Abstract
Background
Data visualization, especially the genome track plots, is crucial for genomics researchers to discover patterns in large-scale sequencing dataset. Although existing tools works well for producing a normal view of the input data, they are not convenient when users want to create customized data representations. Such gap between the visualization and data processing, prevents the users to uncover more hidden structure of the dataset.
Results
We developed CoolBox—an open-source toolkit for visual analysis of genomics data. This user-friendly toolkit is highly compatible with the Python ecosystem and customizable with a well-designed user interface. It can be used in various visualization situations like a Swiss army knife. For example, to produce high-quality genome track plots or fetch commonly used genomic data files with a Python script or command line, to explore genomic data interactively within Jupyter environment or web browser. Moreover, owing to the highly extensible Application Programming Interface design, users can customize their own tracks without difficulty, which greatly facilitate analytical, comparative genomic data visualization tasks.
Conclusions
CoolBox allows users to produce high-quality visualization plots and explore their data in a flexible, programmable and user-friendly way.
Keywords: Genomics, Visualization, Genome browser
Background
With the rapid development of Next-Generation Sequencing (NGS) technologies, more and more genomic assays have been developed to profile the genome from various aspects, such as RNA expression [1], protein-DNA binding [2], chromatin accessibility [3] and 3D structure [4, 5]. By integrating data from such types of different assays or the so-called multi-omics approach, biologists can comprehensively investigate genome dynamics during biological processes. This methodology has been successfully applied to many biological fields, such as neurological diseases [6], development of nervous system [7] and virus infection [8, 9]. Data visualization, especially the genome track like plots, are crucial for exploring or demonstrating some local or global properties of the genomics data.
Many visualization tools have been developed to meet these demands, and these tools can be classified into three categories: (1) Command-line plotting tool [10, 11], (2) Graphical User Interface(GUI) software [12], and (3) Web-based track browser [13–15]. In different situations, each kind of tools has its own advantages and limitations. As for command-line tools, they are convenient for bioinformaticians to produce plots or results easily but require Linux command line skills. GUI tools are friendly to people who are not skilled at programming and command line. Web-based browsers could share visualization results between colleagues. However, they are not efficient in transmission and have relative high latency between the websites and customers. Moreover, for program developers, GUI and web-based tools are not as convenient as command-line tools and plotting packages, which could be locally installed and easily called between stacks. Despite the above tools work well for providing an overview of the input genomic data. However, during actual scientific research, users need a detail comparative and analytical data visualization more than just the basic view of the data. For example, to visualize the differential contact interaction (DCI) of two Hi-C contact matrices [16] or predicted chromatin loops on the matrix [17]. In most cases, bioinformaticians work in programmatic and interactive environments like RStudio, IPython console and Jupyter notebook to complete the data analysis, algorithm development and visualization tasks. However, there is a gap between the data analysis ecosystem and the existing genomic data visualization tools. Researchers spend a lot of time on unnecessary stuffs like file format conversion and environment switching. Therefore, a versatile tool that fills the gap will significantly facilitate the genomics study.
To fill this gap, we developed CoolBox, a versatile toolkit for exploration-driven visualization of genomic data. It combines advantages of existing tools and is highly compatible with the Python scientific ecosystem, highly customizable, easy to use with intuitive interface design and simple installation procedure. It can be used in different scenarios: (1) Python script or another python package for data fetching and plotting; (2) Shell as a command-line plotting tool; (3) Jupyter notebook environment for data fetching, plotting, and exploration; and (4) Web application for exploration and demonstration within the web browser.
Implementation
The plotting system of CoolBox is based on the matplotlib package. A part of the plotting code in the CoolBox is a fork from pyGenomeTracks package. [10] The data stored in bigWig, “.cool” and ”.hic” file format are loaded through pybbi (https://github.com/nvictus/pybbi), cooler [18] and straw [19] packages. Pairwise interaction data in Browser Extensible Data Paired-End (BEDPE) and Pairs format is indexed and randomly accessed using the pairix software (https://github.com/4dn-dcic/pairix). Other text-based genomic feature data format, like Browser Extensible Data (BED), Gene transfer format (GTF), and BedGraph is indexed and random accessed using the tabix [20] software. The widget panel in the GUI is implemented by using the ipywidgets package.
Results and discussion
Flexible and user-friendly API and CLI for producing high-quality genome track plots
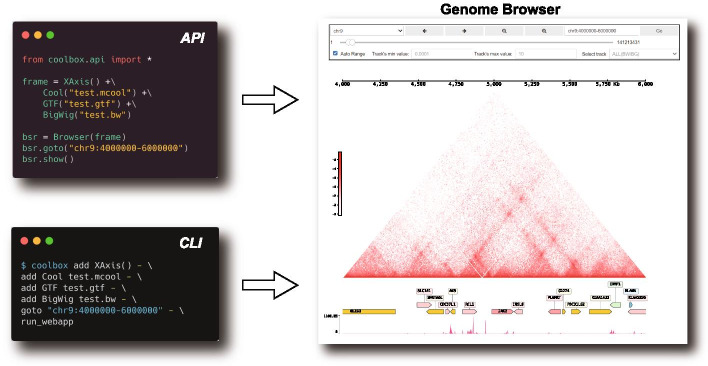
CoolBox provides an Application Programming Interface (API) for Python script or Jupyter environment as well as a Command Line Interface (CLI) for Shell. The interface design is inspired by the popular R package ggplot2 [21]. It allows users to compose their figures with highly intuitive syntax. In CoolBox, users can use the “+” operator in Python or “add” command in Shell to compose low-level track elements to a higher-level figure. For example, to compose track objects of various kinds of genomic data into a single frame and interactively review interested regions in genome browser with few lines of Python codes or Shell commands (Fig. 1).
Fig. 1.
CoolBox has a clear and intuitive syntax to compose genome browser in both API and CLI mode. Inspired by the ggplot2 syntax, figures in CoolBox can be composed and adjusted (color, height, style etc.) from different tracks and features by using the ‘+‘ operator in API or ‘add‘ in CLI, almost every figure composed in the API mode has a paired CLI composing command that produces identical figures. This design facilitates bioinformaticians who work regularly in both environments
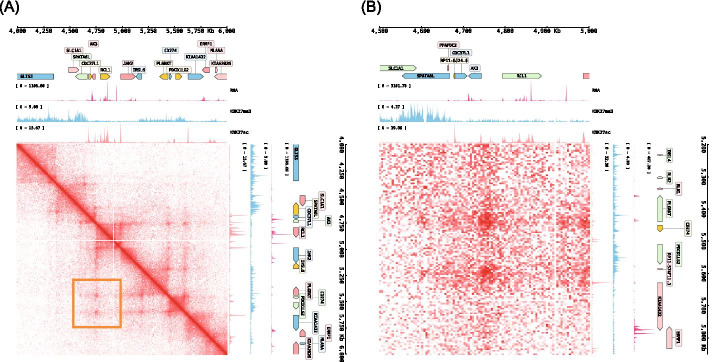
Besides the 1-dimensional viewing mode supported by most other visualization tools, CoolBox supports a joint-view mode that enables users to visualize trans or cis-remote regions in a Hi-C contact matrix (Fig. 2).
Fig. 2.
Joint (2d) view example, CoolBox can compose big figure which put frames around a center contact matrix. This allows to visualize the trans or cis remote (off-diagonal) contact matrix along with genome features. A Joint view on an on-diagonal region. B Joint view on an cis remote region, which shows the magnified detail of the orange box marked loop region that contains two chromatin loops in (A)
Most sets of commonly generated genomic assay data such as RNA-Seq, ChIP-Seq, ATAC-Seq, Hi-C, HiChIP [22] data which stored in BedGraph, bigwig [23], cool [18], .hic [19] and other file formats (see Table 1) can be visualized in CoolBox by different kinds of tracks. Most tracks’ features (color, height, style, etc.) can also be configured in the same way via the API or CLI. In the CoolBox plotting system, the plot contains not only a single layer. Users can put another layer (Coverage) upon the original plot to produce more comprehensive and high-quality figures. Furthermore, figures can be generated in different formats, including PNG, JPEG, PDF, and SVG. More details about the API and CLI are available in the online documents and user manual.
Table 1.
A part of CoolBox builtin tracks for visualizing different kinds of genomics data formats
| Track type | File format | Description |
|---|---|---|
| XAxis | None | Coordinate of the reference genome |
| Spacer | None | For add vertical space between two tracks |
| BigWig | .bigwig | Track for bigWig file, draw the histogram |
| BedGraph | .bedgraph | Track for BedGraph file, draw the histogram |
| BAM | .bam | BAM track for visualize the coverage or alignment |
| BED | .bed | For visualization genome annotation, like refSeq genes and chromatin states |
| GTF | .gtf | Track of GTF file, for visualize gene annotation |
| Arcs | .pairs, .bedpe | Show the chromosome interactions get from ChIA-PET, HiChIP or Hi-C loop data |
| HiCMat | .cool, .mcool, .hic | Show the chromosome contact matrix from Hi-C data |
| Virtual4C | .cool, .mcool, .hic | Virtual 4C track, using Hi-C data to mimic 4C |
| DiScore | .cool, .mcool, .hic | Directional index of Hi-C matrix for detecting TAD |
| InsuScore | .cool, .mcool, .hic | Insulation score of Hi-C matrix for inferring TAD borders |
| HiCDiff | .cool, .mcool, .hic | Show the difference between two contact matrix |
| Selfish | .cool, .mcool, .hic | Apply the selfish algorithm [16] on two contact matrices to detect differential contact interactions |
| SNP | .tsv | Track for show SNPs Manhattan plot |
Interactive exploration and reproducible analysis on genomic data
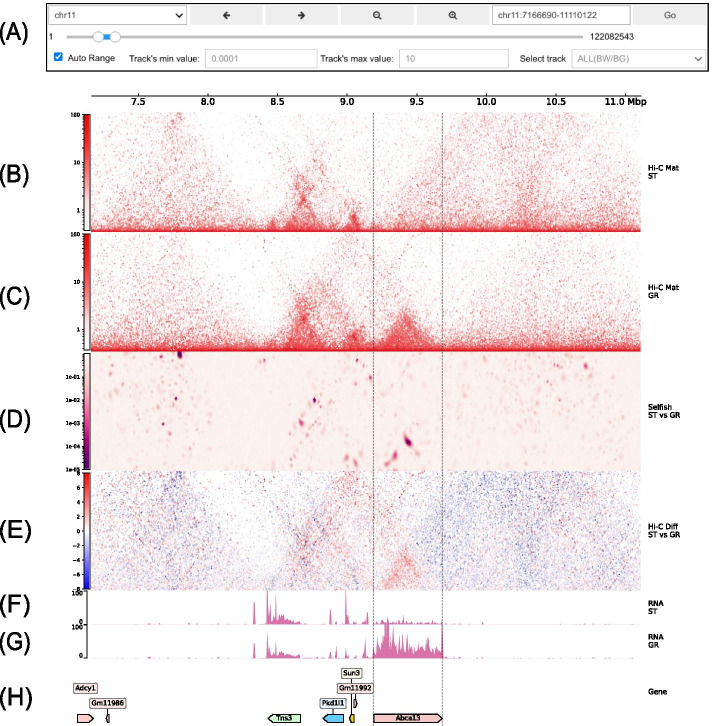
As shown in Fig. 3, CoolBox provides a GUI for interactive data visualization, by which users can explore different genomic regions by operating a simple widget panel and visualize the data within a specific region.
Fig. 3.
A CoolBox figure representing differential interactions of Hi-C contact matrices. Shown Hi-C and RNA-Seq data are produced from the process of hematopoietic differentiation [24]. Its clearly that there is a topological association domain (TAD) formation at the Abca13 gene region and its RNA expression is up-regulated at the same time after the differentiation. A The widgets panel of CoolBox browser, used for zooming, sliding, and locating the genome region. When moving to a new region, the figure draw bellow will be updated automatically. B Hi-C track of short-term hematopoietic stem cell (ST) shows the contact map of ST sample. The color bar indicates the normalized value of the contact map. C Hi-C track of granulocyte (GR). D Differential contact interaction result of the Selfish algorithm [16] on ST and GR Hi-C contact map. The color bar indicates the q-value (BH adjusted p value) produced from the DCI analysis. Darker color means this interaction has a lower q-value; that is to say, two contact maps are more diverse at this location. E Hi-C Diff track. It shows the difference between GR and ST’s z-score normalized contact matrices. The red region of the matrix indicates where GR has a more significant contact frequency compared to ST, and the opposite for blue areas. F BigWig track of ST RNA-Seq data, showing the RNA expression level of ST in this region. G BigWig track of GR RNA-Seq data. H A gene annotation track shows the corresponding genes within this genomic region
Besides, the data and the figures are bound together by Python objects. In this way, users can get the precise data of each track within the current view of the genomic region through the API. This design facilitates comparative visualization and statistical analysis. CoolBox is also a general genomic-file reading package. Data within a particular genome region can be retrieved in a short time, as almost all supported file formats can be indexed and randomly accessed.
Moreover, by leveraging the power of the Jupyter notebook, the visualization result and the entire process can be recorded in the notebook. It is convenient for sharing the visualization result and reproducing the whole analysis by other researchers.
A testing and visualizing framework for new algorithm development
Owing to the user-friendly and highly extensible API design, users can implement their custom tracks without any difficulty, thus enabling seamless cooperation in Python-based algorithm development and scientific research. The algorithm developer can check and visualize the intermediate results produced by their algorithm and adjust parameters simultaneously. In addition, as CoolBox uses an object-oriented programming paradigm in its design, users can reuse each track’s codes by inheritance, including data extraction and drawing-related functions. In most cases, users only need to write algorithm-related core parts. The most tedious part including raw-data reading, preprocessing, and figure drawing are handed over to CoolBox through inheritance (see method section and user manual for implementation details). In this way, bioinformaticians can free themselves from those repetitive procedures and only focuses on the data post-processing.
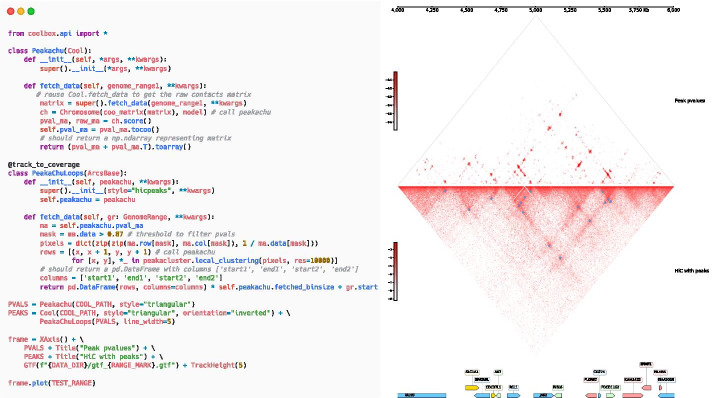
We demonstrate the advantages by implementing a track that visualizes the outputs of the Peakachu algorithm [17], which is a RandomForest based method for detecting loops in the Hi-C contact matrix. As depicted in Fig. 4, the main part of the whole track contains merely 20 lines of Python code. The data fetching and plotting functionality are fully reused by inheriting Cool/ArcsBase Track base class. Furthermore, the custom-defined track is empowered to be used in CLI, API, and browser mode in couple with other built-in tracks. More details include a reproducible code block and can be found in the online documents and user manual.
Fig. 4.
An example to define custom tracks that display Hi-C contact matrix along with peaks detected by Peakachu algorithm [17]. An example of peak prediction result is demonstrated in the right panel. The upper triangular matrix shows the peak p-value output by Peakachu algorithm. The predicted peaks drawn as blue squares upon the original matrix is shown in the lower triangular matrix. The Hi-C matrices and the peaks upon them will be automatically recomputed and updated after the change of genome region. The left panel is the full Runnable python codes used for generating the right panel. The custom track is implemented by following the same intuitive and clear design pattern as other built-in tracks: i.e., reusing the data fetching and plotting functionality as much as possible. For this figure, the functionality of fetching and plotting contact matrix with peaks are totally reused by inheriting Cool/ArcsBase track base class, and the rest of the codes merely calls the computing function of the peachachu package. After the track definition, we can see that the custom track is born to support being used in a ggplot2-like syntax with other tracks, and this capability is also valid in CLI and GUI mode
Comparison with other existing visualization tools
As stated before, there is an urgent need for better visualization tools to accelerate the integration and mining of biological data. Therefore, more and more visualization tools have been developed in recent years. A comparison of features between CoolBox and these tools is listed in Table 2. Most of the visualization tools require a tedious installation process and are operated through the command line. Before visualization, the data needs to be preprocessed through specific steps, and then a static or interactive web interface is generated.
Table 2.
Summary of genomic visualization tools
| Tools | Programming language | API plot | CLI plot | Online access |
|---|---|---|---|---|
| CoolBox | Python | |||
| pyGenomeTracks | Python | |||
| gcMapExplorer | Python | |||
| HiCPlotter | Python | |||
| HiGlass | Python, HTML, CSS, JS | |||
| YueLab Browser | HTML, CSS, JS | |||
| WashU Browser | HTML, CSS, JS | |||
| TADkit | HTML, CSS, JS | |||
| JuiceBox.js | HTML, CSS, JS | |||
| JuiceBox | Java |
| GUI | Input | Installation | Customization | |
|---|---|---|---|---|
| Web and Jupyter | Raw data | Bioconda or PyPI | Python knowledge, very easy | |
| Raw data | PyPI | Python knowledge, easy | ||
| Local | Preprocessed data | PyPI | Python knowledge, easy | |
| Preprocessed data | Manually install | |||
| Web and Jupyter | Preprocessed data, via network | Docker | Web knowledge | |
| Web | Via network | |||
| Web | Via network | |||
| Web | Preprocessed data, via network | Manually install | ||
| Web | Via network | |||
| Local | Raw data | Download | ||
The visualization and data processing of most visualization tools are dissociated, which is not convenient for bioinformaticians whose routine works rely on Python-based scientific computation ecosystem. Except for the CLI mode supported by most visualization tools, the API that the CoolBox has been used internally and exposed follows the same design as the CLI, making switching between these two modes with no pain. More importantly, since the API in CoolBox combines computation and visualization, users can dynamically add different tracks or even custom tracks in the python notebook while processing raw data or developing new methods.
Conclusion
CoolBox is a versatile toolkit for the visualization and exploration of multi-omics data in the Python ecosystem. It provides a user-friendly ggplot2-like syntax for composing various kinds of tracks in CLI, API, GUI and web browser mode. More importantly, its built on a highly extensible plotting system that allows users to implement their custom tracks without wasting time on data fetching and figure plotting procedures. Through the power of Jupyter notebook, it provides a convenient way for bioinformaticians to exploit it’s versatility for better personalized data manipulation and demonstration. It could also increase the reproducibility of genomic data visualization tasks as codes and figures are all organized into the same page.
Availability and requirements
Project name: CoolBox
Project home page: https://github.com/GangCaoLab/CoolBox
Operating system(s): Linux, macOS, Windows WSL
Programming language: Python
Other requirements: All software requirements are listed in https://github.com/GangCaoLab/CoolBox/blob/master/environment.yml
License: GPLv3
Any restrictions to use by non-academics: GPLv3 licensing restrictions apply.
Acknowledgements
We thank everyone who contributed to this project on GitHub. We thank Khaista Rahman for helping to review and improve our English writing.
Abbreviations
- NGS
Next-generation sequencing
- GUI
Graphical user interface
- API
Application programming interface
- CLI
Command line interface
Authors’ contributions
WX: Conceptualization, Investigation, Software design, Software maintain, Software test, Writing—Original Draft Preparation, Writing–Review and Editing. QZ: Conceptualization, Software design, Software maintain, Software test, Writing—Original Draft Preparation, Writing—Review and Editing. DL: Conceptualization, Resources, Writing—Review and Editing. YZ: Conceptualization, Writing—Review and Editing. JD: Conceptualization, Funding Acquisition, Supervision, Writing—Review and Editing. LG: Conceptualization, Funding Acquisition, Investigation, Resources, Supervision, Writing—Review and Editing. GC: Conceptualization, Funding Acquisition, Investigation, Resources, Supervision, Writing—Review and Editing. All the authors have read and approved the final manuscript.
Funding
1. The Key Research and Development Program of Guangdong Province (Grant 2019B020211003 to Gang Cao). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. 2. National Natural Science Foundation of China (NSFC) (Grants 31941014 to Gang Cao). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. 3. National Natural Science Foundation of China (NSFC) (Grants 31941005 to Huanchun Chen). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. 4. National Natural Science Foundation of China (Grants 31702196 to Ke Xiao). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. 5. China Postdoctoral Science Foundation (Grant 2019M662676, to Ke Xiao). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
Sample data designed to demonstrate most features of the software is provided at https://github.com/GangCaoLab/CoolBox/tree/master/tests/test_data.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Guoliang Li, Email: guoliang.li@mail.hzau.edu.cn.
Gang Cao, Email: gcao@mail.hzau.edu.cn.
References
- 1.Morin RD, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh TJ, McDonald H, Varhol R, Jones SJ, Marra MA. Profiling the HeLa s3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45(1):81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- 2.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of stat1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4(8):651. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 3.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for multimodal regulatory analysis and personal epigenomics. Nat Methods. 2013;10(12):1213. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fullwood MJ, Ruan Y. Chip-based methods for the identification of long-range chromatin interactions. J Cell Biochem. 2009;107(1):30–39. doi: 10.1002/jcb.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corces MR, Shcherbina A, Kundu S, Gloudemans MJ, Frésard L, Granja JM, Louie BH, Eulalio T, Shams S, Bagdatli ST, et al. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat Genet. 2020;52(11):1158–68. [DOI] [PMC free article] [PubMed]
- 7.Song M, Pebworth M-P, Yang X, Abnousi A, Fan C, Wen J, Rosen JD, Choudhary MN, Cui X, Jones IR, et al. Cell-type-specific 3d epigenomes in the developing human cortex. Nature. 2020;587(7835):644–649. doi: 10.1038/s41586-020-2825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinz S, Texari L, Hayes MG, Urbanowski M, Chang MW, Givarkes N, Rialdi A, White KM, Albrecht RA, Pache L, et al. Transcription elongation can affect genome 3d structure. Cell. 2018;174(6):1522–1536. doi: 10.1016/j.cell.2018.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao C, Hong P, Huang X, Lin D, Cao G, Wang L, Feng B, Wu P, Shen H, Xu Q, et al. HPV-CCDC106 integration alters local chromosome architecture and hijacks an enhancer by three-dimensional genome structure remodeling in cervical cancer. J Genet Genomics. 2020;47(8):437–450. doi: 10.1016/j.jgg.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Delisle L, Rabbani L, Wolff J, Bhardwaj V, Backofen R, Grüning B, Ramírez F, Manke T. pyGenome Tracks: reproducible plots for multivariate genomic data sets. Bioinformatics. 2020;37:422. doi: 10.1093/bioinformatics/btaa692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akdemir KC, Chin L. Hicplotter integrates genomic data with interaction matrices. Genome Biol. 2015;16(1):198. doi: 10.1186/s13059-015-0767-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar R, Sobhy H, Stenberg P, Lizana L. Genome contact map explorer: a platform for the comparison, interactive visualization and analysis of genome contact maps. Nucleic Acids Res. 2017;45(17):152. doi: 10.1093/nar/gkx644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Song F, Zhang B, Zhang L, Xu J, Kuang D, Li D, Choudhary MN, Li Y, Hu M, et al. The 3d genome browser: a web-based browser for visualizing 3d genome organization and long-range chromatin interactions. Genome Biol. 2018;19(1):1–12. doi: 10.1186/gb-2007-8-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li D, Hsu S, Purushotham D, Sears RL, Wang T. Washu epigenome browser update 2019. Nucleic Acids Res. 2019;47(W1):158–165. doi: 10.1093/nar/gkz348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerpedjiev P, Abdennur N, Lekschas F, McCallum C, Dinkla K, Strobelt H, Luber JM, Ouellette SB, Azhir A, Kumar N, et al. Higlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol. 2018;19(1):1–12. doi: 10.1186/s13059-018-1486-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ardakany AR, Ay F, Lonardi S. Selfish: discovery of differential chromatin interactions via a self-similarity measure. Bioinformatics. 2019;35(14):145–153. doi: 10.1093/bioinformatics/btz362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salameh TJ, Wang X, Song F, Zhang B, Wright SM, Khunsriraksakul C, Ruan Y, Yue F. A supervised learning framework for chromatin loop detection in genome-wide contact maps. Nat Commun. 2020;11(1):1–12. doi: 10.1038/s41467-020-17239-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdennur N, Mirny LA. Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics. 2019;36:311–316. doi: 10.1093/bioinformatics/btz540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durand NC, Shamim MS, Machol I, Rao SS, Huntley MH, Lander ES, Aiden EL. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016;3(1):95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H. Tabix: fast retrieval of sequence features from generic tab-delimited files. Bioinformatics. 2011;27(5):718–719. doi: 10.1093/bioinformatics/btq671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wickham H. ggplot2: elegant graphics for data analysis. J Stat Softw. 2010;35(1):65–88. [Google Scholar]
- 22.Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, Chang HY. Hichip: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016;13(11):919–922. doi: 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. Bigwig and bigbed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26(17):2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang C, Xu Z, Yang S, Sun G, Jia L, Zheng Z, Gu Q, Tao W, Cheng T, Li C, et al. tagHi-C reveals 3d chromatin architecture dynamics during mouse hematopoiesis. Cell Rep. 2020;32(13):108206. doi: 10.1016/j.celrep.2020.108206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sample data designed to demonstrate most features of the software is provided at https://github.com/GangCaoLab/CoolBox/tree/master/tests/test_data.