Abstract
Triple-negative breast cancer (TNBC) is characterized by aggressive clinicopathological features and is associated with a poor prognosis. Identifying patients that are non-responsive to chemotherapy remains a critical goal for effective personalized therapies. In the present study, the predictive value of exosomal microRNAs (miRNAs) was investigated in patients with TNBC. Exosomes were isolated from patients with TNBC undergoing neoadjuvant chemotherapy. Microarray-based miRNA profiles were compared between patients with pathological complete response (pCR; n=12) and non-pCR (n=12). Furthermore, the miRNA profiles of non-pCR patients with breast cancer recurrence were compared with those with no recurrence. A total of 16 differentially expressed exosomal miRNAs were identified between the patients with pCR and non-pCR by microarray analysis. Of these, a combined signature of four miRNAs (miR-4448, miR-2392, miR-2467-3p and miR-4800-3p) could be used to discriminate between pCR and non-pCR patients with TNBC with an area under the curve value of 0.7652. Furthermore, this study found 43 differentially expressed miRNAs between the patients with non-pCR and recurrence and non-pCR patients without recurrence. In network analysis, ‘pathway in cancer’, ‘focal adhesion’ and ‘cell cycle’ were identified as the crucial pathways in patients with non-pCR who also developed recurrence. Several exosomal miRNAs may be useful biomarkers to predict treatment efficacy for TNBC. The present study identified patients who were resistant to standard chemotherapy and therefore more likely to develop breast cancer recurrence.
Keywords: exosome, microRNA, triple-negative breast cancer, neoadjuvant chemotherapy, pathological complete response
Introduction
Triple-negative breast cancer (TNBC), which is characterized at the molecular level as being estrogen receptor (ER)- and progesterone receptor (PR)-negative, and lacking overexpression of human epidermal growth factor-2 (HER2), accounts for 15–20% of all breast cancers and has more aggressive biological features and worse prognosis than other subtypes (1,2). Since there is no targeted therapy for TNBC patients, they typically receive standard chemotherapy, consisting of anthracycline and taxane-containing agents (3,4). Neoadjuvant treatment, which is an established standard therapy for patients with early breast cancer, facilitates evaluation of drug efficacy and allows a more precise determination of prognosis. Patients with TNBC who do not achieve a pathological complete response (pCR) after neoadjuvant chemotherapy (NAC) have worse survival rates than those who achieve pCR (5,6). Since the response is associated with survival benefit, pCR is often used for the clinical evaluation of treatment strategies. In TNBC, the addition of carboplatin to a paclitaxel regimen consistently improved pCR rates compared with paclitaxel alone (7,8). Another neoadjuvant study (the GeparSepto trial) demonstrated that pCR could be increased by exchanging solvent-based paclitaxel with nab-paclitaxel; patients with TNBC were among those who benefited (9). Thus, a lot of effort has been made to find ways of improving the pCR rates to neoadjuvant treatment.
In recent years, several circulating cancer-specific biomarkers have been developed to accurately predict clinical outcome and response to therapy for early breast cancer (10). Exosomes are small membranous vesicles (30–100 nm) containing lipid, proteins, microRNAs (miRNAs) and mRNAs that are secreted by viable cells into the blood circulation (11). The ‘cargo’ in exosomes may contain not only important mediators of intercellular communication, but can also transmit information to the surrounding microenvironment (12,13). miRNAs are non-coding small RNA molecules 19–25 nucleotides in length. They regulate gene expression at the post-transcriptional level by binding to 3′ or 5′ untranslated regions of target messenger RNAs (mRNAs), resulting in inhibition of translation or regulated mRNA degradation (14,15). Deregulation of miRNA has been observed in several types of cancer, including malignancies of the breast (16,17). miRNAs are commonly incorporated into microvesicles or bound to lipoproteins such as HDL or associated with argonaute-2 (Ago2)-containing complexes in the blood (18) Among these complexes, exosome-derived miRNAs are the most appealing for longitudinal observation of disease development, since the membrane encapsulation can maintain structural integrity and protect against degradation by external proteases and other enzymes that are present in biofluids (19).
As described above, exosomal miRNAs represent an informative readout for several tumor types, in terms of detection of the cancer and prediction of clinical outcome (20–22). Additionally, differential expression of subtype-specific miRNAs has also been reported in several studies (23,24). A recent study by Stevic et al demonstrated different exosomal miRNA signatures in HER2-positive and TNBC patients (24). A combined signature consisting of exosomal miR-335, miR-628, and miR-422a could be used to discriminate between TNBC and HER2-positive breast cancer patients with a sensitivity of 68% and a specificity of 81% (24). A deeper understanding of subtype-specific miRNA profiles has provided insight into identifying potential therapeutic strategies and inferring tumor progression, particularly for aggressive TNBC subtypes.
In this study, we investigated the predictive role of exosomal miRNAs in serum samples derived from patients with TNBC before and after NAC. We performed miRNA array analysis in 24 patients with and without pCR to identify which miRNA profile was most closely correlated with the effect of NAC. Furthermore, we determined which patients experienced recurrence among those who received standard chemotherapy and nevertheless did not achieve pCR, since these patients may need more extensive therapy.
Materials and methods
Study population and treatment
First, we retrospectively selected 24 TNBC patients diagnosed with invasive breast cancer, who underwent NAC treatment and subsequent surgery at Kumamoto University Hospital between 2008 and 2016 (Study 1, Fig. S1). This cohort included 12 patients with pCR and 12 patients with non-pCR. TNBC was defined as no expression of ER, PR or HER2. HER2 expression was determined by immunohistochemical (IHC) staining based on the Hercep test. Ki67 was scored as the percentage of nuclear-stained cells out of all cancer cells in the hot spot of the tumor, regardless of the intensity in a ×400 high-power field (Ki67 labeling index). Patient characteristics for Study 1 are shown in Table I.
Table I.
Clinicopathological characteristics in patients with NAC.
| Clinicopathological characteristics | Patients with pCR (n=12) | Patients with non-pCR (n=12) | P-value |
|---|---|---|---|
| Median age at diagnosis, median (range) | 46 (24–63) | 59 (37–74) | 0.04 |
| Menopausal status, n (%) | 0.21 | ||
| Premenopausal | 7 (58) | 3 (25) | |
| Postmenopausal | 5 (42) | 9 (75) | |
| Tumor size, cm, median (range) | 2.5 (1.1–6.4) | 3.2 (1.6–7.8) | 0.34 |
| Nodal status, n (%) | 1.00 | ||
| Negative | 6 (50) | 6 (50) | |
| Positive | 6 (50) | 6 (50) | |
| Clinical T, n (%) | 0.75 | ||
| T1 | 3 (25) | 2 (17) | |
| T2 | 8 (67) | 8 (67) | |
| T3 | 0 (0) | 1 (8) | |
| T4 | 1 (8) | 1 (8) | |
| Stage, n (%) | 0.54 | ||
| 1 | 2 (17) | 2 (17) | |
| 2 | 9 (75) | 7 (58) | |
| 3 | 1 (8) | 3 (25) | |
| Nuclear grade, n (%) | 0.17 | ||
| 1,2 | 5 (42) | 2 (17) | |
| 3 | 7 (58) | 10 (83) | |
| Ki67 labeling index, %, median (range) | 61 (30–97) | 65 (20–100) | 0.75 |
| NAC, n (%) | 1.00 | ||
| FEC followed by DOC | 9 (75) | 8 (67) | |
| PAC followed by FEC | 3 (25) | 3 (25) | |
| FEC | 0 (0) | 1 (8) |
pCR, pathological complete response; NAC, neoadjuvant chemotherapy; FEC, 5-fluorouracil, epirubicin and cyclophosphamide; DOC, docetaxel; PAC, paclitaxel.
Neoadjuvant treatment was assigned to each patient according to their risk on the basis of the clinical parameters, and in accordance with the recommendation of the St. Gallen International Expert Consensus on primary therapy of early breast cancer at the time. For the NAC regimen, all but one patient received eight cycles of anthracycline and taxane-containing agents (Table I); 17 patients received four cycles of 5-fluorouracil 100 mg/m2, epirubicin 100 mg/m2, and cyclophosphamide 500 mg/m2 (FEC), followed by four cycles of docetaxel 75 mg/m2 (DOC), six patients received 12 cycles of paclitaxel 80 mg/m2, followed by four cycles of FEC, one patient received six cycles of FEC. Treatment responses were evaluated according to clinical and pathological features. A pCR was defined based on postoperative specimens as the absence of invasive carcinoma both in breast and axillary lymph nodes.
For Study 2, we selected 16 TNBC patients with non-pCR, comprising eight patients with recurrence and eight without recurrence. The median follow-up period for the patients without recurrence was 81 months (65–121). Patient characteristics before NAC are shown in Table SI. Some of the patients in Study 2 had already been included in Study 1. Serum samples for Study 1 were collected before NAC and those for Study 2 were collected after NAC. We stored the serum samples from all patients at −80°C before use.
Written informed consent was obtained from all subjects for the collection and research use of serum samples. The entire study was approved by the ethics committee of Kumamoto University Graduate School of Medical Sciences.
Isolation of total exosomes from serum
Exosomes in serum samples (500 ml) were extracted using ExoQuick (System Biosciences) according to the manufacturer's instructions. In brief, the reagent was added into the serum sample and incubated at 4°C for at least 30 min to precipitate the exosome pellet. We previously confirmed the identity of the isolated exosomes (25). The size of the exosomes was confirmed using the Nanosight LM10 instrument and presence of an exosomal membrane marker (CD63) was confirmed by western blot analysis (25).
Isolation of total RNA
Total RNA that included the small RNA fraction from exosome pellets was isolated using a miRNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. Cel-miR-39 was used as a spike-in control to confirm the extraction of RNA. The quality of RNA samples and the small RNA fraction was checked using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). The RNA samples were immediately stored at −80°C until use.
MicroRNA microarray analysis
Total RNA was extracted from exosome pellets using the 3D-Gene® RNA extraction reagent (Toray Industries, Inc.). One-half of the amount of extracted total RNA was prepared for miRNA expression analysis using the 3D-Gene® miRNA Labeling kit and the 3D-Gene® Human miRNA Oligo Chip (Toray Industries, Inc.), which was designed to detect 2,565 miRNA sequences registered in miRbase release 21 (http://www.mirbase.org/). The signal values were quantified using the 3D-Gene Scanner 3000 (Toray Industries, Inc.). After subtracting the background values from the signal values, the signals were standardized by the global normalization method, which adjusted the median signal intensity on the chip to 25 (Toray Industries, Inc.). All microarray data obtained in the present study were in agreement with the Minimum Information About a Microarray Experiment guidelines.
Quantitative reverse-transcription polymerase chain reaction (RT-qPCR)
Total RNA extracted from exosomes was reverse transcribed to cDNA using a miScript IIRT kit (Qiagen), according to the manufacturer's protocol. miRNA expression levels were investigated using miScript Primer Assay with a miScript SYBR Green PCR kit according to the manufacturer's protocol (Qiagen). For the normalization of real-time PCR, we selected miR-16-5p and miR-451 because their levels were unchanged across all our datasets, consistent with their use as reference miRNAs in other reports (23,26). Quantification of miRNA expression was performed using the 2−∆∆Ct method.
Survival analyses
The prognostic significance of four selected miRNAs (miR-4448, miR-2392, miR-2467, and miR-4800) was evaluated using the online database, Kaplan-Meier (KM) plotter (http://kmplot.com/analysis/). We analyzed the overall survival of TNBC patients using KM plotter, where they were divided into two groups according to the median expression of each miRNA (high or low). Log rank P-value and hazard ratio (HR) with 95% confidence interval (CI) was calculated.
Statistical analyses
In microarray analysis, candidate miRNAs with differential expression between the two groups (Study 1 and Study 2) were determined based on Student's t-test. Significant miRNAs were defined as those with a P-value <0.05 and at least a 1.5-fold change. The significance of differences in categorized demographic variables was evaluated using the Chi-square test or Fisher's exact test and the nonparametric Mann-Whitney U test. For pCR prediction of miRNAs, we calculated the area under the curve (AUC) using a receiver-operating characteristic (ROC) curve analysis. Logistic regression methods were adopted for univariate analyses to assess the associations of each miRNA with pCR. Odds ratios (ORs) and 95% CIs were calculated. All statistical analyses were carried out using STATA ver.13 (Stata Corp.). All tests were two-sided and P-values <0.05 were considered statistically significant.
Results
Comparison of exosomal miRNA profiles between patients with pCR and non-pCR
First, we retrospectively evaluated the expression of exosomal miRNAs before chemotherapy between patients with pCR and non-pCR (Fig. S1, Study 1). Patient characteristics (n=24) are summarized in Table I. In this cohort, we have adjusted as much as possible for other confounding factors to compare expression of exosomal miRNAs. Therefore, we found no difference in clinicopathological factors other than age between two groups: patients with pCR were younger than those with non-pCR (median age at diagnosis: 46 vs. 59 years, respectively). Most patients received neoadjuvant treatment including a combined anthracycline- and taxane-containing regimen except for one. All patients had tumors with relatively aggressive molecular features with high nuclear grade and Ki67 labeling index (median 65%, range 20–100).
We performed a comprehensive analysis of exosomal miRNAs, in which we identified several miRNAs that were differentially expressed prior to NAC between the patients with pCR and non-pCR. Table II shows only the differentially-expressed miRNAs identified among the 2,556 miRNAs that were analyzed. In patients with pCR, there were five upregulated miRNAs (miR-1273e, miR-4800-3p, miR-2392, miR-2467-3p, and miR-4448) and 11 downregulated miRNAs (miR-3918, miR-4740-3p, miR-3177-5p, miR-1203, miR-874-3p, miR-4419a, miR-637, miR-423-5p, miR-6796-3p, miR-4707-3p, and miR-210-5p); (Table II, Fig. 1). miR-1273e was the most highly upregulated gene (2.38-fold) and miR-3918 was the top downregulated gene (0.27-fold compared to control).
Table II.
Differentially expressed exosomal miRNAs related to patients with pCR compared with non-pCR determined via miRNA array analyses.
| A, Upregulated miRNAs | ||
|---|---|---|
|
| ||
| Mature ID | Fold-change | P-value |
| hsa-miR-1273e | 2.38 | 0.02 |
| hsa-miR-4800-3p | 1.70 | 0.03 |
| hsa-miR-2392 | 1.68 | 0.03 |
| hsa-miR-2467-3p | 1.67 | 0.02 |
| hsa-miR-4448 | 1.54 | 0.02 |
|
| ||
| B, Downregulated miRNAs | ||
|
| ||
| Mature ID | Fold-change | P-value |
|
| ||
| hsa-miR-3918 | 0.27 | 0.03 |
| hsa-miR-4740-3p | 0.38 | 0.03 |
| hsa-miR-3177-5p | 0.40 | 0.03 |
| hsa-miR-1203 | 0.42 | 0.03 |
| hsa-miR-874-3p | 0.45 | <0.01 |
| hsa-miR-4419a | 0.51 | 0.04 |
| hsa-miR-637 | 0.59 | <0.01 |
| hsa-miR-423-5p | 0.60 | 0.05 |
| hsa-miR-6796-3p | 0.61 | 0.04 |
| hsa-miR-4707-3p | 0.63 | 0.01 |
| hsa-miR-210-5p | 0.67 | 0.02 |
Only the miRNAs with statistically significant P-values are shown. miRNA/miR, microRNA; pCR, pathological complete response.
Figure 1.
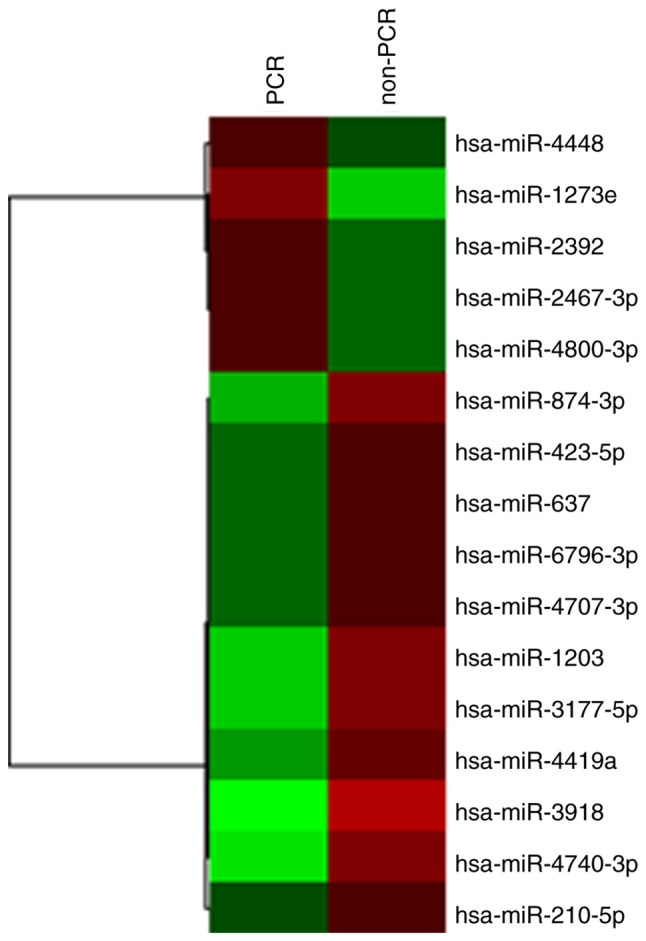
Hierarchal clustering analysis heatmap of patients with pCR compared with those with non-pCR. Red and green represent upregulated and downregulated miRNAs, respectively. The shaded areas show groups with similar expression levels. pCR, pathological complete response; miR, microRNA.
Validation of identified miRNAs
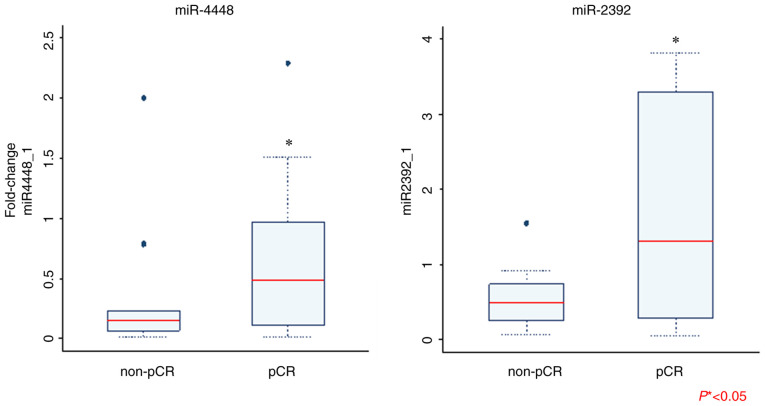
Next, we validated the expression of the key differentially-regulated miRNAs using RT-qPCR. Of the 16 miRNAs, the expression of miR-4448 and miR-2392 was significantly higher in the patients with pCR than in those with non-pCR (Fig. 2). We found no statistically-significant difference in the expression level of other miRNAs between the pCR and non-pCR groups. Following logistic regression analysis for prediction of pCR, we found no significant difference in expression of exosomal miRNAs between the two groups (Table III). We then selected four miRNAs (miR-4448, miR-2392, miR-2467-3p, and miR-4800-3p) with relatively small P-values (P<0.2) in the corresponding logistic regression models. The AUC for prediction of pCR was 0.7652 when these four miRNAs were combined in the analysis; this value is considered to indicate moderate accuracy (Fig. S2).
Figure 2.
Comparison of exosomal miRNA expression between patients with pCR and non-pCR as determined via reverse transcription-quantitative PCR. Only results that are statistically significant are shown. Mann-Whitney U test was performed for data analysis (*P<0.05). pCR, pathological complete response; miRNA/miR, microRNA.
Table III.
Logistic regression analyses for pCR by miRNA expression in exosomes.
| miRNA (high vs. low) | OR (95% CI) | P-value |
|---|---|---|
| hsa-miR-1273e | 2.25 (0.32–15.80) | 0.414 |
| hsa-miR-4448 | 4.50 (0.67–30.20) | 0.122 |
| hsa-miR-2392 | 10.00 (0.96–104.50) | 0.054 |
| hsa-miR-2467-3p | 4.50 (0.67–30.21) | 0.122 |
| hsa-miR-4800-3p | 4.50 (0.67–30.21) | 0.122 |
| hsa-miR-637 | 2.45 (0.46–13.23) | 0.296 |
| hsa-miR-874-3p | 2.45 (0.46–13.23) | 0.296 |
| hsa-miR-18a-5p | 1.69 (0.65–4.38) | 0.282 |
| hsa-miR-423-5p | 1.68 (0.32–8.76) | 0.538 |
| hsa-miR-3177-5p | 2.45 (0.46–13.23) | 0.296 |
| hsa-miR-1203 | 1.68 (0.32–8.76) | 0.538 |
| hsa-miR-4707-3p | 2.45 (0.46–13.23) | 0.296 |
| hsa-miR-3918 | 0.60 (0.11–3.10) | 0.538 |
| hsa-miR-210-5p | 0.41 (0.08–2.19) | 0.296 |
| hsa-miR-4419a | 1.68 (0.32–8.76) | 0.538 |
| hsa-miR-6769-3p | 2.45 (0.46–13.23) | 0.296 |
| hsa-miR-4740-3p | 1.68 (0.32–8.76) | 0.538 |
miRNA/miR, microRNA; pCR, pathological complete response; OR, odds ratio; CI, confidence interval.
Survival analysis of miRNAs identified in TNBC
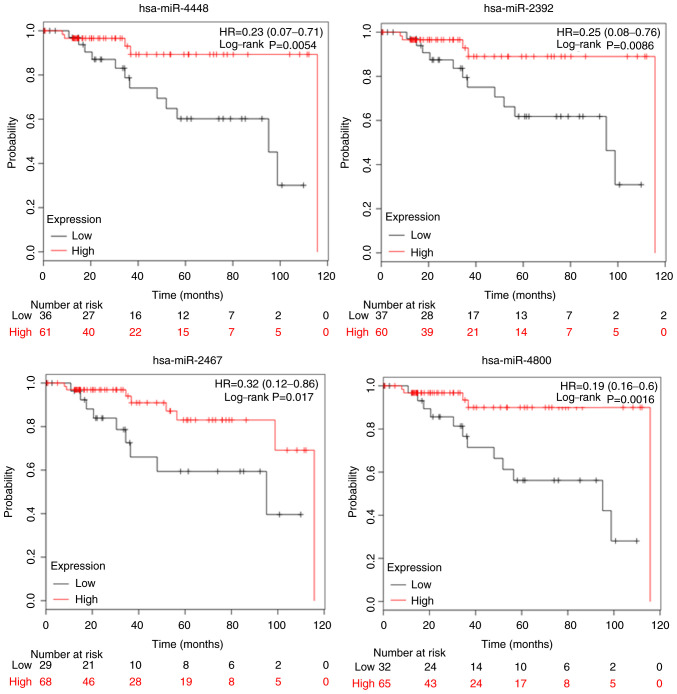
The biological mechanisms-of-action for the miRNAs we identified are not fully described in the published literature. However, to determine whether they may have an impact on survival, we determined whether there were any correlations between the four miRNAs (miR-4448, miR-2392, miR-2467, miR-4800) and overall survival (OS) using the Kaplan-Meier Plotter online database (http://kmplot.com/analysis/). When stratified according to TN subtypes, patients with higher levels of miR-4448 had a longer OS compared with those with lower levels (HR=0.23, 95% CI=0.07–0.71, P=0.0054), as did those with higher levels of miR-2392 (HR=0.25, 95% CI=0.06–0.76, P=0.0086), miR-2467 (HR=0.32, 95% CI=0.12–0.86, P=0.017), and miR-4800 (HR=0.19, 95% CI=0.06–0.6, P=0.0016) (Fig. 3). Based on these results, we suggest that these four miRNAs may have predictive and prognostic utility in TNBC.
Figure 3.
Prognostic value of miR-4448, miR-2392, miR-2467 and miR-4800 expression. Overall survival was evaluated using an online database, Kaplan-Meier plotter. Red lines, high expression of exosomal miRNAs; black lines, low expression. The cut-off values are set at the median for each gene. HR, hazard ratio; miRNA/miR, microRNA.
Identification of miRNAs that are differentially expressed in TNBC patients with non-pCR and breast cancer recurrence
We isolated exosomal miRNA from serum samples of patients who underwent NAC. This cohort consisted of TNBC patients with non-pCR who experienced recurrence as well as those who did not. A total of 16 patients with non-pCR were analyzed (Table SI). We found the clinicopathological factors between the groups with recurrence (n=8) and without recurrence (n=8) were relatively balanced, as the patients with recurrence were more likely to have high levels of Ki67 (median 88 vs. 60%, P=0.064). A hierarchical clustering analysis showed a total of 43 differentially-expressed miRNAs (15 upregulated and 28 downregulated) between non-pCR patients with or without recurrence (Fig. 4 and Table SII). miR-195-5p had the highest fold change (4.43, P=0.02) among the upregulated miRNAs, while miR-548ab was the most downregulated (0.23, P<0.001).
Figure 4.
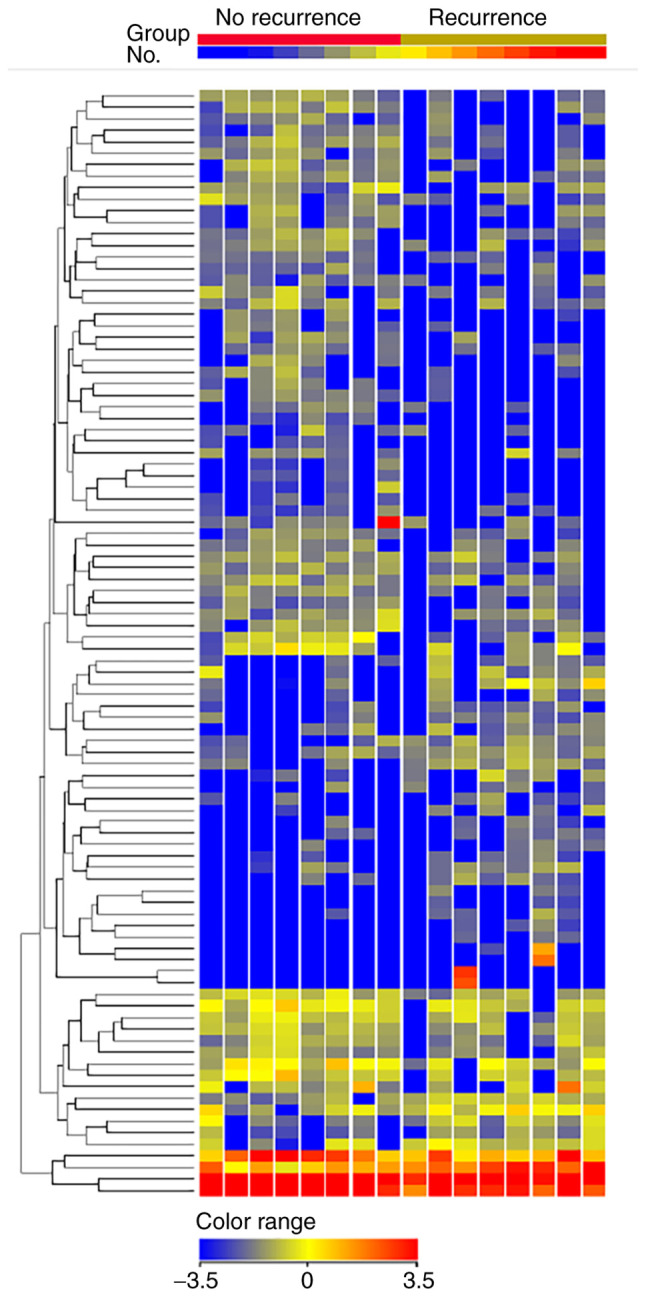
Hierarchal clustering analysis observed among the non-pCR patients with recurrence (n=8) and no recurrence (n=8). Only miRNAs with fold change ≥2.0 are shown. Heatmap colors represent exosomal miRNA expression. The name of each miRNA is omitted in the figure. pCR, pathological complete response; miRNA/miR, microRNA.
Pathway enrichment analysis and prediction of genes that are targets of the differentially-expressed miRNAs
We used the online database Diana Tools mirPath v.3 analysis (http://snf-515788.vm.okeanos.grnet.gr/). to identify pathways associated with the differentially-expressed genes. The top 20 KEGG pathways related to patients with non-pCR and breast cancer recurrence are shown in Table SIII. These included cancer, focal adhesion, cell cycle, and the MAPK signaling pathway, all of which are involved in cancer progression. Subsequently, we obtained a list of putative target genes of differentially expressed miRNAs using the mirTargetLink software (https://ccb-web.cs.uni-saarland.de/mirtargetlink/). Six target genes were controlled by three or more miRNAs (BCL2, CCND1, CDK6, MET, VEGFA, and WEE1; Table IV). Similarly, target genes controlled by two miRNAs observed in the patients with non-pCR and breast cancer recurrence are shown in Table SIV. Among these miRNAs, miR-16-5p and miR-195-5p regulated the most target genes (18 and 17, respectively). Taken together, these pathways and genes may be involved in drug resistance and contribute to a poor prognosis in TNBC.
Table IV.
Target genes controlled by ≥3 miRNAs among the differentially expressed genes in patients with non-pCR and breast cancer recurrence.
| Target gene | miRNAs |
|---|---|
| BCL2 | hsa-miR-195-5p, hsa-miR-16-5p, hsa-miR-34b-3p |
| CCND1 | hsa-miR-106b-5p, hsa-miR-195-5p, hsa-miR-16-5p, hsa-let-7f-5p |
| CDK6 | hsa-miR-195-5p, hsa-miR-16-5p, hsa-miR-34b-3p |
| MET | hsa-miR-31-5p, hsa-miR-34b-3p, hsa-miR-7515 |
| VEGFA | hsa-miR-106b-5p, hsa-miR-195-5p, hsa-miR-16-5p, hsa-miR-34b-3p, had-miR-133a-3p |
| WEE1 | hsa-miR-106b-5p, hsa-miR-195-5p, hsa-miR-16-5p |
miRNA/miR, microRNA; pCR, pathological complete response; CCND1, G1/S-specific cyclin-D1; WEE1, Wee1-like protein kinase.
Discussion
In the present study, we evaluated the exosomal miRNA profiles of TNBC patients in order to identify novel potential biomarkers for pCR to NAC. We suggest that specific exosomal miRNAs identified from pretreatment serum samples may predict treatment efficacy. The combined model of four miRNAs, that is miR-4448, miR-2392, miR-2467-3p, and miR-4800-3p, could clearly discriminate between pCR and non-pCR patients of TNBC. Additionally, we found an association between the expression levels of these genes and overall survival.
Recently, several studies on circulating miRNAs using clinical samples have emerged (27–30). Stevic et al (24). examined the miRNA profiles in circulating exosomes of breast cancer patients before the neoadjuvant GeparSixto trial. The authors demonstrated that the expression of miR-155 and miR-301 in exosomes most significantly predicted pCR in uni- and multivariate analyses. The impact of miR-155 was found consistently through all subtypes, whereas that of miR-301 was found in all cancer subtypes except TNBC. Rodriguez-Martinez et al investigated exosomal miRNA profiles in breast cancer patients with locally-advanced cancer who were undergoing NAC (31). Patients with a complete response to therapy were more likely to have lower levels of miR-21 compared to those with stable disease, although this finding did not reach statistical significance (P=0.060). Another study by Yang et al (32). provided clinical evidence that chemotherapy-elicited exosomal miR-378a-3p and miR-378d were related to chemotherapy response via the EZH2/STAT3 axis.
Here, we showed that, prior to NAC, deregulation of 16 exosomal miRNAs was specifically associated with the pCR patients. To date, the biological activities of the corresponding miRNAs (with the exception of miR-4448 and miR-2392) in breast cancer have not been fully elucidated. A study of spheroid-enriched cells with cancer stem cell properties identified an association between miR-4448 and breast cancer chemoresistance and self-renewal capacity (33). In addition, a study by Hibino et al suggested that miR-4448 acts as a tumor suppressor in gastric and liver cancer cells (34). Another study by Li et al demonstrated that miR-2392 suppresses metastasis and the epithelial-mesenchymal transition by targeting MAML3 and WHSC1 in gastric cancer (35). A study by Fan et al also showed an association of miR-2392 with chemoresistance (36). They reported that mitochondrial miR-2392 regulated chemoresistance in tongue squamous cell carcinoma cells by reprogramming metabolism via downregulation of oxidative phosphorylation and upregulation of glycolysis.
In our study, the above-mentioned miRNAs were upregulated in patients with pCR, indicating a relationship between these genes and treatment efficacy of NAC. In addition, the potential tumor suppressor roles of these miRNAs might explain why higher expression of these miRNAs is associated with longer OS in TNBC (Fig. 3), although the patient cohorts were not adjusted for confounding factors. Additional functional studies investigating how these miRNAs affect the response to breast cancer treatments are now required.
Currently approximately 30% of TNBC patients receive NAC, and the pCR rate following chemotherapy is between 30 and 47% (37,38). Neoadjuvant treatment offers us an opportunity to evaluate the efficacy of drugs in patients who have residual invasive disease after therapy. By analyzing the exosomal miRNA profiles as we outlined here, we can help patients avoid ineffective treatment and consider alternative treatment options. Two recent clinical studies, the CREATE-X (39) and KATHERINE (40) trials, have shown a survival benefit of additional chemotherapy for patients with invasive residual disease after NAC. The CREATE-X trial included patients with both luminal and TN subtypes, who were randomly selected to receive postsurgical treatment either with or without capecitabine (39). Among the patients with TNBC, capecitabine significantly improved 5-year disease-free survival, with survival of 69.8 vs. 56.1% in the control group (HR=0.58; 95% CI: 0.39–0.87). The KATHERINE trial (40) showed that adjuvant ado-trastuzumab-emtansine (T-DM1) significantly improved invasive disease-free survival compared with trastuzumab alone (HR=0.50; 95% CI: 0.39–0.64) in patients with residual disease after neoadjuvant HER2-targeted therapy. Taken together, these results suggest that additional adjuvant chemotherapy may be a treatment option for patients with residual disease after NAC.
As emphasized above, one of the most important issues for patients with an aggressive TN subtype is selection of the right treatment strategy after neoadjuvant therapy. In the present study, we examined the profiles of exosomal miRNAs for patients who did not achieve pCR and eventually experienced tumor recurrence. We found 43 differentially-expressed miRNAs in non-pCR patients with breast cancer recurrence vs. those without recurrence (Table SII). Furthermore, we employed pathway analysis and miRNA target gene prediction in order to identify which pathways might influence treatment efficacy and recurrence. Among the 43 miRNAs, miR-16-5p interfered with the most potential target genes, with miR-195-5p being the second most promiscuous (Table SIV). A recent study by Shen et al (41). revealed that docetaxel treatment caused elevation of miR-195-5p in circulating extracellular vesicles to stimulate cancer stem-like cells, rendering cancer cells resistant to therapy. These miRNAs regulate several target genes that are involved in cancer development, such as those controlling the cell cycle (CCND1, CCND3, CCNE1, CDC42, CDK4, CDK6, and WEE1), proto-oncogenes (KRAS, MYB, RAF1, and RET), and apoptosis (BCL2 and BIRC5). In line with this, several studies have suggested that TNBC is highly sensitive to levels of cell cycle regulators (42–44). A single-cell dynamic study by Asqhar et al demonstrated that the luminal androgen receptor (LAR) subtype of TNBC was more sensitive to CDK4/6 inhibitor compared to basal-like breast cancer both in vitro and in vivo (42). Also, Cretella et al explored the potential of combining the CDK4/6 inhibitor palbociclib with chemotherapy in Rb-positive TNBC cells, in which the sequential treatment inhibited cell proliferation and increased cell death more efficaciously than single treatments (44). Another study suggested that combined PI3Kα and CDK4/6 inhibition significantly increased apoptosis, cell cycle arrest, and tumor immunogenicity, and elicited immunogenic cell death in human TNBC cell lines (43). Use of these agents, which negatively regulate the cell cycle, may provide a novel therapeutic strategy for TNBC.
In the present study, we evaluated differences in the miRNA profiles between patients with pCR and non-pCR to NAC, and also those between non-pCR patients with recurrence and non-recurrence after treatment. We found completely different miRNA profiles between the two studies. This raises the possibility that NAC treatment selectively alters the expression patterns of miRNA, which may reflect the resistance to treatment and the subsequent prognosis. Consequently, we should evaluate changes in expression both before and after NAC for accurate prediction of prognosis. Besides this, our study has several limitations. With the exception of miR-4448 and miR-2392, we were unable to validate the predictive nature of most of the miRNAs identified by the microarray analysis when we moved to PCR-based analysis. This discrepancy may be attributable to the small sample size. Our cohort included only 24 patients, which is too small to establish whether particular associations are specific. Another possible explanation of the discordance may be due to a difference in the technologies used to measure gene expression. For example, the relatively low sensitivity of microarray platforms may have led to false negatives that would therefore not have been evaluated by the PCR-based method. In the present study, we identified several miRNAs such as miR-2467-3p, miR-4419a, and miR-6796-3p that have not previously been studied in the context of cancer; indeed, little is known about their function and mechanism of action. Our previous review of exosomal miRNAs highlighted that there is little overlap between exosomal miRNAs identified in different studies (45). Different methods used for the isolation of exosomes, and/or differences in patient characteristics may have contributed to these inconsistent results. Lastly, there is growing evidence of genetic heterogeneity in TNBC populations, which are divided into several subgroups with different treatment responses and prognoses (46,47). Since TNBC is considered as a single disease in the present study, the obtained results may differ when categorized into subgroups.
Liquid biopsy has emerged as an increasingly important tool since it is a relatively noninvasive method compared to tissue sampling. As we show here, it can be used to detect potential exosomal miRNA biomarkers in TNBC patients undergoing NAC. Indeed, we were able to detect clear differences in miRNA profiles between TNBC patients with pCR and non-pCR. Furthermore, we evaluated the expression of exosomal miRNAs in patients treated with standard chemotherapy and in those with residual cancers. Patients with specific miRNA profiles tended to experience recurrence, and our results pave the way for identifying these patients prior to treatment in order to provide alternative therapies when needed. Further clinical studies focusing on exosomal miRNAs and treatment efficacy are needed to validate our findings.
Supplementary Material
Acknowledgements
The authors would like to thank Mrs Yuki Azakami for their technical support, and Mrs Miki Kawakami for their help with clinical data management (Department of Breast and Endocrine Surgery, Kumamoto University Graduate School of Medical Sciences, Kumamoto, Japan).
Glossary
Abbreviations
- AUC
area under the curve
- CI
confidence interval
- ER
estrogen receptor
- HER2
human epidermal growth factor receptor 2
- HR
hazard ratio
- miRNA
microRNA
- mRNA
messenger RNA
- NAC
neoadjuvant chemotherapy
- OR
odds ratio
- pCR
pathological complete response
- PR
progesterone receptor
- ROC
receiver-operating characteristic
- TNBC
triple-negative breast cancer
Funding Statement
This work was supported by a grant-in-aid (grant no. 18K15324) for scientific research from the Ministry of Education, Science and Culture of Japan.
Funding
This work was supported by a grant-in-aid (grant no. 18K15324) for scientific research from the Ministry of Education, Science and Culture of Japan.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
AS contributed to the conception and design of the study, and performed the experiments and wrote the manuscript. AS and YY confirm the authenticity of all the raw data. YY and HI participated in its design and coordination, and helped to draft the manuscript. MYI contributed to the analysis and interpretation of data. LGY, MT and YF contributed to the acquisition of data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The entire study was approved by the ethics committee of Kumamoto University Graduate School of Medical Sciences (approval nos. 1662 and 1772; Kumamoto, Japan). Written informed consent was obtained from all subjects for the collection and use of serum samples for research. All relevant procedures were performed after obtaining the consent form.
Patient consent for publication
Not applicable.
Competing interests
Yutaka Yamamoto has relevant competing interests, including personal fees from Chugai Pharmaceutical Co., Ltd., AstraZeneca, Kyowa Hakko Kirin Co., Ltd., Pfizer, Inc., Novartis International AG, Essai, Inc., Takeda Pharmaceutical Company, Ltd., Taiho Pharmaceutical Co., Ltd., GE Healthcare Japan, Nippon Kayaku Co., Ltd., Daiichi Sankyo Co., Ltd., Sysmex Corporation and Eli Lilly Japan K.K. The other authors declare that they have no competing interests.
References
- 1.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: A population-based study from the California cancer Registry. Cancer. 2007;109:1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Yang J, Peng L, Sahin AA, Huo L, Ward KC, O'Regan R, Torres MA, Meisel JL. Triple-negative breast cancer has worse overall survival and cause-specific survival than non-triple-negative breast cancer. Breast Cancer Res Treat. 2017;161:279–287. doi: 10.1007/s10549-016-4059-6. [DOI] [PubMed] [Google Scholar]
- 3.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 4.Sharma P. Biology and management of patients with triple-negative breast cancer. Oncologist. 2016;21:1050–1062. doi: 10.1634/theoncologist.2016-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, Bonnefoi H, Cameron D, Gianni L, Valagussa P, et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet. 2014;384:164–172. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 6.Symmans WF, Wei C, Gould R, Yu X, Zhang Y, Liu M, Walls A, Bousamra A, Ramineni M, Sinn B, et al. Long-term prognostic risk after neoadjuvant chemotherapy associated with residual cancer burden and breast cancer subtype. J Clin Oncol. 2017;35:1049–1060. doi: 10.1200/JCO.2015.63.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loibl S, O'Shaughnessy J, Untch M, Sikov WM, Rugo HS, McKee MD, Huober J, Golshan M, von Minckwitz G, Maag D, et al. Addition of the PARP inhibitor veliparib plus carboplatin or carboplatin alone to standard neoadjuvant chemotherapy in triple-negative breast cancer (BrighTNess): A randomised, phase 3 trial. Lancet Oncol. 2018;19:497–509. doi: 10.1016/S1470-2045(18)30111-6. [DOI] [PubMed] [Google Scholar]
- 8.Sikov WM, Berry DA, Perou CM, Singh B, Cirrincione CT, Tolaney SM, Kuzma CS, Pluard TJ, Somlo G, Port ER, et al. Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once-per-week paclitaxel followed by dose-dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage II to III triple-negative breast cancer: CALGB 40603 (Alliance) J Clin Oncol. 2015;33:13–21. doi: 10.1200/JCO.2014.57.0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Untch M, Jackisch C, Schneeweiss A, Conrad B, Aktas B, Denkert C, Eidtmann H, Wiebringhaus H, Kümmel S, Hilfrich J, et al. Nab-paclitaxel versus solvent-based paclitaxel in neoadjuvant chemotherapy for early breast cancer (GeparSepto-GBG 69): A randomised, phase 3 trial. Lancet Oncol. 2016;17:345–356. doi: 10.1016/S1470-2045(15)00542-2. [DOI] [PubMed] [Google Scholar]
- 10.Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 12.Thery C, Zitvogel L, Amigorena S. Exosomes: Composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 13.Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. The biogenesis and functions of exosomes. Traffic. 2002;3:321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 16.Schwarzenbach H. Circulating nucleic acids as biomarkers in breast cancer. Breast Cancer Res. 2013;15:211. doi: 10.1186/bcr3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortez MA, Welsh JW, Calin GA. Circulating microRNAs as noninvasive biomarkers in breast cancer. Recent Results Cancer Res. 2012;195:151–161. doi: 10.1007/978-3-642-28160-0_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang HX, Gires O. Tumor-derived extracellular vesicles in breast cancer: From bench to bedside. Cancer Lett. 2019;460:54–64. doi: 10.1038/s41416-018-0342-0. [DOI] [PubMed] [Google Scholar]
- 20.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hessvik NP, Sandvig K, Llorente A. Exosomal miRNAs as biomarkers for prostate Cancer. Front Genet. 2013;4:36. doi: 10.3389/fgene.2013.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eichelser C, Stuckrath I, Müller V, Milde-Langosch K, Wikman H, Pantel K, Schwarzenbach H. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget. 2014;5:9650–9663. doi: 10.18632/oncotarget.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevic I, Muller V, Weber K, Fasching PA, Karn T, Marmé F, Schem C, Stickeler E, Denkert C, van Mackelenbergh M, et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med. 2018;16:179. doi: 10.1186/s12916-018-1163-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sueta A, Yamamoto Y, Tomiguchi M, Takeshita T, Yamamoto-Ibusuki M, Iwase H. Differential expression of exosomal miRNAs between breast cancer patients with and without recurrence. Oncotarget. 2017;8:69934–69944. doi: 10.18632/oncotarget.19482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Z, Dong J, Wang LE, Ma H, Liu J, Zhao Y, Tang J, Chen X, Dai J, Wei Q, et al. Serum microRNA profiling and breast cancer risk: The use of miR-484/191 as endogenous controls. Carcinogenesis. 2012;33:828–834. doi: 10.1093/carcin/bgs030. [DOI] [PubMed] [Google Scholar]
- 27.Hannafon BN, Ding WQ. Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci. 2013;14:14240–14269. doi: 10.3390/ijms140714240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bach DH, Hong JY, Park HJ, Lee SK. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int J Cancer. 2017;141:220–230. doi: 10.1002/ijc.30669. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi RU, Miyazaki H, Ochiya T. The roles of MicroRNAs in breast cancer. Cancers (Basel) 2015;7:598–616. doi: 10.3390/cancers7020598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bovy N, Blomme B, Freres P, Dederen S, Nivelles O, Lion M, Carnet O, Martial JA, Noël A, Thiry M, et al. Endothelial exosomes contribute to the antitumor response during breast cancer neoadjuvant chemotherapy via microRNA transfer. Oncotarget. 2015;6:10253–10266. doi: 10.18632/oncotarget.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodríguez-Martínez A, de Miguel-Pérez D, Ortega FG, García-Puche JL, Robles-Fernández I, Exposito J, Martorell-Marugan J, Carmona-Sáez P, Garrido-Nava MDC, Rolfo C, et al. Exosomal miRNA profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Res. 2019;21:21. doi: 10.1186/s13058-019-1109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Q, Zhao S, Shi Z, Cao L, Liu J, Pan T, Zhou D, Zhang J. Chemotherapy-elicited exosomal miR-378a-3p and miR-378d promote breast cancer stemness and chemoresistance via the activation of EZH2/STAT3 signaling. J Exp Clin Cancer Res. 2021;40:120. doi: 10.1186/s13046-021-01901-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boo L, Ho WY, Ali NM, Yeap SK, Ky H, Chan KG, Yin WF, Satharasinghe DA, Liew WC, Tan SW, et al. miRNA Transcriptome profiling of spheroid-enriched cells with cancer stem cell properties in human breast MCF-7 cell line. Int J Biol Sci. 2016;12:427–445. doi: 10.7150/ijbs.12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hibino S, Saito Y, Muramatsu T, Otani A, Kasai Y, Kimura M, Saito H. Inhibitors of enhancer of zeste homolog 2 (EZH2) activate tumor-suppressor microRNAs in human cancer cells. Oncogenesis. 2014;3:e104. doi: 10.1038/oncsis.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Li T, Lu Y, Shen G, Guo H, Wu J, Lei C, Du F, Zhou F, Zhao X, et al. miR-2392 suppresses metastasis and epithelial-mesenchymal transition by targeting MAML3 and WHSC1 in gastric cancer. FASEB J. 2017;31:3774–3786. doi: 10.1096/fj.201601140RR. [DOI] [PubMed] [Google Scholar]
- 36.Fan S, Tian T, Chen W, Lv X, Lei X, Zhang H, Sun S, Cai L, Pan G, He L, et al. Mitochondrial miRNA determines chemoresistance by reprogramming metabolism and regulating mitochondrial transcription. Cancer Res. 2019;79:1069–1084. doi: 10.1158/0008-5472.CAN-18-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bagegni NA, Tao Y, Ademuyiwa FO. Clinical outcomes with neoadjuvant versus adjuvant chemotherapy for triple negative breast cancer: A report from the National Cancer Database. PLoS One. 2019;14:e0222358. doi: 10.1371/journal.pone.0222358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Battisti NML, True V, Chaabouni N, Chopra N, Lee K, Shepherd S, Shapira-Rotenberg T, Joshi R, McGrath S, Okines A, et al. Pathological complete response to neoadjuvant systemic therapy in 789 early and locally advanced breast cancer patients: The Royal Marsden experience. Breast Cancer Res Treat. 2020;179:101–111. doi: 10.1007/s10549-019-05444-0. [DOI] [PubMed] [Google Scholar]
- 39.Masuda N, Lee SJ, Ohtani S, Im YH, Lee ES, Yokota I, Kuroi K, Im SA, Park BW, Kim SB, et al. Adjuvant capecitabine for breast cancer after preoperative chemotherapy. N Engl J Med. 2017;376:2147–2159. doi: 10.1056/NEJMoa1612645. [DOI] [PubMed] [Google Scholar]
- 40.von Minckwitz G, Huang CS, Mano MS, Loibl S, Mamounas EP, Untch M, Wolmark N, Rastogi P, Schneeweiss A, Redondo A, et al. Trastuzumab emtansine for residual invasive HER2-Positive breast cancer. N Engl J Med. 2019;380:617–628. doi: 10.1056/NEJMoa1814017. [DOI] [PubMed] [Google Scholar]
- 41.Shen M, Dong C, Ruan X, Yan W, Cao M, Pizzo D, Wu X, Yang L, Liu L, Ren X, Wang SE. Chemotherapy-induced extracellular vesicle miRNAs promote breast cancer stemness by targeting ONECUT2. Cancer Res. 2019;79:3608–3621. doi: 10.1158/0008-5472.CAN-18-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asghar US, Barr AR, Cutts R, Beaney M, Babina I, Sampath D, Giltnane J, Lacap JA, Crocker L, Young A, et al. Single-cell dynamics determines response to CDK4/6 inhibition in triple-negative breast cancer. Clin Cancer Res. 2017;23:5561–5572. doi: 10.1158/1078-0432.CCR-17-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teo ZL, Versaci S, Dushyanthen S, Caramia F, Savas P, Mintoff CP, Zethoven M, Virassamy B, Luen SJ, McArthur GA, et al. Combined CDK4/6 and PI3Kα inhibition is synergistic and immunogenic in triple-negative breast cancer. Cancer Res. 2017;77:6340–6352. doi: 10.1158/0008-5472.CAN-17-2210. [DOI] [PubMed] [Google Scholar]
- 44.Cretella D, Fumarola C, Bonelli M, Alfieri R, La Monica S, Digiacomo G, Cavazzoni A, Galetti M, Generali D, Petronini PG. Pre-treatment with the CDK4/6 inhibitor palbociclib improves the efficacy of paclitaxel in TNBC cells. Sci Rep. 2019;9:13014. doi: 10.1038/s41598-019-49484-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sueta A YY, Iwase H. The role of exosomal microRNAs; focus on clinical applications in breast cancer. Cancer Drug Resist. 2019:847–861. doi: 10.20517/cdr.2019.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, Savage MI, Osborne CK, Hilsenbeck SG, Chang JC, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.