The worldwide pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in more than 4.5 million deaths. Although coronaviruses have a proofreading mechanism to maintain the stability of their long genomic RNAs, mutations emerge continuously, and new variants conferring advantages rapidly become the dominant lineages [1]. Strategies to fight the COVID-19 pandemic using either vaccines or nonpharmaceutical interventions have specifically been threatened by the emergence of SARS-CoV-2 variants of concern (VOCs) [2]. The N501Y mutation of the viral spike (S) protein, which is found in lineages such as B.1.1.7 (Alpha strain), B.1.351 (Beta strain), and P.1 (Gamma strain), increase viral transmission due to the enhanced affinity binding to ACE2 [3]. The E484K/Q mutation in the spike contributes to evasion of antibody neutralization in VOCs, such as B.1.351 and P.1 [4]. The neutralizing titers of the sera from convalescent and vaccinated individuals were reduced against the Beta strain, as reported by us and others [4]. Notably, a novel variant, B.1.617.2 (Delta strain), quickly spread to 135 countries and became a dominant strain in many countries as of 8 Sep 2021. The Delta strain harbors eight mutations in the S protein, including L452R and T478K in the receptor binding domain (RBD). This variant is highly transmissible and causes breakthrough infection in vaccinated people as the protection of vaccines wanes [5, 6]. However, the humoral immune responses of the Delta strain are largely unknown in naive and vaccinated individuals as well as convalescent patients.
In this study, we collected serum samples from 26 convalescent patients who were infected with the SARS-CoV-2 Delta strain and exhibited mild or moderate symptoms during May–June 2021 in Guangzhou, Guangdong, China and hospitalized in the Guangzhou 8th People’s Hospital. Some samples were sequenced to confirm that the COVID-19 pathogen was the Delta strain. Because locally infected people were quite limited in Guangzhou, Guangdong, during May–June 2021, the transmission route study clearly indicated that the rest of them were infected by the SARS-CoV-2 Delta strain. The median age of the Delta strain-infected patients was 45 years (range: 22–83 years), and 42.3% of them were women. According to the Chinese national treatment guidelines, the symptoms were classified as mild (4 patients) or moderate (22 patients), and the median disease duration was 22 days (range: 15–28 days). Nine of the 26 recovered patients were fully vaccinated with the Sinovac-CoronaVac vaccine (Sinovac Life Sciences Co., Ltd), prior to breakthrough infection by the Delta strain. As controls, convalescent serum samples from 22 patients with mild or moderate symptoms who were infected with the early strains (D614 or G614) from March 1 to April 15, 2020, in Guangzhou, Guangdong, were collected. The median age of the early strain-infected patients was 45 years (range: 32–77 years), and 77.3% of them were women. According to the Chinese national treatment guidelines, the symptoms were classified as mild (2 patients) or moderate (20 patients), and the median disease duration was 26 days (range: 17–39 days). None of these patients had been vaccinated. The information regarding all the patients is listed in Table S1.
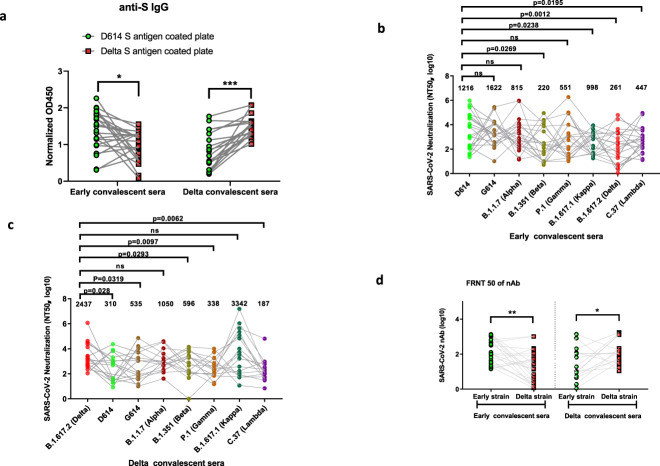
To further evaluate the intensity of the overall humoral immune response in convalescent patients, we first measured the titers of anti-S antibodies in convalescent sera. Recombinant S proteins of both the D614 and Delta strains were coated to capture the antibody in the convalescent sera of samples and thus to compare the total IgG titer and cross-reactivity. After normalization with the baseline from the healthy donor sera, spike-specific IgG was detected in convalescent sera from patients infected with both the early dominant strains (n = 22) and the Delta strain (n = 26) (Fig. S1). Cross-reactivity was detected in anti-spike total IgG when the early convalescent sera were probed with the Delta strain spike protein and vice versa. The results showed that the IgG titers in early convalescent sera to the Delta strain S protein decreased significantly (n = 22, 31.1%, p = 0.0101) (Fig. 1a). Moreover, the titers of IgG specific to the Delta strain S protein in the convalescent sera from the patients infected with the Delta strain (nonvaccinated group) increased significantly (n = 17, 1.79-fold, p = 0.0037) (Fig. 1a).
Fig. 1. The “seesaw effect” between the early strain and Delta strain in the neutralizing titer of respective convalescent sera.
a Normalized OD450 nm values of the anti-IgG to the different S proteins were compared between early convalescent serum (n = 22) and Delta convalescent serum (n = 17) samples. b The NT50 of each early convalescent serum against each pseudotyped virus (n = 22). c NT50 of each Delta convalescent serum against each pseudotyped virus (n = 17). d Each convalescent serum sample was serially diluted tenfold and incubated with 400 FFU of authentic SARS-CoV-2 (D614/B.1.617.2), followed by incubation with Vero E6 cells. The FRNT spots of each well were counted. The FRNT50 of nAbs of each convalescent serum was determined by FRNT and is represented as the IC50. The p value was calculated using one-way ANOVA with Tukey’s multiple comparisons test *p < 0.05, ***p < 0.001. Data are represented as the mean ± SD (n = 17 or 22).
To further evaluate whether the major SARS-CoV-2 variants could escape the cross-neutralization of convalescent sera from the patients infected with the early strains or the Delta strain, we first constructed various S protein-expressing plasmids derived from the D614 virus (Wuhan-Hu-1), G614 (SYSU-HIV), B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.1 (Kappa), B.1.617.2 (Delta), and C.37 (Lambda) strains for packaging in the pseudotyped SARS-CoV-2 S/HIV-1 viruses. The luciferase gene was incorporated into the HIV-1 vector and can be expressed after infection with the pseudotyped virus. We incubated various pseudotyped SARS-CoV-2 S/HIV-1 viruses with convalescent sera from 22 early strain-infected patients, 26 nonvaccinated or vaccinated patients infected with the Delta strain, and 10 healthy donors, followed by the detection of infectivity. In the pseudotyped virus neutralization assay, the average neutralization titer of early-strain convalescent sera dropped significantly for the Beta strain (B.1.351), Kappa strain (B.1.617.1), Delta strain (B.1.617.2), and Lambda strain (C.37) compared to that of the early D614 strain. However, the convalescent sera potently neutralized the Alpha (B.1.1.7) and Gamma (P.1) strains (Fig. 1b). Conversely, we titrated the nonvaccinated Delta convalescent sera against the pseudotyped viruses of SARS-CoV-2 variants. According to our assay, 7.86-fold, 4.56-fold, 4.09-fold and 7.21-fold decreases in the neutralizing antibody (nAb) titer were observed against the D614, G614, Beta (B.1.351), and Gamma (P.1) strains, respectively. However, the neutralizing titer only decreased by 2.32-fold against the Alpha strain (B.1.1.7), with a similar titer against the Kappa strain (B.1.617.1) (Fig. 1c). Interestingly, the convalescent sera from the patients infected with neither the early strains nor the Delta strain could effectively neutralize the Lambda pseudotyped virus, and the neuralization titers were significantly reduced by 2.72-fold and 13.03-fold, respectively (Fig. 1b, c).
To further examine the functional consequences of reduced antibody recognition, we measured the half-maximal inhibitory concentration (IC50) of convalescent sera from the early- or Delta strain-infected patients using an in vitro neutralization assay with authentic D614 or Delta viral isolates. As expected, the titers of nAb correlated most closely with the levels of the pseudotyped virus NT50 for the infecting strain. The neutralization of authentic Delta viruses by D614 sera decreased significantly compared with the neutralization of the authentic D614 strain (n = 22, fold change 7.28; p = 0.0053), while the neutralization of authentic D614 viruses by sera from the Delta strain-infected patients (nonvaccinated group) also largely decreased significantly compared with the neutralization of the authentic D614 strain (n = 17, fold change 6.04; p = 0.0108) (Fig. 1d).
Considering that some Delta strain-infected patients have been fully vaccinated with the Sinovac-CoronaVac vaccine [7], we analyzed the impact of vaccination on the production of nAbs in convalescent sera. Interestingly, we found that complete vaccination not only led to an increase in the production of nAbs against the Delta strain but also increased the titers of nAbs for other variant viruses, such as G614 (SYSU-HIV), B.1.1.7 (Alpha), B.1.351 (Beta), B.1.617.2 (Delta), and C.37 (Lambda) (Fig. S2).
In this report, with studies of nAbs whose production was induced by D614 and B.1.617.2 infection, we demonstrated that natural infection with each SARS-CoV-2 strain induces the generation of antibodies that recognize the infecting strain most strongly. Consistent with recent reports, we observed a significant reduction in the neutralization of Delta viruses by the D614-infected convalescent sera [8–10]. Conversely, the convalescent sera from the Delta strain-infected patients showed higher antibody escape to early dominant strains. Our data illustrate a “seesaw effect” between the early dominant strains and the Delta strain in terms of the neutralizing titer of the respective convalescent sera. Importantly, antibodies whose production was induced by D614 infection were less cross-reactive with other VOCs than those induced by Delta. Our data imply that the mutations in the RBD of the Delta variant likely cause fundamental steric and epitope changes. As most nAbs are highly sensitive to RBD-specific epitopes and have poor cross-reactivity with other Beta coronavirus strains, these mutations lead to the poor cross-reactivity of nAbs from the convalescent sera obtained from patients infected with the early dominant strains and Delta strain. To achieve effective neutralization, researchers could use different BCR germlines during infection with these two virus strains. Unfortunately, as we did not have the opportunity to collect peripheral blood mononuclear cells from recovered patients for B-cell receptor (BCR) sequencing, this hypothesis remains to be verified.
The significant reduction in nAb titers against the Delta strain in convalescent sera of the patients infected with the early strains indicates that these recovered patients are at risk of reinfection with the Delta strain, although they are not as vulnerable as the naive population. In fact, breakthrough infection occurs as the nAb titer wanes following vaccination. In this report, 9 of 26 patients in our study were fully vaccinated with the D614 Sinovac-CoronaVac vaccine. However, although the vaccination did not confer effective protection against the Delta strain in these patients, significantly higher nAb titers against the Alpha, Beta, Delta and Lambda strains were observed in these convalescent patients infected with the Delta strain. This result suggests that breakthrough infection possibly mobilized a memory immune response established by vaccination, resulting in stronger broad-spectrum humoral immunity. The waning of nAb titers after immunization is an inevitable natural process, and thus, the protective ability of the vaccine decreases. If so, our data strongly advocate widespread vaccination. Memory broad-spectrum humoral immunity established by vaccination or breakthrough infection could eventually shield us during our long-term global combat with SARS-CoV-2 variants.
As SARS-CoV-2 viruses are constantly evolving, the danger posed by the variants could still be underestimated. The degree of heterotypic immunity should be an important consideration in the choice of vaccine candidates. In addition to the emergence and global spread of the Delta variant, several other variants have recently emerged. A new variant of interest, the Lambda strain (C.37), is currently spreading in South American countries such as Peru, Chile, Argentina, and Ecuador (https://cov-lineages.org/lineage.html?lineage=C.37). Our results have shown that the Lambda strain possesses mutations that confer immune evasion from Delta strain-specific convalescent sera, with a 13.03-fold reduction in nAb titers. As different BCR germlines could be used during the infection of SARS-CoV-2 variants, a COVID-19 vaccine specifically designed for Delta and Lambda strains is highly warranted, as the current vaccines are all designed based on the S protein of the early dominant strains. Conversely, as the Delta strain-specific vaccine may fail to provide sufficient protection against the early dominant strains, the Alpha strain (B.1.1.7), and the newly emerged Lambda (C.37) strain, multivalent vaccines that mobilize more BCR germlines and therefore induce the production of more broad-spectrum nAbs should also be considered.
Supplementary information
Acknowledgements
This work was supported by the National Special Research Program of China for Important Infectious Diseases (2017ZX10202102 and 2018ZX10302103), the Special 2019-nCoV Project of the National Key Research and Development Program of China (2020YFC0841400), the Emergency Key Program of Guangzhou Laboratory (EKPG21-24), the Special 2019-nCoV Program of the Natural Science Foundation of China (NSFC) (82041002), the Special Research and Development Program of Guangzhou (202008070010), and the Important Key Program of NSFC (81730060) to HZ. This work was also supported by the Zhongnanshan Medical Foundation of Guangdong Province (ZNSA-2021004) and the Emergency Key Program of Guangzhou Laboratory (EKPG21-29) to XT. This work was supported by the National Natural Science Foundation of China (32100743, 82171825) to XH. This work was also supported by the National Natural Science Foundation of China (81971918), the Shenzhen Science and Technology Program (Grant Nos. JSGG20200225150431472 and JCYJ20200109142601702), the Pearl River S&T Nova Program of Guangzhou (201806010118) and the Fundamental Research Funds for the Central Universities, Sun Yat-sen University (2021qntd43) to TP. This work was supported by National Natural Science Foundation of China (32100743, 82171825) to X.H.
Author contributions
Conceptualization, TP, FH, XH, YZ, XT and HZ; methodology, TP, YZ, BL, XH, XT and HZ; validation, XT and HZ; formal analysis, TP, XH, YZ, BL, QS, ZH and HZ; investigation, TP, YZ, XH, XT, FH, ZH and HZ; resources, TP, BL, XH, YZ, CK, XT and HZ; data curation, XH and HZ; writing-original draft, TP, YZ and HZ; writing-reviewing and editing, XH and HZ; supervision, XH, XT and HZ; project administration, TP, XH, XT, and HZ and funding acquisition, TP, XT and HZ.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Ting Pan, Zhongwei Hu, Fengyu Hu, Yiwen Zhang.
Contributor Information
Xin He, Email: hexin59@mail.sysu.edu.cn.
Xiaoping Tang, Email: tangxp@gzhmu.edu.cn.
Hui Zhang, Email: zhangh92@mail.sysu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00776-8.
References
- 1.Robson F, Khan KS, Le TK, Paris C, Demirbag S, Barfuss P, et al. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol Cell. 2020;80:1136–8. doi: 10.1016/j.molcel.2020.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021;27:622–5. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 3.Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford KHD, Dingens AS, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–61. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chia PY, Onget SWX, Chiew CJ, Ang LW, Chavatte J-M, Mak T-M, et al. Virological and serological kinetics of SARS-CoV-2 Delta variant vaccine-breakthrough infections: a multi-center cohort study. 2021. 10.1101/2021.07.28.21261295. [DOI] [PMC free article] [PubMed]
- 6.Mlcochova P, Kemp S, Dhar MS, Papa G, Meng B, Mishra S, et al. SARS-CoV-2 B. 1.617. 2 Delta variant emergence and vaccine breakthrough. 2021. 10.1038/s41586-021-03944-y. Online ahead of print.
- 7.Jara, A, Undurraga EA, González C, Paredes F, Fontecilla T, Jara G, et al. Effectiveness of an Inactivated SARS-CoV-2 Vaccine in Chile. N Engl J Med. 2021. 10.1056/NEJMoa2107715. [DOI] [PMC free article] [PubMed]
- 8.Liu C, Ginn HM, Dejnirattisai W, Supasa P, Wang B, Tuekprakhon A, et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–36. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M, Hofmann-Winkler H, Krüger N, Kempf A, Nehlmeier I, Graichen L, et al. SARS-CoV-2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. 2021;36:109415. doi: 10.1016/j.celrep.2021.109415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–80. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.