Abstract
Background:
To investigate the intergenerational effects of grandmaternal smoking during pregnancy (GMSDP) on the DNA methylation of grandchildren.
Methods:
Data from the Isle of Wight birth cohort with information regarding GMSDP and DNA methylation profiling at the birth of grandchildren (n = 161) were used. Differentially methylated CpG sites related to GMSDP were identified using testing–training screening, analysis of variance and multivariate analysis of covariance. The association between identified CpG sites and expression levels of neighboring genes was tested by linear regression.
Results:
Twenty-three CpG sites were differentially methylated in grandchildren because of GMSDP, and eight of these were associated with expression levels of 13 neighboring genes.
Conclusion:
GMSDP has an intergenerational effect on the DNA methylation profile of grandchildren independent of maternal smoking during pregnancy.
Keywords: : DNA methylation, grandchildren, grandmaternal smoking during pregnancy, intergenerational effect, smoking
Lay abstract
This study aimed to assess how grandmaternal smoking during pregnancy can affect the health of grandchildren. Underlying mechanisms may include epigenetic modifications. To address this topic, the authors investigated the intergenerational effects of grandmaternal smoking during pregnancy on the DNA methylation of grandchildren at birth based on the Isle of Wight birth Cohort. Twenty-three CpG sites were differentially methylated in grandchildren because of grandmaternal smoking during pregnancy, and eight of these were associated with changes in expression levels of 13 neighboring genes. Thus, grandmaternal smoking during pregnancy has an intergenerational effect on the DNA methylation profile of grandchildren independent of maternal smoking during pregnancy.
Maternal smoking during pregnancy (MSDP) carries adverse health effects for the mother and her child and is a significant public health concern around the world [1–4]. Previous reports have shown that MSDP is a risk factor for several adverse perinatal outcomes, including compromised lung function [5], low birth weight [6–8] and preterm birth [9,10], as well as increased susceptibility to various diseases later in life, including childhood asthma [11,12] and obesity [13,14]. However, very few studies have examined the intergenerational effects of grandmaternal smoking during pregnancy (GMSDP) on the health of grandchildren. Specifically, little is known about the epigenetic mechanisms potentially mediating these effects.
The evidence from animal and human studies points to an intergenerational effect of prenatal exposures on the health outcomes of grandchildren. One study in rats reported emphysematous and other structural changes in the lungs of offspring after their pregnant grandmothers were exposed to nicotine [15]. Of four population-based studies that have explored the relationship between GMSDP and asthma in the grandchild, three have found that GMSDP increases the risk of asthma in the grandchildren, independent of the smoking status of the mother [16–18]. However, the Avon Longitudinal Study of Parents and Children found no effect of GMSDP on respiratory outcomes of grandchildren [19].
The potential mechanisms underlying the observed adverse health effects of GMSDP on the health of grandchildren are poorly understood. Current evidence suggests that epigenetic mechanisms, such as DNA methylation (DNAm), may mediate the effect of MSDP on the children's health outcomes. MSDP-induced alterations in DNAm have been observed in the placenta, umbilical cord blood and buccal cells of the offspring [20,21]. In addition, a dose–response effect of MSDP has been reported on the methylation levels of 15 CpG sites located in seven gene regions [22]. It has also been shown that prenatal exposure to tobacco smoke can affect germline reprogramming during embryonic development and result in increased susceptibility to disease during adult life [12]. Together, these pieces of evidence raise the question of whether the underlying mechanisms of the effects of GMSDP on the health of grandchildren may also involve epigenetic mechanisms such as DNAm. In this study, we examined the associations between GMSDP and DNAm profiles of grandchildren using an epigenome-wide association study approach and utilizing data from three generations of the Isle of Wight birth cohort (IOWBC).
Methods
IOWBC & study population
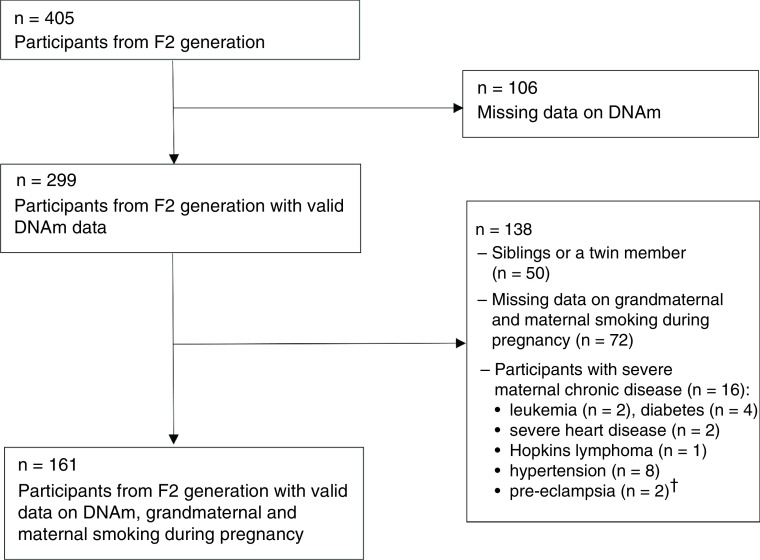
Participants in this study were a subgroup of the ongoing IOWBC [23,24]. This cohort was initially established in 1989 in the UK to observe the natural history of allergic conditions. A total of 1518 pregnant women (F0 generation) were approached at the time of delivery during 1989–1990 and invited to enroll in the study along with all their newborns (F1 generation). Of these, 1456 (94.8%) mother–child pairs consented [25]. Between 2011 and 2018, 231 F1 daughters became pregnant and gave birth to grandchildren (F2 generation, n = 405). This study utilized information from 161 F2 grandchildren with complete data on their DNAm as well as their grandmothers' (F0) and mothers' (F1) smoking behavior. Exclusion criteria included severe clinical disease of the mother (F1), such as type 1 or 2 diabetes, cancer (leukemia, lymphoma), hypertension, pre-eclampsia or severe cardiovascular disease. For mothers with data available on multiple children, only the first child was included in the analysis (Figure 1).
Figure 1. . Flowchart of participants.
†Some participants may have more than one maternal chronic disease.
DNAm: DNA methylation; F2 generation: F1 daughters who gave birth to grandchildren.
Exposure assessment
Information regarding grandmaternal (F0) prenatal smoking status was determined by self-report and serum cotinine levels at the end of pregnancy. A grandmother who had a serum cotinine level greater than 10 μg/l was classified as a smoker [26]. Mothers' (F1) prenatal smoking was measured by self-report at 20 and 28 weeks of pregnancy and by urine cotinine measured at each of the two aforementioned times. Participants with urine cotinine levels ≥2.47 ng/ml were classified as smokers [27]. For both grandmothers and mothers, nonsmoker status was defined as reporting no smoking during pregnancy and negative serum or urine cotinine test. Unfortunately, there is a lack of information regarding smoking cessation of grandmothers or mothers during pregnancy.
Assessment of DNAm in grandchildren
The grandchildren's DNAm data, which were measured using umbilical cord blood (n = 106) or Guthrie card (n = 55), were utilized in this study. High agreement between CpG DNAm from cord blood and neonatal blood on Guthrie cards has been reported before, making Guthrie cards an acceptable source of neonatal blood [28]. Umbilical cord blood samples of the grandchildren were collected after delivery and before the umbilical cord was cut. Dried blood spots on Guthrie cards were collected from a heel prick within 5–8 days after birth as part of the UK newborn screening program. Guthrie cards were stored in the dark at room temperature in individual paper protectors.
The umbilical cord blood samples were centrifuged at 3000 rpm to separate blood cells from plasma and stored at -80°C. Next, DNA was extracted using a standard salting-out procedure [29] with a QIAamp DNA blood mini kit (Qiagen, Hilden, Germany) according to the manufacturer's guidelines. The DNA from dried blood spots on Guthrie cards was extracted using the procedure described by Beyan et al. [30]. The samples were purified using Qiagen QIAamp mini columns and eluted in 100 μl of elution buffer. Next, a Qubit instrument (Invitrogen, MA, USA) was used to measure the concentration of DNA.
Samples were randomized according to their study identifiers when placed on plates. A total of 1 μg of DNA from each subject underwent cytosine to thymine conversion with sodium bisulfite using an EZ-96 DNAm kit (Zymo Research, CA, USA). Genome-wide DNAm was measured using the Infinium HumanMethylation450 BeadChip (Illumina, CA, USA). BeadChips were scanned and processed using a standard protocol based on the random self-assembly of a bead pool on a patterned substrate [31]. The β-values were extracted from image data files using the GenomeStudio software (Illumina) methylation module. The β-values (percent methylation) represented the proportion of intensity of methylated (M) over the sum of methylated and unmethylated (U) allele (β = M/[M+U+c]), where c indicates a constant to prevent dividing by zero [32]. To adjust for different cell type fractions, the proportion of cell types has been estimated in cord blood and Guthrie card using the estimateCellCounts() function in the Bioconductor minfi package, following the Bakulski approach with a cord blood reference panel [33] and Houseman approach [34] with an adult reference panel [35], respectively.
DNAm data preprocessing
To improve DNAm data quality, the Bioconductor Illumina Methylation Analyzer package was used for quantile normalization and type I and II probe peak correction [36]. Furthermore, the ComBat package was used to remove batch effects and other unwanted variations [37]. Additionally, CpG sites with probe SNPs were removed from the list of all CpG sites if the minor allele frequency of probe SNPs was >1%. Probe SNPs are SNPs located within the probe sequence and can cause false interpretation of Illumina microarray results [38].
Gene expression measurement
Gene expression data were available for a subsample of 87 grandchildren with complete GMSDP and DNAm data. Total RNA was extracted from umbilical cord blood using PAXgene RNA kits (PreAnalytiX GmbH, Hombrechtikon, Switzerland). Samples were randomized according to their study identifiers when placed on plates. Total RNA yield and absence of DNA contamination were measured by Qubit 2.0 fluorometer (Qiagen). RNA quality was evaluated using a 2100 Bioanalyzer via RNA 6000 Nano Chips (Agilent Technologies, CA, USA). RNA was reverse-transcribed into cDNA. The cDNA was subsequently used as a template for DNA amplification by real-time PCR with commercially available primer and one-color (Cy3) labeled probes following Microarray-Based Gene Expression Analysis protocol, version 6.0 (Agilent Technologies). For each gene, multiple primers were designed and created to match sense and antisense cDNA. Next, sample cDNA and reference cDNA were hybridized separately on a SurePrint G3 human GE v2 8 × 60K microarray (Agilent Technologies). After hybridization, slides were washed and scanned using a G2565AA microarray scanner system (Agilent Technologies). Scanned images were subsequently analyzed based on fluorescence intensities. Quantitative expression levels were reported after correction for background signal and normalization through Agilent's feature extraction software, version 9.5.
Measurement of covariates & potential confounders
Exposure to secondhand smoke during pregnancy of the mothers (F1) was ascertained by a questionnaire administered to both parents of F2 and was defined as the presence of one of the following: a father who lived in the same residence as the mother and was reported smoking during her pregnancy; other people in the same household who smoked during the mother's pregnancy or regular exposure to secondhand smoke at places other than home, such as in the workplace. The mother's socioeconomic status was assessed by a composite variable based on her education level, income and number of rooms in the house and was categorized into four categories [39]. Information regarding the gestational age and sex of the grandchild was also collected. The age of mothers at conception was not considered a confounder because of the narrow range of maternal ages at first pregnancy.
Statistical analysis
A comparison of main characteristics between the study sample and whole cohort was made using t-test and chi-square test. The grandchildren were classified into four exposure groups according to the different smoking status combinations of the grandmother and mother: both the grandmother and mother smoked during pregnancy (yes/yes group), only the grandmother smoked during pregnancy (yes/no group), only the mother smoked during pregnancy (no/yes group) and neither smoked during pregnancy (no/no group). In addition, the differences between the four groups regarding main characteristics were determined by analysis of variance and chi-square test.
A training–testing screening method has been applied to the cell proportion-adjusted β-values to identify potential CpG sites in the grandchildren's blood that were differentially methylated in relation to their GMSDP/MSDP exposure status [40]. Briefly, training–testing screening is an efficient algorithm that uses a randomly selected two-thirds of the data for training and the other one-third for testing. This approach has been shown to better control type I and II errors compared with the commonly used approaches for multiple testing correction. CpG sites detected in at least 60% of the training–testing pairs (100 randomly selected pairs in total) were considered to be potentially associated with GMSDP/MSDP and were used in subsequent analyses.
Analysis of variance was used to assess the differences in DNAm levels across the four exposure groups (yes/yes, yes/no, no/yes and no/no). To tease out the independent effect of GMSDP from that of MSDP, methylation levels among those who were exposed to only GMSDP (yes/no group) were compared with the unexposed (no/no) group using Tukey's honestly significant difference test.
Additionally, we compared the direction and magnitude of DNAm changes between the grandchildren who were exposed to both GMSDP and MSDP (yes/yes group) and those exposed to GMSDP alone (yes/no group) in relation to the unexposed (no/no) group. Only CpG sites with DNAm changes in the same direction and at least the same magnitude in response to GMSDP and MSDP were selected for further analysis. This is because in order to conclude an effect of exposure to GMSDP alone on the DNAm of the grandchildren, the effect of exposure to both GMSDP and MSDP should be in the same direction and at least as large as exposed to GMSDP alone.
Multivariate analysis of covariance was used to adjust for potential confounders. Cell type-adjusted DNAm level (β-value) was the dependent variable, and GMSDP exposure status, mother's (F1) exposure to secondhand smoke, sex of the grandchild, gestational age of the grandchild, socioeconomic status of the mother and source of the grandchild's blood (Guthrie card or cord blood) were included as independent variables. The magnitude of DNAm changes was evaluated between the GMSDP exposure-only group (yes/no group) and the unexposed (no/no) group. False discovery rate was used to control multiple testing with a cutoff p-value of 0.05.
GMSDP-associated CpG sites were explored for biological relevance by assessing their association with gene expression of the neighboring genes (250 kb upstream and downstream of each CpG) [41]. Corresponding annotated genes were identified using the methylation label file (Infinium MethylationEPIC v1.0 B4 manifest file; Illumina, Inc.), Snipper (https://csg.sph.umich.edu/boehnke/snipper/) [42] and University of California Santa Cruz Genome Browser (https://genome.ucsc.edu/) [43]. Linear regression models were run with gene expression levels as the dependent variables and DNAm levels (β-value), sex, source of blood, MSDP, secondhand smoke exposure during pregnancy and gestational age as the independent variables. The statistical analyses were performed using SAS 9.4 (SAS Institute, NC, USA) and R 3.4.4.
Results
Baseline characteristics
Of the 161 eligible grandmother–mother–grandchild triads, 34 (21.1%) were exposed to both GMSDP and MSDP, 45 (30.0%) were exposed to GMSDP only, 29 (18.0%) were exposed to MSDP only and 53 (32.9%) were not exposed to either (Table 1). The average age of F1 mothers at the time of conception was 23.7 years (range: 18–28 years). The comparison of main characteristics between the analytical samples and whole cohort revealed no significant differences in sex distribution (F2), birth weight (F2), exposure to secondhand smoke during pregnancy (F1) or socioeconomic status of the F1 mother. However, the proportions of participants with smoking grandmothers and mothers were higher among the study sample (Supplementary Table 1).
Table 1. . Characteristics of grandmother–mother–grandchild triads in Isle of Wight birth cohort.
| Characteristics | GMSDP/MSDP groups | |||
|---|---|---|---|---|
| Yes/yes (n = 34) | Yes/no (n = 29) | No/yes (n = 45) | No/no (n = 53) | |
| Sex, n (%) | ||||
| – Male | 17 (50.0) | 20 (44.4) | 21 (72.4) | 32 (60.4) |
| – Female | 17 (50.0) | 25 (55.6) | 8 (27.6) | 21 (39.6) |
| Birth weight, g, mean ± SD† | 3294.8 ± 515.8 | 3315.5 ± 617.7 | 3641.5 ± 414.0 | 3427.6 ± 476.7 |
| Gestational age, week, mean ± SD† | 38.8 ± 2.3 | 39.0 ± 2.2 | 40.2 ± 1.2 | 39.6 ± 1.4 |
| Blood source, n (%) | ||||
| – Cord blood | 21 (61.8) | 30 (66.7) | 18 (62.1) | 37 (69.8) |
| – Guthrie card | 13 (38.2) | 15 (33.3) | 11 (37.9) | 16 (30.2) |
| Exposure to secondhand smoke, n (%) † | ||||
| – No | 5 (15.2) | 6 (13.3) | 11 (37.9) | 37 (69.8) |
| – Yes | 28 (84.9) | 39 (86.7) | 18 (62.1) | 16 (30.2) |
| Socioeconomic status, low to high, n (%) | ||||
| – 1 | 6 (18.8) | 8 (17.8) | 6 (20.7) | 9 (17.0) |
| – 2 | 13 (40.6) | 20 (44.4) | 7 (24.1) | 10 (18.9) |
| – 3 | 7 (21.9) | 6 (13.3) | 11 (37.9) | 12 (22.6) |
| – 4 | 6 (18.8) | 11 (24.4) | 5 (17.2) | 22 (41.5) |
Significant association between variables and GMSDP and/or MSDP.
GMSDP: Grandmaternal smoking during pregnancy; MSDP: Maternal smoking during pregnancy; SD: Standard deviation.
Epigenome-wide association study for differential DNAm associated with GMSDP
Of the 1664 CpG sites selected by training–testing screening as differentially methylated in association with GMSDP and/or MSDP, 27 were still significant after adjusting for multiple testing (Table 2). Of these 27 CpG sites, 25 were significantly differentially methylated in the same direction (i.e., hyper- or hypomethylated) in grandchildren exposed to either GMSDP only or GMSDP + MSDP (ten CpG sites were more methylated and 15 were less methylated). Figure 2 shows two examples of such CpG sites, one with increased methylation and one with decreased methylation due to GMSDP and/or MSDP. After adjusting for potential confounders, 23 CpG sites remained significantly associated with GMSDP (Table 2).
Table 2. . Differentially methylated CpG sites associated with grandmaternal smoking during pregnancy.
| CpG site | Chr | Position | Nearest annotated gene† | UCSC RefGene group | Relation to CpG island | Unadjusted model effect size | Unadjusted p-value | Unadjusted FDR | Adjusted model effect size§ | p-value§ | FDR§ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| cg01718065 | 19 | 51774429 | SIGLECL ¶ | Island | 0.247 | < 0.001 | 0.003 | 0.338 | < 0.001 | 0.001 | |
| cg02296145 | 16 | 1840711 | IGFALS | Body | Island | 0.290 | 0.001 | 0.005 | 0.354 | 0.001 | 0.004 |
| cg02399044 | 12 | 2500229 | CACNA1C | Body | ‐0.263 | < 0.001 | 0.003 | ‐0.295 | 0.001 | 0.004 | |
| cg02473254 | 8 | 2000469 | MYOM2 | Body | N shelf | ‐0.180 | < 0.001 | 0.002 | ‐0.177 | 0.001 | 0.003 |
| cg02615735 | 6 | 31648276 | LY6G5C | TSS200 | N shore | ‐0.260 | 0.003 | 0.007 | ‐0.244 | 0.014 | 0.020 |
| cg03807330 | 11 | 76327232 | LRRC32 ¶ | ‐0.111 | 0.001 | 0.004 | ‐0.078 | 0.045 | 0.049 | ||
| cg06293294 | 11 | 100557954 | FLJ32810 | TSS1500 | Island | 0.177 | 0.001 | 0.005 | 0.159 | 0.123 | 0.123 |
| cg08859138 | 5 | 74532894 | ANKRD31 | TSS200 | Island | 0.460 | < 0.001 | 0.003 | 0.455 | 0.005 | 0.010 |
| cg11596397 | 11 | 75526167 | UVRAG | TSS200 | N shore | 0.138 | 0.002 | 0.005 | 0.142 | 0.038 | 0.045 |
| cg12732436 | 18 | 77752726 | TXNL4A ¶ | S shelf | ‐0.229 | 0.004 | 0.007 | ‐0.287 | 0.007 | 0.012 | |
| cg12799739 | 3 | 97595056 | CRYBG3 | TSS1500 | ‐0.093 | < 0.001 | 0.003 | ‐0.112 | < 0.001 | 0.001 | |
| cg13749070 | 15 | 99451935 | IGF1R | Body | ‐0.235 | 0.001 | 0.005 | ‐0.199 | 0.025 | 0.033 | |
| cg14284055 | 1 | 24439399 | MYOM3 | TSS1500 | ‐0.233 | < 0.001 | 0.003 | ‐0.193 | 0.002 | 0.004 | |
| cg15497724 | 19 | 51774377 | SIGLECL1 ¶ | N shore | 0.176 | 0.001 | 0.005 | 0.218 | 0.001 | 0.004 | |
| cg15517847 | 13 | 41241076 | FOXO1 | TSS1500 | Island | ‐0.190 | 0.003 | 0.007 | ‐0.228 | 0.011 | 0.018 |
| cg16409505 | 14 | 73425357 | DCAF4 | Body | ‐0.126 | < 0.001 | 0.003 | ‐0.098 | 0.071 | 0.074 | |
| cg20056908 | 2 | 85808945 | VAMP8 | 3′UTR | N shelf | ‐0.105 | 0.001 | 0.005 | ‐0.104 | 0.009 | 0.016 |
| cg23072559 | 19 | 55944231 | SHISA7 | 3′UTR | N shore | 0.104 | < 0.001 | 0.003 | 0.100 | 0.014 | 0.020 |
| cg23243463 | 18 | 35146454 | BRUNOL4 | TSS1500 | Island | ‐0.352 | 0.001 | 0.005 | ‐0.420 | 0.005 | 0.010 |
| cg23350744 | 5 | 122422098 | PRDM6 ¶ | N shelf | 0.279 | < 0.001 | 0.003 | 0.272 | 0.006 | 0.012 | |
| cg23670101 | 10 | 46168396 | ANUBL1 | TSS200 | Island | 0.141 | < 0.001 | 0.004 | 0.124 | 0.043 | 0.049 |
| cg24735611 | 1 | 146494928 | LOC728989 | Body | ‐0.107 | < 0.001 | 0.004 | ‐0.081 | 0.032 | 0.040 | |
| cg25256538 | 6 | 159343074 | C6orf99 ¶ | ‐0.188 | 0.001 | 0.004 | ‐0.201 | 0.002 | 0.004 | ||
| cg26961622 | 2 | 20426146 | SDC1 | TSS1500 | S shore | ‐0.155 | < 0.001 | 0.002 | ‐0.159 | 0.002 | 0.004 |
| cg27106230 | 13 | 99229068 | STK24 | First exon; 5′UTR | Island | 0.184 | 0.002 | 0.005 | 0.292 | < 0.001 | 0.002 |
Grandmother–mother–grandchild triad n = 161. Effect size = difference in DNA methylation (cell proportion-adjusted β-value) between grandchildren who were exposed to grandmaternal pregnancy during pregnancy only and those who were not exposed to either grandmaternal smoking during pregnancy or maternal smoking during pregnancy. RefGene group = gene region feature category describing the CpG position.
Only information for the closest corresponding gene displayed.
Adjusted for maternal exposure to secondhand smoke during pregnancy, maternal socioeconomic status, source of grandchild's blood, sex and gestational age.
Based on UCSC Genome Browser (hg19).
Chr: Chromosome; FDR: False discovery rate; N: North; S: South; UCSC: University of California Santa Cruz.
Figure 2. . Relationship between grandmaternal smoking during pregnancy and/or maternal smoking during pregnancy and cell proportion-adjusted methylation levels (β-values) at cg24735611 and cg02296145.
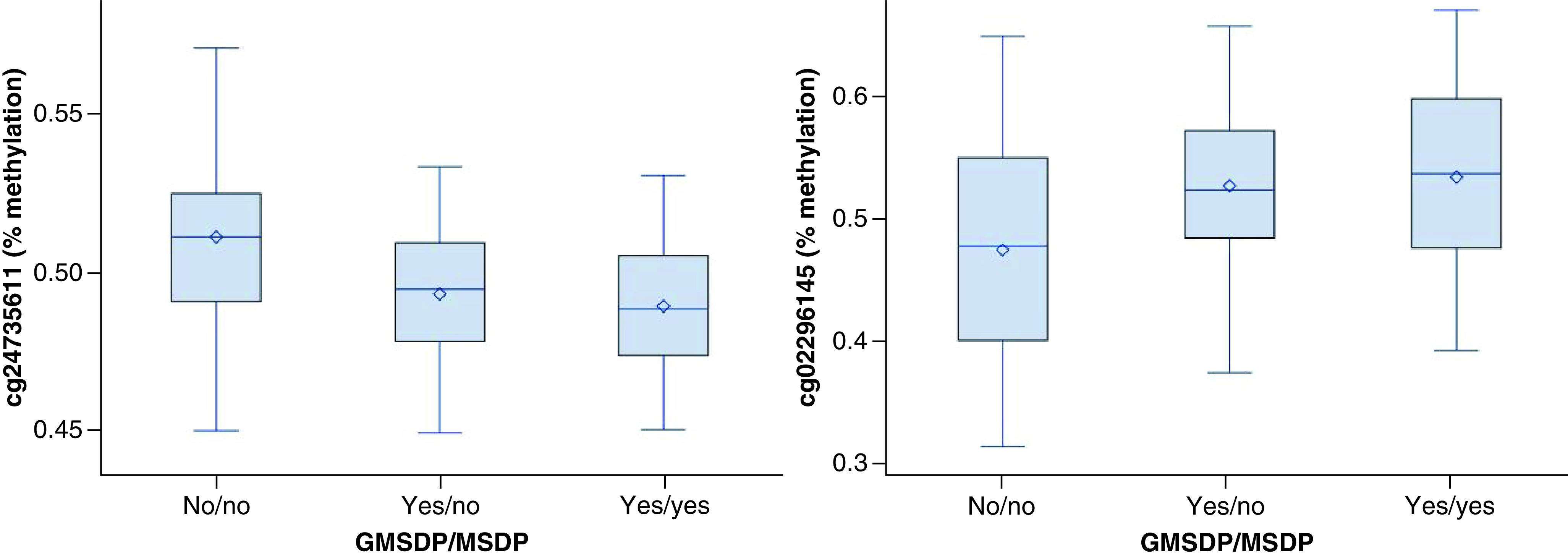
GMSDP: Grandmaternal smoking during pregnancy; MSDP: Maternal smoking during pregnancy.
Association of GMSDP-induced DNAm alterations with gene expression
There were 87 grandchildren with available data on DNAm and gene expression, 33 (37.9%) of whom were exposed to GMSDP. The 23 differentially methylated CpG sites were mapped to 256 genes using a proximity range of ± 250 kb. DNAm levels of eight significantly differentially methylated CpG sites were associated with the expression levels of 13 nearby genes (Table 3 & Supplementary Table 2).
Table 3. . Association between DNA methylation of grandmaternal smoking during pregnancy-associated CpG sites and annotated gene expression level.
| CpG site | Nearest annotated gene | UCSC RefGene group | Relation to CpG island | Gene symbol | Probe name | Estimate | Chi-square | p-value |
|---|---|---|---|---|---|---|---|---|
| cg02296145 | IGFALS | Body | Island | GFER | A_24_P31003 | 0.29 | 4.22 | 0.040 |
| cg02296145 | IGFALS | Body | Island | IFT140 | A_23_P140725 | 0.28 | 4.80 | 0.029 |
| cg02615735 | LY6G5C | TSS200 | N shore | LY6G6F | A_33_P3214334 | 0.88 | 7.20 | 0.007 |
| cg15497724 | SIGLECL1 | – | N shore | CD33 | A_21_P0000094 | 0.73 | 6.11 | 0.013 |
| cg15497724 | SIGLECL1 | – | N shore | SIGLEC12 | A_23_P164596 | 0.55 | 3.90 | 0.048 |
| cg20056908 | VAMP8 | 3′UTR | N shelf | ELMOD3 | A_33_P3235204 | 1.36 | 7.07 | 0.008 |
| cg23072559 | SHISA7 | 3′UTR | N shore | PPP6R1 | A_23_P119448 | ‐1.60 | 3.87 | 0.049 |
| cg23072559 | SHISA7 | 3′UTR | N shore | SSC5D | A_21_P0000127 | ‐1.26 | 5.80 | 0.016 |
| cg23350744 | PRDM6 | – | N shelf | SNX24 | A_33_P3702364 | ‐0.40 | 5.52 | 0.019 |
| cg24735611 | LOC728989 | Body | – | CHD1L | A_23_P45831 | 1.04 | 8.14 | 0.004 |
| cg24735611 | LOC728989 | Body | – | FMO5 | A_33_P3376214 | 1.27 | 5.46 | 0.019 |
| cg27106230 | STK24 | First exon; 5′UTR | Island | FARP1 | A_33_P3238856 | ‐0.57 | 5.55 | 0.019 |
| cg27106230 | STK24 | First exon; 5′UTR | Island | SLC15A1 | A_33_P3373273 | ‐0.75 | 6.58 | 0.010 |
Grandchildren n = 87. RefGene group = gene region feature category describing the CpG position.
N: North; UCSC: University of California Santa Cruz.
Discussion
Very little is known about the intergenerational effects of GMSDP on the health outcomes of grandchildren. Using data from three generations of IOWBC, association of GMSDP with changes in the epigenome of grandchildren has been examined. We identified 23 CpG sites that were differentially methylated in association with GMSDP, of which eight were associated with neighboring gene expression.
These findings indicate that GMSDP has lasting effects that persist across generations. We compared the results with studies focused on the effect of MSDP on DNAm, which have identified over 6000 differentially methylated CpG sites [44,45]. Interestingly, none of the 23 CpG sites identified in this study as being associated with GMSDP (Table 2) have been reported in studies of MSDP. However, when comparing DNAm in grandchildren exposed to both GMSDP and MSDP with unexposed grandchildren in the sample, we found 12 CpG sites that were differentially methylated (Supplementary Table 3) and located on three genes repeatedly reported to be associated with MSDP: MYO1G, AHRR and GFI1. Together, these findings indicate that GMSDP and MSDP affect different epigenetic locations, which may translate into different epigenetic effects. The mechanism of the intergenerational epigenetic transmission remains unclear since there is no evidence to date to support direct germline transmission of DNAm from mother to offspring [46]. One possible explanation of theses findings is that GMSDP exerts a direct effect on F2 gametes located in the developing ovaries of the F1 fetus [44,47,48].
Of the 23 CpG sites that were associated with GMSDP, eight were located in CpG islands, four in north shore, three in north shelf and one in south shore (Table 2). The methylation level of these sites could have a functional impact on nearby genes, some of which are involved in important biological processes [49]. For example, MYOM2 and MYOM3 genes are expressed in skeletal and cardiac muscle [50], whereas IGF1R and IGFALS genes are essential for normal human growth [51–53] and CNS development [54,55]. Mutations in IGF1R and IGFALS genes have been related to short stature, delayed puberty and growth retardation [56,57].
Moreover, methylation levels at eight CpG sites were associated with nearby gene expression levels (Table 3), which indicates a potential functional implication of GMSDP. Among these genes, CD33 encodes a sialic acid-binding immunoglobulin-like lectin that is involved in the regulation of leukocyte functions during the inflammatory and immune response [58,59]. Additionally, studies suggested that the CHD1L gene plays a vital role in nervous system development and neuronal differentiation [60] as well as kidney and urinary tract development [61]. The ELMOD3 gene is associated with hearing impairment [62] and autism spectrum disorder [63]. The FMO5 gene is a key regulator of metabolic aging [64], and absence of the FMO5 protein is associated with high glucose tolerance and insulin sensitivity [65].
The results support an intergenerational effect of GMSDP on DNAm of grandchildren which, in turn, may affect the health of the grandchildren. Of note is that the novel CpG sites identified in this study were not reported in previous studies that assessed the association of MSDP with DNAm of the newborn. This implies that the epigenetic mechanisms involved in MSDP and GMSDP may be different. Finally, these results provide evidence of the potential physiological effects of these changes through gene function changes.
There are several methodological strengths in this study. Data from three generations of IOWBC were utilized to directly observe the intergenerational effect of GMSDP. By comparing grandchildren exposed to different combinations of GMSDP and MSDP, the pure effect of GMSDP on DNAm of grandchildren was identified. In addition, it resulted in high-quality data by implementing state-of-the-art methodology for exposure and outcome assessments.
The study has several limitations, including the small sample size, which may have reduced the power of the study, and the failure to find another available cohort for replication. In addition, the effect of maternal GMSDP on DNAm of grandchildren alone was assessed, but it is important to examine the effect of paternal grandmother smoking as well to have a complete picture of the intergenerational effect of GMSDP. Also, DNAm from both cord blood and Guthrie cards was used. However, the comparability of DNAm between umbilical cord blood and Guthrie cards was examined based on 34 grandchildren in the IOWBC, which showed that around 70% of the CpG sites agreed with regard to DNAm mean between cord blood and Guthrie cards [28]. Additionally, the source of DNAm (cord blood or Guthrie card) was included as a covariate in the multivariate model and was not associated with DNAm levels. Although grandmaternal smoking behavior was assessed by self-report and validated by serum cotinine levels measured at the end of pregnancy, misclassification could still be a possibility. Smoking cessation in early pregnancy may negate the effects on methylation at birth [66]. Unfortunately, due to the lack of cessation information, we were unable to determine whether the effect of GMSDP would be same with grandmaternal smoking cessation during pregnancy. Finally, loss to follow-up could be another limitation considering IOWBC included three generations, but there is no evidence to suggest that the loss to follow-up was not at random.
Conclusion
This study identified novel loci differentially methylated in newborn grandchildren associated with GMSDP independent from MSDP. To our knowledge, this is the first study assessing the intergenerational effect of GMSDP on DNAm patterns of newborn grandchildren. In addition, the study provides preliminary evidence of the potential effects of GMSDP on the health of grandchildren through its effect on the function of several important genes.
Future perspective
Although we demonstrated a potential biological effect of GMSDP on several CpG sites in the blood of grandchildren, the nature and extent of these effects remain largely unknown. It remains unknown whether the differential levels due to GMSDP are biologically and clinically relevant. Future studies are needed to examine whether these changes are related to clinical outcomes in grandchildren.
Summary points.
The authors identified an intergenerational effect of grandmaternal smoking during pregnancy (GMSDP) on the DNA methylation profile of grandchildren at birth independent of maternal smoking during pregnancy.
The training–testing screening method was used to identify the differentially methylated CpG sites related to GMSDP and/or maternal smoking during pregnancy in the peripheral blood of grandchildren.
A total of 23 CpG sites were identified as differentially methylated in association with GMSDP in the newborn after adjusting for confounding and multiple testing.
DNA methylation levels of eight significantly differentially methylated CpG sites were associated with the expression levels of 13 nearby genes, which suggests a functional effect of GMSDP on the health of grandchildren.
Some of these 13 genes have important functional implications, including the CD33, CHD1L and ELMOD3 genes, which are related to leukocyte function regulation and nervous system and kidney development.
The identified CpG sites could potentially serve as blood biomarkers of intergenerational smoking exposure.
Acknowledgments
The authors gratefully acknowledge the valuable effort of all staff at the David Hide Asthma and Allergy Research Centre in maintaining the Isle of Wight birth cohort. The authors also thank the team at the information technology services research computing office at the University of Memphis for their help in supporting high-performance computing. The authors would also like to thank the participants and their families, who have helped them over the last two decades.
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/epi-2021-0433
Author contributions
R Luo tested hypotheses, conducted the analysis and drafted the manuscript. H Zhang contributed to the conception and interpretation of the data. N Mukherjee assisted with data analysis and interpretation of results. W Karmaus assisted with data interpretation and manuscript revision. V Patil was involved in manuscript revision. H Arshad was involved in sample collection, data acquisition, DNA methylation measurement in the Isle of Wight birth cohort and manuscript revision. F Mzayek designed the study, guided the analysis and was involved in the drafting and revision of the manuscript prior to submission. The manuscript has been read and approved by all authors.
Financial & competing interests disclosure
This study was funded by the NIH (grant number: R01 AI091905-01). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate approval from the research ethics committee of the David Hide Asthma and Allergy Research Centre (reference number: 09/H0504/129) as well as the institutional review board at the University of Memphis (FWA00006815; 7 December 2012). In addition, informed consent has been obtained from the participants involved.
Reference
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Gaysina D, Fergusson DM, Leve LD et al. Maternal smoking during pregnancy and offspring conduct problems: evidence from 3 independent genetically sensitive research designs. JAMA Psychiatry 70(9), 956–963 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Zuckerman B, Pearson C et al. Maternal cigarette smoking, metabolic gene polymorphism, and infant birth weight. JAMA 287(2), 195–202 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Stick SM, Burton PR, Gurrin L, Sly PD, Lesouef PN. Effects of maternal smoking during pregnancy and a family history of asthma on respiratory function in newborn infants. Lancet 348(9034), 1060–1064 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Cnattingius S. The epidemiology of smoking during pregnancy: smoking prevalence, maternal characteristics, and pregnancy outcomes. Nicotine Tob. Res. 6(Suppl. 2), S125–S140 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Hanrahan JP, Tager IB, Segal MR et al. The effect of maternal smoking during pregnancy on early infant lung function. Am. Rev. Respir. Dis. 145(5), 1129–1135 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Horta BL, Victora CG, Menezes AM, Halpern R, Barros FC. Low birthweight, preterm births and intrauterine growth retardation in relation to maternal smoking. Paediatr. Perinat. Epidemiol. 11(2), 140–151 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Chiolero A, Bovet P, Paccaud F. Association between maternal smoking and low birth weight in Switzerland: the EDEN study. Swiss Med. Wkly 135(35-36), 525–530 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Pereira PP, Da Mata FA, Figueiredo AC, De Andrade KR, Pereira MG. Maternal active smoking during pregnancy and low birth weight in the Americas: a systematic review and meta-analysis. Nicotine Tob. Res. 19(5), 497–505 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Kyrklund-Blomberg NB, Cnattingius S. Preterm birth and maternal smoking: risks related to gestational age and onset of delivery. Am. J. Obstet. Gynecol. 179(4), 1051–1055 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Kyrklund-Blomberg NB, Granath F, Cnattingius S. Maternal smoking and causes of very preterm birth. Acta Obstet. Gynecol. Scand. 84(6), 572–577 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Jaakkola JJ, Gissler M. Maternal smoking in pregnancy, fetal development, and childhood asthma. Am. J. Public Health 94(1), 136–140 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burke H, Leonardi-Bee J, Hashim A et al. Prenatal and passive smoke exposure and incidence of asthma and wheeze: systematic review and meta-analysis. Pediatrics 129(4), 735–744 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Power C, Jefferis BJ. Fetal environment and subsequent obesity: a study of maternal smoking. Int. J. Epidemiol. 31(2), 413–419 (2002). [PubMed] [Google Scholar]
- 14.Ino T. Maternal smoking during pregnancy and offspring obesity: meta-analysis. Pediatr. Int. 52(1), 94–99 (2010). [DOI] [PubMed] [Google Scholar]; • Describes the risks of grandmaternal nicotine exposure during gestation.
- 15.Maritz GS, Mutemwa M. The effect of grand maternal nicotine exposure during gestation and lactation on lung integrity of the F2 generation. Pediatr. Pulmonol. 49(1), 67–75 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Magnus MC, Haberg SE, Karlstad O, Nafstad P, London SJ, Nystad W. Grandmother's smoking when pregnant with the mother and asthma in the grandchild: the Norwegian Mother and Child Cohort Study. Thorax 70(3), 237–243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lodge CJ, Braback L, Lowe AJ, Dharmage SC, Olsson D, Forsberg B. Grandmaternal smoking increases asthma risk in grandchildren: a nationwide Swedish cohort. Clin. Exp. Allergy 48(2), 167–174 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Li YF, Langholz B, Salam MT, Gilliland FD. Maternal and grandmaternal smoking patterns are associated with early childhood asthma. Chest 127(4), 1232–1241 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Miller LL, Henderson J, Northstone K, Pembrey M, Golding J. Do grandmaternal smoking patterns influence the etiology of childhood asthma? Chest 145(6), 1213–1218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am. J. Respir. Crit. Care Med. 180(5), 462–467 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maccani JZJ, Koestler DC, Houseman EA, Marsit CJ, Kelsey KT. Placental DNA methylation alterations associated with maternal tobacco smoking at the RUNX3 gene are also associated with gestational age. Epigenomics 5(6), 619–630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richmond RC, Simpkin AJ, Woodward G et al. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon Longitudinal Study of Parents and Children (ALSPAC). Hum. Mol. Genet. 24(8), 2201–2217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arshad SH, Hide DW. Effect of environmental factors on the development of allergic disorders in infancy. J. Allergy Clin. Immunol. 90(2), 235–241 (1992). [DOI] [PubMed] [Google Scholar]
- 24.Soto-Ramirez N, Ziyab AH, Karmaus W et al. Epidemiologic methods of assessing asthma and wheezing episodes in longitudinal studies: measures of change and stability. J. Epidemiol. 23(6), 399–410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arshad SH, Holloway JW, Karmaus W et al. Cohort profile: the Isle Of Wight whole population birth cohort (IOWBC). Int. J. Epidemiol. 47(4), 1043–1044i (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verification SSOB. Biochemical verification of tobacco use and cessation. Nicotine Tob. Res. 4(2), 149–159 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Balhara YPS, Jain R. A receiver operated curve-based evaluation of change in sensitivity and specificity of cotinine urinalysis for detecting active tobacco use. J. Cancer Res. Ther 9(1), 84–89 (2013). [DOI] [PubMed] [Google Scholar]; •• Provides justifications for using DNA methylation (DNAm) profiles from umbilical cord blood and Guthrie cards.
- 28.Jiang Y, Wei JF, Zhang HM et al. Epigenome wide comparison of DNA methylation profile between paired umbilical cord blood and neonatal blood on Guthrie cards. Epigenetics 15(5), 454–461 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16(3), 1215 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beyan H, Down TA, Ramagopalan SV et al. Guthrie card methylomics identifies temporally stable epialleles that are present at birth in humans. Genome Res. 22(11), 2138–2145 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bibikova M, Fan J-B. GoldenGate assay for DNA methylation profiling. Methods Mol. Biol. 507, 149–163 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Du P, Zhang X, Huang C-C et al. Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 11, 587 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakulski KM, Feinberg JI, Andrews SV et al. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics 11(5), 354–362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houseman EA, Accomando WP, Koestler DC et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 86 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reinius LE, Acevedo N, Joerink M et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE 7(7), e41361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang D, Yan L, Hu Q et al. IMA: an R package for high-throughput analysis of Illumina's 450K Infinium methylation data. Bioinformatics 28(5), 729–730 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8(1), 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Lehne B, Drong AW, Loh M et al. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 16, 37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogbuanu IU, Karmaus W, Arshad SH, Kurukulaaratchy RJ, Ewart S. Effect of breastfeeding duration on lung function at age 10 years: a prospective birth cohort study. Thorax 64(1), 62–66 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Provides the statistical model for screening in epigenome-wide association studies.
- 40.Ray MA, Tong X, Lockett GA, Zhang H, Karmaus WJ. An efficient approach to screening epigenome-wide data. Biomed. Res. Int. 2016, 2615348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, Wang F, Kranzler HR et al. Identification of methylation quantitative trait loci (mQTLs) influencing promoter DNA methylation of alcohol dependence risk genes. Hum. Genet. 133(9), 1093–1104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welch RP, Willer CJ, SL J., Boehnke M. Snipper: a research tool for extracting and searching biological annotations on genes near SNPs (2013). https://csg.sph.umich.edu/boehnke/snipper/
- 43.Karolchik D, Baertsch R, Diekhans M et al. The UCSC Genome Browser database. Nucleic Acids Res. 31(1), 51–54 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joubert BR, Felix JF, Yousefi P et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am. J. Hum. Genet. 98(4), 680–696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joubert BR, Haberg SE, Nilsen RM et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ. Health Persp. 120(10), 1425–1431 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Byrnes JJ, Babb JA, Scanlan VF, Byrnes EM. Adolescent opioid exposure in female rats: transgenerational effects on morphine analgesia and anxiety-like behavior in adult offspring. Behav. Brain Res. 218(1), 200–205 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes potential mechanisms of the intergenerational epigenetic inheritance.
- 47.Barouki R, Melen E, Herceg Z et al. Epigenetics as a mechanism linking developmental exposures to long-term toxicity. Environ. Int. 114, 77–86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Otterdijk SD, Michels KB. Transgenerational epigenetic inheritance in mammals: how good is the evidence? FASEB J. 30(7), 2457–2465 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Ziller MJ, Gu HC, Muller F et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 500(7463), 477–481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schoenauer R, Lange S, Hirschy A, Ehler E, Perriard JC, Agarkova I. Myomesin 3, a novel structural component of the M-band in striated muscle. J. Mol. Biol. 376(2), 338–351 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Guler HP, Zapf J, Schmid C, Froesch ER. Insulin-like growth factors I and II in healthy man. Estimations of half-lives and production rates. Acta Endocrinol. (Copenh.) 121(6), 753–758 (1989). [DOI] [PubMed] [Google Scholar]
- 52.Iniguez G, Argandona F, Medina P et al. Acid-labile subunit (ALS) gene expression and protein content in human placentas: differences according to birth weight. J. Clin. Endocrinol. Metab. 96(1), 187–191 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Ester WA, Hokken-Koelega AC. Polymorphisms in the IGF1 and IGF1R genes and children born small for gestational age: results of large population studies. Best Pract. Res. Clin. Endocrinol. Metab. 22(3), 415–431 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Beck KD, Powell-Braxton L, Widmer HR, Valverde J, Hefti F. Igf1 gene disruption results in reduced brain size, CNS hypomyelination, and loss of hippocampal granule and striatal parvalbumin-containing neurons. Neuron 14(4), 717–730 (1995). [DOI] [PubMed] [Google Scholar]
- 55.D'Ercole AJ, Ye P, O'Kusky JR. Mutant mouse models of insulin-like growth factor actions in the central nervous system. Neuropeptides 36(2–3), 209–220 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Hess O, Khayat M, Hwa V et al. A novel mutation in IGFALS, c.380T > C (p.L127P), associated with short stature, delayed puberty, osteopenia and hyperinsulinaemia in two siblings: insights into the roles of insulin growth factor-1 (IGF1). Clin. Endocrinol. 79(6), 838–844 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Choi J-H, Kang M, Kim G-H et al. Clinical and functional characteristics of a novel heterozygous mutation of the IGF1R gene and IGF1R haploinsufficiency due to terminal 15q26.2->qter deletion in patients with intrauterine growth retardation and postnatal catch-up growth failure. J. Clin. Endocrinol. Metab. 96(1), e130–e134 (2011). [DOI] [PubMed] [Google Scholar]; •• Discusses the role of CD33 in the immune system.
- 58.Lock K, Zhang J, Lu J, Lee SH, Crocker PR. Expression of CD33-related siglecs on human mononuclear phagocytes, monocyte-derived dendritic cells and plasmacytoid dendritic cells. Immunobiology 209(1–2), 199–207 (2004). [DOI] [PubMed] [Google Scholar]; •• Discusses the role of CD33 in the nervous system.
- 59.Griciuc A, Serrano-Pozo A, Parrado AR et al. Alzheimer's disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron 78(4), 631–643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Discusses the role of CHD1L in neuronal differentiation in embryonic stem cells.
- 60.Dou D, Zhao H, Li Z et al. CHD1L promotes neuronal differentiation in human embryonic stem cells by upregulating PAX6. Stem Cells Dev. 26(22), 1626–1636 (2017). [DOI] [PubMed] [Google Scholar]; •• Discusses the role of CD33 in congenital abnormalities.
- 61.Brockschmidt A, Chung B, Weber S et al. CHD1L: a new candidate gene for congenital anomalies of the kidneys and urinary tract (CAKUT). Nephrol. Dial. Transplant. 27(6), 2355–2364 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Li W, Feng Y, Chen A et al. Elmod3 knockout leads to progressive hearing loss and abnormalities in cochlear hair cell stereocilia. Hum. Mol. Genet. 28(24), 4103–4112 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loi E, Moi L, Blois S et al. ELMOD3-SH2D6 gene fusion as a possible co-star actor in autism spectrum disorder scenario. J. Cell. Mol. Med. 24(2), 2064–2069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malagon SGG, Melidoni AN, Hernandez D et al. The phenotype of a knockout mouse identifies flavin-containing monooxygenase 5 (FMO5) as a regulator of metabolic ageing. Biochem. Pharmacol. 96(3), 267–277 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scott F, Malagon SGG, O'Brien BA et al. Identification of flavin-containing monooxygenase 5 (FMO5) as a regulator of glucose homeostasis and a potential sensor of gut bacteria. Drug Metab. Dispos. 45(9), 982–989 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joubert BR, Haberg SE, Bell DA et al. Maternal smoking and DNA methylation in newborns: in utero effect or epigenetic inheritance? Cancer Epidemiol. Biomarkers Prev. 23(6), 1007–1017 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]