistance. 2020;13:45. doi: 10.2147/IDR.S234353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moradi M, Norouzi A, Taatimoghadam M. Prevalence of bla-CTX-M, bla-SHV, and bla-TEM Genes and Comparison of Antibiotic Resistance Pattern in Extended-spectrum β-lactamase producing and non-producing groups of Klebsiella pneumoniae Isolated from Clinical Samples in Kerman Hospitals. Journal of Fasa University of Medical Sciences. 2016;6(1):120–128. [Google Scholar]
- 7.Moghadam MT, Khoshbayan A, Chegini Z, Farahani I, Shariati A. Bacteriophages, a new therapeutic solution for inhibiting multidrug-resistant bacteria causing wound infection: lesson from animal models and clinical trials. Drug Des Dev Ther. 2020;14:1867. doi: 10.2147/DDDT.S251171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bialvaei AZ, Samadi KH. Colistin, mechanisms and prevalence of resistance. Curr Med Res Opin. 2015;31(4):707–721. doi: 10.1185/03007995.2015.1018989. [DOI] [PubMed] [Google Scholar]
- 9.Kengkla K, Kongpakwattana K, Saokaew S, Apisarnthanarak A, Chaiyakunapruk N. Comparative efficacy and safety of treatment options for MDR and XDR Acinetobacter baumannii infections: a systematic review and network meta-analysis. J Antimicrob Chemother. 2018;73(1):22–32. doi: 10.1093/jac/dkx368. [DOI] [PubMed] [Google Scholar]
- 10.Shariati A, Asadian E, Fallah F, Azimi T, Hashemi A, Sharahi JY, et al. Evaluation of Nano-curcumin effects on expression levels of virulence genes and biofilm production of multidrug-resistant Pseudomonas aeruginosa isolated from burn wound infection in Tehran. Iran Infection Drug Resistance. 2019;12:2223. doi: 10.2147/IDR.S213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohebi S, Hossieni Nave H, Norouzi A, Kandehkar Gharaman M, Taati MM. Detection of extended spectrum beta lactamases on class I integron in escherichia coli isolated from clinical samples. J Mazandaran Univ Med Sci. 2016;26(138):66–76. [Google Scholar]
- 12.Taati Moghadam M, Mirzaei M, Fazel Tehrani Moghaddam M, Babakhani S, Yeganeh O, Asgharzadeh S, et al. The Challenge of Global Emergence of Novel Colistin-Resistant Escherichia coli ST131. Microbial Drug Resistance. 2021. [DOI] [PubMed]
- 13.Gharaibeh MH, Shatnawi SQ. An overview of colistin resistance, mobilized colistin resistance genes dissemination, global responses, and the alternatives to colistin: A review. Veterinary World. 2019;12(11):1735. doi: 10.14202/vetworld.2019.1735-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dosari AS, Norouzi A, Moghadam MT, Satarzadeh N. Antimicrobial activity of Ephedra pachyclada methanol extract on some enteric gram negative bacteria which causes nosocomial infections by agar dilution method. Zahedan J Res Med Sci. 2016;18(11).
- 15.Mattila S, Ruotsalainen P, Jalasvuori M. On-demand isolation of bacteriophages against drug-resistant bacteria for personalized phage therapy. Front Microbiol. 2015;6:1271. doi: 10.3389/fmicb.2015.01271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin S, Huang G, Zhang Y, Jiang B, Yang Z, Dong Z, et al. Phage Abp1 rescues human cells and mice from infection by pan-drug resistant Acinetobacter baumannii. Cell Physiol Biochem. 2017;44(6):2337–2345. doi: 10.1159/000486117. [DOI] [PubMed] [Google Scholar]
- 17.Moghadam MT, Nave HH, Mohebi S, Norouzi A. The evaluation of connection between integrons class I and II and ESBL-producing and Non-ESBL klebsiella pneumoniae isolated from clinical samples, Kerman. 2016.
- 18.Shahbandeh M, Moghadam MT, Mirnejad R, Mirkalantari S, Mirzaei M. The efficacy of AgNO3 nanoparticles alone and conjugated with imipenem for combating extensively drug-resistant pseudomonas aeruginosa. Int J Nanomed. 2020;15:6905–6916. doi: 10.2147/IJN.S260520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74(3):417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirzaei B, Bazgir ZN, Goli HR, Iranpour F, Mohammadi F, Babaei R. Prevalence of multi-drug resistant (MDR) and extensively drug-resistant (XDR) phenotypes of Pseudomonas aeruginosa and Acinetobacter baumannii isolated in clinical samples from Northeast of Iran. BMC Res Notes. 2020;13(1):1–6. doi: 10.1186/s13104-020-05224-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nathwani D, Raman G, Sulham K, Gavaghan M, Menon V. Clinical and economic consequences of hospital-acquired resistant and multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2014;3(1):32. doi: 10.1186/2047-2994-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raman G, Avendano EE, Chan J, Merchant S, Puzniak L. Risk factors for hospitalized patients with resistant or multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2018;7(1):79. doi: 10.1186/s13756-018-0370-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doi Y, Husain S, Potoski BA, McCurry KR, Paterson DL. Extensively drug-resistant Acinetobacter baumannii. Emerg Infect Dis. 2009;15(6):980. doi: 10.3201/eid1506.081006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassetti M, Righi E, Esposito S, Petrosillo N, Nicolini L. Drug treatment for multidrug-resistant Acinetobacter baumannii infections. Future Microbiol. 2008;3:649–660. doi: 10.2217/17460913.3.6.649. [DOI] [PubMed] [Google Scholar]
- 25.Sileshi A, Tenna A, Feyissa M, Shibeshi W. Evaluation of ceftriaxone utilization in medical and emergency wards of Tikur Anbessa specialized hospital: a prospective cross-sectional study. BMC Pharmacol Toxicol. 2016;17(1):1–10. doi: 10.1186/s40360-016-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moghadam MT, Mirzaei M, Moghaddam MFT, Babakhani S, Yeganeh O, Asgharzadeh S, et al. The Challenge of Global Emergence of Novel Colistin-Resistant Escherichia coli ST131. Microbial drug resistance (Larchmont, NY). [DOI] [PubMed]
- 27.Poulikakos P, Tansarli G, Falagas M. Combination antibiotic treatment versus monotherapy for multidrug-resistant, extensively drug-resistant, and pandrug-resistant Acinetobacter infections: a systematic review. Eur J Clin Microbiol Infect Dis. 2014;33(10):1675–1685. doi: 10.1007/s10096-014-2124-9. [DOI] [PubMed] [Google Scholar]
- 28.Tigen ET, Koltka EN, Dogru A, Orhon ZN, Gura M, Vahaboglu H. Impact of the initiation time of colistin treatment for Acinetobacter infections. J Infect Chemother. 2013;19(4):703–708. doi: 10.1007/s10156-013-0549-1. [DOI] [PubMed] [Google Scholar]
- 29.Falagas ME, Rafailidis PI, Ioannidou E, Alexiou VG, Matthaiou DK, Karageorgopoulos DE, et al. Colistin therapy for microbiologically documented multidrug-resistant Gram-negative bacterial infections: a retrospective cohort study of 258 patients. Int J Antimicrob Agents. 2010;35(2):194–199. doi: 10.1016/j.ijantimicag.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Ah Y-M, Kim A-J, Lee J-Y. Colistin resistance in Klebsiella pneumoniae. Int J Antimicrob Agents. 2014;44(1):8–15. doi: 10.1016/j.ijantimicag.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 31.Moubareck CA, Mouftah SF, Pál T, Ghazawi A, Halat DH, Nabi A, et al. Clonal emergence of Klebsiella pneumoniae ST14 co-producing OXA-48-type and NDM carbapenemases with high rate of colistin resistance in Dubai, United Arab Emirates. Int J Antimicrob Agents. 2018;52(1):90–95. doi: 10.1016/j.ijantimicag.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. MBio. 2017;8(3). [DOI] [PMC free article] [PubMed]
- 33.Abd El-Baky RM, Masoud SM, Mohamed DS, Waly NG, Shafik EA, Mohareb DA, et al. Prevalence and some possible mechanisms of colistin resistance among multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa. Infection and Drug Resistance. 2020;13:323. doi: 10.2147/IDR.S238811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rebelo AR, Bortolaia V, Kjeldgaard JS, Pedersen SK, Leekitcharoenphon P, Hansen IM, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eurosurveillance. 2018;23(6):17–00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. MBio. 2019;10(3). [DOI] [PMC free article] [PubMed]
- 36.Al-Kadmy IM, Ibrahim SA, Al-Saryi N, Aziz SN, Besinis A, Hetta HF. Prevalence of genes involved in colistin resistance in acinetobacter baumannii: first report from Iraq. Microb Drug Resist. 2020;26(6):616–622. doi: 10.1089/mdr.2019.0243. [DOI] [PubMed] [Google Scholar]
- 37.Wang C, Feng Y, Liu L, Wei L, Kang M, Zong Z. Identification of novel mobile colistin resistance gene mcr-10. Emerging Microbes Infections. 2020;9(1):508–516. doi: 10.1080/22221751.2020.1732231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lei T, Zhang J, Jiang F, He M, Zeng H, Chen M, et al. First detection of the plasmid-mediated colistin resistance gene mcr-1 in virulent Vibrio parahaemolyticus. Int J Food Microbiol. 2019;308:108290. doi: 10.1016/j.ijfoodmicro.2019.108290. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Liu S, Fu J, Yin J, Zhao J, Zhong C, et al. Co-occurrence of colistin and meropenem resistance determinants in a Stenotrophomonas strain isolated from sewage water. Microb Drug Resist. 2019;25(3):317–325. doi: 10.1089/mdr.2018.0418. [DOI] [PubMed] [Google Scholar]
- 40.Lin J, Xu C, Fang R, Cao J, Zhang X, Zhao Y, et al. Resistance and heteroresistance to colistin in Pseudomonas aeruginosa isolates from Wenzhou. China Antimicrobial agents and chemotherapy. 2019;63(10):e00556–e619. doi: 10.1128/AAC.00556-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ortiz dlTV, Ortega A, Buñuel F, Pérez-Vázquez M, Marcos B, Oteo J, Detection of the high-risk clone ST131 of Escherichia coli carrying the colistin resistance gene mcr-1 and causing acute peritonitis. Int J Antimicrobial Agents. 2017;49(1):115. doi: 10.1016/j.ijantimicag.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 42.Attia H, Szubin R, Yassin AS, Monk JM, Aziz RK. Draft genome sequences of four metallo-beta-lactamase-producing multidrug-resistant Klebsiella pneumoniae clinical isolates, including two colistin-resistant strains, from Cairo, Egypt. Microbiology resource announcements. 2019;8(7). [DOI] [PMC free article] [PubMed]
- 43.Ahmed SS, Alp E, Hopman J, Voss A. Global epidemiology on colistin resistant Acinetobacter baumannii. Journal of Infectious Diseases & Therapy. 2016.
- 44.El-Sayed Ahmed MAE-G, Zhong L-L, Shen C, Yang Y, Doi Y, Tian G-B. Colistin and its role in the Era of antibiotic resistance: an extended review (2000–2019). Emerging Microbes & Infections. 2020;9(1):868–85. [DOI] [PMC free article] [PubMed]
- 45.Kaye KS, Pogue JM, Tran TB, Nation RL, Li J. Agents of last resort: polymyxin resistance. Infect Dis Clin North Am. 2016;30(2):391. doi: 10.1016/j.idc.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 46.Falagas ME, Rafailidis PI, Matthaiou DK. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist Updates. 2010;13(4–5):132–138. doi: 10.1016/j.drup.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 47.Zhong L-L, Zhou Q, Tan C-Y, Roberts AP, El-Sayed MAE-G. Multiplex loop-mediated isothermal amplification (multi-LAMP) assay for rapid detection of mcr-1 to mcr-5 in colistin-resistant bacteria. Infection and drug resistance. 2019;12:1877. [DOI] [PMC free article] [PubMed]
- 48.Madec J-Y, Haenni M. Antimicrobial resistance plasmid reservoir in food and food-producing animals. Plasmid. 2018;99:72–81. doi: 10.1016/j.plasmid.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 49.Andersson DI, Hughes D, Kubicek-Sutherland JZ. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist Updates. 2016;26:43–57. doi: 10.1016/j.drup.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 50.Yang Q, Li M, Spiller OB, Andrey DO, Hinchliffe P, Li H, et al. Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms. Nat Commun. 2017;8(1):1–12. doi: 10.1038/s41467-017-02149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moffatt JH, Harper M, Adler B, Nation RL, Li J, Boyce JD. Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55(6):3022–3024. doi: 10.1128/AAC.01732-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherry N, Howden B. Emerging Gram negative resistance to last-line antimicrobial agents fosfomycin, colistin and ceftazidime-avibactam–epidemiology, laboratory detection and treatment implications. Expert Rev Anti Infect Ther. 2018;16(4):289–306. doi: 10.1080/14787210.2018.1453807. [DOI] [PubMed] [Google Scholar]
- 53.Soon RL, Nation RL, Cockram S, Moffatt JH, Harper M, Adler B, et al. Different surface charge of colistin-susceptible and-resistant Acinetobacter baumannii cells measured with zeta potential as a function of growth phase and colistin treatment. J Antimicrob Chemother. 2011;66(1):126–133. doi: 10.1093/jac/dkq422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olaitan AO, Morand S, Rolain J-M. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poirel L, Madec JY, Lupo A, Schink AK, Kieffer N, Nordmann P, et al. Antimicrobial resistance in Escherichia coli. Antimicrobial Resistance in Bacteria from Livestock and Companion Animals. 2018:289–316. [DOI] [PubMed]
- 56.Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willett J. Bacterial two-component systems share a common mechanism to regulate signaling and specificity. 2012.
- 58.Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, et al. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother. 2011;55(7):3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wösten MM, Kox LF, Chamnongpol S, Soncini FC, Groisman EA. A signal transduction system that responds to extracellular iron. Cell. 2000;103(1):113–125. doi: 10.1016/S0092-8674(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 60.Huang J, Li C, Song J, Velkov T, Wang L, Zhu Y, et al. Regulating polymyxin resistance in Gram-negative bacteria: roles of two-component systems PhoPQ and PmrAB. Future Microbiol. 2020;15(6):445–459. doi: 10.2217/fmb-2019-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baron S, Hadjadj L, Rolain J-M, Olaitan AO. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents. 2016;48(6):583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 62.Llobet E, Tomas JM, Bengoechea JA. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology. 2008;154(12):3877–3886. doi: 10.1099/mic.0.2008/022301-0. [DOI] [PubMed] [Google Scholar]
- 63.Magnet S, Courvalin P, Lambert T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother. 2001;45(12):3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin M-F, Lin Y-Y, Lan C-Y. Contribution of EmrAB efflux pumps to colistin resistance in Acinetobacter baumannii. J Microbiol. 2017;55(2):130–136. doi: 10.1007/s12275-017-6408-5. [DOI] [PubMed] [Google Scholar]
- 65.Sundaramoorthy NS, Suresh P, Ganesan SS, GaneshPrasad A, Nagarajan S. Restoring colistin sensitivity in colistin-resistant E. coli: Combinatorial use of MarR inhibitor with efflux pump inhibitor. Sci Reports. 2019;9(1):1–13. doi: 10.1038/s41598-019-56325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bojkovic J, Richie DL, Six DA, Rath CM, Sawyer WS, Hu Q, et al. Characterization of an Acinetobacter baumannii lptD deletion strain: permeability defects and response to inhibition of lipopolysaccharide and fatty acid biosynthesis. J Bacteriol. 2016;198(4):731–741. doi: 10.1128/JB.00639-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hood MI, Becker KW, Roux CM, Dunman PM, Skaar EP. Genetic determinants of intrinsic colistin tolerance in Acinetobacter baumannii. Infect Immun. 2013;81(2):542–551. doi: 10.1128/IAI.00704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 69.Nhu NTK, Riordan DW, Nhu TDH, Thanh DP, Thwaites G, Lan NPH, et al. The induction and identification of novel Colistin resistance mutations in Acinetobacter baumannii and their implications. Sci Rep. 2016;6(1):1–8. doi: 10.1038/srep28291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mlynarcik P, Kolar M. Molecular mechanisms of polymyxin resistance and detection of mcr genes. Biomed Pap Med Fac Palacky Univ Olomouc. 2019;163(1):28–38. doi: 10.5507/bp.2018.070. [DOI] [PubMed] [Google Scholar]
- 71.Mills G, Dumigan A, Kidd T, Hobley L, Bengoechea JA. Identification and characterization of two Klebsiella pneumoniae lpxL lipid A late acyltransferases and their role in virulence. Infection and immunity. 2017;85(9). [DOI] [PMC free article] [PubMed]
- 72.Panta PR, Kumar S, Stafford CF, Billiot CE, Douglass MV, Herrera CM, et al. A DedA family membrane protein is required for Burkholderia thailandensis colistin resistance. Front Microbiol. 2019;10:2532. doi: 10.3389/fmicb.2019.02532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun J, Zhang H, Liu Y-H, Feng Y. Towards understanding MCR-like colistin resistance. Trends Microbiol. 2018;26(9):794–808. doi: 10.1016/j.tim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 74.Burrowes B, Harper DR, Anderson J, McConville M, Enright MC. Bacteriophage therapy: potential uses in the control of antibiotic-resistant pathogens. Expert Rev Anti Infect Ther. 2011;9(9):775–785. doi: 10.1586/eri.11.90. [DOI] [PubMed] [Google Scholar]
- 75.Abedon ST, Thomas-Abedon C, Thomas A, Mazure H. Bacteriophage prehistory: is or is not Hankin, 1896, a phage reference? Bacteriophage. 2011;1(3):174–178. doi: 10.4161/bact.1.3.16591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lepelletier D, Berthelot P, Lucet J-C, Fournier S, Jarlier V, Grandbastien B, et al. French recommendations for the prevention of ‘emerging extensively drug-resistant bacteria’(eXDR) cross-transmission. J Hosp Infect. 2015;90(3):186–195. doi: 10.1016/j.jhin.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 77.Nagel TE, Chan BK, De Vos D, El-Shibiny A, Kang'ethe EK, Makumi A, et al. The developing world urgently needs phages to combat pathogenic bacteria. Front Microbiol. 2016;7:882. doi: 10.3389/fmicb.2016.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pereira C, Moreirinha C, Teles L, Rocha RJ, Calado R, Romalde JL, et al. Application of phage therapy during bivalve depuration improves Escherichia coli decontamination. Food Microbiol. 2017;61:102–112. doi: 10.1016/j.fm.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 79.Rozema EA, Stephens TP, Bach SJ, Okine EK, Johnson RP, Stanford K, et al. Oral and rectal administration of bacteriophages for control of Escherichia coli O157: H7 in feedlot cattle. J Food Prot. 2009;72(2):241–250. doi: 10.4315/0362-028X-72.2.241. [DOI] [PubMed] [Google Scholar]
- 80.De Smet J, Hendrix H, Blasdel BG, Danis-Wlodarczyk K, Lavigne R. Pseudomonas predators: understanding and exploiting phage–host interactions. Nat Rev Microbiol. 2017;15(9):517. doi: 10.1038/nrmicro.2017.61. [DOI] [PubMed] [Google Scholar]
- 81.Tran TAT, Struck DK, Young R. Periplasmic domains define holin-antiholin interactions in T4 lysis inhibition. J Bacteriol. 2005;187(19):6631–6640. doi: 10.1128/JB.187.19.6631-6640.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chegini Z, Khoshbayan A, Moghadam MT, Farahani I, Jazireian P, Shariati A. Bacteriophage therapy against Pseudomonas aeruginosa biofilms: a review. Ann Clin Microbiol Antimicrob. 2020;19(1):1–17. doi: 10.1186/s12941-020-00389-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nobrega FL, Costa AR, Kluskens LD, Azeredo J. Revisiting phage therapy: new applications for old resources. Trends Microbiol. 2015;23(4):185–191. doi: 10.1016/j.tim.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 84.Jeannot K, Bolard A, Plesiat P. Resistance to polymyxins in Gram-negative organisms. Int J Antimicrob Agents. 2017;49(5):526–535. doi: 10.1016/j.ijantimicag.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 85.Antoniadou A, Kontopidou F, Poulakou G, Koratzanis E, Galani I, Papadomichelakis E, et al. Colistin-resistant isolates of Klebsiella pneumoniae emerging in intensive care unit patients: first report of a multiclonal cluster. J Antimicrob Chemother. 2007;59(4):786–790. doi: 10.1093/jac/dkl562. [DOI] [PubMed] [Google Scholar]
- 86.Ebrahimi S, Sisakhtpour B, Mirzaei A, Karbasizadeh V, Moghim S. Efficacy of Isolated Bacteriophage Against Biofilm Embedded Colistin-Resistant Acinetobacter baumannii. 2020.
- 87.Hao G, Chen AI, Liu M, Zhou H, Egan M, Yang X, et al. Colistin resistance-mediated bacterial surface modification sensitizes phage infection. Antimicrobial Agents and Chemotherapy. 2019;63(12). [DOI] [PMC free article] [PubMed]
- 88.Shokri D, Soleimani-Delfan A, Fatemi SM. Assessment of phage cocktails with extended host range activity against antibiotic resistant strains of Pseudomonas aeruginosa. Comp Clin Pathol. 2017;26(2):417–422. doi: 10.1007/s00580-016-2394-y. [DOI] [Google Scholar]
- 89.Abdelkader K, Gutiérrez D, Grimon D, Ruas-Madiedo P, Lood C, Lavigne R, et al. Lysin LysMK34 of Acinetobacter baumannii bacteriophage PMK34 has a turgor pressure-dependent intrinsic antibacterial activity and reverts colistin resistance. Applied and Environmental Microbiology. 2020;86(19). [DOI] [PMC free article] [PubMed]
- 90.Manohar P, Tamhankar AJ, Lundborg CS, Nachimuthu R. Therapeutic characterization and efficacy of bacteriophage cocktails infecting Escherichia coli, Klebsiella pneumoniae, and Enterobacter species. Front Microbiol. 2019;10:574. doi: 10.3389/fmicb.2019.00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Blasco L, Ambroa A, Trastoy R, Bleriot I, Moscoso M, Fernández-Garcia L, et al. In vitro and in vivo efficacy of combinations of colistin and different endolysins against clinical strains of multi-drug resistant pathogens. Sci Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-64145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thummeepak R, Kitti T, Kunthalert D, Sitthisak S. Enhanced antibacterial activity of Acinetobacter baumannii bacteriophage ØABP-01 endolysin (LysABP-01) in combination with colistin. Front Microbiol. 2016;7:1402. doi: 10.3389/fmicb.2016.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blasco L, Ambroa A, Trastoy R, Perez-Nadales E, Fernández-Cuenca F, Torre-Cisneros J, et al. In vitro and in vivo efficacy of the combination of colistin and endolysins against clinical strains of Multi-Drug Resistant (MDR) pathogens. bioRxiv. 2019:662460. [DOI] [PMC free article] [PubMed]
- 94.Bernasconi OJ, Donà V, Tinguely R, Endimiani A. In vitro activity of three commercial bacteriophage cocktails against multidrug-resistant Escherichia coli and Proteus spp strains of human and non-human origin. J Global Antimicrobial Resistance. 2017;8:179–185. doi: 10.1016/j.jgar.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 95.Aslam S, Courtwright AM, Koval C, Lehman SM, Morales S, Furr CLL, et al. Early clinical experience of bacteriophage therapy in 3 lung transplant recipients. Am J Transplant. 2019;19(9):2631–2639. doi: 10.1111/ajt.15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vukotic G, Obradovic M, Novovic K, Di Luca M, Jovcic B, Fira D, et al. Characterization, antibiofilm, and depolymerizing activity of two phages active on carbapenem-resistant acinetobacter baumannii. Front Med. 2020;7:426. doi: 10.3389/fmed.2020.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schirmeier E, Zimmermann P, Hofmann V, Biebl M, Gerstmans H, Maervoet VE, et al. Inhibitory and bactericidal effect of Artilysin® Art-175 against colistin-resistant mcr-1-positive Escherichia coli isolates. Int J Antimicrob Agents. 2018;51(3):528–529. doi: 10.1016/j.ijantimicag.2017.08.027. [DOI] [PubMed] [Google Scholar]
- 98.Defraine V, Schuermans J, Grymonprez B, Govers SK, Aertsen A, Fauvart M, et al. Efficacy of artilysin Art-175 against resistant and persistent Acinetobacter baumannii. Antimicrob Agents Chemother. 2016;60(6):3480–3488. doi: 10.1128/AAC.00285-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.