Abstract
Background
Tumor metastasis is a major factor for poor prognosis of hepatocellular carcinoma (HCC), but the relationship between ubiquitination and metastasis need to be studied more systematically. We analyzed the ubiquitinome of HCC in this study to have a more comprehensive insight into human HCC metastasis.
Methods
The protein ubiquitination levels in 15 HCC specimens with vascular invasion and 15 without vascular invasion were detected by ubiquitinome. Proteins with significantly different ubiquitination levels between HCCs with and without vascular invasion were used to predict E3 ubiquitin ligases associated with tumor metastasis. The topological network of protein substrates and corresponding E3 ubiquitin ligases was constructed to identify the key E3 ubiquitin ligase. Besides, the growth, migration and invasion ability of LM3 and HUH7 hepatoma cell lines with and without SYVN1 expression interference were measured by cell proliferation assay, subcutaneous tumor assay, umphal vein endothelium tube formation assay, transwell migration and invasion assays. Finally, the interacting proteins of SYVN1 were screened and verified by protein interaction omics, immunofluorescence, and immunoprecipitation. Ubiquitin levels of related protein substrates in LM3 and HUH7 cells were compared in negative control, SYVN1 knockdown, and SYVN1 overexpression groups.
Results
In this study, our whole‐cell proteomic dataset and ubiquitinomic dataset contained approximately 5600 proteins and 12,000 ubiquitinated sites. We discovered increased ubiquitinated sites with shorter ubiquitin chains during the progression of HCC metastasis. In addition, proteomic and ubiquitinomic analyses revealed that high expression of E3 ubiquitin‐protein ligase SYVN1 is related with tumor metastasis. Furthermore, we found that SYVN1 interacted with heat shock protein 90 (HSP90) and impacted the ubiquitination of eukaryotic elongation factor 2 kinase (EEF2K).
Conclusions
The ubiquitination profiles of HCC with and without vascular invasion were significantly different. SYVN1 was the most important E3 ubiquitin‐protein ligase responsible for this phenomenon, and it was related with tumor metastasis and growth. Therefore, SYVN1 might be a potential therapeutic target for HCC.
Keywords: cancer, E3 ubiquitin‐protein ligase, eukaryotic elongation factor 2 kinase, heat shock protein 90, hepatocellular carcinoma, liver, metastasis, proteomics, synoviolin, ubiquitin
We constructed one of the largest ubiquitinome dataset obtained from human cancer tissues to date using the high‐resolution mass spectrometry and immunoenrichment of diGly‐modified peptides. We identified a E3 ligase, SYVN1, which potentially participated in the metastasis of HCC. We studied the interaction omics of SYVN1, and found it interacted with HSP90 and impacted the ubiquitination of EEF2K.
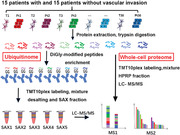
Abbreviations
- ACN
acetonitrile
- AFP
alpha fetal protein
- AGC
automatic gain control
- DAPI
4‐6‐diamidino‐2‐phenylindole
- EEF2K
eukaryotic elongation factor 2 kinase
- FA
formic acid
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HSP90
heat shock protein 90
- HSPs
Heat shock proteins
- PR‐619
2,6‐diamino‐5‐thiocyanatopyridin‐3‐yl thiocyanate
- SAX
strong anion exchange
- SYVN1
synoviolin
- TFA
trifluoroacetic acid
1. BACKGROUND
Hepatocellular carcinoma (HCC) is the dominant subtype of liver cancer and a common malignant tumor worldwide. The incidence of liver cancer has rapidly increased by 2%‐3% annually from 2007 to 2016 [1]. The 5‐year relative survival rate of liver cancer (18%) is one of the lowest among all cancers [1]. Vascular invasion, including macro‐ and micro‐invasion, was found to be an aggressive biological behavior and a prerequisite for systemic tumor dissemination [2, 3]. The angiogenic, invasive, and metastatic potency of cancer vary widely, and integrative multi‐omics analyses could help identify the subtypes of HCC [4]. These phenotypic differences might result from the complicated, multi‐protein and highly precisely regulated molecular machines that are subjected to posttranslational modification [5].
Ubiquitination is the conjugation of protein substrates with the 76‐amino acid protein ubiquitin. Ubiquitination touches upon all aspects of eukaryotic biology [6] and plays important roles in many essential cellular processes, including protein‐protein interactions, protein trafficking, and transcriptional modification [7, 8]. Ubiquitination involves multistep enzymatic reactions sequentially catalyzed by Ub‐activating enzyme (E1), Ub‐conjugating enzyme (E2), and Ub‐ligated protein (E3) [9, 10]. Gallo et al. [11] described how various cancers take advantage of the misregulated expression of the members of the ubiquitination cascade for proliferation, survival, and metastasis. Of the three ubiquitylating enzymes, E3 ubiquitin ligases are of particular concern. Acting as the specific substrate‐recognizing element, E3s play an essential role in cancer development and associated tightly with the clinical prognosis of cancer [12]. For instance, the E3 ubiquitin ligase tripartite motif‐containing 7 (TRIM7) induced Lys48‐linked polyubiquitination of Src protein, which further led to the suppression of its downstream mechanistic target of rapamycin complex 1‐ ribosomal protein S6 kinase (mTORC1‐S6K1) signaling pathway and acted as a tumor suppressor in HCC [13].
Synoviolin (SYVN1), an evolutionarily conserved endoplasmic reticulum‐resident E3 ligase, is the central component of a complex facilitating degradation of misfolded proteins during the ubiquitin‐proteasome‐dependent process of endoplasmic reticulum‐associated degradation [14, 15]. SYVN1 participates in biological events in several cancer types by regulating the ubiquitination and degradation of P53 [16], insulin‐like growth factor‐I receptor [17], and Sirtuin 2 [18]. Heat shock proteins (HSPs), which could help cancer cells survive and obtain an invasive phenotype, always function as molecular chaperones. A study demonstrated that HSP70 picked up and subsequently escorted mutant Blimp‐1 proteins to SYVN1, resulting in the ubiquitination‐dependent degradation of Blimp‐1 [19]. Various E3 ligases are responsible for the regulation of many cancer‐related kinases, such as Akt and eukaryotic elongation factor 2 kinase (EEF2K) [20, 21]. However, the mechanisms by which SYVN1 affects kinases are unclear.
In this study, we combined the whole‐cell proteome and ubiquitinome for analysis of HCC tumorigenesis and metastasis. Besides, we systematically studied the interaction proteome of SYVN1 and investigated the potential roles and mechanisms of SYVN1 in HCC metastasis.
2. MATERIALS AND METHODS
2.1. Clinical tissue sample collection
Clinical tissue samples were collected from 15 HCC patients with and 15 patients without vascular invasion who underwent surgical resection between October 12th, 2017 and July 2, 2018 at the First Affiliated Hospital of Zhejiang University. Clinical tissue samples were snap‐frozen in liquid nitrogen and stored at ‐80°C until use. All patients did not receive any preoperative anticancer treatments, and no parasitic eggs were found in the liver through computed tomography. Vascular invasion was classified as macrovascular invasion in 6 patients with the invasion identified by gross examination and as microvascular invasion in 9 patients with the invasion only visible under a microscope using the reported criteria [22, 23].
The ethical committee of the First Affiliated Hospital of Zhejiang University approved this study. Informed consent was obtained from all patients, allowing the use of their data in researches.
2.2. Cell culture and animal experiments
HUH7, LM3, 293T, and human umbilical vein endothelial cells (HUVECs) from the National Collection of Authenticated Cultures (Shanghai, China) were cultured in Dulbecco's modified eagle medium (DMEM) or modification of minimum essential medium (α‐MEM) containing 10% dialyzed fetal bovine serum and tested for mycoplasma contamination prior to the experiments. Specific pathogen‐free BALB/c male nude mice (4 weeks old) were obtained from Zhejiang Laboratory Animal Center (Hangzhou, Zhejiang, China) and housed according to rules for raising laboratory animals released by Zhejiang University in 2009. The experimental procedures were reviewed and approved by the Ethical Committee of Zhejiang University and were performed according to the Guide for the Care and Use of Laboratory Animals released by Zhejiang University in 2009. Nude mice were randomly divided into three groups with 7 mice in each group, and a subcutaneous tumor‐bearing nude mouse model was established by inoculating 5 × 106 lentiviral shNC‐, sh1SYVN1‐, or sh2SYVN1‐infected LM3 and 1 × 107 HUH7 cells in 100 μL phosphate buffer solution (PBS) into the subcutaneous tissue of the right hind limbs of nude mice. Their body weight and tumor volume were measured every two days. Tumor volume was calculated as (length × width2) / 2. When approximately half of the nude mice displayed cachexia (significant weight loss), the mice were euthanized and their tumors were weighed and photographed.
2.3. Protein extraction and digestion
Before the frozen samples were ground into powder using a bead‐shocker at a speed of 5 m/s for 30‐s pulses, we added freshly prepared 4°C lysis buffer [8 mol/L urea, 150 mmol/L NaCl, 50 mmol/L Tris‐HCl (pH = 8.0), 1 × phosphorylase inhibitor (Cat#P1049, Beyotime, Shanghai, China), 1 × ethylene diamine tetraacetic acid, 50 μmol/L (2,6‐diamino‐5‐thiocyanatopyridin‐3‐yl) thiocyanate (PR‐619) (Cat#S7130, Selleck, Houston, TX, USA), 1 × protease inhibitor] and ceramic beads to each sample. The protein concentration of the supernatant was estimated using the bicinchoninic acid (BCA) protein assay (Cat#23227, Thermo Scientific, Waltham, MA, USA). Proteins were reduced with 10 mmol/L dithiothreitol for 45 min at 30°C and subsequently carbamidomethylated with 30 mmol/L iodoacetamide for 30 min at room temperature in the dark. Next, the liquid was replaced with 50 mmol/L ammonium bicarbonate using Zeba Spin Desalting Columns (Cat#89892, Thermo Scientific). All the samples were digested overnight at 37°C with a trypsin‐to‐substrate ratio of 1:40 (wt/wt) and shaken at 150 rpm. After digestion, peptides were acidified with trifluoroacetic acid (TFA) to a final concentration of 1% (pH ≤ 3.0). Acidified peptides were centrifuged at 1800 × g for 10‐20 min at 4°C to remove lipids, and the supernatant was desalted using a 500 mg preconditioned C18 Sep‐Pak SPE cartridge (Cat#WAT043395, Waters, Milford, MA, USA). C18 cartridges were conditioned with 4 mL acetonitrile (ACN) followed by one wash with 4 mL of the mixture of 50% ACN and 0.1% TFA, and two washes with 0.1% TFA. The sample was loaded onto the conditioned C18 cartridge, washed thrice with 0.1% TFA, and finally eluted with 50% ACN and 0.1% FA. Desalted peptides were lyophilized over 48 h in a vacuum lyophilizer (LABCONCO, Kansas City, MO, USA). The amount of peptides was accurately estimated by using Pierce Quantitative Colorimetric Peptide Assay (Cat#23227, Thermo Scientific), and 7 mg peptides from each sample were needed for our study.
2.4. Chemical cross‐linking of anti‐diGly‐modified peptides antibody
For antibody cross‐linking, we washed the antibody‐bound beads, which were obtained from the PTMScan ubiquitin remnant motif kit (Cat#5562, Cell Signaling Technology, Boston, MA, USA), thrice with 1 mL cold antibody cross‐linking wash buffer (100 mmol/L sodium borate, pH = 9.0). After washing, beads were resuspended in 1 mL antibody cross‐linking buffer (20 mmol/L dimethyl pimelimidate in antibody cross‐linking wash buffer) and incubated at room temperature for 30 min with gentle end‐over‐end rotation. To stop the cross‐linking reaction, we washed the beads twice with blocking buffer (200 mmol/L ethanolamine, pH = 8.0) and incubated the beads in 1 mL blocking buffer for 2 h at 4°C with end‐over‐end rotation. To test the cross‐linking efficiency, we eluted pre‐ and post‐cross‐linking antibody from the same amount of beads using 0.15% TFA and completely dried the eluates. The cross‐linking antibody beads were washed thrice with 1 mL cold 1 × PBS and stored for several weeks at 4°C.
2.5. Enrichment of diGly‐modified peptides
Seven milligrams of dried peptides were resuspended in cold 1 × immunoaffinity purification (IAP) buffer and incubated with 50 μg washed cross‐linked antibody beads in an immunoprecipitation (IP) tube for 2 h at 4°C with end‐over‐end rotation. Subsequently, the IP tube was centrifuged at 100 × g for 30 s at 4°C to gather beads. Beads were washed thrice with ice‐cold 1 × IAP buffer and twice with ice‐cold mass spectrum (MS) water. diGly‐modified peptides (also known as K‐ε‐GG peptides) were eluted by adding 50 μL 0.15% TFA twice. The eluate was dried completely by vacuum centrifugation and then used in the following steps.
2.6. Tandem mass tag (TMT) isobaric labeling
Immediately before use, we equilibrated the TMT Label Reagents (Cat#90111, Thermo Scientific) to room temperature. To 0.8‐mg vials, we added 84 μL anhydrous acetonitrile and dissolved the reagent for 5 min with occasional vortexing. Each post‐IP peptide sample was dissolved in 20 μL 100 mmol/L tetraethylammonium bromide (pH = 8.0), and 8 μL TMT Label Reagent was carefully added. The reaction was incubated for 1 h at room temperature. Subsequently, 2 μL 5% hydroxylamine was added to each sample, and the sample was incubated for 15 min to quench the reaction. The supernatant was desalted using a homemade C18 stage tip and dried by vacuum centrifugation.
2.7. Pipette tip‐based strong anion exchange (SAX) fractionation of diGly‐modified peptides
SAX chromatography with tip columns was performed using Empore Anion‐SR Extraction Disks (Cat#98060402298, 3M, Saint Paul, MN, USA). Wash solutions were applied sequentially, including 80 μL ACN, 80 μL SAX B, 80 μL SAX C twice, and 80 μL SAX A twice. The combined TMT10plex‐labeled diGly‐modified peptides were resuspended in 80 μL SAX A and loaded on the preconditioning SAX tips. The flow‐through was labeled as SAX fraction 0. We eluted the loaded SAX tips with 80 μL SAX buffers 1, 2, 3, 4, and 5 in sequence, and eluates were labeled as SAX fractions 1, 2, 3, 4, and 5, respectively. The formulations of SAX reagents are listed in Supplementary Table S1. Before vacuum centrifugation, SAX fraction 0 was combined with fraction 1.
2.8. Liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) analysis
All peptide samples were dried by vacuum centrifugation and resuspended in 2% ACN and 0.1% formic acid (FA). Then, the peptides were separated by nanoLC‐MS/MS using an UltiMate 3000 RSLCnano system (Thermo Scientific) and analyzed by Q Exactive HF‐X (Thermo Scientific). Solvent A contained 2% ACN and 0.1% FA, and solvent B contained 98% ACN and 0.1% FA. Gradient elution was performed at 32°C using linear gradients of 120 min at a flow rate of 400 nL/min: 1‐4 min with 3% (v/v) of solvent B, 4‐6 min with 3%‐5% solvent B, 6‐70 min with 5%‐15% solvent B, 70‐90 min with 15%‐30% solvent B, 90‐100 min with 30%‐80% solvent B, 100‐110 min with 80% solvent B, and 110‐120 min with 3% solvent A. MS spectra were acquired using 120,000 resolutions with a mass range of 300‐1500 m/z and an automatic gain control (AGC) target of 3E6. MS2 spectra were acquired using 45,000 resolutions, and higher energy collision induces dissociation fragmentation was performed with a collision energy of approximately 32% normalized collisional energy. In addition, we used a method of data‐dependent top 20 on MS2 with an AGC target of 2E5. The isolation window was 1.0 m/z. Charge exclusion was ≤ 2 and ≥ 7, and dynamic exclusion was set to 30 s. The fixed first mass was 100 m/z. Peptide match was preferred, and the exclude isotope feature was on.
2.9. Immunoprecipitation and Western blotting
Cells and tissue samples were lysed with urea lysis buffer or nonidet P‐40, and protein concentrations were quantified. The extracts were incubated with rabbit polyclonal anti‐ubiquitin (Cat#3933, Cell Signaling Technology), rabbit monoclonal anti‐SYVN1 (Cat#14773, Cell Signaling Technology), rabbit polyclonal anti‐actin (Cat#ab8227, Abcam, Cambridge, UK), mouse monoclonal anti‐HSP90 (Cat#sc‐13119, Santa, Dallas, TX, USA), rabbit polyclonal anti‐EEF2K (Cat#3692S, Cell Signaling Technology), mouse monoclonal anti‐Flag (Cat#sc‐51590, Santa), or mouse monoclonal anti‐CD31 (Cat#3528, Cell Signaling Technology) at 4°C for 2 h. Then, protein A/G beads were added to samples and incubated for 2 h. After incubation, protein A/G beads were washed four times with ice‐cold 1 × PBS and heated in boiling water for 5 min with loading buffer. Then, samples were separated by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) on a 4%‐15% gradient gel (Cat#4561085, Bio‐Rad, Hercules, CA, USA). A polyvinylidene fluoride membrane was incubated with the indicated antibodies after transfer and subsequently incubated with corresponding second antibodies and detected using a chemiluminescence kit (Cat#P0018M, Beyotime).
2.10. Cell proliferation
We constructed lentiviruses containing two shRNA sequences of SYVN1 (shSYVN1) and negative control shRNA (shNC) and transfected LM3 and HUH7 cells to obtain stable knockdown cell lines of SYVN1. Both shNC‐ and shSYVN1‐transfected cells were inoculated into a 96‐well cell culture plate. Cell proliferation was detected every 12 h using the cell counting kit 8 (CCK‐8) kit (Cat#CK04, DOJINDO, Shanghai, China) according to specification.
2.11. Tube formation
Briefly, 1.5 × 104 HUVECs were transferred to 96‐well plates coated with 10 mg/mL Matrigel and cultured for 12 h with conditioned media collected from LM3 and HUH7 cells, which was diluted 1:1 with normal cell culture media. Mesh areas of cellular networks were quantified using ImagePro‐Plus software (https://www.mediacy.com/, Media Cybernetics, Rockville, MD, USA).
2.12. Cell migration and invasion
LM3 and HUH7 cells infected with shSYVN1 or shNC lentivirus were suspended in a medium without serum and transferred to Matrigel‐uncoated or Matrigel‐coated upper chambers of a Transwell insert. Medium without serum was added to the lower chamber for 12 h and then replaced with medium containing 20% FBS for another 24‐48 h, and the cells were incubated at 37°C in 5% CO2. The average number of cells that migrated or invaded through the membrane in five fields was photographed and counted per chamber.
2.13. Immunohistochemistry
Frozen sections of liver cancer and subcutaneous tumor tissues of nude mice were fixed with 4% paraformaldehyde and stained with anti‐SYVN1 antibody (1:1000 dilution). Images were captured using a digital automatic slide scanner (Hamamatsu, Iwata, Japan) and quantified using ImagePro‐Plus.
2.14. Immunofluorescence
LM3 and HUH7 cells were cultured in 24‐well plates overnight at 37°C in 5% CO2. The attached slides with cells were fixed, membrane broken, sealed, and incubated with anti‐SYVN1 (1:1000 dilution) and anti‐HSP90 (1:1000 dilution) primary antibodies overnight at 4°C followed by Alexa Fluor 555‐conjugated donkey anti‐rabbit IgG antibody and Alexa Fluor 488‐conjugated donkey anti‐mouse IgG antibody for 2 h at room temperature. Then, 4‐6‐diamidino‐2‐phenylindole (DAPI) was used to stain nuclei.
2.15. Plasmid transfection and lentiviral infection
Briefly, 293T cells were transfected with an HA‐tagged ubiquitin plasmid containing a point mutation [wild type (WT), K6, K11, K27, K29, K33, K48, and K63] and SYVN1 plasmid according to the manufacturer's instructions (Cat#TF20121201, NEOFECT, San Francisco, CA, USA). LM3 and HUH7 cells were infected with shSYVN1 (target sequences: 5’‐GCTCACGCCTACTACCTCAAA‐3’ and 5’‐GGAGACTGCCACTACAGTTGT‐3’) or shNC (target sequence: 5’‐TTCTCCGAACGTGTCACGT‐3’) lentivirus according to the manufacturer's instructions (Genomeditech, Shanghai, China). The infection efficiency was observed using a fluorescence microscope.
2.16. Real‐time PCR
Total RNA from LM3 and HUH7 cells was extracted using a rapid total RNA extraction kit (Cat#220011, Fastagen Biotech, Shanghai, China) according to the manufacturer's instructions. mRNA was quantified using a reverse transcription kit (Cat#RR036A, Takara, Kyoto, Japan) and fast qPCR mix (Cat#RR820A, Takara) according to the manufacturer's instructions. Primer sequences were as follows: basic fibroblast growth factor (bFGF), forward 5′‐AGAAGAGCGACCCTCACATCA‐3′, reverse 5′‐ ACTGCCCAGTTCGTTTCAGTG‐3′; granulocyte‐colony stimulating factor (GCSF), forward 5′‐GCTGCTTGAGCCAACTCCATA‐3′, reverse 5′‐GAACGCGGTACGACACCTC‐3′; platelet‐derived growth factor AA (PDGFAA), forward 5′‐CCAGCGACTCCTGGAGATAGA‐3′, reverse 5′‐CGTCCTGGTCTTGCAGACAG‐3′; PDGFBB, forward 5′‐CCAGGTGAGAAAGATCGAGATTG‐3′, reverse 5′‐ATGCGTGTGCTTGAATTTCCG‐3′; placental growth factor (PLGF), forward 5′‐GAACGGCTCGTCAGAGGTG‐3′, reverse 5′‐ACAGTGCAGATTCTCATCGCC‐3′; vascular endothelial growth factor (VEGF), forward 5′‐AGGGCAGAATCATCACGAAGT‐3′, reverse 5′‐AGGGTCTCGATTGGATGGCA‐3′; β‐actin, forward 5′‐CATGTACGTTGCTATCCAGGC‐3′, reverse 5′‐CTCCTTAATGTCACGCACGAT‐3′.
2.17. Peptide‐protein identification and quantification
MaxQuant (version 1.6.2.10, https://www.maxquant.org) was used for protein identification and quantification. The human UniProtKB database (October 2018) and viral hepatitis type B (HBV)‐b/‐c database (October 2018) were utilized as the search database, while the automatic reverse database and known contaminants were used for decoy searches. Variable modifications included oxidation (M) (+15.99491 Da) and acetylation (protein N‐term) (+42.01056 Da) for both ubiquitinome and whole‐cell proteome but GlyGly(K)_10plex_TMT (+343.20586 Da) for only ubiquitinome. Carbamidomethyl (C) (+57.02146 Da) was set as the fixed modification, and the max number of modifications per peptide was set as 5. Trypsin was set as the specificity of digestion, and the maximum number of missed cleavage sites was 2. We used 20 parts per million (ppm) in the first search ion tolerance and 4.5 ppm in the main search ion tolerance. Both peptide and protein identification were performed at false discovery rate (FDR) < 1%. Except for the abovementioned settings, the default parameters of MaxQuant were adopted.
2.18. Functional annotation enrichment analysis
To identify the top enriched pathways during the occurrence and development of HCC, we ranked the diGly sites according to t‐statistics and fold change. Proteins containing diGly sites with fold changes >2 and P < 0.05 were selected for functional annotation enrichment analysis in a database for annotation, visualization and integrated discovery (DAVID) (https://david.ncifcrf.gov/) [24, 25]. Fisher's exact test and Hochberg's FDR method were chosen for significance determinations. The heat map was generated using the heatmap3 package in R software.
2.19. E3 enrichment analysis
To identify the E3s that contributed to changes in ubiquitination between different groups, we ran some searches using substrate proteins containing obviously changed ubiquitination sites on UbiBrowser (http://ubibrowser.ncpsb.org) [26]. The four most likely E3s from each search result and their corresponding substrate proteins were imported into Cytoscape (https://cytoscape.org/).
2.20. Statistical analyses
R framework (version 3.5.1, https://www.r‐project.org/), Perseus (https://maxquant.net/perseus/) [27], and GraphPad Prism (https://www.graphpad.com/, GraphPad Software, San Diego, CA, USA) were utilized to analyze all statistical and bioinformatics data. The P value threshold was set to 0.05, and the relative fold change threshold was set to ±1 after log2 transformation. Boxplots were generated using GraphPad or the ggplot2 package of R environment. The lower hinge, upper hinge, lower whisker, upper whisker, and bar in the box represented the 25% percentile, 75% percentile, 1.5 times less than the lower interquartile range (IQR) hinges, 1.5 times more than the upper IQR hinges, and the median, respectively. The Pearson correlation between different proteins was calculated using R software (“cor” function in the basic package). Principal component analysis (PCA) was performed using R software (“principal” function in psych package). Heatmap3 was used to plot the heat map and clustering for quantitative changes in ubiquitination sites. Transcriptome data were obtained from the GEPIA database [28].
2.21. Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository [29] with the dataset identifiers PXD011895 and PXD018593. All other data that can support our findings are available in the article and supplemental information.
3. RESULTS
3.1. Ubiquitinome and whole‐cell proteome in HCC
Clinicopathological characteristics of the 30 HCC patients are summarized (Table 1 and Supplementary Table S2). We discovered that SAX combined with TMT10plex labeling was an efficient method for mapping endogenous ubiquitination (Figure 1). By applying the method from Udeshi et al. [30], we cross‐linked the antibody with beads before IP which prevented antibody loss and improved IP efficiency (Supplementary Figure S1A). Consequently, diGly‐modified peptides accounted for approximately 50% of the total identified peptides in almost every fraction (Supplementary Figure S1B), and greater than 60% diGly‐modified peptides were separated into only one fraction (Supplementary Figure S1C). Other findings of the identified diGly‐modified peptides and proteins included the distribution of charge, intensity, molecular weight, MS/MS count, sequence length, and number of diGly‐modified sites (Supplementary Figures S1D‐I). Remarkably, we successfully identified a total of 11,913 diGly‐modified sites (9800 in HCC with vascular invasion, 7241 in HCC without vascular invasion) and 4317 diGly‐modified proteins (3909 in HCC with vascular invasion, 3259 in HCC without vascular invasion) from HCC (Table 2). PCA and heat maps showed the difference between HCC with and without vascular invasion in both the ubiquitinome and whole‐cell proteome (Figures 2A and 2B, Supplementary Figures S2A and S2B). Briefly, 7532 diGly‐modified sites and 3465 diGly‐modified proteins were identified in at least 10 HCCs (Figure 2C). Although extensive overlaps of both diGly‐modified sites and diGly‐modified proteins were observed between HCC with and without vascular invasion, we discovered a small portion of diGly‐modified proteins that only existed in one group (Supplementary Figure S2C). We assumed that these diGly‐modified proteins might have a potential impact on the occurrence of vascular invasion in HCC.
TABLE 1.
Baseline clinical characteristics of the 30 HCC patients
| Vascular invasion | ||||
|---|---|---|---|---|
| Characteristic | Whole cohort | Present | Absent | P value |
| Total (cases) | 30 | 15 | 15 | |
| Age (years; mean ± SD) | 56.9 ± 11.2 | 54.4 ± 10.0 | 59.4 ± 11.7 | 0.234 |
| Etiology [cases (%)] | 1.000 | |||
| HBV | 17 (56.7) | 8 (26.7) | 9 (30.0) | |
| HBV + alcohol | 13 (43.3) | 7 (23.3) | 6 (20.0) | |
| Gender [cases (%)] | 1.000 | |||
| Male | 28 (93.3) | 14 (46.7) | 14 (46.7) | |
| Female | 2 (6.7) | 1 (3.3) | 1 (3.3) | |
| Tumor size (cm; mean ± SD) | 7.0 ± 2.7 | 8.4 ± 3.1 | 5.6 ±1.3 | 0.004 |
| AFP (IU/mL; mean ± SD) | 9400 ± 20856 | 17593 ± 26768 | 1208 ± 4384 | 0.039 |
| Edmondson grade [cases (%)] | 0.709 | |||
| I‐II | 18 (60.0) | 8 (26.7) | 10 (33.3) | |
| III‐IV | 12 (40.0) | 7 (23.3) | 5 (16.7) | |
Abbreviations: HCC, hepatocellular carcinoma; HBV, hepatitis B virus; AFP, alpha fetal protein; SD, standard deviation.
FIGURE 1.
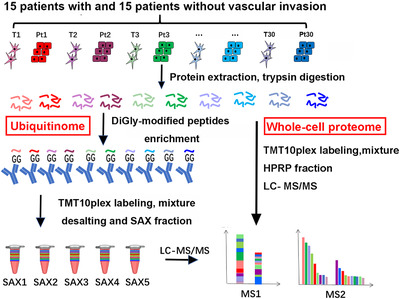
The workflow of systemic ubiquitinome and whole‐cell proteome analyses of HCC. Abbreviations: HCC, hepatocellular carcinoma; T, tumor tissue; Pt, peritumoral tissue; SAX, strong anion‐ exchange; TMT, Tandem mass tag; HPRP, high performance reverse chromatography; LC‐MS, liquid chromatography‐mass spectrometry
TABLE 2.
The identified diGly‐modified sites and proteins in proteome
| HCC sample | diGly‐modified sites | diGly‐modified proteins | WCP proteins |
|---|---|---|---|
| Total | 11,913 | 4317 | 5573 |
| With vascular invasion | 9800 | 3909 | 5382 |
| Without vascular invasion | 7241 | 3259 | 5227 |
Abbreviations: HCC, hepatocellular carcinoma; WCP, whole‐cell proteome.
FIGURE 2.
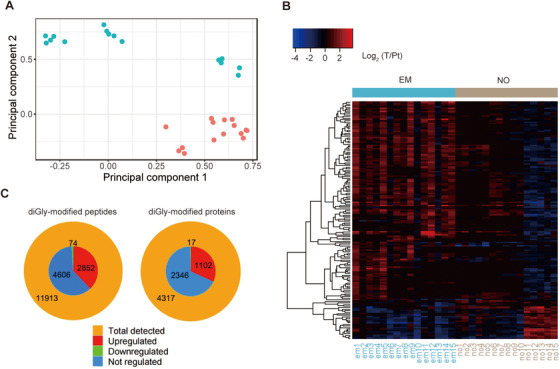
Quantitative profiling of ubiquitinome and whole‐cell proteome in HCC. (A) Principal component analysis (PCA) analysis of HCC whole‐cell proteome. Red dots represent HCC samples with vascular invasion, and green dots represent HCC samples without vascular invasion. (B) Heat map shows the regulation pattern of proteins in HCC with and without vascular invasion in the whole‐cell proteome. (C) The outer circle represents the number of all identified diGly‐modified sites and proteins in at least 1 HCC sample. The inner circle represents the number of quantified diGly‐modified sites and proteins in at least 10 HCC samples. Upregulated or downregulated peptides or proteins refer to those with a fold change > 1.5 and P < 0.05. Abbreviations: HCC, hepatocellular carcinoma; EM, HCC with vascular invasion; NO, HCC without vascular invasion; T, tumor tissue; Pt, peritumoral tissue
3.2. Increased ubiquitination sites with shorter ubiquitin chains in HCC with vascular invasion
Compared with peritumoral tissues, we observed a significant upregulation of diGly‐modified sites in tumor tissues, and this phenomenon was more obvious in HCC with vascular invasion than in HCC without vascular invasion (Figure 3A and Supplementary Figure S3A). Further analysis revealed that the percentages of significantly changed (fold change > 2) diGly‐modified sites increased from 15.6% in HCC without vascular invasion to 18.0% in HCC with vascular invasion (Figure 3B). Regarding ubiquitin itself, diGly‐modified sites (such as K63) were increased in cancer tissues in both HCC with and without vascular invasion, and the upregulation of diGly‐modified sites in ubiquitin itself was more observable in HCC without vascular invasion (Figure 3C and Supplementary Figure S3B). This phenomenon was validated by Western blotting (Figure 3D and Supplementary Figure S3C). The combination of these two discoveries prompted us to conclude that HCC formation was accompanied with an increase in ubiquitination sites and longer ubiquitin chains. The diGly‐modified sites for ubiquitin reflect the length of the ubiquitin chain, while the total diGly‐modified sites reflect the quantity of the ubiquitination. Comparing the relationship between the length of ubiquitin chain and the quantity of ubiquitination, we can recognize polyubiquitination and monoubiquitination in samples. Therefore, we speculate that vascular invasion and HCC metastasis were followed by increased ubiquitination sites and shorter ubiquitin chains (Figures 3E and 3F).
FIGURE 3.
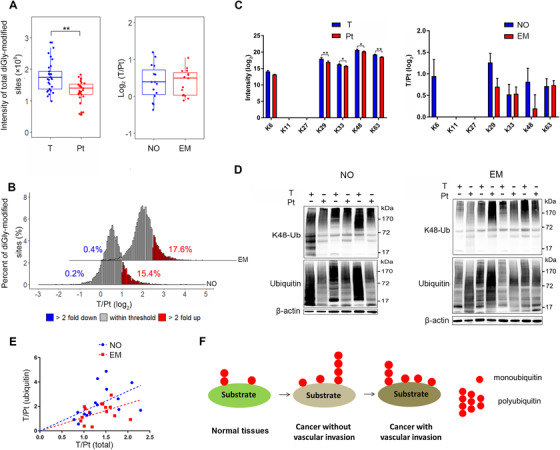
Ubiquitination sites are increased while ubiquitin chains are shortened in HCC with vascular invasion. (A) The distribution of total diGly‐modified sites in tumor and peritumoral tissues (left) and the ratio of total diGly‐modified sites in tumor tissues to those in peritumoral tissues in EM and NO groups (right). The lower, median, and upper lines in each box correspond to 25%, 50%, and 75%, respectively. (B) diGly‐modified site changes between HCC with and without vascular invasion. The proportions of diGly‐modified sites that are at least 2‐fold decreased or increased in abundance are marked in blue or red, respectively. (C) The intensity of ubiquitinated lysine residues on ubiquitin itself (left: tumor tissues vs. peritumoral tissues; right: HCC with vascular invasion vs. HCC without vascular invasion). Error bars represent the standard error of the mean. (D) Lysates from tumor and peritumoral tissues in HCC with and without vascular invasion were subjected to ubiquitin and K48‐Ub detection by SDS‐PAGE. (E) Intensity ratio of diGly‐modified sites in ubiquitin itself to total diGly‐modified sites in each patient. (F) Pattern diagram shows the dynamic change in ubiquitination in HCC genesis and metastasis. ** P < 0.01. Abbreviations: HCC, hepatocellular carcinoma; K48‐Ub, K48‐ubiquitin; EM, HCC with vascular invasion; NO, HCC without vascular invasion; T, tumor tissue; Pt, peritumoral tissue
3.3. E3 ubiquitin ligase SYVN1 was associated with tumor metastasis
Protein ubiquitination levels are regulated by two opposing systems: ubiquitination and deubiquitination. E3 ubiquitin ligase is a major functional enzyme of the ubiquitination system, which mainly transfers ubiquitin from E2 to the substance. For a comprehensive understanding of the ubiquitination modification in tumor development and metastasis, we focused on several diGly‐modified sites with significant changes. Comparing HCC with vascular invasion to HCC without vascular invasion, the top upregulated diGly‐modified sites were K255 of thioredoxin reductase 1 (TXNRD1), K17 of Ras association domain family member 4 (RASSF4), K375 of epoxide hydrolase 1 (EPHX1), K257 of sulfotransferase family 1E member 1 (SULT1E1), K447 of EEF2K, K64 of mothers against decapentaplegic homolog 5 (SMAD5), K230 of serine palmitoyltransferase long chain base subunit 1 (SPTLC1), and K254 of pyridoxal phosphate binding protein (PLPBP); the top downregulated diGly‐modified sites were K1059 of structural maintenance of chromosomes 3 (SMC3), K64 of high mobility group nucleosomal binding domain 2 (HMGN2), K207 of leucine‐rich repeat containing 59 (LRRC59), K164 of tryptophan 2,3‐dioxygenase (TDO2), K685 of the ankyrin repeat and KH domain containing 1 (ANKHD1), K420 of sodium voltage‐gated channel alpha subunit 7 (SCN7A), K7 of sequestosome 1 (SQSTM1), and K51 of cytidine deaminase (CDA) (Figures 4A and 4B). We predicted that SYVN1 was the most important E3 ubiquitin ligase increasing vascular invasion in HCC based on differentially ubiquitinated proteins between HCC with and without vascular invasion (Figure 4C). We also compared tumor tissues with peritumoral tissues and predicted that the E3 ubiquitin ligase was critical in the occurrence of HCC (Supplementary Figures S4A‐C).
FIGURE 4.
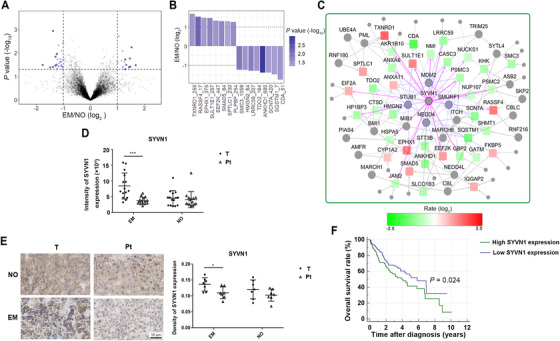
The E3 ubiquitin ligase SYVN1 plays an important role in HCC with vascular invasion. (A) Differentially regulated diGly‐modified sites in HCC with vascular invasion versus HCC without vascular invasion. Blue dots represent fold change ≥ 2 and P < 0.05. (B) The most significantly regulated diGly‐modified sites (the number after protein name indicates the position of lysine residues) in HCC with vascular invasion. (C) Prediction of E3 ligases from proteins containing significantly regulated diGly‐modified sites in HCC with vascular invasion versus HCC without vascular invasion. The circles represent E3 ligases. The squares represent substrates. Red represents upregulation. Green represents downregulation. The size of a colored circle or square represents the amount of regulated diGly‐modified sites. (D) SYVN1 protein expression in tumor tissues and peritumoral tissues quantified by whole‐cell proteome. Error bars represent the standard deviation. (E) SYVN1 protein expression in tumor tissues and peritumoral tissues quantified by immunohistochemistry. Error bars represent the standard deviation. (F) Kaplan‐Meier survival overall survival curves of HCC patients with high and low SYVN1 mRNA expression using data from TCGA. * P < 0.05, ** P < 0.01, *** P < 0.001. Abbreviations: HCC, hepatocellular carcinoma; EM, HCC with vascular invasion; NO, HCC without vascular invasion; T, tumor tissue; Pt, peritumoral tissue.
In‐depth inspection of our whole‐cell proteomic data showed that SYVN1 expression was increased in tumor tissues compared with peritumoral tissues in HCC with vascular invasion, but no differences were found for HCC without vascular invasion (Figure 4D). Similar findings were observed on immunohistochemical analysis (Figure 4E). Transcriptome data from the GEPIA database also showed that SYVN1 was highly expressed in most tumor tissues, including HCC (Supplementary Figure S4D). Overall survival curves of HCC patients from the TCGA database indicated that high SYVN1 expression was associated with poor prognosis in HCC (Figure 4F). In summary, high SYVN1 expression in HCC tissues may affect prognosis by promoting tumor metastasis. To further understand the role of ubiquitination in HCC, we performed functional annotation analysis using proteins containing significantly regulated diGly‐modified sites in HCC. Three pathways were found enriched: regulation of cellular response to heat, proteasome, and ubiquitin binding (Supplementary Figure S4E).
3.4. SYVN1 promoted cancer cell proliferation in serum‐deficient cases
To clarify the role of SYVN1 in HCC metastasis, we first determined its effect on cancer cell proliferation. The results showed that low SYVN1 expression did not affect cell proliferation in LM3 and HUH7 cell lines under serum‐deficient condition (Figure 5A). Surprisingly, when those cell lines were inoculated subcutaneously in nude mice, and grown 3‐4 weeks, the volume and weight of subcutaneous tumors derived from cells with low SYVN1 expression were significantly smaller than those from cells with normal SYVN1 expression (Figures 5B‐D and Supplementary Figure S5A). SYVN1 expression was decreased in SYVN1‐knockdown cell xenografts (Figure 5E). The effects of SYVN1 on the proliferation of LM3 and HUH7 cell lines were different in in vivo and in vitro experiments, suggesting that the promoting effect of SYVN1 was conditional due to the complex microenvironment in vivo. In serum‐free conditions, low SYVN1 expression inhibited the proliferation of LM3 and HUH7 cell lines (Figure 5F).
FIGURE 5.
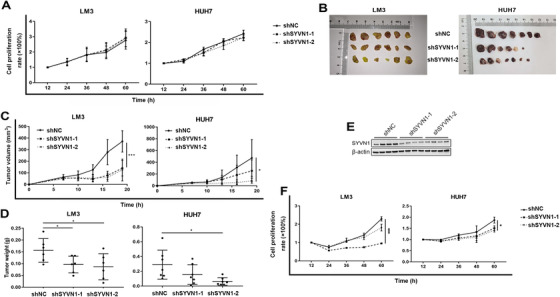
SYVN1 affects cell proliferation in serum‐deficient conditions. (A) Cell proliferation under serum‐sufficient conditions was detected in LM3 and HUH7 cells at different time points using a CCK‐8 kit. Error bars represent the standard error of the mean from two or three biological replicates with eight technical replicates each. (B‐D) Stable shNC‐ and shSYVN1‐transfected LM3 and HUH7 cells were injected subcutaneously into nude mice, and tumor lesions, volume, and weight were measured. Error bars represent the standard deviation. (E) Lysates from subcutaneous tumors formed by stable shNC‐ and shSYVN1‐transfected LM3 cells were subjected to SDS‐PAGE and Western blotting to detect the expression of SYVN1. (F) Cell proliferation under serum‐free conditions was detected at different time points using a CCK‐8 kit. Error bars represent the standard error of the mean from two or three biological replicates with eight technical replicates each. * P < 0.05, ** P < 0.01, *** P < 0.001. Abbreviations: NC, negative control
3.5. SYVN1 affected cell migration and angiogenesis
Cancer cell migration and invasion in the tumor microenvironment under the condition of serum and nutrient deficiency are crucial to tumor metastasis. We found that when LM3 and HUH7 cells in a serum‐deficient environment were stimulated by serum, the migration and invasion capacities of shSYVN1‐transfected cells were decreased (Figures 6A and 6B, Supplementary Figures S5B and S5C). The ability of cancer cells to promote angiogenesis affects tumor growth and metastasis. We found that the mesh area of HUVECs cultured in the medium from shSYVN1‐transfected LM3 and HUH7 cells was significantly smaller than that noted in the shNC group (Figures 6C and 6D). We also examined the mRNA expression of angiogenesis‐related molecules in LM3 and HUH7 cells. In the LM3 cell line, bFGF, PDGFBB, and PLGF mRNA levels in cells with low SYVN1 expression were significantly reduced compared with the shNC group; in the HUH7 cell line, bFGF, GCSF, PDGFAA, and VEGF mRNA levels in cells with low SYVN1 expression were significantly reduced (Figure 6E).
FIGURE 6.
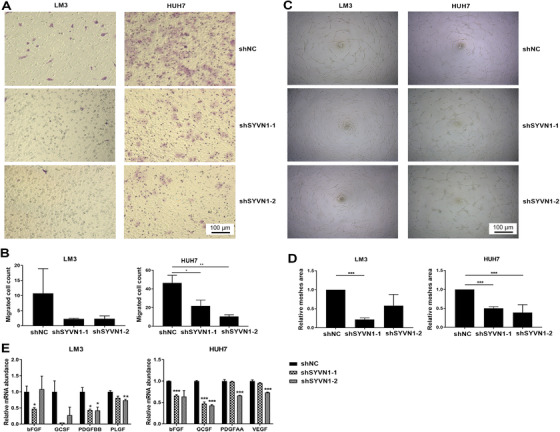
SYVN1 enhances cell migration and angiogenesis. (A‐B) LM3 and HUH7 cells infected with lentiviral shNC or shSYVN1 were subjected to a transwell migration assay. (C‐D) HUVECs were cultured on Matrigel‐coated plates with conditioned media from LM3 and HUH7 cells infected with lentiviral shNC or shSYVN1 for 12 h. The relative mesh areas to shNC group were calculated. (E) mRNA expression levels of genes involved in angiogenesis were measured by qPCR in LM3 and HUH7 cells infected with lentiviral shNC or shSYVN1. Error bars represent the standard deviation. * P < 0.05, ** P < 0.01, *** P < 0.001. Abbreviations: NC, negative control
3.6. SYVN1 interacted with HSP90 and increased EEF2K ubiquitination
To determine the mechanism of the influence of SYVN1 on HCC metastasis, we preliminarily screened various molecules interacting with SYVN1 using MS in both LM3 and HUH7 cell lines. The results showed that HSP90AA1, HSP90AB1, and HSP90B1 exhibited a very high probability of interacting with SYVN1 (Figure 7A and Supplementary Figure S6A). In addition, HSP90AA1, HSP90AB1, and HSP90B1 protein levels were highly correlated with SYVN1 protein levels in our whole‐cell proteomic data (Figure 7B and Supplementary Figure S6B). Co‐IP and laser confocal experiments also confirmed the interaction between HSP90 family members and SYVN1 (Figures 7C and 7D, Supplementary Figure S6C). HSP90 is used in the diagnosis of liver cancer and plays an important role in the development of liver cancer. CDC37 and STIP1 are the molecular chaperones of HSP90 which are used to indicate the activity of HSP90. In the whole‐cell proteomic data, HSP90 family proteins and their co‐chaperones exhibited significantly increased expression in tumor tissues than in corresponding peritumoral tissues in HCC with vascular invasion, but this difference was not significant in HCC without vascular invasion (Figure 7E and Supplementary Figure S6D). Thus, we considered that the HSP90 family potentially promoted HCC metastasis. Transcriptome data from the GEPIA database also indicated that these proteins were highly expressed in tumor tissues compared with the normal tissues, but the differences in HSP90AA1, HSP90B1, and CDC37 were not significant (Supplementary Figure S6E). However, IP and Western blotting experiments showed that SYVN1 did not affect the ubiquitination of the HSP90 family (Supplementary Figures S6F‐G). The relationship between SYVN1 and HSP90 may be more complex, and SYVN1 may influence liver cancer metastasis via the ubiquitination of other proteins.
FIGURE 7.
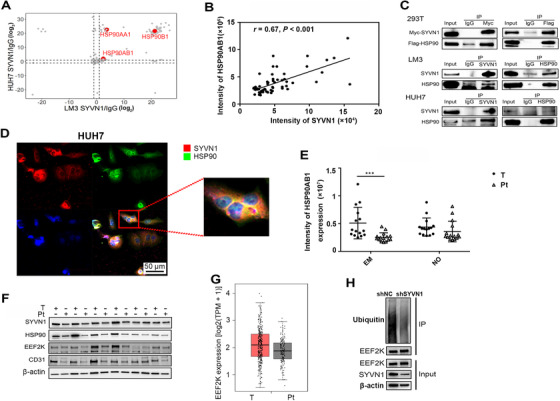
SYVN1 interacts with HSP90 and affects EEF2K ubiquitination. (A) Lysates from LM3 and HUH7 cells were subjected to SYVN1 and IgG pull‐down followed by protein detection using LC‐MS. The data were derived from three biological replicates. (B) Pearson's correlation plots for the protein intensities of HSP90AB1 and SYVN1 in 30 patients as detected by whole‐cell proteome analysis. (C) Lysates from 293T, LM3, and HUH7 cells were subjected to SYVN1 and HSP90 pull‐down followed by SDS‐PAGE and Western blotting. (D) HUH7 cells were fixed and stained using antibodies against SYVN1 (red), HSP90 (green), and DAPI (blue). (E) HSP90AB1 expression in tumor tissues and peritumoral tissues from whole‐cell proteome analysis. (F) EEF2K protein expression in tumor tissues and peritumoral tissues quantified by Western blotting. (G) The protein levels of EEF2K in HCC and normal tissues acquired from GEPIA. (H) Lysates from LM3 cells infected with lentiviral shNC or shSYVN1 were subjected to EEF2K pull‐down followed by SDS‐PAGE and Western blotting. Error bars represent the standard deviation. * P < 0.05, ** P < 0.01, *** P < 0.001. Abbreviations: HCC, hepatocellular carcinoma; EM, HCC with vascular invasion; NO, HCC without vascular invasion; T, tumor tissue; Pt, peritumoral tissue
EEF2K plays an important role in angiogenesis and tumor metastasis. In our ubiquitinome data, the ubiquitination of EEF2K was significantly increased in HCC with vascular invasion than in HCC without vascular invasion (Figure 4C). We used UbiBrowser to predict the E3 ubiquitin ligase of EEF2K and found that SYVN1 had the highest score (Figure 4C). Western blotting experiments showed that EEF2K, CD31 (a marker of vascular endothelial cells), and SYVN1 were highly expressed in tumor tissues (Figure 7F). Transcriptome data from the GEPIA database also indicated increased EEF2K expression in tumor tissues compared with normal tissues (Figure 7G). Therefore, we hypothesized that SYVN1 plays a role in promoting liver cancer metastasis via the ubiquitination of EEF2K. Finally, IP and Western blotting experiments revealed that SYVN1 increased EEF2K ubiquitination (Figure 7H). Surprisingly, HSP90 inhibition did not affect the expression of SYVN1 and EEF2K (Supplementary Figure S6H), the relationship between the three might be complicated.
4. DISCUSSION
In the present study, more than 10,000 protein ubiquitination modification sites were identified in HCC samples using ubiquitinome analysis. HCC tumor tissues had higher ubiquitination modification level than peritumoral tissues. The ubiquitination modification levels of EEF2K, TXNRD1, RASSF4, and other proteins were significantly increased in HCC with vascular invasion. SYVN1 promoted the proliferation of hepatoma cell lines both in vitro and in vivo. SYVN1 promoted angiogenesis and enhanced the migration and invasion of hepatoma cell lines. SYVN1 promoted the ubiquitination of EEF2K.
Ubiquitinome profiles are significantly different between tumor tissues and peritumoral tissues. The whole‐cell proteome has been well investigated in both HCC [31, 32]. Besides, ubiquitinome in HCC cell lines with different metastasis potentials have been investigated, and Ku80 was confirmed to be closely associated with invasion and migration of HCC cells [33]. Therefore, we profiled the global ubiquitination using a method for quantitative proteomics of diGly‐modified peptides and performed an integrative analysis of the ubiquitinome and whole‐cell proteome in patients with HCC to have a deeper understanding of HCC metastasis. Quantitative proteomics indicated that many proteins exhibited dysregulated ubiquitination; in general, ubiquitination was upregulated. Ubiquitination plays an important role in tumors, particularly in cell cycle pathways. For example, the polyubiquitination of P53 leads to its degradation and results in the reduction of tumor‐suppressive function [34]. The globally increased ubiquitination level in cancer tissues might be the result of increased cancer cell metabolism given their faster protein turnover rate. The total number of diGly‐modified sites was upregulated, but the diGly‐modified sites of ubiquitin itself were downregulated in HCC with vascular invasion compared with HCC without vascular invasion. We hypothesized that this phenomenon might have resulted from the upregulation of monoubiquitination, which remains to be proven by further research. Monoubiquitination is widespread in cells and has biological significance [35]. In addition, tumor‐suppressor genes, such as p53, forkhead box protein O (FOXO), and phosphatase and tensin (PTEN), depend on monoubiquitination for their correct localization and consequently proper tumor‐suppressor function [36]. The upregulation of polyubiquitination and monoubiquitination during tumorigenesis and the increase in global monoubiquitination during metastasis are beneficial to cancer cell survival by ensuring their rapid and efficient metabolism because the assembly of ubiquitin chains requires a significant amount of energy and monoubiquitination is sufficient for most functions.
E3 ubiquitin ligase SYVN1 plays an important role in HCC metastasis. Significant differences in the ubiquitination of some proteins, such as EEF2K, between HCC with and without vascular invasion suggest that the ubiquitination of these proteins contributes to HCC metastasis. E3 ubiquitin ligase is a key enzyme in the ubiquitination system that determines the specific recognition of substrate proteins. Using UbiBrowser, we predicted the E3 ubiquitin ligases of these proteins whose ubiquitination was significantly regulated and constructed the network diagram to identify the core enzyme SYVN1. In addition, SYVN1 was verified to be highly expressed in HCC with vascular invasion but not in HCC without vascular invasion. The changes of substrates and E3 ubiquitin ligases invariably suggested that SYVN1 played an indispensable role in HCC metastasis. Subsequently, we performed functional analysis on SYVN1 in HCC cell lines. Interestingly, SYVN1 did not affect the proliferation of HCC cells cultured in vitro but significantly promoted the growth of HCC cells in nude mice. Considering the differences in serum, oxygen, and carbon dioxide concentrations in in vivo and in vitro culture environments, we hypothesized that the influence of SYVN1 on HCC cell proliferation was restricted by one of the above factors. The proliferation‐promoting effect of SYVN1 in HCC cells in a low serum environment suggested that SYVN1 enhanced the tolerance of cancer cells to nutrient deficiency, thus, promoting tumor malignancy. This notion might be similar to the role of LLGL scribble cell polarity complex component 2 (LLGL2) protein in breast cancer in promoting leucine uptake under conditions of nutritional deficiency [37]. Interestingly, SYVN1 affects the proliferation of Hep3B cells both in vitro and in vivo [38], and the contradictory results need further study. In addition, the vascular metastasis of tumor requires abundant blood vessels and a strong migration and invasion ability of tumor cells in the tumor microenvironment. Our experimental data demonstrated that SYVN1 promoted the vascular metastasis of tumor based on all of the above aspects, which was consistent with ubiquitinome and whole‐cell proteome results.
SYVN1 enhanced the ubiquitination of EEF2K. To clarify the mechanism of SYVN1, we screened and confirmed its interaction with HSP90. HSP90 is a member of the heat shock protein family, and many HSP90 inhibitors have been developed to treat cancer [39, 40]. Therapy targeting HSP90 is used for cholangiocarcinoma [41], non–small cell lung cancer [42], acute lymphoblastic leukemias [43], and prostate cancer [44]. Unfortunately, we did not identify any effect of SYVN1 on HSP90 ubiquitination. The interaction patterns between these two molecules might be complex. To identify the substrates of SYVN1, we shifted focus to another molecule, namely, EEF2K. EEF2K ubiquitination was increased in HCC with vascular invasion, and SYVN1 was predicted to be its E3 based on UbiBrower analysis. Furthermore, EEF2K was reported to protect cancer cells from energy depletion or nutrient deprivation and promote angiogenesis [45, 46, 47], which was consistent with our experimental results. Inhibition or genetic deletion of EEF2K was also reported to promote tumor growth [48, 49]. We confirmed that SYVN1 affected the ubiquitination of EEF2K. Interestingly, the interaction between HSP90 and EEF2K has been previously reported [50, 51]. However, the relationships among SYVN1, HSP90, and EEF2K and the role of EEF2K in HCC metastasis were not investigated in this study.
5. CONCLUSIONS
We integrated quantitative proteomics of ubiquitination and the whole‐cell proteome in HCC, recognized robust changes in the ubiquitin‐proteasome system in the process of carcinogenesis and metastasis, and demonstrated that SYVN1 could be used as a new drug target for future cancer therapies.
DECLARATIONS
AUTHOR CONTRIBUTIONS
FJ, ZJ, MZ and ZS performed the majority of experiments. FJ and ZJ performed the majority of data and statistical analysis. FJ and MZ conceived and designed experiments. ZJ, FJ, ZS and MZ wrote and edited the manuscript. ZS and LL directed the study. HZ, ZJ, MZ and ZX collected HCC tissues and clinical patient information. XO and LZ assisted sample processing of LC‐MS.
CONSENT FOR PUBLICATION
Written informed consent for publication was obtained from all participants.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The experimental protocol was established, according to the ethical guidelines of the Helsinki Declaration and was approved by the Human Ethics Committee of the First Affiliated Hospital of Zhejiang University. Written informed consent was obtained from individual or guardian participants.
CONFLICT OF INTEREST
The authors declare no competing interests.
Supporting information
Supporting information
Supporting information
ACKNOWLEDGMENTS
We thank Jing Jiang for mass spectrometry technical support. We also thank Danhua Zhu and Xiaopeng Yu for helpful discussions. We thank Haiyang Xie and Leiming Chen for their assistance with sample collection. This work was supported by the National Key Research and Development Program (2017YFC1200100), the National Natural Science Foundation of China (81400589), Chinese National Science and Technology Major Project of the 13th Five‐year plan (2017ZX10202202‐001‐008).
Ji F, Zhou M, Sun Z, Jiang Z, Zhu H, Xie Z, et al. Integrative proteomics reveals the role of E3 ubiquitin ligase SYVN1 in hepatocellular carcinoma metastasis. Cancer Commun. 2021;41:1007–1023. 10.1002/cac2.12192
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2. Rodríguez‐Perálvarez M, Luong TV, Andreana L, Meyer T, Dhillon AP, Burroughs AK. A systematic review of microvascular invasion in hepatocellular carcinoma: diagnostic and prognostic variability. Ann Surg Oncol. 2013;20(1):325–39. 10.1245/s10434-012-2513-1 [DOI] [PubMed] [Google Scholar]
- 3. Thuluvath PJ. Vascular invasion is the most important predictor of survival in HCC, but how do we find it? J Clin Gastroenterol. 2009;43(2):101–2. 10.1097/MCG.0b013e318191e64f [DOI] [PubMed] [Google Scholar]
- 4. Wu Y, Liu Z, Xu X. Molecular subtyping of hepatocellular carcinoma: A step toward precision medicine. Cancer Commun (Lond). 2020;40(12):681–93. 10.1002/cac2.12115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yin J, Zhu JM, Shen XZ. The role and therapeutic implications of RING‐finger E3 ubiquitin ligases in hepatocellular carcinoma. Int J Cancer. 2015;136(2):249–57. 10.1002/ijc.28717 [DOI] [PubMed] [Google Scholar]
- 6. Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2(3):169–78. 10.1038/35056563 [DOI] [PubMed] [Google Scholar]
- 7. Ciechanover A, Heller H, Elias S, Haas AL, Hershko A. ATP‐dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc Natl Acad Sci U S A. 1980;77(3):1365–8. 10.1073/pnas.77.3.1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kwon YT, Ciechanover A. The Ubiquitin Code in the Ubiquitin‐Proteasome System and Autophagy. Trends Biochem Sci. 2017;42(11):873–86. 10.1016/j.tibs.2017.09.002 [DOI] [PubMed] [Google Scholar]
- 9. Haas AL, Warms JV, Hershko A, Rose IA. Ubiquitin‐activating enzyme. Mechanism and role in protein‐ubiquitin conjugation. J Biol Chem. 1982;257(5):2543–8. [PubMed] [Google Scholar]
- 10. Hershko A. Ubiquitin‐mediated protein degradation. J Biol Chem. 1988;263(30):15237–40. [PubMed] [Google Scholar]
- 11. Gallo LH, Ko J, Donoghue DJ. The importance of regulatory ubiquitination in cancer and metastasis. Cell Cycle. 2017;16(7):634–48. 10.1080/15384101.2017.1288326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8(8):645–54. 10.1593/neo.06376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu L, Qin C, Li T, Ma X, Qiu Y, Lin Y, et al. The E3 ubiquitin ligase TRIM7 suppressed hepatocellular carcinoma progression by directly targeting Src protein. Cell Death Differ. 2020;27(6):1819–31. 10.1038/s41418-019-0464-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tan X, He X, Fan Z. Upregulation of HRD1 promotes cell migration and invasion in colon cancer. Mol Cell Biochem. 2019;454(1‐2):1–9. 10.1007/s11010-018-3447-0 [DOI] [PubMed] [Google Scholar]
- 15. Schulz J, Avci D, Queisser MA, Gutschmidt A, Dreher LS, Fenech EJ, et al. Conserved cytoplasmic domains promote Hrd1 ubiquitin ligase complex formation for ER‐associated degradation (ERAD). J Cell Sci. 2017;130(19):3322–35. 10.1242/jcs.206847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamasaki S, Yagishita N, Nishioka K, Nakajima T. The roles of synoviolin in crosstalk between endoplasmic reticulum stress‐induced apoptosis and p53 pathway. Cell Cycle. 2007;6(11):1319–23. 10.4161/cc.6.11.4277 [DOI] [PubMed] [Google Scholar]
- 17. Xu YM, Wang HJ, Chen F, Guo WH, Wang YY, Li HY, et al. HRD1 suppresses the growth and metastasis of breast cancer cells by promoting IGF‐1R degradation. Oncotarget. 2015;6(40):42854–67. 10.18632/oncotarget.5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu L, Yu L, Zeng C, Long H, Duan G, Yin G, et al. E3 Ubiquitin Ligase HRD1 Promotes Lung Tumorigenesis by Promoting Sirtuin 2 Ubiquitination and Degradation. Mol Cell Biol. 2020;40(7):e00257–19. Published 2020 Mar 16. 10.1128/MCB.00257-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang WF, Yan L, Liu Z, Liu LX, Lin J, Liu ZY, et al. HSP70‐Hrd1 axis precludes the oncorepressor potential of N‐terminal misfolded Blimp‐1s in lymphoma cells. Nat Commun. 2017;8(1):363. Published 2017 Aug 25. 10.1038/s41467-017-00476-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, et al. The Skp2‐SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149(5):1098–1111. 10.1016/j.cell.2012.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arora S, Yang JM, Hait WN. Identification of the ubiquitin‐proteasome pathway in the regulation of the stability of eukaryotic elongation factor‐2 kinase. Cancer Res. 2005;65(9):3806–10. 10.1158/0008-5472.CAN-04-4036 [DOI] [PubMed] [Google Scholar]
- 22. Mínguez B, Hoshida Y, Villanueva A, Toffanin S, Cabellos L, Thung S, et al. Gene‐expression signature of vascular invasion in hepatocellular carcinoma. J Hepatol. 2011;55(6):1325–31. 10.1016/j.jhep.2011.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roayaie S, Blume IN, Thung SN, Guido M, Fiel MI, Hiotis S, et al. A system of classifying microvascular invasion to predict outcome after resection in patients with hepatocellular carcinoma. Gastroenterology. 2009;137(3):850–5. 10.1053/j.gastro.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 25. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Y, Xie P, Lu L, Wang J, Diao L, Liu Z, et al. An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase‐substrate interaction network. Nat Commun. 2017;8(1):347. Published 2017 Aug 24. 10.1038/s41467-017-00299-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13(9):731–40. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- 28. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98‐W102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma J, Chen T, Wu S, Yang C, Bai M, Shu K, et al. iProX: an integrated proteome resource. Nucleic Acids Res. 2019;47(D1):D1211–7. 10.1093/nar/gky869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Udeshi ND, Mertins P, Svinkina T, Carr SA. Large‐scale identification of ubiquitination sites by mass spectrometry. Nat Protoc. 2013;8(10):1950–60. 10.1038/nprot.2013.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang RC, Neo JC, Lo SL, Tan GS, Seow TK, Chung MC. Proteome database of hepatocellular carcinoma. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;771(1‐2):303–28. 10.1016/s1570-0232(02)00041-7 [DOI] [PubMed] [Google Scholar]
- 32. Ding SJ, Li Y, Shao XX, Zhou H, Zeng R, Tang ZY, et al. Proteome analysis of hepatocellular carcinoma cell strains, MHCC97‐H and MHCC97‐L, with different metastasis potentials. Proteomics. 2004;4(4):982–94. 10.1002/pmic.200300653 [DOI] [PubMed] [Google Scholar]
- 33. Sun Y, Zheng X, Yuan H, Chen G, Ouyang J, Liu J, et al. Proteomic analyses reveal divergent ubiquitylation patterns in hepatocellula carcinoma cell lines with different metastasis potential. J Proteomics. 2020;225:103834. 10.1016/j.jprot.2020.103834 [DOI] [PubMed] [Google Scholar]
- 34. Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin‐like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6(10):776–88. 10.1038/nrc1994 [DOI] [PubMed] [Google Scholar]
- 35. Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125(4):703–17. 10.1016/j.cell.2006.04.029 [DOI] [PubMed] [Google Scholar]
- 36. Salmena L, Pandolfi PP. Changing venues for tumour suppression: balancing destruction and localization by monoubiquitylation. Nat Rev Cancer. 2007;7(6):409–13. 10.1038/nrc2145 [DOI] [PubMed] [Google Scholar]
- 37. Saito Y, Li L, Coyaud E, Luna A, Sander C, Raught B, et al. LLGL2 rescues nutrient stress by promoting leucine uptake in ER+ breast cancer. Nature. 2019;569(7755):275–9. 10.1038/s41586-019-1126-2 [DOI] [PubMed] [Google Scholar]
- 38. Liu L, Long H, Wu Y, Li H, Dong L, Zhong JL, et al. HRD1‐mediated PTEN degradation promotes cell proliferation and hepatocellular carcinoma progression. Cell Signal. 2018;50:90–9. 10.1016/j.cellsig.2018.06.011 [DOI] [PubMed] [Google Scholar]
- 39. Kryeziu K, Bruun J, Guren TK, Sveen A, Lothe RA. Combination therapies with HSP90 inhibitors against colorectal cancer. Biochim Biophys Acta Rev Cancer. 2019;1871(2):240‐247. 10.1016/j.bbcan.2019.01.002 [DOI] [PubMed] [Google Scholar]
- 40. Franke J, Eichner S, Zeilinger C, Kirschning A. Targeting heat‐shock‐protein 90 (Hsp90) by natural products: geldanamycin, a show case in cancer therapy. Nat Prod Rep. 2013;30(10):1299–1323. 10.1039/c3np70012g [DOI] [PubMed] [Google Scholar]
- 41. Lampis A, Carotenuto P, Vlachogiannis G, Cascione L, Hedayat S, Burke R, et al. MIR21 Drives Resistance to Heat Shock Protein 90 Inhibition in Cholangiocarcinoma. Gastroenterology. 2018;154(4):1066–1079.e5. 10.1053/j.gastro.2017.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sang J, Acquaviva J, Friedland JC, Smith DL, Sequeira M, Zhang C, et al. Targeted inhibition of the molecular chaperone Hsp90 overcomes ALK inhibitor resistance in non‐small cell lung cancer. Cancer Discov. 2013;3(4):430–43. 10.1158/2159-8290.CD-12-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kucine N, Marubayashi S, Bhagwat N, Papalexi E, Koppikar P, Martin MS, et al. Tumor‐specific HSP90 inhibition as a therapeutic approach in JAK‐mutant acute lymphoblastic leukemias. Blood. 2015;126(22):2479–83. 10.1182/blood-2015-03-635821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lamoureux F, Thomas C, Yin MJ, Fazli L, Zoubeidi A, Gleave ME. Suppression of heat shock protein 27 using OGX‐427 induces endoplasmic reticulum stress and potentiates heat shock protein 90 inhibitors to delay castrate‐resistant prostate cancer [published correction appears in Eur Urol. 2016 Jul;70(1):e27‐e28]. Eur Urol. 2014;66(1):145–55. 10.1016/j.eururo.2013.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kenney JW, Moore CE, Wang X, Proud CG. Eukaryotic elongation factor 2 kinase, an unusual enzyme with multiple roles. Adv Biol Regul. 2014;55:15–27. 10.1016/j.jbior.2014.04.003 [DOI] [PubMed] [Google Scholar]
- 46. Wang X, Xie J, Proud CG. Eukaryotic Elongation Factor 2 Kinase (eEF2K) in Cancer. Cancers (Basel). 2017;9(12):162. Published 2017 Nov 27. 10.3390/cancers9120162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou Y, Li Y, Xu S, Lu J, Zhu Z, Chen S, et al. Eukaryotic elongation factor 2 kinase promotes angiogenesis in hepatocellular carcinoma via PI3K/Akt and STAT3. Int J Cancer. 2020;146(5):1383–95. 10.1002/ijc.32560 [DOI] [PubMed] [Google Scholar]
- 48. Faller WJ, Jackson TJ, Knight JR, Ridgway RA, Jamieson T, Karim SA, et al. mTORC1‐mediated translational elongation limits intestinal tumour initiation and growth. Nature. 2015;517(7535):497–500. 10.1038/nature13896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang X, Xie J, da Mota SR, Moore CE, Proud CG. Regulated stability of eukaryotic elongation factor 2 kinase requires intrinsic but not ongoing activity. Biochem J. 2015;467(2):321–31. 10.1042/BJ20150089 [DOI] [PubMed] [Google Scholar]
- 50. Yang J, Yang JM, Iannone M, Shih WJ, Lin Y, Hait WN. Disruption of the EF‐2 kinase/Hsp90 protein complex: a possible mechanism to inhibit glioblastoma by geldanamycin. Cancer Res. 2001;61(10):4010–16. [PubMed] [Google Scholar]
- 51. Xie J, Van Damme P, Fang D, Proud CG. Ablation of elongation factor 2 kinase enhances heat‐shock protein 90 chaperone expression and protects cells under proteotoxic stress. J Biol Chem. 2019;294(18):7169–76. 10.1074/jbc.AC119.008036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository [29] with the dataset identifiers PXD011895 and PXD018593. All other data that can support our findings are available in the article and supplemental information.


