Zhang et al. demonstrate EYA2 as a novel therapeutic target in glioblastoma stem cells with localization to the centrosome to promote mitotic spindle function and self-renewal. Inhibitors of EYA2 tyrosine phosphatase activity display selective therapeutic benefit for glioblastoma.
Abstract
Glioblastoma ranks among the most lethal of primary brain malignancies, with glioblastoma stem cells (GSCs) at the apex of tumor cellular hierarchies. Here, to discover novel therapeutic GSC targets, we interrogated gene expression profiles from GSCs, differentiated glioblastoma cells (DGCs), and neural stem cells (NSCs), revealing EYA2 as preferentially expressed by GSCs. Targeting EYA2 impaired GSC maintenance and induced cell cycle arrest, apoptosis, and loss of self-renewal. EYA2 displayed novel localization to centrosomes in GSCs, and EYA2 tyrosine (Tyr) phosphatase activity was essential for proper mitotic spindle assembly and survival of GSCs. Inhibition of the EYA2 Tyr phosphatase activity, via genetic or pharmacological means, mimicked EYA2 loss in GSCs in vitro and extended the survival of tumor-bearing mice. Supporting the clinical relevance of these findings, EYA2 portends poor patient prognosis in glioblastoma. Collectively, our data indicate that EYA2 phosphatase function plays selective critical roles in the growth and survival of GSCs, potentially offering a high therapeutic index for EYA2 inhibitors.
Introduction
Glioblastoma (World Health Organization grade IV glioma) is the most prevalent and malignant primary brain tumor, with a median survival of 12–15 mo (Ostrom et al., 2017; Weller et al., 2015). Standard-of-care treatment for glioblastoma includes maximal surgical resection, followed by concurrent radiotherapy and chemotherapy with the oral methylator temozolomide (Stupp et al., 2009). Although other therapies, such as tumor treating fields and bevacizumab, are approved for glioblastoma, current therapy remains palliative (Stupp et al., 2017). As one of the first tumors investigated by The Cancer Genome Atlas (TCGA; Cancer Genome Atlas Research Network, 2008), glioblastoma ranks among the most deeply characterized of human cancers, yet this molecular understanding has translated poorly into meaningful survival benefit for glioblastoma patients.
Glioblastoma was previously designated as glioblastoma multiforme, reflecting inter- and intra-patient heterogeneity with diversity in genetic aberrations and regional transcriptional program variations at the single cell level (Darmanis et al., 2017; Meyer et al., 2015; Patel et al., 2014). Cellular hierarchies in glioblastoma partially recapitulate the hierarchies observed during central nervous system development. Cells with stemlike properties, called glioblastoma stem cells (GSCs), stand at the apex of these complex and dynamic hierarchies (Lathia et al., 2015). Although the precise identification and cellular origins of GSCs remain under investigation, glioblastoma represents a solid tumor for which cancer stem cells can be reliably studied (Bao et al., 2006a; Galli et al., 2004; Singh et al., 2004). GSCs are functionally defined by their ability to self-renew, sustain proliferation, and propagate tumors upon transplantation (Galli et al., 2004; Singh et al., 2004). GSCs reside in specific supportive locations, including perivascular and hypoxia niches, which provide essential cues to sustain proliferation, angiogenesis, and invasion (Bao et al., 2006b; Calabrese et al., 2007; Jin et al., 2017). GSCs contribute to tumor recurrence after therapy due to preferential activation of DNA damage response pathways and aberrant checkpoint activation (Bao et al., 2006a).
Collectively, targeting GSCs could provide novel therapeutic strategies to improve patient outcome. Based on this background, we sought genetic targets that were specifically expressed by GSCs relative to their nontransformed counterparts (i.e., neural stem cells [NSCs]) and differentiated progeny (i.e., differentiated glioblastoma cells [DGCs]) that may serve as novel drug targets for the disease.
Results
GSCs preferentially express EYA2 (eyes absent homologue 2)
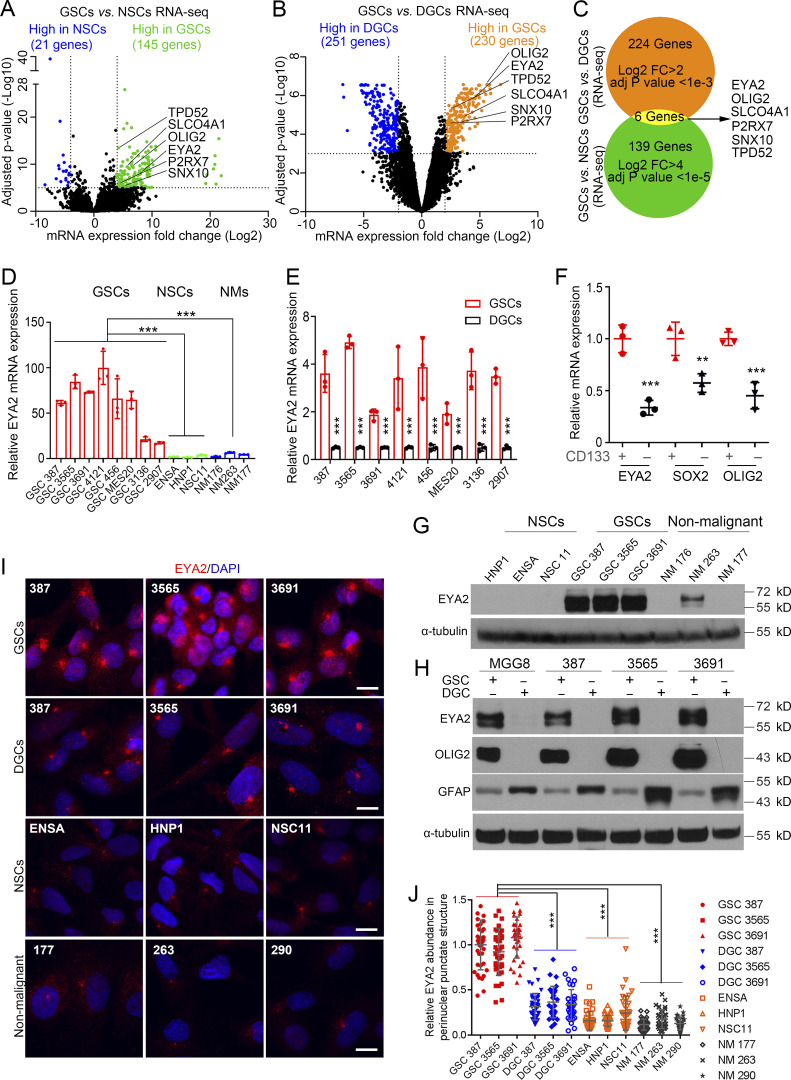
Differentially expressed genes (DEGs) provide a rich resource to discover regulators of cellular phenotypes. We reasoned that DEGs associated with defining cancer stemness in glioblastoma may yield key GSC dependencies amenable to targeting. Therefore, we took a two-stage approach by interrogating DEGs comparing GSCs to nontransformed NSCs (Fig. 1 A) and comparing GSCs to differentiated tumor cells (DGCs; Fig. 1 B). Although several biological processes were enriched in the DEGs expressed by GSCs from these comparisons (Fig. S1, A and B), DEGs that fulfilled both criteria were limited to six genes: EYA2, OLIG2, SLCO4A1, P2RX7, SNX10, and TPD52 (Fig. 1 C). Validating our approach, OLIG2 is a transcriptional regulator that maintains GSC proliferation and resistance to radiation (Mehta et al., 2011; Trépant et al., 2015). As EYA2 ranked the highest in the differential expression levels among the genes, we selected EYA2 for further study. EYA2 belongs to the EYA family, which contains four members with conserved domain structures that contain a tyrosine (Tyr) phosphatase domain and transactivation domain, as well as an associated serine/threonine phosphatase domain through an interaction with protein phosphatase 2A (PP2A; Zhang et al., 2018). To determine the epigenetic regulation of EYA2 in GSCs, we leveraged a cohort of matched, patient-derived GSCs and DGCs that we analyzed for active chromatin using histone 3 lysine 27 acetyl (H3K27ac) chromatin immunoprecipitation followed by deep sequencing, which revealed extensive preferential activated chromatin consistent with enhancer activity close to the EYA2 gene promoter region in GSCs (Fig. S1 C).
Figure 1.
GSCs preferentially express EYA2 relative to normal NSCs and differentiated tumor cells. (A) Volcano plot of DEGs in 38 GSCs and 5 NSCs (GSE119834). Genes highly expressed in GSCs were labeled as green, and genes highly expressed in NSCs were labeled as blue. As indicated by the dotted lines, cutoff was set at fold change (FC) >4, adjusted (adj) P value <1e-5. (B) Volcano plot of three GSCs and their corresponding DGCs (GSE54791). Genes highly expressed in GSCs were labeled as orange, and genes highly expressed in DGCs were labeled as blue. As indicated by the dotted lines, cutoff was set at fold change >2, adjusted P value <1e-3. (C) Venn diagram visualizes the overlap in genes highly expressed in GSCs based on the comparison between GSCs and NSCs (green) and the comparison between GSCs and DGCs (orange). The overlap is shown in yellow. (D) EYA2 mRNA in GSCs, NSCs, and NMs. Data are presented as mean ± SD. The P values were calculated by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (E) EYA2 mRNA levels in GSCs and corresponding DGCs. Data are presented as mean ± SD. The P values were calculated by two-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (F) mRNA levels of EYA2, SOX2, and OLIG2 in CD133+ versus CD133− tumor cells sorted from xenografted tumors derived from GSC 387 expressing mCherry. Data are presented as mean ± SD. The P values were calculated by two-way ANOVA with Tukey’s multiple comparisons. **, P < 0.01; ***, P < 0.001. (G) Immunoblotting of EYA2 protein in GSCs, NSCs, and NMs. α-Tubulin was used as loading control. (H) Immunoblotting of EYA2 protein in GSCs and corresponding DGCs. OLIG2 was used as GSC marker while GFAP was used as DGC marker. α-Tubulin was used as loading control. (I) Immunofluorescence staining of EYA2 in GSCs, NSCs, DGCs, and NMs. EYA2 is shown in red, DAPI in blue. Scale bars represent 10 µm. (J) Quantification of EYA2 staining in the perinuclear punctate structure in GSCs, NSCs, DGCs, and NMs. Data are presented as mean ± SD. The P values were calculated by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. D, E, and G–J are representative of three independent experiments. Three mice were used in F. At least 30 cells per GSC model were quantified in J.
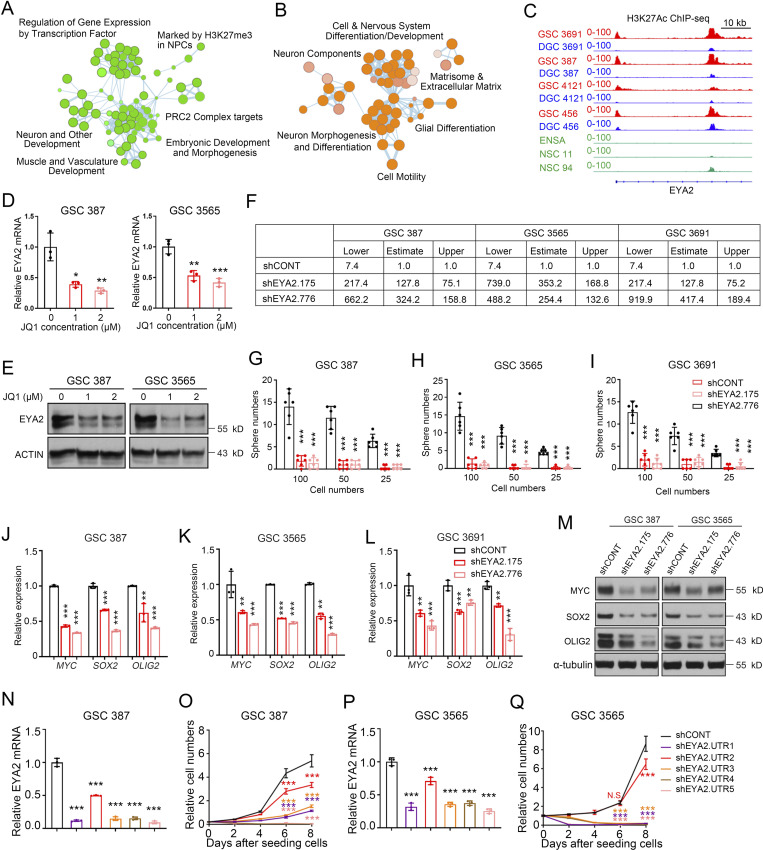
Figure S1.
EYA2 is required for GSC maintenance. (A and B) Pathway enrichment bubble plots of DEGs preferentially expressed in GSCs from the comparison between GSCs and NSCs (A) and the comparison between GSCs and DGCs (B). NPC, neural progenitor cell. (C) H3K27Ac immunoprecipitation followed by deep sequencing (ChIP-seq) profiles in GSCs (red) and their corresponding DGCs (blue) and NSCs (green) are shown around the promoter region of EYA2. (D and E) EYA2 mRNA (D) and protein (E) levels in different GSCs after treatment with JQ1. Data are presented as mean ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons. *, P < 0.05; **, P < 0.01; ***, P < 0.001. ACTIN was used as loading control. (F) Confidence intervals of stem cell frequencies in different GSCs transduced with shCONT or shEYA2. (G–I) Sphere numbers of different GSCs transduced with shCONT or shEYA2. Data are presented as mean ± SD. Significance was determined by two-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (J–L) qPCR analysis of MYC, SOX2, and OLIG2 expression in GSCs transduced with shCONT or shEYA2. Data are presented as mean ± SD. Significance was determined by two-way ANOVA with Tukey’s multiple comparisons. **, P < 0.01; ***, P < 0.001. (M) Immunoblotting of MYC, SOX2, and OLIG2 expression in GSCs transduced with shCONT or shEYA2. α-Tubulin was used as loading control. (N) Relative EYA2 mRNA measured by qPCR in GSC 387 transduced with shCONT or different shRNAs (shEYA2.UTRs) targeting the 3′ UTR region of EYA2 mRNA. Data are presented as mean ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (O) CellTiter-Glo assay to measure the growth of GSC 387 following transduction with shCONT or shEYA2.UTRs. Data are presented as mean ± SD. Significance was determined by two-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (P) Relative EYA2 mRNA measured by qPCR in GSC 3565 transduced with shCONT or different shRNAs (shEYA2.UTRs) targeting the 3′ UTR region of EYA2 mRNA. Data are presented as mean ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (Q) CellTiter-Glo assay to measure the growth of GSC 3565 following transduction with shCONT or shEYA2.UTRs. Data are presented as mean ± SD. Significance was determined by two-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. All data except A and B are representative of three independent experiments.
The BRD4 inhibitor, JQ1, disrupts enhancers to block gene expression (Lovén et al., 2013). To interrogate the functions of the GSC-specific enhancer proximal to the EYA2 promoter, patient-derived GSCs were treated with JQ1, inducing a reduction of EYA2 expression at both the mRNA and protein levels (Fig. S1, D and E), suggesting that EYA2 is epigenetically up-regulated in GSCs. To confirm preferential GSC expression of EYA2, we examined its comparative expression in a panel of patient-derived GSCs and nonneoplastic neural cells, including NSCs and nonmalignant brain cultures (NMs) derived from epilepsy surgical resections. EYA2 mRNA expression was consistently higher in GSCs compared with neural cells or DGCs (Fig. 1, D and E), which was confirmed by elevated EYA2 expression in xenografted tumor cells flow sorted by CD133 surface expression (Fig. 1 F), a well-known cancer stem cell marker. As expected, EYA2 expression was up-regulated in GSCs compared with neural cells or DGCs at the protein level (Fig. 1, G and H).
EYA2 localizes both to the nucleus, where it serves as a coactivator to the sine oculis homeobox (SIX) family of transcription factors and a phosphatase to regulate the response to DNA damage (Cook et al., 2009; Krishnan et al., 2009), and in the cytoplasm, where it acts to enhance migration and invasion (Pandey et al., 2010; Zhou et al., 2018). In addition to the expected nuclear staining, EYA2 was detectable in a perinuclear punctate structure in GSCs, as revealed by immunofluorescent staining, whereas weaker EYA2 signals from such structures were observed in differentiated tumor cells and normal brain cells (Fig. 1, I and J). Collectively, these data indicate that EYA2 is preferentially expressed in GSCs with a novel perinuclear location.
EYA2 maintains GSC proliferation and self-renewal
EYA2 up-regulation in GSCs suggested potential dependency of GSCs on EYA2. To interrogate the functional roles of EYA2 in GSCs, we targeted EYA2 expression using two nonoverlapping shRNAs against EYA2 (designated shEYA2.175 and shEYA2.776 based on the start site of each shRNA sequence) compared with a control shRNA with a sequence that does not target any sequence in the mammalian genome (shCONT). As measured by a luminescence-based cell viability assay, disruption of EYA2 expression impaired GSC growth (Fig. 2 A). Extreme Limiting Dilution Assay, a surrogate readout of self-renewal, showed that EYA2 silencing reduced GSC self-renewal (Fig. 2 B; and Fig. S1, F–I) and expression of key transcription factors essential for GSC self-renewal (MYC, SOX2, and OLIG2) at the mRNA and protein levels (Fig. S1, J–M).
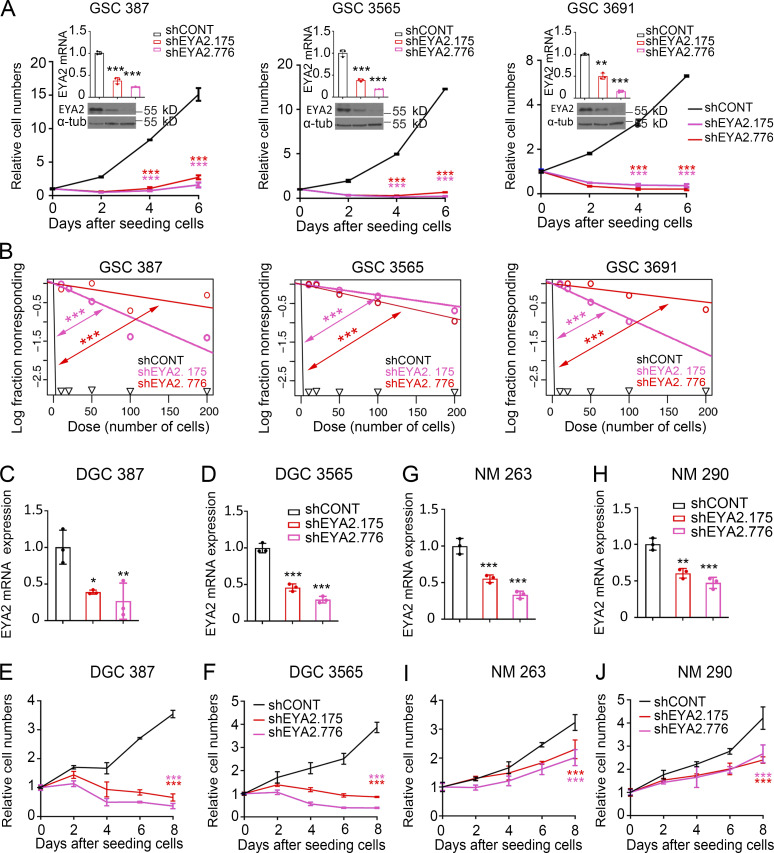
Figure 2.
EYA2 is necessary for GSC maintenance. (A) CellTiter-Glo assays to measure the GSC proliferation after EYA2 knockdown. Knockdown efficiency of EYA2 was examined by qPCR and Western blot. Data are presented as the mean ± SD. The P values were calculated in qPCR assays by one-way ANOVA with Tukey’s multiple comparisons. The P values were calculated in CellTiter-Glo assays by two-way ANOVA with Tukey’s multiple comparisons. **, P < 0.01; ***, P < 0.001. α-Tubulin (α-tub) was used as loading control for immunoblots. (B) Limiting dilution assays with different GSCs transduced by shCONT or shEYA2. ***, P < 0.001 by χ2 test for pairwise differences. (C and D) Knockdown efficiency of EYA2 examined by qPCR and Western blot in DGC 387 (C) and DGC 3565 (D). α-Tubulin was used as loading control. Data are presented as mean ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (E and F) CellTiter-Glo assays to measure cell growth of DGC 387 (E) and DGC 3565 (F) after EYA2 knockdown. Data are presented as mean ± SD. Significance was determined by two-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (G and H) Knockdown efficiency of EYA2 examined by qPCR and Western blot in NM 263 (G) and NM 290 (H). α-Tubulin was used as loading control. Data are presented as mean ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons. **, P < 0.01; ***, P < 0.001. (I and J) CellTiter-Glo assays to measure cell growth of NM 263 (I) and NM 290 (J) after EYA2 knockdown. Data are presented as mean ± SD. Significance was determined by two-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. All data are representative of three independent experiments.
To interrogate the functional importance of EYA2 to GSCs and rule out off-target effects, several additional shRNAs targeting EYA2 were tested; every shRNA that diminished EYA2 levels caused growth defects in GSCs, with the extent of the defect largely in proportion to the efficiency of reduction in EYA2 expression (Fig. S1, N–Q). Targeting EYA2 also moderately reduced the proliferation of DGCs, suggesting a role for EYA2 in both GSCs and DGCs, but showed much lower potency in nonneoplastic cells (Fig. 2, C–J). These data demonstrate a dependency of GSCs and DGCs on EYA2 compared with nonneoplastic brain cells. To our knowledge, these results are the first to demonstrate the essentiality of EYA2 to GSC maintenance.
EYA2 is required for GSC cell cycle progression and survival
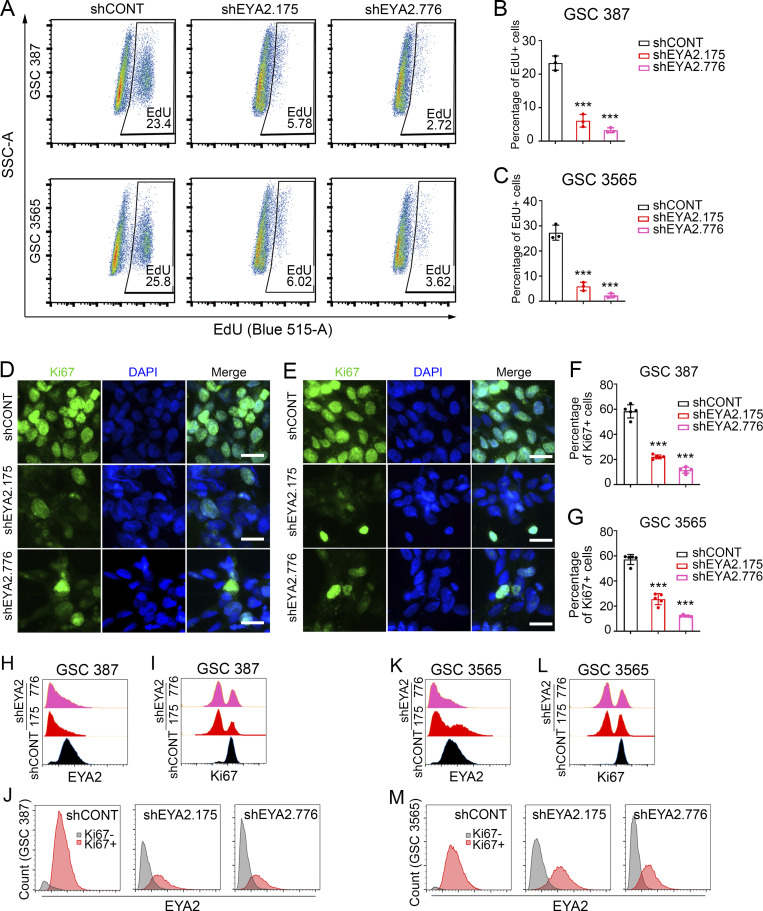
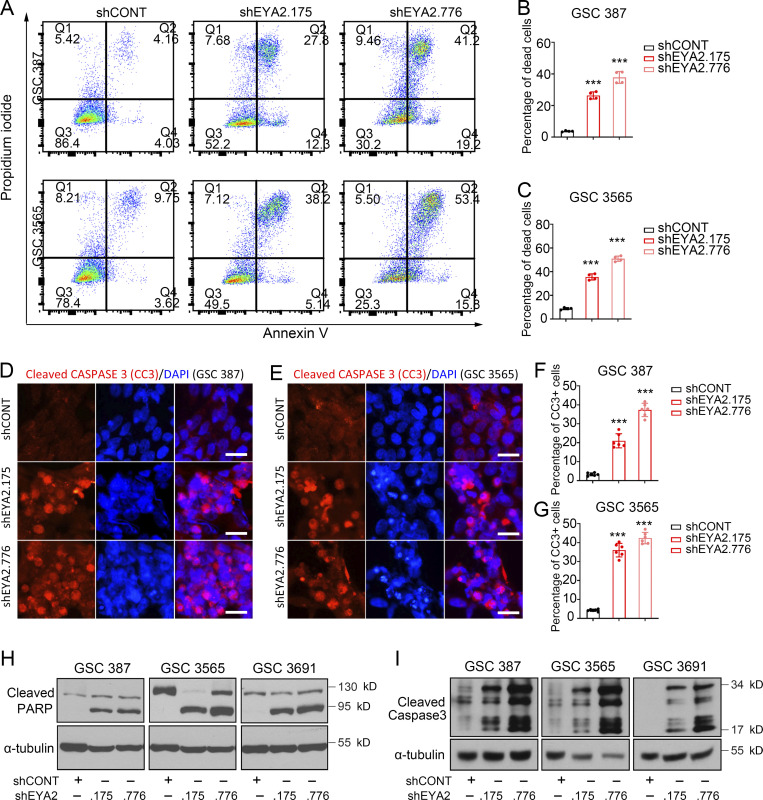
To dissect the cellular effects of EYA2 on GSCs, we interrogated cell cycle progression and induction of apoptosis following silencing of EYA2. GSCs transduced with either shEYA2 or shCONT were pulse-labeled with 5-ethynyl-2-deoxyuridine (EdU) to assess the fraction of actively proliferating cells. Flow cytometric (FACS) analysis revealed a strong reduction in the percentage of EdU+ cells upon targeting EYA2 in patient-derived GSCs (Fig. 3, A–C), which was confirmed using Ki67 staining (Fig. 3, D–G). FACS analysis of EYA2 and Ki67 levels in patient-derived GSCs upon EYA2 knockdown further supported the critical roles of EYA2 in GSC proliferation (Fig. 3, H–M). EYA2 loss also induced apoptosis in GSCs, which was confirmed with three complementary assays: annexin V–propidium iodide (PI) staining quantified by FACS analysis (Fig. 4, A–C); cleaved CASPASE 3 immunofluorescence (Fig. 4, D–G); and immunoblotting to detect cleaved poly-ADP ribose polymerase (PARP) and CASPASE 3 (Fig. 4, H and I). Collectively, these results demonstrate that EYA2 is required for GSC cell cycle progression and survival.
Figure 3.
EYA2 knockdown impairs GSC proliferation. (A) Flow cytometry of EdU-incorporated GSCs transduced with shCONT or shEYA2. (B and C) Quantification of the EdU+ population in GSC 387 (B) or GSC 3565 (C). Data are presented as mean ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (D and E) Ki67 staining in GSC 387 (D) or GSC 3565 (E) transduced with shCONT or shEYA2. Ki67 is shown in green, DAPI in blue. Scale bars represent 20 µm. (F and G) Quantification of Ki67+ cells in GSC 387 (F) or GSC 3565 (G) transduced with shCONT or shEYA2. Data are presented as mean ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (H and I) EYA2 (H) and Ki67 (I) protein levels measured by FACS in GSC 387 transduced with shEYA2 or shCONT. (J) EYA2 expression in Ki67+ versus Ki67− populations of GSC 387 transduced with shEYA2 or shCONT. (K and L) EYA2 (K) and Ki67 (L) protein levels measured by FACS in GSC 387 transduced with shEYA2 or shCONT. (M) EYA2 expression in Ki67+ versus Ki67− populations of GSC 387 transduced with shEYA2 or shCONT. A and D–M are representatives of at least three independent experiments. Five different 60× images per arm were used in F and G. B and C are from three independent experiments. SSC-A, side scatter-A.
Figure 4.
Disrupting EYA2 expression causes GSC apoptosis. (A) Flow cytometry of annexin V/PI-stained GSCs transduced with shCONT or shEYA2. (B and C) Quantification of dead cells labeled as annexin V/PI double-positive in GSC 387 (B) and GSC 3565 (C) transduced with shCONT or shEYA2. Data are presented as mean ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (D and E) Immunofluorescence staining of cleaved CASPASE 3 (CC3) in GSC 387 (D) and GSC 3565 (E) transduced with shCONT or shEYA2. CC3 is shown in red, DAPI in blue. Scale bars represent 20 µm. (F and G) Quantification of CC3+ cells in GSC 387 (F) and GSC 3565 (G) transduced with shCONT or shEYA2. Data are presented as mean ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (H and I) Western blot of PARP (H) and cleaved CASPASE 3 (I) in GSCs transduced with shCONT or shEYA2. α-Tubulin was used as loading control. A and D–I are representatives of three independent experiments. B and C are representatives of four independent experiments. Six different 60× images per arm were used in F and G.
EYA2 phosphatase function is required for GSC maintenance
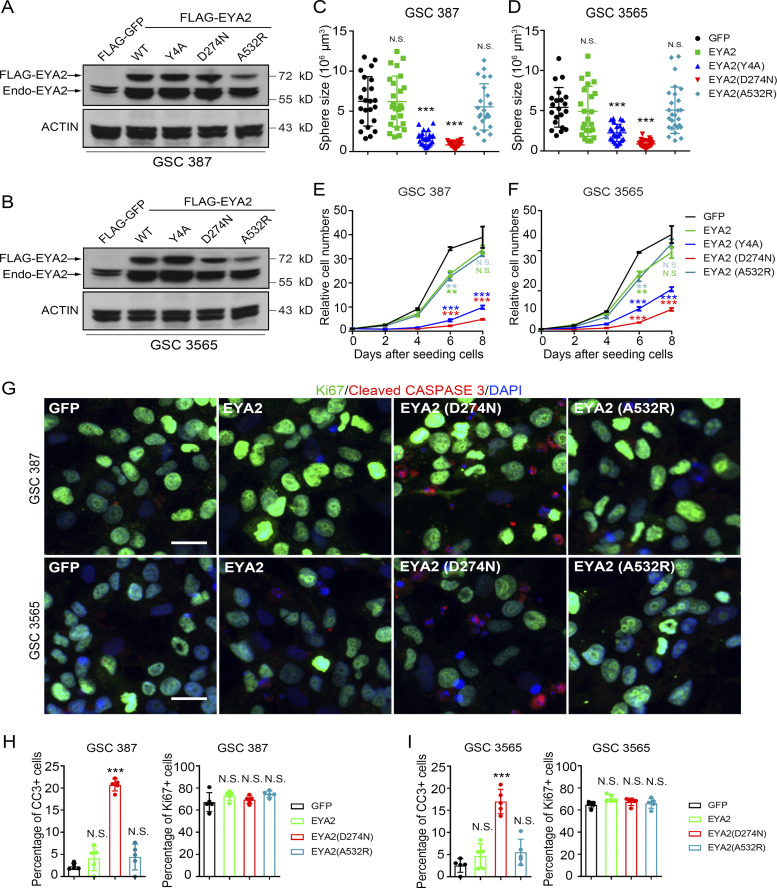
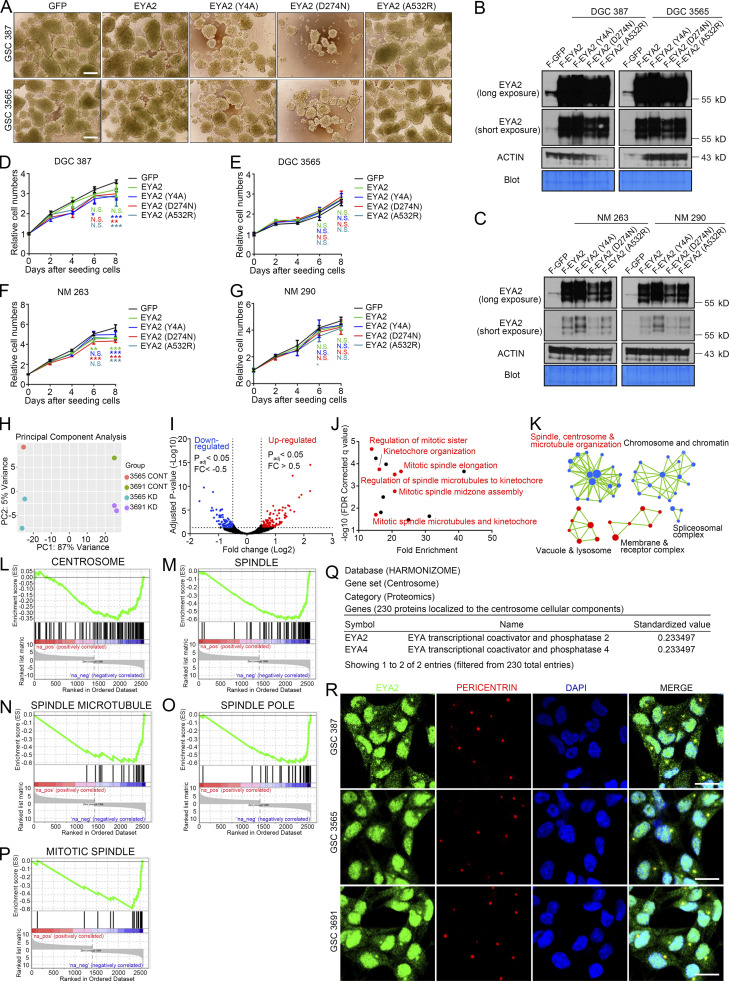
EYA2 is a multifunctional protein with discrete domains that transactivate the SIX transcription factors or function as an intrinsic phosphatase for selected Tyr (Tadjuidje and Hegde, 2013) or, through an interaction with PP2A, for serine/threonine (Zhang et al., 2018). To dissect the specific EYA2 function in GSCs, we leveraged distinct EYA2 mutants to selectively disrupt interactions with SIX family members (EYA2A532R; Patrick et al., 2013) or inhibit Tyr phosphatase function (EYA2D274N; Pandey et al., 2010). We also added an EYA2 mutant that was based on mutations in other EYA family members shown to disrupt the associated serine/threonine phosphatase function of EYA (EYA2Y4A; Okabe et al., 2009). GSCs were transduced with FLAG-GFP, WT FLAG-EYA2, or one of the EYA2 mutants (Fig. 5, A and B), then underwent evaluation for sphere formation and proliferation. GSCs transduced with FLAG-GFP, WT EYA2, or a mutant defective in binding SIX family members and associated transactivating function (EYA2A532R) displayed similar sphere sizes (considered a surrogate of proliferation) and cell number (Fig. 5, C–F; and Fig. S2 A). In contrast, GSCs transduced with EYA2 mutants targeting Tyr phosphatase function (EYA2D274N) or, to a lesser degree, that have been shown to disrupt associated serine/threonine phosphatase function in other EYA family members (EYA2Y4A) inhibited sphere size and cell numbers over time (Fig. 5, C–F; and Fig. S2 A). Supporting a specific EYA2 function in GSCs, overexpression of WT EYA2 or EYA2 mutants caused little to no change in cellular growth in DGCs or in the nonmalignant neural cells derived from epilepsy resections (NMs), despite far greater levels of overexpression in these conditions than in GSCs (Fig. S2, B–G), suggesting that the phosphatase activity of EYA2 plays a unique role in GSCs.
Figure 5.
EYA2 phosphatase is required for GSC maintenance. (A and B) Western blot for EYA2 in GSC 387 (A) and GSC 3565 (B) overexpressing FLAG-tagged WT or mutant EYA2. “Endo-EYA2” indicates endogenous EYA2. ACTIN was used as loading control. (C and D) Sphere size of GSC 387 (C) or GSC 3565 (D) overexpressing WT or mutant EYA2. Data are presented as mean ± SD. The P values were calculated by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (E and F) CellTiter-Glo assay to measure the cell growth with GSC 387 (E) or GSC 3565 (F) overexpressing WT or mutant EYA2. Data are presented as mean ± SD. The P values were calculated by two-way ANOVA with Tukey’s multiple comparisons. **, P < 0.01; ***, P < 0.001. (G) Immunofluorescence staining of Ki67 and cleaved CASPASE 3 (CC3) in GSC 387 and GSC 3565 overexpressing WT or mutant EYA2. Ki67 is shown in green, CC3 in red, and DAPI in blue. Scale bar represents 20 µm. (H and I) Quantification of CC3+ cells (left) and Ki67+ cells (right) in GSC 387 (H) or GSC 3565 (I) overexpressing WT or mutant EYA2. Data are presented as mean ± SD. The P values were calculated by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. All data are representative of three independent experiments. Five different 60× images per arm were used in H and I.
Figure S2.
EYA2 is associated with spindle formation in GSCs, but its phosphatase activities are not required for non-GSC cells. (A) Sphere size of GSC 387 or GSC 3565 overexpressing WT or mutant EYA2. Scale bar represents 200 µm. (B) Immunoblotting of EYA2 protein in DGC 387 or DGC 3565 overexpressing WT or different EYA2 mutants. ACTIN and the Coomassie Blue–stained PVDF membrane were used as loading control. (C) Immunoblotting of EYA2 protein in NM 263 or NM 290 overexpressing WT or different EYA2 mutants. ACTIN and the Coomassie Blue–stained PVDF membrane were used as loading control. (D–G) CellTiter-Glo assay to measure the growth of DGC 387 (D), DGC 3565 (E), NM 263 (F), and NM 290 (G) overexpressing GFP, WT EYA2, and different EYA2 mutants. Data are presented as mean ± SD. Significance was determined by two-way ANOVA with Tukey’s multiple comparisons. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (H) Principal component analysis with the RNA-seq datasets obtained from GSC 3565 and GSC 3691 transduced with shCONT (CONT) or shEYA2 (KD). (I) Volcano plot to show the common changes in transcriptional profiles between GSC 3565 and GSC 3691 upon EYA2 knockdown. Genes with adjusted (adj) P value <0.05, fold change (FC) >0.5 were labeled as red (up-regulated), and genes with adjusted P value <0.05, fold change less than −0.5 were labeled as blue (down-regulated). (J) GO analysis of the down-regulated genes in GSCs upon EYA2 knockdown. (K) Network analysis of the DEGs after EYA2 knockdown in GSCs. Pre-ranked genes (adjusted P value <0.05) based on fold change were processed to GSEA. GO_biological process was selected as the targeted gene sets. (L–P) Enrichment plots for gene sets associated with centrosome and spindle. (Q) Localization of EYA2 and EYA4 to centrosome revealed by Harmonizome proteomics database. (R) Co-staining of EYA2 and centrosome marker, PERICENTRIN, in different GSC 387, GSC 3565, and GSC 3691. EYA2 is shown in green, PERICENTRIN in red, and DAPI in blue. Scale bars represent 20 µm. A–G and R are representative of three independent experiments. FDR, false discovery rate.
As expression of the Tyr phosphatase–defective EYA2 mutant caused the most potent suppression of GSC growth (Fig. 5, C–F) and the Tyr phosphatase activity of EYA2 is targetable, we focused on the impact of the EYA2 Tyr phosphatase activity in GSCs moving forward. Of note, we confirmed that the EYA2D274N mutant lacked phosphatase activity, but we were unable to detect dominant negative effects (data not shown), likely due to limitations with the phosphatase activity assay. EYA2 loss of function decreased proliferation and increased apoptosis in GSCs, so we interrogated the effects of EYA2 mutants on Ki67 and cleaved CASPASE 3 levels, revealing that GSCs transduced with WT EYA2 or a mutant with deficient transactivation displayed baseline levels of proliferation and apoptosis, whereas transduction with the EYA2 mutant deficient in Tyr phosphatase activity induced apoptosis (Fig. 5, G–I). Sustained Ki67 staining in GSCs expressing EYA2 mutants suggested that distinct EYA2 functions contribute together to regulate GSC proliferation. Concordant with the differential levels of EYA2 in GSCs versus DGCs and NSCs, EYA2 Tyr phosphatase function was particularly important in GSC maintenance, providing a potential therapeutic index for targeting EYA2.
EYA2 is associated with centrosome and mitotic spindle control
To interrogate the molecular mediators of EYA2 in GSC maintenance, we performed RNA sequencing (RNA-seq) on two patient-derived GSCs transduced with either shEYA2 or shCONT. Unsupervised principal component analysis grouped the GSCs separately, but targeting EYA2 expression induced transcriptional changes shared by the GSCs (Fig. S2, H and I). Gene ontology (GO) analysis of the shared DEGs in GSCs after EYA2 knockdown revealed enrichment of transcriptional programs related to mitosis and mitotic spindle control (Fig. S2 J), which was confirmed by gene set enrichment analysis (GSEA; Fig. S2, K–P). Interrogating the Harmonizome database, a collection of information about genes and proteins from 114 datasets of multiple cell types provided by 66 online resources (Rouillard et al., 2016), suggested that EYA2 as well as its related family member EYA4 were components of the centrosome (Fig. S2 Q), consistent with the perinuclear punctate EYA2 localization in GSCs (Fig. 1 I). Collectively, these data suggest dual functions for EYA2 in GSCs that likely entail transcriptional as well as nontranscriptional functions through localization outside of the nucleus.
EYA2 localizes to GSC centrosomes
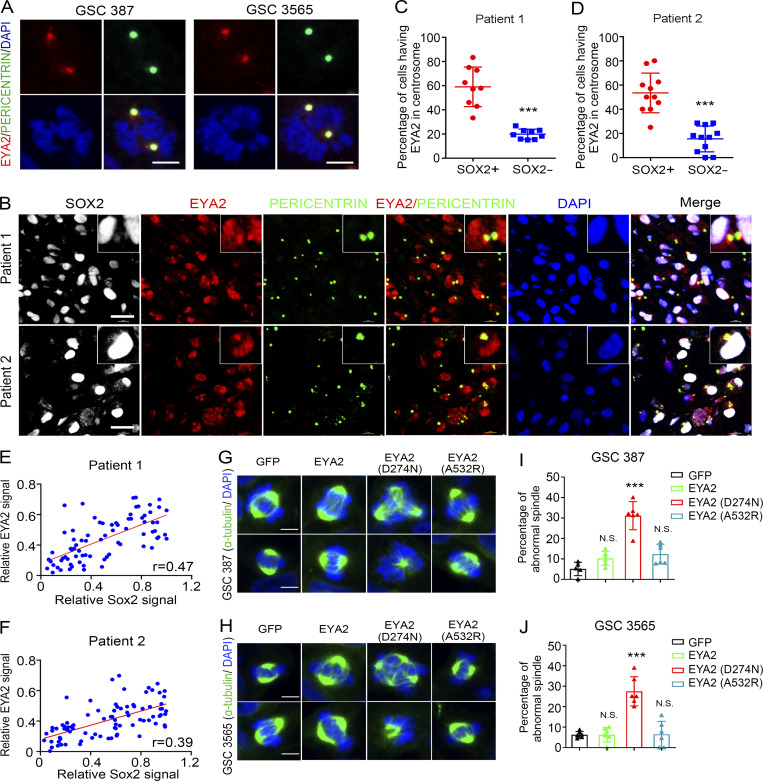
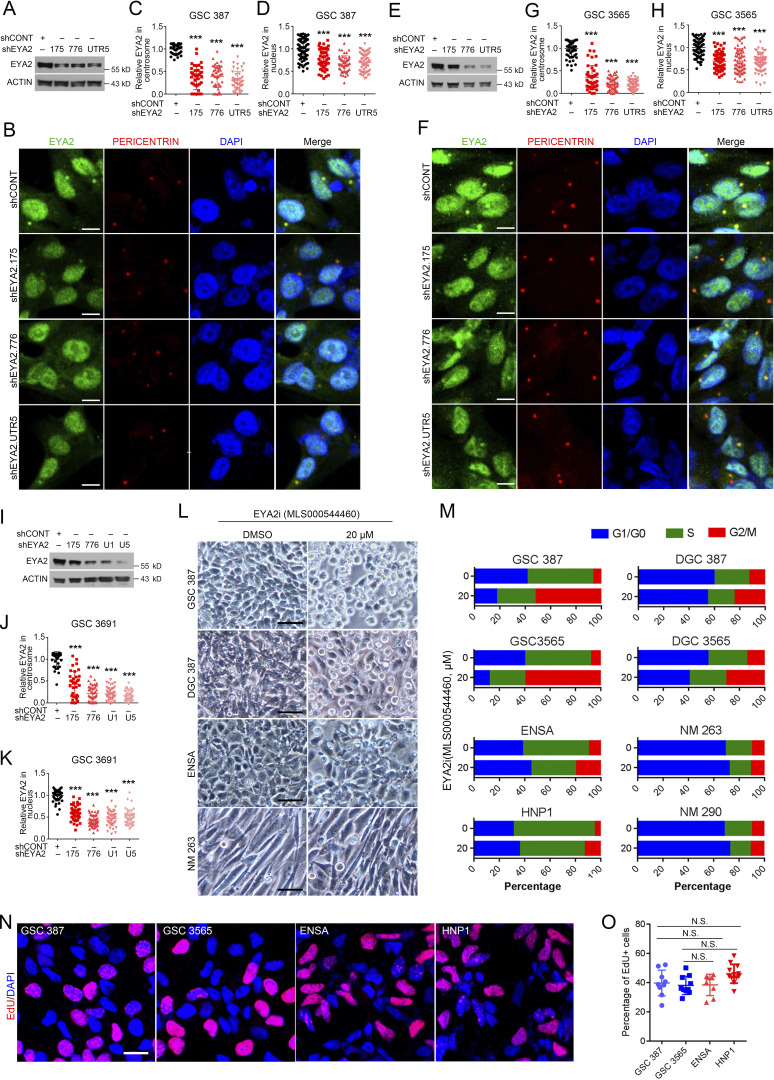
Given the perinuclear localization, we sought to determine if EYA2 localizes to centrosomes in GSCs. GSCs were co-stained with EYA2 and the centrosome marker PERICENTRIN. While EYA2 staining was observed in the nucleus as expected, we observed an additional novel localization of EYA2 to the perinuclear cytoplasm in puncta that were PERICENTRIN-positive (Fig. S2 R). During the cell cycle, centrosomes are duplicated and then separate from each other, which is critical for proper spindle formation and cell cycle progression (Tanenbaum and Medema, 2010). In cycling cells, EYA2 signals were duplicated along with the centrosomes in GSCs (Fig. 6 A), suggesting a functional association of EYA2 with centrosomes in GSCs. Supporting EYA2 localization to the centrosome, EYA2 knockdown in three patient-derived GSCs induced a loss of EYA2 signal not only in the nucleus but also in the centrosome (Fig. S3, A–K).
Figure 6.
EYA2 localizes to GSC centrosomes. (A) Co-staining of EYA2 and PERICENTRIN in cycling cells of GSC 387 (left) and GSC 3565 (right). EYA2 was labeled as red, PERICENTRIN as green, and DAPI as blue. Scale bars represent 5 µm. (B) Immunofluorescence staining of SOX2, EYA2, and PERICENTRIN in two different glioblastoma patient surgical specimens. SOX2 is shown in gray, EYA2 in red, PERICENTRIN in green, and DAPI in blue. Scale bars represent 20 µm. (C and D) Quantification of EYA2 localization to the centrosome in SOX2+ or SOX2− cells in glioblastoma specimens from patient 1 (C) or patient 2 (D). Data are presented as mean ± SD. ***, P < 0.001. The P values were calculated by unpaired t test. (E and F) Correlation between EYA2 and SOX2 expression in glioblastoma patient samples. (G and H) Immunofluorescence staining of α-tubulin in GSC 387 (G) and GSC 3565 (H) overexpressing GFP, WT EYA2, or different EYA2 mutants. Scale bars represent 5 µm. (I and J) Quantification of the aberrant spindles found in GSC 387 (I) and GSC 3565 (J) expressing WT or mutant EYA2. Data are presented as mean ± SD. The P values were calculated by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. Data are representatives of three independent experiments except B–F. Tumor samples from two glioblastoma patients were used in B, and 9 or 11 different images were quantified in C and D, respectively. 77 or 84 cells from 10 different 120× images per patient were measured in E and F, respectively. Six different 80× images were quantified in I and J.
Figure S3.
EYA2 localizes to centrosomes, and EYA2 Tyr phosphatase inhibitor treatment induces selective G2/M arrest in GSCs. (A) Immunoblot of EYA2 in GSC 387 transduced with shCONT or shRNAs targeting EYA2. ACTIN was used as loading control. (B) Co-staining of EYA2 and PERICENTRIN in GSC 387 transduced with shCONT or shRNAs targeting EYA2. EYA2 is shown in green, PERICENTRIN in red, and DAPI in blue. Scale bars represent 10 µm. (C and D) Quantification of EYA2 staining in centrosome (C) or nucleus (D) of GSC 387 transduced with shCONT or shRNAs targeting EYA2. Data are presented as mean ± SD. ***, P < 0.001. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison. (E) Immunoblot of EYA2 in GSC 3565 transduced with shCONT or shRNAs targeting EYA2. ACTIN was used as loading control. (F) Co-staining of EYA2 and PERICENTRIN in GSC 3565 transduced with shCONT or shRNAs targeting EYA2. EYA2 is shown in green, PERICENTRIN in red, and DAPI in blue. Scale bars represent 10 µm. (G and H) Quantification of EYA2 staining in centrosome (G) or nucleus (H) of GSC 3565 transduced with shCONT or shRNAs targeting EYA2. Data are presented as mean ± SD. ***, P < 0.001. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison. (I) Immunoblot of EYA2 in GSC 3691 transduced with shCONT or shRNAs targeting EYA2. ACTIN was used as loading control. (J and K) Quantification of EYA2 staining in centrosome (J) or nucleus (K) of GSC 3691 transduced with shCONT or shRNAs targeting EYA2. Data are presented as mean ± SD. ***, P < 0.001. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison. (L) The morphology of GSCs, DGCs, NSCs, and NMs treated by DMSO or 20 µM EYA2 Tyr phosphatase inhibitor (EYA2i; MLS000544460). Scale bars represent 100 µm. (M) Flow cytometry–based cell cycle analysis in GSCs, DGCs, NSCs, and nonmalignant brain cultures treated with the EYA2 Tyr phosphatase inhibitor (MLS000544460) at the indicated concentration. (N) EdU staining in GSCs and NSCs after 2-h co-culture with 10 µM EdU. EdU is shown in red and DAPI in blue. Scale bar represents 20 µm. (O) Quantification of EdU+ cells in GSCs and NSCs. Data are presented as mean ± SD. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison. All data are representative of three independent experiments. At least 30 cells from five different 120× images were quantified in C and D, G and H, and J and K. Nine different 120× images were quantified in O.
To validate the localization of EYA2 to the centrosome in GSCs in the absence of culture, human glioblastoma patient surgical samples underwent immunofluorescence staining for EYA2, PERICENTRIN, and the GSC marker SOX2. SOX2+ cells showed increased EYA2 staining in centrosomes compared with SOX2− cells (Fig. 6, B–D). Strong correlation between SOX2 staining and EYA2 staining was observed in patient surgical specimens (Fig. 6, E and F), supporting high EYA2 expression in neoplastic tumor cells. These data underscore the relevance of this finding to human disease and suggest a potential role for EYA2 in mitotic spindle function within GSCs.
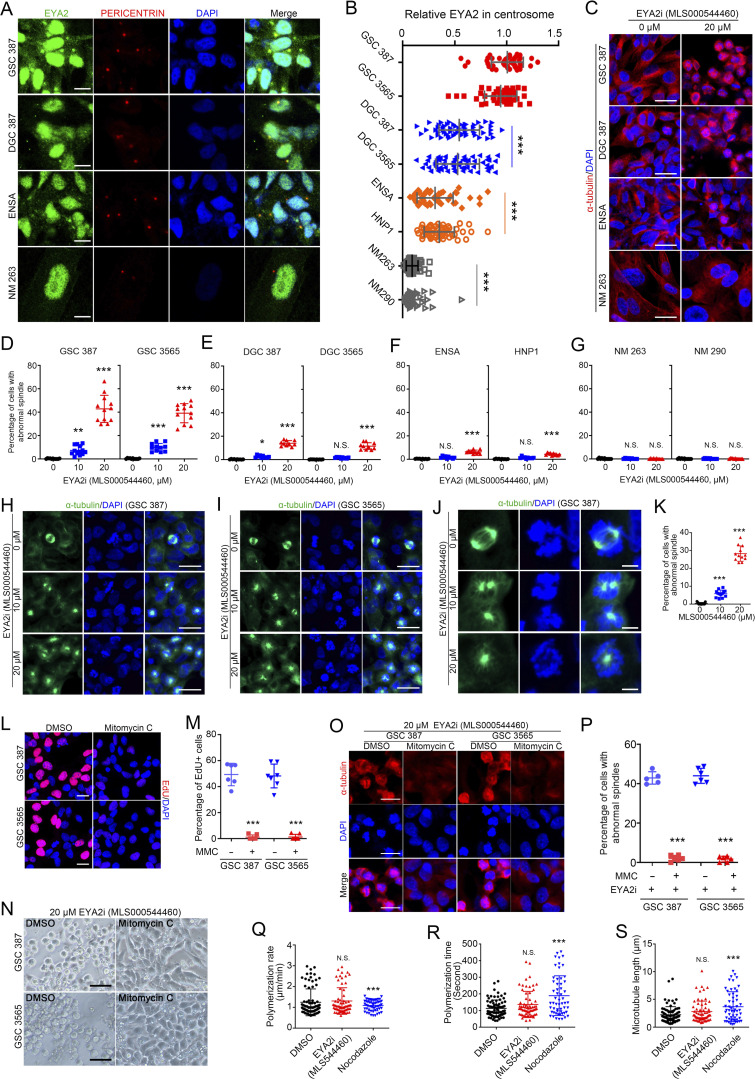
To interrogate the function of EYA2 in spindle formation in GSCs, GSCs were transduced with WT or mutant EYA2, then stained with α-tubulin to localize the spindle. GSCs expressing FLAG-tagged GFP, WT EYA2, or the EYA2A532R mutant (Patrick et al., 2013) displayed normal spindle formation, whereas EYA2 mutant deficient for Tyr phosphatase activity (EYA2D274N; Pandey et al., 2010) caused aberrant spindle formation (Fig. 6, G–J). These data demonstrate that EYA2 and its associated Tyr phosphatase activity is critical for proper mitotic spindle formation and GSC survival.
Pharmacologic inhibition of EYA2 Tyr phosphatase results in defective mitotic spindle formation in GSCs
To lay the foundation for potential therapeutic translation of our findings, we hypothesized that the effect of the Tyr phosphatase mutation on EYA2 function in GSC viability and on mitotic spindles could leverage pharmacologic inhibition of the Tyr phosphatase EYA2 activity against GSCs. To this end, we used a highly specific allosteric inhibitor of the EYA2 Tyr phosphatase (MLS000544460) that we recently developed (Anantharajan et al., 2019; Krueger et al., 2013, 2014). GSCs were much more sensitive to the EYA2 Tyr phosphatase inhibitor compared with DGCs and normal brain cells with a typical morphology of G2/M phase arrest (Fig. S3 L), which was confirmed by flow cytometric cell cycle analysis (Fig. S3 M). GSCs and NSCs displayed similar rates of cell proliferation measured by EdU incorporation (Fig. S3, N and O), ruling out increased cellular proliferation as an explanation for the increased sensitivity of GSCs to the EYA2 Tyr phosphatase inhibitor. Instead, the sensitivities of different cells to the EYA2 Tyr phosphatase inhibitor strongly correlated with the abundance of centrosomal EYA2 revealed by co-staining of EYA2 and PERICENTRIN (Fig. S4, A and B). The greater sensitivity of GSCs to inhibition of EYA2 Tyr phosphatase activity was confirmed by the increase of cells with aberrant α-tubulin staining (Fig. S4, C–G), suggesting impaired spindle assembly.
Figure S4.
EYA2 Tyr phosphatase inhibitor blocks spindle formation in GSCs in a cell cycle–dependent manner. (A) Co-staining of EYA2 and PERICENTRIN in GSC, DGC, NSC, and nonmalignant cells (NMs). EYA2 is shown in green, PERICENTRIN in red, and DAPI in blue. Scale bars represent 10 µm. (B) Quantification of EYA2 staining in the centrosomes of GSCs, NSCs, DGCs, and NMs. Data are presented as mean ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (C) α-Tubulin staining in GSC, DGC, NSC, and NM treated with DMSO or 20 µM EYA2 Tyr phosphatase inhibitor (EYA2i; MLS000544460) for 24 h. α-Tubulin is shown in red and DAPI in blue. Scale bars represent 20 µm. (D–G) Quantification of the cells with abnormal spindles in GSCs (D), DGCs (E), NSCs (F), and NMs (G) after treatment with EYA2 Tyr phosphatase inhibitor (MLS000544460) for 24 h at indicated concentrations. n ≥ 10. Data are presented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison. (H and I) Immunofluorescent staining of α-tubulin in GSC 387 (H) or GSC 3565 (I) treated with EYA2 Tyr phosphatase inhibitor (EYA2i; MLS000544460). α-Tubulin is shown in green and DAPI in blue. Scale bars represent 20 µm. (J) Immunofluorescent staining of α-tubulin in GSC 387 treated with EYA2 Tyr phosphatase inhibitor (EYA2i; MLS000544460). α-Tubulin is shown in green and DAPI in blue. Scale bars represent 5 µm. (K) Quantification of α-tubulin–positive cells in GSC 387 after treatment with EYA2 Tyr phosphatase inhibitor (MLS000544460). Data are presented as mean ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (L) EdU staining in GSCs treated with DMSO or 10 µg/ml mitomycin C. EdU is shown in red and DAPI in blue. Scale bars represent 20 µm. (M) Quantification of EdU+ cells in GSCs treated with DMSO or 10 µg/ml mitomycin C. Data are presented as mean ± SD. ***, P < 0.001. Statistical significance was determined by unpaired t test. (N) Morphology of DMSO or 10 µg/ml mitomycin C–treated GSCs upon treatment with 20 µM EYA2 Tyr phosphatase inhibitor (MLS000544460). Scale bars represent 100 µm. (O) α-Tubulin staining in DMSO or 10 µg/ml mitomycin C–treated GSCs upon treatment with 20 µM EYA2 Tyr phosphatase inhibitor (MLS000544460). α-Tubulin is shown in red, DAPI in blue. Scale bars represent 20 µm. (P) Quantification of the cells having abnormal spindles in DMSO or 10 µg/ml mitomycin C–treated GSCs upon treatment with 20 µM EYA2 Tyr phosphatase inhibitor (MLS000544460). n ≥ 5. Data are presented as mean ± SD. ***, P < 0.001. Statistical significance was determined by unpaired t test. (Q–S) Quantification of the polymerization rate (Q), polymerization time (R) of the purified tubulin, and the length of the microtubule (S) in the presence of DMSO, 20 µM EYA2 Tyr phosphatase inhibitor (MLS000544460), or 500 nM Nocodazole. Data are presented as mean ± SD. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison. ***, P < 0.001. All data are representative of three independent experiments. At least 40 cells from five (GSC 387, DGC 387, and ENSA) or 12 (NM 263) 120× images were quantified in B. 12 60× images per arm were quantified in D and K, and 10 60× images per arm were quantified in E–G. At least six 60× images per arm were quantified in M and P.
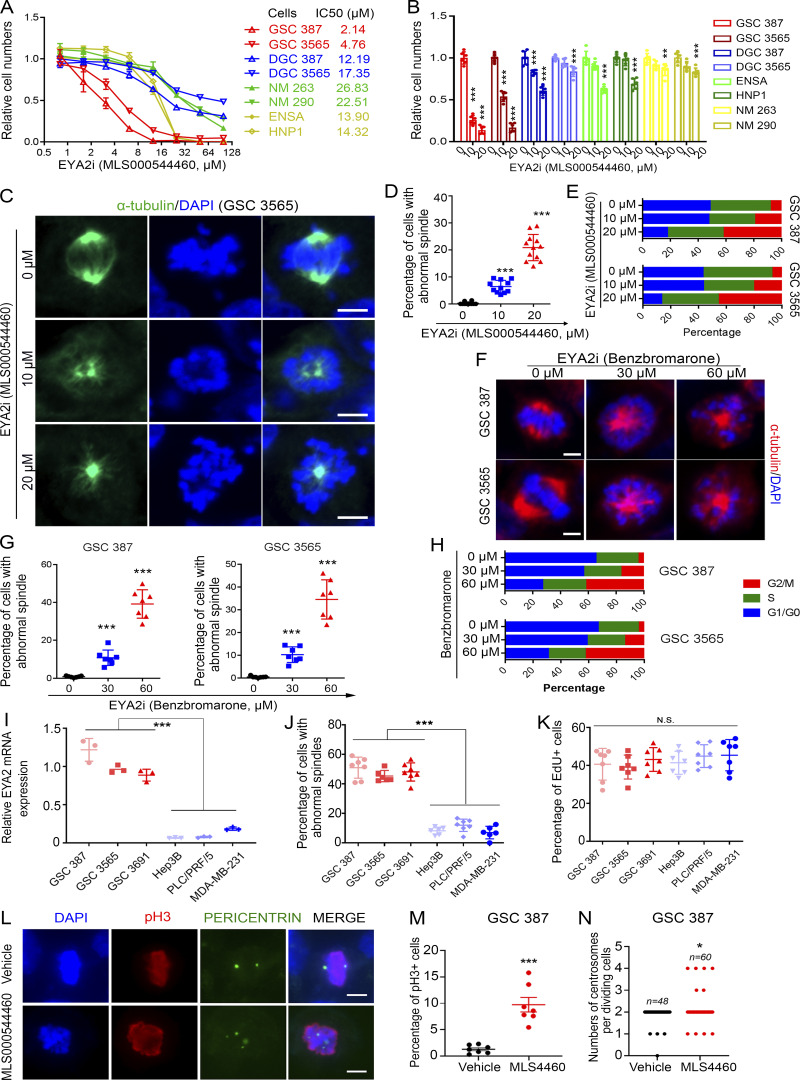
Supporting a potential therapeutic index, GSC proliferation was inhibited by the EYA2-specific Tyr phosphatase inhibitor (Krueger et al., 2014) at concentrations 4–10-fold lower than normal brain cells and DGCs (Fig. 7, A and B). EYA2 Tyr phosphatase inhibitor treatment of two patient-derived GSCs induced loss of bipolar spindle formation (Fig. 7, C and D; and Fig. S4, H–L) along with a G2/M cell cycle arrest (Fig. 7 E). In support of this finding, treatment with mitomycin C, which efficiently blocked cell cycle progression (Fig. S4, M and N), desensitized GSCs to the EYA2 Tyr phosphatase inhibitor (Fig. S4, O and P).
Figure 7.
Inhibitor of EYA2 Tyr phosphatase blocks spindle formation in GSCs. (A) Dose- response curves of different cells to EYA2 Tyr phosphatase inhibitor, MLS000544460. The half-maximal inhibitory concentration (IC50) of EYA2 Tyr phosphatase inhibitor in different cells is shown to the right. Data are presented as the mean ± SD. (B) Relative cell numbers after treating different cell models with EYA2 Tyr phosphatase inhibitor (MLS000544460). Data are presented as the mean ± SD. **, P < 0.01; ***, P < 0.001. (C) Immunofluorescence staining of α-tubulin in GSC 3565 treated with EYA2 Tyr phosphatase inhibitor (EYA2i; MLS000544460). α-tubulin was labeled as green, DAPI as blue. Scale bars represent 5 µm. (D) Quantification of cells with abnormal spindle in GSC 3565 after treatment with EYA2 Tyr phosphatase inhibitor (MLS000544460). Data are presented as the mean ± SD. The P values were calculated by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (E) Flow cytometry–based cell cycle analysis following PI staining in GSC 387 or GSC 3565 treated with either DMSO or EYA2 Tyr phosphatase inhibitor (MLS000544460) with indicated concentrations. (F) Morphologies of spindles in GSC 387 or GSC 3565 treated with EYA2 Tyr phosphatase inhibitor (benzbromarone) at indicated concentrations. α-Tubulin is in red and DAPI in blue. Scale bars represent 5 µm. (G) Quantification of GSC 387 (left) or GSC 3565 (right) with abnormal spindles after treatment with EYA2 Tyr phosphatase inhibitor (benzbromarone) at indicated concentrations. Data are presented as mean ± SD. The P values were calculated by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (H) Flow cytometry–based cell cycle analysis following PI staining in GSC 387 or GSC 3565 treated with either DMSO or EYA2 Tyr phosphatase inhibitor (benzbromarone) at indicated concentrations. (I) EYA2 mRNA expression in GSCs, liver cancer cells (Hep3B and PLC/PRF/5), and breast cancer cells (MDA-MB-231) measured by real-time qPCR. Data are presented as the mean ± SD. ***, P < 0.001. (J) Quantification of cells having abnormal spindles in GSCs, breast cancer cells, and liver cancer cells after treatment with 20 µM of EYA2 Tyr phosphatase inhibitor for 24 h. Data are presented as mean ± SD. ***, P < 0.001. Statistical significance was determined by one-way ANOVA with Tukey’s correction for multiple comparisons. (K) Statistics of EdU+ cell in GSCs, breast cancer cells, and liver cancer cells. Data are presented as mean ± SD. Statistical significance was determined by one-way ANOVA with Tukey’s correction for multiple comparisons. (L) PERICENTRIN and phospho-histone 3 (pH3) staining in GSC 387 treated by MLS000544460 or vehicle. PERICENTRIN is in green, pH3 in red, and DAPI in blue. Scale bars represent 10 µm. (M and N) Quantification of pH3+ cells (M) and the pH3+ cells having abnormal centrosome foci numbers (N) in GSC 387 treated with either MLS000544460 (indicated as MLS4460) or vehicle. Abnormal PERICENTRIN foci were defined as 1, 3, or >4 foci per GSC. Data are presented as mean ± SD. Statistical significance was determined by t test. *, P < 0.05; ***, P < 0.001. A–K are representatives of three independent experiments. L–N are representative figures of four independent experiments using either GSC 387 treated with 4460 (2×) or GSC 3565 and GSC 3691 treated with 9987 (1× each). 12 or 7 different 120× images were quantified in D and G, respectively. At least six 120× images were quantified in B, and seven 80× images were quantified in E.
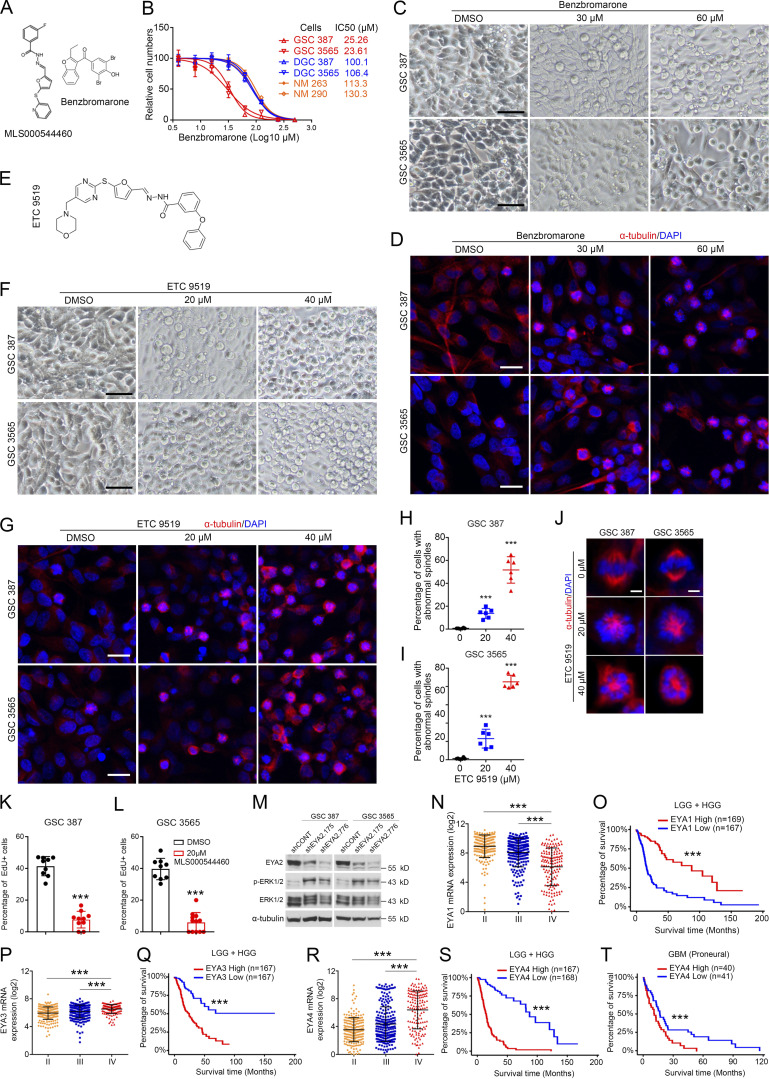
To exclude the possibility that the inhibitor of EYA2 Tyr phosphatase activity directly impaired microtubule dynamics, we used total internal reflection fluorescence microscopy to live image purified tubulin polymerization (Gell et al., 2010). The EYA2 Tyr phosphatase inhibitor did not disrupt microtubule polymerization rates, polymerization time, or microtubule length, in contrast to a known inhibitor of microtubules, nocodazole (Fig. S4, Q–S; Vasquez et al., 1997). The crystal structure of an analogue of this inhibitor has recently been determined, revealing the mechanistic details of inhibition and demonstrating its on-target actions (Anantharajan et al., 2019). To confirm the on-target effect of this class of inhibitors and the functional importance of the EYA2 Tyr phosphatase activity in spindle formation in GSCs, we used another general EYA inhibitor, benzbromarone, which has a different structure from MLS000544460 (Fig. S5 A) but also inhibits EYA Tyr phosphatase activity (Tadjuidje et al., 2012). Confirming our findings with MLS000544460, GSCs were selectively sensitive to benzbromarone compared with other types of cells (Fig. S5 B). In parallel to the observed effects of MLS000544460 on GSCs, benzbromarone blocked spindle formation and induced G2M arrest (Fig. 7, F–H; and Fig. S5, C and D).
Figure S5.
Different EYA2 Tyr phosphatase inhibitors consistently block spindle formation in GSCs. (A) Molecular structures of MLS000544460 and benzbromarone. (B) The half-maximal inhibitory concentration (IC50) of benzbromarone in GSCs, DGCs, and NMs. Data are presented as mean ± SD. (C) Morphology of GSC 387 and 3565 after treatment with benzbromarone at indicated concentrations. Scale bars represent 100 µm. (D) α-Tubulin staining in GSC 387 and 3565 after treatment with benzbromarone at indicated concentrations. α-Tubulin is shown in red and DAPI in blue. Scale bars represent 20 µm. (E) Molecular structure of ETC 9519. (F) Morphology of GSC 387 or GSC 3565 treated with ETC 9519 at indicated concentrations. Scale bars represent 100 µm. (G) α-Tubulin staining in GSC 387 and 3565 after treatment with ETC 9519 at indicated concentrations. α-Tubulin is shown in red and DAPI in blue. Scale bars represent 20 µm. (H and I) Quantification of GSC 387 (H) or GSC 3565 (I) with abnormal spindles after treatment with ETC 9519 at indicated concentrations. Data are presented as mean ± SD. ***, P < 0.001. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison. (J) Morphologies of spindles in GSC 387 or GSC 3565 treated with ETC 9519 at indicated concentrations. α-Tubulin is shown in red and DAPI in blue. Scale bars represent 5 µm. (K and L) Statistics of EdU labeling in GSC 387 (K) or 3565 (L) after MLS000544460 treatment. Data are presented as mean ± SD. ***, P < 0.001. Statistical significance was determined by t test. (M) Representative immunoblot of EYA2, ERK1/2, and phosphorylated ERK1/2 (p-ERK1/2) in GSC 387 (left) and GSC 3565 (right) transduced with either shCONT or shEYA2. α-Tubulin was used as loading control. (N) EYA1 mRNA levels in different grades of glioma in TCGA datasets. Data are presented as mean ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (O) Survival curves of low-grade glioma (LGG) and high-grade glioma (HGG) patients with high or low EYA1 expression in TCGA datasets. Significance values were determined by Mantel-Cox log-rank test. ***, P < 0.001. (P) EYA3 mRNA levels in different grades of glioma in TCGA datasets. Data are presented as mean ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (Q) Survival curves of low-grade glioma (LGG) and high-grade glioma (HGG) patients with high or low EYA3 expression in TCGA datasets. Significance values were determined by Mantel-Cox log-rank test. ***, P < 0.001. (R) EYA4 mRNA levels in different grades of glioma in TCGA datasets. Data are presented as mean ± SD. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (S and T) Survival curves of low-grade glioma (LGG) and high-grade glioma patients (S) or glioblastoma patients with proneural subtype (T) based on EYA4 expression in TCGA datasets. Significance values were determined by Mantel-Cox log-rank test. ***, P < 0.001. B–M are representative of three independent experiments. Six 60× images were quantified in H and I. Nine 60× images were quantified in K and L.
Finally, we developed a novel analogue of MLS000544460, designated as ETC 9519 (please see Materials and methods for the synthesis of ETC 9519), which showed similar phenotypes to both MLS000544460 and benzbromarone (Fig. S5, E–J). Consistent with the observation of preferential expression of EYA2 in GSCs compared with liver cancer cells or breast cancer cells (Fig. 7 I), blocking EYA2 Tyr phosphatase activity only caused abnormal spindles in GSCs but not in liver cancer cells or breast cancer cells despite similar rates of proliferation (Fig. 7, J and K), which further supports the importance of the EYA2 Tyr phosphatase in spindle formation in GSCs as well as suggests that the EYA2 Tyr phosphatase inhibitor is on target. MLS000544460 caused abnormal centrosome foci numbers in dividing GSCs, serving as a potential mechanism of aberrant spindles caused by targeting EYA2 Tyr phosphatase activity in GSCs (Fig. 7, L–N). Taken together, these data support EYA2 Tyr phosphatase activity as a therapeutic target in GSCs and suggest a potential novel function for EYA2 in mitotic spindle assembly and proper chromosome segregation.
Targeting EYA2 expression suppresses in vivo glioblastoma growth
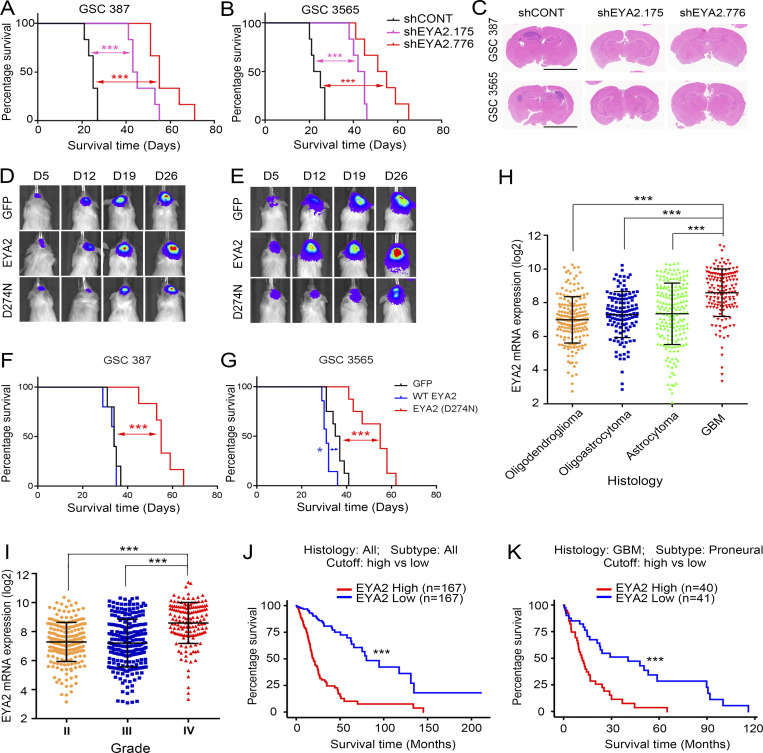
As in vivo tumor growth is the gold standard for assessing tumor initiation, we interrogated the impact of EYA2 silencing on orthotopic tumor formation. Viable GSCs transduced with either shCONT or one of two nonoverlapping shEYA2s were intracranially implanted in immunocompromised mice. Tumor-bearing mice with GSCs transduced with shEYA2s survived longer than those transduced with shCONT (Fig. 8, A and B). Histological analysis of tumor-bearing brains collected 20 d after GSC implantation revealed large tumor masses in mice bearing shCONT GSCs, whereas tumors were not readily apparent in brains bearing GSCs transduced with shEYA2s (Fig. 8 C). To interrogate the functional importance of the EYA2 Tyr phosphatase activity for in vivo GSC growth, two patient-derived GSCs transduced with either GFP or WT EYA2 or with the Tyr phosphatase-dead EYA2 mutant (D274N) were implanted orthotopically in immunodeficient mice. Consistent with in vitro observations, tumor latency was longer in mice bearing GSCs transduced with the Tyr phosphatase-dead EYA2 (D274N) compared with mice bearing GSCs transduced with GFP or WT EYA2 (Fig. 8, D and E). Mice harboring cells from two different patient-derived GSCs transduced with the D274N EYA2 mutant survived longer than mice harboring the same GSCs transduced with GFP or WT EYA2 (Fig. 8, F and G), suggesting Tyr phosphatase activity of EYA2 as a potential target in vivo.
Figure 8.
Targeting EYA2 expression suppresses tumor growth in vivo. (A and B) Kaplan–Meier survival curves of immunocompromised mice bearing GSC 387 (A) or GSC 3565 (B) transduced with either shCONT or shEYA2. n = 6. The P values were calculated by Mantel-Cox log-rank test. ***, P < 0.001. (C) H&E staining of the brain sections from NGS mice bearing GSC 387 (A) or GSC 3565 (B) transduced with shCONT or shEYA2. Scale bars represent 5 mm. (D and E) Representative bioluminescence images of mice bearing GSC 387 (D) or GSC 3565 (E) expressing GFP, WT EYA2, and EYA2 (D274N). (F and G) Kaplan–Meier survival curves of immunocompromised mice bearing GSC 387 (F; n = 5) or GSC 3565 (G; n ≥ 7) expressing GFP, WT EYA2, and EYA2 (D274N). The P values were calculated by Mantel-Cox log-rank test. *, P < 0.05; ***, P < 0.001. (H and I) EYA2 mRNA levels in different glioblastoma (H) or different grades (I) in TCGA of glioma. Data are presented as mean ± SD. The P values were calculated by one-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (J and K) Survival curves of glioma patients (J) or glioblastoma patients of proneural subtype (K) in TCGA based on EYA2 mRNA expression. The P values were calculated by Mantel-Cox log-rank test. ***, P < 0.001.
To determine whether our findings are applicable to glioblastoma patients, in silico analysis of TCGA glioma datasets was examined, revealing that EYA2 expression was increased in glioblastoma relative to other brain tumor histologies (Fig. 8 H) and that EYA2 expression correlated with increased tumor grade (Fig. 8 I). Across all tumor grades, EYA2 expression informed poor survival (Fig. 8 J). EYA2 expression was not prognostic in all glioblastomas, but high EYA2 expression informed poor prognosis in glioblastoma patients with the proneural subtype (Fig. 8 K). Collectively, these data suggest that EYA2 may be a key clinical target for glioblastoma.
Pharmacologic inhibition of EYA2 Tyr phosphatase suppresses glioblastoma growth
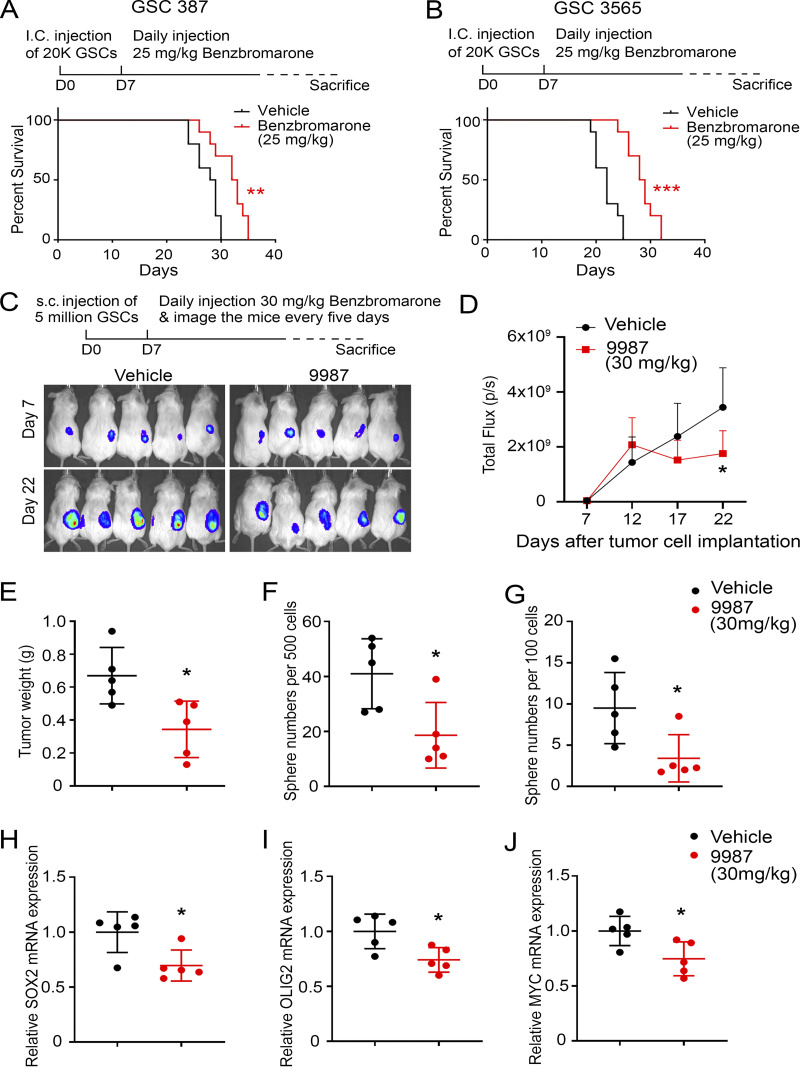
The genetic data in tumor models, coupled with our in vitro data that demonstrate that GSCs were much more sensitive to EYA2 Tyr phosphatase inhibition compared with DGCs or nonmalignant cells (NSCs and NMs; Fig. 7 A; Fig. S3, L and M; and Fig. S4, C–G), strongly suggested that pharmacologic EYA2 Tyr phosphatase inhibition may be efficacious in vivo. Therefore, GSCs were intracranially implanted into immunocompromised mice, and treatment with the EYA2 Tyr phosphatase inhibitor was initiated 7 d after tumor cell implantation to mimic established tumors. As pharmacokinetic and pharmacodynamic properties of MLS000544460 are not optimal, we tested benzbromarone as it has previously been used in vivo. Oral benzbromarone treatment prolonged the survival of mice bearing two different patient-derived GSCs compared with vehicle-treated mice (Fig. 9, A and B). Supporting EYA2 as a druggable target in vivo, treatment with our recently characterized EYA2 inhibitor, NCGC00249987 (Anantharajan et al., 2019), a newer analogue of 4460, reduced the growth of subcutaneously implanted GSCs compared with control-treated mice (Fig. 9, C–E). GSCs were implanted subcutaneously in this case to enable characterization of stem cells, as GSCs are difficult to obtain from the brain microenvironment after transplantation.
Figure 9.
Pharmacologic inhibition of EYA2 Tyr phosphatase suppresses glioblastoma growth. (A and B) Kaplan–Meier survival curves of immunocompromised mice bearing GSC 387 (A) or GSC 3565 (B) treated with benzbromarone or vehicle. The P values were calculated by Mantel-Cox log-rank test. **, P < 0.01; ***, P < 0.001. I.C., intracranial. (C) Bioluminescence images of mice bearing GSC 3565 treated per os with either vehicle or 30 mg/kg 9987. (D) Quantification of the bioluminescence signals from either 9987-treated mice or vehicle-treated mice. Data are presented as mean ± SD. *, P < 0.05. The P values were calculated by two-way ANOVA with Tukey’s correction for multiple comparisons. (E) The weight of tumors dissected from tumor-bearing mice 25 d after tumor cell implantation. Data are presented as mean ± SD. *, P < 0.05. The P values were calculated by unpaired t test. (F and G) The number of spheres generated from either 500 (F) or 100 (G) FACS-sorted tumor cells. Data are presented as mean ± SD. *, P < 0.05. The P values were calculated by unpaired t test. (H–J) mRNA levels of SOX2 (H), OLIG2 (I), and MYC (J) measured by qPCR in tumor cells sorted from either vehicle-treated mice or 9987-treated mice. Data are presented as mean ± SD. *, P < 0.05. The P values were calculated by unpaired t test. 10 mice per arm were used for A and B, and five mice per arm for C–J.
To determine whether NCGC00249987 (referred to as 9987) alters stem cell characteristics in vivo, tumor cells were transduced with mCherry to enable selective enrichment of tumor cells, which were sorted via FACS upon sacrifice of tumor-bearing mice. FACS-sorted mCherry-positive cells were then plated for sphere formation assays and analyzed for expression of stem cell markers. In vivo 9987 treatment reduced ex vivo sphere formation of tumor cells compared with cells from control-treated tumors, demonstrating that 9987 suppresses tumor cell self-renewal (Fig. 9, F and G). Likewise, ex vivo expression levels of stemness genes, such as SOX2, OLIG2, and MYC, were reduced in tumor cells treated with 9987 compared with tumor cells taken from control-treated mice (Fig. 9, H–J).
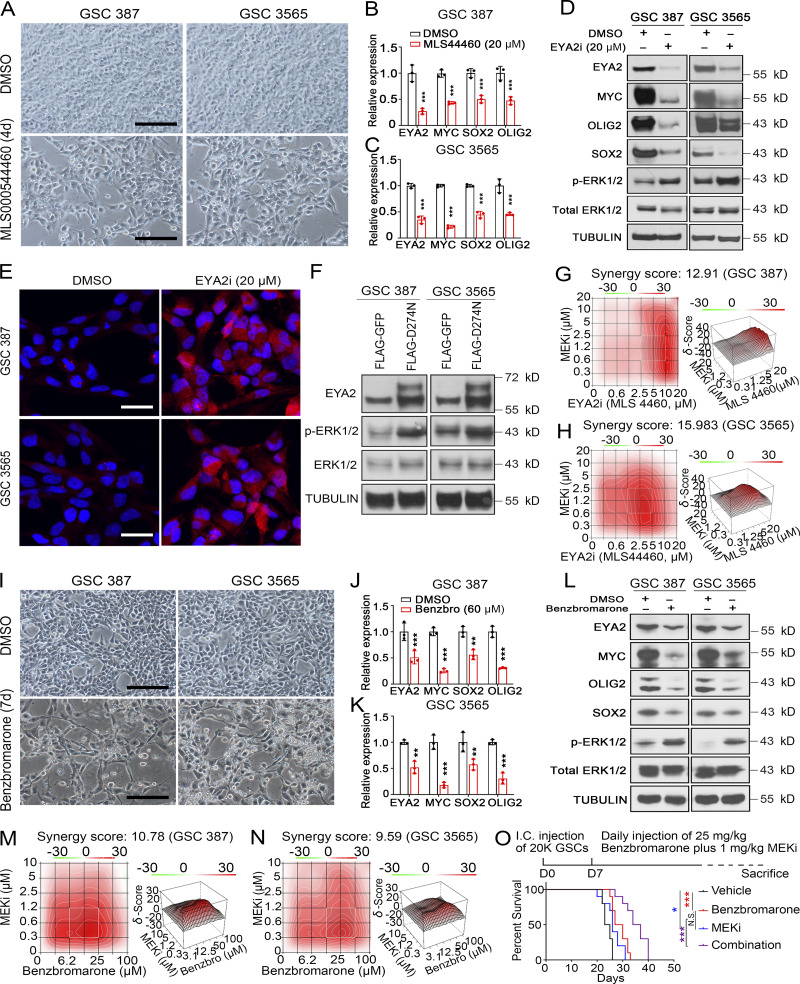
While prolonged survival of tumor-bearing mice was observed with inhibition of the EYA2 Tyr phosphatase activity (Fig. 9, A and B), mice in all groups eventually succumbed to tumor growth, which was consistent with the in vitro observation that a population of cells in GSCs survived treatment with the EYA2 Tyr phosphatase inhibitor MLS000544460 (Fig. 10 A). Supporting an on-target effect of MLS000544460, EYA2 expression was decreased in cells that survived treatment with the inhibitor (Fig. 10, B–D). The surviving cells were relatively quiescent (Fig. S5 K and Fig. S5 L) and expressed lower levels of MYC, SOX2, and OLIG2, suggesting the loss of stemness may be what leads to the loss of EYA2 in response to MLS000544460 treatment (Fig. 10, B–D). Treatment-induced dormancy can be accompanied by reactivation of ERK signaling (Kurppa et al., 2020). As revealed by both immunoblot and immunofluorescent staining, phosphorylated ERK1/2 was up-regulated in cells that survived EYA2 Tyr phosphatase inhibition (Fig. 10, D and E), suggesting compensatory activation of the ERK/MAPK pathway in response to EYA2 Tyr phosphatase inhibition. Supporting these observations, up-regulated ERK1/2 phosphorylation was also observed in GSCs with EYA2 knockdown or expressing the EYA2D274N mutant (Fig. 10 F and Fig. S5 M). These data suggested that dual treatment with EYA2 and MEK1/2 inhibitors may synergize to kill GSCs. Indeed, strong synergistic effects between the different EYA2 Tyr phosphatase inhibitors and a MEK1/2 inhibitor were observed in GSCs (Fig. 10, G and H). Supporting these findings, treatment with benzbromarone also induced similar effects in GSCs (Fig. 10, I–N). These data suggest that targeting the EYA2 Tyr phosphatase together with suppression of ERK/MAPK signaling may improve tumor control, which we demonstrated, as dual in vivo treatment with benzbromarone and MEK1/2 inhibitor further prolonged the life span of the tumor-bearing mice (Fig. 10 O).
Figure 10.
Dual inhibition of EYA2 Tyr phosphatase and MEK/ERK causes synergistic decrease in glioblastoma growth. (A) Brightfield images of cells treated with EYA2 Tyr phosphatase inhibitor (MLS000544460) or vehicle. Scale bars represent 200 µm. (B and C) mRNA levels of indicated genes in GSC 387 (B) or 3565 (C) treated with EYA2 Tyr phosphatase inhibitor MLS000544460 (indicated as MLS44460) or vehicle. Data are presented as mean ± SD. The P values were calculated by two-way ANOVA with Tukey’s multiple comparisons. ***, P < 0.001. (D) Immunoblot of indicated proteins in GSCs treated with EYA2 Tyr phosphatase inhibitor (EYA2i; MLS000544460) or vehicle. p-ERK, phosphorylated ERK. α-Tubulin was used as loading control. (E) Immunostaining of phosphorylated ERK1/2 in GSCs treated with EYA2 Tyr phosphatase inhibitor (MLS000544460) or vehicle. p-ERK1/2 is in red and DAPI in blue. Scale bars represent 20 µm. (F) Immunoblot of indicated proteins in GSCs expressing GFP or EYA2D274N. α-Tubulin was used as loading control. (G and H) Synergy indices of MLS000544460 (indicated as MLS44460 or MLS4460) and MEK1/2 inhibitor (MEKi) in GSC 387 (G) or GSC 3565 (H) analyzed by R package synergyfinder. (I) Whitefield images of cells treated with benzbromarone or vehicle. Scale bars represent 200 µm. (J and K) mRNA levels of indicated genes in GSC 387 (J) or 3565 (K) treated with benzbromarone (Benzbro) or vehicle. Data are presented as the mean ± SD. The P values were calculated by two-way ANOVA with Tukey’s multiple comparisons. **, P < 0.01; ***, P < 0.001. (L) Immunoblot of indicated proteins in GSCs treated with benzbromarone or vehicle. α-Tubulin was used as loading control. (M and N) Synergy indices of benzbromarone and MEK1/2 inhibitor in GSC 387 (M) or GSC 3565 (N) analyzed by R package synergyfinder. (O) Kaplan–Meier survival curves of immunocompromised mice bearing GSC 387 treated with vehicle, benzbromarone, MEK1/2 inhibitor, or both benzbromarone and MEK1/2 inhibitor. The P values were calculated by Mantel-Cox log-rank test. *, P < 0.05; ***, P < 0.001. I.C., intracranial. A–N are representatives of three independent experiments. 10 mice per arm were used for O.
Discussion
The universal lethality of glioblastoma requires novel therapeutic approaches to improve patient outcome. While anti-angiogenic and onco-immunology paradigms have transformed the clinical care of patients with many cancer types, outcomes in glioblastoma have been largely disappointing. The most effective nonsurgical therapies for glioblastoma patients continue to be cytotoxic agents that induce DNA damage, including ionizing radiation and alkylating chemotherapies, to which GSCs display relative resistance (Bao et al., 2006a). Given that nests of GSCs in areas of proliferation have been associated with poor patient outcome (Pallini et al., 2008), targeting mitosis represents a potential therapeutic paradigm that might be useful for glioblastoma. A genetic screen in GSCs identified the mitotic regulator BUGZ as an important target, but the lack of enzymatic function of BUGZ amenable to therapeutic targeting limits the translational potential to date (Toledo et al., 2014). General targeting of mitotic machinery confers significant side effects (Zhou and Giannakakou, 2005, 2005; Tischer and Gergely, 2019); thus, identification of novel means to target mitotic machinery, particularly those enhanced in GSCs, may be an approach to inhibit the disease with less toxic side effects. Here, we leveraged expression analysis to identify molecules that were enriched in neoplasia and associated with stemlike features, providing a tightly focused group of targets led by EYA2.
EYA2 is a member of the EYA family of proteins (EYA1–EYA4), which are important for myogenesis (Grifone et al., 2007), neurogenesis (Eisner et al., 2015), thymus (Xu et al., 2002), kidney, and ear development (Xu et al., 1999). As multifunctional proteins, EYAs contain domains associated with both Tyr and threonine phosphatases, although only EYA Tyr phosphatase activity is intrinsic to the molecule, whereas we recently demonstrated that EYA threonine phosphatase activity is due to an interaction with PP2A (Zhang et al., 2018). EYA proteins also act as transcriptional cofactors to SIX family members to activate target gene expression important for multiple biological processes (Tadjuidje and Hegde, 2013; Zhou et al., 2018). Expression of most EYA family members decreases after completion of development (Zhou et al., 2018), although low levels of expression are maintained in some tissues (Tadjuidje and Hegde, 2013). Reexpression or overexpression of EYA2 is observed in several cancers, including ovarian cancer (Zhang et al., 2005), brain cancer (Wen et al., 2017), lung cancer (Li et al., 2017), and hematopoietic cancers (Ono et al., 2017). Ectopic expression of EYA2 in breast cancer cells increases proliferation, transformation, and migration (Farabaugh et al., 2012; Pandey et al., 2010). EYA2 expression correlates with poor prognosis in ovarian and breast cancer patients (Zhang et al., 2005) and is required for Six1-mediated breast cancer metastasis (Farabaugh et al., 2012; Patrick et al., 2013), revealing EYA2 as a potential clinical target in cancer therapy.
The EYAs have been extensively characterized for their roles as transcriptional coactivators to drive gene expression, and most identified molecular interactors of EYAs to date are nuclear (Cook et al., 2009; Yuan et al., 2014; Zhou et al., 2018), including nontranscriptional targets such as H2AX (Cook et al., 2009; Krishnan et al., 2009). However, potential cytoplasmic roles for EYA family members, through their associated phosphatase activities, have been described, both in modulating migration and invasion (Pandey et al., 2010) and also in regulating innate and adaptive immunity (Okabe et al., 2009; Vartuli et al., 2018), although the direct targets of these activities remain largely elusive. While EYA2 overexpression has not been previously described in glioblastoma, up-regulation of EYA1 and SIX1 has been observed in A2B5+ glioma progenitor cells, in which the function of EYA1 was suggested to be as a transcriptional partner to SIX1 during gliomagenesis (Auvergne et al., 2013). In contrast, our results uncover a novel, likely cytoplasmic function of the EYA2 Tyr phosphatase in regulating the mitotic spindle, a function that is critical to GSC maintenance. EYA1 serves as a Tyr phosphatase required for Cyclin D1 expression in breast cancer cells and has been implicated in breast cancer cell proliferation, migration, and invasion (Wu et al., 2013), although the role of its Tyr phosphatase activity in proliferation is controversial (Pandey et al., 2010). In breast cancer cells, EYA2 dephosphorylates ERβY36 to promote tumor growth (Yuan et al., 2014). Additional targets of EYA Tyr phosphatase activity include H2AX and WDR1 (Cook et al., 2009; Mentel et al., 2018; Yuan et al., 2014). Upon DNA damage, EYA1-3 dephosphorylates the phosphorylated Y142 site on histone H2AX, which triggers DNA repair instead of apoptosis (Cook et al., 2009; Krishnan et al., 2009), suggesting that EYA Tyr phosphatase inhibition could be used effectively in combination with chemotherapy (Robin et al., 2012). In glioma, while expression of EYA2-4 increases with tumor grade and informs poor prognosis, EYA1 expression decreases with tumor grade and correlates with a better prognosis (Fig. S5, N–T). In addition, among the EYAs, only EYA2 and EYA4 have been detected at the centrosome in the Harmonizome proteomic database (Fig. S2 Q), suggesting a potentially distinct role for these two EYA family members.
GSCs can be both quiescent (Deleyrolle et al., 2011) and highly proliferative, in contrast to NSCs, which are usually quiescent (Tirosh et al., 2016). As dysregulation of the centrosome and chromosome segregation impairs mitosis (Weaver et al., 2007; Williams et al., 2008; Yang et al., 2008), the selective dependency of GSCs on EYA2 function could derive from its high levels causing differential mitotic control, resulting in maintenance of genome integrity. Given the importance of the EYA2 Tyr phosphatase in GSC maintenance, targeting this activity may inhibit glioblastoma growth. Designing specific inhibitors of protein Tyr phosphatases has proven difficult, largely due to the lack of specificity of inhibitors, because the over 100 protein Tyr phosphatases in the human genome often share similar active site structures (Barr, 2010; Tadjuidje and Hegde, 2013). However, the EYAs belong to the HAD family of phosphatases that use an Asp as active site residues instead of Cys as in most cellular phosphatases, raising the possibility of identifying inhibitors of EYAs with some specificity (Tadjuidje and Hegde, 2013; Zhou et al., 2018). Using high-throughput screening approaches, Hegde and colleagues identified benzbromarone-related compounds that target the Tyr phosphatase activity of EYA2 and EYA3 (Pandey et al., 2013; Tadjuidje et al., 2012), which we demonstrate can inhibit glioblastoma growth in vivo. Our group identified a series of N-arylidenebenzohydrazide–containing compounds highly specific to EYA2. These compounds are unable to inhibit related EYA family members despite highly conserved active sites. Biochemical and crystallographic data demonstrate that our novel inhibitors act through an allosteric mechanism (Krueger et al., 2013, 2014), inhibiting Mg2+ access specifically to EYA2 by binding to a pocket behind the active site (Anantharajan et al., 2019). Due to the allosteric mechanism of action of these EYA2 Tyr phosphatase inhibitors and the unique role of EYA2 in glioblastoma, use of such inhibitors may enable highly specific targeting of the most therapeutically resistant cells in glioblastoma, without the unwanted side effects normally conferred by phosphatase inhibitors. However, improvement of their in vivo properties will be required to test their efficacy against glioblastoma. In vitro data indicated that ERK1/2 phosphorylation was up-regulated upon treatment with different EYA2 Tyr phosphatase inhibitors. Consistently, combined treatment with both MEK1/2 inhibitor and different EYA2 Tyr phosphatase inhibitors synergized in killing GSCs in vitro and prolonged the survival of glioblastoma-bearing mice.
In summary, targeting mitotic control offers a potential cellular mechanism for improved cancer therapy, though general inhibitors of this process may present significant toxicity. As EYA2 can be considered broadly an oncofetal gene with reactivation after development in multiple cancer types, the therapeutic index of EYA2-targeting strategies is likely high. This property of EYA2, along with the allosteric nature of the inhibitors developed by our group, suggests that targeting EYA2 in glioblastoma is likely to be highly specific. While critical molecular targets of the EYA2 phosphatase in GSCs remain to be determined, our discovery of a novel potential function of phosphatase control of the mitotic spindle offers a directly translatable therapeutic paradigm for a lethal malignancy.
Materials and methods
Derivation of GSCs, nonmalignant brain cultures, and NSCs
Patient-derived GSC 387 and GSC 3691 were generated in our laboratory. GSC 3565 was derived with a human glioblastoma specimen from University Hospitals-Cleveland. NMs (NM 176, NM 177, NM 263, and NM 290) were derived from human epilepsy resection specimens from Cleveland Clinic Foundation. NSC11s are human-induced pluripotent cell-derived NSCs (ALSTEM). ENSAs are human embryonic stem-derived neural progenitor cells (Millipore). HNP1 Human Neural Progenitors (ArunA Biomedical) were derived from the hESC WA09 line. All NSCs were cultured in Neurobasal media (Invitrogen) supplemented with B27 (Invitrogen), epidermal growth factor, and basic fibroblast growth factor (20 ng/ml each; R&D Systems). Short Tandem Repeat analyses were performed to authenticate the identity of each tumor model used in this article on at least a yearly basis. Mycoplasma testing was performed by qPCR cellular supernatants at least twice a year. The use of human patient samples in this study has been approved by the institutional review board of the University of California, San Diego, and patients provided informed consent.
In vivo tumorigenesis and drug treatment
In the experiments measuring the survival curves of mice bearing GSCs transduced with shRNAs, intracranial xenograft assays were performed by transplanting 5 × 104 GSCs into the right cerebral cortex of NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ; The Jackson Laboratory) mice at a depth of 3.5 mm. 2 × 104 cells were transplanted into the right cerebral cortex of NSG mice via intracranial injection in survival experiments on mice bearing GSCs expressing GFP, WT EYA2, or EYA2D274N or on mice treated with small-molecule inhibitors. For the GSC-bearing mice treated with 9987, 5 × 106 cells were transplanted into the flank via subcutaneous injection. In all experiments involving inhibitor treatment, the small molecules were administrated daily via oral gavage and were started on day 7 after GSC transplantation. The doses of the small molecules used in this study can be found in the figures and figure legends.
All mouse experiments were performed under an animal protocol approved by the Institutional Animal Care and Use Committee in the University of California, San Diego. At least five 4–6-week-old healthy male and female NSG mice per arm were randomly selected and used in this study. Five mice per arm were used to generate the survival curves of mice bearing GSCs transduced with either shCONT or shEYA2. More than seven mice per arm were used to generate survival curves of mice bearing GSCs overexpressing GFP, EYA2, or EYA2D274N. 10 mice per arm were used in experiments involving small-molecule treatment. All mice were maintained in a specific pathogen–free facility in 12-h light/12-h dark cycle at the University of California, San Diego. Animals were monitored daily during the experiments until neurological signs or signs of morbidity were observed, at which point they were sacrificed. Neurological signs or signs of morbidity include hunched posture, gait changes, lethargy, and weight loss. Brains were collected and fixed in 4% formaldehyde. H&E staining was performed on sections from the fixed tissues for histological analysis.
Tumor dissociation and glioma stem cell culture
Xenografted tumors, which were dissociated using a papain dissociation system according to the manufacturer’s instructions, were used as renewal source of the GSCs in this study. The dissociated cells were then cultured in Neurobasal medium (Invitrogen) supplemented with 2% B27 (Invitrogen), 1% L-glutamine (Invitrogen), 1% sodium pyruvate (Invitrogen), 1% penicillin/streptomycin (Invitrogen), 10 ng/ml basic fibroblast growth factor (R&D Systems), and 10 ng/ml EGF (R&D Systems) for at least 6 h to recover surface antigens expression. The cells were then labeled with CD133/2(293C)-Allophycocyanin antibody kit (130098826; Miltenyi Biotec), and the CD133+ cells were sorted and analyzed by flow cytometry. The sorted CD133+ cells were cultured in Neurobasal medium containing B27, 20 ng/ml of both EGF (R&D Systems) and fibroblast growth factor (R&D Systems) for a short period before treatment and analyzed. GSC phenotypes were then validated by expression of stem cell markers (SOX2 and OLIG2) and functional assays of self-renewal. Corresponding differentiated glioma cells were obtained by culturing the GSCs in DMEM medium supplemented with 10% FBS for at least 2 wk.
Proliferation and neurosphere formation assay
CellTiter-Glo assays were performed in 96-well plates by plating 103 cells per well with at least three replicates. Relative cell numbers were then measured at desired time points by CellTiter-Glo (Promega) assay based on the manufacturer’s instructions. All data were normalized to the data of day 0 and presented as mean ± SD to show the relative proliferation rates after different treatments. Sphere formation capacity of GSCs after desired treatments was measured by in vitro limiting dilution assay. Briefly, different numbers of GSC cells (200, 100, 50, 25, and 10) were plated into individual wells in 96-well plates with at least eight replicates. The numbers of spheres in each well were counted 7 d later, and the frequencies of stem cells were estimated by extreme limiting dilution analysis using software available at http://bioinf.wehi.edu.au/software/elda, as previously described (Hu and Smyth, 2009).
Immunofluorescent staining
All cells were plated on Matrigel-coated round coverslips (Electron Microscopy Sciences). After desired treatments, the cells were fixed with 4% paraformaldehyde at room temperature (rt) for 10 min, followed by permeabilizations with PBS containing 0.1% Tween 20 for 10 min and blocking with 5% normal goat serum in 0.1% PBST for 20 min at rt. After that, cells were incubated with anti–α-tubulin (Cat# T6074; 1:2,000; Sigma-Aldrich), anti-EYA2 (Cat# HPA027024; 1:200; Sigma-Aldrich), anti-Ki67 (Cat# M7240; 1:100; DAKO), anti–Cleaved CASPASE 3 (Cat# 9664; 1:500; CST), anti-SOX2 (Cat# AB5603; 1:200; Millipore), anti-PERICENTRIN (Cat# ab28144; 1:1,000; Abcam), anti–γ-tubulin (Cat# T3195; 1:1,000; Sigma-Aldrich), and anti–phospho-HISTONE 3 (Cat# H0412; 1:1,000; Sigma-Aldrich) antibodies overnight at 4°C. After that, the cells on the coverslip were washed three times with PBS, followed by incubation with secondary antibodies conjugated with desired dyes (1:500; Invitrogen) supplemented with 1 µg/ml DAPI for 2 h at rt. After washing three times with PBS, slides were then mounted with Fluoromount-G (SouthernBiotech) or Fluoroshield with DAPI (Cat# F6057; Sigma-Aldrich) and imaged with fluorescence microscope or confocal. For the Ki67 staining in GSCs overexpressing GFP, WT EYA2, or EYA2D274N, a goat anti-mouse secondary antibody conjugated with Alexa Fluor 647 was used. The signals from Alexa Fluor 647 were pseudo-colored in green for the figures.
Microtubule dynamics assays
Porcine brain α-tubulin was purified as previously described (Waterman-Storer, 2001). In vitro microtubule dynamics assays were performed as previously described (Fees and Moore, 2019), with DMSO, 20 µM EYA2 Tyr phosphatase inhibitor (MLS000544460), or 500 nM Nocodazole (Cat# M1404; Sigma-Aldrich) added to the final reaction. Each condition was tested in three separate experiments, with α-tubulin concentrations ranging from 6 to 10 µM. Total Internal Reflection Fluorescence images were acquired at 3-s intervals using a Nikon Ti-E microscope equipped with a 1.49 numerical aperture 100× CFI160 Apochromat objective, total internal reflection fluorescence illuminator, OBIS 488-nm and Sapphire 561-nm lasers (Coherent), an ORCA-Flash 4.0 LT sCMOS camera (Hammamatsu Photonics), and NIS Elements software (Nikon). Images were analyzed using a custom-made MATLAB program to identify and segment individual microtubules and generate maximum-intensity projections for each time point (Fees and Moore, 2018). Kymographs of image series were used to calculate microtubule length, polymerization rate, and polymerization time before catastrophe.
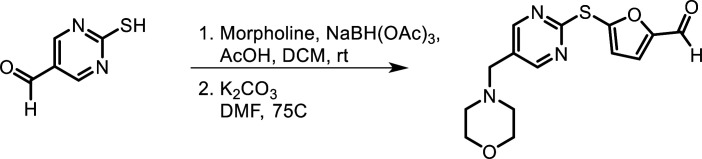
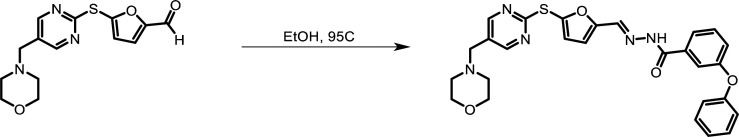
Synthesis of ETC 9519
(Scheme 1).
Morpholine (43.68 µl, 0.499 mmol, 1.0 equivalent [equiv.]) and 2-mercaptopyrimidine-5-carbaldehyde (140.0 mg, 0.999 mmol, 2.0 equiv.) were dissolved in anhydrous dichloromethane (DCM), and then acetic acid (28.59 µl, 0.499 mmol, 1.0 equiv.) was added. The reaction was stirred for 1 h at rt under nitrogen atmosphere. Sodium triacetoxyborohydride (317.5 mg, 1.498 mmol, 3.0 equiv.) was added, and the reaction was stirred for another 2.5 h. When the reaction was completed, the resultant yellow suspension was filtered, and the filtrate was collected and concentrated under reduced pressure. It was redissolved in methanol, stirred at ambient for 2 h, and concentrated under reduced pressure to afford crude 5-(morpholinomethyl)pyrimidine-2-thiol (210.2 mg, 1.201 mmol) as bright yellow solid, which was used for the next step without purification.
A solution of 5-(morpholinomethyl)pyrimidine-2-thiol (210.2 mg, 1.201 mmol, 1.0 equiv.), 5-bromofuran-2-carbaldehyde (329.96 mg, 1.562 mmol, 1.3 equiv.), and potassium carbonate (501.7 mg, 3.604 mmol, 3.0 equiv.) in anhydrous dimethylformamide (DMF; 6.0 ml, 0.2 M) was stirred for 18 h at 75°C under nitrogen atmosphere. Upon completion, the reaction mixture was passed through a cake of celite, and the filtrate collected was concentrated under reduced pressure. The crude material was purified using reverse phase chromatography (C18, eluent 40–60% acetonitrile [0.1% formic acid]/water [0.1% formic acid] to afford 5-((5-(morpholinomethyl)pyrimidin-2-yl)thio)furan-2-carbaldehyde; 78.1 mg, 0.256 mmol) as pale orange solid.
1H NMR (DMSO-d6) δ 9.65 (s, 1H), 8.59 (s, 2H), 7.66 (d, J = 3.60 Hz, 1H), 7.26 (d, J = 3.60 Hz, 1H), 3.55 (t, J = 4.80 Hz, 4H), 3.45 (s, 2H), 2.34 (t, J = 4.80 Hz, 4H).
(Scheme 2).
A solution of 5-((5-(morpholinomethyl)pyrimidin-2-yl)thio)furan-2-carbaldehyde (78.1 mg, 0.256 mmol, 1.0 equiv.) and 3-phenoxybenzohydrazide (58.28 mg, 0.256 mmol, 1.0 equiv.) in anhydrous ethanol (2.6 ml, 0.1 M) was stirred for 2 h at 95°C under nitrogen atmosphere. Upon completion, excess solvent was removed under reduced pressure and partitioned between DCM and water. The organic phase was separated, washed with aqueous sodium chloride, dried over sodium sulfate, and concentrated to dryness. The residue was purified using reverse phase chromatography (C18, eluent 60–80% acetonitrile [0.1% formic acid]/water [0.1% formic acid]). Upon removal of solvents, the solid was sonicated in hexanes and decanted to afford (E)-N'-((5-((5-(morpholinomethyl)pyrimidin-2-yl)thio)furan-2-yl)methylene)-3-phenoxybenzohydrazide (80.56 mg, 0.156 mmol) as pale orange solid.
1H NMR (DMSO-d6) δ 11.92 (s, 1H),), 8.57 (s, 2H), 8.32 (s, 1H), 7.67 − 7.69 (m, 1H), 7.52 − 7.57 (m, 2H), 7.41 − 7.45 (m, 2H), 7.17 − 7.24 (m, 2H), 7.06 − 7.13 (m, 4H), 3.54 (t, J = 4.40 Hz, 4H), 3.44 (s, 2H), 2.34 (t, J = 4.40 Hz, 4H). LCMS (ESI) m/z 516 (MH+) with a purity of 98.0%.
Flow cytometry–based cell cycle analysis, EdU, and annexin V staining
For the cell cycle analysis, two million cells were harvested, trypsinized, and washed with ice-cold PBS twice. The washed cells were fixed with ice-cold 70% ethanol for 2 h after desired treatment and then washed with PBS twice. Cells were then suspended in 400 µl PBS containing 0.1% Triton X-100, 100 µg/ml DNase-free RNase, and 50 µg/ml PI and kept in 4°C for overnight. Stained cells were quantified with a BD Fortessa analyzer. FlowJo software was used to generate flow cytometry plots.
EdU staining was performed with the Click-iT Plus EdU Flow Cytometry Assay Kit (Cat# C10632; Invitrogen) according to the manufacturer’s instruction. Briefly, GSCs cultured in complete Neurobasal medium were incubated with 10 µM EdU for 2 h. Then, the cells were collected and washed with PBS supplemented with 1% BSA followed by fixation with the provided reagent at rt for 15 min. The fixed cells were washed with PBS supplemented with 1% BSA and suspended in permeabilization buffer for 15 min by avoiding light. The cell suspension was mixed with Click-iT plus reaction cocktail and was incubated at rt for 30 min by avoiding light. The stained cells were washed with provided reagent and then suspended with saponin-based permeabilization buffer. After incubating with DAPI (1 µg/ml), the cell suspension was processed to flow cytometry analysis with a BD Fortessa analyzer.
Annexin V staining was conducted based on the manufacturer’s instruction provided by Alexa Fluor 488 Annexin V/Dead Cell Apoptosis Kit (Cat# V13241; Invitrogen). Briefly, cells after desired treatment were harvested and washed with ice-cold PBS. Appropriate numbers of the washed cells were suspended in 100 µl Annexin-binding buffer supplemented with Alexa Fluor 488 annexin V and PI and were kept at rt for 15-min incubation by avoiding light. After that, 400 µl Annexin-binding buffer was added to each sample and was mixed gently. The stained cells were kept on ice and processed to flow analysis immediately.
Western blotting
Cells after desired treatment were collected and suspended in radio-immunoprecipitation assay buffer supplemented with protease inhibitors and phosphatase inhibitors. The suspended cells were kept on ice for 30 min. The lysates were centrifuged at 12,000 rpm for 10 min at 4°C, and the supernatant was collected and processed to Bradford assay to measure the protein concentration. The protein samples were processed for immediate use by mixing with SDS Laemmli loading buffer and boiling for 5 min, or else the protein samples were split into small amounts and then stored at −80°C. Equal amounts of protein were used for electrophoresis with PAGE gels. The polyvinylidene difluoride (PVDF) membranes with proteins were blocked with TBS and 0.1% polysorbate 20 supplemented with 3.5% BSA at rt for 30 min, which was followed by incubation with primary antibodies overnight at 4°C. Anti-EYA2 (Cat# HPA027024; 1:500; Sigma-Aldrich), anti–Cleaved CASPASE 3 (Cat# 9664; 1:1,000; CST), anti-PARP (Cat# 9532; 1:1,000; CST), anti-SOX2 (Cat# AB5603; 1:1,000; Millipore), anti-OLIG2 (Cat# MABN50; 1:1,000; Millipore), anti-MYC (Cat# sc-764; 1:500), anti-ACTIN (Cat# A5316; 1:1,000; Sigma-Aldrich), and anti–α-tubulin (Cat# T6074; 1:10,000; Sigma-Aldrich) antibodies were used as primary antibodies. The membranes were developed by SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific) and Autoradiography Film (Denville Scientific). The developed films were scanned with Epson Perfection V600 Photo.
RNA-seq analysis
RNA-seq data for GSC versus NSC comparisons was downloaded from Gene Expression Omnibus (GEO) using the accession no. GSE119834 (Mack et al., 2019). 38 GSC and 5 NSC samples were used for this analysis. Reads were trimmed using Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) and cutadapt (http://cutadapt.readthedocs.io/en/stable/guide.html). Trimmed reads were quantified using Salmon in the quasi mapping-based mode using –seqBias, –gcBias, and –validateMappings flags. TXimport was used to import data into DESeq2 for differential expression analysis using the R programming language. Differential gene expression cutoffs were set at log2 mRNA expression fold change >4 (or less than −4) and with an adjusted P value of <1e-5. Volcano plots were generated using GraphPad Prism. GSEA was performed through the online GSEA web portal (https://www.gsea-msigdb.org/gsea/index.jsp). Pathway enrichment bubble plots were generated using the Bader Lab Enrichment Map Application and Cytoscape (http://www.cytoscape.org).
RNA-seq data for GSC versus DGC comparisons were downloaded from GEO using the accession no. GSE54791 (Suvà et al., 2014). Three paired GSC and DGC samples were used for this analysis. RNA-seq counts-per-million data were analyzed, and differential expression was calculated using the DESeq2 in R. Differential gene expression cutoffs were set at log2 mRNA expression fold change >2 (or less than −2) and with an adjusted P value of <1e-3. Volcano plots were generated using GraphPad Prism. GSEA was performed as described above. Gene set overlaps were calculated using GeneVenn (http://www.bioinformatics.org/gvenn/).
Total RNA from GSCs transduced with shCONT or shEYA2 were collected with miRNeasy mini kit (Qiagen) and processed to library construction with TruSeq RNA library Prep Kit (Illumina). The libraries were applied to paired sequencing with HiSeq4000 in Institute for Genomic Medicine, University of California, San Diego. The Fastq files of the RNA-seq data were processed to read quality control by running “FastQC” (Galaxy version 0.72) in Galaxy (https://usegalaxy.org) and then to remove the adapters by running “Trim Galore!” (Galaxy version 0.4.3.1). The trimmed reads were mapped to human genome 38 by “HISAT2” (Galaxy version 2.1.0+galaxy 4), and the mapped reads were then counted with “featureCounts” (Galaxy version 1.6.3+galaxy 2). The counts were processed to DEG analysis with R package DESeq2 (Version 1.14.1). Volcano plots were generated using GraphPad Prism. Down-regulated genes upon EYA2 knockdown were processed to GO analysis (http://geneontology.org/). Pre-ranked genes by fold change were processed to GSEA (Broad institute).
The normalization of the RNA-seq data was done using DESeq2, which was only one step on the user end, although there are multiple steps involved on the back end. Briefly, a pseudo-reference sample was created for each gene across all samples, and then ratios were calculated between the samples and pseudo reference for each gene across all samples. The median value of all ratios in one sample was taken as the normalization factor for the given sample. The normalized count values were generated by dividing each raw count value by the normalization factor of each sample. The normalized counts were then processed to desired applications like DEG analysis.
RNA and real-time quantitative (q)PCR
Total cellular RNA was isolated by use of Trizol reagent (Cat# 15596018; Invitrogen) based on the manufacturer’s instruction and dissolved in RNase-free water. By use of High Capacity of cDNA Reverse Transcription kit (Cat# 4368814; Life Technologies), cDNA was synthesized from 1 µg total RNA. Real-time qPCR was then performed to measure the relative mRNA expression by using Applied Biosystems 7900HT cycler or Bio-Rad CFX 9600 with SYBR-Green PCR Master Mix (Cat# A25778; Life Technologies) or Green Hi-ROX qPCR Kit (Cat# QS2050; Alkali Scientific).
Plasmids and lentiviral transduction
Lentiviral clones expressing two or three nonoverlapping shRNAs directed against human EYA2 (TRCN0000051947 and TRCN0000051946) or a nontargeting control shRNA was obtained from Sigma-Aldrich. shRNAs targeting 3′ untranslated region (UTR) of human EYA2 were designed with hairpin design tool in GPP Web Portal (https://portals.broadinstitute.org/gpp/public/seq/search; Board Institute). Annealed oligos were then inserted into PLKO.1 backbone (Cat# 10878; Addgene) as instructed. 293FT cells were transfected with the transfer plasmids together with the packaging vectors pCMV-dR8.2 dvpr and pCI-VSVG using a standard calcium phosphate transfection method in DMEM plus 10% FBS. 12 h later after transfection, culture medium was changed to complete Neurobasal medium. Medium containing virus particles was collected and centrifuged at 3,000 rpm for 10 min at 4°C. The supernatant was filtered with a 0.45-µm filter for immediate use or stock in −80°C.
Patient database bioinformatics
TCGA glioblastoma dataset (Bowman et al., 2017) was interrogated to determine the clinical relevance of patients with low-grade glioma or glioblastoma. Expression levels of genes of interest were categorized into two groups with the top quarter or bottom quarter as cutoff. The survival time and status for patients in both groups were analyzed using the Kaplan-Meier statistical test via log-rank test.
Quantification and statistical analysis
Student’s t test was used to assess statistical significance of studies having only two groups. One-way ANOVA was used to determine statistical significance in studies having more than one group. For quantification having subgroups in each group, two-way ANOVA was used to assess statistical significance. χ2 test was used for pairwise differences in assessing stem population frequency in the limited dilution assays. Kaplan-Meier survival curves were generated by use of Graphpad Prism 6 software, and log-rank tests were performed to assess statistical significance between groups. All experiments were performed at least three times independently. All data are presented as mean ± SD with *, P < 0.05; **, P < 0.01; and ***, P < 0.001. No statistical methods or criteria were used to estimate sample size or to include or exclude samples. The investigators were not blinded to the group allocation during the experiments.
Online supplemental material
Fig. S1 shows the GO analysis of DEGs preferentially expressed in GSCs compared with NSCs or DGCs. It also shows the H3K27Ac profiles in GSCs, NSCs, and DGCs around the EYA2 promoter region. Stemness after EYA2 knockdown in GSCs was examined, and the cellular growth was also measured upon EYA2 knockdown with additional shRNAs. Fig. S2 shows the effects of EYA2 mutants in GSCs or DGCs. It also shows the correlation between EYA2 and spindles in GSCs. Fig. S3 shows the co-staining of EYA2 and PERICENTRIN upon EYA2 knockdown in GSCs. It also shows the percentage of cells in the various cell cycle phases upon EYA2 Tyr phosphatase inhibitor treatment (or vehicle control) in GSCs, NSCs, DGCs, and NMs. Fig. S4 shows the increased centrosomal localization of EYA2 in GSCs and the abnormal spindles caused by the EYA2 Tyr phosphatase inhibitor. Fig. S5 shows that different EYA2 Tyr phosphatase inhibitors similarly cause spindle phenotypes. It also shows the expression of other EYAs in glioblastoma and the correlation between EYA expression and prognosis in gliomas or glioblastoma.
Acknowledgments
We appreciate the Histology Core at University of California, San Diego for its work on histological experiments and analysis and the Flow Cytometry Shared Resource from the University of Colorado Cancer Center for its work on cell cycle analysis (P30CA046934). We also appreciate the technical help of Alli Siclair.
The work was supported by National Institutes of Health grants (B.C. Prager: CA217066; R.C. Gimple: CA217065; L.J.Y. Kim: CA203101; S. Bao: CA184090, NS091080, NS099175; H.L. Ford: CA224867, CA221282, NS108396; and J.N. Rich: CA197718, CA238662, NS103434). P. Hamerlik is supported by the Danish Cancer Society/KBVU Foundation, Knæk Cancer, and NovoNordisk Foundation. We thank the Singapore Ministry of Health, National Medical Research Council for support under its open fund individual research grant (OFIRG17may050, NMRC/OFIRG/0051/2017).
Authors’ contributions: Conception and design: G. Zhang, Z. Dong, S. Bao, P. Hamerlik, H.L. Ford, J.N. Rich; development of methodology: G. Zhang, Z. Dong, Q. Wu, T.H. Keller, G. Lin, C. Kang, J.K. Moore, R. Zhao, H.L. Ford, J.N. Rich; acquisition of data (provided animals, acquired patients, provided facilities, etc.): G. Zhang, Z. Dong, Q. Wu, A. Wolin, J.Z. Shen, L. Jiang, L. Zhao, X. Wang, L. Zhang, L.M. Wood, D. Lv, J.K. Moore, T.H. Keller, G. Lin, C. Kang; analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): G. Zhang, Z. Dong, R.C. Gimple, Z. Qiu, B.C. Prager, L.J.Y. Kim, H.L. Ford, J.N. Rich; writing, reviewing, and/or revision of the manuscript: G. Zhang, Z. Dong, R.C. Gimple, Z. Qiu, B.C. Prager, L.J.Y. Kim, J.Z. Shen, L. Jiang, X. Wang, R.L. Anderson, R. Zhao, S. Bao, P. Hamerlik, H.L. Ford, J.N. Rich; administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): H.L. Ford, J.N. Rich.
Data availability
All RNA-seq data files are deposited in the GEO repository under accession no. GSE135306.
References
- Anantharajan, J., Zhou H., Zhang L., Hotz T., Vincent M.Y., Blevins M.A., Jansson A.E., Kuan J.W.L., Ng E.Y., Yeo Y.K., et al. 2019. Structural and Functional Analyses of an Allosteric EYA2 Phosphatase Inhibitor That Has On-Target Effects in Human Lung Cancer Cells. Mol. Cancer Ther. 18:1484–1496. 10.1158/1535-7163.MCT-18-1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auvergne, R.M., Sim F.J., Wang S., Chandler-Militello D., Burch J., Al Fanek Y., Davis D., Benraiss A., Walter K., Achanta P., et al. 2013. Transcriptional differences between normal and glioma-derived glial progenitor cells identify a core set of dysregulated genes. Cell Rep. 3:2127–2141. 10.1016/j.celrep.2013.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, S., Wu Q., McLendon R.E., Hao Y., Shi Q., Hjelmeland A.B., Dewhirst M.W., Bigner D.D., and Rich J.N.. 2006a. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 444:756–760. 10.1038/nature05236 [DOI] [PubMed] [Google Scholar]
- Bao, S., Wu Q., Sathornsumetee S., Hao Y., Li Z., Hjelmeland A.B., Shi Q., McLendon R.E., Bigner D.D., and Rich J.N.. 2006b. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 66:7843–7848. 10.1158/0008-5472.CAN-06-1010 [DOI] [PubMed] [Google Scholar]
- Barr, A.J. 2010. Protein tyrosine phosphatases as drug targets: strategies and challenges of inhibitor development. Future Med. Chem. 2:1563–1576. 10.4155/fmc.10.241 [DOI] [PubMed] [Google Scholar]
- Bowman, R.L., Wang Q., Carro A., Verhaak R.G.W., and Squatrito M.. 2017. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro-oncol. 19:139–141. 10.1093/neuonc/now247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese, C., Poppleton H., Kocak M., Hogg T.L., Fuller C., Hamner B., Oh E.Y., Gaber M.W., Finklestein D., Allen M., et al. 2007. A perivascular niche for brain tumor stem cells. Cancer Cell. 11:69–82. 10.1016/j.ccr.2006.11.020 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network . 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 455:1061–1068. 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, P.J., Ju B.G., Telese F., Wang X., Glass C.K., and Rosenfeld M.G.. 2009. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 458:591–596. 10.1038/nature07849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis, S., Sloan S.A., Croote D., Mignardi M., Chernikova S., Samghababi P., Zhang Y., Neff N., Kowarsky M., Caneda C., et al. 2017. Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma. Cell Rep. 21:1399–1410. 10.1016/j.celrep.2017.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleyrolle, L.P., Harding A., Cato K., Siebzehnrubl F.A., Rahman M., Azari H., Olson S., Gabrielli B., Osborne G., Vescovi A., and Reynolds B.A.. 2011. Evidence for label-retaining tumour-initiating cells in human glioblastoma. Brain. 134:1331–1343. 10.1093/brain/awr081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisner, A., Pazyra-Murphy M.F., Durresi E., Zhou P., Zhao X., Chadwick E.C., Xu P.X., Hillman R.T., Scott M.P., Greenberg M.E., and Segal R.A.. 2015. The Eya1 phosphatase promotes Shh signaling during hindbrain development and oncogenesis. Dev. Cell. 33:22–35. 10.1016/j.devcel.2015.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh, S.M., Micalizzi D.S., Jedlicka P., Zhao R., and Ford H.L.. 2012. Eya2 is required to mediate the pro-metastatic functions of Six1 via the induction of TGF-β signaling, epithelial-mesenchymal transition, and cancer stem cell properties. Oncogene. 31:552–562. 10.1038/onc.2011.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fees, C.P., and Moore J.K.. 2018. Regulation of microtubule dynamic instability by the carboxy-terminal tail of β-tubulin. Life Sci. Alliance. 1:e201800054. 10.26508/lsa.201800054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fees, C.P., and Moore J.K.. 2019. A unified model for microtubule rescue. Mol. Biol. Cell. 30:753–765. 10.1091/mbc.E18-08-0541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli, R., Binda E., Orfanelli U., Cipelletti B., Gritti A., De Vitis S., Fiocco R., Foroni C., Dimeco F., and Vescovi A.. 2004. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 64:7011–7021. 10.1158/0008-5472.CAN-04-1364 [DOI] [PubMed] [Google Scholar]
- Gell, C., Bormuth V., Brouhard G.J., Cohen D.N., Diez S., Friel C.T., Helenius J., Nitzsche B., Petzold H., Ribbe J., et al. 2010. Microtubule Dynamics Reconstituted In Vitro and Imaged by Single-Molecule Fluorescence Microscopy. In Methods in Cell Biology. Wilson L., and Correia J.J., editors. Academic Press, Burlington, MA. 221–245. [DOI] [PubMed] [Google Scholar]
- Grifone, R., Demignon J., Giordani J., Niro C., Souil E., Bertin F., Laclef C., Xu P.-X., and Maire P.. 2007. Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev. Biol. 302:602–616. 10.1016/j.ydbio.2006.08.059 [DOI] [PubMed] [Google Scholar]
- Hu, Y., and Smyth G.K.. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 347:70–78. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Jin, X., Kim L.J.Y., Wu Q., Wallace L.C., Prager B.C., Sanvoranart T., Gimple R.C., Wang X., Mack S.C., Miller T.E., et al. 2017. Targeting glioma stem cells through combined BMI1 and EZH2 inhibition. Nat. Med. 23:1352–1361. 10.1038/nm.4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan, N., Jeong D.G., Jung S.-K., Ryu S.E., Xiao A., Allis C.D., Kim S.J., and Tonks N.K.. 2009. Dephosphorylation of the C-terminal tyrosyl residue of the DNA damage-related histone H2A.X is mediated by the protein phosphatase eyes absent. J. Biol. Chem. 284:16066–16070. 10.1074/jbc.C900032200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger, A.B., Dehdashti S.J., Southall N., Marugan J.J., Ferrer M., Li X., Ford H.L., Zheng W., and Zhao R.. 2013. Identification of a selective small-molecule inhibitor series targeting the eyes absent 2 (Eya2) phosphatase activity. J. Biomol. Screen. 18:85–96. 10.1177/1087057112453936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger, A.B., Drasin D.J., Lea W.A., Patrick A.N., Patnaik S., Backos D.S., Matheson C.J., Hu X., Barnaeva E., Holliday M.J., et al. 2014. Allosteric inhibitors of the Eya2 phosphatase are selective and inhibit Eya2-mediated cell migration. J. Biol. Chem. 289:16349–16361. 10.1074/jbc.M114.566729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurppa, K.J., Liu Y., To C., Zhang T., Fan M., Vajdi A., Knelson E.H., Xie Y., Lim K., Cejas P., et al. 2020. Treatment-Induced Tumor Dormancy through YAP-Mediated Transcriptional Reprogramming of the Apoptotic Pathway. Cancer Cell. 37:104–122.e12. 10.1016/j.ccell.2019.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia, J.D., Mack S.C., Mulkearns-Hubert E.E., Valentim C.L.L., and Rich J.N.. 2015. Cancer stem cells in glioblastoma. Genes Dev. 29:1203–1217. 10.1101/gad.261982.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Qiu R., Qiu X., and Tian T.. 2017. EYA2 promotes lung cancer cell proliferation by downregulating the expression of PTEN. Oncotarget. 8:110837–110848. 10.18632/oncotarget.22860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén, J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., and Young R.A.. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 153:320–334. 10.1016/j.cell.2013.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack, S.C., Singh I., Wang X., Hirsch R., Wu Q., Villagomez R., Bernatchez J.A., Zhu Z., Gimple R.C., Kim L.J.Y., et al. 2019. Chromatin landscapes reveal developmentally encoded transcriptional states that define human glioblastoma. J. Exp. Med. 216:1071–1090. 10.1084/jem.20190196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, S., Huillard E., Kesari S., Maire C.L., Golebiowski D., Harrington E.P., Alberta J.A., Kane M.F., Theisen M., Ligon K.L., et al. 2011. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer Cell. 19:359–371. 10.1016/j.ccr.2011.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentel, M., Ionescu A.E., Puscalau-Girtu I., Helm M.S., Badea R.A., Rizzoli S.O., and Szedlacsek S.E.. 2018. WDR1 is a novel EYA3 substrate and its dephosphorylation induces modifications of the cellular actin cytoskeleton. Sci. Rep. 8:2910. 10.1038/s41598-018-21155-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, M., Reimand J., Lan X., Head R., Zhu X., Kushida M., Bayani J., Pressey J.C., Lionel A.C., Clarke I.D., et al. 2015. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc. Natl. Acad. Sci. USA. 112:851–856. 10.1073/pnas.1320611111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe, Y., Sano T., and Nagata S.. 2009. Regulation of the innate immune response by threonine-phosphatase of Eyes absent. Nature. 460:520–524. 10.1038/nature08138 [DOI] [PubMed] [Google Scholar]
- Ono, R., Masuya M., Ishii S., Katayama N., and Nosaka T.. 2017. Eya2, a Target Activated by Plzf, Is Critical for PLZF-RARA-Induced Leukemogenesis. Mol. Cell. Biol. 37:e00585-16. 10.1128/MCB.00585-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom, Q.T., Gittleman H., Liao P., Vecchione-Koval T., Wolinsky Y., Kruchko C., and Barnholtz-Sloan J.S.. 2017. CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010-2014. Neuro-oncol. 19(suppl_5):v1–v88. 10.1093/neuonc/nox158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallini, R., Ricci-Vitiani L., Banna G.L., Signore M., Lombardi D., Todaro M., Stassi G., Martini M., Maira G., Larocca L.M., and De Maria R.. 2008. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin. Cancer Res. 14:8205–8212. 10.1158/1078-0432.CCR-08-0644 [DOI] [PubMed] [Google Scholar]
- Pandey, R.N., Rani R., Yeo E.J., Spencer M., Hu S., Lang R.A., and Hegde R.S.. 2010. The Eyes Absent phosphatase-transactivator proteins promote proliferation, transformation, migration, and invasion of tumor cells. Oncogene. 29:3715–3722. 10.1038/onc.2010.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, R.N., Wang T.S., Tadjuidje E., McDonald M.G., Rettie A.E., and Hegde R.S.. 2013. Structure-activity relationships of benzbromarone metabolites and derivatives as EYA inhibitory anti-angiogenic agents. PLoS One. 8:e84582. 10.1371/journal.pone.0084582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L., et al. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 344:1396–1401. 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick, A.N., Cabrera J.H., Smith A.L., Chen X.S., Ford H.L., and Zhao R.. 2013. Structure-function analyses of the human SIX1-EYA2 complex reveal insights into metastasis and BOR syndrome. Nat. Struct. Mol. Biol. 20:447–453. 10.1038/nsmb.2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin, T.P., Smith A., McKinsey E., Reaves L., Jedlicka P., and Ford H.L.. 2012. EWS/FLI1 regulates EYA3 in Ewing sarcoma via modulation of miRNA-708, resulting in increased cell survival and chemoresistance. Mol. Cancer Res. 10:1098–1108. 10.1158/1541-7786.MCR-12-0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillard, A.D., Gundersen G.W., Fernandez N.F., Wang Z., Monteiro C.D., McDermott M.G., and Ma’ayan A.. 2016. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford). 2016:baw100. 10.1093/database/baw100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., and Dirks P.B.. 2004. Identification of human brain tumour initiating cells. Nature. 432:396–401. 10.1038/nature03128 [DOI] [PubMed] [Google Scholar]
- Stupp, R., Hegi M.E., Mason W.P., van den Bent M.J., Taphoorn M.J.B., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Belanger K., et al. National Cancer Institute of Canada Clinical Trials Group . 2009. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 10:459–466. 10.1016/S1470-2045(09)70025-7 [DOI] [PubMed] [Google Scholar]
- Stupp, R., Taillibert S., Kanner A., Read W., Steinberg D., Lhermitte B., Toms S., Idbaih A., Ahluwalia M.S., Fink K., et al. 2017. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: A randomized clinical trial. JAMA. 318:2306–2316. 10.1001/jama.2017.18718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvà, M.L., Rheinbay E., Gillespie S.M., Patel A.P., Wakimoto H., Rabkin S.D., Riggi N., Chi A.S., Cahill D.P., Nahed B.V., et al. 2014. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 157:580–594. 10.1016/j.cell.2014.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadjuidje, E., and Hegde R.S.. 2013. The Eyes Absent proteins in development and disease. Cell. Mol. Life Sci. 70:1897–1913. 10.1007/s00018-012-1144-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadjuidje, E., Wang T.S., Pandey R.N., Sumanas S., Lang R.A., and Hegde R.S.. 2012. The EYA tyrosine phosphatase activity is pro-angiogenic and is inhibited by benzbromarone. PLoS One. 7:e34806. 10.1371/journal.pone.0034806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum, M.E., and Medema R.H.. 2010. Mechanisms of centrosome separation and bipolar spindle assembly. Dev. Cell. 19:797–806. 10.1016/j.devcel.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Tirosh, I., Venteicher A.S., Hebert C., Escalante L.E., Patel A.P., Yizhak K., Fisher J.M., Rodman C., Mount C., Filbin M.G., et al. 2016. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 539:309–313. 10.1038/nature20123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer, J., and Gergely F.. 2019. Anti-mitotic therapies in cancer. J. Cell Biol. 218:10–11. 10.1083/jcb.201808077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo, C.M., Herman J.A., Olsen J.B., Ding Y., Corrin P., Girard E.J., Olson J.M., Emili A., DeLuca J.G., and Paddison P.J.. 2014. BuGZ is required for Bub3 stability, Bub1 kinetochore function, and chromosome alignment. Dev. Cell. 28:282–294. 10.1016/j.devcel.2013.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trépant, A.-L., Bouchart C., Rorive S., Sauvage S., Decaestecker C., Demetter P., and Salmon I.. 2015. Identification of OLIG2 as the most specific glioblastoma stem cell marker starting from comparative analysis of data from similar DNA chip microarray platforms. Tumour Biol. 36:1943–1953. 10.1007/s13277-014-2800-5 [DOI] [PubMed] [Google Scholar]
- Vartuli, R.L., Zhou H., Zhang L., Powers R.K., Klarquist J., Rudra P., Vincent M.Y., Ghosh D., Costello J.C., Kedl R.M., et al. 2018. Eya3 promotes breast tumor-associated immune suppression via threonine phosphatase-mediated PD-L1 upregulation. J. Clin. Invest. 128:2535–2550. 10.1172/JCI96784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez, R.J., Howell B., Yvon A.M., Wadsworth P., and Cassimeris L.. 1997. Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol. Biol. Cell. 8:973–985. 10.1091/mbc.8.6.973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer, C.M. 2001. Microtubule/organelle motility assays. Curr. Protoc. Cell Biol. Chapter 13:13.1.1-13.1.21. 10.1002/0471143030.cb1301s00 [DOI] [PubMed] [Google Scholar]
- Weaver, B.A.A., Silk A.D., Montagna C., Verdier-Pinard P., and Cleveland D.W.. 2007. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 11:25–36. 10.1016/j.ccr.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Weller, M., Wick W., Aldape K., Brada M., Berger M., Pfister S.M., Nishikawa R., Rosenthal M., Wen P.Y., Stupp R., and Reifenberger G.. 2015. Glioma. Nat. Rev. Dis. Primers. 1:15017. 10.1038/nrdp.2015.17 [DOI] [PubMed] [Google Scholar]
- Wen, Z., Liang C., Pan Q., and Wang Y.. 2017. Eya2 overexpression promotes the invasion of human astrocytoma through the regulation of ERK/MMP9 signaling. Int. J. Mol. Med. 40:1315–1322. 10.3892/ijmm.2017.3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B.R., Prabhu V.R., Hunter K.E., Glazier C.M., Whittaker C.A., Housman D.E., and Amon A.. 2008. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 322:703–709. 10.1126/science.1160058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K., Li Z., Cai S., Tian L., Chen K., Wang J., Hu J., Sun Y., Li X., Ertel A., and Pestell R.G.. 2013. EYA1 phosphatase function is essential to drive breast cancer cell proliferation through cyclin D1. Cancer Res. 73:4488–4499. 10.1158/0008-5472.CAN-12-4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P.-X., Adams J., Peters H., Brown M.C., Heaney S., and Maas R.. 1999. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat. Genet. 23:113–117. 10.1038/12722 [DOI] [PubMed] [Google Scholar]
- Xu, P.-X., Zheng W., Laclef C., Maire P., Maas R.L., Peters H., and Xu X.. 2002. Eya1 is required for the morphogenesis of mammalian thymus, parathyroid and thyroid. Development. 129:3033–3044. 10.1242/dev.129.13.3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., Loncarek J., Khodjakov A., and Rieder C.L.. 2008. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat. Cell Biol. 10:748–751. 10.1038/ncb1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, B., Cheng L., Chiang H.-C., Xu X., Han Y., Su H., Wang L., Zhang B., Lin J., Li X., et al. 2014. A phosphotyrosine switch determines the antitumor activity of ERβ. J. Clin. Invest. 124:3378–3390. 10.1172/JCI74085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Yang N., Huang J., Buckanovich R.J., Liang S., Barchetti A., Vezzani C., O’Brien-Jenkins A., Wang J., Ward M.R., et al. 2005. Transcriptional coactivator Drosophila eyes absent homologue 2 is up-regulated in epithelial ovarian cancer and promotes tumor growth. Cancer Res. 65:925–932. [PubMed] [Google Scholar]
- Zhang, L., Zhou H., Li X., Vartuli R.L., Rowse M., Xing Y., Rudra P., Ghosh D., Zhao R., and Ford H.L.. 2018. Eya3 partners with PP2A to induce c-Myc stabilization and tumor progression. Nat. Commun. 9:1047. 10.1038/s41467-018-03327-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., and Giannakakou P.. 2005. Targeting microtubules for cancer chemotherapy. Curr. Med. Chem. Anticancer Agents. 5:65–71. 10.2174/1568011053352569 [DOI] [PubMed] [Google Scholar]
- Zhou, H., Zhang L., Vartuli R.L., Ford H.L., and Zhao R.. 2018. The Eya phosphatase: Its unique role in cancer. Int. J. Biochem. Cell Biol. 96:165–170. 10.1016/j.biocel.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All RNA-seq data files are deposited in the GEO repository under accession no. GSE135306.