Abstract
Development of combination therapy to decrease side effects of chemotherapeutic drugs and increase their utilization rate in combination with gene editing is a key research topic in tumor treatment. The present study aimed to investigate the effect of cationic microbubbles (CMBs) carrying paclitaxel (PTX) and C-erbB-2 knockout plasmid on the endometrial cancer cell line HEC-1A and to determine how C-erbB-2 regulates the function of endometrial cancer cells. Cells were treated with CMB, PTX, PTX-CMBs, cationic plasmid-carrying or cationic PTX-carrying plasmid groups. After verifying the most effective combination of PTX-CMBs and plasmids, HEC-1A cells were transfected. Reverse transcription-quantitative (RT-q)PCR and western blotting were used to measure C-erbB-2 and protein expression. After verifying C-erbB-2 knockout, invasion, healing, clone formation and proliferation of HEC-1A cells were assessed. Simultaneously, expression levels of the genes for P21, P27, mammalian target of rapamycin (mTOR), and Bcl-2 associated death promoter (Bad) were measured by RT-qPCR. Compared with the PTX group, CMBs significantly enhanced the absorption efficiency of PTX by HEC-1A cells. C-erbB-2 knockout had an inhibitory effect on the proliferation, migration and invasion of HEC-1A cells; cell proliferation and invasion of the group carrying PTX and plasmids simultaneously were significantly weakened. The C-erbB-2-knockout group exhibited increased expression of P21 and P27. Simultaneously loading PTX and plasmid may be novel combination therapy with great potential. C-erbB-2 may regulate the proliferation of HEC-1A cells by downregulating expression of P21 and P27.
Keywords: chemotherapy-gene combination therapy, chemotherapy drug, gene editing, tumor therapy, ultrasound cationic microbubbles
Introduction
Endometrial cancer (EC) is the sixth most commonly diagnosed cancer worldwide (1). It has been estimated that the incidence of EC in China will increase twofold by 2030 to reach >120,000 cases (2).
Treatments for EC include radiotherapy, chemotherapy, surgery, hormone and molecular targeted therapy and immune-checkpoint inhibitors (3). The prognosis of EC is poor, so developing safe and efficacious treatments is key to improve patient outcomes.
Paclitaxel (PTX) is an anti-cancer drug isolated and extracted from yew trees that is used in the treatment of breast and ovarian cancer, EC and other types of disease (4,5). PTX kills dividing cancer cells by stabilizing microtubules of mitotic spindles (6) but PTX is also highly toxic to healthy cells in the human body (7). Improving the anti-tumor effects of drugs and decreasing their systemic side effects is important.
Human epidermal growth factor receptor-2, also known as C-erbB-2, is a transmembrane tyrosine kinase receptor (8). C-erbB-2 is expressed at low levels in the epithelial cells of most organs in healthy human tissue and at slightly higher levels in fetal tissue (9,10). Studies have shown that C-erbB-2 is overexpressed in various types of tumor, such as EC, breast cancer, ovarian cancer, stomach and lung cancer (11–14) and its overexpression is associated with proliferation of tumor cells. Zhou et al (15) note that high expression of C-erbB-2 is closely related to the prognosis of ovarian cancer; Li et al (16) suggest that C-erbB-2 is a potential therapeutic target for lung cancer and Erickson et al (17) state that C-erbB-2 is also an effective potential therapeutic target in endometrial cancer. Therefore, directly knocking out C-erbB-2 to explore the effect on the endometrium is a feasible method
Gene-level editing is based on clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) technology. CRISPR technology was first used in Escherichia coli (18,19). CRISPR enables correction of errors in the genome and gene regulation in cells and organisms to be performed rapidly, cheaply and with relative ease (20). Guide (g)RNA matches a desired target gene and Cas9 (an endonuclease) causes a double-stranded DNA break, thereby allowing modifications to the genome (21). The CRISPR/Cas9 system is the most powerful gene-editing method worldwide (22) but its safe and efficient use in the human body is a major challenge (23).
Ultrasound non-invasively controls the release of drugs and carriers wrapped in or around gas-filled microbubbles (MBs) (24). Ultrasound also induces cavitation effects, which can produce transient pores in cell membranes. This increases cell permeability and enhances the efficiency of drug delivery (25,26). The surface of cationic (C) gas-filled MBs has a positive charge, allowing effective combination with negatively charged plasmid DNA to increase the loading rate of plasmids (27). Hence, CMBs are used to deliver drugs or genes.
In the present study, the EC cell line HEC-1A was cultured in vitro. CMB were used to carry PTX and the CRISPR/Cas9 gene-targeted editing system under ultrasonic irradiation. The present study aimed to explore the interference effect of C-erbB-2 knockout by CRISPR/Cas9-PTX-CMB on endometrial cancer cells.
Materials and methods
Cell culture
Wuhan Procell Life Technology (Wuhan, China) provided the human EC cell line (HEC-1A). The HEC-1A cell line was supplemented with 10% fetal bovine serum (Biosharp Life Sciences) and 5% penicillin-streptomycin (Biosharp Life Sciences). Cells were cultured (37°C; 5% CO2) in complete medium [DMEM (Gibco; Thermo Fisher Scientific, Inc.) + 10% fetal bovine serum (Biosharp Life Sciences) + 5% penicillin-streptomycin (Biosharp Life Sciences)] lacking bacteria, yeasts, fungi or mycoplasma.
Construction of primers and plasmids
The sequences of three candidate gRNAs targeting C-erbB-2 were selected. For gRNA1, target 1 was 5′-TCATCGCTCACAACCAAGTG-3′ and target 2 was 5′-CAGGGGTGGTATTGTTCAGC-3′. For gRNA2, target 1 was 5′-TCATCGCTCACAACCAAGTG-3′ and target 2 was 5′-CGGGTCTCCATTGTCTAGCA-3′. For gRNA3, target 1 was 5′-CGCTCACAACCAAGTGAGGC-3′ and target 2 was 5′-ACAGGGGTGGTATTGTTCAG-3′.
These sequences were cloned into the pGE-5 plasmid encoding Cas9 and the gRNA scaffold. The empty pGE-5 plasmid (Shanghai GenePharma Co., Ltd.) was designed as a negative control. Synthesis was performed by Shanghai GenePharma Co., Ltd. The pGE-5 plasmid contained enhanced green fluorescent protein, ampicillin, puromycin-resistance genes and other markers, to aid selection of the best gRNA sequence and screen out successfully transfected cells.
Screening for the best targeting C-erbB-2-knockout gRNA
HEC-1A cells were inoculated in a 6-well plate (4×105/well) and large petri dish (1×106/dish). Then, 2.5 ml complete medium [DMEM (Gibco; Thermo Fisher Scientific, Inc.) + 10% fetal bovine serum (Biosharp Life Sciences) + 5% penicillin-streptomycin (Biosharp Life Sciences)] was added to each well of the 6-well plate and 10 ml complete medium was added to the large dish in a 37°C incubator. Culture was performed for 24 h until cells adhered to the wall and confluence reached 70%. Cells were washed once with phosphate-buffered saline (PBS) before transfection. A total of 2.5 ml Opti-MEM Reduced-Serum Medium (Gibco; Thermo Fisher Scientific, Inc.) was added each well of the 6-well plate and 5 ml Opti-MEM Reduced-Serum Medium (Gibco; Thermo Fisher Scientific, Inc.) was added to the large dish, followed by incubation at 37°C for 2 h. Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) was employed to transfect different CRISPR/Cas9 plasmids into HEC-1A cells according to the manufacturer's instructions. Lipofectamine® 3000 6 µl or 18 µl (Thermo Fisher Scientific, Inc.) and gRNA plasmid 2 µg or 9 µg and P3000 4 µl or 18 µl (Thermo Fisher Scientific, Inc.) were mixed gently and added to a 6-well plate (Lipofectamine® 3000 6 µl+ gRNA plasmid 2 µg + P3000 4 µl) or a large dish (Lipofectamine® 3000 18 µl + gRNA plasmid 9 µg + P3000 18 µl) for cell growth after standing at room temperature for 15 minutes. The cells were placed in a 37°C incubator with 5% CO2 for transfection for 6 h and then the medium in the 6-well plate and the large dish was changed to complete medium, the cells continued to be cultured in an incubator at 37°C with 5% CO2, Images were captured using a fluorescence microscope (cat. no. BX51; Olympus Corporation) after 24 h and 48 h. After capturing the 48 h image, RNA from cells in the 6-well plate was collected for reverse transcription-quantitative (RT-q)PCR. The total protein from cells in the large dish was collected for western blotting to measure the expression of C-erbB-2 protein and verify the knockout efficiency of each group of plasmids.
CMB construction
Stearoyl phosphatidylcholine (Avanti Polar Lipids, Inc.), (2,3-dioleoyl-propyl) trimethylammonium chloride (Avanti Polar Lipids) and polyoxyethylene stearate (Shanghai Trustin Chemical Co., Ltd.) were weighed accurately and used at a molar ratio of 70:16:14. Reagents were dissolved in chloroform and the organic solvent was removed by rotary evaporation under reduced pressure following uniform mixing. The mixture was placed in an ultrasonic water bath at 60°C for 15 min to obtain a lipid suspension. The suspension was divided into vials, filled with sulfur hexafluoride (SF6) gas and shaken mechanically for 1 min to obtain CMB.
Construction of PTX-loaded CMB
Stearoyl phosphatidylcholine, trimethyl ammonium chloride and polyoxyethylene stearate were weighed accurately and used at a molar ratio of 70:16:14. Reagents were dissolved in chloroform. PTX dissolved in methanol (1 mg/ml) was added. Following uniform mixing, the organic solvent was removed by rotary evaporation under reduced pressure. The lipid film containing PTX was added to a buffer solution. A PTX-containing lipid suspension was obtained following placement in an ultrasonic bath at 60°C for 15 min. After dispensing in vials, the suspension was filled with SF6 gas and shaken mechanically for 1 min to obtain membrane shells containing PTX (i.e., PTX-CMB).
Physical measurement of CMB and PTX-CMB
CMB were observed under an optical microscope to detect their morphology and distribution. The particle size and surface potential were detected by a laser particle size and surface potential detector (Malvern Instruments).
Combination and identification of CRISPR/Cas9 with CMB
CMB and PTX-CMB (both 160 ml) were collected and 20 ml 3,3′-dioctadecyloxacarbocyanine perchlorate fluorescent dye (Dio; 7.561 mM) was added. After mixing and incubation at room temperature for 15 min, 40 mg plasmid was added and a Cy3 nucleic-acid labeling kit (Label IT® Tracker™ Intracellular Nucleic Acid Localization kit; Mirus Bio, LLC) was used for labeling. CMB, PTX-CMB and plasmids were mixed thoroughly and incubated at 4°C for 20 min. The binding of MB and plasmids was observed under a confocal microscope (magnification, ×600).
MB transfection
To prepare MB-plasmid mixture, 10 µg plasmid and 1.14×109 MB was added to a sterile centrifuge tube, followed by gentle blowing with a pipette tip to aid mixing. The mixture was allowed to stand for 20 min at 4°C. For cell preparation, 2×105 cells were added to a 6-well plate with a sterile 22×22 mm cover glass at the bottom and then placed in a 37°C incubator for 24 h. After 24 h, the original medium [DMEM (Gibco; Thermo Fisher Scientific, Inc.) + 10% fetal bovine serum (Biosharp Life Sciences) + 5% penicillin-streptomycin (Biosharp Life Sciences)] was aspirated and washed with PBS. Tweezers were used to place the coverslip on the cover of the 6-well plate with the cell surface facing the liquid surface. Complete medium (as aforementioned) was added to each well of the 6-well plate until the liquid overflowed. The MB-plasmid mixture was added to the upper layer and placed on the 6-hole plate cover. A small ultrasonic instrument was used to sonicate MBs and cells at the bottom of the 6-well plate. The sonication conditions were 1 MHz at 0.75 W/cm2 for 30 sec. The CMB and PTX-CMB group were sonicated under identical conditions. For the group carrying PTX and CRISPR/Cas9 transfection (CRISPR/Cas9-PTX-CMB), the cells were cultured at 37°C for 24 h, and a Puro drug sieve was used for 7-day screening. The expanded culture was used for subsequent experiments.
RT-qPCR
An RNA extraction kit (Qiagen GmbH) was used to extract RNA from HEC-1A cells in the CMB, PTX, PTX-CMB, CRISPR/Cas9-CMB and CRISPR/Cas9-PTX-CMB groups. RT-qPCR was performed using a RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.; 42°C for 60 min, then 25°C for 5 min, then 42°C for 60 min and 70°C for 5 min.) and PowerUp SYBR Green Master Mix (Applied Biosystems) to determine cellular mRNA levels of C-erbB-2, mechanistic target of rapamycin (mTOR), Bcl-2 associated death promoter (Bad), P27 and P21 in each group. The primer sequence of each gene is as follows: C-erbB-2: F 5′-ACCCAGCTCTTTGAGGACAA-3′ R 5′-ATCGTGTCCTGGTAGCAGAG-3′; mTOR: F 5′-CCTGCCTTTGTCATGCCTTT-3′ R 5′-CTGGGTTTGGATCAGGGTCT-3′; Bad: F 5′-GAAGACTCCAGCTCTGCAGA-3′ R 5′-CATCCCTTCGTCGTCCTCC-3′; P27: F 5′-AGGAACTCGACTCAGACGTG-3′ R 5′-TATTTGGAGGCACAGCAGGA-3′; P21: F 5′-GCCCAGTGGACAGCGAGCAG-3′ R 5′-GCCGGCGTTTGGAGTGGTAGA-3′; β-actin (Reference gene): F 5′-AAGGATTCCTATGTGGGCGAC-3′ R 5′-CGTACAGGGATAGCACAGCC-3′. Thermal cycling conditions were: 50°C for 2 min, 95°C for 2 min, then 40 cycles of 95°C for 15 sec, 60°C for 15 sec, and 72°C for 1 min. The method of quantification was the 2−ΔΔCq method (28).
Western blotting
RIPA (Beyotime Institute of Biotechnology) lysis solution [200 µl; containing 2 µl PMSF (Beyotime Institute of Biotechnology) + 2 µl Cocktail (Cell Signaling Technology, Inc.)] was added to the cell pellets of each group (CMB, PTX, PTX-CMB, CRISPR/Cas9-CMB and CRISPR/Cas9-PTX-CMB groups) to lyse the cells. The cells were dispersed and mixed well and then placed on ice for 30 min lysis. Following centrifugation at 12,000 × g for 10 min at 4°C, the supernatant was aspirated. The protein concentration was determined by a bicinchoninic acid kit (Beyotime Institute of Biotechnology). Total protein (30 µg/well) was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 8% gels and transferred to polyvinylidene fluoride (PVDF) membranes. After fixing with anhydrous methanol at room temperature for 10 sec, PVDF membranes were washed with TBST(0.05% Tween-20) and blocked with TBST (0.05% Tween-20) containing 3% bovine serum albumin(Beyotime Institute of Biotechnology) at room temperature for 1 h. PVDF membranes were incubated with anti-C-erbB-2 monoclonal antibody (Abcam; cat. no. ab16901, 1:1,000) and anti-β-actin-monoclonal antibody (Cell Signaling Technology, Inc.; cat. no. 4970s, 1:1,000) overnight at 4°C, washed three times with TBST, and incubated with horseradish peroxidase-labeled secondary antibody (Abcam; cat. no. ab205719, 1:10,000) and β-actin secondary antibody(Cell Signaling Technology, Inc.; cat. no. 7074P2, 1:2,000) for 1 h at room temperature on a shaker. at room temperature for 1 h on a shaker. After washing three times with TBST, bands were visualized using chemiluminescent reagents (A and B solution; Immobilon Western; Sigma-Aldrich; Merck KGaA) in a dark room and analyzed using ImageJ (National Institutes of Health; v1.50i).
Cell scratching and healing
Each cell group (CMB, PTX, PTX-CMB, CRISPR/Cas9-CMB and CRISPR/Cas9-PTX-CMB groups) was cultured in a Medium petri dish containing complete culture medium as aforementioned for 24 h at 37°C and 5% CO2. The cells were trypsinized and centrifuged at 80 × g for 3 min at room temperature. After aspirating the supernatant, 1 ml serum-free medium [DMEM (Gibco; Thermo Fisher Scientific, Inc.) + 5% penicillin-streptomycin (Biosharp Life Sciences)] was used to resuspend the cells. Then, 190 µl serum-free medium was added to 10 µl resuspended cells, followed by thorough mixing. Then, 10 µl cell suspension was removed for counting. The density of HEC-1A cells was maintained at 7×105/ml. Next, 70 µl cell suspension was placed in the wound-healing inserts (ibidi GmbH) and cultured in a 37°C incubator until the cells in the chambers on both sides of the wound-healing inserts covered the field of view. The scratch cell was pulled out vertically and the cells washed gently with PBS. Then, 1 ml serum-free medium was added and culturing allowed to continue. Images were recorded under a light microscope (magnification, ×100) every 4 h and stopped when the cells began to fuse. ImageJ (National Institutes of Health, v1.50i) was used to measure the healing area of each group of cells.
Clone formation
From each group (CMB, PTX, PTX-CMB, CRISPR/Cas9-CMB and CRISPR/Cas9-PTX-CMB groups) cells in the logarithmic growth phase were collected and inoculated in a 6-well plate at 1×103 cells/well. Then, 2 ml complete medium was added and changed every 3 days. Cells were cultured for 7 days in an incubator at 37°C. The culture was stopped when a clonal population of cells was visible to the naked eye. The medium was aspirated, washed with PBS and cells were fixed at room temperature with 4% paraformaldehyde for 20 min. After aspirating the 4% paraformaldehyde, 1 ml 10% Giemsa stain was added to each well at room temperature for 5 min and images were captured. In each group, the number of clones was manually counted in four randomly selected fields of view under a light microscope (magnification, ×40); >50 cells was considered as one colony.
Cell invasion
Matrigel was spread at 37°C on a 24-well plate chamber (membrane pore size: 8 µm) and aspirated after 1 h. Opti-MEM Reduced-Serum Medium (Gibco; Thermo Fisher Scientific, Inc.) was used to resuspend 1.5×105 cells in each group (CMB, PTX, PTX-CMB, CRISPR/Cas9-CMB and CRISPR/Cas9-PTX-CMB groups) into the upper chamber and complete medium was added to the lower chamber. Following 36 h culture at 37°C, the medium in the upper chamber was removed and washed three times with PBS. After removing the medium in the lower chamber, cells were fixed at room temperature with 4% paraformaldehyde for 30 min. Giemsa solution (10%; 600 µl) was added for staining at room temperature for 10 min. A total of 4 randomly selected fields of view were observed under a light microscope(×200) and images were captured.
Cell Counting Kit (CCK)-8 assay
CCK-8 (Beyotime Institute of Biotechnology) was used to measure the viability and proliferation of cells. A total of 2×103 cells/well of each group (CMB, PTX, PTX-CMB, CRISPR/Cas9-CMB and CRISPR/Cas9-PTX-CMB groups) were inoculated into 96-well plates. Three replicate wells were set up for each group and each time point. There were 75 wells in total, 15 holes in each group and 3 holes were used per group per day). After the cells were cultured at 37°C for 24 h, 3-wells were selected for each group. The cells were washed once with PBS, then 10 µl of CCK-8 reagent was added and the cells were cultured at 37°C for 2 h. Then, a microplate reader (Gene Company Limited) was used to measure the absorbance of the cells at 450 nm this was recorded as Day 1. After the cells were cultured at 37°C for 48 h, 3-wells were selected for each group again, rinsed with PBS, CCK-8 reagent added and cultured at 37°C for 2 h. Then, a microplate reader (Gene Company Limited) was used to measure the absorbance of the cells at 450 nm and recorded as Day 2. These steps were repeated until the fifth day of measurement.
Statistical analysis
The cell invasion and clone formation were repeated four times, the other experiments were repeated three times. Data are presented as the mean ± standard deviation. Statistical analysis were performed using SPSS 24.0 (IBM Corp.). Differences between multiple groups were assessed using independent samples Kruskal-Wallis test followed by Dunn's post hoc test for multiple comparisons. P<0.05 was considered to indicate a statistically significant difference.
Results
gRNA3 sequence has the highest efficiency in knocking out C-erbB-2
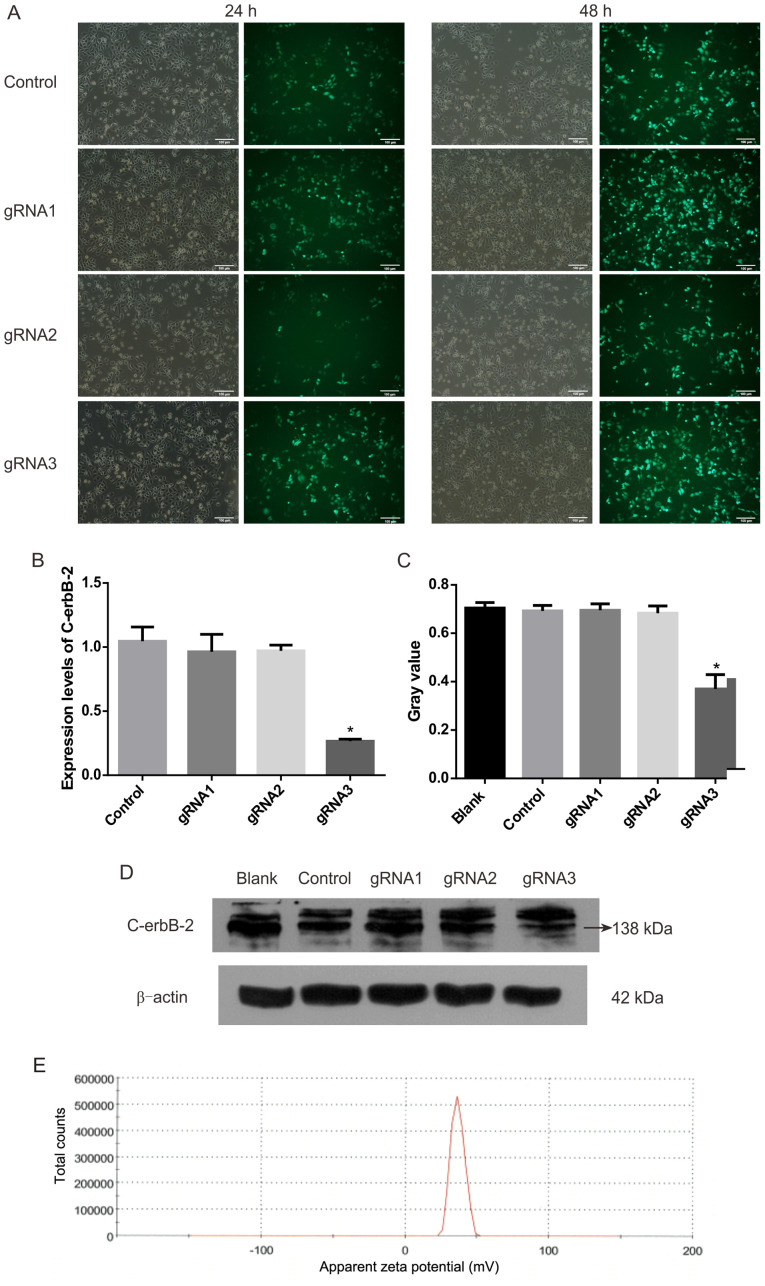
After three gRNA sequences were transfected into HEC-1A cells, the expression levels of C-erbB-2 and protein significantly decreased (Fig. 1). gRNA3 was the best sequence to knock out C-erbB-2 among the three gRNA sequences tested.
Figure 1.
Plasmid screening fluorescence map. (A) Observation and comparison of HEC-1A cells transfected with plasmids at 24 and 48 h under a fluorescence microscope. (B) Expression of C-erbB-2 mRNA was detected by reverse transcription-quantitative PCR following plasmid transfection. *P<0.05 vs. control. (C) Grayscale value of C-erbB-2 protein. (D) C-erbB-2 protein bands of plasmid-transfected cells. n=4. *P<0.05 vs. blank. (E) The Zeta potential of the CMB (mV). g, guide.
CMBs and PTX-CMBs are positively charged on the surface
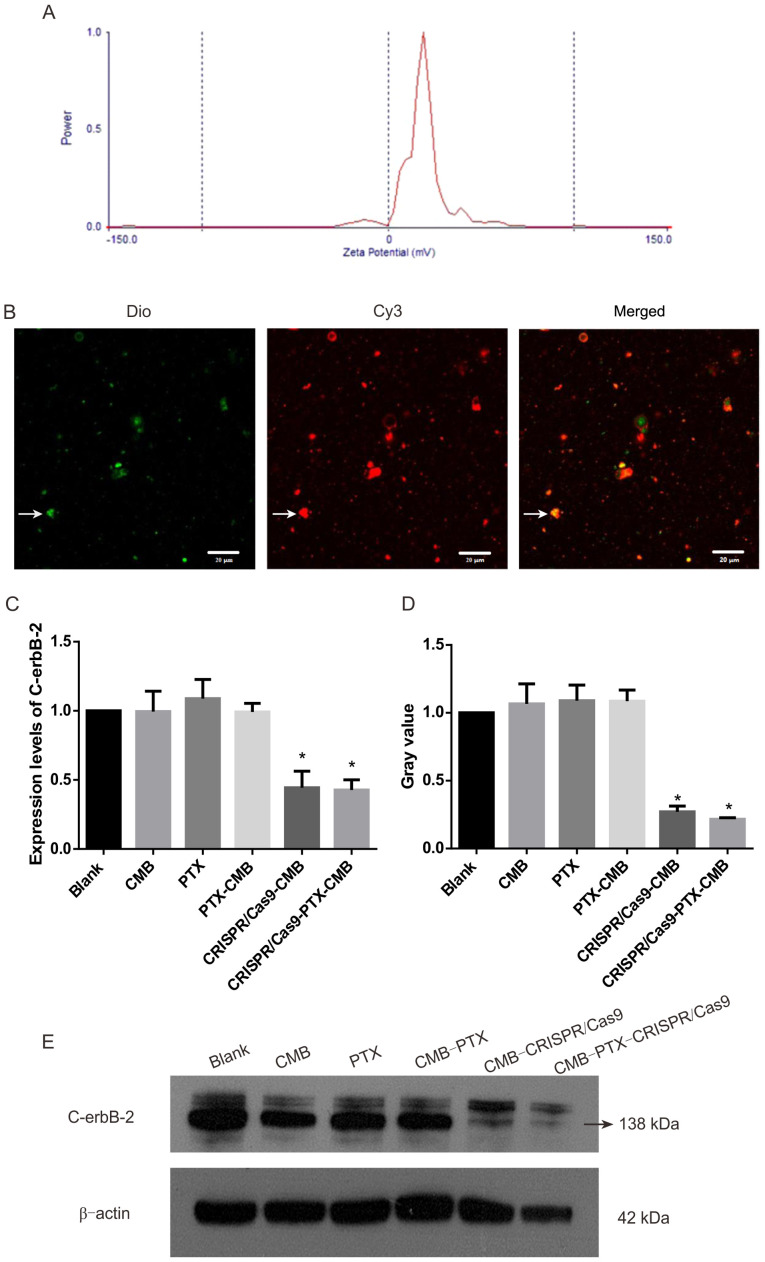
The particle size of CMBs was 1.213 µm and that of PTX-CMBs was 1.970 µm. The surface potential of CMBs was 36.70 mV (Fig. 1E) and that of PTX-CMBs was 20.41 mV (Fig. 2A). The concentration of CMBs and PTX-CMBs was calculated using a cell counter (CMBs, 28.38×109/ml; PTX-CMBs, 7.84×108/ml). Compared with CMB, the positive surface potential of PTX-CMBS decreased slightly and the particle size increased.
Figure 2.
MB and plasmid binding map and verification of knockout of the stable strain of C-erbB-2. (A) The Zeta potential of the PTX-CMB (mV). (B) CMBs bound to plasmids. White arrows indicate microvesicles. (C) Expression of C-erbB-2 mRNA following treatment was measured by reverse transcription-quantitative PCR. (D) C-erbb-2 protein levels in each treatment group. (E) Western blotting determined C-erbB-2 protein expression levels in each treatment group. n=3. *P<0.05 vs. blank. CMB, cationic microbubble; PTX, paclitaxel; Dio, 3,3′-dioctadecyloxacarbocyanine perchlorate.
Preparation and in vitro identification of CRISPR/Cas9-CMB
CMBs (160 µl) and Cy3-labeled Cas9 plasmids (40 µg) were incubated for 20 min. Under a fluorescence microscope, green MBs stained with Dio and Cy3-labeled Cas9 plasmids were observed (Fig. 2B). These data confirmed the effective combination of CMBs and plasmids.
CRISPR/Cas9-CMB and CRISPR/Cas9-PTX-CMB groups can effectively knock out C-erbB-2
Expression of C-erbB-2 protein in the CRISPR/Cas9-CMB and CRISPR/Cas9-PTX-CMB group decreased significantly compared with that in the other groups (Fig. 2D-E). This confirmed the effectiveness of transfection and establishment of a C-erbB-2 knockout line. Compared with the blank group, RT-qPCR demonstrated significant gene knockout in the CRISPR/Cas9-CMB and CRISPR/Cas9-PTX-CMB groups (Fig. 2C).
Knockout of C-erbB-2 in vitro can effectively inhibit cell invasion
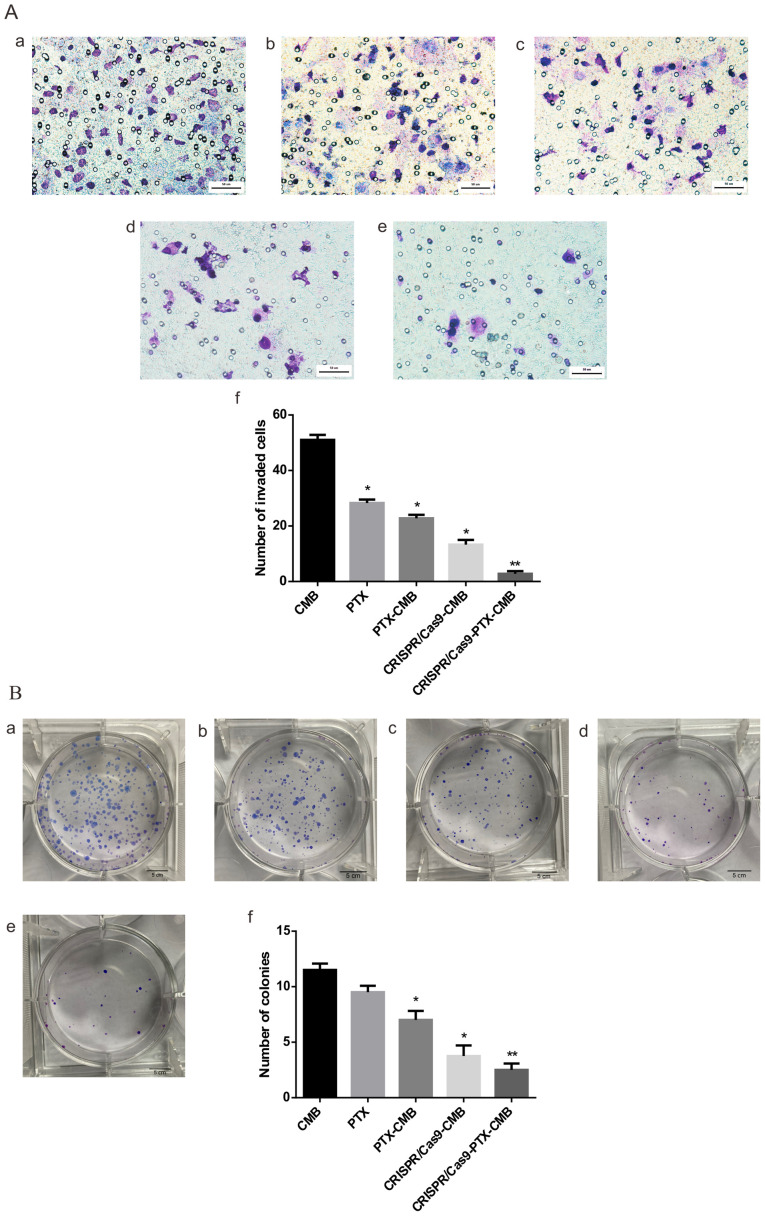
Compared with the CMB group, the number of invaded cells in the PTX, PTX-CMB, CRISPR/Cas9-CMB and CRISPR/Cas9-PTX-CMB groups significantly decreased (Fig. 3A-a-e). The CRISPR/Cas9-PTX-CMBs group had the lowest number of invading cells (Fig. 3A-f). These data confirmed that the invasion ability of cells treated with PTX or gene knockout was effectively inhibited, and combined drug/gene therapy exhibited the most notable inhibitory effect.
Figure 3.
Cell invasion and colony formation assay. (A) Cell invasion. (B) Colony formation. a, CMB; b, PTX; c, PTX-CMB; d, CRISPR/Cas9; e, CRISPR/Cas9-PTX-CMB; f, quantification. n=4. *P<0.05, **P<0.01 vs. CMB. CMB, cationic microbubble; PTX, paclitaxel.
Knockout of C-erbB-2 in vitro can effectively inhibit cell clone formation
Compared with the CMB group, CRISPR/Cas9-CMB and CRISPR/Cas9-PTX-CMB groups had fewer cell clones, with the lowest number observed in the CRISPR/Cas9-PTX-CMB group (Fig. 3B-a-f). The PTX-CMB group exhibited fewer cell clones than the PTX group. CRISPR/Cas9-PTX-CMBs exhibited fewer cell clones than the CRISPR/Cas9-CMB group. The results of cell clone formation experiments showed that PTX and CRISPR/Cas9 knockout of C-erbb-2 exhibited an inhibitory effect on HEC-1A cells, while CRISPR/Cas9-PTX-CMB exhibited the greatest inhibitory effect.
Knockout of C-erbB-2 in vitro can effectively inhibit cell migration
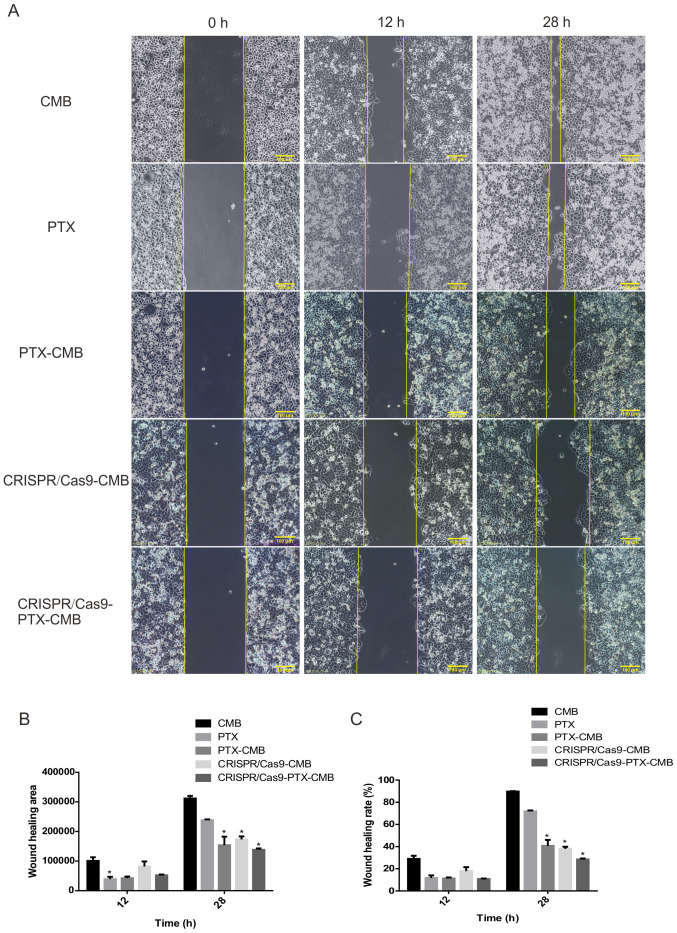
At 48 h, compared with the CMB group, the wound healing rate of the CMB-PTX, CRISPR/Cas9-CMB and CRISPR/Cas9-PTX-CMB groups was inhibited (Fig. 4B and C). The cell scratch experiment demonstrated that the proliferation of cells in the CRISPR/Cas9-PTX-CMB was significantly inhibited.
Figure 4.
Wound healing. (A) Comparison of wound healing at 12 and 28 h. CRISPR/Cas9-PTX-CMB group had the lowest wound healing (B) area and (C) rate. n=3. *P<0.05 vs. CMB. CMB, cationic microbubble; PTX, paclitaxel.
Knockout of C-erbB-2 in vitro can effectively inhibit cell proliferation
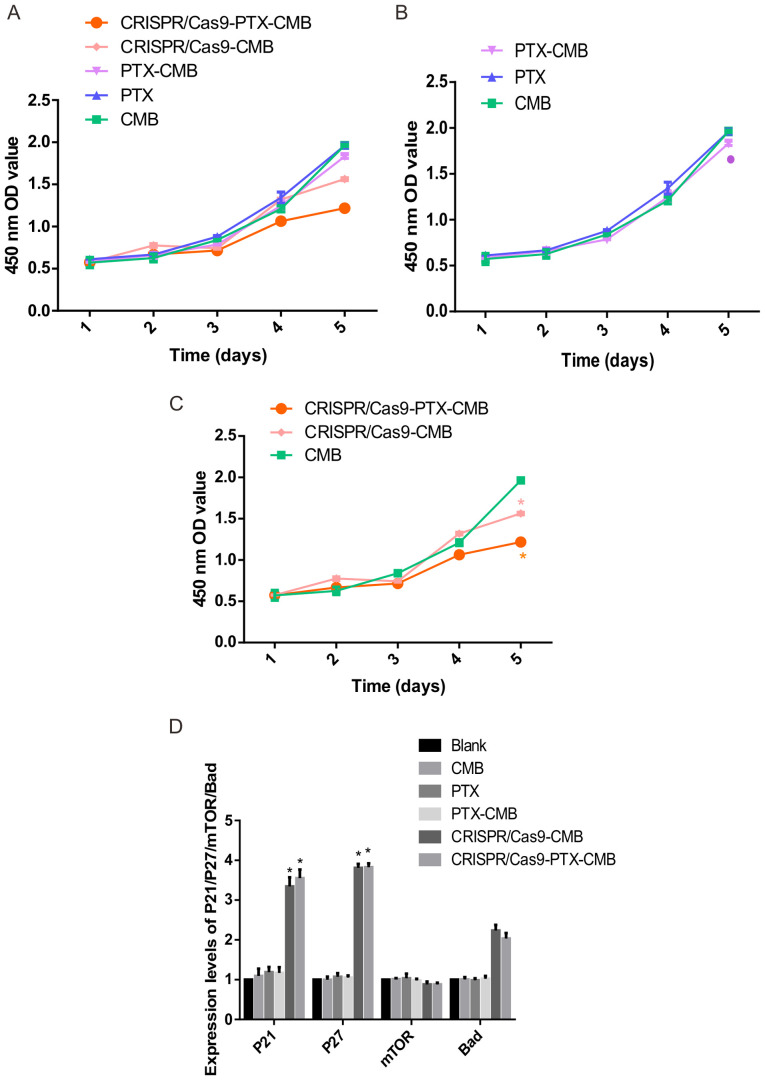
Absorbance values at 450 nm were measured to assess proliferation (Fig. 5A). On day 5, compared with the CMB group, the absorbance value of the cells in the PTX-CMB group decreased (Fig. 5B). At the same time, compared with the CMB group, the absorbance values of the CRISPR/Cas9-CMB and CRISPR/Cas9-PTX-CMB groups were significantly decreased (Fig. 5C). These results suggested that proliferation of cells treated with drug-gene combination was inhibited.
Figure 5.
Cell proliferation and detection of expression levels of each gene. (A) Cell proliferation in all groups. (B) Cell proliferation in the drug group. *P<0.1 vs. CMB. (C) Cell proliferation in C-erbB-2 knockout group. *P<0.05 vs. CMB (D) Expression of P21, P27, mTOR and Bad. n=3. *P<0.05 vs. blank. CMB, cationic microbubble; PTX, paclitaxel; OD, optical density.
Knockout of C-erbB-2 gene in vitro can increase the expression level of P21 and P27 genes
To assess the effect of knockout of C-erbB-2 cell line constructed by CMB burst transfection on the expression of its downstream genes, RT-qPCR was performed. Compared with the CMB, PTX, PTX-CMB, CRISPR/Cas9-CMB and CRISPR/Cas9-PTX-CMB groups, the expression of P21 and P27 was increased significantly (Fig. 5D). Hence, C-erbB-2 may participate in the occurrence and progression of EC by regulating expression of P27 and P21.
Discussion
In recent years, CRISPR/Cas9 systems have been developed, including use of lentiviruses, lipid nanoparticles, artificial viruses, and non-viral method (29,30). All of these systems have drawbacks; adeno-associated virus (AAV)-mediated Cas9 delivery may cause inadvertent interruption of the expression of important genes. In addition, the long-term existence of a virus-mediated drug delivery system will increase the accumulation of off-target cleavage, therefore, virus-mediated methods are usually limited in vitro. As to non-viral-mediated physical methods such as microscopic injection and electroporation; microscopic injection requires high labor costs and harsh experimental conditions and electroporation requires specific plans for different types of cells, which makes the operation complicated. (31,32). The present study described an efficient delivery system using CMB and ultrasound. CMB has a positive charge on its surface and is fat-soluble (33,34), so it has advantages when loading DNA materials such as plasmids and carrying PTX drugs (24,35). CMB not only retains the physical and chemical properties of ordinary MBs but also significantly improves the ability to carry plasmid DNA due to its positive charge, which improves the efficiency of ultrasound targeted transfection, confirming the efficiency of CMB as a delivery system (36). Ultrasound targeted MBs destruction (UTMD) is an emerging method for delivering target genes into cells or living animal organs. It has become a research hotspot due to its advantages of simplicity, non-invasiveness, targeting and reproducibility (37–39). Low-frequency ultrasound has little effect on the biological interaction between healthy cells and normal tissue and is safe. The low-frequency ultrasound principle is to combine non-invasive MBs with drugs, DNA or RNA vectors (such as adenovirus, plasmids or nanoparticles) (37,38). Low frequency and high mechanical index ultrasound are used to destroy MBs and release drugs, adenovirus, plasmids or nanoparticles to specific areas or organs (40). At the same time, it produces a non-invasive cavitation effect: Multiple reversible pores with diameters of hundreds of nanometers appear in the cell membrane. Non-invasive cavitation effect can further improve the transmission efficiency (41,42).
Cell function experiments showed that HEC-1A cells treated with CRISPR/Cas9-CMBs or CRISPR/Cas9-PTX-CMBs knockout C-erbB-2, The rate of proliferation, healing, as well as cloning, migration and invasion ability of HEC-1A cells were weakened. These data demonstrated in vitro proliferation of the EC cell line HEC-1A after knocking out C-erbB-2. The group CRISPR/Cas9-PTX-CMB simultaneously weakened the proliferation and invasion ability of HEC-1A cells most robustly. The present study demonstrated the possibility of delivery of genes or drugs based on CMB-targeted destruction. At the same drug concentration, compared with the PTX group, the anti-tumor effect of CMBs following release of PTX was improved. Combination therapy is expected to overcome the limitations of traditional treatments that rely on only one therapy, including the adverse reactions and toxic effects caused by ineffective increase of drug doses, off-target effects of gene editing and the effects of drugs or surgery on normal tissue and organs (43,44). Gene therapy combining chemotherapeutic agents and genetic material has become a promising combination therapy strategy due to its synergistic effect and ability to decrease chemotherapeutic dose without affecting anti-tumor activity (45).
The present data suggested that the mechanism of C-erbB-2 gene regulation of EC may involve regulation of expression of P27 and P21. P21 and P27 are tumor suppressor genes that effectively inhibit the proliferation and division of tumor cells and are also known as negative cell cycle regulators (46). Although the tumor-suppressive function of P21 is one of the most studied aspects of this protein in cancer, the role of P21 in phenotypic plasticity and its carcinogenic/anti-apoptotic function depends on the subcellular localization of P21. P21 can be an oncogenic protein or a tumor suppressor, depending on its localization in the cytoplasm or the nucleus, respectively (47,48). Huang et al (49) found that nuclear p21 inhibits, but cytoplasmic p21 promotes, cell migration and invasion abilities. The present data suggested that the high expression of P21 and P27 in EC primarily serves a role in suppressing tumors, which also indicated that C-erbB-2 may be downregulated by P21. The expression of P27 serves a role in promoting proliferation of tumor cells (50). Bad is a mitochondrial pro-apoptotic factor, the primary function of which is to promote cell apoptosis (51). In the present experiment, it remains unknown whether C-erbB-2 regulates expression of Bad and affects the occurrence and progress of EC. The mTOR pathway is a central signaling pathway that controls metabolic processes, such as protein synthesis, growth and metabolism (52). It supports proliferation by controlling cell growth and metabolism. Preclinical studies have shown that inhibiting mTOR results in anti-tumor activity (53,54). However, after knocking out C-erbB-2 gene in the present experiment, Changes in mTOR gene were not statistically significant. The present cell function experiments demonstrated that proliferation of the CRISPR/Cas9-CMB and CRISPR/Cas9-PTX-CMB decreased. Therefore, it was hypothesized that C-erbB-2 downregulates P21 and P27 to promote the proliferation of HEC-1A cells. It was also speculated that the targeted release of PTX may decrease its systemic toxic and side effects when used against EC. This should be validated in larger studies involving animal models.
EC (HEC-1A) cells and CMBs were used as carriers for drug and gene delivery. HEC-1A cells treated with CRISPR/Cas9-CMBs or CRISPR/Cas-9PTX-CMBs knocked out C-erbB-2. The proliferation, healing, as well as cloning, migration and invasion ability of HEC-1A cells were weakened. CMB-assisted transfection method based on ultrasound may aid development of novel treatment methods against EC.
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the Hainan Key Program of Research and Development (grant no. ZDYF2019123).
Funding
The present study was supported by the Hainan Key Program of Research and Development (grant no. ZDYF2019123).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SP and JC performed experiments and data analysis and wrote the manuscript. SB designed the study, revised the manuscript and obtained funding. SP and JC confirm the authenticity of all the raw data, All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lu KH, Broaddus RR. Endometrial Cancer. N Engl J Med. 2020;383:2053–2064. doi: 10.1056/NEJMra1514010. [DOI] [PubMed] [Google Scholar]
- 2.Njoku K, Abiola J, Russell J, Crosbie EJ. Endometrial cancer prevention in high-risk women. Best Pract Res Clin Obstet Gynaecol. 2020;65:66–78. doi: 10.1016/j.bpobgyn.2019.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Aoki Y, Kanao H, Wang X, Yunokawa M, Omatsu K, Fusegi A, Takeshima N. Adjuvant treatment of endometrial cancer today. Jpn J Clin Oncol. 2020;50:753–765. doi: 10.1093/jjco/hyaa071. [DOI] [PubMed] [Google Scholar]
- 4.Habibi Jouybari M, Hosseini S, Mahboobnia K, Boloursaz LA, Moradi M, Irani M. Simultaneous controlled release of 5-FU, DOX and PTX from chitosan/PLA/5-FU/g-C3N4-DOX/g-C3N4-PTX triaxial nanofibers for breast cancer treatment in vitro. Colloids Surf B Biointerfaces. 2019;179:495–504. doi: 10.1016/j.colsurfb.2019.04.026. [DOI] [PubMed] [Google Scholar]
- 5.Zhou X, Cao C, Li N, Yuan S. SYL3C aptamer-anchored microemulsion co-loading β-elemene and PTX enhances the treatment of colorectal cancer. Drug Deliv. 2019;26:886–897. doi: 10.1080/10717544.2019.1660733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebeid K, Meng X, Thiel KW, Do AV, Geary SM, Morris AS, Pham EL, Wongrakpanich A, Chhonker YS, Murry DJ, et al. Synthetically lethal nanoparticles for treatment of endometrial cancer. Nat Nanotechnol. 2018;13:72–81. doi: 10.1038/s41565-017-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinkic C, Kruse A, Zygmunt M, Schuetz F, Brucker J, Rom J, Sohn C, Fluhr H. Influence of Paclitaxel and Heparin on Vitality, Proliferation and Cytokine Production of Endometrial Cancer Cells. Geburtshilfe Frauenheilkd. 2017;77:1104–1110. doi: 10.1055/s-0043-119289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q, Liu F, Wang B, Li Z, Zhou D, Yang Q, Dong J, Li J. HER-2 expression in biopsy and surgical specimen on prognosis of osteosarcoma: A systematic review and meta-analysis of 16 studies. Medicine (Baltimore) 2016;95:e3661. doi: 10.1097/MD.0000000000003661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Press MF, Cordon-Cardo C, Slamon DJ. Expression of the HER-2/neu proto-oncogene in normal human adult and fetal tissues. Oncogene. 1990;5:953–962. [PubMed] [Google Scholar]
- 10.Mori S, Akiyama T, Yamada Y, Morishita Y, Sugawara I, Toyoshima K, Yamamoto T. C-erbB-2 gene product, a membrane protein commonly expressed on human fetal epithelial cells. Lab Invest. 1989;61:93–97. [PubMed] [Google Scholar]
- 11.Hua S, Chuanbo F, Zhonglin W, Shuangjiu Z, Xinwen Z. The expression and prognostic significance of Topo-II and c-erbB-2 in breast cancer. Minerva Med. 2020 Jul 17; doi: 10.23736/S0026-4806.20.06637-9. (Epub ahead of print). doi: 10.23736/S0026-4806.20.06637-9. [DOI] [PubMed] [Google Scholar]
- 12.Wang K, Liu J, Duan Y, Wu J, Dongye S, Wang Y, Liu Z, Han G. C-erbB-2 expression is related with pathological progression of gastric cancer: Results of a non-radioactive in situ hybridization. Int J Clin Exp Pathol. 2017;10:9649–9653. [PMC free article] [PubMed] [Google Scholar]
- 13.Canoz O, Ozkan M, Arsav V, Er O, Coskun HS, Soyuer S, Altinbas M. The role of c-erbB-2 expression on the survival of patients with small-cell lung cancer. Lung. 2006;184:267–272. doi: 10.1007/s00408-005-2591-y. [DOI] [PubMed] [Google Scholar]
- 14.Xiao W, Dong X, Zhao H, Han S, Nie R, Zhang X, An R. Expression of MIF and c-erbB-2 in endometrial cancer. Mol Med Rep. 2016;13:3828–3834. doi: 10.3892/mmr.2016.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Q, Hou CN, Yang HJ, He Z, Zuo MZ. Distinct expression and prognostic value of members of the epidermal growth factor receptor family in ovarian cancer. Cancer Manag Res. 2018;10:6937–6948. doi: 10.2147/CMAR.S183769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li BT, Ross DS, Aisner DL, Chaft JE, Hsu M, Kako SL, Kris MG, Varella-Garcia M, Arcila ME. HER2 Amplification and HER2 Mutation Are Distinct Molecular Targets in Lung Cancers. J Thorac Oncol. 2016;11:414–419. doi: 10.1016/j.jtho.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson BK, Zeybek B, Santin AD, Fader AN. Targeting human epidermal growth factor receptor 2 (HER2) in gynecologic malignancies. Curr Opin Obstet Gynecol. 2020;32:57–64. doi: 10.1097/GCO.0000000000000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho S, Shin J, Cho BK. Applications of CRISPR/Cas System to Bacterial Metabolic Engineering. Int J Mol Sci. 2018;19:1089. doi: 10.3390/ijms19041089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hryhorowicz M, Lipiński D, Zeyland J, Słomski R. CRISPR/Cas9 Immune System as a Tool for Genome Engineering. Arch Immunol Ther Exp (Warsz) 2017;65:233–240. doi: 10.1007/s00005-016-0427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhan T, Rindtorff N, Betge J, Ebert MP, Boutros M. CRISPR/Cas9 for cancer research and therapy. Semin Cancer Biol. 2019;55:106–119. doi: 10.1016/j.semcancer.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Gupta D, Bhattacharjee O, Mandal D, Sen MK, Dey D, Dasgupta A, Kazi TA, Gupta R, Sinharoy S, Acharya K, et al. CRISPR-Cas9 system: A new-fangled dawn in gene editing. Life Sci. 2019;232:116636. doi: 10.1016/j.lfs.2019.116636. [DOI] [PubMed] [Google Scholar]
- 23.Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release. 2017;266:17–26. doi: 10.1016/j.jconrel.2017.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang F, Li Y, Liufu C, Wang Y, Chen Z. Preparation of Cationic Lipid-coated Ultrasound Contrast Agents and Noninvasive Gene Transfection Via Ultrasound-targeted Microbubble Destruction. Curr Pharm Des. 2018;24:3587–3595. doi: 10.2174/1381612824666181011120031. [DOI] [PubMed] [Google Scholar]
- 25.Delalande A, Bastié C, Pigeon L, Manta S, Lebertre M, Mignet N, Midoux P, Pichon C. Cationic gas-filled microbubbles for ultrasound-based nucleic acids delivery. Biosci Rep. 2017;37:BSR20160619. doi: 10.1042/BSR20160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Q, Deng Q, Hu B, Wang YJ, Chen JL, Cui JJ, Cao S, Song HN. Ultrasound combined with targeted cationic microbubble-mediated angiogenesis gene transfection improves ischemic heart function. Exp Ther Med. 2017;13:2293–2303. doi: 10.3892/etm.2017.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manta S, Renault G, Delalande A, Couture O, Lagoutte I, Seguin J, Lager F, Houzé P, Midoux P, Bessodes M, et al. Cationic microbubbles and antibiotic-free miniplasmid for sustained ultrasound-mediated transgene expression in liver. J Control Release. 2017;262:170–181. doi: 10.1016/j.jconrel.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 28.Arocho A, Chen B, Ladanyi M, Pan Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol Pathol. 2006;15:56–61. doi: 10.1097/00019606-200603000-00009. [DOI] [PubMed] [Google Scholar]
- 29.Xu X, Wan T, Xin H, Li D, Pan H, Wu J, Ping Y. Delivery of CRISPR/Cas9 for therapeutic genome editing. J Gene Med. 2019;21:e3107. doi: 10.1002/jgm.3107. [DOI] [PubMed] [Google Scholar]
- 30.Lino CA, Harper JC, Carney JP, Timlin JA. Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 2018;25:1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen M, Mao A, Xu M, Weng Q, Mao J, Ji J. CRISPR-Cas9 for cancer therapy: Opportunities and challenges. Cancer Lett. 2019;447:48–55. doi: 10.1016/j.canlet.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 32.Yip BH. Recent Advances in CRISPR/Cas9 Delivery Strategies. Biomolecules. 2020;10:839. doi: 10.3390/biom10060839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Li X, Liu L, Liu B, Wang F, Chen C. Tissue Targeting and Ultrasound-Targeted Microbubble Destruction Delivery of Plasmid DNA and Transfection In Vitro. Cell Mol Bioeng. 2019;13:99–112. doi: 10.1007/s12195-019-00597-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kooiman K, Roovers S, Langeveld SAG, Kleven RT, Dewitte H, O'Reilly MA, Escoffre JM, Bouakaz A, Verweij MD, Hynynen K, et al. Ultrasound-Responsive Cavitation Nuclei for Therapy and Drug Delivery. Ultrasound Med Biol. 2020;46:1296–1325. doi: 10.1016/j.ultrasmedbio.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu X, Guo J, He C, Geng H, Yu G, Li J, Zheng H, Ji X, Yan F. Ultrasound triggered image-guided drug delivery to inhibit vascular reconstruction via paclitaxel-loaded microbubbles. Sci Rep. 2016;6:21683. doi: 10.1038/srep21683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang D, Yang L, Tian H, Fu B, Chen W, Liu K, Jia Z, Jiang S, Han X, Sun L. [Enhancement of gene transfection efficiency and therapeutic effect of ultrasound-targeted microbubble destruction in vivo with cationic microbubble] Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2018;32:228–236. doi: 10.7507/1002-1892.201706058. (In Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du M, Chen Z, Chen Y, Li Y. Ultrasound-Targeted Delivery Technology: A Novel Strategy for Tumor- Targeted Therapy. Curr Drug Targets. 2019;20:220–231. doi: 10.2174/1389450119666180731095441. [DOI] [PubMed] [Google Scholar]
- 38.de Leon A, Perera R, Nittayacharn P, Cooley M, Jung O, Exner AA. Ultrasound Contrast Agents and Delivery Systems in Cancer Detection and Therapy. Adv Cancer Res. 2018;139:57–84. doi: 10.1016/bs.acr.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Endo-Takahashi Y, Negishi Y. Microbubbles and Nanobubbles with Ultrasound for Systemic Gene Delivery. Pharmaceutics. 2020;12:964. doi: 10.3390/pharmaceutics12100964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian L, Thapa B, Hong J, Zhang Y, Zhu M, Chu M, Yao J, Xu D. The present and future role of ultrasound targeted microbubble destruction in preclinical studies of cardiac gene therapy. J Thorac Dis. 2018;10:1099–1111. doi: 10.21037/jtd.2018.01.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szablowski JO, Bar-Zion A, Shapiro MG. Achieving Spatial and Molecular Specificity with Ultrasound-Targeted Biomolecular Nanotherapeutics. Acc Chem Res. 2019;52:2427–2434. doi: 10.1021/acs.accounts.9b00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou LQ, Li P, Cui XW, Dietrich CF. Ultrasound nanotheranostics in fighting cancer: Advances and prospects. Cancer Lett. 2020;470:204–219. doi: 10.1016/j.canlet.2019.11.034. [DOI] [PubMed] [Google Scholar]
- 43.Wu J, Chen J, Feng Y, Zhang S, Lin L, Guo Z, Sun P, Xu C, Tian H, Chen X. An immune cocktail therapy to realize multiple boosting of the cancer-immunity cycle by combination of drug/gene delivery nanoparticles. Sci Adv. 2020;6:eabc7828. doi: 10.1126/sciadv.abc7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu P, Liu X, Cheng Y, Zhong S, Shi X, Wang S, Liu M, Ding J, Zhou W. Core-Shell Nanosystems for Self-Activated Drug-Gene Combinations against Triple-Negative Breast Cancer. ACS Appl Mater Interfaces. 2020;12:53654–53664. doi: 10.1021/acsami.0c15089. [DOI] [PubMed] [Google Scholar]
- 45.Han H, Kim D, Jang Y, Seo M, Kim K, Lee JB, Kim H. Focused ultrasound-triggered chemo-gene therapy with multifunctional nanocomplex for enhancing therapeutic efficacy. J Control Release. 2020;322:346–356. doi: 10.1016/j.jconrel.2020.03.041. [DOI] [PubMed] [Google Scholar]
- 46.Zhou X, Yang Y, Ma P, Wang N, Yang D, Tu Q, Sun B, Xiang T, Zhao X, Hou Z, et al. TRIM44 is indispensable for glioma cell proliferation and cell cycle progression through AKT/p21/p27 signaling pathway. J Neurooncol. 2019;145:211–222. doi: 10.1007/s11060-019-03301-0. [DOI] [PubMed] [Google Scholar]
- 47.Shamloo B, Usluer S. p21 in Cancer Research. Cancers (Basel) 2019;11:1178. doi: 10.3390/cancers11081178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez-Cupello C, Dam M, Serini L, Wang S, Lindgren D, Englund E, Kjellman P, Axelson H, García-Mariscal A, Madsen CD. The STRIPAK Complex Regulates Response to Chemotherapy Through p21 and p27. Front Cell Dev Biol. 2020;8:146. doi: 10.3389/fcell.2020.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y, Wang W, Chen Y, Huang Y, Zhang J, He S, Tan Y, Qiang F, Li A, Røe OD, et al. The opposite prognostic significance of nuclear and cytoplasmic p21 expression in resectable gastric cancer patients. J Gastroenterol. 2014;49:1441–1452. doi: 10.1007/s00535-013-0900-4. [DOI] [PubMed] [Google Scholar]
- 50.Razavipour SF, Harikumar KB, Slingerland JM. p27 as a Transcriptional Regulator: New Roles in Development and Cancer. Cancer Res. 2020;80:3451–3458. doi: 10.1158/0008-5472.CAN-19-3663. [DOI] [PubMed] [Google Scholar]
- 51.Lu P, Redd Bowman KE, Brown SM, Joklik-Mcleod M, Vander Mause ER, Nguyen HTN, Lim CS. p53-Bad: A Novel Tumor Suppressor/Proapoptotic Factor Hybrid Directed to the Mitochondria for Ovarian Cancer Gene Therapy. Mol Pharm. 2019;16:3386–3398. doi: 10.1021/acs.molpharmaceut.9b00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murugan AK. mTOR: Role in cancer, metastasis and drug resistance. Semin Cancer Biol. 2019;59:92–111. doi: 10.1016/j.semcancer.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 53.Hua H, Kong Q, Zhang H, Wang J, Luo T, Jiang Y. Targeting mTOR for cancer therapy. J Hematol Oncol. 2019;12:71. doi: 10.1186/s13045-019-0754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Magaway C, Kim E, Jacinto E. Targeting mTOR and Metabolism in Cancer: Lessons and Innovations. Cells. 2019;8:1584. doi: 10.3390/cells8121584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.