Abstract
V(D)J recombination in differentiating lymphocytes is a highly regulated process in terms of both cell lineage and the stage of cell development. Transgenic and knockout mouse studies have demonstrated that transcriptional enhancers from antigen receptor genes play an important role in this regulation by activating cis-recombination events. A striking example is the T-cell receptor β-chain (TCRβ) gene enhancer (Eβ), which in the mouse consists of at least seven nuclear factor binding motifs (βE1 to βE7). Here, using a well-characterized transgenic recombination substrate approach, we define the sequences within Eβ required for recombination enhancer activity. The Eβ core is comprised of a limited set of motifs (βE3 and βE4) and an additional previously uncharacterized 20-bp sequence 3′ of the βE4 motif. This core element confers cell lineage- and stage-specific recombination within the transgenic substrates, although it cannot bypass the suppressive effects resulting from transgene integration in heterochromatic centromeres. Strikingly, the core enhancer is heavily occupied by nuclear factors in immature thymocytes, as shown by in vivo footprinting analyses. A larger enhancer fragment including the βE1 through βE4 motifs but not the 3′ sequences, although active in inducing germ line transcription within the transgenic array, did not retain the Eβ recombinational activity. Our results emphasize the multifunctionality of the TCRβ enhancer and shed some light on the molecular mechanisms by which transcriptional enhancers and associated nuclear factors may impact on cis recombination, gene expression, and lymphoid cell differentiation.
Immunoglobulin (Ig) and T-cell receptor (TCR) genes are assembled from separate variable (V), diversity (D), or joining (J) gene segments in a process known as V(D)J recombination (4, 44, 55). Normally, V(D)J recombination is restricted to, and required for, early B and T lymphocyte development. It depends on a unique activity, called the V(D)J recombinase, which targets recombination signal sequences (RSSs; consisting of a heptamer, a spacer of 12 or 23 bp, and a nonamer) flanking the rearranging sides of all V, D, and J segments. Rearrangement events primarily involve pairs of segments with RSSs of asymmetrical spacer length (12/23 rule).
The functional core of the V(D)J recombinase consists of the lymphoid-restricted RAG-1 and RAG-2 gene products which recognize, pair off, and cleave the RSSs from two rearranging segments, a step also involving architectural proteins from the high-mobility-group family (22, 61). These cleavages consist of DNA double-strand breaks (DSBs) introduced precisely at the junction between the heptamer and adjacent coding sequences, yielding two distinct products: the hairpin-sealed coding ends (CEs) and the phosphorylated, blunt-ended signal ends (SEs). Eventually, the cleaved products are assembled together, a process which requires, in addition to the RAG factors, components of the general DNA DSB repair machinery (26, 54). The two CEs are assembled to form a coding joint (CJ) on the rearranged chromosome following hairpin opening and possible deletion and/or addition of nucleotides at the free extremities. The SEs are fused without further processing, yielding a signal joint (SJ) usually contained within a circular piece of extrachromosomal DNA.
V(D)J recombination is strictly controlled with respect to the lymphoid cell lineage, stage of cell differentiation, and allele usage. For example, there is a strong bias for TCR gene rearrangement to occur only in T cells (and for Ig gene rearrangement to occur only in B cells). Also, during αβ thymic cell development, TCRβ rearrangement initiates in CD4− CD8− double-negative (DN) cells prior to TCRα rearrangement which occurs during the transition from DN to CD4+ CD8+ double-positive (DP) cells (37). Moreover, during TCRβ rearrangement, Dβ-to-Jβ recombination usually precedes complete Vβ-to-DJβ recombination (in DN CD44+ CD25+ and DN CD44− CD25+ thymocytes, respectively). Finally, Vβ-to-DJβ joining is subject to allelic exclusion, a process whereby productive rearrangement (e.g., those encoding a TCR β chain) on one allele precludes further Vβ recombination. It has been hypothesized that these levels of control reflect the ability of the individual loci and/or segments to serve as substrates for the recombinase, a concept known as V(D)J recombinational accessibility (63, 67). Recent evidence has revealed that recombinational accessibility, as tested by specific DSB formation, is a regulated property of lymphocyte chromatin (69). However, the molecular mechanism(s) and structural basis underlying these phenomena are not yet understood. Proposed activating factors include transcription across the unrearranged (germ line) region, the down-modulation of CpG methylation, and/or other epigenetic changes in chromatin (e.g., level of histone acetylation) affecting nucleosome positioning (13, 20, 54).
In studies using transgenic and knockout mouse techniques, it has been demonstrated that transcriptional cis-regulatory elements affect the recombination potential of adjacent gene segments (29, 63, 67). One striking example has come from studies on the TCRβ gene locus and the associated enhancer. This large locus (∼500 kb in the mouse) (Fig. 1A) carries two homologous regions, each containing one Dβ segment (Dβ1 or Dβ2), six functional Jβ segments (Jβ1.1 to 6 or Jβ2.1 to 6; one additional pseudo-J is present in each Jβ cluster), and one constant (C) region gene (Cβ1 or Cβ2). Most of the Vβ genes are located 5′ of the Dβ-Jβ clusters, except for one (Vβ14) oriented in the opposite transcriptional direction 3′ of Cβ2. A single transcriptional enhancer (Eβ), contained within a 560 bp HpaI-NcoI DNA fragment (Eβ560), has been found 5.9 kb downstream of the Cβ2 exons (38, 50). Eβ560 activates V(D)J recombination within a transgenic substrate in early developing thymocytes from both embryonic and adult mice (12, 56). Moreover, deletion of the endogenous Eβ560 fragment by using gene targeting in embryonic stem cells and production of mutant mice results in a drastic inhibition of cis rearrangement of the mutated TCRβ alleles (5, 6), indicating that the fragment contains regulatory sequences that are absolutely required for V(D)J recombination to proceed normally at that locus. Finally, detailed analyses of Dβ and Jβ SE intermediates as well as Dβ-to-Jβ CJ and SJ products in thymocytes from mice homozygous for the targeted deletion (Eβ−/− mice) lead to the surprising finding that loss of accessibility alone may not fully account for the defect in recombination, suggesting an unsuspected role for Eβ during the recombination reaction (30).
FIG. 1.
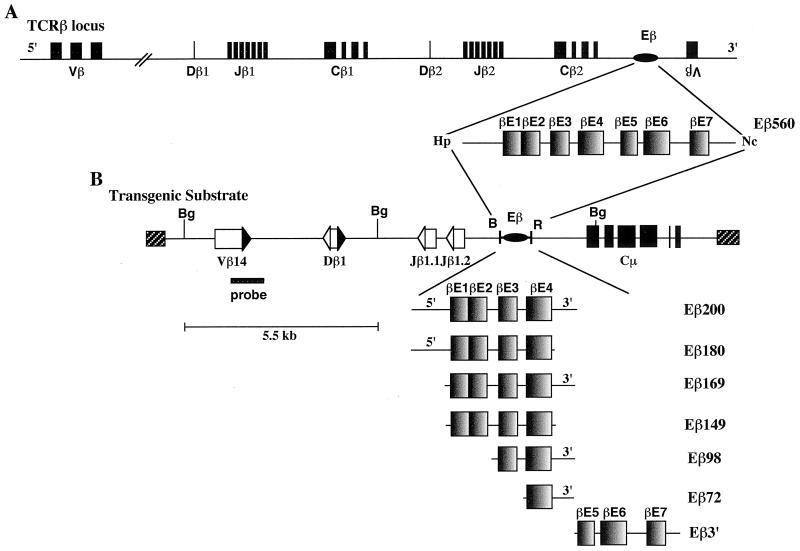
Structural organization of Eβ and truncated Eβ variants used in this study. (A) Location within the TCRβ locus and structure of the 560-bp HpaI-NcoI DNA fragment containing Eβ (not drawn to scale). The TCRβ V, D, and J segments as well as the Cβ exons are represented by shaded boxes, and Eβ is represented by an oval. 5′ and 3′ indicate transcriptional orientations of these various elements within the TCRβ locus, with the exception of the single Vβ14 gene located downstream of Eβ, which lies in the opposite direction. An enlargement of the 560-bp enhancer (Eβ560) shows the several nuclear factor binding sites (βE1 to βE7) previously identified by EMSA within this fragment (71). (B) Partial restriction endonuclease map of the microinjected inserts and structure of the Eβ variants. The TCRβ V, D, and J segments are represented by open boxes, their flanking RSSs are represented by shaded (23-bp spacer) or open (12- or 13-bp spacer) triangles, the IgH Cμ exons are represented by shaded boxes, the cosmid sequences are represented by hatched boxes, and Eβ is represented by an oval (these various elements are not drawn to scale). Restriction endonuclease sites are indicated as B (BamHI), Bg (BglII), Hp (HpaI), Nc (NcoI), and R (EcoRI). The structural organization of each Eβ variant is shown, including the 80- and 30-bp sequences which, in some variants, flank the 5′ and/or 3′ sides of the βE1 and βE4 motifs, respectively (delineation of βE motifs according to Takeda et al. [71]). Locations of the Vβ14-hybridizing probe used in Southern analysis of BglII-restricted genomic DNA from the transgenic mice and of the 5.5-kb BglII-hybridizing fragment from the unrearranged substrate are indicated.
Studies of mouse and human Eβ, using in vitro footprinting analysis and electrophoretic mobility shift assays (EMSA), have identified multiple nuclear factor binding sites along this element which are conserved between the two species, supporting their functional importance (25, 58, 71). Several such motifs, including GATA- and helix-loop-helix (HLH)-binding E-box motifs, a cyclic AMP response element-like sequence, and Ets and core-binding factor (CBF) sites, are shared by other lymphoid gene enhancers and have been shown to be required for Eβ activity (25, 31, 42, 59, 70, 71). Accordingly, transcription factors known to bind discrete enhancer motifs were also found to activate reporter genes placed under the control of related Eβ sequences, including notably GATA-3 (47) and Ets as well as CBF (36, 59, 70, 78). In addition, screening of cDNA expression libraries with oligonucleotide probes spanning Eβ or related sequences contributed to the identification of novel factors with homology to POU domain proteins (51) or to CACCC box binding proteins (73). Finally, signal transduction pathways involving raf and ras were proposed to play a role in Eβ-mediated transcriptional activation (79). Despite all of these studies however, Eβ structural and functional organization remains less well understood compared to regulatory elements from other lymphoid genes such as the Ig heavy-chain (IgH) intronic enhancer or the TCRα enhancer (23, 53). Notably, in contrast to these other elements, a minimal core Eβ sequence has not yet been unequivocally defined (25, 58, 71). Moreover, the functional importance of individual motifs and their associated factors to act nonredundantly as positive or negative elements is still unclear; such motifs include GATA and Ets (31, 47, 59, 70).
In this study, we used transgenic mice to better characterize the function of the Eβ element in vivo, focusing especially on its role in the activation of germ line transcription and V(D)J recombination. Notably, we have defined a core TCRβ enhancer for recombination which is comprised of two known nuclear factor binding motifs as well as additional, previously unidentified sequences which we show are absolutely required to support cis rearrangement of adjacent TCRβ gene segments.
MATERIALS AND METHODS
Construction of recombination substrates.
All recombination substrates were derived from the construct VβDβJβCμB*R*. VβDβJβCμB*R* is identical to the VβDβJβCμ construct (19) except that it lacks the BamHI and EcoRI restriction sites located within exon 2 of the Cμ gene and 5.3 kb further downstream, respectively. Both sites were eliminated following BamHI and EcoRI partial endonucleolytic cleavages, fill-in of the extremities, and religation. This allowed direct cloning of the several Eβ variants, using the adjacent BamHI and EcoRI sites located 130 bp 3′ of the Jβ1.2 gene segment in VβDβJβCμB*R*. The Eβ variants were produced by PCR amplification, using a 4.95-kb Eβ-containing genomic fragment subcloned into pGEM-4Z (Promega France, Charbonnières, France) as a template (6), as well as appropriate forward and reverse primers flanked by BamHI and EcoRI sites, respectively. With position 1 assigned to the first nucleotide in the HpaI site from the 560bp HpaI-NcoI Eβ-containing fragment (38), the various truncated variants of Eβ used in this study were as follows: Eβ200 (+124 to +324), Eβ180 (+124 to +304), Eβ169 (+155 to +324), Eβ149 (+155 to +304), Eβ98 (+226 to +324), Eβ72 (+252 to +324), and Eβ3′ (+300 to +560). Each Eβ fragment inserted into the recombination substrate was verified by nucleotide sequencing.
Production of transgenic mice.
Linearization, purification, microinjection of the individual DNA inserts into fertilized (C57BL/6 × CBA/J)F2 oocytes, identification of the transgenic founders, and production and maintenance of transgenic mouse lines were performed as described previously (12, 18, 19). The characteristics of the transgenic animals in lines 1003 (carrying an enhancerless recombination substrate) and Eβ2 (carrying the Eβ560-driven substrate, hereafter referred to as the Eβ560 line) have been reported previously (12, 19). Nontransgenic littermates in the Eβ560 line were used as the source of wild-type (WT) genomic DNA and total RNA. All mice were housed in a specific-pathogen-free animal facility in accordance with institutional guidelines. Mice were sacrificed for analysis between 4 and 6 weeks of age.
Southern blot analyses.
Preparation of genomic DNA, restriction enzyme digests, agarose gel electrophoresis, DNA blotting onto nylon membranes (Hybond-N+; Amersham, Les Ulis, France), preparation and [α-32P]dCTP labeling of the Vβ14 probe, and hybridization procedures were performed as described previously (12, 19). Images were generated from hybridized filters by use of a phosphorimager (BAS 1000; Fuji, Raytest France S.A.R.L., Courbevoie, France) and quantitated with MacBAS software.
DNA and RNA PCR assays.
Genomic DNA and total RNA were simultaneously extracted from transgenic cell populations (∼5 × 105 cells), using TRIzol (GIBCO BRL, Cergy Pontoise, France) as recommended by the manufacturer. RNA samples were treated with RNase-free DNase I (Pharmacia, Orsay, France) and were converted to cDNA by reverse transcription (RT), using the SuperScript II reverse transcriptase (GIBCO BRL). Analysis of transgenic specific V(D)J rearrangements and Jβ germ line transcripts by, respectively, DNA long-range PCR (LR-PCR and RNA RT)-PCRs were performed as described previously (12, 18). LR-PCRs were performed for 25 cycles of 1 min at 94°C, 30 s at 56°C, and 2 min at 72°C; RT-PCRs were performed for 28 cycles of 30 s at 94°C, 30 s at 58°C, and 30 s at 72°C. Oligonucleotide primers for amplification of fragments encompassing substrate Dβ-to-Jβ and Vβ-to-DJβ rearrangements and substrate Jβ1-IgH Cμ exons, as well as fragments encompassing β-actin, CD14, and Cβ2 exons, were as described previously (12, 30). Sequences of forward and reverse primers for amplification of fragments encompassing endogenous Dβ1-to-Jβ1.1/Jβ1.2 rearrangements were 5-CTGGTGGTTTCTTCCAGCCCT-3 and 5-CCTTCCTCTGATTACCAGAAC-3′. PCR amplification of endogenous Vκ-to-Jκ2 rearrangements was performed as described by Schlissel and Baltimore (62). After amplification, PCR products were electrophoresed through 1% agarose–0.5% NuSieve gels, transferred to nylon membranes (Hybond-N+; Amersham), and hybridized with [γ-32P]ATP-labeled locus-specific oligonucleotide probes internal to the corresponding primers. Images were generated and quantitated as described above.
Antibodies and fluorescence-activated cell sorting (FACS) purification of lymphocytes.
Fluorescein isothiocyanate (FITC)- and phycoerythrin-conjugated monoclonal antibodies (MAbs) against CD25 (7D4), CD44 (Pgp-1), CD3ɛ (145-2C11), and B220 (RA3-6B2) were purchased from PharMingen (San Diego, Calif.). Lymphocyte preparation and cell staining with saturating levels of MAbs were performed as described previously (10, 18). Cell sorting of peripheral T and B cells was accomplished with anti-CD3ɛ and anti-B220 MAbs. For cell sorting of DN thymic cell populations, an initial depletion of CD4+ and/or CD8+ cells was performed with anti-CD4- and anti-CD8-containing supernatants and rabbit complement, as described previously (33), before staining with anti-CD44 and anti-CD25 MAbs. Cell sorting was performed with a FACStar Plus (Becton Dickinson, Mountain View, Calif.).
Fluorescence in situ hybridization (FISH) analysis.
Metaphase spreads were prepared from transgenic splenic lymphocytes stimulated with concanavalin A (ConA) at 37°C for 72 h and cultured with 5-bromodeoxyuridine (60 μg/ml) added for the final 6 h of culture to ensure chromosomal R banding. Chromosome spreads were hybridized with a biotinylated 10-kb EcoRI fragment containing the Cμ exons (19), using standard protocols (49, 57). Biotinylation of the Cμ probe was carried out by nick translation using biotin-16-dUTP (Boehringer Mannheim, Meylan, France) as recommended by the manufacturer. Before hybridization, the biotinylated probe was annealed for 45 min at 37°C with a 200-fold excess of murine Cot-1 DNA (GIBCO-BRL) in order to compete with nonspecific repetitive sequences; 200 ng of the probe (at 10 μg/ml in the hybridization solution) was used per slide. The hybridized probe was detected with FITC-conjugated avidin (Vector Laboratories, Burlingame, Calif.). Chromosomes were counterstained with propidium iodide diluted in antifade solution (pH 11.0) as described previously (43). For each chromosomal spread, a total of 30 metaphase cells were analyzed. Over 85% of the cells showed, in addition to the two hybridization signals detectable at the distal end of chromosomes 12 (corresponding to the endogenous IgH loci), a specific signal corresponding to the transgenic site.
In vivo footprinting analyses.
The in vivo genomic footprint analysis was performed by the dimethyl sulfate (DMS)/ligation-mediated PCR (LM-PCR) technique (52) essentially as described by Algarté et al. (1). Briefly, thymocytes from 4- to 6-week-old WT or RAG-1-deficient (RAG-1−/−) mice (68) were harvested and subjected to DMS treatment, and the DNA was extracted. In parallel, DNA from cells of the same thymuses was first extracted, deproteinized, and then subjected to DMS treatment (naked DNA). The resulting DMS-treated DNA samples were cleaved by pipiridine and subjected to LM-PCR. Specific oligonucleotide primers, encompassing the Eβ sequence, were used to carry out a primer extension reaction for both top and bottom strands. The resulting double-stranded DNA was ligated to the LM-PCR unidirectional linker consisting of two strands of dissimilar length: LM-PCR.1 (5′-GGGGTGACCC GGGAGATCTGAATTC-3′) and LM-PCR.2 (5′-CTAGACTTAAG-3′). The linker-ligated DNA was subsequently subjected to PCR using Eβ-specific oligonucleotide primers internal to that used for the primer extension step and primer LM-PCR.1 and a final two cycles of PCR with a third [γ-32P]ATP-labeled Eβ-specific oligonucleotide primer. The resulting labeled PCR products were resolved on 5% Hydrolink Long Ranger sequencing gels (TEBU, Le Perray en Yvelines, France) and exposed to X-ray film. The primers used for the primer extension, PCR, and labeling steps were, respectively, as follows: bottom strand, βE1.1 (5′-GACCGATTCCATCAAAGAG-3′), βE1.2 (5′-GCAACTGAAGAGATGCATTCCTGGG-3′) and βE1.3 (5′-GAGATGCATTCCTGGGACTTTTCGGTTCC-3′); top strand, βE2.1 (5′-TTAGAGACCCTCCTCTTGG-3′), βE2.2 (5′-GGTGATAGCTAGAGGCTGAGGTAGA-3′), and βE2.3 (5′-AGAGGCTGAGGTAGAAAGGGCTGCATGAG-3′).
RESULTS
Experimental design.
The Eβ560 mouse DNA fragment (Eβ560 [Fig. 1]) has been shown by in vitro footprinting and EMSA to include seven nuclear factor-binding motifs, designated βE1 to βE7 (71). The aim of this study was to define which sequences within Eβ560 are involved in targeting TCRβ variable segments for cis recombination and expression in vivo. Thus, we used a well-characterized transgenic minilocus system in which substrate rearrangements are strictly dependent on the presence of a transcriptional enhancer (19, 66, 74). Briefly, the minilocus (Fig. 1B) is comprised of the unrearranged Vβ14, Dβ1, Jβ1.1, and Jβ1.2 gene segments (and flanking RSSs) in the same transcriptional orientation such that substrate Dβ-to-Jβ and Vβ-to-(D)Jβ rearrangements result in deletion of the corresponding intervening sequences; within these transgenes, the former rearrangements can occur in both T and B cells, whereas the latter are highly restricted to T cells. The TCRβ region is linked to a downstream DNA fragment containing the IgH Cμ gene, thus conferring a hybrid structure to the minilocus that facilitates molecular analyses of the integrated transgenes. Truncated versions of Eβ were inserted between the TCRβ and IgH regions in the minilocus, a location in which the Eβ 560 fragment was previously shown to activate cis recombination and transcription (12, 56). In preliminary experiments using this transgenic approach, we found that a 200-bp fragment (Eβ200) comprised of sequences upstream of the βE5 motif (including the βE1 to βE4 motifs and 80 and 30 bp of 5′ and 3′ flanking sequences, respectively [Fig. 1B]) induces transgene rearrangements, whereas a downstream fragment containing the βE5, βE6, and βE7 motifs (Eβ3′) is essentially inactive (G. Bouvier and P. Ferrier, unpublished data). The present study further dissects the recombinational activity of the 5′ region of Eβ, using several truncated versions of Eβ200 as schematized at the bottom of Fig. 1B.
Recombination activity of the transgenic substrates.
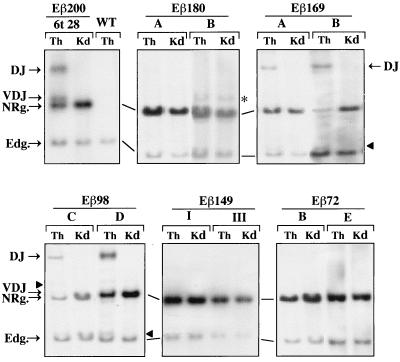
The linearized recombination substrates were used to produce transgenic mice, utilizing standard protocols. The resulting transgenic founders were backcrossed onto WT (C57BL/6 × CBA/J)F1 mice to derive transgenic lines. At least three independent transgenic lines shown to contain from 1 to 18 copies of the intact transgene were established for each construct (Table 1). Substrate rearrangements in the individual lines were first analyzed by Southern blot assay using BglII-restricted genomic DNA from transgenic thymuses and a Vβ14-specific hybridization probe to allow for the identification of site-specific rearrangements within the integrated transgenes, as previously described (19) (see the legends to Fig. 1 and 2). DNA from a nonrearranging tissue (kidney) was used as a negative control. In the case of the Eβ98 substrate, two additional founders (Eβ98 D and Eβ98 E) were sacrificed and similarly analyzed. Overall, these analyses demonstrated substantial differences in the efficiency of V(D)J recombination among the various transgenes, as judged by the presence or absence of thymus-specific hybridizing bands, the sizes of which correspond to predicted rearranged fragments within the transgenic substrates (Fig. 2; results from all transgenic lines are reported in Table 1). Specifically, substrate Dβ-to-Jβ rearrangement was clearly detected in thymus DNA carrying the Eβ200 construct (in three of four lines), the Eβ169 construct (in all four lines), and the Eβ98 construct (in three of five lines) but not in thymus DNA carrying the Eβ180, Eβ149, or Eβ72 construct (three lines in the first two cases; four lines in the latter). When absent from thymus DNA, substrate Dβ-to-Jβ rearrangement was also not detected in DNA from other lymphoid tissues such as the lymph nodes, spleen, and bone marrow (data not shown). Generally, substrate Vβ-to-(D)Jβ rearrangement was barely visible by this assay, including in mice exhibiting substrate Dβ-to-Jβ joints (Fig. 2). This may be due to the fact that Vβ-to-(D)Jβ joining in T-lineage cells is frequently less efficient within this recombination transgene (12, 18, 19, 56), coupled to the related sizes of the unrearranged and Vβ-to-(D)Jβ rearranged hybridizing fragments (Fig. 2, legend; also see the results from PCR analyses reported below).
TABLE 1.
Transgenic mouse lines
| Enhancer fragmenta | Transgenic mouse linesb | Copy no.c | DJ/V(D)J rearrangementd | Jβ1 germ line transcriptione |
|---|---|---|---|---|
| Eβ200 | 3t 15 | 6 | + | + |
| 1t 30 | 7 | + | + | |
| 6t 28 | 8 | + | + | |
| 4t 26 | 1 | − | − | |
| Eβ180 | 5A | 14 | − | + |
| 5B | 5 | − | + | |
| 5C | 13 | − | + | |
| Eβ169 | 3A | 9 | + | + |
| 3B | 2 | + | + | |
| 3C | 2 | + | + | |
| 3D | 1 | + | + | |
| Eβ149 | I | 18 | +/− | +/− |
| II | 9 | − | + | |
| III | 3 | − | + | |
| Eβ98 | 98A | 1 | − | − |
| 98B | 3 | − | − | |
| 98C | 3 | + | + | |
| 98D | + | + | ||
| 98E | + | + | ||
| Eβ72 | 72A | 10 | − | − |
| 72B | 5 | − | +/− | |
| 72D | 10 | − | +/− | |
| 72E | 7 | − | +/− |
Variants of Eβ within the microinjected inserts.
Transgenic mouse lines were established from independent founders and maintained by crosses with normal (C57BL/6 × CBA/J)F1 mice; transgenic animals of the various lines were shown to carry mostly intact copies of the transgene, integrated in a head-to-tail configuration at a single site. Mice 98D and 98E represent transgenic founders for which transgenic lines have not been established.
DNA copy number of microinjected constructs per diploid genome, determined by phosphorimager densitometric analysis of Vβ14-hybridized Southern blots of transgenic kidney DNA and comparison between fragments containing either the transgene or the endogenous Vβ14 gene segment.
Determined by Southern blotting and DNA LR-PCR analysis as detailed in Results. (−), lack of detectable rearrangement by Southern analysis or <1% by LR-PCR; +, presence of DJ rearrangement in Southern assays and of DJ and V(D)J rearrangements in LR-PCR assays; +/−, presence of DJ and/or V(D)J rearrangements in LR-PCR assays at <6% of those found in Eβ200 transgenic mice.
Determined by RNA RT-PCR analysis as detailed in Results. −, lack of detectable Jβ1 transcripts; + and +/−, presence of Jβ1 transcripts at ≥18 and ≤2%, respectively, of those found in Eβ200 transgenic mice.
FIG. 2.
Southern blot analysis of the microinjected constructs in transgenic mouse tissues. Ten micrograms of genomic DNA from thymus (Th) and kidney (Kd) of 4-week-old transgenic mice in the indicated lines and from the thymus of a WT animal was digested with BglII and assayed by Southern blot analysis for hybridization to the [α-32P]dCTP-labeled Vβ14 probe. In the unrearranged substrate, the probe labels a 5.5-kb BglII fragment. Because of deletion of the intervening sequences following V(D)J recombination within the transgene, the probe is expected to label additional fragments of predicted sizes depending on the type of rearrangement (19). The positions of the BglII fragments containing the endogenous Vβ14 gene (Edg., 3.7 kb), the unrearranged substrate (NRg., 5.5 kb), and the Dβ-to-Jβ (DJ) and Vβ-to-DJβ (VDJ) rearrangements are indicated. The predicted sizes of both the DJ and VDJ fragments may vary depending on the length of the inserted Eβ variant (in the range of 128 bp), with the largest Vβ14-hybridizing DJ (9.74-kb) and VDJ (5.74-kb) fragments being seen in the Eβ 200 thymus. Arrowheads indicate substrate rearrangements of lesser abundance which most likely correspond to other types of recombinase-mediated joints, as described previously (19). The asterisk indicates a 6.1-kb hybridizing fragment present in both thymus and kidney DNA from transgenic mouse Eβ180 B, which most likely corresponds to a truncated copy of the transgene.
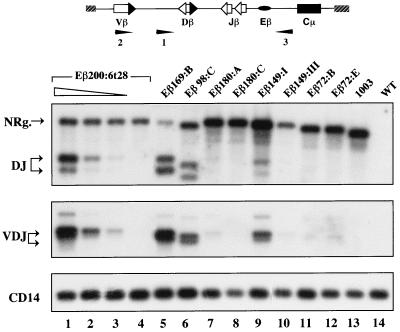
To further analyze V(D)J recombination within the various transgenes and confirm the differential activities of the corresponding Eβ variants in activating rearrangement, we used specific LR-PCR assays as described previously (12). Depending on the oligonucleotide primers used, the assays allow for amplification of transgene fragments carrying either the unrearranged Dβ1, Jβ1.1, and Jβ1.2 segments as well as the rearranged Dβ1-to-Jβ1.1 and Dβ1-to-Jβ1.2 products or the rearranged Vβ14-to-(D)Jβ1.1 and Vβ14-to-(D)Jβ1.2 products, respectively. Representative data are shown in Fig. 3, top and middle panels; overall results are reported in Table 1. Significantly, amplified fragments containing Dβ-to-Jβ or Vβ-to-(D)Jβ rearrangements were readily detected in thymus DNA from transgenic mice in the same Eβ200, Eβ169, and Eβ98 lines that showed substrate recombination in Southern analyses. Conversely, transgene rearrangements were barely visible in thymus DNA from transgenic mice in the Eβ180, Eβ149, and Eβ72 lines (within this group, maximum levels of transgene rearrangements were observed in the 18-copy line Eβ149:I [Fig. 3]). By comparing thymus DNA from transgenic mice of these three types of lines to serial dilutions of thymus DNA from a rearranging Eβ200:6t28 transgenic mouse, we estimated that there was an overall 100-fold reduction in the levels of Dβ-to-Jβ and Vβ-to-(D)Jβ recombination per copy number in the Eβ180, Eβ149, and Eβ72 lines [an 18-fold reduction for Vβ-to-(D)Jβ rearrangement in the particular line Eβ149:I; see the legend to Fig. 3 for calculation]. A similar analysis and calculation indicated that substrate rearrangements in the Eβ200 and Eβ169 lines were roughly equivalent (when copy number is taken into account, the highest level of rearrangement is actually found in line Eβ169:B), whereas they were reduced two- to threefold in the Eβ98 lines compared to the other two. Collectively, these results strongly suggest that when observed, the recombinational activity is not due to the mere polymerization of nuclear factor binding motifs in the transgenic array (the synergistic effect of multimerization of transcription binding sites has been well documented in transfection studies [for example, see reference 15]). Instead, we conclude that a short (98-bp) region of Eβ, comprised of the βE3 and βE4 motifs as well as an additional 30 bp on the 3′ side of βE4, is sufficient to promote V(D)J rearrangement within the integrated minilocus.
FIG. 3.
LR-PCR analysis of the microinjected constructs in transgenic mouse tissues. Fifty nanograms of genomic DNA from thymus (lanes 5 to 13) and kidney (lane 4) of 4-week-old transgenic mice in the indicated lines and from a WT thymus (lane 14) was tested by PCR using substrate-specific oligonucleotide primers (scheme at the top) and a Jβ1.2-specific oligonucleotide probe, as described previously (12). The transgenic mouse in line 1003 carries six copies of the enhancerless minilocus (19). Top panel, hybridization images of DNA LR-PCR-amplified products using a forward primer located upstream of the Dβ1 segment (primer 1) and a reverse primer located on the 5′ side of the IgH region (primer 3) in the transgenic construct to allow the simultaneous amplification of substrate DNA fragments containing either unrearranged (NRg.) or rearranged Dβ and Jβ segments (DJ). Middle panel, hybridization images of DNA LR-PCR-amplified products using a forward primer located in the Vβ14 gene (primer 2) and the reverse primer 3 to allow the amplification of substrate fragments containing rearranged Vβ and Jβ segments (VDJ); the faint band visible in the rearranged samples corresponds to DNA fragments carrying a Vβ-to-Dβ partial rearrangement, as described previously (12). Bottom panel, amplification products of the nonrearranging CD14 gene, used here to control for sample loading. Lanes 1 to 3 show a titration of transgenic thymus DNA in the rearranging line Eβ200:6t28. Input DNA was kept constant (50 ng) by using decreasing amounts of thymus DNA and increasing amounts of kidney DNA. Lane 1, undiluted thymus; lanes 2 and 3, 1-to-10 and 1-to-100 dilutions, respectively. Quantification of the levels of substrate rearrangement per transgene copy in the individual lines was performed by phosphorimager scanning of the hybridizing images and correction for the amount of loaded DNA and the copy number per diploid genome. Phosphorimager analysis of the CD14 hybridization indicated that sample loading varied no more than 2.5-fold.
Transcription activity of the transgenic substrates.
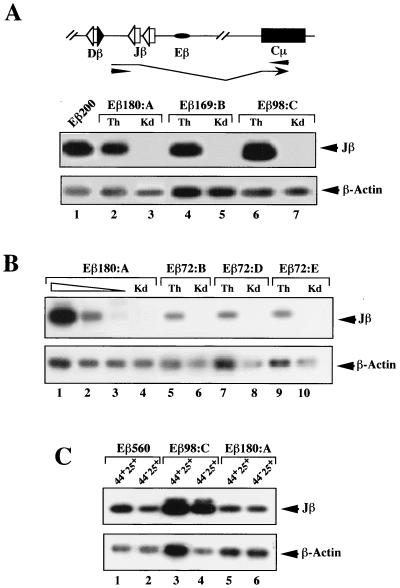
A host of studies, including transgenic analyses of various forms of recombination substrates, have shown that transcription of germ line Ig and TCR gene loci and/or segments tightly correlates with their activation for V(D)J recombination during lymphoid cell development (29, 67). However, a few examples which support a possible dissociation between transcriptional and recombinational activities have been reported (2, 18, 35, 40, 56). Using a previously described RT-PCR assay (12), we analyzed germ line transcription through the unrearranged Jβ segments in the various integrated transgenes. High levels of germ line Jβ transcripts were found in thymus RNA from the rearranging Eβ200, Eβ169, and Eβ98 as well as nonrearranging Eβ180 and Eβ149 transgenic mice (Fig. 4A and Table 1). Among these lines, levels of Jβ transcripts in thymus RNA varied no more than threefold according to quantification analysis (Fig. 4 and data not shown). However, when accounting for the number of nonrearranged copies of the transgene in the different lines (calculated following densitometric scanning of Southern blots of thymus DNA similar to those in Fig. 2), we found that levels of Jβ transcripts from the nonrearranging lines were generally the lowest. For example, Jβ transcription in line Eβ180:A was calculated to represent ∼20% of that found in the Eβ200:6t28 line, whereas levels of transcription in the other two rearranging mouse lines shown in Fig. 4A were somewhat higher (see figure legend for details). Thus, the difference between the levels of germ line transcription between line Eβ180:A and the rearranging lines is relatively modest. A somewhat different picture was found in assays using thymus RNA from the nonrearranging Eβ72 transgenic animals. Although Jβ transcripts could generally be detected, they were produced at significantly lower levels, exhibiting variations between transgenic animals in the same litter, including some that had no detectable Jβ transcription (Fig. 4B, Table 1, and data not shown). Compared to serial dilutions of thymus cDNA from a nonrearranging Eβ180:A transgenic mouse, we estimated the level of Jβ transcripts per transgene copy to be decreased 10- to 25-fold, depending on the Eβ72 line studied (i.e., 0.8 to 2% of Jβ transcription in the Eβ200:6t28 line [Table 1]). These data indicate that all of the truncated Eβ variants tested in this study except Eβ72 are efficient in stimulating Jβ transcription within the integrated transgene.
FIG. 4.
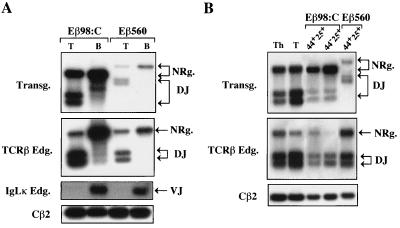
Germ line transcription of the transgenic substrates. An RT-PCR assay (schematized at the top) was used to specifically detect germ line Jβ transcripts from the transgenic substrates (12), starting with 10 ng of total RNA from transgenic thymus (Th) and kidney (Kd) (A and B, lanes 1 to 7 and 4 to 10, respectively) or with total RNA from FACS-purified DN CD44+ CD25+ (44+25+) and CD44− CD25+ (44−25+) thymocytes (C) in the indicated lines. In panel A, lanes 1 shows a transgenic thymus cDNA in the rearranging line Eβ200:6t28. In panel B, lanes 1 to 3 show a titration of transgenic thymus cDNA in the nonrearranging line Eβ180:A. Input cDNA was kept constant. Lane 1, thymus cDNA undiluted; lanes 2 and 3, thymus cDNA diluted 1 to 10 and 1 to 50 in kidney cDNA. Analysis of β-actin transcripts was performed in parallel to control for sample loading. Because primers which crossed intron-exon boundaries were used, contaminating DNA could not account for the hybridizing amplified products. Quantifications were carried out as described for Fig. 3. For the lines shown in panel A, for example, the percentages of Jβ transcripts calculated for the individual lines, after correction for differences in sample loading and the amount of unrearranged substrate, were as follows: Eβ200:6t28, 100%; Eβ180:A, 20%; Eβ169:B, 118%; and Eβ98:C, 145%.
According to the above data, the Eβ180 subfragment activates germ line transcription but not V(D)J recombination within the transgenic array. To verify whether transcription is activated in early-developing thymocytes, we purified DN CD44+ CD25+ and CD44− CD25+ thymocytes from a transgenic mouse of line Eβ180:A by FACS and analyzed their RNA for the presence of Jβ transcripts. Purified thymocytes from transgenic mice of lines Eβ98:C and Eβ560 were tested in parallel (DN CD25+ thymocytes in the Eβ560 line were previously shown to carry transgene recombination [12]). Thus, Jβ transcripts were detected in both CD44+ CD25+ and CD44− CD25+ DN subpopulations from all tested mice (Fig. 4C). In separate analyses using LR-PCR and genomic DNA from the same sorted cells, we have confirmed that substrate Dβ-to-Jβ rearrangements are not found in the Eβ180:A transgenic mouse, in contrast to the Eβ98:C and Eβ560 mice (data not shown). We conclude that substrate Jβ transcription is activated in the Eβ180 thymocytes at a developmental stage where TCRβ gene recombination normally takes place.
Effect of transgene integration on enhancer activity.
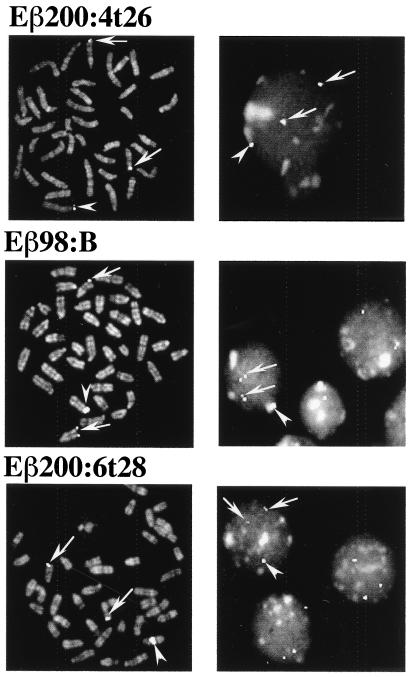
The integration of transgenes into the mouse germ line following pronuclear injection of DNA usually results in an array of tandemly repeated, head-to-tail copies at a single chromosomal site. Depending on the site of chromosomal integration, side effects which generally have a negative effect on transgene expression can result (48). For example, it has been suggested that silencing of transgene expression could be a consequence of the spreading of centromeric heterochromatin within the nearby integrated transgenic array, a mechanism analogous to position effect variegation (PEV) in drosophila (7, 16, 21). Among the several Eβ reporter transgenes which we found to rearrange and/or be expressed in a majority of independent transgenic lines, there were a few in which both transgene recombination and transcription could not be detected (e.g., Table 1, lines Eβ200:4t26, Eβ98:A, and Eβ98:B). Since the copy number was stable and there was no evidence of structural alteration of the transgenes within these lines (R. K. Tripathi, M.-G. Mattei, and P. Ferrier, unpublished data), it seemed possible that the transgenic arrays had been silenced for expression following integration within or close to chromosomal centromeres. To test this hypothesis, we performed FISH analysis on spread chromosomes from ConA-stimulated splenocytes in transgenic animals of the nonrearranging Eβ200:4t26 and Eβ98:B lines, using as a probe the IgH Cμ DNA fragment (∼10 kb) present on the 3′ side of the recombination substrates. The rearranging lines Eβ200:6t28 and Eβ98:C were similarly analyzed. Under these conditions, the probe is expected to hybridize to the endogenous IgH loci, thus labeling the telomeric region of the two homologous chromosomes 12, as well as to the transgenic array, leading to an additional spot located on an unpredictable chromosome. In agreement with our hypothesis, we found that the transgenic arrays in lines Eβ200:4t26 and Eβ98:B are located within the centromeric region of, respectively, a large and a medium-sized chromosome (Fig. 5, left panels). Significantly, in both cases, a signal (corresponding presumably to the transgenic substrates) is associated with heterochromatin visible as condensed areas in interphase nuclei (Fig. 5, right panels). In marked contrast, the transgenic signal in line Eβ200:6t28 is located in the middle part of a medium-sized chromosome, distant from any possible effect of centromeric heterochromatin (bottom panels); a similar image of chromosomal midarm integration was observed in transgenic splenocytes from line Eβ98:C (data not shown). As expected, in all cases, fluorescent signals are visible at the distal end of both chromosomes 12, corresponding to the endogenous IgH loci.
FIG. 5.
Localization of IgH Cμ sequences by chromosomal FISH analysis to metaphase chromosomes (left panels) and interphase nuclei (right panels) from ConA-stimulated splenocytes of hemizygous transgenic mice. The top and middle panels are from the nonrearranging lines Eβ200:4t26 and Eβ98:B; the bottom panels are from the rearranging line Eβ200:6t28. The biotinylated IgH Cμ probe was revealed with avidin-FITC. Arrows indicate locations of the endogenous IgH Cμ locus at the distal end of chromosomes 12; arrowheads indicate locations of transgenic areas in the various lines. Chromosomes and nuclei were counterstained with propidium iodide.
Eβ98 confers lineage and temporal specificity to transgene recombination.
The minimal TCRβ enhancer fragment that was found to activate transcription and recombination within the transgenic substrates integrated at sites distant from centromeric heterochromatin was Eβ98. To test whether this element does so in a lineage- and development stage-restricted manner, we analyzed Dβ-to-Jβ rearrangement by LR-PCR using genomic DNA prepared from sorted populations of T and B peripheral lymphocytes or DN CD44+ CD25+ and DN CD44− CD25+ thymocytes from transgenic mice of line Eβ98:C. Genomic DNA from the corresponding cells purified from Eβ560 transgenic mice was similarly tested. Previously, we found that transgene rearrangement in this line is T-cell specific and occurs early during T-cell ontogeny and differentiation (12). Significantly, fragments containing substrate Dβ-to-Jβ rearrangements were found to predominate in purified T compared to B cells from the Eβ98:C mouse, similar to the recombination profiles observed with a transgenic mouse from line Eβ560 (Fig. 6A, top panel). Additional LR-PCR assays to analyze fragments containing endogenous Dβ-to-Jβ and Vκ-to-Jκ rearrangements confirmed the T- and B-lineage origin of the sorted cells (Fig. 6A, middle panels). Moreover, Dβ-to-Jβ recombination products were detected in purified DN CD25+ thymocytes from both Eβ98 and Eβ560 mice, again with profiles related to those observed for endogenous TCRβ gene rearrangement (Fig. 6B). Together with our finding of germ line Jβ transcripts in DN thymocytes from the Eβ98:C mice (see above), these data strongly suggest that Eβ98 represents a true TCRβ core enhancer for recombination, as it appears to confer appropriate cell lineage and temporal specificity.
FIG. 6.
(A) Lineage and (B) developmental stage specificity of transgene rearrangement in line Eβ98:C. Lymphoid cells were purified by cell sorting from individual transgenic mice of lines Eβ98:C and Eβ560, as indicated. The sorted cells were >98.5% pure as determined by FACS reanalysis. Genomic DNA prepared from the indicated purified cell populations was analyzed by LR-PCR to detect transgene Dβ-to-Jβ rearrangements (A and B, top panels), endogenous Dβ-to-Jβ rearrangements (A and B, second panels from the top), and endogenous Vκ-to-Jκ rearrangements (A, third panel from the top). Amplification of fragments containing endogenous Cβ2 sequences was carried out to control for sample loading (A and B, bottom panel). Unrearranged and rearranged TCRβ-hybridizing fragments are labeled as in Fig. 3A. VJ, VκJκ-containing fragment. The sorted cells analyzed were peripheral T and B cells (T and B, respectively) (A) and DN CD44+ CD25+ and CD44− CD25+ thymocytes (44+25+ and 44−25+, respectively) (B). In panel B, DNA from thymocytes (Th) and peripheral T cells (T) of a transgenic mouse in line Eβ98:C were used as controls.
Developing thymocytes show nuclear factor occupancy within Eβ98.
The Eβ98 core enhancer defined in this study contains the previously identified βE3 and βE4 nuclear factor binding sites as well as additional sequences 3′ of βE4. βE3 contains a typical E-box motif, while βE4 has been shown to be comprised of a composite Ets-CBF binding site (36, 70, 71, 78). Moreover, examination of the βE4 3′ flanking sequences by using the TRANSFAC database of transcription factor binding sites (76, 77) revealed that this region also contains an E-box motif (Fig. 7). To test whether the Eβ98 element is bound by nuclear factors in vivo, we analyzed enhancer occupancy within the endogenous TCRβ locus by genomic footprinting using LM-PCR (52). This technique can reveal nuclear factor binding in chromosomal DNA following comparison of LM-PCR products between DNA which is treated in vivo with the membrane-permeable DNA-methylating agent DMS and that treated in vitro after extraction (thus generating the complete pattern of G residues across the given region). This is visualized as either protected G's or hypersensitive G's and A's for the in vivo versus in vitro DMS-treated DNA. The LM-PCR strategy was adapted to analyze the 250-nucleotide (nt) region spanning from 5′ of βE1 to 3′ of βE7. In addition to WT thymocytes, we analyzed thymocytes from RAG−/− mice. Whereas the former are comprised of mostly DP cells carrying a productively rearranged TCRβ locus, the latter are made up of DN cells arrested in development at a stage where TCRβ gene rearrangement normally takes place (e.g., CD25+) (37). The endogenous locus was chosen for this analysis instead of the transgenic loci because of technical difficulties encountered in specifically amplifying the Eβ region from the flanking A-T-rich IgH sequences in the integrated substrates (R. K. Tripathi, D. Payet, S. Spicuglia, W. M. Hempel, and P. Ferrier, unpublished data).
FIG. 7.
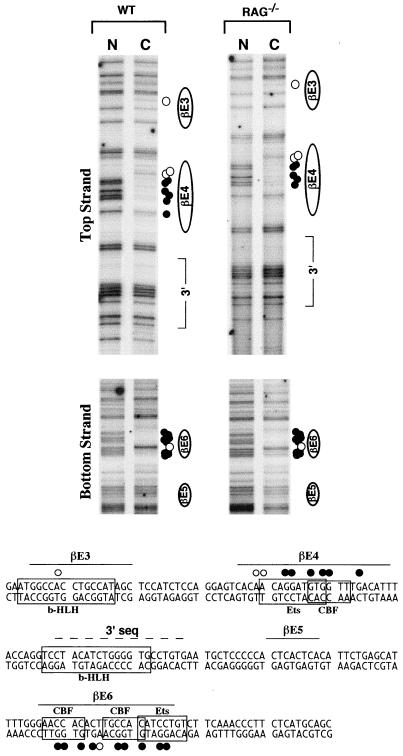
Analysis of in vivo occupancy of Eβ in primary thymocytes. Thymocyte DNA from a (WT) mouse and from 10 RAG−/− mice was methylated with DMS either as naked DNA in vitro (N) or as chromosomal DNA in intact cells in vivo (C). Methylated DNA samples were treated with piperidine and subjected to LM-PCR using Eβ-specific primers. The top and bottom panels show footprints for the top and bottom strands, respectively. The corresponding sequence is shown below. Protected guanines are indicated by filled circles, while hypersensitive adenines are indicated by open circles to the right of the panels and above the top strand and below the bottom strand of the sequence shown. Positions of known protein binding sites within the analyzed Eβ sequences, as well as the location of the element defined in this study 3′ of the βE4 motif, are indicated. Only the sequences showing in vivo occupancy within the 250-nt Eβ region studied are shown.
Comparison of the LM-PCR products between in vitro and in vivo DMS-treated DNA across the 250-nt Eβ region demonstrated one strong footprint for both top and bottom strands (Fig. 7). For the top strand, there was strong occupancy at βE4 visualized as a series of protected G's and several hypersensitive A's for both WT and RAG−/− thymocytes (Fig. 7, top panel). As there are no G's in the βE4 motif on the bottom strand, there was no apparent footprint in this region (data not shown). On this strand, however, there was evidence of strong occupancy of βE6, outside the Eβ98 region, visualized as a series of protected G's flanking a hypersensitive A (Fig. 7, lower panel). In addition to the strong occupancy observed at βE4 and βE6, there was also evidence for occupancy at βE3, suggested by the presence of a single hypersensitive A within this element on the top strand. Finally, there was no evidence of nuclear factor binding throughout the rest of Eβ, including the sequence 3′ of βE4 (Fig. 7 and data not shown). These results demonstrate that the βE3 and βE4 sites within the Eβ98 core are occupied in vivo, in agreement with our functional data. In addition, the results strongly suggest that (i) nuclear factor occupancy at Eβ is established in DN cells (as represented by RAG−/− thymocytes) and (ii) this occupancy is not significantly modified in more developed DP cells (as represented by WT thymocytes).
DISCUSSION
Previous genetic analyses demonstrate a crucial role of Eβ in regulating TCRβ gene recombination and, in addition, suggest a possible dual function for the control of both chromatin structure and V(D)J recombination at the TCRβ gene locus (5, 6, 12, 30, 56). These functions are most likely mediated by the combined action of nuclear factors bound to Eβ in developing T cells. Provocatively, several of the identified Eβ-binding factors also interact with Ig and other TCR enhancers (39, 42), which were similarly demonstrated to affect V(D)J recombination at their own loci (29, 67). Despite these findings, however, gene-targeting studies have so far failed to reveal an indisputable role for the relevant factors in regulating V(D)J recombinational accessibility (14). This may be due to either a redundancy between members of a given transcription factor family or a pleiotropic role of the inactivated factor in cell differentiation and/or survival. The definition of minimal cis-regulatory elements required to modulate the recombination of linked gene segments may, in addition to providing molecular clues as to the mechanism(s) involved, constitute an alternative approach to address the issue of which trans-acting factors are critical for this process.
We have shown that a 98-bp domain from the 560-bp Eβ-containing fragment, comprised of two well-defined nuclear factor binding motifs (βE3 and βE4) and a 30-bp flanking sequence 3′ of βE4, is sufficient to promote germ line transcription and V(D)J recombinase-mediated rearrangements within an enhancer-dependent recombination substrate integrated into the mouse genome in several transgenic mouse lines. An overlapping 72-bp region that lacks the upstream βE3 motif was found to be uniformly inactive when tested in this system, whereas longer Eβ subfragments containing additional βE1 and βE2 motifs but lacking most of the sequences 3′ of βE4 (e.g., Eβ180 and Eβ149; both subfragments contain only 10 bp 3′ of βE4) were relatively efficient in activating germ line transcription but not recombination. These data indicate that the Eβ98 domain represents a TCRβ core enhancer for V(D)J recombination and point toward a novel Eβ sequence that plays a critical role in this process. The implications of these results, in terms of the molecular mechanisms and nuclear factors involved, are discussed below.
The V(D)J recombination-enhancing Eβ subfragments identified in this study, including the Eβ98 core, promoted rearrangement and germ line transcription from the integrated transgenes in several independent transgenic mouse lines, in conformity with the correlation between the two activities generally observed at antigen receptor gene loci (20, 67). In a few instances, however, we found that some Eβ subfragments (e.g., Eβ200 and Eβ98) were not active after transgene integration within or close to a chromosomal centromere. Most likely, the subfragments could not antagonize the repressive effect of centromeric heterochromatin structures. In a study investigating the function of two enhancer elements, the 5′ HS2 enhancer from the human β-globin locus and the metallothionein enhancer, it was found that instead of regulating the level of expression of a linked reporter cassette, both enhancers act in cis to protect constructs from repression by flanking chromatin and to suppress PEV (72). Strikingly, however, the strength of this effect was shown to depend on the site of integration, indicating that as in PEV, chromatin varies in its ability to repress activity in a given gene unit. These data support a model in which enhancers (and their bound factors) directly interact with repressive chromatin structures to permit and stabilize expression rather than to increase the rate of transcription. While Eβ activity of the subfragments is suppressed by centromeric heterochromatin, the possibility remains that it can overcome the repressive effects of chromatin regions characteristic of highly regulated loci, such as the TCRβ locus, as opposed to those characteristic of housekeeping genes, somewhat analogous to the situation just described. We anticipate a similar mode of action for other recombination enhancers; already, the Ig μ enhancer core has been shown to establish factor access in chromatin independent of transcriptional stimulation (34).
Unexpectedly, early-programmed Jβ transcription (e.g., in DN CD25+ thymocytes) was detected within transgenes carrying the rearrangement-defective Eβ180 subfragment, suggesting that this element could also confer chromatin accessibility to the transgenic templates in a significant proportion of cells undergoing TCRβ gene rearrangement. How then could the V(D)J recombinase apparently ignore the homologous gene segments in the particular transgenes? A trivial explanation would be that significant differences in accessibility still exist between rearranging and nonrearranging loci, despite the modest difference in germ line transcription, and/or that a higher threshold of chromatin access is required to permit recombination as opposed to germ line transcription. Alternatively, germ line transcription in the Eβ180-containing transgenes may be qualitatively inadequate, for example, not being initiated at the promoter upstream of Dβ1, an element which has been shown by gene targeting to be required for rearrangement of this gene segment (75). Finally, in line with our recent report of unresolved DSBs at RSSs from enhancer-deleted TCRβ alleles, it is also possible that specific nucleoprotein complexes organized at Eβ regulatory elements may affect the V(D)J recombination process at a step beyond that of chromatin access (30). Of note, two potential functions of Eβ, increasing chromatin accessibility and affecting directly the course of the recombination reaction, may not be mutually exclusive. If this were the case, it would provide an explanation for the apparent paradox of Jβ transcription in the absence of recombination in the Eβ180 transgenic mouse lines. Current analyses of recombination intermediates and germ line transcripts in lymphoid cells from transgenic mouse models, including some established on a RAG−/− background, may help us to distinguish among these possibilities. In any case, our data add to the increasing experimental evidence concerning transgenic and endogenous TCR loci, suggesting that transcription per se is not sufficient to permit rearrangement of the given gene segments (5, 18, 45, 56).
It is believed that all lymphoid cells are derived from a common lymphoid cell progenitor (65). As they differentiate, the developing lymphocytes progressively lose their multilineage potential. In the rearranging Eβ98:C line, transgene Dβ-to-Jβ rearrangements were found to predominate in T as opposed to B lymphocytes. Accordingly, these products were readily observed in DN CD44+ CD25+ thymocytes, a subpopulation which contains αβ (and γδ) T-cell precursors but can no longer give rise to B cells (65). Overall, transgene rearrangement profiles within the Eβ98:C line (as well as within the Eβ560 line [Fig. 6 and reference 12]) mimic those at the endogenous TCRβ locus (11, 24, 46). Coupled to the fact that the reporter transgenes in the Eβ98:C and Eβ560 lines are located on different chromosomes (Tripathi, Mattei, and Ferrier, unpublished data) thus presumably within independent genomic contexts, these data strongly suggest that the regulatory elements within the Eβ core act positively in regulating cis recombination, independently of the action of adjacent motifs outside the core domain. Interestingly, also in a study using a transgenic approach, it was found that rearrangements within substrates carrying DNA fragments containing the full-length TCRα enhancer (Eα) are appropriately timed (e.g., with respect to those at the endogenous TCR-Jα locus) and strictly limited to the αβ T lineage during thymocyte differentiation (12, 41). In contrast, those under the control of a shorter core Eα fragment initiate slightly earlier during ontogeny and occur in both αβ and γδ T cells (60). Because some nuclear factor binding motifs are shared by the Eβ and Eα cores, it is tempting to speculate, based on our results as well as those of Roberts et al. (60), that nucleoprotein complexes comprised of the corresponding factors could enhance cis recombination in early T cells, this activity at the TCR-Jα locus being delayed by the combinatory effect of factors bound outside the Eα core. Contrary to our results, however, equivalent levels of Dβ-to-Jβ junctions in both T and B cells within Eβ560-containing transgenes have been reported (56). We note that among the four types of transgenic mice analyzed, two were of high copy number (25 and 60 copies), a fact which could lead to the dysregulation of cell lineage specificity in V(D)J recombination (9). One way to reconcile these data would be that Eβ560 has the capacity to enhance cis recombination in both T and B cells, but in the latter case less efficiently and only occasionally, an assumption which would parallel the differential transcriptional efficiency of related fragments following transfection into lymphoid T- and B-cell lines (25, 71).
The Eβ98 subfragment identified here as a core enhancer of V(D)J recombination exhibits several interesting characteristics. Extensive (90%) homology is found between the mouse and human sequences over the entire fragment, including the 3′ flanking sequences shown to be strictly required for the activity, whereas overall homology is less conserved (<70%) outside this particular domain (58; Tripathi, Payet, et al., unpublished data). In addition, within this subfragment, the βE3 and βE4 sites that have been shown to interact with nuclear factors from early developing thymocytes following EMSA and in vitro footprinting analyses (71; Tripathi, Payet, et al., unpublished data) as well as in vivo genomic footprinting (this study) overlap with, respectively, a bHLH-binding E-box motif and the Ets-CBF composite motif, both of which are commonly found in antigen receptor gene enhancers (17, 23, 59). Notably, a direct role for bHLH factors on the regulation of V(D)J recombination in T lymphocytes is supported by several studies (3, 64), including, in the case of the TCRβ locus, the finding that enforced expression of Id3 (which inhibits many bHLH transcription factors) in cultured human thymocytes prevents the appearance of cells with Dβ-to-Jβ rearrangements (28). Interestingly, another typical E box is also present within the most 3′ 20-bp flanking sequences of Eβ98 (bottom of Fig. 7). Although we have not been able to definitively confirm binding at this site by using conventional DNA-protein interaction assays, we have obtained evidence suggesting that when present, the 3′ sequences modify binding at the βE4 site, leading to the formation of a distinct nucleoprotein complex(es) (Tripathi, Payet, et al., unpublished data). One attractive possibility would be that the E box within the 3′ sequence, along with another site(s) (βE3 and/or βE4), contributes to a bipartite DNA motif that nucleates a multimeric complex in developing T cells, as recently described in a study of nucleoprotein structures involving the LIM-only protein Lmo2 (27). Alternatively, the 3′ sequence may help to stabilize the interaction of cofactors which bind to the minimal enhancer. Intriguingly, within the TCRα enhancer has been found a sequence which, in the absence of demonstrable factor binding, is required for the formation of a ternary complex involving a coactivator (ALY) and a bound factor (8). Further analyses in our lab will focus on a deeper characterization of the molecular complexes identified at Eβ98 and their putative role in regulating V(D)J cis recombination.
Indirectly, our results suggest that the Eβ motifs outside of Eβ98 may play a role during early T-cell differentiation distinct from activating recombination and/or at a latter stage of T-cell life, for example, during T-cell activation. In particular, this may be the case for the motif in βE1 which was proposed to represent the main GATA-3 factor binding site within Eβ (31). Indeed, expression of GATA-3 has been found to be down-modulated during periods of TCR gene recombination in thymocytes (32), making it unlikely that this factor could contribute directly to regulation of the TCRβ rearrangement process. More intriguing, however, is the function of the second Ets- and CBF-interacting element within βE6 which, like that in βE4, is heavily occupied in early thymocytes (Fig. 7). These elements have been shown to be responsible for the inducibility of the TCRβ enhancer in response to phorbol ester treatment, which mimics the signals for a range of cellular changes associated with T-cell differentiation and activation (58, 59). It is, therefore, possible that each of the two motifs plays a unique role in enhancer function, the βE4 site being more closely involved with modulating recombinational accessibility. It is equally possible, however, that these two elements act cooperatively to ensure the chromatin-opening activity of Eβ, the βE6 site being, for example, required for maximal efficiency as proposed for other types of cis-regulatory elements (21). These issues are currently being addressed by inserting Eβ subfragments at the endogenous TCRβ locus.
ACKNOWLEDGMENTS
We thank M. Algarté, J. Imbert, and P. Rameil for advice on in vivo genomic footprinting assays, C. Beziers La Fosse for preparing the artwork, N. Brun-Roubereau for helping in FACS sorting, and A. Loussif, M. Pontier, and G. Warcollier for maintaining the mouse colonies.
This work was supported by institutional grants from INSERM and CNRS and by specific grants from the Association pour la Recherche sur le Cancer, the Commission of the European Communities, the Fondation Princesse Grace de Monaco, the Ligue Nationale Contre le Cancer, and Rhone-Poulenc Pharmaceuticals (to P.F.). R.K.T. was a fellow of the Ministère des Affaires Etrangères and is now a fellow of the Fondation pour la Recherche Médicale. W.M.H. is the recipient of a Foreign Associate Scientist position from the CNRS.
REFERENCES
- 1.Algarté M, Lecine P, Costello R, Plet A, Olive D, Imbert J. In vivo regulation of interleukin-2 receptor α gene transcription by the coordinated binding of constitutive and inducible factors in human primary T cells. EMBO J. 1995;14:5060–5072. doi: 10.1002/j.1460-2075.1995.tb00188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez J D, Anderson S J, Loh D Y. V(D)J recombination and allelic exclusion of a TCR β-chain minilocus occurs in the absence of a functional promoter. J Immunol. 1995;155:1191–1202. [PubMed] [Google Scholar]
- 3.Bain G, Romanow W J, Albers K, Havran W L, Murre C. Positive and negative regulation of V(D)J recombination by the E2A proteins. J Exp Med. 1999;189:289–300. doi: 10.1084/jem.189.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogue M A, Roth D B. Mechanism of V(D)J recombination. Curr Biol. 1996;8:175–180. doi: 10.1016/s0952-7915(96)80055-0. [DOI] [PubMed] [Google Scholar]
- 5.Bories J C, Demengeot J, Davidson L, Alt F W. Gene-targeted deletion and replacement mutations of the T-cell receptor β-chain enhancer: the role of enhancer elements in controlling V(D)J recombinational accessibility. Proc Natl Acad Sci USA. 1996;93:7871–7876. doi: 10.1073/pnas.93.15.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouvier G, Watrin F, Naspetti M, Verthuy C, Naquet P, Ferrier P. Deletion of the mouse T-cell receptor β gene enhancer blocks αβ T-cell development. Proc Natl Acad Sci USA. 1996;93:7877–7881. doi: 10.1073/pnas.93.15.7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer O, Zhao J C, Cohen J L, Depetris D, Yagello M, Lejeune L, Bruel S, Mattéi M G, Klatzmann D. Position-dependent variegation of a CD4 minigene with targeted expression to mature CD4+ T cells. J Immunol. 1997;159:3383–3390. [PubMed] [Google Scholar]
- 8.Bruhn L, Munnerlyn A, Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 9.Bucchini D, Reynaud C-A, Ripoche M-A, Grimal H, Jami J, Weill J C. Rearrangement of a chicken immunoglobulin gene occurs in the lymphoid lineage of transgenic mice. Nature. 1987;326:409–411. doi: 10.1038/326409a0. [DOI] [PubMed] [Google Scholar]
- 10.Capone M, Curnow J, Bouvier G, Ferrier P, Horvat B. T cell development in TCRαβ transgenic mice: analysis using V(D)J recombination substrates. J Immunol. 1995;10:5165–5172. [PubMed] [Google Scholar]
- 11.Capone M, Hockett R D, Jr, Zlotnik A. Kinetics of T cell receptor β, γ, and δ rearrangements during adult thymic development: T cell receptor rearrangements are present in CD44+CD25+ pro-T thymocytes. Proc Natl Acad Sci USA. 1998;95:12522–12527. doi: 10.1073/pnas.95.21.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capone M, Watrin F, Fernex C, Horvat B, Krippl B, Scollay R, Ferrier P. TCRβ and TCRα gene enhancers confer tissue- and stage-specificity on V(D)J recombination events. EMBO J. 1993;12:4335–4346. doi: 10.1002/j.1460-2075.1993.tb06118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cedar H, Bergman Y. Developmental regulation of immune system gene rearrangement. Curr Opin Immunol. 1999;11:64–69. doi: 10.1016/s0952-7915(99)80012-0. [DOI] [PubMed] [Google Scholar]
- 14.Clevers H, Ferrier P. Transcriptional control during T-cell development. Curr Opin Immunol. 1998;10:166–171. doi: 10.1016/s0952-7915(98)80245-8. [DOI] [PubMed] [Google Scholar]
- 15.Dang W, Nikolajczyk B S, Sen R. Exploring functional redundancy in the immunoglobulin μ heavy-chain gene enhancer. Mol Cell Biol. 1998;18:6870–6878. doi: 10.1128/mcb.18.11.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobie K W, Lee M, Fantes J A, Graham E, Clark A J, Springbett A, Lathe R, McClenaghan M. Variegated transgene expression in mouse mammary gland is determined by the transgene integration locus. Proc Natl Acad Sci USA. 1996;93:6659–6664. doi: 10.1073/pnas.93.13.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erman B, Cortes M, Nikolajczyk B S, Speck N A, Sen R. ETS-core binding factor: a common composite motif in antigen receptor gene enhancers. Mol Cell Biol. 1998;18:1322–1330. doi: 10.1128/mcb.18.3.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernex C, Capone M, Ferrier P. The V(D)J recombinational and transcriptional activities of the immunoglobulin heavy-chain intronic enhancer can be mediated through distinct protein-binding sites in a transgenic substrate. Mol Cell Biol. 1995;15:3217–3226. doi: 10.1128/mcb.15.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrier P, Krippl B, Blackwell T K, Furley A, Suh H, Winoto A, Cook W D, Hood L, Costantini F, Alt F W. Separate elements control DJ and VDJ rearrangement in a transgenic recombination substrate. EMBO J. 1990;9:117–125. doi: 10.1002/j.1460-2075.1990.tb08087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrier P, Krippl B, Furley A J, Blackwell T K, Suh H, Mendelsohn M, Winoto A, Cook W D, Hood L, Costantini F. Control of VDJ recombinase activity. Cold Spring Harbor Symp Quant Biol. 1989;54:191–202. doi: 10.1101/sqb.1989.054.01.024. [DOI] [PubMed] [Google Scholar]
- 21.Festenstein R, Tolaini M, Corbella P, Mamalaki C, Parrington J, Fox M, Miliou A, Jones M, Kioussis D. Locus control region function and heterochromatin-induced position effect variegation. Science. 1996;271:1123–1125. doi: 10.1126/science.271.5252.1123. [DOI] [PubMed] [Google Scholar]
- 22.Gellert M. Recent advances in understanding V(D)J recombination. Adv Immunol. 1997;64:39–64. doi: 10.1016/s0065-2776(08)60886-x. [DOI] [PubMed] [Google Scholar]
- 23.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCRα enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 24.Godfrey D I, Kennedy J, Mombaerts P, Tonegawa S, Zlotnik A. Onset of TCR-β gene rearrangement and role of TCR-β expression during CD3−CD4−CD8− thymocyte differentiation. J Immunol. 1994;152:4783–4792. [PubMed] [Google Scholar]
- 25.Gottschalk L R, Leiden J M. Identification and functional characterization of the human T-cell receptor β gene transcriptional enhancer: common nuclear proteins interact with the transcriptional regulatory elements of the T-cell receptor α and β genes. Mol Cell Biol. 1990;10:5486–5495. doi: 10.1128/mcb.10.10.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grawunder U, West R B, Lieber M R. Antigen receptor gene rearrangement. Curr Opin Immunol. 1998;10:172–173. doi: 10.1016/s0952-7915(98)80246-x. [DOI] [PubMed] [Google Scholar]
- 27.Grutz G G, Bucher K, Lavenir I, Larson T, Larson R, Rabbitts T H. The oncogenic T cell LIM-protein Lmo2 forms part of a DNA-binding complex specifically in immature T cells. EMBO J. 1998;17:4594–4605. doi: 10.1093/emboj/17.16.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heemskerk M H, Blom B, Nolan G, Stegmann A P, Bakker A Q, Weijer K, Res P C, Spits H. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J Exp Med. 1997;186:1597–1602. doi: 10.1084/jem.186.9.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hempel W M, Leduc I, Mathieu N, Tripathi R K, Ferrier P. Accessibility control of V(D)J recombination: lessons from gene targeting. Adv Immunol. 1998;69:309–352. doi: 10.1016/s0065-2776(08)60610-0. [DOI] [PubMed] [Google Scholar]
- 30.Hempel W M, Stanhope-Baker P, Mathieu N, Huang F, Schlissel M S, Ferrier P. Enhancer control of V(D)J recombination at the TCRβ locus: differential effects on DNA cleavage and joining. Genes Dev. 1998;12:2305–2317. doi: 10.1101/gad.12.15.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henderson A J, McDouglas S, Leiden J M, Calame K L. GATA elements are necessary for the activity and tissue specificity of the T-cell receptor beta-chain transcriptional enhancer. Mol Cell Biol. 1994;14:4286–4294. doi: 10.1128/mcb.14.6.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendriks R W, Nawijn M C, Engel J D, van Doorninck H, Grosveld F, Karis A. Expression of the transcription factor GATA-3 is required for the development of the earliest T cell progenitors and correlates with stages of cellular proliferation in the thymus. Eur J Immunol. 1999;29:1912–1918. doi: 10.1002/(SICI)1521-4141(199906)29:06<1912::AID-IMMU1912>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 33.Ismaili J, Antica M, Wu L. CD4 and CD8 expression and T cell antigen receptor gene rearrangement in early intrathymic precursor cells. Eur J Immunol. 1996;26:731–737. doi: 10.1002/eji.1830260402. [DOI] [PubMed] [Google Scholar]
- 34.Jenuwein T, Forrester W C, Qiu R-G, Grosschedl R. The immunoglobulin μ enhancer core establishes local factor access in nuclear chromatin independent of transcriptional stimulation. Genes Dev. 1993;7:2016–2032. doi: 10.1101/gad.7.10.2016. [DOI] [PubMed] [Google Scholar]
- 35.Kallenbach S, Babinet C, Pournin S, Cavelier P, Goodhardt M, Rougeon F. The intronic immunoglobulin κ gene enhancer acts independently on rearrangement and transcription. Eur J Immunol. 1993;23:1917–1921. doi: 10.1002/eji.1830230828. [DOI] [PubMed] [Google Scholar]
- 36.Kim W Y, Sieweke M, Ogawa E, Wee H J, Englmeier U, Graf T, Ito Y. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J. 1999;18:1609–1620. doi: 10.1093/emboj/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kisielow P, Von Boehmer H. Development and selection of T cells: facts and puzzles. Adv Immunol. 1995;58:87–208. doi: 10.1016/s0065-2776(08)60620-3. [DOI] [PubMed] [Google Scholar]
- 38.Krimpenfort P, De Jong R, Uematsu Y, Dembic Z, Ryser S, Von Boehmer H, Steinmetz M, Berns A. Transcription of T cell receptor β-chain genes is controlled by a downstream regulatory element. EMBO J. 1988;7:745–750. doi: 10.1002/j.1460-2075.1988.tb02871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuo C T, Leiden J M. Transcriptional regulation of T lymphocyte development and function. Annu Rev Immunol. 1999;17:149–197. doi: 10.1146/annurev.immunol.17.1.149. [DOI] [PubMed] [Google Scholar]
- 40.Lauster R, Reynaud C-A, Martensson L, Peter A, Bucchini D, Jami J, Weill J C. Promoter, enhancer and silencer elements regulate rearrangement of an immunoglobulin transgene. EMBO J. 1993;12:4615–4623. doi: 10.1002/j.1460-2075.1993.tb06150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lauzurica P, Krangel M S. Temporal and lineage-specific control of T cell receptor αandδ enhancers. J Exp Med. 1994;179:1913–1921. doi: 10.1084/jem.179.6.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leiden J M. Transcriptional regulation of T cell receptor genes. Annu Rev Immunol. 1993;11:539–570. doi: 10.1146/annurev.iy.11.040193.002543. [DOI] [PubMed] [Google Scholar]
- 43.Lemieux N, Dutrillaux B, Viegas-Pequignot E. A simple method for simultaneous R- or G-banding and fluorescence in situ hybridization of small single-copy genes. Cytogenet Cell Genet. 1992;59:311–312. doi: 10.1159/000133277. [DOI] [PubMed] [Google Scholar]
- 44.Lewis S M. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:29–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 45.Livak F, Schatz D G. Alternative splicing of rearranged T cell receptor δ sequences to the constant region of the α locus. Proc Natl Acad Sci USA. 1998;95:5694–5699. doi: 10.1073/pnas.95.10.5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Livak F, Tourigny M, Schatz D G, Petrie H T. Characterization of TCR gene rearrangements during adult murine T cell development. J Immunol. 1999;162:2575–2580. [PubMed] [Google Scholar]
- 47.Marine J, Winoto A. The human enhancer binding GATA-3 binds to several T-cell receptor regulatory elements. Proc Natl Acad Sci USA. 1991;88:7284–7288. doi: 10.1073/pnas.88.16.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin D I K, Whitelaw E. The vagaries of variegating transgenes. Bioessays. 1996;18:919–923. doi: 10.1002/bies.950181111. [DOI] [PubMed] [Google Scholar]
- 49.Matsuda Y, Harada Y N, Natsuume-Sakai S, Lee K, Shiomi T, Chapman V M. Location of the mouse complement factor H gene (cfh) by FISH analysis and replication R-banding. Cytogenet Cell Genet. 1992;61:282–285. doi: 10.1159/000133423. [DOI] [PubMed] [Google Scholar]
- 50.McDougall S, Peterson C L, Calame K. A transcription enhancer 3′ of Cβ2 in the T cell receptor β locus. Science. 1988;241:205–208. doi: 10.1126/science.2968651. [DOI] [PubMed] [Google Scholar]
- 51.Messier H, Brickner H, Gaikwad J, Fotedar A. A novel POU domain protein which binds to the T-cell receptor β enhancer. Mol Cell Biol. 1993;11:5450–5460. doi: 10.1128/mcb.13.9.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mueller P R, Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 53.Nelsen B, Tian G, Erman B, Gregoire J, Maki R, Graves B, Sen R. Regulation of lymphoid-specific immunoglobulin μ heavy chain gene enhancer by ETS-domain proteins. Science. 1993;261:82–86. doi: 10.1126/science.8316859. [DOI] [PubMed] [Google Scholar]
- 54.Oettinger M A. V(D)J recombination: on the cutting edge. Curr Opin Cell Biol. 1999;11:325–329. doi: 10.1016/S0955-0674(99)80044-1. [DOI] [PubMed] [Google Scholar]
- 55.Okada A, Alt F W. Mechanisms that control antigen receptor variable region gene assembly. Semin Immunol. 1994;6:185–196. doi: 10.1006/smim.1994.1024. [DOI] [PubMed] [Google Scholar]
- 56.Okada A, Mendelsohn M, Alt F W. Differential activation of transcription versus recombination of transgenic T cell receptor β variable region gene segments in B and T lineage cells. J Exp Med. 1994;180:261–272. doi: 10.1084/jem.180.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pinkel D, Straume T, Gray J W. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prosser H M, Lake R A, Wotton D, Owen M J. Identification and functional analysis of the transcriptional enhancer of the human T cell receptor β gene. Eur J Immunol. 1991;21:161–166. doi: 10.1002/eji.1830210124. [DOI] [PubMed] [Google Scholar]
- 59.Prosser H M, Wotton D, Gegonne A, Ghysdael J, Wang S, Speck N A, Owen M J. A phorbol ester response element within the human T-cell receptor β-chain enhancer. Proc Natl Acad Sci USA. 1992;89:9934–9938. doi: 10.1073/pnas.89.20.9934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roberts J L, Lauzurica P, Krangel M S. Development regulation of VDJ recombination by the core fragment of the T cell receptor α enhancer. J Exp Med. 1997;185:131–140. doi: 10.1084/jem.185.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schatz D G. V(D)J recombination moves in vitro. Semin Immunol. 1997;9:149–159. doi: 10.1006/smim.1997.0068. [DOI] [PubMed] [Google Scholar]
- 62.Schlissel M S, Baltimore D. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 1989;58:1001–1007. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
- 63.Schlissel M S, Stanhope-Baker P. Accessibility and the developmental regulation of V(D)J recombination. Semin Immunol. 1997;9:161–170. doi: 10.1006/smim.1997.0066. [DOI] [PubMed] [Google Scholar]
- 64.Schlissel M S, Voronova A, Baltimore D. Helix-loop-helix transcription factor E47 activates germ-line immunoglobulin heavy-chain gene transcription and rearrangement in a pre-T-cell line. Genes Dev. 1991;5:1367–1376. doi: 10.1101/gad.5.8.1367. [DOI] [PubMed] [Google Scholar]
- 65.Shortman K, Wu L. Early T lymphocyte progenitors. Annu Rev Immunol. 1996;14:29–47. doi: 10.1146/annurev.immunol.14.1.29. [DOI] [PubMed] [Google Scholar]
- 66.Sleckman B P, Bassing C H, Bardon C G, Okada A, Khor B, Bories J C, Monroe R, Alt F W. Accessibility control of variable region gene assembly during T-cell development. Immunol Rev. 1998;165:121–130. doi: 10.1111/j.1600-065x.1998.tb01235.x. [DOI] [PubMed] [Google Scholar]
- 67.Sleckman B P, Gorman J R, Alt F W. Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu Rev Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- 68.Spanopoulou E, Roman C A, Corcoran L M, Schlissel M S, Silver D P, Nemazee D, Nussenzweig M C, Shinton S A, Hardy R R, Baltimore D. Functional immunoglobulin transgenes guide ordered B-cell differentiation in RAG-1-deficient mice. Genes Dev. 1994;8:1030–1042. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- 69.Stanhope-Baker P, Hudson K M, Shaffer A L, Constantinescu A, Schlissel M S. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 1996;85:887–897. doi: 10.1016/s0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- 70.Sun W, Graves B J, Speck N A. Transactivation of the Moloney murine leukemia virus and T-cell receptor β-chain enhancers by cbf and ets requires intact binding sites for both proteins. J Virol. 1995;69:4941–4949. doi: 10.1128/jvi.69.8.4941-4949.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takeda J, Cheng A, Mauxion F, Nelson C A, Newberry R D, Sha W C, Sen R, Loh D Y. Functional analysis of the murine T-cell receptor β enhancer and characteristics of its DNA-binding proteins. Mol Cell Biol. 1990;10:5027–5035. doi: 10.1128/mcb.10.10.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walters M C, Magis W, Fiering S, Eidemiller T, Scalzo D, Groudine M, Martin D I K. Transcriptional enhancers act in cis to suppress position-effect variegation. Genes Dev. 1996;10:185–195. doi: 10.1101/gad.10.2.185. [DOI] [PubMed] [Google Scholar]
- 73.Wang Y K, Kobori J A, Hood L. The htβ gene encodes a novel CACCC box-binding protein that regulates T-cell receptor gene expression. Mol Cell Biol. 1993;13:5691–5701. doi: 10.1128/mcb.13.9.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watrin F, Fernex C, Capone M, Horvat B, Caillol D, Ferrier P. Transgenic mouse models to study VDJ recombination. In: Bluethmann H, Ohashi P, editors. Analysis of the immune system of mice utilizing transgenesis and targeting mutagenesis. Orlando, Fla: Harcourt Brace Janovich; 1994. pp. 1–14. [Google Scholar]
- 75.Whitehurst C E, Chattopadhyay S, Chen J. Control of V(D)J recombinational accessibility of the Dβ1 gene segment at the TCRβ locus by a germline promoter. Immunity. 1999;10:313–322. doi: 10.1016/s1074-7613(00)80031-x. [DOI] [PubMed] [Google Scholar]
- 76.Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wingender E, Kel A E, Kel O V, Karas H, Heinemeyer T, Dietze P, Knuppel R, Romaschenko A G, Kolchanov N A. TRANSFAC, TRRD and COMPEL: towards a federated database system on transcriptional regulation. Nucleic Acids Res. 1997;25:265–268. doi: 10.1093/nar/25.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wotton D, Ghysdael J, Wang S, Speck N A, Owen M J. Cooperative binding of Ets-1 and core binding factor to DNA. Mol Cell Biol. 1994;14:840–850. doi: 10.1128/mcb.14.1.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wotton D, Ways D K, Parker P J, Owen M J. Activity of both Raf and Ras is necessary for activation of transcription of the human T cell receptor β gene by protein kinase C, Ras plays multiple roles. J Biol Chem. 1993;268:17975–17982. [PubMed] [Google Scholar]