Abstract
Aberrant connectivity of large-scale brain networks has been observed among individuals with alcohol use disorders (AUDs) as well as in those at risk, suggesting deficits in neural communication between brain regions in the liability to develop AUD. Electroencephalographical (EEG) coherence, which measures the degree of synchrony between brain regions, may be a useful measure of connectivity patterns in neural networks for studying the genetics of AUD. In 8,810 individuals (6,644 of European and 2,166 of African ancestry) from the Collaborative Study on the Genetics of Alcoholism (COGA), we performed a Multi-Trait Analyses of Genome-Wide Association Studies (MTAG) on parietal resting-state theta (3-7 Hz) EEG coherence, which previously have been associated with AUD. We also examined developmental effects of GWAS findings on trajectories of neural connectivity in a longitudinal subsample of 2,316 adolescent/young adult offspring from COGA families (ages 12-30) and examined the functional and clinical significance of GWAS variants. Six correlated single nucleotide polymorphisms located in a brain-expressed lincRNA (ENSG00000266213) on chromosome 18q23 were associated with posterior interhemispheric low theta EEG coherence (3-5 Hz). These same variants were also associated with alcohol use behavior and posterior corpus callosum volume, both in a subset of COGA and in the UK Biobank. Analyses in the subsample of COGA offspring indicated that the association of rs12954372 with low theta EEG coherence occurred only in females, most prominently between ages 25-30 (p<2x10−9). Converging data provide support for the role of genetic variants on chromosome 18q23 in regulating neural connectivity and alcohol use behavior, potentially via dysregulated myelination. While findings were less robust, genome-wide associations were also observed with rs151174000 and parieto-frontal low theta coherence, rs14429078 and parieto-occipital interhemispheric high theta coherence, and rs116445911 with centro-parietal low theta coherence. These novel genetic findings highlight the utility of the endophenotype approach in enhancing our understanding of mechanisms underlying addiction susceptibility.
Keywords: African-ancestry, European-ancestry, EEG, Connectivity, Alcohol Dependence, Alcohol Consumption
Introduction
Differences in the connectivity of large scale brain networks (“the connectome”) have been observed among individuals with neuropsychiatric disorders, including alcohol use disorders1 (AUD). While the development of MRI-based resting-state functional connectivity has profoundly improved our understanding of the spatial organization of human brain connectivity, it does not provide the real-time temporal resolution required to observe neural activity on the order of milliseconds at which neural communication for most relevant sensory, motor and cognitive phenomena in the brain occur that is provided by EEG functional connectivity2. One measure of EEG functional connectivity is EEG coherence, which measures the degree of synchrony in oscillatory activity between two brain regions, where increased coherence indicates functional integration between these brain regions and decreased coherence reflects unrelated neural activity3,4. EEG coherence provides detailed frequency specific measures of neural connectivity, enabled by the millisecond sampling rate of EEG recording. An advantage of EEG connectivity is the ability to assess local and distal connectivity patterns as a function of frequency band, as aspects of neural function and connectivity patterns between brain regions are specific to EEG frequency2 . EEG coherence is a notable measure of functional connectivity that has been studied extensively3,4. Further, EEG coherence phenotypes are highly heritable5 and have demonstrated important utility in genetic studies of neuropsychiatric disorders, including AUD6,7. For example, highly significant linkage and association for theta coherence at parietal-occipital leads were found8,9 in the same GABRA2 variants associated with AD10. Given its exquisite temporal resolution, EEG coherence is a promising neural endophenotype, which alongside clinical and fMRI data, with its powerful spatial resolution, can aid in the discovery of novel genetic variants for neuropsychiatric disorders and further our understanding of neural and molecular mechanisms of AUD. Despite the promise of EEG coherence as an endophenotype for AUD or other neuropsychiatric disorders, we know of no prior GWAS (Genome-Wide Association Study) of resting state EEG coherence, and only a single previous GWAS of theta inter-trial phase coherence during the P300 window conducted by Malone and colleagues, which identified a region on chromosome 2 (~TEKT4)11. Previous GWAS have examined other EEG measures, including EEG power and P300 amplitude12,13. Our group also conducted an earlier GWAS of event-related theta oscillations during the P300 response, which indicated important variants in KCNJ614; however this result was not confirmed in Malone et al11 with a closely related phenotype. In 2018, Smit et al.15 performed the largest GWAS to date of resting-state EEG oscillatory power across a range of frequencies and identified novel variants in chromosome 3p21.1 for alpha EEG power. In addition, GABRA2 significantly affected beta power, consistent with the known relation between GABAA interneuron activity and beta oscillations16. More recently, a GWAS of resting-state beta EEG power conducted in individuals of African ancestry implicated variants on chromosome 3q267.
EEG coherence is a function of thalamocortical and cortico-cortical synchronization, involving both short axonal connections of neighboring pyramidal cells, and long axonal propagation via intracortical association pathways (i.e., inter- and intra-cortical pathways4). EEG coherence is known to change dynamically across the lifespan, with some studies showing normative increases in childhood17, adolescence18 and early adulthood19 and decreases in later adulthood20. Since these measures are well suited for detecting relatively subtle structural brain abnormalities, coherence has been particularly useful in the study of normal brain development, learning disorders, and neuropsychiatric disorders such as AUD21. Differences in resting state interhemispheric coherence have been observed in individuals with AUD when compared to unaffected individuals across most frequency bands, with findings most prominent in parietally coupled theta (3-7 Hz), alpha (8-12 Hz), and beta (13-28 Hz) bands5. In a subsample of the Collaborative Study on the Genetics of Alcoholism (COGA), individuals with DSM-IV alcohol dependence (AD) manifested increased resting EEG interhemispheric theta coherence8,9, indicating altered cortico-cortical functional connectivity. Recent work22 identified resting state theta EEG coherence networks that were correlated with resting state executive control networks detected with functional magnetic resonance imaging (fMRI) in prior AD studies; this demonstrates that there is a correspondence between resting state fMRI connectivity and resting-state EEG activity. When compared to light drinkers, individuals with AUD have reduced cortical grey matter23,24 and white matter volumes, and reduced myelination25,26. In addition, individuals with AUD have a smaller corpus callosum25,27, and show disruptions of the integrity of white matter tracts in the corpus callosum on the micro-structural level28, possibly via disruptions in myelination29. Taken together, these findings suggest corpus callosal interhemispheric connections may be disturbed in AUD.
To determine if genetic variants associated with theta EEG coherence would point to genetic factors important for AUD, we conducted a multi-trait GWAS of eight measures of parietal theta EEG coherence in 8,810 individuals from the Collaborative Study on the Genetics of Alcoholism (COGA). We aimed to characterize these genetic findings with respect to alcohol use behavior (maximum drinks in a 24-hour period and DSM-5 symptom count) and related aspects of brain structure and function shown to be aberrant in AUD (using MRI and EEG data) both in a subset of COGA subjects as well as in replication samples (UK Biobank MRI subsample30 and Psychiatric Genomics Consortium-Substance Use Disorder31). In addition, we also examine differential expression of implicated genetic variants in various brain regions in individuals in the general population. Because of prior evidence indicating that the effects of genetic variants on neural oscillations may differ throughout development32, we examined developmental effects of the most significant theta EEG coherence GWAS findings on EEG coherence throughout adolescence and young adulthood in a subset of individuals followed longitudinally, including the assessment of gender differences in these associations.
SAMPLE & METHODS
The sample included alcohol dependent (AD) probands and their family members recruited and assessed through seven participating COGA sites, as well as a community sample assessed without regards to their AD status, as described previously30. Experimental protocols were approved by each site's institutional review board, and informed consent was obtained from all participants. Participants were administered the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA), a poly-diagnostic interview33 that assesses DSM-IV psychiatric disorders, including AD. Individuals below age 18 were administered an adolescent SSAGA. The analytic sample consisted of all participants assessed by the SSAGA with EEG coherence and GWAS data available: 8,810 individuals (6,644 of European ancestry and 2,166 of African ancestry) residing in the United States. Demographic characteristics of the full sample (all individuals with genotypic data) and the analytic sample (all individuals with genotypic data and EEG data) are comparable (Supplemental Table 1). 26.3% of the analytic sample met criteria for DSM-IV AD.
EEG Recording & Processing has been detailed previously19. Briefly, resting (eyes-closed) EEG was recorded for 4.25 min (256 seconds) at a rate of 256 samples/second; the entire time-series was analyzed by Fourier transform methods. EEG procedures were identical at all collection sites. This study examined eight EEG inter- and intra- hemispheric parietally coupled coherence phenotypes (depicted in Supplemental Figure 5; namely, theta (low theta: 3-5 Hz and high theta: 5-7 Hz) between interhemispheric parietal-occipital bipolar pairs (P3-O1—P4-O2), neighboring interhemispheric centro-parietal (C4-P3—C3-P3) bipolar pairs, and right and left intrahemispheric fronto-parietal (P8-P4—F8-F4, P7-P3—F7-F3) bipolar pairs. Because of differences in low (3-5 Hz) and high (5-7 Hz) theta bands suggested in this study (correlations ranging from 0.60-0.80), and by previous work in COGA34 and other studies 35-37, we examined low and high theta bands separately to reduce phenotypic heterogeneity. Data on the full or combined theta band (3-7 Hz) is presented in Supplemental Materials. Use of bipolar electrode pairs effectively results in a low resolution spatial filter that reduces volume conduction effects38. Further details regarding the EEG coherence data processing has been detailed previously5 and is provided in the Supplemental Material (EEG Coherence Recording & Data Reduction).
Genotyping, Imputation and Quality Control.
Genotyping of 3,654 individuals of AA and 7,382 individuals of EA was performed using the Illumina 2.5M array (Illumina, San Diego, CA, USA), the Illumina OmniExpress39, the Illumina 1M array, or the Affymetrix Smokescreen array40. SNPs with a genotyping rate <98%, or that violated Hardy-Weinberg equilibrium (p<10−6), or with minor allele frequency (MAF) less than 3% were excluded from analyses. Mendelian inconsistencies were removed38, after which data were imputed to 1000 genomes (EUR and AFR, Phase 3, b37, October 2014; build hg19) using SHAPEIT41 and IMPUTE242. Following imputation, genotype probabilities ≥ 0.90 were changed to genotypes. Mendelian errors in the imputed SNPs were reviewed and resolved as described previously7,43. SNPs with an imputation information score < 0.30 or MAF < 0.03 were excluded from subsequent analysis. Further details are in Supplemental Information.
Statistical analysis.
Genetic analysis was conducted in GWAF (Genome-Wide Association analyses with Family)41 on 6,832,792 SNPs using a linear mixed model (LMM) incorporating a genetic relationship matrix to control for the relatedness in the family sample41-42. Principal components (PCs) derived from GWAS data were used to assign ancestry in the genotyped sample, and families were classified as European ancestry (EA) or African ancestry (AA) according to the ancestry of the greatest proportion of family members. Analyses were conducted separately in the families of AA and EA, using identical phenotypic definitions, covariates, SNP QC standards, MAF thresholds and imputation protocols. Subsequently, MTAG44 (multi- trait analyses of GWAS) was run to include summary statistics from all eight theta EEG coherences phenotypes, both low and high theta for the four pairs described in the EEG Recording & Processing section. MTAG is a multi-trait approach that leverages the common genetic information across multiple correlated traits to boost the power of each individual trait44. MTAG is also powerful at disentangling spurious sources of genetic correlation, such as due to overlapping samples, using Linkage Disequilibrium Score Regression45. Meta-analysis across the EA and AA samples was performed using inverse-variance weighting and genomic control in METAL46. Sex and log-transformed age (at the time of EEG recording) were included as covariates in the model, as each of these variables were associated with theta EEG (p<0.001).
The first three PCs (PC1-PC3) computed from SNPRelate42 and genotype array, birth cohort (prior to 1930, 1930-1949, 1950-1969, and 1970 and after) were also included as covariates to reduce the risk of false-positives owing to population stratification. Established thresholds for genomewide significance (p< 5 x 10−8) were used. Genome-wide Complex Trait Analysis (GCTA)47 was used to determine SNP heritability of parietal coupled theta EEG coherences in the analytic sample. We performed MAGMA48 competitive gene-set analyses using the summary statistics from the GWAS of theta EEG coherence using FUMA v1.2.8 (Functional Mapping and Annotation of Genome-Wide Association Studies). The Genotype-Tissue Expression (GTEx v649) database was used to obtain gene expression levels in 10 brain tissues. In addition, we examined the most significantly associated variant for low theta EEG using HUGIn (Hi-C Unifying Genomic Interrogator50), assessing DNA-DNA interactions which can localize a GWAS finding to a specific gene that may be nearby or hundreds of kb away using data generated by Schmitt et al.51 Search parameters included the two available brain tissues (dorsolateral prefrontal cortex and hippocampus) in neural progenitor and embryonic stem cell lines.
Alcohol Consumption and Problems.
We asked whether variants that met genome-wide significance with theta EEG coherences were also associated with alcohol use behavior in the larger COGA sample in which GWAS AUD data were available (even if EEG was not; 8,038 EA individuals and 3,654 AA individuals). Measures included maximum number of drinks consumed in a 24-hour period (maxdrinks) and DSM-5 AUD criterion count (0-11) over the lifetime. See Supplemental Table 1 for analytic sample details.
Next, we examined if genome-wide significant variants were associated with alcohol drinker status (never, previous, current) and alcohol intake frequency (average units of wine, beer, and spirits per week) in the UK Biobank, the largest GWAS of alcohol consumption published to date30 (N: 337,000 population-based individuals of European ancestry from a volunteer cohort in the UK aged 40-69 years old30). We also examined if GWAS variants were associated with AD in the Psychiatric Genomics Consortium’s meta-analysis of DSM-IV AD31, the largest GWAS of alcohol use disorder conducted to date (N: 14,904 AD cases and 37,944 controls from 28 studies; European, N: 46,568; African; N: 6,280).
We examined association of genome-wide significant SNPs with longitudinal trajectories of theta EEG coherence in adolescent and young adult offspring ages 12-31 from COGA families (N: 2,316, EA: 1,265, AA: 1,051). Association trajectory modeling was carried out as described previously32. Briefly, a local linear regression model was fitted to the data, with additive effects of the genome-wide significant SNP as the predictor and low theta EEG coherence measure as the outcome. Additional covariates included sex, family history of AUD, 3 ancestral PCs, and sex × genotype interaction. Additionally, a sensitivity model was examined, in which we controlled for the participant’s maximum number of drinks consumed in a typical week over the last 12 months.
Post-Hoc Analyses
In both a subset of the UK Biobank (N: 3,722), and in 60 representative participants from COGA’s adolescent and young adult offspring on whom MRI data were available52 (Supplemental Table 1), we examined if genome-wide significant regions were also associated with posterior corpus callosum volume. Genetic association with posterior corpus callosum volume was tested using linear regression, adjusted for gender, age, total brain volume, PCs 1-3, and genotype array.
P-values were adjusted using the Pnorm procedure53, which accounts for both the LD structure of the SNPs and the sampling of relatives. Pnorm uses the multivariate normal distribution approximation to evaluate the significance of each test adjusting for simultaneous testing. Given differences in patterns of LD, different thresholds for EA and AA populations were established. In all subsequent analyses, a Bonferroni test-correction was utilized.
RESULTS
GWAS of Theta EEG Coherence
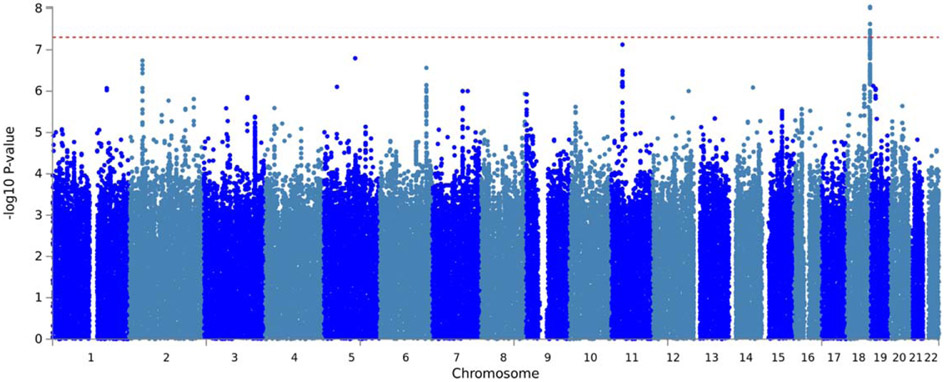
A meta-analysis of the COGA EA and AA revealed six SNPs in high LD (pair-wise r2>0.9 and 0.7, D’>0.9 and 0.8 in EAs and AAs respectively) were associated with low theta EEG coherence at p<5x10−8 (Table 1, Figure 1, Supplemental Figure 1, q-q plot in Supplemental Figure 6). They are located in AC107892 (ENSG00000266213) a long intergenic non-coding RNA (lincRNA) on 18q23 (Chr 18, 75894847-75900263, hg38). The most significant SNP was rs12954372 (Meta-analyzed p = 9.15E-09; EA p: 1.51x10−5 and AA p: 1.04x10−5; Supplemental Figure 1). The minor allele (A) was associated with increased posterior interhemispheric low theta EEG coherence (Meta-analyzed Z-score: 5.75). rs12954372 was also associated with other highly correlated (r2>0.70) posterior-related coherence measures (Supplemental Table 2) in COGA families, including high theta (5-7 Hz) and the full theta band (3-7 Hz), but these findings did not reach genome-wide significance.
Table 1.
Chromosome 18 variants demonstrating association with interhemispheric posterior low theta EEG coherence at P3-O1—P4-O2.
| SNP | Base-pair Position |
Effect Allele |
Minor Allele Frequency |
Effect Size (Z-score) |
Meta p-value |
|---|---|---|---|---|---|
| rs12954372 | 75900263 | A | 0.212 | 5.746 | 9.15E-09 |
| rs12965943 | 75900225 | G | 0.202 | 5.735 | 9.75E-09 |
| rs12957925 | 75900192 | A | 0.203 | 5.582 | 2.38E-08 |
| rs12455089 | 75899708 | G | 0.209 | 5.521 | 3.38E-08 |
| rs12456531 | 75899668 | C | 0.209 | 5.502 | 3.76E-08 |
| rs8088204 | 75901055 | T | 0.211 | 5.485 | 4.14E-08 |
| rs12457297 | 75893680 | A | 0.210 | 5.451 | 5.01E-08 |
| rs8085845 | 75896982 | T | 0.209 | 5.448 | 5.10E-08 |
| rs12959612 | 75894165 | T | 0.208 | 5.438 | 5.38E-08 |
| rs12455126 | 75893273 | G | 0.210 | 5.434 | 5.50E-08 |
| rs12966809 | 75894211 | C | 0.210 | 5.415 | 6.13E-08 |
| rs924888 | 75900840 | A | 0.208 | 5.412 | 6.25E-08 |
| rs34622862 | 75896444 | T | 0.209 | 5.401 | 6.61E-08 |
| rs8098981 | 75896607 | T | 0.209 | 5.401 | 6.61E-08 |
| rs12966267 | 75900507 | C | 0.209 | 5.400 | 6.66E-08 |
| rs924887 | 75900912 | C | 0.208 | 5.397 | 6.77E-08 |
| rs4799267 | 75896178 | G | 0.209 | 5.384 | 7.29E-08 |
| rs4799268 | 75896184 | A | 0.209 | 5.384 | 7.29E-08 |
| rs4799266 | 75895923 | A | 0.209 | 5.370 | 7.88E-08 |
| rs12965699 | 75895741 | A | 0.209 | 5.363 | 8.20E-08 |
| rs924890 | 75900657 | T | 0.208 | 5.362 | 8.23E-08 |
| rs67507908 | 75891447 | G | 0.212 | 5.339 | 9.34E-08 |
| rs111340774 | 75894847 | T | 0.208 | 5.332 | 9.71E-08 |
Figure 1. Manhattan plot displaying genome-wide meta-analysis association results for interhemispheric low theta coherence (P3-O1—P4-O2).
Genome-wide association results for low theta EEG coherence in the genome-wide meta-analysis of individuals of European and African ancestry. Y axis denotes the −log10 (P-value) for association. X axis is the physical position of the SNPs across the genome. Note: Red line indicates the threshold of genome-wide significance (P<5 × 10–8).
Genome-wide SNPs included five non-coding RNA exonic variants, one variant downstream of lincRNA AC107892 (ENSG00000266213) and one intergenic variant. In the GTEX data, AC107892 is expressed at low levels across brain tissue, particularly in the substantia nigra (Supplemental Figure 3). In the HUGIn database, interaction of rs12954372 was observed with LINC01029 in dorsolateral prefrontal cortex tissue (p<0.01) and SALL3 (p<0.001) in hippocampus tissue (Supplemental Figure 4), SALL3 in neural progenitor cells (p<0.00001), and ATP9B and MBP in embryonic stem cells (p<0.00001). HaploReg V4.1 indicated that rs12954372 alters regulatory motifs in some cell types, including H1 and H9 derived neuronal progenitor cells, in the ROADMAP Epigenomics data.
Genome-wide associations were also observed with rs151174000 and parieto-frontal (P8-P4--F8-F4) low theta coherence (meta-analyzed p = 3.26E-08), rs144290781 with parieto-occipital (P3-O1--P4-O2) interhemispheric high theta coherence (meta-analyzed p = 3.15E-08), and rs116445911 with centro-parietal (C4-P4--C3-P3) low theta coherence (meta-analyzed p = 4.82E-08). While several additional SNPs in high linkage disequilibrium with rs116445911 were also implicated at subthreshold levels in centro-parietal low theta coherence GWAS (Supplemental Table 3) for both rs144290781 and rs151174000, only a single variant was implicated, suggesting that these associations may be spurious.
MAGMA identified no individual gene that was significantly associated with low theta coherences, although subthreshold (<5x10−4) associations were observed with: EPS8L3, HAAO, BCHE, ASF1A, MCM9, and CDH.
Alcohol Use Behavior
In the analytic sample, individuals with increased parietal-occipital theta EEG coherences had an increased number of maxdrinks (B: 2.03, p=0.03) and individuals with increased parietal-frontal low theta EEG coherence had a decreased number of maxdrinks (Beta= 2.11, p=0.03) and were less likely to meet criteria for DSM-5 AUD (B: 3.52, p<0.001). All six variants associated with parieto-occipital low theta (3-5 Hz) EEG coherence showed nominal positive associations with maxdrinks and DSM-5 AUD symptom count in both EA and AA COGA samples (Table 2) and with alcohol drinker status and alcohol intake frequency in the UK Biobank (Table 3). [Note, this excludes rs8088204 which was not assessed in the UK Biobank.] Given the high LD observed among these six SNPs, p-values were adjusted for the number of tests (1.2 for EAs and 2.1 for AAs) as estimated using the pnorm procedure53 and four alcohol phenotypes examined. Associations of rs12965943 and rs8088204 survived multiple test-correction (0.05/# of independent tests x four phenotypes= 0.021 for EAs and 0.012 for AAs) for both maxdrinks and DSM-5 AUD symptom count, and only among AA participants. Associations of rs12957925 and rs12457297 with maxdrinks among AA participants also survived a multiple test correction. All six SNPs were significantly (i.e., p<0.0001) associated with alcohol drinker status and alcohol intake frequency in the UK Biobank (Table 3), and withstood a multiple test correction; the minor allele was associated with increased AUD risk. None of the SNPs were significantly associated with DSM-IV AD in the PGC (Table 3).
Table 2.
Associations of Chromosome 18 SNPs with maximum drinks consumed in a 24-hour period (maxdrinks) and DSM-5 AUD symptom count in COGA families
| Maxdrinks (EA, N: 7,289) |
Maxdrinks (AA, N: 3,076) |
AUD Severity (EA, N: 7,169) |
AUD Severity (AA, N: 3,026) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | A1 | beta | p-value | beta | p-value | beta | p-value | beta | p-value |
| rs12954372 | A | 0.03 | 0.047 | 0.08 | 0.006 | 0.03 | 0.036 | 0.07 | 0.013 |
| rs12965943 | G | 0.03 | 0.039 | 0.10 | 0.002* | 0.03 | 0.037 | 0.08 | 0.003* |
| rs12957925 | A | 0.03 | 0.038 | 0.10 | 0.002* | 0.03 | 0.036 | 0.08 | 0.005 |
| rs12455089 | G | 0.03 | 0.034 | 0.09 | 0.005 | 0.03 | 0.031 | 0.08 | 0.005 |
| rs12456531 | C | 0.03 | 0.034 | 0.09 | 0.005 | 0.03 | 0.031 | 0.07 | 0.006 |
| rs8088204 | T | 0.03 | 0.023 | 0.10 | 0.001* | 0.03 | 0.029 | 0.09 | 0.001* |
Withstands multiple test correction: 0.05/# of independent tests x four phenotypes= 0.01 for EAs and 0.005 for AAs. Note: Given the high LD observed among these six SNPs, p-values were adjusted for the number of tests (1.2 for EAs and 2.1 for AAs) as estimated using the pnorm procedure53.
Table 3.
Associations of chromosome 18 SNPs with alcohol consumption measures in the UK 796 Biobank and alcohol dependence in the Psychiatric Genomics Consortium
| UK Biobank Alcohol Drinker Status: Never (EA, N: 337,000) |
UK Biobank Alcohol Use Frequency (EA, N: 337,000) |
PGC DSM-IV Alcohol Dependence (EA, N: 46,568) |
PGC DSM-IV Alcohol Dependence (AA, N: 6,280) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | A1 | beta | p-value | beta | p-value | OR | p-value | OR | p-value |
| rs12954372 | A | 0.0017 | 3.7e-4* | 0.015 | 2.7e-4* | 0.960 | 1.1E-01 | 0.937 | 4.3E-01 |
| rs12965943 | G | 0.0017 | 2.9e-4* | 0.015 | 2.1e-4* | 0.960 | 1.1E-01 | 0.890 | 6.3E-02 |
| rs12957925 | A | 0.0017 | 4.8e-4* | 0.015 | 2.4e-4* | 0.960 | 1.1E-01 | 0.908 | 1.2E-01 |
| rs12455089 | G | 0.0017 | 5.0e-4* | 0.015 | 1.9e-4* | 0.959 | 1.0E-01 | 0.942 | 3.2E-01 |
| rs12456531 | C | 0.0017 | 3.7e-4* | 0.015 | 1.4e-4* | 0.960 | 1.1E-01 | 0.940 | 3.1E-01 |
| rs8088204 | T | NA | NA | NA | NA | 0.961 | 1.2E-01 | 0.920 | 1.6E-01 |
Given the high LD observed among these six SNPs, p-values were adjusted for the number of tests (1.2 for EAs and 2.1 for AAs) as estimated using the pnorm procedure53. Associations of rs12965943 and rs8088204 survived multiple test-correction (0.05/# of independent tests x four phenotypes= 0.021 for EAs and 0.012 for AAs) for both phenotypes, and only among AA participants. Association of rs12957925 with maxdrinks among AA participants also survived a multiple test correction. All six SNPs were significantly (i.e., p<0.0001) associated with alcohol drinker status and alcohol intake frequency in the UK Biobank (Table 3). All associations withstood a multiple test correction; the minor allele was associated with increased AUD risk. None of the SNPs were significantly associated with DSM-IV AD in the PGC (Table 3).
Post-Hoc Analyses
We examined association of rs12954372 with developmental trajectories of low theta EEG coherence in a longitudinal subsample of the discovery sample, including 2,316 adolescent/young adult offspring from COGA families. Significant effects of rs12954372 were driven by associations among females after age 18 (Supplemental Figure 2); the most significant p-value was observed for the association of rs12954372 and low theta P3-O1—P4-O2 coherence among females between ages 25-30 (2x10−9). Results from the sensitivity analysis, in which we controlled for the participant’s maximum number of drinks consumed in a typical week over the last 12 months, remained largely unchanged.
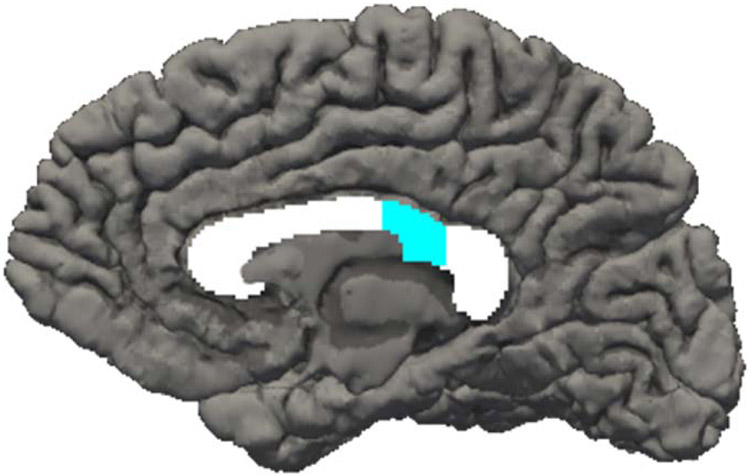
An association of rs12954372 with posterior corpus callosum volume measured via MRI was found in the UK Biobank sample (N = 3,722, B: 0.03; p: 0.042); the same association was found in a subset of young adult offspring from COGA families in COGA’s prospective study on whom MRI data was available (N = 60, B: 0.34; p: 0.038, Figure 2). In the MRI subsample of COGA’s prospective study, posterior interhemispheric low theta coherence was associated with increased maxdrinks (N:50, B:0.51, p<0.0001), but not DSM-IV AD (N= 49, B: 0.12, p: 0.415) or posterior corpus callosum volume (N: 49, B: 0.18, p: 0.230).
Figure 2.
Increased posterior corpus callosum volume was observed among rs12954372 A-allele carriers (B: 0.34, p: 0.038, adjusted for gender, age, total brain volume, PC1-PC3, and genotype array) in COGA participants (Total MRI N=60, A-allele carriers N=7)
DISCUSSION
Although previous studies have reported differences in EEG coherence among individuals diagnosed with AD and related neuropsychiatric conditions, no comprehensive genomic studies have been published prior to this first GWAS of EEG connectivity. We report a genome-wide significant signal in a 5.4 kb intergenic region on chromosome 18q23. The most significant SNP, rs12954372, located in a brain-expressed lincRNA AC107892 was positively associated with not only low theta (3-5 Hz) EEG coherence, but also with alcohol use behaviors (e.g., maxdrinks), and corpus callosum volume in the COGA sample, and with alcohol intake frequency and corpus callosum volume in an independent sample.
18q23, Neural Connectivity, and Neurocognitive Dysfunction
While little is known about the function of AC107892, it was previously associated with schizophrenia54 and educational attainment55, and is expressed throughout the substantia nigra (Supplemental Figure 3). There are also several syndromes and disorders associated with structural variation in this region of potentially relevance to neural connectivity; e.g., 18q23 deletion syndrome is characterized by diverse neurological and psychiatric features, including developmental delays, epilepsy, and autism spectrum disorders (ASD). Relatedly, there several structural variations (e.g., deletions) observed in this region among individuals with ASD56 and distal 18q deletion syndrome which is commonly characterized by impaired myelin production, neurological problems, delayed development, learning disabilities, seizures, hyperactivity, mood disorders and features of ASD that impair communication and social interaction. LincRNAs are emerging as key transcriptional and post-translational regulators that have been shown to be important for normal development and in disease pathophysiology57, including in psychiatric58 and substance-abuse disorders59. Recent studies show their involvement in cis-regulation of neighboring genes58 and regulation of gene expression60, with highest expression in brain.
Supplemental figure 4 graphically illustrates the known genes located further upstream and downstream of the 18q23 GWAS signal found to interact (via HUGIn50) with rs12954372; including LINC01029 and SALL3 throughout different brain tissues, and ATP9B and MBP in embryonic stem cells. LINC01029 was recently associated with schizophrenia and nicotine dependence61, SALL3 with digit length ratio62 and modestly with AUD in the Genetic Analysis Workshop 14 dataset63 (includes a subset of COGA), ATP9B with prostate cancer64 and frontotemporal lobar degeneration65, and NFATC1 with Crohn’s and Inflammatory Bowel disease66, and creatinine levels67. While little is known about the function of most of these genes, much is known regarding MBP (Myelin Basic Protein) and its important role in neural connectivity. Myelin increases efficient neural signal transmission throughout the brain, and it is thought to contribute to the enhanced brain-regional connectivity, processing speed, and related cognitive function throughout the lifespan68. Subtle abnormalities of myelination can lead to persistent cortical network dysfunction68, which may provide one potential link to dysfunction in neurocognitive and psychiatric phenotypes.
As noted above, GABRA2 has been linked to resting-state beta oscillatory power in both an earlier iteration of the COGA study16, as well as in independent samples15. We note that GABRA2 SNPs were nominally associated with low theta coherence (p<0.05) in the current study, however this p-value did not withstand multiple test correction.
While findings were less robust, genome-wide associations were also observed among intergenic chromosome 18 variant rs151174000 with parieto-frontal low theta coherence, chromosome 1 variant rs14429078 with parieto-occipital interhemispheric high theta coherence, and chromosome 4 variant rs116445911 with centro-parietal low theta coherence. While not much is known about these variants, rs116445911 is associated with the expression of SLC9B1 in brain69, and SLC9B1 has been associated with schizophrenia70, another neuropsychiatric disorder characterized by dysregulated neural connectivity71. As noted above, only a single variant was implicated for both rs144290781 and rs151174000, suggesting that these associations may be spurious.
18q23, Neural Connectivity and Alcohol Use/Disorders
In COGA, individuals with increased parietal-occipital theta EEG coherence had an increased number of maxdrinks. There was evidence of association between several variants meeting genome-wide criteria for low theta (3-5 Hz) parietal-occipital theta EEG coherence with maxdrinks and AUD severity (p<0.05 in EAs and p<0.01 in AAs), suggesting a potential role for low theta EEG coherence variants in alcohol use behavior in the COGA sample, particularly among participants of African ancestry. Further, all six SNPs meeting genome-wide criteria for low parietal-occipital theta EEG coherence in COGA were also associated with alcohol use and intake frequency in the UK Biobank, a population of European ancestry. Taken together, these data suggest a role of 18q23 in alcohol use behavior, however whether this also extends to alcohol use disorder (DSM-IV or DSM-5) is less clear. We also note that previous work from our group implicated high theta coherence as most significant for alcohol dependence in a sample of non-COGA participants as well as for significant genetic association in a COGA sample of linkage families, densely affected with alcohol dependence9, whereas the current study’s findings focus on genetic associations with low theta (3-5 Hz) coherence with alcohol use behavior. As noted above, while low and high theta are highly correlated, there is evidence of distinctions between the two sub-frequency bands. While both the prior and current genetic samples were derived from COGA, the current analytic sample differs from the previous analytic sample in several important ways that may impact the association of low and high theta with alcohol use behavior. For example, the previous genetic study was limited to participants of European ancestry, and had larger proportions of males, older individuals, and subjects more severely affected with AUD. The current sample also includes participants of African ancestry, more females and younger individuals, and those less severely affected with AUD. Given research indicating that ancestry72, sex73, age74,75 and disorder severity76 each introduce important heterogeneity in genetic association findings of complex traits, as well as in the EEG parameters themselves77-80, it is highly plausible that findings based on these three samples will differ.
18q23 and Corpus Callosum Volume
Interestingly, variants in 18q23 were also associated with posterior corpus callosum volume in the UK Biobank and among subjects from COGA who were also assessed with MRI. This suggests that 18q23 may contribute to variability in interhemispheric connectivity between the posterior right and left hemispheres of the brain, although the current study does not provide evidence of the direct relation of EEG coherence and corpus callosum measures (possibly due to the small MRI sample size). Chronic alcohol use is toxic to specific areas in the brain, including the corpus callosum, and impacts myelination throughout the brain81. More specifically, reductions in corpus callosum volume and microstructural degradations29 have been observed among individuals with AUD. Further, a significant loss of axonal and myelin proteins, and lower white matter integrity following chronic alcohol exposure has also been observed in the corpus callosum82,83. Diffusion tensor imaging studies of white-matter abnormalities in alcoholics highlight a disruption of myelin microstructure in prefrontal areas and the corpus callosum52 that appears distinct from callosal atrophy seen with Alzheimer’s disease29. In its most severe form, chronic alcohol abuse can lead to Marchiafava-Bignami disease, a neurologic disorder with corpus callosum demyelination84. The commissural fibers of the corpus callosum play a dominant role in the maintenance of interhemispheric connectivity, which was assessed in our study by EEG coherence. Although further evidence is needed to support the link between corpus callosum measures and EEG coherence, these findings may suggest that the variability in interhemispheric connectivity in AUD results from a specific loss of interhemispheric commissural fibers that enable communication between the cerebral hemispheres projecting through the corpus callosum. Given the intriguing associations among variants in 18q23, EEG coherences and AUD, large, comprehensive in-depth studies, are needed to evaluate the associations of EEG coherences, structural brain imaging phenotypes, and neuropsychiatric disorders.
18q23, Neural Connectivity and Development
We also investigated the association of rs12954372 with developmental trajectories of parietal-occipital EEG theta coherence in the COGA adolescent/young adult subsample to further our understanding of the developmental influences of salient genetic risk factors emerging from the coherence GWASs and their connection to alcohol use behavior. Interestingly, significant effects were only observed in females and were driven by associations observed after age 18 (Supplemental Figure 2).
The brain undergoes its greatest growth and development in the first years of life, with a second phase beginning in adolescence characterized by increased sexual differentiation as well as growth of white matter tracts and synaptic pruning in gray matter, leading to anatomical and functional maturation85. This second phase of development is most profound in regions of the brain involved in higher order cognitive functions (e.g., top-down control functions, such as inhibition and other aspects of executive function), as well as affective functions (e.g., emotional and reward processing; 86,87).
Although both clinical and experimental studies have demonstrated the greater vulnerability of female adolescents to the neurotoxic/neuroinflammatory effects of alcohol88; neuroimaging studies have shown that female adolescent binge drinkers and those with adolescent onset AD exhibit greater reduction than males in the prefrontal cortex and larger white fiber disruptions, particularly in corpus callosum89. Results from this study show that coherence differences associated with rs12954372 predate significant alcohol use, and sensitivity analysis was unable to find a significant effect of early drinking on coherence measures. This suggests that the differences in parietal-occipital theta EEG coherence observed in this study are more likely markers for risk, rather than a consequence of alcohol use.
Limitations and Future Directions
The most notable limitation is the relatively modest sample size in the primary GWAS. Nevertheless, this is the only GWAS of EEG coherence conducted to date. In addition, several highly correlated SNPs with genome-wide significance were associated with low theta EEG coherence and more modestly with alcohol related behaviors and posterior corpus callosum volume in our primary sample. Importantly, they were also associated with alcohol consumption and posterior corpus callosum volume in an independent sample. This region of 18q23 has been previously associated with other neuropsychiatric traits of potential relevance to neural connectivity and alcohol use behavior in both human and animal data. Given the modest sample size for GWAS and lack of direct replication sample available for theta EEG coherences, and nominal associations observed in eQTL analyses, all findings must be replicated in larger samples. Recent GWAS of alcohol use behaviors31,90 have not, however, found variants in this region of chromosome 18 to be genome-wide significant. In addition, it will be important for future work to combine the advantages of the superior spatial resolution of MRI and the superior temporal resolution of EEG to better understand functional connectivity. Finally, future studies should examine how findings from this work could potentially translate into useful targets for neurofeedback among individuals with AUD, and those at risk, given the success of neurofeedback as a therapeutic tool, alongside other behavioral and pharmacological therapies91.
Conclusions
This study found a novel association between SNPs in a brain-expressed lincRNA (AC107892) on 18q23 and theta EEG coherence in a sample of related individuals of EA and AA. Further, theta EEG coherence genome-wide associated variants were associated with alcohol use behavior both in the discovery sample and an independent replication sample. Converging data provide support for the role of genetic variants within 18q23 in regulating neural connectivity and alcohol use behavior, potentially via dysregulation and/or demyelination. This study also demonstrates the utility of the endophenotype approach; genetic findings of EEG coherence have provided an underlying biological hypothesis (i.e., neural connectivity) that can enhance our understanding of functional cerebral circuits and one potential mechanism underlying a predisposition to AUD and related behaviors. Our findings also indicate important sex and developmental differences, underscoring the need for us to examine these effects throughout the lifespan.
Supplementary Material
Acknowledgements
This research is supported by K01DA037914 (JLM), K02DA32573 (AA), K01AA024152 (JES), K02AA018755 (DMD), and U10AA008401 (BP). COGA: The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes eleven different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, J. Rice, K. Bucholz, A. Agrawal); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield, A. Brooks); Department of Biomedical and Health Informatics, The Children’s Hospital of Philadelphia; Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia PA (L. Almasy), Virginia Commonwealth University (D. Dick), Icahn School of Medicine at Mount Sinai (A. Goate), and Howard University (R. Taylor). Other COGA collaborators include: L. Bauer (University of Connecticut); J. McClintick, L. Wetherill, X. Xuei, Y. Liu, D. Lai, S. O’Connor, M. Plawecki, S. Lourens (Indiana University); G. Chan (University of Iowa; University of Connecticut); J. Meyers, D. Chorlian, C. Kamarajan, A. Pandey, J. Zhang (SUNY Downstate); J.-C. Wang, M. Kapoor, S. Bertelsen (Icahn School of Medicine at Mount Sinai); A. Anokhin, V. McCutcheon, S. Saccone (Washington University); J. Salvatore, F. Aliev, B. Cho (Virginia Commonwealth University); and Mark Kos (University of Texas Rio Grande Valley). A. Parsian and H. Chen are the NIAAA Staff Collaborators. We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. We also thank T.B.B. for his assistance with this manuscript. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
Footnotes
Disclosures: This material is original research, has not been previously published and has not been submitted for publication elsewhere while under consideration. Unrelated to this work, coauthor Arpana Agrawal received peer-reviewed funding, travel and an honorarium from ABMRF (end 12/2012) which receives support from the brewing industry. We have no other conflicts of interest to disclose.
References
- 1.Filbey FM The American Journal of Drug and Alcohol Abuse An introduction to & quot;The addiction connectome: brain connectivity in drug and alcohol addiction" Am J Drug Alcohol Abus. 39, 341–342 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Uhlhaas PJ & Singer W Neural Synchrony in Brain Disorders: Relevance for Cognitive Dysfunctions and Pathophysiology. Neuron 52, 155–168 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Nunez PL et al. EEG coherency: I: statistics, reference electrode, volume conduction, Laplacians, cortical imaging, and interpretation at multiple scales. Electroencephalogr. Clin. Neurophysiol 103, 499–515 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Srinivasan R, Winter WR, Ding J & Nunez PL EEG and MEG coherence: Measures of functional connectivity at distinct spatial scales of neocortical dynamics. J. Neurosci. Methods 166, 41–52 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chorlian DB et al. Heritability of EEG coherence in a large sib-pair population. Biol. Psychol 75, 260–6 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porjesz B et al. The utility of neurophysiological markers in the study of alcoholism. Clin. Neurophysiol 116, 993–1018 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Meyers JL et al. An endophenotype approach to the genetics of alcohol dependence: a genome wide association study of fast beta EEG in families of African ancestry. Mol. Psychiatry (2017). doi: 10.1038/mp.2016.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porjesz B & Rangaswamy M Neurophysiological Endophenotypes, CNS Disinhibition, and Risk for Alcohol Dependence and Related Disorders. Sci. World J 7, 131–141 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rangaswamy M & Porjesz B Uncovering genes for cognitive (dys)function and predisposition for alcoholism spectrum disorders: a review of human brain oscillations as effective endophenotypes. Brain Res. 1235, 153–71 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edenberg HJ et al. Variations in GABRA2, Encoding the α2 Subunit of the GABAA Receptor, Are Associated with Alcohol Dependence and with Brain Oscillations. Am. J. Hum. Genet 74, 705–714 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malone SM, McGue M & Iacono WG What can time-frequency and phase coherence measures tell us about the genetic basis of P3 amplitude? Int. J. Psychophysiol 115, 40–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iacono WG, Malone SM, Vaidyanathan U & Vrieze SI Genome-wide scans of genetic variants for psychophysiological endophenotypes: A methodological overview. Psychophysiology 51, 1207–1224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vrieze SI et al. Genetic associations of nonsynonymous exonic variants with psychophysiological endophenotypes. Psychophysiology 51, 1300–1308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang SJ et al. Family-based genome-wide association study of frontal θ oscillations identifies potassium channel gene KCNJ6. Genes. Brain. Behav 11, 712–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smit DJA et al. Genome-wide association analysis links multiple psychiatric liability genes to oscillatory brain activity. Hum. Brain Mapp 39, 4183–4195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porjesz B et al. Linkage disequilibrium between the beta frequency of the human EEG and a GABAA receptor gene locus. Proc. Natl. Acad. Sci. U. S. A 99, 3729–33 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whedon M, Perry NB, Calkins SD & Bell MA Changes in frontal EEG coherence across infancy predict cognitive abilities at age 3: The mediating role of attentional control. Dev. Psychol 52, 1341–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segalowitz SJ, Santesso DL & Jetha MK Electrophysiological changes during adolescence: A review. Brain Cogn. 72, 86–100 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Chorlian DB, Rangaswamy M & Porjesz B EEG coherence: topography and frequency structure. Exp. brain Res 198, 59–83 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Duffy FH, Mcanulty GB & Albert MS Effects of age upon interhemispheric EEG coherence in normal adults. Neurobiol. Aging 17, 587–599 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Rangaswamy M & Porjesz B Understanding alcohol use disorders with neuroelectrophysiology. Handb. Clin. Neurol 125, 383–414 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardenas VA, Price M & Fein G EEG coherence related to fMRI resting state synchrony in long-term abstinent alcoholics. Neuroimage. Clin 17, 481–490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fein G et al. Cortical gray matter loss in treatment-naïve alcohol dependent individuals. Alcohol. Clin. Exp. Res 26, 558–64 (2002). [PMC free article] [PubMed] [Google Scholar]
- 24.Jernigan TL et al. Reduced cerebral grey matter observed in alcoholics using magnetic resonance imaging. Alcohol. Clin. Exp. Res 15, 418–27 (1991). [DOI] [PubMed] [Google Scholar]
- 25.Hommer DW, Momenan R, Kaiser E & Rawlings RR Evidence for a Gender-Related Effect of Alcoholism on Brain Volumes. Am. J. Psychiatry 158, 198–204 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Pfefferbaum A et al. Brain gray and white matter volume loss accelerates with aging in chronic alcoholics: a quantitative MRI study. Alcohol. Clin. Exp. Res 16, 1078–89 (1992). [DOI] [PubMed] [Google Scholar]
- 27.Pfefferbaum A, Lim KO, Desmond JE & Sullivan EV Thinning of the corpus callosum in older alcoholic men: a magnetic resonance imaging study. Alcohol. Clin. Exp. Res 20, 752–7 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Pfefferbaum A & Sullivan EV Microstructural but Not Macrostructural Disruption of White Matter in Women with Chronic Alcoholism. Neuroimage 15, 708–718 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Pitel A-L, Chanraud S, Sullivan EV & Pfefferbaum A Callosal microstructural abnormalities in Alzheimer’s disease and alcoholism: same phenotype, different mechanisms. Psychiatry Res. Neuroimaging 184, 49–56 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clarke T-K et al. Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117). Mol. Psychiatry 22, 1376–1384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walters RK et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat. Neurosci 21, 1656–1669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chorlian DB et al. Genetic correlates of the development of theta event related oscillations in adolescents and young adults. Int. J. Psychophysiol 115, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bucholz KK et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J. Stud. Alcohol 55, 149–58 (1994). [DOI] [PubMed] [Google Scholar]
- 34.Chorlian DB, Rangaswamy M & Porjesz B EEG coherence: topography and frequency structure. Exp. Brain Res 198, 59–83 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Travis F Temporal and spatial characteristics of meditation EEG. Psychol. Trauma Theory, Res. Pract. Policy (2019). doi: 10.1037/tra0000488 [DOI] [PubMed] [Google Scholar]
- 36.Ahnaou A, Huysmans H, Jacobs T & Drinkenburg WHIM Cortical EEG oscillations and network connectivity as efficacy indices for assessing drugs with cognition enhancing potential. Neuropharmacology 86, 362–377 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Passynkova N, Neubauer H & Scheich H Spatial organization of EEG coherence during listening to consonant and dissonant chords. Neurosci. Lett 412, 6–11 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Cook IA, O’Hara R, Uijtdehaage SH, Mandelkern M & Leuchter AF Assessing the accuracy of topographic EEG mapping for determining local brain function. Electroencephalogr. Clin. Neurophysiol 107, 408–14 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Wang J-C et al. A genome-wide association study of alcohol-dependence symptom counts in extended pedigrees identifies C15orf53. Mol. Psychiatry 18, 1218–1224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baurley JW, Edlund CK, Pardamean CI, Conti DV & Bergen AW Smokescreen: a targeted genotyping array for addiction research. BMC Genomics 17, 145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delaneau O, Howie B, Cox AJ, Zagury J-F & Marchini J Haplotype estimation using sequencing reads. Am. J. Hum. Genet 93, 687–96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das S et al. Next-generation genotype imputation service and methods. Nat. Genet 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wetherill L et al. Association of substance dependence phenotypes in the COGA sample. Addict. Biol 20, 617–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turley P et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet 50, 229–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willer CJ, Li Y & Abecasis GR METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet 42, 565–569 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Leeuw CA, Mooij JM, Heskes T & Posthuma D MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carithers LJ & Moore HM The Genotype-Tissue Expression (GTEx) Project. Biopreserv. Biobank (2015). doi: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin JS et al. HUGIn: Hi-C Unifying Genomic Interrogator. Bioinformatics 33, 3793–3795 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmitt AD et al. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Rep. 17, 2042–2059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pandey AK et al. Lower Prefrontal and Hippocampal Volume and Diffusion Tensor Imaging Differences Reflect Structural and Functional Abnormalities in Abstinent Individuals with Alcohol Use Disorder. Alcohol. Clin. Exp. Res (2018). doi: 10.1111/acer.13854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z Direct assessment of multiple testing correction in case-control association studies with related individuals. Genet. Epidemiol 35, 70–9 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Fanous AH et al. Genome-Wide Association Study of Clinical Dimensions of Schizophrenia: Polygenic Effect on Disorganized Symptoms. Am. J. Psychiatry 169, 1309–1317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okbay A et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vorstman JAS et al. Autism genetics: opportunities and challenges for clinical translation. Nat. Rev. Genet 18, 362–376 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Farris SP, Arasappan D, Hunicke-Smith S, Harris RA & Mayfield RD Transcriptome organization for chronic alcohol abuse in human brain. Mol. Psychiatry 20, 1438–1447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zuo L et al. Long noncoding RNAs in psychiatric disorders. Psychiatr. Genet 26, 109–116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sartor GC, St. Laurent G & Wahlestedt C The Emerging Role of Non-Coding RNAs in Drug Addiction. Front. Genet 3, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spadaro PA et al. Long Noncoding RNA-Directed Epigenetic Regulation of Gene Expression Is Associated With Anxiety-like Behavior in Mice. Biol. Psychiatry 78, 848–859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen J et al. Genetic Relationship between Schizophrenia and Nicotine Dependence. Sci. Rep 6, 25671 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Warrington NM et al. Genome-wide association study identifies nine novel loci for 2D:4D finger ratio, a putative retrospective biomarker of testosterone exposure in utero. Hum. Mol. Genet 27, 2025–2038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen H et al. Inferring Alcoholism SNPs and Regulatory Chemical Compounds Based on Ensemble Bayesian Network. Comb. Chem. High Throughput Screen 20, 107–115 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Eeles R et al. The genetic epidemiology of prostate cancer and its clinical implications. Nat. Rev. Urol 11, 18–31 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Pottier C et al. Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal lobar degeneration and GRN mutations: a genome-wide association study. Lancet Neurol. 17, 548–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu JZ et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet 47, 979–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanai M et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet 50, 390–400 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Poggi G et al. Cortical network dysfunction caused by a subtle defect of myelination. Glia 64, 2025–40 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin JS et al. HUGIn: Hi-C Unifying Genomic Interrogator. Bioinformatics 33, 3793–3795 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Z et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat. Genet 49, 1576–1583 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Kam JWY, Bolbecker AR, O’Donnell BF, Hetrick WP & Brenner CA Resting state EEG power and coherence abnormalities in bipolar disorder and schizophrenia. J. Psychiatr. Res 47, 1893–901 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peterson RE et al. Genome-wide Association Studies in Ancestrally Diverse Populations: Opportunities, Methods, Pitfalls, and Recommendations. Cell 179, 589–603 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khramtsova EA, Davis LK & Stranger BE The role of sex in the genomics of human complex traits. Nat. Rev. Genet 20, 173–190 (2019). [DOI] [PubMed] [Google Scholar]
- 74.Meyers JL et al. Association of Polygenic Liability for Alcohol Dependence and EEG Connectivity in Adolescence and Young Adulthood. Brain Sci. 9, 280 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cousminer DL et al. Genome-wide association study of sexual maturation in males and females highlights a role for body mass and menarche loci in male puberty. Hum. Mol. Genet 23, 4452–4464 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.CONVERGE consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523, 588–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barry RJ et al. Age and gender effects in EEG coherence: II. Boys with attention deficit/hyperactivity disorder. Clin. Neurophysiol 116, 977–84 (2005). [DOI] [PubMed] [Google Scholar]
- 78.Barry RJ, Clarke AR, McCarthy R & Selikowitz M Age and gender effects in EEG coherence: III. Girls with attention-deficit/hyperactivity disorder. Clin. Neurophysiol 117, 243–251 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Cardenas VA, Price M & Fein G EEG coherence related to fMRI resting state synchrony in long-term abstinent alcoholics. NeuroImage Clin. 17, 481–490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marosi E et al. Sex Differences in Eeg Coherence in Normal Children. Int. J. Neurosci 72, 115–121 (1993). [DOI] [PubMed] [Google Scholar]
- 81.Costin BN & Miles MF Molecular and neurologic responses to chronic alcohol use. Handb. Clin. Neurol 125, 157–71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pfefferbaum A, Adalsteinsson E & Sullivan EV Supratentorial Profile of White Matter Microstructural Integrity in Recovering Alcoholic Men and Women. Biol. Psychiatry 59, 364–372 (2006). [DOI] [PubMed] [Google Scholar]
- 83.McQueeny T et al. Altered White Matter Integrity in Adolescent Binge Drinkers. Alcohol. Clin. Exp. Res 33, 1278–1285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tozakidou M, Stippich C & Fischmann A Teaching NeuroImages: Radiologic findings in Marchiafava-Bignami disease. Neurology 77, e67–e67 (2011). [DOI] [PubMed] [Google Scholar]
- 85.Sowell ER et al. Mapping cortical change across the human life span. Nat. Neurosci 6, 309–315 (2003). [DOI] [PubMed] [Google Scholar]
- 86.Sowell ER, Delis D, Stiles J & Jernigan TL Improved memory functioning and frontal lobe maturation between childhood and adolescence: a structural MRI study. J. Int. Neuropsychol. Soc 7, 312–22 (2001). [DOI] [PubMed] [Google Scholar]
- 87.Sowell ER, Thompson PM & Toga AW Mapping Changes in the Human Cortex throughout the Span of Life. Neurosci. 10, 372–392 (2004). [DOI] [PubMed] [Google Scholar]
- 88.Alfonso-Loeches S, Pascual M & Guerri C Gender differences in alcohol-induced neurotoxicity and brain damage. Toxicology 311, 27–34 (2013). [DOI] [PubMed] [Google Scholar]
- 89.Welch KA, Carson A & Lawrie SM Brain Structure in Adolescents and Young Adults with Alcohol Problems: Systematic Review of Imaging Studies. Alcohol Alcohol. 48, 433–444 (2013). [DOI] [PubMed] [Google Scholar]
- 90.Liu M et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet 51, 237–244 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sokhadze TM, Cannon RL & Trudeau DL EEG biofeedback as a treatment for substance use disorders: review, rating of efficacy, and recommendations for further research. Appl. Psychophysiol. Biofeedback 33, 1–28 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.